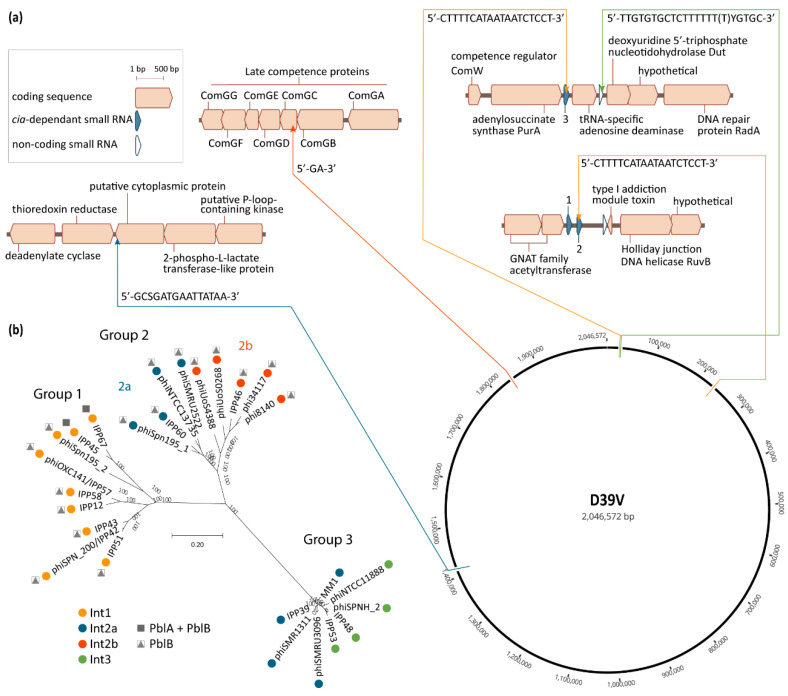
Figure 2.
Pneumococcal temperate phage groups and their chromosomal integration loci. (a) Chromosomal integration sites of pneumococcal phages. Thin arrows indicate the precise locus of the phage attachment sites, in relation to their immediate genetic context and their location on the circular representation of the D39V (CP027540.1) genome. Direction of the arrows reflects the orientation of phage integration in the chromosome respective to the integrase being located at the 5′ end of the integrated phage, as is the convention. Color of the arrows and connecting lines refer to the integrase type in panel (b). The nucleotide sequence of each core attachment site is indicated at each integration locus, as previously defined by Romero et al. [26] (Int1) and Gindreau et al. [24] (Int2a, including modifications reflecting single nucleotide polymorphisms in the genomes used in panel (b)), or as determined by the authors by aligning the left and right phage–chromosome junctions with the corresponding chromosomal site in the genome of D39V, in the case of Int3 and Int2b; (b) phylogenetic tree of a selection of pneumococcal temperate phages for which the genetic context could be determined. The multiple alignment of phage genomes was performed using MUSCLE v.3.8 [60]. The phylogenetic tree was constructed using FastTree v.2.1.10 [61] and visualized in MEGA-X v10.1.8 [62]. Colored dots represent the integrase type according to the key, and refer to the similarly-colored arrows pointing to the integration sites in panel (a). All Int1-encoding phages depicted here, except for IPP67, integrate in csRNA3. The presence of genes encoding the putative virulence factors PblA and PblB is indicated. Accession numbers of the sequences used are listed in Appendix A, Table A1.