Abstract
Changes in the genetic material can lead to serious human health defects, as mutations in somatic cells may cause cancer and can contribute to other chronic diseases. Genotoxic events can appear at both the DNA, chromosomal or (during mitosis) whole genome level. The study of mechanisms leading to genotoxicity is crucially important, as well as the detection of potentially genotoxic compounds. We consider the current state of the art and describe here the main endpoints applied in standard human in vitro models as well as new advanced 3D models that are closer to the in vivo situation. We performed a literature review of in vitro studies published from 2000–2020 (August) dedicated to the genotoxicity of nanomaterials (NMs) in new models. Methods suitable for detection of genotoxicity of NMs will be presented with a focus on advances in miniaturization, organ-on-a-chip and high throughput methods.
Keywords: nanomaterials, genotoxicity, in vitro 3D models, high throughput methods, miniaturization
1. Introduction
Nanomaterials (NMs) are materials with unique properties and size with at least one dimension between 1 and 100 nm, as defined by the European Commission (Regulation (EC) No. 1907/2006) [1]. They have already been in use for several decades, and are being utilized in almost every industrial sector. Due to constantly growing commercial applications and their presence in the majority of consumer products, NMs are being extensively investigated for their safety. A main concern with NM exposure is their genotoxic potential. Genotoxicity describes the property of chemical or physical agents that are able to alter the genetic information. Genotoxic events can be transient (repairable damage), or can lead to permanent changes (mutations) in the amount or structure of the genetic material in a cell. When a mutation is present in a germ cell, it is inherited to the next generation, and can cause a genetic disorder. In somatic cells, a mutation in a critical gene may lead to cancer. Thus, every mutagen is considered to be potentially carcinogenic.
A strategy for genotoxicity testing needs to cover all necessary endpoints: gene mutations, chromosomal aberrations (clastogenicity) and larger genome damage leading to aneuploidy. All these endpoints can be covered by the so-called two-test strategy, covering gene mutations, chromosomal aberrations (clastogenicity) and changes in number of chromosomes (aneuploidy). Because of the size-related special physicochemical properties of NMs, many standard test methods are not suitable for the toxicity testing of NMs or the test protocols have to be optimized. For assessing gene mutations caused by NMs, the Ames test is not suitable owing to the small size of bacteria (comparable with NMs) as well as the bacterial cell wall which is different from the human/mammalian cell membrane [2,3,4]. Thus, in the test strategy the mammalian gene mutation test is recommended together with the micronucleus assay [4,5]. Additionally, many other assays exist addressing intermediate endpoints, such as various types of DNA damage and novel markers as omics or epigenetic changes [6]. These markers can give supporting evidence about the safety of tested compounds, including NMs [7].
Standard in vitro genotoxicity tests are typically based on a single cell type. However, a key observation in in vivo genotoxicity studies conducted with NMs is that the permanent changes induced in DNA are often the result of secondary genotoxicity associated with inflammation [8,9,10,11]. Thus, there has been a strong effort in recent years to develop more complex, in vivo-like in vitro models based on 3D structures either of a single cell type or as co-cultures of two or more cell types. The application of these models in toxicology is believed to provide reliable data that are more relevant for evaluating genotoxicity in humans than standard 2D models. Several of these models are already well advanced, and new protocols have been established for example for skin, lung and liver tissue models [4,6]. The advanced 3D models are technically more difficult to perform and more time-consuming; however, they can better address exposure scenarios, such as lung models reflecting inhalation exposure, skin models for dermal exposure, and liver models for systemic or oral exposure. Another advantage of such advanced models is that most 3D models are based on human cells, enhancing the relevance of the results for assessing potential human genotoxicity. As there are huge numbers of NMs to be tested, research in in vitro toxicology focuses on miniaturization and development of robust high throughput methods. An overview of standard and novel genotoxicity tests, advanced models and high throughput methods for application with NMs is provided below [12,13,14].
2. Methods
Our approach was to describe the main endpoints applied in new advanced in vitro 3D models, organ-on-chip and high throughput methods suitable for detection of genotoxicity of NMs. An electronic literature search was conducted through PubMed databases to identify articles dealing with genotoxicity of NMs and advanced in vitro models and high throughput methods from year 2000–2020 (August). The following keyword combinations were used: “genotoxicity & 3D in vitro model”, “genotoxicity & advanced in vitro model”, “genotoxicity & high throughput”, “genotoxicity & high throughput & nanomaterials“, “genotoxicity & high throughput & nanoparticles”, “genotoxicity & organ on chip”, “genotoxicity & 3D models & nanoparticles”, “genotoxicity & 3D models & nanomaterials”. As publication type, both research articles as well as reviews were considered.
3. Results of Literature Search
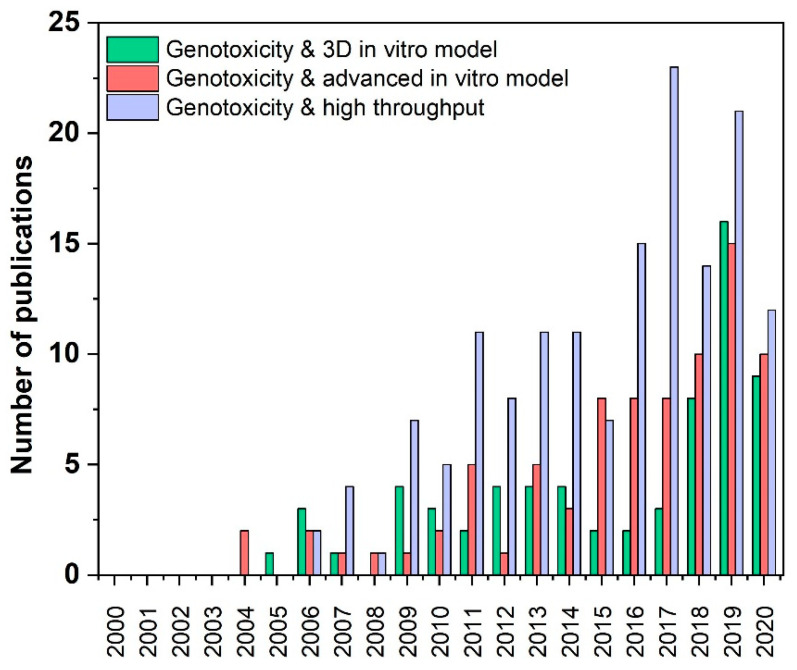
The literature search resulted in 59 papers for “genotoxicity & 3D in vitro model”, 73 papers for “genotoxicity & advanced in vitro model”, 134 papers for “genotoxicity & high throughput”, eight papers for “genotoxicity & high throughput & nanomaterials” [15,16,17,18,19,20,21] and six papers for “genotoxicity & high throughput & nanoparticles” (4 are the same as for “genotoxicity & high throughput & nanomaterials”) [16,17,18,19,21,22], seven papers for “genotoxicity & organ on chip“ [23,24,25,26,27,28,29], seven papers for “genotoxicity & 3D models & nanoparticles” [30,31,32,33,34,35,36], 11 papers for “genotoxicity & 3D models & nanomaterials” [30,31,33,36,37,38,39,40,41] for the period of 20 years (2000–2020), whereby the year 2020 was considered only from January to August. (Table 1). 265 (88.0%) out the overall 301 studies have been published in the last ten years and 91 (30.2%) out the overall studies in 2019 and 2020 (Table 1). The accelerated publication of papers on these topics over the last few years clearly shows that the importance of the fields of research is increasing (Figure 1). This is partly due to the development of alternative methods to animal testing, as required by REACH (Registration, Evaluation, Authorisation and Restriction of Chemicals) [42], and to the ever-increasing implementation of the 3Rs (Replace, Reduce, Refine) principle [43].
Table 1.
Number of publications during the last 20 years in the field of advanced in vitro 3D models, organ-on-chip and high throughput methods suitable for detection of genotoxicity of nanomaterials. An electronic literature search was conducted through PubMed databases to identify articles dealing with genotoxicity of nanomaterials and advanced in vitro models and high throughput methods from year 2000–2020 (August). The listed keyword combinations were used.
| Keywords of Literature Search | Number of Publications Per Time Period | ||
|---|---|---|---|
| 2000–2020 | 2010–2020 | 2019–2020 | |
| genotoxicity and 3D models in vitro | 59 | 47 | 23 |
| genotoxicity and advanced in vitro model | 73 | 63 | 23 |
| genotoxicity and & high throughput | 134 | 120 | 32 |
| genotoxicity and organ on chip | 7 | 7 * | 2 |
| genotoxicity and 3D models & nanoparticles | 7 | 7 ** | 1 |
| genotoxicity and 3D models & nanomaterials | 11 | 11 *** | 5 |
| genotoxicity and high throughput & nanoparticles | 6 | 6 *** | 4 |
| genotoxicity and high throughput & nanomaterials | 8 | 8 *** | 4 |
* first paper published in 2013; ** first paper published in 2014; *** first paper published in 2011.
Figure 1.
Number of publications during the last 20 years in the field of advanced in vitro 3D models, organ-on-chip and high throughput methods suitable for detection of genotoxicity of nanomaterials. An electronic literature search was conducted through PubMed databases to identify articles dealing with genotoxicity of nanomaterials and advanced in vitro models and high throughput methods from year 2000–2020 (August). The number of publications is assigned to the individual years.
If one differentiates again all identified publications on the topic of “genotoxicity & 3D models & nanomaterials/nanoparticles/organ on chip”, with regard to review articles and research studies and studies mentioned twice, it becomes clear that much less is known and so far only ten studies have been published dealing with in vitro 3D models and organ-on-chip systems for the detection of genotoxicity of NMs. A detailed overview of the study designs is shown in Table 2.
Table 2.
Overview of the in vitro studies published in the field genotoxicity of nanomaterials in combination with a 3D model, organ-on-chip and high throughput methods. All publications found via PubMed literature search to the following keyword combinations are listed in this table. “genotoxicity & 3D models & nanoparticles”, “genotoxicity & 3D models & nanomaterials” and “genotoxicity & organ on chip”. ECL: electrochemiluminescent (no nanomaterial tested); CFA: colony forming assay; MNA: micronucleus assay; IFL: immune fluorescence; SWCNT: single walled carbon nanotube; MWCNT: multi walled carbon nanotube; UFCB: ultrafine carbon black; ASB: crocidolite asbestos; ZnO: zinc oxide; TiO2: titanium dioxide; Ag: silver; LD: live/dead; n.sp.: not specified; LDH: lactate dehydrogenase; ATP: adenosine tri-phosphate; SiO2: silica; ZrO2: zirconium dioxide; TEM: transmission electron microscopy; RPD: relative population doubling analysis; NM: nanomaterial.
| Human Cell Origin | Cell Model | Culture Dimension | Endpoints | Test Methods | NM Tested | Year | Ref |
|---|---|---|---|---|---|---|---|
| Nasal mucosa | mini organ culture | 3D | cytotoxicity, genotoxicity | comet assay, caspase–3 ELISA, ROS assay, TEM | ZnO | 2011 | [44] |
| lung | EpiAirway™ | 3D | cytotoxicity, genotoxicity | comet assay, LDH assay, ATP assay | Ag, SiO2, ZrO2 | 2017 | [33] |
| lung | small airway epithelium | 2D | cytotoxicity, genotoxicity | apoptosis assay, CFA, p53 downregulation, IFL, γH2AX assay, spheroid formation assay | SWCNT, MWCNT, UFCB, ASB | 2019 | [39] |
| skin | EpiDerm™, TK6 | 2D & 3D | cytotoxicity, genotoxicity | MNA, TEM, RPD analysis | SiO2 | 2016 | [32] |
| liver | HepG2 | 2D & 3D | cytotoxicity, genotoxicity | comet assay, LD staining, alamarBlue™ assay | Ag, ZnO, TiO2 | 2020 | [34] |
| liver | HepG2 | 3D | cytotoxicity, genotoxicity, liver functionality | MNA, cytokine secretion | Ag, TiO2 | 2020 | [38] |
| liver | HepG2, HepaRG | 3D | cytotoxicity, genotoxicity, liver functionality | MNA, TB assay, albumin level, urea level | ZnO | 2020 | [37] |
| liver | micro tissue | 3D | cytotoxicity, genotoxicity, liver functionality | comet assay, LD staining, adenylate kinase assay, cytokine secretion, comet assay, albumin ELISA, CYP3A4 activity, lipid peroxidation assay | Ag, ZnO, MWCNT, TiO2 | 2014 | [31] |
| liver, blood, breast | HepG2, TK6, MCF7 | 2D | genotoxicity | comet assay, Epi-comet assay | - | 2017 | [24] |
| liver lung kidney, intestine | n. sp. | 2D | genotoxicity | ECL fluidic chip LC-MS/MS | - | 2015 | [25] |
4. General Mechanisms of Genotoxicity
The genotoxicity of NMs and mechanisms leading to transient or permanent genetic changes have been intensively investigated [8,9,10,11,12,13]. Recent studies show that NM genotoxicity can result from two main mechanisms; primary (direct or indirect) or secondary genotoxicity. For certain NMs, one of these mechanisms might apply; however, for some NMs both mechanisms can occur simultaneously after NM exposure.
Primary direct genotoxicity is caused by direct interaction of NMs with the genome, and requires physical contact of NMs with DNA in the nucleus. This interaction could lead to DNA damage such as DNA breaks and other DNA lesions, large DNA malformation, or chromosomal damage. The primary indirect mechanism arises from NM-induced reactive oxygen species (ROS), or from toxic ions released from dissolution of NMs and production of intracellular ROS via the Fenton-type reaction [5,12]. Oxidative stress is considered a key mechanism in primary indirect genotoxicity. Free radicals may interact with cellular biomolecules including DNA, leading to potentially serious consequences. ROS may attack the DNA causing purine or pyrimidine oxidation lesions and strand breaks. The damage can be repaired but can also result in gene mutations and even larger chromosomal damage. NMs can promote DNA damage also via other molecules that either have the capacity to interact with DNA or interfere with DNA replication and cell division. NMs can interact for example with protein kinases responsible for regulation of cell cycle events, such as DNA replication and cell division [11].
For understanding whether primary genotoxicity is direct or indirect, it is important to study the mechanism of uptake and whether NMs can enter the nucleus. The smallest NMs (with a size of only a few nm) could penetrate the nucleus via nuclear pores. However, some studies show the presence of larger NMs in the nucleus, indicating that there could be other pathways for nuclear uptake, for example intracellular processes resembling endocytosis [45]. NMs can enter the nucleus also during mitosis when the nuclear membrane is dissolved. Once in the nucleus, NMs might directly interact with DNA, or with DNA organized in chromosomal structures, depending on the stage of the cell cycle (direct genotoxicity). During interphase, NMs could interact or bind with DNA molecules and interfere with DNA replication and transcription of DNA into RNA. NMs can also interfere with the spindle apparatus, the centrioles and associated proteins and disturb the process of mitosis, thus leading to the formation of micronuclei that could be manifested as clastogenicity or aneugenicity [46].
Secondary genotoxicity is considered to be the main mechanism of genotoxicity of NMs. It is mediated via ROS produced by inflammatory cells. NMs can trigger an oxidative burst caused by activation of phagocytes. This inflammation is an initial defense mechanism against invasion of microorganisms or foreign materials, involved in clearance. In case clearance of e.g., inhaled NMs fails, it can cause a chronic immune cell response. This type of secondary genotoxicity is not possible to study with standard in vitro approaches, and has until recently been investigated only in vivo following chronic inflammation caused by activation of immune cells, such as macrophages or neutrophils [8,47]. An in vitro approach to studying secondary genotoxicity has many challenges, as standard monoculture systems cannot be used to assess the ability of NMs to induce genotoxicity via inflammation. Advances in new models thus focus on co-culture of target cells with cells from the immune system.
Several studies show that genotoxicity of NMs not only depends on dose and exposure duration, but also on their size, surface properties, chemical composition as well as on shape [48,49,50,51].
Huk et al. investigated three silver (Ag) NMs with same surface properties but different sizes and found that Ag NMs genotoxicity depends on NM size [52]. Comparative in vitro genotoxicity studies of zinc oxide (ZnO) nano- and macroparticles cells also came to the conclusion that size matters [53]. Guo et al. summarized their studies, that coatings have less effect on the relative genotoxicity of Ag NMs than the particles size [54]. With both the micronucleus and mouse lymphoma assay, the smaller the Ag NMs, the greater the cyto- and genotoxicity [54]. Also other studies investigated the contribution of nano- and micro-sized fractions (titanium dioxide (TiO2) [55], gold (Au) [56], cobalt chrome [57], polystyrene [58], silica [59], Ag [54,60,61], Kaolin [62], ZnO [32], copper (Co) [63]. Using the mini-gel comet assay Lebedová et al. studied the size-dependent genotoxicity of Ag, Au and platinum (Pt) NMs [64]. The authors detected size- and material-dependent DNA strand breaks [64].
In a study with Ag NMs of same size the authors found that genotoxicity and mutagenicity of nano-Ag depends on their surface properties and charge [65]. Similarly Gabelova et al. showed that surface chemistry affects genotoxicity of iron oxide NMs [66].
Another study compared the effect of size and surface functionalization of Au NMs on genotoxicity [67]. Two core sizes (5 and 20 nm) and three functionalizations (negative, positive, and neutral surface charges) were examined by Vales et al. [67]. Surface charge-dependent effects have been identified [67]. Further studies found that silica oxide (SiO2)-coated nanosized TiO2 induced less DNA damage than uncoated [68]. This finding may be associated with abilities that reduce the formation of free radicals mediated by TiO2 NMs [69], change NMs’ agglomeration stage [70] and influence the interaction with biological components [71]. A relationship between genotoxicity and surface composition has also been demonstrated for many other metal and metal oxide particles [61,63,71,72,73,74,75] as well as for biopolymer particles [76].
Shape-dependent genotoxicity has also been studied in vitro [77,78]. Different shapes (bipyramids, rods, platelets) of TiO2 NMs have been compared to commercial TiO2 NMs [78]. Also sphere- and rod-shaped mesoporous SiO2 NMs have been screened regarding their genotoxic potential [77].
Besides shape, surface properties and size, also the chemical composition plays an important role in NM genotoxicity [61,79]. Studies on genotoxicity of metal NMs (Zn, TiO2, Ag, iron oxide, etc.) suggest this [61,79].
The interrelationship between particle shape, size, chemical composition and toxicological effects has been explored by Yang et al. [80]. Compared with ZnO NMs, carbon nanotubes were moderately cytotoxic but induced more DNA damage determined by the comet assay [80]. This comparative study demonstrates that chemical composition plays a major role in cytotoxicity and the size in genotoxicity [80].
Among different types of metal or metal oxide nanoparticles, the genotoxic potential of carbon-based materials such as SWCNT [81], multi wall carbon nanotube (MWCN) [82,83] or graphene [84,85,86], have also been evaluated in vitro. High-aspect-ratio MWCNTs were found to be more toxic than the low-aspect-ratio MWCNTs [83]. Studies on interactions of graphene oxide and graphene nanoplatelets with the in vitro intestinal barrier model resulted in no oxidative damage induction [87]. Whereas MWCNTs induced significant DNA damage [88,89], an increase in 8 nitroguanine [88,90] and an increase in micronucleused cells [88,91].
Transformation processes in the environment, including chemical, physical and biological processes could affect the toxic potential of NMs. So called aging processes may change the surface properties as well as chemical composition and size of the NMs. Few studies are available investigating the effect of aging on the genotoxic potential of NMs [92,93,94,95,96,97] assuming an influence on genotoxicity, but too little data are available to be able to conclude.
5. Nanomaterials–Characteristics in Cell Culture Media and Assay Interference
NMs can be divided into broad categories of materials, such as metals (silver, gold, copper), metal oxides (titanium dioxide, iron oxide, zinc oxide), carbon based NMs (single- or multi-walled carbon nano tubes), different types of polymers and further advanced NMs such as complex, hybrid, multi-component or multi-structure NMs. NMs not only vary in their chemical composition, but also in size, size distribution, shape, surface characteristics, surface area etc. [98]. In vitro studies between different laboratories often contradict one another, due to an inadequate NM characterization under the study conditions. For the most cellular assays, NMs stock solutions are diluted in cell culture medium (CCM). In a second step this suspension is applied to the cultured cells in vitro. When NMs are suspended in CCM or other biological fluids, they change their particle surface, the associated particle-particle and particle-cell interactions [99,100]. This is dependent on the characteristics of the primary NM (composition, size, shape, surface chemistry) and the properties of the surrounding fluid (pH, ionic strength, protein content, temperature) [16,17]. CCM containing fetal bovine serum leads to a formation of so-called protein coronas, which change the physicochemical nature and toxicological behavior of the NMs so enormously, that they cannot be compared any longer to untreated NM stock solutions [99]. Thus, the characterization of NMs under test conditions is indispensable to understand their impact on cellular systems and to interpret the test result accurately [101,102,103]. Typically, the size and size distribution of NMs are determined by light scattering techniques such as dynamic light scattering (DLS) and nanoparticle tracking analysis (NTA). On the other hand, shape and size (distribution) can be directly visualized by microscopic techniques, such as transmission electron microscopy (TEM), scanning electron microscopy (SEM) and atomic force microscopy (AFM) [104]. The surface charge of NMs can be characterized by the determination of the Zeta potential, which is a method to measure the electrostatic potential at the electrical double layer, which surrounds NMs in suspension [105]. Field flow fractionation (FFF) is a powerful sizing technique and in combination with UV- and multi-angle light scattering (MALS)-detectors it is suitable for separation of different particle sizes in a low concentration range. When combined with inductively coupled plasma-mass spectrometry (ICP–MS), this technique allows determination of the (ion) composition and concentration of NM test solutions [106]. Moreover, characterization regarding interference of NMs with the applied assay system is highly recommended. It has been shown that some NMs interfere with fluorescence/absorption in widely used assays such as alamarBlue™, Neutral Red or WST-1, which can lead to false positive cytotoxicity results unless interference test is included in the test [107,108].
6. Cytotoxicity Testing before Genotoxicity Studies
Before genotoxicity testing can be performed, it is necessary to know the cytotoxicity of the tested NMs and to establish the LC50 (lethal concentration- at which 50% of cells will die) in order to choose the appropriate range of concentrations before genotoxicity testing. The dose range depends on the type of genotoxicity test applied to the NMs. For the gene mutation and micronucleus assays, the concentration range should cover non-cytotoxic concentrations and up to 50 ± 5% cell death. For assays that are detecting DNA breaks, NMs are tested only at non-cytotoxic concentrations, as DNA breaks can also be induced indirectly as a consequence of cytotoxicity.
DNA fragmentation is a step in the cell signaling cascade for induction of apoptotic cell death, downstream of caspase 3 activation and cleavage and activation of the DNA fragmentation factor DFF, and many apoptotic assays are based on detection of DNA fragmentation [109]. As DNA breaks are coupled to the cell death process, false positive results and classification of cytotoxic compounds as genotoxic can occur if cytotoxicity is not an integrated part of the genotoxicity testing. However, there is no consensus when it comes to the threshold value for cytotoxicity. Strictly, cytotoxicity is stated from 80% viability (20% cytotoxicity compared to negative control), but others consider that the cut-off for cytotoxicity in genotoxicity testing should be as low as around 50% viability [110,111].
Cytotoxicity can be tested by different assays [112,113]. The most significant and meaningful tests are based on the ability of cells to proliferate or to survive and form colonies, i.e., viability. These tests are interference-free, and the read-out is based directly on cell survival. The plating efficiency, relative growth activity and colony forming efficiency assays contain all previously described characteristics [22]. Plating efficiency and relative growth are integrated parts of OECD Test Guidelines 476 and 490 [22] (for mammalian gene mutation tests (OECDTG 476, 1997; OECD TG 490, 2015) [114,115]. Alternative measures of cytotoxicity (rather than viability) can be based on membrane integrity and application of membrane impermeable dyes, allowing staining only of cells with damaged cell membrane, such as the Trypan blue exclusion test. Another common method is dual staining with the fluorescent dyes propidium iodide for dead cells and fluorescein diacetate for live cells, and analysis by microscopy or flow cytometer. Dead or dying cells can be detected by leakage of intracellular substances, as lactate dehydrogenase (LDH), or by other colorimetric or fluorometric methods, such as staining with dyes requiring metabolic activation for detection. A common and convenient test is the alamarBlue™ assay, based on reduction of Resazurin to the highly fluorescent Resorufin, or the similar MTT test. Results of cytotoxicity assays that are not based on cell survival should be interpreted with caution; the effects they measure may be reversible, such as damage to the membrane, and so might not indicate cell death. It is important, when selecting a cytotoxicity test for application in genotoxicity testing, that the same exposure conditions should apply to both tests, and that the tests should be performed in parallel. If genotoxicity is measured after, e.g., three and 24 h exposure, as is common for the comet assay, then cytotoxicity should also be measured after three and 24 h’ exposure. The alamarBlue™ assay is a convenient and reliable cytotoxicity assay to be used in combination with the comet assay, which has been tested and found to be applicable with several different NMs [6,26,36].
7. Biomarkers for Genotoxicity
Genotoxicity biomarkers include a battery of assays that cover DNA damage, gene mutations and chromosomal damage as sensitive genotoxicity endpoints. In spite of the limited data and the lack of information concerning NM safety, the number of produced NMs is in constant increase: thus in several past and on-going European projects, such as NanoTEST, NanoGENOTOX, NanoReg, HISENTS, RiskGONE and others, efforts have been made to understand the NMs mode of action and to develop or adapt validated OECD test methods used on chemicals for use on NMs. The biomarkers of genotoxicity have been extensively applied to nanotoxicology risk assessment.
For the risk assessment of NMs in vitro, two test strategy is recommended: (a) mammalian gene mutation test, and (b) test for chromosomal damage.
7.1. Mammalian Gene Mutation Test
The most commonly used assays for gene mutation of NMs are based on Tk (thymidine kinase) or Hprt (hypoxanthine phosphoribosyltransferase) genes as described in OECD Test Guidelines (TGs) (OECD TG 476, 2015 and OECD 490, 2015) [114,115], with small adaptations especially regarding NM characterization and exposure [41]. V79 cells were used for detecting Hprt gene mutants after exposure with silver, titanium dioxide, carbon nanotubes, and other NMs [13,31,32,33,116]. The mouse lymphoma (MLA) assay detects a wide spectrum of genetic damage, including gene deletion, as well as epigenetic silencing of the functional Tk allele due to promoter hypermethylation [117]. Several NMs, such as different shapes of silver and iron oxide, were investigated with this assay [66].
7.2. Chomosomal Damage
Clastogenicity and aneugenicity are important genotoxicity endpoints that identify agents causing structural chromosome or chromatid breaks, dicentrics and other abnormal chromosomes, including loss of chromosomes. These could be detected by chromosomal aberration tests. As this test is time consuming, the most common test to detect chromosomal damage is the in vitro micronucleus assay that detects micronuclei in the cytoplasm of interphase cells [118]. Micronuclei are formed from chromosome, chromatid fragments or whole chromosomes that lag behind in cell division forming single or multiple micronuclei in the cytoplasm. The micronucleus assay detects both structural chromosome damage (clastogenic effect) as well as numerical chromosome alterations (aneugenic effect) [119]. They can be measured by visual or automated scoring after staining slides. The OECD TG 487 (2016) [120] describes the cytokinesis-block micronucleus (CBMN) assay in different cell models, including human or other mammalian cells and cell lines. Cytochalasin B is an agent that is used to block cytokinesis, and thus to prevent separation of daughter cells after mitosis, leading to the formation of binucleated cells. This assay is often used for NM testing; however, exposure with NMs should occur before cytochalasin B is added to allow uptake of NMs by cells [15,31,121].
7.3. DNA Damage
Detection of DNA damage (single and double strand breaks and specific DNA lesions) belongs to the group of so-called indicative tests that are also used in hazard assessment of NMs [16,122,123]. DNA strand breaks can be evaluated by the comet assay which is the method of choice for measuring DNA damage in cellular DNA [124]. This assay (also known as single cell gel electrophoresis) is a simple, robust, reliable and user-friendly method, widely used for many years for genotoxicity testing of chemicals, and it is the most commonly applied method to test genotoxicity of NMs [17]. The high sensitivity of the assay, detecting from about 100 up to several thousand breaks per cell, allows detection also of weak genotoxic agents. However, the high sensitivity of the assay may contribute to variability, so to ensure high reproducibility of results, it is important to keep standardized conditions and a constant experimental design.
While the basic comet assay detects strand breaks, a common modification by incorporating digestion with a lesion-specific endonuclease after the lysis step allows detection of damaged bases. Formamidopyrimidine DNA glycosylase (Fpg) has been particularly useful; its primary substrate is the oxidized base 8-oxoGua, and it has therefore been employed to measure the effects of oxidative stress on DNA [125].
DNA double strand breaks (DSBs) and DNA damage foci are DNA damage response biomarkers [126]; local accumulations or modifications of DNA damage response proteins that form at the sites of DNA DSBs can be visualized through microscopic imaging following immunocyto- or -histochemical detection or fluorescent protein tagging [127]. DSBs, among the most severe forms of DNA damage, are triggered by various genotoxic insults and induced directly by a number of physical and chemical agents including NMs [128]. γH2AX is one of the DNA damage response proteins that accumulate and/or are modified in the vicinity of a chromosomal DNA DSB to form microscopically visible, subnuclear foci, contributing to their repair [127]. A number of studies have proposed C-termini phosphorylated histone protein, γH2AX, as a potential biomarker of DNA DSBs caused by genotoxicants [67,129,130,131]. This method is widely used in different fields including in vitro toxicology testing of environmental pollutants [132]. Initially this biomarker was used for identifying ionizing radiation effects [133]. In the last years γH2AX phosphorylation technique has been used in several studies to measure DSB caused by different NMs [15] for instance carbon nanotubes [134,135], zinc oxide [136], gold [137], silica [138], polystyrene [139] and titanium dioxide NMs [50] showing that this technique can be a useful tool to assess the genotoxic potential of NMs [15].
8. Carcinogenicity
Crucial for safety assessment of chemicals and NMs is their carcinogenic potential. The observation that a compound is genotoxic implies that it may be potentially carcinogenic. An additional aspect concerning carcinogenicity is the existence of non-genotoxic carcinogens, which induce their effect through secondary mechanisms, such as oxidative stress or other inflammatory responses [140,141,142]. Until recently, the only recognized test for carcinogenicity was by animal testing, such as the 2-year rodent assay. As an ethical response to the importance of reducing the use of animals in safety assessment of chemicals, tests have been developed as alternative methods for detection of carcinogenic potential. An additional important aspect, particularly due to the large numbers of new chemicals and engineered NMs, is the high cost attributed to the animal testing. A ban on in vivo testing of cosmetics has also increased dependence on in vitro tests.
The cell transformation assay (CTA) is an in vitro approach that makes us of the phenotypic transformation of cells as a marker of carcinogenicity [143,144]. Cells transformed in vitro have been shown to induce tumors when injected into immunosuppressed animals. Cells used in CTA are often derived from rodent embryos, such as the Syrian hamster embryo (SHE), mouse BALBc 3T3, Bhas42 and C3H/10T cells. The CTA assays have been met with some reservations and are still not accepted for regulatory purposes. The main argument has been the lack of understanding of the mechanisms behind the transformation in addition to the subjective nature of the assessment of the transformations. A previous study on the Syrian hamster embryo cell transformation assay also reported a high rate of false positives and limitations in the ability to distinguish between rodent and human carcinogens [145]. Recent works have, however, shown that cell transformation assays are useful tools to predict carcinogens [146]. In particular, the CTAs can be useful to identify non-genotoxic carcinogens and should be included as an integral part of a battery of in vitro tests in order to predict carcinogenic potential of chemicals and also NMs [147]. Fontana et al. investigated four different amorphous non-genotoxic silica particles with the CTA and indicated one non-genotoxic carcinogenic out of two pyrogenic and two precipitated silica materials [147].
9. New Endpoints of Genotoxicity
Among new endpoints in the study of NM genotoxicity, gene expression and epigenetics have to be taken into account. NM exposure has been reported to induce de-regulation of genes involved in several biological processes, including the DNA damage response and DNA repair, cell cycle progression, oxidative stress and inflammatory responses [148,149,150,151,152,153,154,155]. It is relevant to point out that many of these processes have been shown to lead to secondary genotoxicity [156]. Changes in the regulation of gene expression can occur in response to molecular signaling activated within or between the cells after interaction with chemicals and/or NMs.
Another important class of mechanisms affecting gene expression involves epigenetics, i.e., the study of molecules and mechanisms that can perpetuate alternative gene activity states in the context of the same DNA sequence [157]. During the last decade, epigenetics has become one of the most important areas in the study of biological sciences. Some epigenetic responses have been suggested as possible biomarkers of exposure or disease risk, although the mechanistic links between the endpoints and the outcomes still need to be discovered [158]. Epigenetic responses include, among others, DNA methylation, non-coding small single-stranded RNAs termed microRNAs (miRNAs) and histone modifications. Several metallic, non-metallic and carbon-based NMs have been reported to affect epigenetic mechanisms [53,54,56]. DNA methylation is an important mechanism for the maintenance of cell- and tissue-specific gene expression. The methylated status of the DNA is defined by the addition of a methyl group to the fifth position of the cytosine residue in the dinucleotide sequence CpG [158]. Global hypomethylation has been linked to chromosome instability and oncogenesis, while changes in the methylation status of gene promoter regions entail an alteration of the expression of the associated gene. Changes in the methylation of the DNA, both global and gene-specific, have been reported as an effect of exposure to diverse NMs including silver, silicon dioxide, and silica and carbon nanotubes [52,54,57]. miRNAs are involved in the regulation of gene expression at post-transcriptional level, by affecting the stability of mRNA, or targeting them for degradation. Altered miRNA expression has been related to pathologies including cancer, but also cardiovascular, developmental and neurological diseases. Changes in miRNA expression induced by gold, titanium dioxide and iron nanoparticle exposure have been repeatedly reported in in vitro and in vivo studies [155,156]. Histone modifications include several mechanisms, e.g., acetylation, methylation of lysine residues, phosphorylation, ubiquitination and ATP-ribosylation. These alterations result in chromatin remodeling that consequently influences gene transcription. This is the least studied and understood mechanism in epigenetics, although some studies have reported histone interactions with NMs, such as cadmium telluride quantum dots, soft NMs used as scaffolds for biological and medical applications, and gold NMs [156].
Although genetic and epigenetic changes induced by exposure to NMs have been reported, the causal relationship with the onset of diseases is still poorly understood. There is a need for mechanistic studies associating the molecular responses with the functional effects, e.g., linking the alterations of gene expression with cellular behavior, such as cell migration and invasion, and DNA damage repair capacity. In vitro research on NMs and epigenetics has mainly involved short-term studies [158,159]. Long-term studies with chronic exposures to low doses of NMs are needed to better mimic real human exposure, and to clarify genetic and epigenetic modifications.
Standard molecular biology techniques such as quantitative real time PCR (qPCR), sodium bisulphite sequencing, and pyrosequencing are commonly used to investigate DNA methylation and miRNA expression. These are relevant methods in the study of molecular mechanisms underlying the functional effects. They can for example be coupled with other techniques and genome editing methods such as small interfering RNA (siRNA) or the most recent CRISPR-cas9 to investigate the role of specific genes by selectively altering their expression. However, they have limitations in terms of time, and being labor-intensive and low-throughput [154], although methods development such as high throughput real-time PCR platforms may provide promising systems for the simultaneous processing of large numbers of samples [160]. Besides, a modified version of the comet assay is under development for high throughput screening (HTS) of DNA methylation alterations, and has the potential for testing NMs [161].
10. High Throughput Methods
HTS methods are defined as the use of automated tools to facilitate rapid execution of a large number and variety of biological assays that may include several test substances in each assay [162]. 134 publications have been found applying the keywords “genotoxicity & high throughput“ [163,164,165,166,167,168], although only 14 of them related to NMs (Table 1).
In a recent publication developed under EC FP7 NanoREG project, several scientists agreed that the rapid growth of NM production, and the current low throughput, time consuming and laborious approaches for evaluating genotoxicity of the NMs should lead to the development of rapid, efficient and high throughput genotoxicity testing strategies for safety/risk assessment of NMs [15]. HTS methods are needed to allow toxicity testing of large numbers of materials in a timely manner and with savings in terms of labor costs [169].
In a recent review, several existing genotoxicity testing methods which are amenable to HTS/high content screening (HCS) approaches were identified [15]. HTS was introduced in the pharmaceutical and chemical industries as a rapid way of evaluating unwanted effects of novel compounds. HTS in vitro facilitates the hazard ranking of NMs through the generation of a database with all reported effects on biological and environmental systems; thus novel NMs can be prioritized for in vivo testing [170]. Ideally, this screening should allow for mechanistic profiling to better inform on hazard identification and to improve risk assessment strategies [171].
Regulatory in vitro genotoxicity testing exhibits shortcomings in specificity and mode of action (MoA) information [172]. Examples of HTS methods for genotoxicity testing of NMs are described in the following:
10.1. Comet Assay
Recent miniaturized versions of the assay- with 12 mini-gels per slide [173], 96 mini-gels on a GelBond film [174,175] a special 96-well multichamber plate (MCP) [176] -allow increased throughput and analysis of numerous types or modifications of NMs in a time- and cost-effective manner. Numerous studies have been published to date on genotoxicity of NMs with the semi-HTP version of the comet assay [53,177,178,179]. To improve the very time-consuming DNA damage evaluation by manual microscopic fluorescence-analysis, several automatic systems for image analysis have been developed to help increase the throughput of the assay for high screening capacity; for instance the fully automated slide-scanning platform Metafer and the MetaCyte CometScan software. Using such automated evaluating systems, the analysis duration is reduced from hour-range to minute-range. Versatile systems conveniently automate a wide area of image analysis applications in microscopy for life sciences and can be adapted to other assays when needed.
10.2. In Vitro Micronucleus Assay
Different approaches have been proposed until now to increase the speed of the micronucleus assay in vitro. Classically, the long and tedious visual scoring of slides has been changed by using automated platforms scoring different set of slides in each run [50,60,72,73,74]. The micronucleus assay using HTS/HCS platforms has been widely used in NM research; it requires less test material than conventional test methods, and has a greater compatibility with high throughput screening instrumentation [75,76,77,78,180,181,182]. The standard protocol involves (1) the lysis of membranes by a non-ionic detergent; (2) the use of one or more nucleic acid dyes that can permit discrimination between the liberated nuclei and micronuclei, according to their DNA-dye-associated fluorescence intensities. A high throughput change in the micronucleus assay includes the use of flow cytometry for the separation of micronuclei and nuclei [183,184,185,186,187]. Further modifications use 96-well plates in conjunction with a robotic auto-sampling device. Different commercial automatic scoring devices are now available [178].
10.3. γH2AX Assay
Various computer-based approaches have developed the conventional γH2AX assay into a highly efficient technique, which is therefore also suitable for high throughput [82,83]. In addition, this second generation of the γH2AX assay allows the analysis of further parameters in a cell population. Supported by image analysis software tools, foci are identified and quantitative foci parameters are calculated [188,189,190,191,192]. The successful application of this method with iron oxide NMs has been demonstrated by Harris et al. [193] analyzing the H2AX phosphorylation by applying a high content in silico platform.
10.4. ToxTracker Assay
The ToxTracker assay (mechanism-based reporter assay based on embryonic stem cells that uses GFP-tagged biomarkers for detecting DNA damage, oxidative stress and general cellular stress) is a tool that could prove useful in the field of NM toxicology allowing for high throughput screening [18,194].
For an even more high throughput approach, (epi-)genome wide analyses, e.g., based on microarray technology (gene or miRNA microarrays), and next generation sequencing such as massive parallel sequencing, allow investigation of changes across the whole (epi-)genome, and can be used, e.g., to screen NM exposure effects on gene expression and epigenetic endpoints such as miRNA alterations. Besides the higher costs of these techniques, difficulties include analysis of the huge amount of data produced, and the evaluation of confounding factors. Powerful bioinformatics tools are needed for data analysis and integration, but genome-wide investigations on the epigenome and transcriptome, coupled with statistical modeling are a relevant approach for the investigation of associations and causal relationships between NM exposure, molecular effects and adverse health outcomes [154].
11. New Advanced In Vitro Models (3D, Organ-on-a-Chip)
The development of advanced in vitro models for nanotoxicity assessment is greatly encouraged by the principles of 3Rs, which are embedded in international legislation and regulations on the use of animals for scientific purposes [43]. The goal of 3Rs is to avoid animal experiments whenever it is possible (replace), to limit the number of animals (reduce) and their potential suffering to a minimum (refine). New advanced models such as three-dimensional (3D) in vitro models or organ-on-a-chip technologies (OOC) have the potential to serve as alternative methods to replace animal testing and close the gap between in vivo and two-dimensional (2D) in vitro models [195].
When comparing complex organs with cells cultured in standard 2D monolayers, 2D cultures do not have the ability to represent the functionality of an organ, whereas cells cultured in 3D resemble the organ structure better, due to their more “in vivo-like” behavior for key parameters such as cell viability, proliferation, differentiation, morphology, gene and protein expression and function [195,196]. For genotoxicity assessment, robust protocols for 3D models have been established for skin, airways and liver tissue equivalents [10,197]. Many of the 3D cell culture systems applied in genotoxicity testing (of NMs) have been spheroids, such as liver spheroids constructed from primary hepatocytes, HepG2 hepatocellular carcinoma cells or the HepaRG cell line applied to the comet assay [14,31,34,198,199] and micronucleus assay [200]. Human 3D airway models usually consist of a functional and differentiated respiratory epithelium with cilia and mucus and are cultured on the physiological relevant air-liquid-interface (ALI) [19]. The comet assay has been established with commercially available human reconstructed 3D airway models (MucilAir™, Epithelix and EpiAirway, MatTek) as well as a model consisting of the bronchial epithelial cell line BEAS2B and tumor lung epithelial cell line A549 [4,93]. These 3D models for liver and airways show great promise, but are at an early stage of development and need to be further improved for genotoxicity assessment of NMs. For human skin models, two methods have been developed and used for genotoxicity assessment: the reconstructed skin micronucleus test (RSMN) and the reconstructed skin comet assay (RS comet assay). Reconstructed skin tissues are commercially available (EpiDerm™, Phenion® FT, EpiSkin™) and have been shown to have the same metabolic properties as native skin [94,95]. Wills et al. [32] performed the RSMN with silica nanoparticles and the EpiDerm™ model and came to the conclusion that robust exposure characterization and uptake assessment methods are crucial to interpret nano(geno)toxicity studies successfully. However, 3D tissue-based assays definitely provide a more realistic test system to study the genotoxic potential of NMs, compared to 2D test systems [10,32,34].
Even advanced 3D tissue models fail to mimic an organ in some aspects such as spatiotemporal gradients of chemicals and nutrients and mechano-stimulated environments. The incorporation of microfluidic networks in 3D models constitutes the most recent innovation in microfluidics, called organ-on-a-chip technologies (OOC). OOC models are aimed at modeling in vitro the micro-physiological conditions prevalent in the body as closely as possible, thus avoiding the typical disadvantages of conventional cell models [201]. A major challenge in the cultivation of complex, 3D organ models under physiologically relevant conditions is the maintenance of their function over a long period of time [195]. Bhatia and Ingber [202] described OOCs as systems for the cultivation of living cells in continuously perfused chambers to model physiological functions. OOC have been developed for nearly every organ of the human body for different applications, particularly for drug testing approaches [203]. Besides mimicking one organ under physiological conditions, chip-based in vitro models mimicking organ–organ interactions have also been developed, for example the intestine-liver interaction [204,205]. Additionally, chip-based multi-organ in vitro models are described in literature, using primary cells or cell lines as 2D culture or 3D spheroids to mimic the organs and their interactions on chip [206,207,208,209,210].
For nanotoxicity assessment, a placenta-on-a-chip [211] and a lung-on-a-chip [212] have recently been published, though neither included the endpoint of genotoxicity [107,108]. Researchers developed microfabricated chip technologies for the screening of cytotoxicity and genotoxicity of NMs [213,214,215]. Vecchio et al. coupled the cytokinesis block micronucleus assay with micro-array based cell sorting to analyze the genotoxic effect of NMs on human primary lymphocyte subtypes [215]. In the same year, CometChip Technology was published [123]. The widely used comet assay has been transferred to a microarray-based approach, a high throughput platform, which helps to overcome the limitations of the traditional performed comet assay such as low-throughput and poor reproducibility [123]. Thus, chip technologies can help to transfer important methods to a high throughput level [123] and to combine 3D models with more physiologic features.
12. Conclusions and Future Perspectives
Due to constantly growing commercial applications and their presence in the majority of consumer products, NMs are being extensively investigated for their safety. A main concern with NM exposure is their genotoxic potential. Therefore, the study of mechanisms potentially leading to genotoxicity is crucially important. Besides the general mechanisms of genotoxicity (primary direct genotoxicity and secondary genotoxicity) and their verification processes, we have described the additional studies which are needed for a correct implementation of genotoxicity studies. Before genotoxicity testing, it is necessary to determine the LC50 (lethal concentration) in order to define the appropriate dose range for genotoxicity studies. Furthermore, for the interpretation of the results a characterization of the NMs under physiological conditions is essential for the correct interpretation of the biologic data. Due to the limited data concerning NMs safety and the increasing number of yearly produced NMs, different European projects, such as NanoTEST, NanoGENOTOX, NanoReg, HISENTS, RiskGONE and others, made efforts to understand the mode of action of NMs and to develop, or adapt validated OECD chemical test methods for use on NMs. Genotoxicity biomarkers include a battery of assays that were discussed in this chapter. We described the main endpoints applied in standard in vitro models as well as new advanced 3D models that are closer to the in vivo situation. New genotoxicity endpoints were discussed, including genetic and epigenetic changes. We have discussed 3D models and their application for genotoxicity studies as well as the state-of-the-art on adaptation of genotoxicity assays to HTS technologies. The development of advanced in vitro models for nanotoxicity assessment is greatly encouraged by the principles of 3Rs (replace, reduce, refine), which are embedded in international legislation and regulations on the use of animals for scientific purposes. Additionally, their incorporation in microfluidic systems (organ-on-a-chip technologies) are very challenging approaches. Existing approaches of organ-on-a-chip and HTS methods are presented and discussed, such as the use of automated tools to facilitate rapid execution of a large number of comet assays, in vitro micronucleus assays or γH2AX assays.
The need for HTS and rapid automated procedures to perform genotoxicity studies is growing, both in the pharmaceutical and in the chemical industry for approval of new nanoscale materials. A high number of international experts are working on the optimization of the procedures described in this chapter, with the aim of approving the new advanced cell models as new animal replacement models and including the automated culture and analysis procedures in new OECD Test Guidelines.
Acknowledgments
The authors thank Andrew Collins (NorGenoTec AS, Norway) for helpful discussions and kindly editing the manuscript with English corrections.
Author Contributions
Conceptualization, Y.K., E.R.-P. and M.D.; writing—original draft preparation, Y.K., E.R.-P., M.D.; writing—review and editing, Y.K., E.R.-P., E.M., M.H., N.E.Y., E.M.L. and M.D.; supervision, M.D.; funding acquisition, Y.K. and M.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the European Commission under the Horizon 2020 programme (grant agreement No. 862296/Sabydoma; grant agreement No. 814425/RiskGONE; grant agreement No. 814572/NanoSolveIT; grant agreement No. 857381/VISION), hCOMET project (COST Action, CA 15132), by the Research Council Norway (RCN) and the Federal Ministry of Education and Research (BMBF) via the ERA-NET EuroNanoMed II/III projects INNOCENT (RCN 271075), CELLUX (RCN 310571) and GOTTARG (RCN 310565) as well as TRANSCAN-2 project NExT (BMBF 01KT1910) and also via national projects NanoBioReal (RCN 288768) and NanoCELL (BMBF 03XP196F).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Commission Recommendation of 18 October 2011 on the definiton of a nanomaterial, (2011/696/EU) Off. J. Eur. Union. 2011 [Google Scholar]
- 2.Azqueta A., Dusinska M. The use of the comet assay for the evaluation of the genotoxicity of nanomaterials. Front. Genet. 2015;6:239. doi: 10.3389/fgene.2015.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clift M.J.D., Raemy D.O., Endes C., Ali Z., Lehmann A.D., Brandenberger C., Petri-Fink A., Wick P., Parak W.J., Gehr P., et al. Can the Ames test provide an insight into nano-object mutagenicity? Investigating the interaction between nano-objects and bacteria. Nanotoxicology. 2013;7:1373–1385. doi: 10.3109/17435390.2012.741725. [DOI] [PubMed] [Google Scholar]
- 4.Doak S.H., Manshian B., Jenkins G.J.S., Singh N. In vitro genotoxicity testing strategy for nanomaterials and the adaptation of current OECD guidelines. Mutat. Res. 2012;745:104–111. doi: 10.1016/j.mrgentox.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.OECD . Genotoxicity of Manufactured Nanomaterials: Report of the OECD Expert Meeting. Volume 472. Elsevier; Amsterdam, The Netherlands: 2014. pp. 1–37. [Google Scholar]
- 6.Ventura C., Sousa-Uva A., Lavinha J., Silva M.J. Conventional and novel “omics”-based approaches to the study of carbon nanotubes pulmonary toxicity. Environ. Mol. Mutagen. 2018;59:334–362. doi: 10.1002/em.22177. [DOI] [PubMed] [Google Scholar]
- 7.Lan J., Gou N., Gao C., He M., Gu A.Z. Comparative and mechanistic genotoxicity assessment of nanomaterials via a quantitative toxicogenomics approach across multiple species. Environ. Sci. Technol. 2014;48:12937–12945. doi: 10.1021/es503065q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans S.J., Clift M.J.D., Singh N., De Oliveira Mallia J., Burgum M., Wills J.W., Wilkinson T.S., Jenkins G.J.S., Doak S.H. Critical review of the current and future challenges associated with advanced in vitro systems towards the study of nanoparticle (secondary) genotoxicity. Mutagenesis. 2017;32:233–241. doi: 10.1093/mutage/gew054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfuhler S., Downs T.R., Allemang A.J., Shan Y., Crosby M.E. Weak silica nanomaterial-induced genotoxicity can be explained by indirect DNA damage as shown by the OGG1-modified comet assay and genomic analysis. Mutagenesis. 2017;32:5–12. doi: 10.1093/mutage/gew064. [DOI] [PubMed] [Google Scholar]
- 10.Pfuhler S., van Benthem J., Curren R., Doak S.H., Dusinska M., Hayashi M., Heflich R.H., Kidd D., Kirkland D., Luan Y., et al. Use of in vitro 3D tissue models in genotoxicity testing: Strategic fit, validation status and way forward. Report of the working group from the 7th International Workshop on Genotoxicity Testing (IWGT) Mutat. Res. Toxicol. Environ. Mutagen. 2020;850–851:503135. doi: 10.1016/j.mrgentox.2020.503135. [DOI] [PubMed] [Google Scholar]
- 11.Magdolenova Z., Drlickova M., Henjum K., Rundén-Pran E., Tulinska J., Bilanicova D., Pojana G., Kazimirova A., Barancokova M., Kuricova M., et al. Coating-dependent induction of cytotoxicity and genotoxicity of iron oxide nanoparticles. Nanotoxicology. 2015;9:44–56. doi: 10.3109/17435390.2013.847505. [DOI] [PubMed] [Google Scholar]
- 12.Doak S.H., Dusinska M. NanoGenotoxicology: Present and the future. Mutagenesis. 2017;32:1–4. doi: 10.1093/mutage/gew066. [DOI] [PubMed] [Google Scholar]
- 13.Nelson B.C., Wright C.W., Ibuki Y., Moreno-Villanueva M., Karlsson H.L., Hendriks G., Sims C.M., Singh N., Doak S.H. Emerging metrology for high-throughput nanomaterial genotoxicology. Mutagenesis. 2017;32:215–232. doi: 10.1093/mutage/gew037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elje E., Hesler M., Rundén-Pran E., Mann P., Mariussen E., Wagner S., Dusinska M., Kohl Y. The comet assay applied to HepG2 liver spheroids. Mutat. Res. Toxicol. Environ. Mutagen. 2019:0–1. doi: 10.1016/j.mrgentox.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Collins A.R., Annangi B., Rubio L., Marcos R., Dorn M., Merker C., Estrela-Lopis I., Cimpan M.R., Ibrahim M., Cimpan E., et al. High throughput toxicity screening and intracellular detection of nanomaterials. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2017;9 doi: 10.1002/wnan.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moreno-Villanueva M., Eltze T., Dressler D., Bernhardt J., Hirsch C., Wick P., von Scheven G., Lex K., Bürkle A. The automated FADU-assay, a potential high-throughput in vitro method for early screening of DNA breakage. ALTEX. 2011;28:295–303. doi: 10.14573/altex.2011.4.295. [DOI] [PubMed] [Google Scholar]
- 17.Sramkova M., Kozics K., Masanova V., Uhnakova I., Razga F., Nemethova V., Mazancova P., Kapka-Skrzypczak L., Kruszewski M., Novotova M., et al. Kidney nanotoxicity studied in human renal proximal tubule epithelial cell line TH1. Mutat. Res. Toxicol. Environ. Mutagen. 2019 doi: 10.1016/J.MRGENTOX.2019.01.012. [DOI] [PubMed] [Google Scholar]
- 18.Brown D.M., Danielsen P.H., Derr R., Moelijker N., Fowler P., Stone V., Hendriks G., Møller P., Kermanizadeh A. The mechanism-based toxicity screening of particles with use in the food and nutrition sector via the ToxTracker reporter system. Toxicol. In Vitro. 2019;61:104594. doi: 10.1016/j.tiv.2019.104594. [DOI] [PubMed] [Google Scholar]
- 19.Hufnagel M., Schoch S., Wall J., Strauch B.M., Hartwig A. Toxicity and Gene Expression Profiling of Copper- and Titanium-Based Nanoparticles Using Air-Liquid Interface Exposure. Chem. Res. Toxicol. 2020;33:1237–1249. doi: 10.1021/acs.chemrestox.9b00489. [DOI] [PubMed] [Google Scholar]
- 20.Bajpayee M., Kumar A., Dhawan A. Genotoxicity Assessment. Humana; New York, NY, USA: 2019. The Comet Assay: Assessment of In Vitro and In Vivo DNA Damage; pp. 237–257. [DOI] [PubMed] [Google Scholar]
- 21.Gao C.-H., Mortimer M., Zhang M., Holden P.A., Cai P., Wu S., Xin Y., Wu Y., Huang Q. Impact of metal oxide nanoparticles on in vitro DNA amplification. PeerJ. 2019;7:e7228. doi: 10.7717/peerj.7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El Yamani N., Collins A.R., Rundén-Pran E., Fjellsbø L.M., Shaposhnikov S., Zienolddiny S., Dusinska M. In vitro genotoxicity testing of four reference metal nanomaterials, titanium dioxide, zinc oxide, cerium oxide and silver: Towards reliable hazard assessment. Mutagenesis. 2017;32:117–126. doi: 10.1093/mutage/gew060. [DOI] [PubMed] [Google Scholar]
- 23.Alhadrami H.A. Biosensors: Classifications, medical applications, and future prospective. Biotechnol. Appl. Biochem. 2018;65:497–508. doi: 10.1002/bab.1621. [DOI] [PubMed] [Google Scholar]
- 24.Townsend T.A., Parrish M.C., Engelward B.P., Manjanatha M.G. The development and validation of EpiComet-Chip, a modified high-throughput comet assay for the assessment of DNA methylation status. Environ. Mol. Mutagen. 2017;58:508–521. doi: 10.1002/em.22101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wasalathanthri D.P., Li D., Song D., Zheng Z., Choudhary D., Jansson I., Lu X., Schenkman J.B., Rusling J.F. Elucidating Organ-Specific Metabolic Toxicity Chemistry from Electrochemiluminescent Enzyme/DNA Arrays and Bioreactor Bead-LC-MS/MS. Chem. Sci. 2015;6:2457–2468. doi: 10.1039/C4SC03401E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akagi J., Hall C.J., Crosier K.E., Cooper J.M., Crosier P.S., Wlodkowic D. OpenSource lab-on-a-chip physiometer for accelerated zebrafish embryo biotests. Curr. Protoc. Cytom. 2014;67:9.44.1–9.44.16. doi: 10.1002/0471142956.cy0944s67. [DOI] [PubMed] [Google Scholar]
- 27.Klepeisz P., Sagmeister S., Haudek-Prinz V., Pichlbauer M., Grasl-Kraupp B., Gerner C. Phenobarbital induces alterations in the proteome of hepatocytes and mesenchymal cells of rat livers. PLoS ONE. 2013;8:e76137. doi: 10.1371/journal.pone.0076137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardwick R.N., Betts C.J., Whritenour J., Sura R., Thamsen M., Kaufman E.H., Fabre K. Drug-induced skin toxicity: Gaps in preclinical testing cascade as opportunities for complex in vitro models and assays. Lab Chip. 2020;20:199–214. doi: 10.1039/C9LC00519F. [DOI] [PubMed] [Google Scholar]
- 29.Israel J.W., Chappell G.A., Simon J.M., Pott S., Safi A., Lewis L., Cotney P., Boulos H.S., Bodnar W., Lieb J.D., et al. Tissue- and strain-specific effects of a genotoxic carcinogen 1,3-butadiene on chromatin and transcription. Mamm. Genome. 2018;29:153–167. doi: 10.1007/s00335-018-9739-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y., Xu S., Wu T., Hu K., Chen S., Xu A., Wu L. Assessment of Genotoxic Effects by Constructing a 3D Cellular System with Highly Sensitive Mutagenic Human-Hamster Hybrid Cells. Chem. Res. Toxicol. 2018;31:594–600. doi: 10.1021/acs.chemrestox.8b00069. [DOI] [PubMed] [Google Scholar]
- 31.Kermanizadeh A., Løhr M., Roursgaard M., Messner S., Gunness P., Kelm J.M., Møller P., Stone V., Loft S. Hepatic toxicology following single and multiple exposure of engineered nanomaterials utilising a novel primary human 3D liver microtissue model. Part. Fibre Toxicol. 2014;11:1–15. doi: 10.1186/s12989-014-0056-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wills J.W., Hondow N., Thomas A.D., Chapman K.E., Fish D., Maffeis T.G., Penny M.W., Brown R.A., Jenkins G.J.S., Brown A.P., et al. Genetic toxicity assessment of engineered nanoparticles using a 3D in vitro skin model (EpiDermTM) Part. Fibre Toxicol. 2016;13:1–21. doi: 10.1186/s12989-016-0161-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haase A., Dommershausen N., Schulz M., Landsiedel R., Reichardt P., Krause B.-C., Tentschert J., Luch A. Genotoxicity testing of different surface-functionalized SiO2, ZrO2 and silver nanomaterials in 3D human bronchial models. Arch. Toxicol. 2017;91:3991–4007. doi: 10.1007/s00204-017-2015-9. [DOI] [PubMed] [Google Scholar]
- 34.Elje E., Mariussen E., Moriones O.H., Bastús N.G., Puntes V., Kohl Y., Dusinska M., Rundén-Pran E. Hepato(Geno)toxicity assessment of nanoparticles in a hepg2 liver spheroid model. Nanomaterials. 2020;10:545. doi: 10.3390/nano10030545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shoeb M., Kodali V.K., Farris B.Y., Bishop L.M., Meighan T.G., Salmen R., Eye T., Friend S., Schwegler-Berry D., Roberts J.R., et al. Oxidative Stress, DNA Methylation, and Telomere Length Changes in Peripheral Blood Mononuclear Cells after Pulmonary Exposure to Metal-Rich Welding Nanoparticles. NanoImpact. 2017;5:61–69. doi: 10.1016/j.impact.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Efeoglu E., Maher M.A., Casey A., Byrne H.J. Toxicological assessment of nanomaterials: The role of in vitro Raman microspectroscopic analysis. Anal. Bioanal. Chem. 2018;410:1631–1646. doi: 10.1007/s00216-017-0812-x. [DOI] [PubMed] [Google Scholar]
- 37.Conway G.E., Shah U.-K., Llewellyn S., Cervena T., Evans S.J., Al Ali A.S., Jenkins G.J., Clift M.J.D., Doak S.H. Adaptation of the in vitro micronucleus assay for genotoxicity testing using 3D liver models supporting longer-term exposure durations. Mutagenesis. 2020 doi: 10.1093/mutage/geaa018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Llewellyn S.V., Conway G.E., Shah U.-K., Evans S.J., Jenkins G.J.S., Clift M.J.D., Doak S.H. Advanced 3D Liver Models for In vitro Genotoxicity Testing Following Long-Term Nanomaterial Exposure. J. Vis. Exp. 2020 doi: 10.3791/61141. [DOI] [PubMed] [Google Scholar]
- 39.Kiratipaiboon C., Stueckle T.A., Ghosh R., Rojanasakul L.W., Chen Y.C., Dinu C.Z., Rojanasakul Y. Acquisition of Cancer Stem Cell-like Properties in Human Small Airway Epithelial Cells after a Long-term Exposure to Carbon Nanomaterials. Environ. Sci. Nano. 2019;6:2152–2170. doi: 10.1039/C9EN00183B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirsch-Volders M., Decordier I., Elhajouji A., Plas G., Aardema M.J., Fenech M. In vitro genotoxicity testing using the micronucleus assay in cell lines, human lymphocytes and 3D human skin models. Mutagenesis. 2011;26:177–184. doi: 10.1093/mutage/geq068. [DOI] [PubMed] [Google Scholar]
- 41.Dusinska M., Mariussen E., Rundén-Pran E., Hudecova A.M., Elje E., Kazimirova A., El Yamani N., Dommershausen N., Tharmann J., Fieblinger D., et al. Nanotxicity. Volume 1894. Humana Press; New York, NY, USA: 2019. In vitro approaches for assessing the genotoxicity of nanomaterials; pp. 83–122. (Methods in Molecular Biology Book Series). [DOI] [PubMed] [Google Scholar]
- 42.Regulation (EC) No 1907/2006 of the European Parliament and of the Councilconcerning the Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH), establishing a European Chemicals Agency, amending Directive 1999/45/EC and repealing Council Regulation (EEC) No 793/93 and Commission Regulation (EC) No 1488/94 as well as Council Directive 76/769/EEC and Commission Directives 91/155/EEC, 93/67/EEC, 93/105/EC and 2000/21/EC. Off. J. Eur. Union. 2006 [Google Scholar]
- 43.Russell W., Burch R. The Principles of Humane Experimental Technique. Methuen; London, UK: 1959. pp. 1–238. [Google Scholar]
- 44.Hackenberg S., Zimmermann F.-Z., Scherzed A., Friehs G., Froelich K., Ginzkey C., Koehler C., Burghartz M., Hagen R., Kleinsasser N. Repetitive exposure to zinc oxide nanoparticles induces dna damage in human nasal mucosa mini organ cultures. Environ. Mol. Mutagen. 2011;52:582–589. doi: 10.1002/em.20661. [DOI] [PubMed] [Google Scholar]
- 45.Kazimirova A., El Yamani N., Rubio L., García-Rodríguez A., Barancokova M., Marcos R., Dusinska M. Effects of titanium dioxide nanoparticles on the Hprt gene mutations in V79 hamster cells. Nanomaterials. 2020;10:465. doi: 10.3390/nano10030465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kisin E.R., Murray A.R., Sargent L., Lowry D., Chirila M., Siegrist K.J., Schwegler-Berry D., Leonard S., Castranova V., Fadeel B., et al. Genotoxicity of carbon nanofibers: Are they potentially more or less dangerous than carbon nanotubes or asbestos? Toxicol. Appl. Pharmacol. 2011;252:1–10. doi: 10.1016/j.taap.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartek J., Mistrik M., Bartkova J. Long-distance inflammatory and genotoxic impact of cancer in vivo. Proc. Natl. Acad. Sci. USA. 2010;107:17861–17862. doi: 10.1073/pnas.1013093107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fubini B., Ghiazza M., Fenoglio I. Physico-chemical features of engineered nanoparticles relevant to their toxicity. Nanotoxicology. 2010;4:347–363. doi: 10.3109/17435390.2010.509519. [DOI] [PubMed] [Google Scholar]
- 49.Danielsen P.H., Knudsen K.B., Štrancar J., Umek P., Koklič T., Garvas M., Vanhala E., Savukoski S., Ding Y., Madsen A.M., et al. Effects of physicochemical properties of TiO2 nanomaterials for pulmonary inflammation, acute phase response and alveolar proteinosis in intratracheally exposed mice. Toxicol. Appl. Pharmacol. 2020;386:114830. doi: 10.1016/j.taap.2019.114830. [DOI] [PubMed] [Google Scholar]
- 50.Ling C., An H., Li L., Wang J., Lu T., Wang H., Hu Y., Song G., Liu S. Genotoxicity Evaluation of Titanium Dioxide Nanoparticles In Vitro: A Systematic Review of the Literature and Meta-analysis. Biol. Trace Elem. Res. 2020 doi: 10.1007/s12011-020-02311-8. [DOI] [PubMed] [Google Scholar]
- 51.Magdolenova Z., Collins A., Kumar A., Dhawan A., Stone V., Dusinska M. Mechanisms of genotoxicity. A review of in vitro and in vivo studies with engineered nanoparticles. Nanotoxicology. 2014;8:233–278. doi: 10.3109/17435390.2013.773464. [DOI] [PubMed] [Google Scholar]
- 52.Huk A., Izak-Nau E., Reidy B., Boyles M., Duschl A., Lynch I., Dušinska M. Is the toxic potential of nanosilver dependent on its size? Part. Fibre Toxicol. 2014;11:65. doi: 10.1186/s12989-014-0065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kononenko V., Repar N., Marušič N., Drašler B., Romih T., Hočevar S., Drobne D. Comparative in vitro genotoxicity study of ZnO nanoparticles, ZnO macroparticles and ZnCl2 to MDCK kidney cells: Size matters. Toxicol. In Vitro. 2017;40:256–263. doi: 10.1016/j.tiv.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 54.Guo X., Li Y., Yan J., Ingle T., Jones M.Y., Mei N., Boudreau M.D., Cunningham C.K., Abbas M., Paredes A.M., et al. Size- and coating-dependent cytotoxicity and genotoxicity of silver nanoparticles evaluated using in vitro standard assays. Nanotoxicology. 2016;10:1373–1384. doi: 10.1080/17435390.2016.1214764. [DOI] [PubMed] [Google Scholar]
- 55.Proquin H., Rodríguez-Ibarra C., Moonen C.G.J., Urrutia Ortega I.M., Briedé J.J., de Kok T.M., van Loveren H., Chirino Y.I. Titanium dioxide food additive (E171) induces ROS formation and genotoxicity: Contribution of micro and nano-sized fractions. Mutagenesis. 2017;32:139–149. doi: 10.1093/mutage/gew051. [DOI] [PubMed] [Google Scholar]
- 56.Xia Q., Li H., Liu Y., Zhang S., Feng Q., Xiao K. The effect of particle size on the genotoxicity of gold nanoparticles. J. Biomed. Mater. Res. A. 2017;105:710–719. doi: 10.1002/jbm.a.35944. [DOI] [PubMed] [Google Scholar]
- 57.Raghunathan V.K., Devey M., Hawkins S., Hails L., Davis S.A., Mann S., Chang I.T., Ingham E., Malhas A., Vaux D.J., et al. Influence of particle size and reactive oxygen species on cobalt chrome nanoparticle-mediated genotoxicity. Biomaterials. 2013;34:3559–3570. doi: 10.1016/j.biomaterials.2013.01.085. [DOI] [PubMed] [Google Scholar]
- 58.Hesler M., Aengenheister L., Ellinger B., Drexel R., Straskraba S., Jost C., Wagner S., Meier F., von Briesen H., Büchel C., et al. Multi-endpoint toxicological assessment of polystyrene nano- and microparticles in different biological models in vitro. Toxicol. In Vitro. 2019;61:104610. doi: 10.1016/j.tiv.2019.104610. [DOI] [PubMed] [Google Scholar]
- 59.Zhou F., Liao F., Chen L., Liu Y., Wang W., Feng S. The size-dependent genotoxicity and oxidative stress of silica nanoparticles on endothelial cells. Environ. Sci. Pollut. Res. Int. 2019;26:1911–1920. doi: 10.1007/s11356-018-3695-2. [DOI] [PubMed] [Google Scholar]
- 60.Butler K.S., Peeler D.J., Casey B.J., Dair B.J., Elespuru R.K. Silver nanoparticles: Correlating nanoparticle size and cellular uptake with genotoxicity. Mutagenesis. 2015;30:577–591. doi: 10.1093/mutage/gev020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodriguez-Garraus A., Azqueta A., Vettorazzi A., López de Cerain A. Genotoxicity of Silver Nanoparticles. Nanomaterials. 2020;10:251. doi: 10.3390/nano10020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kawanishi M., Yoneda R., Totsuka Y., Yagi T. Genotoxicity of micro- and nano-particles of kaolin in human primary dermal keratinocytes and fibroblasts. Genes Environ. 2020;42:16. doi: 10.1186/s41021-020-00155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Naz S., Gul A., Zia M. Toxicity of copper oxide nanoparticles: A review study. IET Nanobiotechnology. 2020;14:1–13. doi: 10.1049/iet-nbt.2019.0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lebedová J., Hedberg Y.S., Odnevall Wallinder I., Karlsson H.L. Size-dependent genotoxicity of silver, gold and platinum nanoparticles studied using the mini-gel comet assay and micronucleus scoring with flow cytometry. Mutagenesis. 2018;33:77–85. doi: 10.1093/mutage/gex027. [DOI] [PubMed] [Google Scholar]
- 65.Huk A., Izak-Nau E., el Yamani N., Uggerud H., Vadset M., Zasonska B., Duschl A., Dusinska M. Impact of nanosilver on various DNA lesions and HPRT gene mutations—effects of charge and surface coating. Part. Fibre Toxicol. 2015;12:25. doi: 10.1186/s12989-015-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gábelová A., El Yamani N., Alonso T.I., Buliaková B., Srančíková A., Bábelová A., Pran E.R., Fjellsbø L.M., Elje E., Yazdani M., et al. Fibrous shape underlies the mutagenic and carcinogenic potential of nanosilver while surface chemistry affects the biosafety of iron oxide nanoparticles. Mutagenesis. 2017;32:193–202. doi: 10.1093/mutage/gew045. [DOI] [PubMed] [Google Scholar]
- 67.Vales G., Suhonen S., Siivola K.M., Savolainen K.M., Catalán J., Norppa H. Genotoxicity and Cytotoxicity of Gold Nanoparticles In Vitro: Role of Surface Functionalization and Particle Size. Nanomaterials. 2020;10:271. doi: 10.3390/nano10020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Falck G., Lindberg H., Suhonen S., Vippola M., Vanhala E., Catalán J., Savolainen K., Norppa H. Genotoxic effects of nanosized and fine TiO2. Hum. Exp. Toxicol. 2009;28:339–352. doi: 10.1177/0960327109105163. [DOI] [PubMed] [Google Scholar]
- 69.Vales G., Rubio L., Marcos R. Long-term exposures to low doses of titanium dioxide nanoparticles induce cell transformation, but not genotoxic damage in BEAS-2B cells. Nanotoxicology. 2015;9:568–578. doi: 10.3109/17435390.2014.957252. [DOI] [PubMed] [Google Scholar]
- 70.Osman I.F., Baumgartner A., Cemeli E., Fletcher J.N., Anderson D. Genotoxicity and cytotoxicity of zinc oxide and titanium dioxide in HEp-2 cells. Nanomedicine. 2010;5:1193–1203. doi: 10.2217/nnm.10.52. [DOI] [PubMed] [Google Scholar]
- 71.Charles S., Jomini S., Fessard V., Bigorgne-Vizade E., Rousselle C., Michel C. Assessment of the in vitro genotoxicity of TiO2nanoparticles in a regulatory context. Nanotoxicology. 2018;12:357–374. doi: 10.1080/17435390.2018.1451567. [DOI] [PubMed] [Google Scholar]
- 72.Ghosh S., Ghosh I., Chakrabarti M., Mukherjee A. Genotoxicity and biocompatibility of superparamagnetic iron oxide nanoparticles: Influence of surface modification on biodistribution, retention, DNA damage and oxidative stress. Food Chem. Toxicol. 2020;136 doi: 10.1016/j.fct.2019.110989. [DOI] [PubMed] [Google Scholar]
- 73.Pöttler M., Staicu A., Zaloga J., Unterweger H., Weigel B., Schreiber E., Hofmann S., Wiest I., Jeschke U., Alexiou C., et al. Genotoxicity of superparamagnetic iron oxide nanoparticles in granulosa cells. Int. J. Mol. Sci. 2015;16:26280–26290. doi: 10.3390/ijms161125960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang W., Jiang P., Chen W., Zheng B., Mao Z., Antipov A., Correia M., Larsen E.H., Gao C. Genotoxicity of copper oxide nanoparticles with different surface chemistry on rat bone marrow mesenchymal stem cells. J. Nanosci. Nanotechnol. 2016;16:5489–5497. doi: 10.1166/jnn.2016.11753. [DOI] [PubMed] [Google Scholar]
- 75.Laban B., Ralević U., Petrović S., Leskovac A., Vasić-Anićijević D., Marković M., Vasić V. Green synthesis and characterization of nontoxic L-methionine capped silver and gold nanoparticles. J. Inorg. Biochem. 2020;204 doi: 10.1016/j.jinorgbio.2019.110958. [DOI] [PubMed] [Google Scholar]
- 76.Platel A., Carpentier R., Becart E., Mordacq G., Betbeder D., Nesslany F. Influence of the surface charge of PLGA nanoparticles on their in vitro genotoxicity, cytotoxicity, ROS production and endocytosis. J. Appl. Toxicol. 2016;36:434–444. doi: 10.1002/jat.3247. [DOI] [PubMed] [Google Scholar]
- 77.Niu M., Zhong H., Shao H., Hong D., Ma T., Xu K., Chen X., Han J., Sun J. Shape-dependent genotoxicity of mesoporous silica nanoparticles and cellular mechanisms. J. Nanosci. Nanotechnol. 2016;16:2313–2318. doi: 10.1166/jnn.2016.10928. [DOI] [PubMed] [Google Scholar]
- 78.Gea M., Bonetta S., Iannarelli L., Giovannozzi A.M., Maurino V., Bonetta S., Hodoroaba V.D., Armato C., Rossi A.M., Schilirò T. Shape-engineered titanium dioxide nanoparticles (TiO2–NPs): Cytotoxicity and genotoxicity in bronchial epithelial cells. Food Chem. Toxicol. 2019;127:89–100. doi: 10.1016/j.fct.2019.02.043. [DOI] [PubMed] [Google Scholar]
- 79.Cowie H., Magdolenova Z., Saunders M., Drlickova M., Correia Carreira S., Halamoda Kenzaoi B., Gombau L., Guadagnini R., Lorenzo Y., Walker L., et al. Suitability of human and mammalian cells of different origin for the assessment of genotoxicity of metal and polymeric engineered nanoparticles. Nanotoxicology. 2015;9:57–65. doi: 10.3109/17435390.2014.940407. [DOI] [PubMed] [Google Scholar]
- 80.Yang H., Liu C., Yang D., Zhang H., Xi Z. Comparative study of cytotoxicity, oxidative stress and genotoxicity induced by four typical nanomaterials: The role of particle size, shape and composition. J. Appl. Toxicol. 2009;29:69–78. doi: 10.1002/jat.1385. [DOI] [PubMed] [Google Scholar]
- 81.Kisin E.R., Murray A.R., Keane M.J., Shi X.C., Schwegler-Berry D., Gorelik O., Arepalli S., Castranova V., Wallace W.E., Kagan V.E., et al. Single-walled carbon nanotubes: Geno- and cytotoxic effects in lung fibroblast V79 cells. J. Toxicol. Environ. Heal. Part A Curr. Issues. 2007;70:2071–2079. doi: 10.1080/15287390701601251. [DOI] [PubMed] [Google Scholar]
- 82.Catalán J., Siivola K.M., Nymark P., Lindberg H., Suhonen S., Järventaus H., Koivisto A.J., Moreno C., Vanhala E., Wolff H., et al. In vitro and in vivo genotoxic effects of straight versus tangled multi-walled carbon nanotubes. Nanotoxicology. 2016;10:794–806. doi: 10.3109/17435390.2015.1132345. [DOI] [PubMed] [Google Scholar]
- 83.Kim J.S., Lee K., Lee Y.H., Cho H.S., Kim K.H., Choi K.H., Lee S.H., Song K.S., Kang C.S., Yu I.J. Aspect ratio has no effect on genotoxicity of multi-wall carbon nanotubes. Arch. Toxicol. 2011;85:775–786. doi: 10.1007/s00204-010-0574-0. [DOI] [PubMed] [Google Scholar]
- 84.Xu L., Zhao J., Wang Z. Genotoxic response and damage recovery of macrophages to graphene quantum dots. Sci. Total Environ. 2019;664:536–545. doi: 10.1016/j.scitotenv.2019.01.356. [DOI] [PubMed] [Google Scholar]
- 85.Burgum M.J., Clift M.J.D., Evans S.J., Hondow N., Miller M., Lopez S.B., Williams A., Tarat A., Jenkins G.J., Doak S.H. In Vitro Primary-Indirect Genotoxicity in Bronchial Epithelial Cells Promoted by Industrially Relevant Few-Layer Graphene. Small. 2020:e2002551. doi: 10.1002/smll.202002551. [DOI] [PubMed] [Google Scholar]
- 86.Syama S., Mohanan P. V Safety and biocompatibility of graphene: A new generation nanomaterial for biomedical application. Int. J. Biol. Macromol. 2016;86:546–555. doi: 10.1016/j.ijbiomac.2016.01.116. [DOI] [PubMed] [Google Scholar]
- 87.Domenech J., Hernández A., Demir E., Marcos R., Cortés C. Interactions of graphene oxide and graphene nanoplatelets with the in vitro Caco-2/HT29 model of intestinal barrier. Sci. Rep. 2020;10:2793. doi: 10.1038/s41598-020-59755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Samadian H., Salami M.S., Jaymand M., Azarnezhad A., Najafi M., Barabadi H., Ahmadi A. Genotoxicity assessment of carbon-based nanomaterials; Have their unique physicochemical properties made them double-edged swords? Mutat. Res. Mutat. Res. 2020;783:108296. doi: 10.1016/j.mrrev.2020.108296. [DOI] [PubMed] [Google Scholar]
- 89.Panek A., Błażewicz M., Frączek-Szczypta A., Adamczyk J., Wiltowska-Zuber J., Paluszkiewicz C. Applications of Comet Assay for the Evaluation of Genotoxicity and DNA Repair Efficiency in Nanomaterials Research. Acta Phys. Pol. A. 2018;133:280–282. doi: 10.12693/APhysPolA.133.280. [DOI] [Google Scholar]
- 90.Guo F., Ma N., Horibe Y., Kawanishi S., Murata M., Hiraku Y. Nitrative DNA damage induced by multi-walled carbon nanotube via endocytosis in human lung epithelial cells. Toxicol. Appl. Pharmacol. 2012;260:183–192. doi: 10.1016/j.taap.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 91.Lindberg H.K., Falck G.C.M., Suhonen S., Vippola M., Vanhala E., Catalán J., Savolainen K., Norppa H. Genotoxicity of nanomaterials: DNA damage and micronuclei induced by carbon nanotubes and graphite nanofibres in human bronchial epithelial cells in vitro. Toxicol. Lett. 2009;186:166–173. doi: 10.1016/j.toxlet.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 92.Wang J., Wang J., Liu Y., Nie Y., Si B., Wang T., Waqas A., Zhao G., Wang M., Xu A. Aging-independent and size-dependent genotoxic response induced by titanium dioxide nanoparticles in mammalian cells. J. Environ. Sci. 2019;85:94–106. doi: 10.1016/j.jes.2019.04.024. [DOI] [PubMed] [Google Scholar]
- 93.Foltête A.S., Masfaraud J.F., Bigorgne E., Nahmani J., Chaurand P., Botta C., Labille J., Rose J., Férard J.F., Cotelle S. Environmental impact of sunscreen nanomaterials: Ecotoxicity and genotoxicity of altered TiO2 nanocomposites on Vicia faba. Environ. Pollut. 2011;159:2515–2522. doi: 10.1016/j.envpol.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 94.Pereira R., Rocha-Santos T.A.P., Antunes F.E., Rasteiro M.G., Ribeiro R., Gonçalves F., Soares A.M.V.M., Lopes I. Screening evaluation of the ecotoxicity and genotoxicity of soils contaminated with organic and inorganic nanoparticles: The role of ageing. J. Hazard. Mater. 2011;194:345–354. doi: 10.1016/j.jhazmat.2011.07.112. [DOI] [PubMed] [Google Scholar]
- 95.Senapati V.A., Kansara K., Shanker R., Dhawan A., Kumar A. Monitoring characteristics and genotoxic effects of engineered nanoparticle-protein corona. Mutagenesis. 2017;32:479–490. doi: 10.1093/mutage/gex028. [DOI] [PubMed] [Google Scholar]
- 96.Dubey P., Matai I., Kumar S.U., Sachdev A., Bhushan B., Gopinath P. Perturbation of cellular mechanistic system by silver nanoparticle toxicity: Cytotoxic, genotoxic and epigenetic potentials. Adv. Colloid Interface Sci. 2015;221:4–21. doi: 10.1016/j.cis.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 97.Xu L., Xu M., Wang R., Yin Y., Lynch I., Liu S. The Crucial Role of Environmental Coronas in Determining the Biological Effects of Engineered Nanomaterials. Small. 2020;16 doi: 10.1002/smll.202003691. [DOI] [PubMed] [Google Scholar]
- 98.Rasmussen K., Rauscher H., Mech A., Riego Sintes J., Gilliland D., González M., Kearns P., Moss K., Visser M., Groenewold M., et al. Physico-chemical properties of manufactured nanomaterials—Characterisation and relevant methods. An outlook based on the OECD Testing Programme. Regul. Toxicol. Pharmacol. 2018;92:8–28. doi: 10.1016/j.yrtph.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hansen U., Thünemann A.F. Characterization of Silver Nanoparticles in Cell Culture Medium Containing Fetal Bovine Serum. Langmuir. 2015;31:6842–6852. doi: 10.1021/acs.langmuir.5b00687. [DOI] [PubMed] [Google Scholar]
- 100.Precupas A., Gheorghe D., Botea-Petcu A., Leonties A.R., Sandu R., Popa V.T., Mariussen E., Naouale E.Y., Rundén-Pran E., Dumit V., et al. Thermodynamic Parameters at Bio-Nano Interface and Nanomaterial Toxicity: A Case Study on BSA Interaction with ZnO, SiO2, and TiO2. Chem. Res. Toxicol. 2020;33:2054–2071. doi: 10.1021/acs.chemrestox.9b00468. [DOI] [PubMed] [Google Scholar]
- 101.Savage D.T., Hilt J.Z., Dziubla T.D. Nanotoxicity. Humana Press; New York, NY, USA: 2019. In Vitro Methods for Assessing Nanoparticle Toxicity; pp. 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.DeLoid G.M., Cohen J.M., Pyrgiotakis G., Demokritou P. An integrated dispersion preparation, characterization and in vitro dosimetry methodology for engineered nanomaterials. Nat Protoc. 2017;12:335–371. doi: 10.1038/nprot.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pulido-Reyes G., Leganes F., Fernández-Piñas F., Rosal R. Bio-nano interface and environment: A critical review. Environ. Toxicol. Chem. 2017;36:3181–3193. doi: 10.1002/etc.3924. [DOI] [PubMed] [Google Scholar]
- 104.Moore T.L., Rodriguez-Lorenzo L., Hirsch V., Balog S., Urban D., Jud C., Rothen-Rutishauser B., Lattuada M., Petri-Fink A. Nanoparticle colloidal stability in cell culture media and impact on cellular interactions. Chem. Soc. Rev. 2015;44:6287–6305. doi: 10.1039/C4CS00487F. [DOI] [PubMed] [Google Scholar]
- 105.Clogston J.D., Patri A.K. Characterization of Nanoparticles Intended for Drug Delivery. Volume 697. Humana Press; New York, NY, USA: 2011. Zeta Potential Measurement; pp. 63–70. Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- 106.Bednar A.J., Poda A.R., Mitrano D.M., Kennedy A.J., Gray E.P., Ranville J.F., Hayes C.A., Crocker F.H., Steevens J.A. Comparison of on-line detectors for field flow fractionation analysis of nanomaterials. Talanta. 2013;104:140–148. doi: 10.1016/j.talanta.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 107.Almutary A., Sanderson B.J.S. The MTT and Crystal Violet Assays: Potential Confounders in Nanoparticle Toxicity Testing. Int. J. Toxicol. 2016;35:454–462. doi: 10.1177/1091581816648906. [DOI] [PubMed] [Google Scholar]
- 108.Oostingh G.J., Casals E., Italiani P., Colognato R., Stritzinger R., Ponti J., Pfaller T., Kohl Y., Ooms D., Favilli F., et al. Problems and challenges in the development and validation of human cell-based assays to determine nanoparticle-induced immunomodulatory effects. Part. Fibre Toxicol. 2011;8:8. doi: 10.1186/1743-8977-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Majtnerová P., Roušar T. An overview of apoptosis assays detecting DNA fragmentation. Mol. Biol. Rep. 2018;45:1469–1478. doi: 10.1007/s11033-018-4258-9. [DOI] [PubMed] [Google Scholar]
- 110.Koppen G., Azqueta A., Pourrut B., Brunborg G., Collins A.R., Langie S.A.S. The next three decades of the comet assay: A report of the 11th International Comet Assay Workshop. Mutagenesis. 2017;32:397–408. doi: 10.1093/mutage/gex002. [DOI] [PubMed] [Google Scholar]
- 111.Reisinger K., Blatz V., Brinkmann J., Downs T.R., Fischer A., Henkler F., Hoffmann S., Krul C., Liebsch M., Luch A., et al. Validation of the 3D Skin Comet assay using full thickness skin models: Transferability and reproducibility. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2018;827:27–41. doi: 10.1016/j.mrgentox.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 112.Riss T., Niles A., Moravec R., Karassina N., Vidugiriene J. Cytotoxicity Assays: In Vitro Methods to Measure Dead Cells. In: Markossian S., Sittampalam G.S., Grossman A., Brimacombe K., Arkin M., Auld. D., Austin C.P., Baell J., Caaveiro J.M.M., Chung T.D.Y., et al., editors. Assay Guidance Manual. The Assay Guidance Manual, Eli Lilly & Company and the National Center for Advancing Translational Sciences; Bethesda, MD, USA: 2004. [PubMed] [Google Scholar]
- 113.ISO 10993-5:2009—Biological Evaluation of Medical Devices—Part 5: Tests for In Vitro Cytotoxicity. [(accessed on 27 August 2020)]; Available online: https://www.iso.org/standard/36406.html.
- 114.OECD . Test No. 476: In vitro Mammalian Cell Gene Mutation Test. OECD Publishing for the Testing Chemicals Section; Paris, France: 1997. [Google Scholar]
- 115.OECD . Test No. 490: In Vitro Mammalian Cell Gene Mutation Tests Using the Thymidine Kinase Gene. OECD; Paris, France: 2015. [Google Scholar]
- 116.Rubio L., El Yamani N., Kazimirova A., Dusinska M., Marcos R. Multi-walled carbon nanotubes (NM401) induce ROS-mediated HPRT mutations in Chinese hamster lung fibroblasts. Environ. Res. 2016;146:185–190. doi: 10.1016/j.envres.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 117.Cheng T.F., Patton G.W., Muldoon-Jacobs K. Can the L5178Y Tk+/- mouse lymphoma assay detect epigenetic silencing? Food Chem. Toxicol. 2013;59:187–190. doi: 10.1016/j.fct.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 118.Shibai-Ogata A., Kakinuma C., Hioki T., Kasahara T. Evaluation of high-throughput screening for in vitro micronucleus test using fluorescence-based cell imaging. Mutagenesis. 2011;26:709–719. doi: 10.1093/mutage/ger037. [DOI] [PubMed] [Google Scholar]
- 119.Kirsch-Volders M., Plas G., Elhajouji A., Lukamowicz M., Gonzalez L., Vande Loock K., Decordier I. The in vitro MN assay in 2011: Origin and fate, biological significance, protocols, high throughput methodologies and toxicological relevance. Arch. Toxicol. 2011;85:873–899. doi: 10.1007/s00204-011-0691-4. [DOI] [PubMed] [Google Scholar]
- 120.OECD . Test No. 487: In Vitro Mammalian Cell Micronucleus Test. OECD; Paris, France: 2016. OECD Guidelines for the Testing of Chemicals, Section 4. [Google Scholar]
- 121.García-Rodríguez A., Kazantseva L., Vila L., Rubio L., Velázquez A., Ramírez M.J., Marcos R., Hernández A. Micronuclei detection by flow cytometry as a high-throughput approach for the genotoxicity testing of nanomaterials. Nanomaterials. 2019;9:1677. doi: 10.3390/nano9121677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Di Bucchianico S., Cappellini F., Le Bihanic F., Zhang Y., Dreij K., Karlsson H.L. Genotoxicity of TiO2 nanoparticles assessed by mini-gel comet assay and micronucleus scoring with flow cytometry. Mutagenesis. 2017;32:127–137. doi: 10.1093/mutage/gew030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Watson C., Ge J., Cohen J., Pyrgiotakis G., Engelward B.P., Demokritou P. High-throughput screening platform for engineered nanoparticle-mediated genotoxicity using cometchip technology. ACS Nano. 2014;8:2118–2133. doi: 10.1021/nn404871p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Collins A., El Yamani N., Dusinska M. Sensitive detection of DNA oxidation damage induced by nanomaterials. Free Radic. Biol. Med. 2017;107:69–76. doi: 10.1016/j.freeradbiomed.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 125.Collins A.R. The use of bacterial repair endonucleases in the comet assay. In: Gautier J.C., editor. Drug Safety Evaluation. Volume 1641. Humana Press; New York, NY, USA: 2010. Methods in Molecular Biology. [DOI] [Google Scholar]
- 126.Mah L.J., El-Osta A., Karagiannis T.C. γh2AX: A sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24:679–686. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- 127.Rothkamm K., Barnard S., Moquet J., Ellender M., Rana Z., Burdak-Rothkamm S. DNA damage foci: Meaning and significance. Environ. Mol. Mutagen. 2015;56:491–504. doi: 10.1002/em.21944. [DOI] [PubMed] [Google Scholar]
- 128.Wan R., Mo Y., Tong R., Gao M., Zhang Q. Nanotoxicity. Volume 1894. Humana Press; New York, NY, USA: 2019. Determination of phosphorylated histone H2AX in nanoparticle-induced genotoxic studies; pp. 145–159. Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- 129.Elespuru R., Pfuhler S., Aardema M.J., Chen T., Doak S.H., Doherty A., Farabaugh C.S., Kenny J., Manjanatha M., Mahadevan B., et al. Genotoxicity assessment of nanomaterials: Recommendations on best practices, assays, and methods. Toxicol. Sci. 2018;164:391–416. doi: 10.1093/toxsci/kfy100. [DOI] [PubMed] [Google Scholar]
- 130.Bin-Jumah M.N., Al-Abdan M., Al-Basher G., Alarifi S. Molecular Mechanism of Cytotoxicity, Genotoxicity, and Anticancer Potential of Green Gold Nanoparticles on Human Liver Normal and Cancerous Cells. Dose Response. 2020;18:1559325820912154. doi: 10.1177/1559325820912154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.El-Hussein A., Hamblin M.R. ROS generation and DNA damage with photo-inactivation mediated by silver nanoparticles in lung cancer cell line. IET Nanobiotechnol. 2017;11:173–178. doi: 10.1049/iet-nbt.2015.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Audebert M., Dolo L., Perdu E., Cravedi J.-P., Zalko D. Use of the γH2AX assay for assessing the genotoxicity of bisphenol A and bisphenol F in human cell lines. Arch. Toxicol. 2011;85:1463–1473. doi: 10.1007/s00204-011-0721-2. [DOI] [PubMed] [Google Scholar]
- 133.Bonner W.M., Redon C.E., Dickey J.S., Nakamura A.J., Sedelnikova O.A., Solier S., Pommier Y. γH2AX and cancer. Nat. Rev. Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhu L., Chang D.W., Dai L., Hong Y. DNA damage induced by multiwalled carbon nanotubes in mouse embryonic stem cells. Nano Lett. 2007;7:3592–3597. doi: 10.1021/nl071303v. [DOI] [PubMed] [Google Scholar]
- 135.Cveticanin J., Joksic G., Leskovac A., Petrovic S., Sobot A.V., Neskovic O. Using carbon nanotubes to induce micronuclei and double strand breaks of the DNA in human cells. Nanotechnology. 2010;21:015102. doi: 10.1088/0957-4484/21/1/015102. [DOI] [PubMed] [Google Scholar]
- 136.Valdiglesias V., Costa C., Kiliç G., Costa S., Pásaro E., Laffon B., Teixeira J.P. Neuronal cytotoxicity and genotoxicity induced by zinc oxide nanoparticles. Environ. Int. 2013;55:92–100. doi: 10.1016/j.envint.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 137.Chattopadhyay N., Cai Z., Kwon Y.L., Lechtman E., Pignol J.P., Reilly R.M. Molecularly targeted gold nanoparticles enhance the radiation response of breast cancer cells and tumor xenografts to X-radiation. Breast Cancer Res. Treat. 2013;137:81–91. doi: 10.1007/s10549-012-2338-4. [DOI] [PubMed] [Google Scholar]
- 138.Msiska Z., Pacurari M., Mishra A., Leonard S.S., Castranova V., Vallyathan V. DNA Double-Strand Breaks by Asbestos, Silica, and Titanium Dioxide. Am. J. Respir. Cell Mol. Biol. 2010;43:210–219. doi: 10.1165/rcmb.2009-0062OC. [DOI] [PubMed] [Google Scholar]
- 139.Paget V., Dekali S., Kortulewski T., Grall R., Gamez C., Blazy K., Aguerre-Chariol O., Chevillard S., Braun A., Rat P., et al. Specific uptake and genotoxicity induced by polystyrene nanobeads with distinct surface chemistry on human lung epithelial cells and macrophages. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0123297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hernández L.G., van Steeg H., Luijten M., van Benthem J. Mechanisms of non-genotoxic carcinogens and importance of a weight of evidence approach. Mutat. Res. Rev. Mutat. Res. 2009;682:94–109. doi: 10.1016/j.mrrev.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 141.Trouiller B., Reliene R., Westbrook A., Solaimani P., Schiestl R.H. Titanium dioxide nanoparticles induce DNA damage and genetic instability in vivo in mice. Cancer Res. 2009;69:8784–8789. doi: 10.1158/0008-5472.CAN-09-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Toyooka T., Amano T., Ibuki Y. Titanium dioxide particles phosphorylate histone H2AX independent of ROS production. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2012;742:84–91. doi: 10.1016/j.mrgentox.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 143.Kerckaert G.A., LeBoeuf R.A., Isfort R.J. Use of the Syrian hamster embryo cell transformation assay for determining the carcinogenic potential of heavy metal compounds. Fundam. Appl. Toxicol. 1996;34:67–72. doi: 10.1006/faat.1996.0176. [DOI] [PubMed] [Google Scholar]
- 144.LeBoeuf R.A., Kerckaert G.A., Aardema M.J., Gibson D.P., Brauninger R., Isfort R.J. The pH 6.7 Syrian hamster embryo cell transformation assay for assessing the carcinogenic potential of chemicals. Mutat. Res. 1996;356:85–127. doi: 10.1016/0027-5107(95)00199-9. [DOI] [PubMed] [Google Scholar]
- 145.Mauthe R.J., Gibson D.P., Bunch R.T., Custer L., Syrian Hamster Embryo Assay Working Group Response to Alternative models for carcinogenicity testing: Weight of evidence across models. Sam Cohen, Toxicologic Pathology (2001) 29(suppl.): 183–190. Toxicol. Pathol. 2002;30:292–293. doi: 10.1080/019262302753559641. [DOI] [PubMed] [Google Scholar]
- 146.Creton S., Aardema M.J., Carmichael P.L., Harvey J.S., Martin F.L., Newbold R.F., O’Donovan M.R., Pant K., Poth A., Sakai A., et al. Cell transformation assays for prediction of carcinogenic potential: State of the science and future research needs. Mutagenesis. 2012;27:93–101. doi: 10.1093/mutage/ger053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fontana C., Kirsch A., Seidel C., Marpeaux L., Darne C., Gaté L., Remy A., Guichard Y. In vitro cell transformation induced by synthetic amorphous silica nanoparticles. Mutat. Res. 2017;823:22–27. doi: 10.1016/j.mrgentox.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 148.Cui Y., Liu H., Ze Y., Zengli Z., Hu Y., Cheng Z., Cheng J., Hu R., Gao G., Wang L., et al. Gene expression in liver injury caused by long-term exposure to titanium dioxide nanoparticles in mice. Toxicol. Sci. 2012;128 doi: 10.1093/TOXSCI/KFS153. [DOI] [PubMed] [Google Scholar]
- 149.Dawson M.A., Kouzarides T. Cancer epigenetics: From mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 150.Gao G., Ze Y., Li B., Zhao X., Zhang T., Sheng L., Hu R., Gui S., Sang X., Sun Q., et al. Ovarian dysfunction and gene-expressed characteristics of female mice caused by long-term exposure to titanium dioxide nanoparticles. J. Hazard. Mater. 2012;243:19–27. doi: 10.1016/j.jhazmat.2012.08.049. [DOI] [PubMed] [Google Scholar]
- 151.Gojova A., Lee J.-T., Jung H.S., Guo B., Barakat A.I., Kennedy I.M. Effect of cerium oxide nanoparticles on inflammation in vascular endothelial cells. Inhal. Toxicol. 2009;21(Suppl. 1):123–130. doi: 10.1080/08958370902942582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Moos P.J., Olszewski K., Honeggar M., Cassidy P., Leachman S., Woessner D., Cutler N.S., Veranth J.M. Responses of human cells to ZnO nanoparticles: A gene transcription study. Metallomics. 2011;3:1199–1211. doi: 10.1039/c1mt00061f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Park E.-J., Choi J., Park Y.-K., Park K. Oxidative stress induced by cerium oxide nanoparticles in cultured BEAS-2B cells. Toxicology. 2008;245:90–100. doi: 10.1016/j.tox.2007.12.022. [DOI] [PubMed] [Google Scholar]
- 154.Smolkova B., El Yamani N., Collins A.R., Gutleb A.C., Dusinska M. Nanoparticles in food. Epigenetic changes induced by nanomaterials and possible impact on health. Food Chem. Toxicol. 2015;77:64–73. doi: 10.1016/j.fct.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 155.Rossner P., Vrbova K., Rossnerova A., Zavodna T., Milcova A., Klema J., Vecera Z., Mikuska P., Coufalik P., Capka L., et al. Gene Expression and Epigenetic Changes in Mice Following Inhalation of Copper(II) Oxide Nanoparticles. Nanomaterials. 2020;10:550. doi: 10.3390/nano10030550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Smolkova B., Dusinska M., Gabelova A. Nanomedicine and epigenome. Possible health risks. Food Chem. Toxicol. 2017;109:780–796. doi: 10.1016/j.fct.2017.07.020. [DOI] [PubMed] [Google Scholar]
- 157.Cavalli G., Heard E. Advances in epigenetics link genetics to the environment and disease. Nature. 2019;571:489–499. doi: 10.1038/s41586-019-1411-0. [DOI] [PubMed] [Google Scholar]
- 158.Smolkova B., Dusinska M., Gabelova A. Epigenetic Effects of Nanomaterials. Encycl. Environ. Heal. 2019:678–685. doi: 10.1016/B978-0-12-409548-9.11044-9. [DOI] [Google Scholar]
- 159.Stoccoro A., Karlsson H.L., Coppedè F., Migliore L. Epigenetic effects of nano-sized materials. Toxicology. 2013;313:3–14. doi: 10.1016/j.tox.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 160.Lamas A., Franco C.M., Regal P., Miranda J.M., Vázquez B., Cepeda A. Polymerase Chain Reaction for Biomedical Applications. InTech; London, UK: 2016. High-Throughput Platforms in Real-Time PCR and Applications. Chapter 2. [DOI] [Google Scholar]
- 161.Dusinska M., Tulinska J., El Yamani N., Kuricova M., Liskova A., Rollerova E., Rundén-Pran E., Smolkova B. Immunotoxicity, genotoxicity and epigenetic toxicity of nanomaterials: New strategies for toxicity testing? Food Chem. Toxicol. 2017;109:797–811. doi: 10.1016/j.fct.2017.08.030. [DOI] [PubMed] [Google Scholar]
- 162.Nel A., Xia T., Meng H., Wang X., Lin S., Ji Z., Zhang H. Nanomaterial toxicity testing in the 21st century: Use of a predictive toxicological approach and high-throughput screening. Acc. Chem. Res. 2013;46:607–621. doi: 10.1021/ar300022h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Seo J.-E., Tryndyak V., Wu Q., Dreval K., Pogribny I., Bryant M., Zhou T., Robison T.W., Mei N., Guo X. Quantitative comparison of in vitro genotoxicity between metabolically competent HepaRG cells and HepG2 cells using the high-throughput high-content CometChip assay. Arch. Toxicol. 2019;93:1433–1448. doi: 10.1007/s00204-019-02406-9. [DOI] [PubMed] [Google Scholar]
- 164.Mack M., Schweinlin K., Mirsberger N., Zubel T., Bürkle A. Automated screening for oxidative or methylation-induced DNA damage in human cells. ALTEX. 2020 doi: 10.14573/altex.2001221. [DOI] [PubMed] [Google Scholar]
- 165.Baltazar M.T., Cable S., Carmichael P.L., Cubberley R., Cull T., Delagrange M., Dent M.P., Hatherell S., Houghton J., Kukic P., et al. A Next-Generation Risk Assessment Case Study for Coumarin in Cosmetic Products. Toxicol. Sci. 2020;176:236–252. doi: 10.1093/toxsci/kfaa048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Johann S., Goßen M., Behnisch P.A., Hollert H., Seiler T.-B. Combining Different In Vitro Bioassays to Evaluate Genotoxicity of Water-Accommodated Fractions from Petroleum Products. Toxics. 2020;8:45. doi: 10.3390/toxics8020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Seo J.-E., Wu Q., Bryant M., Ren L., Shi Q., Robison T.W., Mei N., Manjanatha M.G., Guo X. Performance of high-throughput CometChip assay using primary human hepatocytes: A comparison of DNA damage responses with in vitro human hepatoma cell lines. Arch. Toxicol. 2020;94:2207–2224. doi: 10.1007/s00204-020-02736-z. [DOI] [PubMed] [Google Scholar]
- 168.Takeiri A., Matsuzaki K., Motoyama S., Yano M., Harada A., Katoh C., Tanaka K., Mishima M. High-content imaging analyses of γH2AX-foci and micronuclei in TK6 cells elucidated genotoxicity of chemicals and their clastogenic/aneugenic mode of action. Genes Environ. Off. J. Jpn. Environ. Mutagen Soc. 2019;41:4. doi: 10.1186/s41021-019-0117-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Damoiseaux R., George S., Li M., Pokhrel S., Ji Z., France B., Xia T., Suarez E., Rallo R., Mädler L., et al. No time to lose—high throughput screening to assess nanomaterial safety. Nanoscale. 2011;3:1345. doi: 10.1039/c0nr00618a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.George S., Xia T., Rallo R., Zhao Y., Ji Z., Lin S., Wang X., Zhang H., France B., Schoenfeld D., et al. Use of a high-throughput screening approach coupled with in vivo zebrafish embryo screening to develop hazard ranking for engineered nanomaterials. ACS Nano. 2011;5:1805–1817. doi: 10.1021/nn102734s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Moller P., Moller L., Godschalk R.W.L., Jones G.D.D. Assessment and reduction of comet assay variation in relation to DNA damage: Studies from the European Comet Assay Validation Group. Mutagenesis. 2010;25:109–111. doi: 10.1093/mutage/gep067. [DOI] [PubMed] [Google Scholar]
- 172.Wilde S., Dambowsky M., Hempt C., Sutter A., Queisser N. Classification of in vitro genotoxicants using a novel multiplexed biomarker assay compared to the flow cytometric micronucleus test. Environ. Mol. Mutagen. 2017;58:662–677. doi: 10.1002/em.22130. [DOI] [PubMed] [Google Scholar]
- 173.Shaposhnikov S., Azqueta A., Henriksson S., Meier S., Gaivão I., Huskisson N.H., Smart A., Brunborg G., Nilsson M., Collins A.R. Twelve-gel slide format optimised for comet assay and fluorescent in situ hybridisation. Toxicol. Lett. 2010;195:31–34. doi: 10.1016/j.toxlet.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 174.Gutzkow K.B., Langleite T.M., Meier S., Graupner A., Collins A.R., Brunborg G. High-throughput comet assay using 96 minigels. Mutagenesis. 2013;28:333–340. doi: 10.1093/mutage/get012. [DOI] [PubMed] [Google Scholar]
- 175.Azqueta A., Gutzkow K.B., Priestley C.C., Meier S., Walker J.S., Brunborg G., Collins A.R. A comparative performance test of standard, medium- and high-throughput comet assays. Toxicol. In Vitro. 2013;27:768–773. doi: 10.1016/j.tiv.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 176.Stang A., Witte I. Performance of the comet assay in a high-throughput version. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2009;675:5–10. doi: 10.1016/j.mrgentox.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 177.Huk A., Collins A.R., El Yamani N., Porredon C., Azqueta A., De Lapuente J., Dusinska M. Critical factors to be considered when testing nanomaterials for genotoxicity with the comet assay. Mutagenesis. 2015;30:85–88. doi: 10.1093/mutage/geu077. [DOI] [PubMed] [Google Scholar]
- 178.García-Rodríguez A., Rubio L., Vila L., Xamena N., Velázquez A., Marcos R., Hernández A. The comet assay as a tool to detect the genotoxic potential of nanomaterials. Nanomaterials. 2019;9:1385. doi: 10.3390/nano9101385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Rubio L., Annangi B., Vila L., Hernández A., Marcos R. Antioxidant and anti-genotoxic properties of cerium oxide nanoparticles in a pulmonary-like cell system. Arch. Toxicol. 2016;90:269–278. doi: 10.1007/s00204-015-1468-y. [DOI] [PubMed] [Google Scholar]
- 180.Fenech M., Kirsch-Volders M., Rossnerova A., Sram R., Romm H., Bolognesi C., Ramakumar A., Soussaline F., Schunck C., Elhajouji A., et al. HUMN project initiative and review of validation, quality control and prospects for further development of automated micronucleus assays using image cytometry systems. Int. J. Hyg. Environ. Health. 2013;216:541–552. doi: 10.1016/j.ijheh.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 181.Roemer E., Zenzen V., Conroy L.L., Luedemann K., Dempsey R., Schunck C., Sticken E.T. Automation of the in vitro micronucleus and chromosome aberration assay for the assessment of the genotoxicity of the particulate and gas-vapor phase of cigarette smoke. Toxicol. Mech. Methods. 2015;25:320–333. doi: 10.3109/15376516.2015.1037413. [DOI] [PubMed] [Google Scholar]
- 182.Decordier I., Papine A., Plas G., Roesems S., Vande Loock K., Moreno-Palomo J., Cemeli E., Anderson D., Fucic A., Marcos R., et al. Automated image analysis of cytokinesis-blocked micronuclei: An adapted protocol and a validated scoring procedure for biomonitoring. Mutagenesis. 2009;24:85–93. doi: 10.1093/mutage/gen057. [DOI] [PubMed] [Google Scholar]
- 183.Nüsse M., Kramer J. Flow cytometric analysis of micronuclei found in cells after irradiation. Cytometry. 1984;5:20–25. doi: 10.1002/cyto.990050105. [DOI] [PubMed] [Google Scholar]
- 184.Avlasevich S., Bryce S., De Boeck M., Elhajouji A., Van Goethem F., Lynch A., Nicolette J., Shi J., Dertinger S. Flow cytometric analysis of micronuclei in mammalian cell cultures: Past, present and future. Mutagenesis. 2011;26:147–152. doi: 10.1093/mutage/geq058. [DOI] [PubMed] [Google Scholar]
- 185.Bryce S.M., Avlasevich S.L., Bemis J.C., Phonethepswath S., Dertinger S.D. Miniaturized flow cytometric in vitro micronucleus assay represents an efficient tool for comprehensively characterizing genotoxicity dose-response relationships. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2010;703:191–199. doi: 10.1016/j.mrgentox.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bryce S.M., Avlasevich S.L., Bemis J.C., Tate M., Walmsley R.M., Saad F., Van Dijck K., De Boeck M., Van Goethem F., Lukamowicz-Rajska M., et al. Flow cytometric 96-well microplate-based in vitro micronucleus assay with human TK6 cells: Protocol optimization and transferability assessment. Environ. Mol. Mutagen. 2013;54:180–194. doi: 10.1002/em.21760. [DOI] [PubMed] [Google Scholar]
- 187.Bryce S.M., Bemis J.C., Avlasevich S.L., Dertinger S.D. In vitro micronucleus assay scored by flow cytometry provides a comprehensive evaluation of cytogenetic damage and cytotoxicity. Mutat. Res. 2007;630:78–91. doi: 10.1016/j.mrgentox.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Cai Z., Vallis K.A., Reilly R.M. Computational analysis of the number, area and density of gamma-H2AX foci in breast cancer cells exposed to (111)In-DTPA-hEGF or gamma-rays using Image-J software. Int. J. Radiat. Biol. 2009;85:262–271. doi: 10.1080/09553000902748757. [DOI] [PubMed] [Google Scholar]
- 189.Hou Y.-N., Lavaf A., Huang D., Peters S., Huq R., Friedrich V., Rosenstein B.S., Kao J. Development of an Automated γ-H2AX Immunocytochemistry Assay. Radiat. Res. 2009;171:360–367. doi: 10.1667/RR1349.1. [DOI] [PubMed] [Google Scholar]
- 190.Roch-Lefèvre S., Mandina T., Voisin P., Gaëtan G., Mesa J.E.G., Valente M., Bonnesoeur P., García O., Voisin P., Roy L. Quantification of γ-H2AX Foci in Human Lymphocytes: A Method for Biological Dosimetry after Ionizing Radiation Exposure. Radiat. Res. 2010;174:185–194. doi: 10.1667/RR1775.1. [DOI] [PubMed] [Google Scholar]
- 191.Bhogal N., Jalali F., Bristow R.G. Microscopic imaging of DNA repair foci in irradiated normal tissues. Int. J. Radiat. Biol. 2009;85:732–746. doi: 10.1080/09553000902785791. [DOI] [PubMed] [Google Scholar]
- 192.Ivashkevich A.N., Martin O.A., Smith A.J., Redon C.E., Bonner W.M., Martin R.F., Lobachevsky P.N. γH2AX foci as a measure of DNA damage: A computational approach to automatic analysis. Mutat. Res. 2011;711:49–60. doi: 10.1016/j.mrfmmm.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Harris G., Palosaari T., Magdolenova Z., Mennecozzi M., Gineste J.M., Saavedra L., Milcamps A., Huk A., Collins A.R., Dusinska M., et al. Iron oxide nanoparticle toxicity testing using high-throughput analysis and high-content imaging. Nanotoxicology. 2015;9(Suppl. 1):87–94. doi: 10.3109/17435390.2013.816797. [DOI] [PubMed] [Google Scholar]
- 194.Karlsson H.L., Gliga A.R., Calléja F.M.G.R., Gonçalves C.S.A.G., Wallinder I.O., Vrieling H., Fadeel B., Hendriks G. Mechanism-based genotoxicity screening of metal oxide nanoparticles using the ToxTracker panel of reporter cell lines. Part. Fibre Toxicol. 2014;11:41. doi: 10.1186/s12989-014-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Alépée N. State-of-the-art of 3D cultures (organs-on-a-chip) in safety testing and pathophysiology. ALTEX. 2014:441–477. doi: 10.14573/altex1406111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Chen L., Li N., Liu Y., Faquet B., Alépée N., Ding C., Eilstein J., Zhong L., Peng Z., Ma J., et al. A new 3D model for genotoxicity assessment: EpiSkinTM Micronucleus Assay. Mutagenesis. 2020:1–11. doi: 10.1093/mutage/geaa003. [DOI] [PubMed] [Google Scholar]
- 197.Tahara H., Sadamoto K., Yamagiwa Y., Nemoto S., Kurata M. Investigation of comet assays under conditions mimicking ocular instillation administration in a three-dimensional reconstructed human corneal epithelial model. Cutan. Ocul. Toxicol. 2019;38:375–383. doi: 10.1080/15569527.2019.1634580. [DOI] [PubMed] [Google Scholar]
- 198.Mandon M., Huet S., Dubreil E., Fessard V., Le Hégarat L. Three-dimensional HepaRG spheroids as a liver model to study human genotoxicity in vitro with the single cell gel electrophoresis assay. Sci. Rep. 2019;9:10548. doi: 10.1038/s41598-019-47114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Štampar M., Tomc J., Filipič M., Žegura B. Development of in vitro 3D cell model from hepatocellular carcinoma (HepG2) cell line and its application for genotoxicity testing. Arch. Toxicol. 2019;93:3321–3333. doi: 10.1007/s00204-019-02576-6. [DOI] [PubMed] [Google Scholar]
- 200.Shah U.-K., Mallia J.d.O., Singh N., Chapman K.E., Doak S.H., Jenkins G.J.S. A three-dimensional in vitro HepG2 cells liver spheroid model for genotoxicity studies. Mutat. Res. Toxicol. Environ. Mutagen. 2018;825:51–58. doi: 10.1016/j.mrgentox.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 201.van Duinen V., Trietsch S.J., Joore J., Vulto P., Hankemeier T. Microfluidic 3D cell culture: From tools to tissue models. Curr. Opin. Biotechnol. 2015;35:118–126. doi: 10.1016/j.copbio.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 202.Bhatia S.N., Ingber D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 203.Huh D., Leslie D.C., Matthews B.D., Fraser J.P., Jurek S., Hamilton G.A., Thorneloe K.S., McAlexander M.A., Ingber D.E. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci. Transl. Med. 2012;4 doi: 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Bricks T., Paullier P., Legendre A., Fleury M.-J., Zeller P., Merlier F., Anton P.M., Leclerc E. Development of a new microfluidic platform integrating co-cultures of intestinal and liver cell lines. Toxicol. In Vitro. 2014;28:885–895. doi: 10.1016/j.tiv.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 205.Gordon S., Daneshian M., Bouwstra J., Caloni F., Constant S., Davies D.E., Dandekar G., Guzman C.A., Fabian E., Haltner E., et al. Non-animal models of epithelial barriers (skin, intestine and lung) in research, industrial applications and regulatory toxicology. ALTEX. 2015;32:327–378. doi: 10.14573/altex.1510051. [DOI] [PubMed] [Google Scholar]
- 206.Maschmeyer I., Lorenz A.K., Schimek K., Hasenberg T., Ramme A.P., Hübner J., Lindner M., Drewell C., Bauer S., Thomas A., et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip. 2015;15:2688–2699. doi: 10.1039/C5LC00392J. [DOI] [PubMed] [Google Scholar]
- 207.Maschmeyer I., Hasenberg T., Jaenicke A., Lindner M., Lorenz A.K., Zech J., Garbe L.A., Sonntag F., Hayden P., Ayehunie S., et al. Chip-based human liver-intestine and liver-skin co-cultures—A first step toward systemic repeated dose substance testing in vitro. Eur. J. Pharm. Biopharm. 2015;95:77–87. doi: 10.1016/j.ejpb.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Materne E.-M., Maschmeyer I., Lorenz A.K., Horland R., Schimek K.M.S., Busek M., Sonntag F., Lauster R., Marx U. The multi-organ chip—A microfluidic platform for long-term multi-tissue coculture. J. Vis. Exp. 2015:e52526. doi: 10.3791/52526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Kim H.J., Ingber D.E. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr. Biol. 2013;5:1130–1140. doi: 10.1039/c3ib40126j. [DOI] [PubMed] [Google Scholar]
- 210.Kim H.J., Huh D., Hamilton G., Ingber D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12:2165–2174. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 211.Yin F., Zhu Y., Zhang M., Yu H., Chen W., Qin J. A 3D human placenta-on-a-chip model to probe nanoparticle exposure at the placental barrier. Toxicol. In Vitro. 2019;54:105–113. doi: 10.1016/j.tiv.2018.08.014. [DOI] [PubMed] [Google Scholar]
- 212.Zhang M., Xu C., Jiang L., Qin J. A 3D human lung-on-a-chip model for nanotoxicity testing. Toxicol. Res. 2018;7:1048–1060. doi: 10.1039/C8TX00156A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Dong H., Sun H. A Microchip for Integrated Single-Cell Gene Expression Profiling and Genotoxicity Detection. Sensors. 2016;16:1489. doi: 10.3390/s16091489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Dong H., Sun H., Zheng J. A microchip for integrated single-cell genotoxicity assay. Talanta. 2016;161:804–811. doi: 10.1016/j.talanta.2016.09.040. [DOI] [PubMed] [Google Scholar]
- 215.Vecchio G., Fenech M., Pompa P.P., Voelcker N.H. Lab-on-a-chip-based high-throughput screening of the genotoxicity of engineered nanomaterials. Small. 2014;10:2721–2734. doi: 10.1002/smll.201303359. [DOI] [PubMed] [Google Scholar]