Abstract
Corals harbor a great diversity of symbiotic microorganisms that play pivotal roles in host nutrition, reproduction, and development. Changes in the ocean environment, such as increasing exposure to artificial light at night (ALAN), may alter these relationships and result in a decline in coral health. In this study, we examined the microbiome associated with gravid specimens of the reef-building coral Acropora digitifera. We also assessed the temporal effects of ALAN on the coral-associated microbial community using high-throughput sequencing of the 16S rRNA gene V4 hypervariable region. The A. digitifera microbial community was dominated by phyla Proteobacteria, Firmicutes, and Bacteroidetes. Exposure to ALAN had no large-scale effect on the coral microbiome, although taxa affiliated with Rhodobacteraceae, Caulobacteraceae, Burkholderiaceae, Lachnospiraceae, and Ruminococcaceae were significantly enriched in corals subjected to ALAN. We further noted an increase in the relative abundance of the family Endozoicomonadaceae (Endozoicomonas) as the spawning period approached, regardless of light treatment. These findings highlight the stability of the A. digitifera microbial community under short-term artificial light pollution and provide initial insights into the response of the collective holobiont to ALAN.
Keywords: 16S rRNA gene, acroporid, coral-associated microbes, ecological light pollution, Endozoicomonadaceae, holobiont
1. Introduction
Corals play vital roles in marine ecosystems. The reefs they construct protect shorelines from wave impacts and provide habitats for diverse marine organisms [1,2]. Corals are considered holobionts or metaorganisms because of their association with photosynthetic algae (Symbiodiniaceae dinoflagellates), fungi, viruses, and prokaryotic microbes (bacteria and archaea) [3]. These symbionts reside in the corals’ surface mucopolysaccharide layer (SML), tissues, gastrovascular cavity, and skeleton [4,5,6,7,8]. These communities can vary across life stages of the coral host [9,10]. Coral-associated prokaryotic microbes are taxonomically and functionally diverse [11,12]. They are key for maintaining holobiont health as they contribute to carbon cycling, sulfur cycling, phosphorus fixation, metal homeostasis, organic remediation, the production of antibiotics, and secondary metabolite production [13,14,15]. However, environmental stressors, such as thermal anomalies, ocean acidification, eutrophication, salinity changes, chemical pollution, and sedimentation, can cause changes in the coral microbiome that disrupt these functions [16,17,18,19,20]. The loss of core microbiome functions may eventually lead to disease or death of the animal host [21].
Artificial light pollution at night (ALAN), which is also known as light pollution or photo-pollution, is an emerging stressor affecting nearshore environments and coastal zones [22,23,24]. In 2010, it was estimated that 22.2% of the world’s shorelines were already exposed to nightly light pollution [25]. The extent and intensity of ALAN impact is expected to increase as more coastal areas are developed [24,26]. The persistence of artificial and bright illumination that masks or adds to natural moonlight and starlight is a growing concern for many marine organisms, particularly corals [22,25].
Corals depend on numerous environmental cues (e.g., sea temperature, day length, wind or current patterns, and lunar cycles) to coordinate physiological processes such as metabolism, calcification, and reproduction [27,28,29]. The exposure of corals to light pollution at night has been shown to have detrimental effects on these processes. One major effect of ALAN exposure is the desynchronization of gamete release in corals [30]. This is likely linked to changes in the expression of genes related to cell cycle progression, cell proliferation, survival, and growth [31]. ALAN exposure has also been shown to have deleterious effects on coral-dinoflagellate symbiosis, resulting in severe oxidative stress, reduced symbiont cell density, lower chlorophyll concentration, and decreased maximum quantum yield of photosystem II [32,33,34]. Metabolic activity of the coral animal and its microalgal and prokaryotic symbionts are tightly intertwined [35]. Hence, artificial light exposure that affects overall holobiont metabolism may also influence the activity and abundance of closely associated microbial community members, with the potential enhancement of probiotic bacteria that can help the coral adapt to new environmental conditions. However, no studies have explored the effect of ALAN on the coral microbiome. Here, we examined the microbiome associated with gravid specimens of the reef-building coral, Acropora digitifera, and assessed the temporal effects of ALAN on the coral-associated microbial community. We hypothesized that the microbial assemblage would display temporal variability in response to ALAN exposure.
2. Materials and Methods
2.1. Coral Collection and Ex Situ Light Pollution Experiment
This work was part of a larger study on the effect of ALAN on coral gametogenesis and spawning. Hence, collection and experimentation were based on the known timing of gamete development and spawning for acroporids in the Bolinao-Anda Reef Complex in Pangasinan, Northwestern Philippines, as described by previous studies [36,37]. Forty-five colonies of A. digitifera (>30 cm in diameter) were collected from the nonlit Caniogan Reef (16°17.633′ N, 120°00.873′ E) in Anda, Pangasinan, at 4 to 5-m depths and transported to the outdoor hatchery of the Bolinao Marine Laboratory of University of the Philippines. The presence of gametes in collected corals was verified through histological analysis [36,38]. Coral sampling was conducted with permission from the Philippine Department of Agriculture–Bureau of Fisheries and Aquatic Resources with Gratuitous Permit No. 0169-19. Colonies were acclimatized in 447-L, opaque, blue plastic tanks (142 cm × 85 cm × 38 cm) with flowthrough sand-filtered seawater and natural daylight and moonlight for 21 days. Tanks were maintained at a temperature of 27.47 ± 0.47 °C (mean ± SD), salinity of 30.77 ± 0.21, dissolved oxygen (DO) of 2.36 ± 1.52 mg/L, and pH of 7.95 ± 0.07. The recorded average temperature at the reef from January to March was 28.51 ± 0.78 °C. Shade nets kept the average mid-day photosynthetically active radiation (PAR) in the tanks at 437.38 ± 363.26 µmol quanta m−2 s−1, which is within range of light levels recorded on the reef (517.47 ± 276.15 µmol quanta m−2 s−1). After the acclimatization period, coral colonies were subjected to nightly exposure to cold white LED (5329 K) with a major blue peak from 420–480 nm (“cold white” treatment) or to warm white LED (2719 K) with a major red peak from 580–620 nm (“warm white” treatment). Nighttime PAR in the tanks ranged from 0.5–0.75 µmol quanta m−2 s−1, which approximates ALAN values in populated nearshore environments, as reported by other studies [32,33,34]. Controls were subjected to natural moonlight (ambient treatment) with an average PAR of 0.01 ± 0.0 µmol quanta m−2 s−1. Each treatment was represented by three replicate tanks with five coral colonies each. Light treatments were conducted every day for up to two months from 26 January to 30 March, 2019. Lights were automatically turned on at sundown and turned off at sunrise by a photocell. Black curtains were placed between the tanks to prevent light contamination between treatments. Curtains were closed at sundown and opened at sunrise. Temperature, salinity, DO, and pH were monitored every day using a YSI Pro2030 multi-parameter meter (YSI Inc., Yellow Springs, OH, USA) and Mettler Toledo SevenGoTM Duo SG78 (Mettler Toledo, Zurich, Switzerland) at 0900H and 1600H (Table S1). PAR was measured using a LI-COR LI-193 light meter (Li-Cor, Inc., Lincoln, NE, USA).
2.2. Sampling and DNA Extraction
Only a subset of colonies that were used in this experiment were sampled for microbial analysis. Coral fragments (1 inch in length) were sampled from the central portion of three colonies after the acclimatization period immediately before the commencement of light treatments (T1; 25 January 2019). Three colonies were sampled from each treatment after one month of exposure (T2; 25 February 2019), and three more colonies were sampled from each treatment after two months of exposure immediately before spawning (T3; 30 March 2019). Coral fragments were flash-frozen in liquid nitrogen and stored at −80 °C prior to DNA extraction. Entire coral fragments were crushed using a mortar and pestle, and total DNA was extracted using a modified cetyltrimethylammonium bromide (CTAB) method [39]. DNA was resuspended in nuclease-free water to a final volume of 30 μL and stored at −20 °C. The quality of extracted DNA was checked by agarose gel electrophoresis using 1% agarose in 1X Tris/Borate/EDTA buffer at 120 V for 20 min. DNA concentration was determined using a NanoDrop 2000c spectrophotometer (Thermo Scientific, Waltham, MA, USA).
2.3. 16S rRNA Gene Sequencing and Analysis
Total genomic DNA was submitted to Macrogen, Inc., Seoul, South Korea for 16S rRNA gene sequencing. The prokaryotic 16S rRNA gene V4 hypervariable region was amplified from 10 ng of DNA using the Herculase II Fusion DNA Polymerase Nextera XT Index Kit V2 (Agilent Technologies, Santa Clara, CA, USA) with primers 515F (5′-GTG CCA GCM GCC GCG GTA A-3′) and 806R (5′ GGA CTA CHV GGG TWT CTA AT-3′) [40]. Sequencing was done on the Illumina MiSeq platform with 300-bp paired-end reads. Raw sequence data were deposited in the NCBI Sequence Read Archive and can be accessed under BioProject accession number PRJNA647288. Raw data are also available at Figshare (https://doi.org/10.6084/m9.figshare.12675374.v2). Microbial community analysis was conducted using the Quantitative Insights Into Microbial Ecology 2 (QIIME2) package (2020.2) ([41], https://docs.qiime2.org/2020.2/). Quality control was carried out by denoising the sequences using the DADA2 package [42] for the correction of amplicon errors following these parameters: --p-trim-left-f 19 --p-trim-left-r 20 --p-trunc-len-f 290 --p-trunc-len-r 250. SILVA version 132 ([43]; https://arb-silva.de) was used for taxonomic classification at a 97% sequence similarity cut-off. Sequences of mitochondrial or chloroplast origin and those with <10 counts in all libraries were removed from the final set of amplicon sequence variants (ASVs).
2.4. Data Analyses and Visualization
Alpha diversity metrics (observed ASVs, Shannon, and Inverse Simpson) were computed using ASV counts rarefied to the smallest sample size (28,061 reads). Normality of alpha diversity data was verified using the Shapiro-Wilk test. Global differences in alpha diversity values were determined using the Kruskal-Wallis test. Variation in community composition amongst treatments was visualized using principal coordinates analysis (PCoA) based on the Bray-Curtis dissimilarity matrix. Dissimilarity in microbial community composition amongst samples was determined by permutational multivariate analysis of variance (PERMANOVA) using the adonis function in the vegan package [44] with default settings (Bray-Curtis distance and 999 permutations). To account for random effects from the repeated sampling of coral colonies, we set strata = colonyID. p-values were adjusted using the Benjamini-Hochberg method. Differentially abundant ASVs between pairwise combinations of treatments and timepoints were identified using ALDEx2 [45,46] on center log ratio-transformed data at an effect size threshold greater than |3|. Bacterial indicator taxa, or ASVs that were significantly associated with specific timepoints or light treatments, were identified using the indicspecies package [47] based on the relative abundance of ASVs in the rarefied dataset. All statistical analyses and data visualizations were done using vegan [44], phyloseq [48], and ggplot2 [49] packages and were performed on Rstudio version 1.2.1335 ([50]; https://rstudio.com).
3. Results
3.1. Prokaryotic Microbial Community Composition of A. digitifera
The sequencing of the 16S rRNA V4 region of the A. digitifera-associated microbiome yielded a total of 2,544,965 reads from 21 libraries (Table S2). A total of 1,225,029 sequences passed the filtering steps. Classification using the SILVA database returned 1063 ASVs affiliated with 24 prokaryotic phyla, 39 classes, 89 orders, and 147 families. Rarefaction curves of observed ASVs approached an asymptote for all coral samples, indicating that the sequencing depth was sufficient to cover all the possible ASVs in each sample (Figure S1).
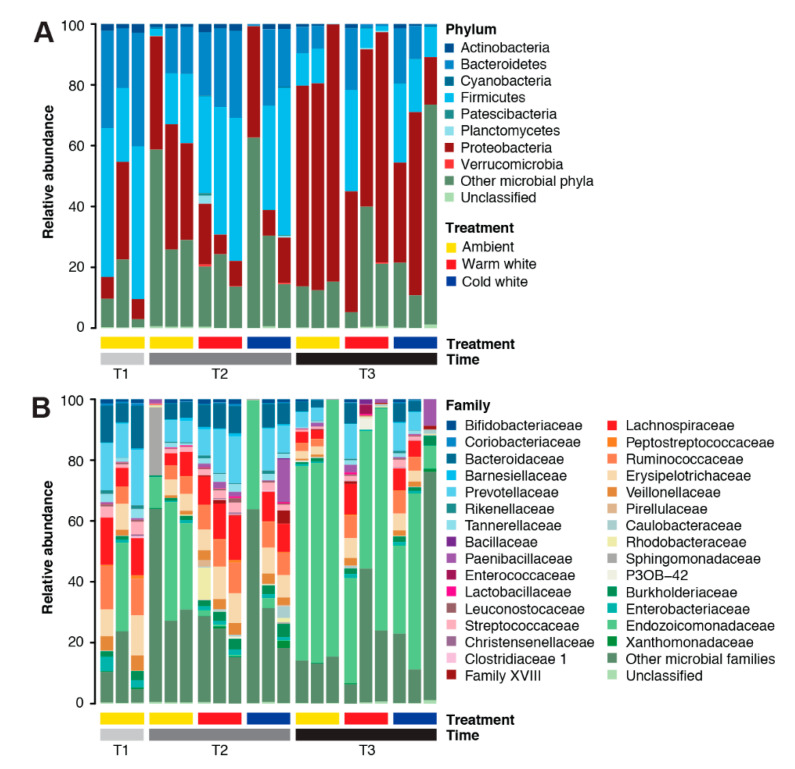
The A. digitifera microbiome was dominated by members of the phyla Proteobacteria (35.03%), Firmicutes (25.71%), Bacteroidetes (16.44%), Actinobacteria (1.34%), Planctomycetes (0.24%), Verrucomicrobia (0.08%), and Cyanobacteria (0.07%) (Figure 1A). Dominant families in the A. digitifera microbiome included Endozoicomonadaceae (28.70%), Prevotellaceae (8.39%), Lachnospiraceae (6.97%), Ruminococcaceae (5.89%), Bacteroidaceae (5.64%), Erysipelotrichiaceae (5.63%), Veillonellaceae (2.03%), Burkholderiaceae (1.78%), Paenibacillaceae (1.69%), Streptococcaceae (1.62%), Tannerellaceae (1.29%), and Enterobacteriaceae (1.11%) (Figure 1B). Archaeal phyla, such as Euryarchaeota and Thaumarchaeota, were also detected but at very low counts. It should be noted that the microbial symbiont communities associated with A. digitifera were highly variable even for colonies subjected to the same treatments and sampled at the same timepoints.
Figure 1.
Microbial community composition of Acropora digitifera. Relative abundance of microbial taxa at (A) phylum and (B) family levels in corals subjected to various light treatments (ambient, warm white, and cold white) for different durations (T1, January; T2, February; and T3, March). Phyla or families representing <0.1% of the total community are represented as “Other microbial phyla” and “Other microbial families”, respectively.
3.2. Effect of Time and Light Treatment on Microbial Community Structure
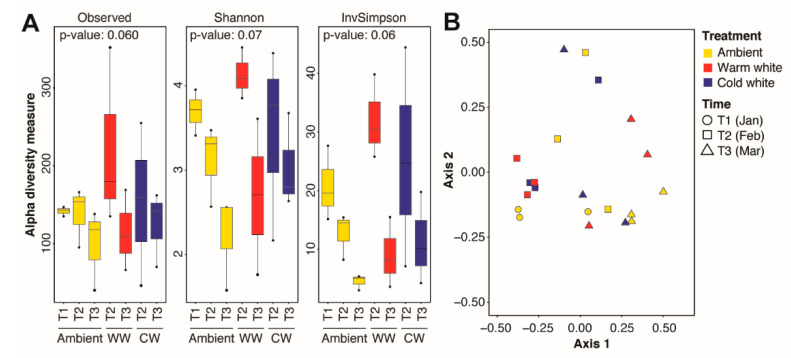
Significant changes in microbial community diversity were observed between corals sampled at different timepoints, regardless of light treatments (Figure S2A). This was also reflected in a shift in the microbial community structure as the spawning period approached (PERMANOVA, Timepoints: R2 = 0.2299, p = 0.01; Table 1), with a significant difference between T2 and T3 (PERMANOVA, T2 × T3: R2 = 0.1658, p = 0.04; Table 1). In contrast, no significant differences in alpha diversity metrics were observed between corals subjected to different light regimes (Figure S2B) or between corals subjected to different light treatments for varying durations (Figure 2A). The microbial community structure also remained similar across light treatments (PERMANOVA, Treatments: R2 = 0.0864, p = 1.00; Table 1) or across treatments at different timepoints (PERMANOVA, Timepoints × Treatments: R2 = 0.0737, p = 0.64; Table 1; Figure 2B).
Table 1.
Comparison of Acropora digitifera microbial communities at the amplicon sequence variant (ASV) level between different light treatments and timepoints. Benjamini-Hochberg-adjusted p-values in bold denote statistical significance at p < 0.05. PERMANOVA: permutational multivariate analysis of variance.
| Groups | PERMANOVA | |
|---|---|---|
| R 2 | p-Value | |
| Global comparisons | ||
| Timepoints | 0.2299 | 0.01 |
| Treatments | 0.0864 | 1.00 |
| Timepoints × Treatments | 0.0737 | 0.64 |
| Comparisons between timepoints | ||
| T1 × T2 | 0.1449 | 0.25 |
| T1 × T3 | 0.2133 | 0.13 |
| T2 × T3 | 0.1658 | 0.04 |
Figure 2.
Alpha and beta diversity of the Acropora digitifera microbiome. (A) Observed amplicon sequence variants (ASVs), Shannon, and inverse Simpson indices for coral samples subjected to various light treatments (ambient moonlight, Ambient; warm white, WW; and cold white, CW) for different durations. Kruskal-Wallis test p-values for comparisons are shown. (B) Principal coordinates analysis of microbial communities in corals exposed nightly to ambient moonlight, warm white light, or cold white light for different durations.
3.3. Microbial Taxa Significantly Associated with Time and Light Treatments
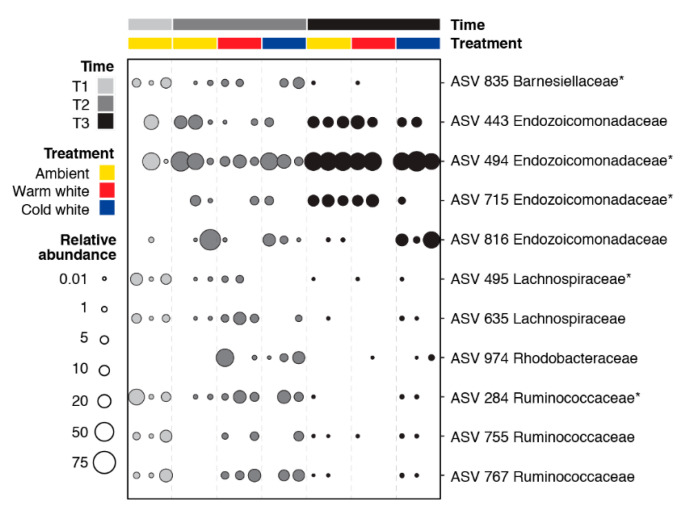
Pairwise comparisons of all time and treatment combinations using an ALDEx2 analysis with an effect size greater than |3| revealed 11 differentially abundant ASVs (Figure 3). Differentially abundant ASVs were affiliated with Barnesiellaceae (one ASV), Endozoicomonadaceae (four ASVs), Lachnospiraceae (two ASVs), Rhodobacteraceae (one ASV), and Ruminococcaceae (three ASVs). ASVs 495, 635, and 755 were relatively more abundant in the T1 samples. ASVs 284, 767, and 835 were abundant in T1 ambient and T2 light-treated samples. ASV 974 was enriched in the light-treated samples at T2. Endozoicomonadaceae ASVs (494, 443, 715, and 816) showed higher relative abundance at T2 and T3.
Figure 3.
Bubble plot showing the relative abundance of 11 differentially abundant ASVs in corals exposed to different light treatments and collected at different timepoints (T1, January; T2, February; and T3, March). Differentially abundant ASVs were identified using ALDEx2 with an effect size greater than |3|. ASVs with asterisks were also identified as bacterial indicator taxa using the indicspecies package (Figure S3).
An indicator species analysis was also conducted as a complementary method to identify ASVs significantly associated with specific timepoints or treatments (Figure S3). This method considers the abundance and frequency of occurrence of ASVs in samples from different conditions [51]. Thirty-three ASVs, including several members of Lachnospiraceae and Ruminococcaceae, were significantly associated with corals sampled at T1, while two ASVs affiliated with Endozoicomonadaceae were associated with corals at T3. Five differentially abundant ASVs, including Barnesiellaceae (ASV 835), Endozoicomonadaceae (ASVs 494 and 715), Lachnospiraceae (ASV 495), and Ruminococcaceae (ASV 284), overlapped with ASVs identified as bacterial indicator taxa (Figure 3).
4. Discussion
4.1. The A. digitifera Microbiome Is Unaffected by Short-Term Light Pollution
The A. digitifera microbiome was dominated by phyla Proteobacteria, Firmicutes, and Bacteroidetes, which agrees with previous investigations on the microbiomes of adult conspecific coral species [17,52]. The microbial community structure remained stable despite nightly exposure to either warm white or cold white light. Only a few microbial taxa showed significant changes in relative abundance. However, the microbial community structure did change over time, regardless of light treatment. This shift could be related to gamete maturation as the spawning period approached or, alternatively, to changes in water quality conditions in the tanks.
The ability of the coral microbiome to tolerate stressors, such as temperature and eutrophication, has also been reported in other studies [16,17]. The compositional stability of the coral microbiome may be attributed to the physiological plasticity of each individual microbial taxon that allows it to buffer environmental change [53]. A stable microbial community ensures the continued provision of metabolites to the host and prevention of the unexpected proliferation of opportunistic or foreign bacteria that may be detrimental to the holobiont [6,54,55]. Moreover, changes in the relative abundance of some microbial taxa in the light pollution treatment is in agreement with the coral probiotic hypothesis, wherein the relative abundance of microbial species changes in a manner that allows the coral holobiont to adapt to new environmental conditions [56].
4.2. Microbial Taxa That Respond to ALAN
Although the overall microbial community structure remained the same in A. digitifera subjected to nightly light pollution, we observed an increase in the relative abundance of aerobic anoxygenic photoheterotrophic bacteria (AAPB), including members of Rhodobacteraceae and Caulobacteraceae, under the warm white and cold white light treatments, respectively. AAPB are chlorophototrophic members of the phylum Proteobacteria that contain bacteriochlorophyll a and use light as an alternative energy source [57,58]. Anoxygenic photosynthesis is a phototrophic process where light energy is captured and converted to ATP without the production of oxygen [59]. Members of the AAPB family can also utilize various organic and inorganic compounds, perform sulfur oxidation, carbon monoxide oxidation, and produce secondary metabolites [60]. Light directly affects and stimulates the growth rates and abundance of natural populations of marine AAPB, which explains their abundance in the corals experiencing light pollution at night [61,62]. Anoxygenic phototrophs have previously been detected in the skeletons of the scleractinian corals Montipora monasteriata, Porites cylindrica, and Isopora palifera [63,64].
Burkholderiaceae, Lachnospiraceae, Ruminococcaceae, and Paenibacillaceae were also enriched in A. digitifera fragments subjected to ALAN. It is worth noting that these microbial taxa are major taxonomic groups found in the human gut and in fecal microbiota [65,66,67,68,69], as well as in the fish gut microbiome [70,71]. The milkfish aquaculture zone and human settlements found in proximity to the Bolinao Marine Laboratory [72] are possible sources of these bacterial groups. Some members of Paenibacillaceae, although present in our coral samples, are known to be soil-associated [73] and may have been transported by coastal runoff [5].
4.3. Microbial Taxa Associated with Symbiodiniaceae Are Enriched under ALAN
Light pollution has been shown to affect the activity of phototrophic symbiotic dinoflagellates (Symbiodiniaceae) of corals [32,33,34]. As Symbiodiniaceae are closely associated with nitrogen-fixing bacteria (diazotrophs) [6,7,35], it is possible that ALAN exposure may also affect the abundance of these particular microorganisms. In our study, we found that members of Lachnospiraceae, Rhodobacterales, and Caulobacterales, which include known diazotrophs [74,75,76,77,78,79], were relatively more abundant in corals subjected to light at night. These bacterial groups have also been found to co-occur with dinoflagellate symbionts of corals [80,81]. It is possible that the higher abundance of symbiont-associated microbes in the corals under ALAN treatment is linked to the greater abundance and activity of the dinoflagellate symbionts.
4.4. Endozoicomonadaceae Abundance Increased as Spawning Period Approached
Taxa affiliated with the proteobacterial family Endozoicomonadaceae increased in abundance in A. digitifera coral fragments over the course of the experiment. It is likely that this increase is related to the progression of gametogenesis, as Endozoicomonas is found at low abundance in the microbial community of nongravid A. digitifera adults [17,52] or in larvae and juveniles [9] but is detected at high abundance in gravid A. digitifera colonies and in the egg-sperm bundles of A. digitifera [9] and A. tenuis [10]. Endozoicomonadaceae (Endozoicomonas) may therefore be vertically transmitted in acroporid corals [10]. Endozoicomonas are reported to produce quorum-sensing molecules and antimicrobial compounds [82,83], which may prevent the proliferation of non-native microbes that can harm the coral holobiont. We posit that the abundance of this bacteria in gravid corals and in their gamete bundles may serve to protect the eggs and sperm in the water column until fertilization occurs [84,85]. However, as our experimental tanks received seawater directly from the reef flat, we are unable to rule out the possibility that changes in the seawater conditions (i.e., nutrient levels and temperature) over the course of the experiment may have also contributed to the observed increase in Endozoicomonas. Thus, further studies are needed to explore the temporal dynamics of Endozoicomonas in the microbiome of A. digitifera.
5. Conclusions
One of the most understudied threats to corals is ALAN, which is an inevitable consequence of coastal development and a growing concern due to its potential negative impacts on coral reproduction. This study revealed that the overall coral microbial community structure remained stable under nightly exposure to light pollution. We did, however, observe that bacteria associated with the phototrophic symbionts of the coral, as well as those that could themselves utilize light for energy production, increased in abundance under ALAN exposure. We also noted a significant increase in the abundance of Endozoicomonas in the A. digitifera microbiome as the spawning period approached. Further studies are needed to determine whether continued exposure to ALAN, especially in combination with other stressors, affects the collective physiology and health of the coral holobiont.
Acknowledgments
We sincerely thank Renato Adolfo, Fernando Castrence, Francis Kenith Adolfo, Robert Valenzuela, and Ronald De Guzman for field and hatchery assistance.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/10/1566/s1: Figure S1: Rarefaction curves for all samples based on number of ASVs identified by sequencing of the 16S rRNA V4 region. Figure S2: Alpha diversity of the Acropora digitifera microbiome. Figure S3: Bacterial indicator taxa for timepoints and light treatments. Table S1: Environmental conditions in experimental tanks from January to March 2019. Table S2: Information on Acropora digitifera colonies used in the study.
Author Contributions
P.C.C., C.C., I.A., Y.R. and O.L. conceptualized the study. P.C.C., C.C., J.I.P.B., M.A.L.N., C.L.D.C., I.A., Y.R., O.L. and S.L.G.S. conducted the experiment. J.I.P.B. and M.A.L.N. performed the microbial community analysis. J.I.P.B., M.A.L.N., C.L.D.C., S.L.G.S., P.C.C. and C.C. analyzed the data and generated the tables and figures. All authors contributed to the drafting of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Department of Science and Technology Philippine Council for Agriculture, Aquatic and Natural Resources Research and Development (QMSR-MRRD-MEC-295-1449) to P.C.C. and C.C.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- 1.Ménard A., Turgeon K., Roche D.G., Binning S.A., Kramer D.L. Shelters and their use by fishes on fringing coral reefs. PLoS ONE. 2012;7:e38450. doi: 10.1371/journal.pone.0038450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodríguez-Zaragoza F.A., Arias-González J.E. Coral biodiversity and bio-construction in the northern sector of the mesoamerican reef system. Front. Mar. Sci. 2015;2:13. doi: 10.3389/fmars.2015.00013. [DOI] [Google Scholar]
- 3.Peixoto R.S., Rosado P.M., Leite D.C.D.A., Rosado A.S., Bourne D.G. Beneficial microorganisms for corals (BMC): Proposed mechanisms for coral health and resilience. Front. Microbiol. 2017;8:341. doi: 10.3389/fmicb.2017.00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sweet M., Croquer A., Bythell J. Bacterial assemblages differ between compartments within the coral holobiont. Coral Reefs. 2011;30:39–52. doi: 10.1007/s00338-010-0695-1. [DOI] [Google Scholar]
- 5.Li J., Chen Q., Long L.-J., Dong J.-D., Yang J., Zhang S. Bacterial dynamics within the mucus, tissue and skeleton of the coral Porites lutea during different seasons. Sci. Rep. 2014;4:1–8. doi: 10.1038/srep07320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ainsworth T.D., Krause L., Bridge T., Torda G., Raina J.-B., Zakrzewski M., Gates R.D., Padilla-Gamiño J.L., Spalding H.L., Smith C. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 2015;9:2261–2274. doi: 10.1038/ismej.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourne D.G., Morrow K.M., Webster N.S. Insights into the coral microbiome: Underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 2016;70:317–3400. doi: 10.1146/annurev-micro-102215-095440. [DOI] [PubMed] [Google Scholar]
- 8.Ricci F., Marcelino V.R., Blackall L.L., Kühl M., Medina M., Verbruggen H. Beneath the surface: Community assembly and functions of the coral skeleton microbiome. Microbiome. 2019;7:159. doi: 10.1186/s40168-019-0762-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernasconi R., Stat M., Koenders A., Paparini A., Bunce M., Huggett M.J. Establishment of coral-bacteria symbioses reveal changes in the core bacterial community with host ontogeny. Front. Microbiol. 2019;10:1529. doi: 10.3389/fmicb.2019.01529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damjanovic K., Menéndez P., Blackall L.L., van Oppen M.J. Early life stages of a common broadcast spawning coral associate with specific bacterial communities despite lack of internalized bacteria. Microb. Ecol. 2020;79:706–719. doi: 10.1007/s00248-019-01428-1. [DOI] [PubMed] [Google Scholar]
- 11.Krediet C.J., Ritchie K.B., Paul V.J., Teplitski M. Coral-associated micro-organisms and their roles in promoting coral health and thwarting diseases. Proc. R. Soc. B Biol. Sci. 2013;280:20122328. doi: 10.1098/rspb.2012.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hernandez-Agreda A., Leggat W., Bongaerts P., Herrera C., Ainsworth T.D. Rethinking the coral microbiome: Simplicity exists within a diverse microbial biosphere. mBio. 2018;9:e00812-18. doi: 10.1128/mBio.00812-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olson N., Ainsworth T., Gates R., Takabayashi M. Diazotrophic bacteria associated with Hawaiian Montipora corals: Diversity and abundance in correlation with symbiotic dinoflagellates. J. Exp. Mar. Biol. Ecol. 2009;371:140–146. doi: 10.1016/j.jembe.2009.01.012. [DOI] [Google Scholar]
- 14.Sharp K.H., Sneed J., Ritchie K., Mcdaniel L., Paul V.J. Induction of larval settlement in the reef coral Porites astreoides by a cultivated marine Roseobacter strain. Biol. Bull. 2015;228:98–107. doi: 10.1086/BBLv228n2p98. [DOI] [PubMed] [Google Scholar]
- 15.Welsh R.M., Zaneveld J.R., Rosales S.M., Payet J.P., Burkepile D.E., Thurber R.V. Bacterial predation in a marine host-associated microbiome. ISME J. 2016;10:1540–1544. doi: 10.1038/ismej.2015.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garren M., Raymundo L., Guest J., Harvell C.D., Azam F. Resilience of coral-associated bacterial communities exposed to fish farm effluent. PLoS ONE. 2009;4:e7319. doi: 10.1371/journal.pone.0007319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gajigan A.P., Diaz L.A., Conaco C. Resilience of the prokaryotic microbial community of Acropora digitifera to elevated temperature. MicrobiologyOpen. 2017;6:e00478. doi: 10.1002/mbo3.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDevitt-Irwin J.M., Baum J.K., Garren M., Vega Thurber R.L. Responses of coral-associated bacterial communities to local and global stressors. Front. Mar. Sci. 2017;4:262. doi: 10.3389/fmars.2017.00262. [DOI] [Google Scholar]
- 19.Grottoli A.G., Dalcin Martins P., Wilkins M.J., Johnston M.D., Warner M.E., Cai W.-J., Melman T.F., Hoadley K.D., Pettay D.T., Levas S. Coral physiology and microbiome dynamics under combined warming and ocean acidification. PLoS ONE. 2018;13:e0191156. doi: 10.1371/journal.pone.0191156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ziegler M., Grupstra C.G., Barreto M.M., Eaton M., BaOmar J., Zubier K., Al-Sofyani A., Turki A.J., Ormond R., Voolstra C.R. Coral bacterial community structure responds to environmental change in a host-specific manner. Nat. Commun. 2019;10:1–11. doi: 10.1038/s41467-019-10969-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egan S., Gardiner M. Microbial dysbiosis: Rethinking disease in marine ecosystems. Front. Microbiol. 2016;7:991. doi: 10.3389/fmicb.2016.00991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Depledge M.H., Godard-Codding C.A., Bowen R.E. Light pollution in the sea. Mar. Pollut. Bull. 2010;9:1383–1385. doi: 10.1016/j.marpolbul.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 23.Davies T.W., Coleman M., Griffith K.M., Jenkins S.R. Night-time lighting alters the composition of marine epifaunal communities. Biol. Lett. 2015;11:20150080. doi: 10.1098/rsbl.2015.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kyba C.C., Kuester T., De Miguel A.S., Baugh K., Jechow A., Hölker F., Bennie J., Elvidge C.D., Gaston K.J., Guanter L. Artificially lit surface of Earth at night increasing in radiance and extent. Sci. Adv. 2017;3:e1701528. doi: 10.1126/sciadv.1701528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies T.W., Duffy J.P., Bennie J., Gaston K.J. The nature, extent, and ecological implications of marine light pollution. Front. Ecol. Environ. 2014;12:347–355. doi: 10.1890/130281. [DOI] [Google Scholar]
- 26.Garratt M.J., Jenkins S.R., Davies T.W. Mapping the consequences of artificial light at night for intertidal ecosystems. Sci. Total Environ. 2019;691:760–768. doi: 10.1016/j.scitotenv.2019.07.156. [DOI] [PubMed] [Google Scholar]
- 27.Babcock R., Bull G., Harrison P.L., Heyward A.J., Oliver J., Wallace C., Willis B. Synchronous spawnings of 105 scleractinian coral species on the Great Barrier Reef. Mar. Biol. 1986;90:379–394. doi: 10.1007/BF00428562. [DOI] [Google Scholar]
- 28.Moya A., Tambutté S., Tambutté E., Zoccola D., Caminiti N., Allemand D. Study of calcification during a daily cycle of the coral Stylophora pistillata: Implications for light-enhanced calcification. J. Exp. Biol. 2006;209:3413–3419. doi: 10.1242/jeb.02382. [DOI] [PubMed] [Google Scholar]
- 29.Baird A.H., Guest J.R., Willis B.L. Systematic and biogeographical patterns in the reproductive biology of scleractinian corals. Annu. Rev. Ecol. Evol. Syst. 2009;40:551–571. doi: 10.1146/annurev.ecolsys.110308.120220. [DOI] [Google Scholar]
- 30.Kaniewska P., Alon S., Karako-Lampert S., Hoegh-Guldberg O., Levy O. Signaling cascades and the importance of moonlight in coral broadcast mass spawning. eLife. 2015;4:e09991. doi: 10.7554/eLife.09991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenberg Y., Doniger T., Levy O. Sustainability of coral reefs are affected by ecological light pollution in the Gulf of Aqaba/Eilat. Commun. Biol. 2019;2:1–9. doi: 10.1038/s42003-019-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ayalon I., de Barros Marangoni L.F., Benichou J.I., Avisar D., Levy O. Red Sea corals under Artificial Light Pollution at Night (ALAN) undergo oxidative stress and photosynthetic impairment. Glob. Chang. Biol. 2019;25:4194–4207. doi: 10.1111/gcb.14795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levy O., de Barros Marangoni L.F., Cohen J.I., Rottier C., Béraud E., Grover R., Ferrier-Pagès C. Artificial light at night (ALAN) alters the physiology and biochemistry of symbiotic reef building corals. Environ. Pollut. 2020;266:114987. doi: 10.1016/j.envpol.2020.114987. [DOI] [PubMed] [Google Scholar]
- 34.Tamir R., Eyal G., Cohen I., Loya Y. Effects of light pollution on the early life stages of the most abundant northern red sea coral. Microorganisms. 2020;8:193. doi: 10.3390/microorganisms8020193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson J.R., Rivera H.E., Closek C.J., Medina M. Microbes in the coral holobiont: Partners through evolution, development, and ecological interactions. Front. Cell. Infect. Microbiol. 2015;4:176. doi: 10.3389/fcimb.2014.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomez E.J., Jamodiong E.A., Maboloc E.A., Ligson C.A., Tabalanza T.D., Villanueva R.D., Cabaitan P.C. Gametogenesis and reproductive pattern of the reef-building coral Acropora millepora in northwestern Philippines. Invertebr. Reprod. Dev. 2018;62:202–208. doi: 10.1080/07924259.2018.1496155. [DOI] [Google Scholar]
- 37.Jamodiong E.A., Maboloc E.A., Leriorato J.C., Tañedo M.C.S., Diaz L.A., Tabalanza T.D., Cabaitan P.C., Villanueva R.D. Coral spawning and spawn-slick observation in the Philippines. Mar. Biodivers. 2018;48:2187–2192. doi: 10.1007/s12526-017-0680-9. [DOI] [Google Scholar]
- 38.Jamodiong E.A., Maboloc E.A., Villanueva R.D., Cabaitan P.C. Gametogenesis and Inter-annual Variability in the Spawning Pattern of Acropora hyacinthus in Northwestern Philippines. Zool. Stud. 2018;57:e46. doi: 10.6620/ZS.2018.57-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winnepenninckx B., Backeljau T., De R.W. Extraction of high molecular weight DNA from molluscs. Trends Genet. TIG. 1993;9:407. doi: 10.1016/0168-9525(93)90102-n. [DOI] [PubMed] [Google Scholar]
- 40.Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Lozupone C.A., Turnbaugh P.J., Fierer N., Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolyen E., Rideout J.R., Dillon M., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P.R., O’Hara R.B., Simpson G.L., Solymos P., et al. Vegan: Community ecology package. R Package Version 2.5-5. [(accessed on 25 May 2020)]; Available online: https://CRAN.R-project.org/package=vegan.
- 45.Fernandes A.D., Macklaim J.M., Linn T.G., Reid G., Gloor G.B. ANOVA-like differential expression (ALDEx) analysis for mixed population RNA-Seq. PLoS ONE. 2013;8:e67019. doi: 10.1371/journal.pone.0067019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gloor G.B., Reid G. Compositional analysis: A valid approach to analyze microbiome high-throughput sequencing data. Can. J. Microbiol. 2016;62:692–703. doi: 10.1139/cjm-2015-0821. [DOI] [PubMed] [Google Scholar]
- 47.De Cáceres M., Legendre P., Moretti M. Improving indicator species analysis by combining groups of sites. Oikos. 2010;119:1674–1684. doi: 10.1111/j.1600-0706.2010.18334.x. [DOI] [Google Scholar]
- 48.McMurdie P.J., Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wickham H., Chang W., Henry L., Pedersen T., Takahsahi K., Wilke C., Woo K., Yutani H., Dunnington D. Create elegant data visualisations using the grammar of graphics. R Package Version 3.3.0. [(accessed on 25 May 2020)]; Available online: https://ggplot2.tidyverse.org.
- 50.R Team RStudio: Integrated development for R. Version 1.2.1335. [(accessed on 20 March 2020)]; Available online: https://rstudio.com.
- 51.De Cáceres M., Legendre P. Associations between species and groups of sites: Indices and statistical inference. Ecology. 2009;90:3566–3574. doi: 10.1890/08-1823.1. [DOI] [PubMed] [Google Scholar]
- 52.Rizal A., Akbarsyah N., Kdyp P., Permana R., Andhikawati A. Molecular diversity of the bacterial community associated with Acropora digitifera (Dana, 1846) corals on Rancabuaya coastline, Garut District, Indonesia. World Sci. News. 2020;144:384–396. [Google Scholar]
- 53.Shade A., Peter H., Allison S.D., Baho D., Berga M., Bürgmann H., Huber D.H., Langenheder S., Lennon J.T., Martiny J.B. Fundamentals of microbial community resistance and resilience. Front. Microbiol. 2012;3:417. doi: 10.3389/fmicb.2012.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bäckhed F., Ley R.E., Sonnenburg J.L., Peterson D.A., Gordon J.I. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 55.Kamada N., Chen G.Y., Inohara N., Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reshef L., Koren O., Loya Y., Zilber-Rosenberg I., Rosenberg E. The coral probiotic hypothesis. Environ. Microbiol. 2006;8:2068–2073. doi: 10.1111/j.1462-2920.2006.01148.x. [DOI] [PubMed] [Google Scholar]
- 57.Kolber Z.S., Gerald F., Lang A.S., Beatty J.T., Blankenship R.E., VanDover C.L., Vetriani C., Koblizek M., Rathgeber C., Falkowski P.G. Contribution of aerobic photoheterotrophic bacteria to the carbon cycle in the ocean. Science. 2001;292:2492–2495. doi: 10.1126/science.1059707. [DOI] [PubMed] [Google Scholar]
- 58.Liu Y., Zheng Q., Lin W., Jiao N. Characteristics and evolutionary analysis of photosynthetic gene clusters on extrachromosomal replicons: From streamlined plasmids to chromids. MSystems. 2019;4:e00319–e00358. doi: 10.1128/mSystems.00358-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yurkov V., Csotonyi J.T. New light on aerobic anoxygenic phototrophs. In: Hunter C.N., Daldal F., Thurnauer M.C., Beatty J.T., editors. The Purple Phototrophic Bacteria. Volume 28. Springer; Dordrecht, The Netherlands: 2009. pp. 31–55. Advances in Photosynthesis and Respiration. [Google Scholar]
- 60.Pujalte M.J., Lucena T., Ruvira M.A., Arahal D.R., Macián M.C. The family rhodobacteraceae. In: Rosenberg E., DeLong E.F., Lory S., Stackebrandt E., Thompson F., editors. The Prokaryotes. 4th ed. Springer; Berlin/Heidelberg, Germany: 2014. pp. 439–512. [Google Scholar]
- 61.Ferrera I., Sánchez O., Kolářová E., Koblížek M., Gasol J.M. Light enhances the growth rates of natural populations of aerobic anoxygenic phototrophic bacteria. ISME J. 2017;11:2391–2393. doi: 10.1038/ismej.2017.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Piwosz K., Vrdoljak A., Frenken T., González-Olalla J.M., Šantić D., McKay R.M., Spilling K., Guttman L., Znachor P., Mujakić I. Light and primary production shape bacterial activity and community composition of aerobic anoxygenic phototrophic bacteria in a microcosm experiment. mSphere. 2020;5:e00354-20. doi: 10.1128/mSphere.00354-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Magnusson S.H., Fine M., Kühl M. Light microclimate of endolithic phototrophs in the scleractinian corals Montipora monasteriata and Porites cylindrica. Mar. Ecol. Prog. Ser. 2007;332:119–128. doi: 10.3354/meps332119. [DOI] [Google Scholar]
- 64.Yang S.H., Lee S.T., Huang C.R., Tseng C.H., Chiang P.W., Chen C.P., Chen H.J., Tang S.L. Prevalence of potential nitrogen-fixing, green sulfur bacteria in the skeleton of reef-building coral Isopora palifera. Limnol. Oceanogr. 2016;61:1078–1086. doi: 10.1002/lno.10277. [DOI] [Google Scholar]
- 65.Tap J., Mondot S., Levenez F., Pelletier E., Caron C., Furet J.P., Ugarte E., Muñoz-Tamayo R., Paslier D.L., Nalin R. Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 2009;11:2574–2584. doi: 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- 66.McLellan S.L., Newton R.J., Vandewalle J.L., Shanks O.C., Huse S.M., Eren A.M., Sogin M.L. Sewage reflects the distribution of human faecal Lachnospiraceae. Environ. Microbiol. 2013;15:2213–2227. doi: 10.1111/1462-2920.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eren A.M., Sogin M.L., Morrison H.G., Vineis J.H., Fisher J.C., Newton R.J., McLellan S.L. A single genus in the gut microbiome reflects host preference and specificity. ISME J. 2015;9:90–100. doi: 10.1038/ismej.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Newton R.J., McLellan S.L., Dila D.K., Vineis J.H., Morrison H.G., Eren A.M., Sogin M.L. Sewage reflects the microbiomes of human populations. mBio. 2015;6:e02574-14. doi: 10.1128/mBio.02574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alam M.T., Amos G.C., Murphy A.R., Murch S., Wellington E.M., Arasaradnam R.P. Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathog. 2020;12:1–8. doi: 10.1186/s13099-019-0341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dehler C.E., Secombes C.J., Martin S.A. Environmental and physiological factors shape the gut microbiota of Atlantic salmon parr (Salmo salar L.) Aquaculture. 2017;467:149–157. doi: 10.1016/j.aquaculture.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chapagain P., Arivett B., Cleveland B.M., Walker D.M., Salem M. Analysis of the fecal microbiota of fast-and slow-growing rainbow trout (Oncorhynchus mykiss) BMC Genom. 2019;20:788. doi: 10.1186/s12864-019-6175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.San Diego-McGlone M.L., Azanza R.V., Villanoy C.L., Jacinto G.S. Eutrophic waters, algal bloom and fish kill in fish farming areas in Bolinao, Pangasinan, Philippines. Mar. Pollut. Bull. 2008;57:295–301. doi: 10.1016/j.marpolbul.2008.03.028. [DOI] [PubMed] [Google Scholar]
- 73.Grady E.N., MacDonald J., Liu L., Richman A., Yuan Z.-C. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Factories. 2016;15:203. doi: 10.1186/s12934-016-0603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Igai K., Itakura M., Nishijima S., Tsurumaru H., Suda W., Tsutaya T., Tomitsuka E., Tadokoro K., Baba J., Odani S. Nitrogen fixation and nifH diversity in human gut microbiota. Sci. Rep. 2016;6:1–11. doi: 10.1038/srep31942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sarria-Guzmán Y., Chávez-Romero Y., Gómez-Acata S., Montes-Molina J.A., Morales-Salazar E., Dendooven L., Navarro-Noya Y.E. Bacterial communities associated with different Anthurium andraeanum L. plant tissues. Microbes Environ. 2016;31:321–328. doi: 10.1264/jsme2.ME16099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tsoy O.V., Ravcheev D.A., Čuklina J., Gelfand M.S. Nitrogen fixation and molecular oxygen: Comparative genomic reconstruction of transcription regulation in Alphaproteobacteria. Front. Microbiol. 2016;7:1343. doi: 10.3389/fmicb.2016.01343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Angel R., Panhölzl C., Gabriel R., Herbold C., Wanek W., Richter A., Eichorst S.A., Woebken D. Application of stable-isotope labelling techniques for the detection of active diazotrophs. Environ. Microbiol. 2018;20:44–61. doi: 10.1111/1462-2920.13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lesser M.P., Morrow K.M., Pankey S.M., Noonan S.H. Diazotroph diversity and nitrogen fixation in the coral Stylophora pistillata from the Great Barrier Reef. ISME J. 2018;12:813–824. doi: 10.1038/s41396-017-0008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang L., English M.K., Tomas F., Mueller R.S. Recovery and Community Succession of the Zostera marina Rhizobiome after Transplantation. bioRxiv. 2020 doi: 10.1101/2020.04.20.052357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bernasconi R., Stat M., Koenders A., Huggett M.J. Global networks of Symbiodinium-bacteria within the coral holobiont. Microb. Ecol. 2019;77:794–807. doi: 10.1007/s00248-018-1255-4. [DOI] [PubMed] [Google Scholar]
- 81.Quigley K.M., Alvarez Roa C., Torda G., Bourne D.G., Willis B.L. Co-dynamics of Symbiodiniaceae and bacterial populations during the first year of symbiosis with Acropora tenuis juveniles. MicrobiologyOpen. 2020;9:e959. doi: 10.1002/mbo3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Waters C.M., Bassler B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 83.Bourne D., Iida Y., Uthicke S., Smith-Keune C. Changes in coral-associated microbial communities during a bleaching event. ISME J. 2008;2:350–363. doi: 10.1038/ismej.2007.112. [DOI] [PubMed] [Google Scholar]
- 84.Shnit-Orland M., Kushmaro A. Coral mucus-associated bacteria: A possible first line of defense. FEMS Microbiol. Ecol. 2009;67:371–380. doi: 10.1111/j.1574-6941.2008.00644.x. [DOI] [PubMed] [Google Scholar]
- 85.Leite D.C., Leão P., Garrido A.G., Lins U., Santos H.F., Pires D.O., Castro C.B., van Elsas J.D., Zilberberg C., Rosado A.S. Broadcast spawning coral Mussismilia hispida can vertically transfer its associated bacterial core. Front. Microbiol. 2017;8:176. doi: 10.3389/fmicb.2017.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.