Abstract
Simple Summary
Early diagnosis of breast cancer greatly increases the chance of cure and survival from the disease. The mammogram is widely used for early detection of breast cancer, but its effectiveness and accuracy have been a concern for a long time as well as its inability in detecting small cancers, especially in women with dense breast tissues. Therefore, it is an unmet clinical need to develop a simple, convenient test to overcome the shortcomings of mammography. Liquid biopsy, which is based on the analysis of body fluids, has attracted much attention in the search for cancer biomarkers. Recent advances in analytical techniques have gradually made it possible to detect breast cancer early through a biomarker analysis of blood, nipple aspirate fluid, sweat, urine, tears, or the breath. We envision that a simple blood or breath test holds great promise as a biomarker for early detection of breast cancer in the near future.
Abstract
Breast cancer is the most common cancer in women worldwide. Accurate early diagnosis of breast cancer is critical in the management of the disease. Although mammogram screening has been widely used for breast cancer screening, high false-positive and false-negative rates and radiation from mammography have always been a concern. Over the last 20 years, the emergence of “omics” strategies has resulted in significant advances in the search for non-invasive biomarkers for breast cancer diagnosis at an early stage. Circulating carcinoma antigens, circulating tumor cells, circulating cell-free tumor nucleic acids (DNA or RNA), circulating microRNAs, and circulating extracellular vesicles in the peripheral blood, nipple aspirate fluid, sweat, urine, and tears, as well as volatile organic compounds in the breath, have emerged as potential non-invasive diagnostic biomarkers to supplement current clinical approaches to earlier detection of breast cancer. In this review, we summarize the current progress of research in these areas.
Keywords: breast cancer, biomarker, diagnosis, detection, blood, body fluid
1. Introduction
Breast cancer is the most commonly diagnosed cancer in women worldwide. The incidence and mortality rates for female breast cancer far exceeded those for other cancers [1]. Although the incidence rate of breast cancer has risen, the mortality rates have steadily fallen due to early diagnosis and better treatments [2]. Early detection of breast cancer often leads to better outcomes. According to Cancer Australia’s National Cancer Control Indicators, the relative survival for females diagnosed with early-stage breast cancers at diagnosis was much higher than that for those with advanced breast cancers. Survival for early-stage breast cancer (stage 1) remained at 100% at 1, 3, and 5 years from diagnosis. However, survival for metastatic breast cancer (stage 4) reduced to 69% at 1 year, 47% at 3 years, and 32% at 5 years from diagnosis [3]. Therefore, detection of breast cancer at an early stage plays a pivotal role in reducing the mortality.
Mammogram screening has been commonly used for early detection of breast cancer in many high-income countries. However, the benefits and limitations of mammography have been a heated international debate for decades [4,5,6,7,8,9]. The main concerns with mammography are radiation and overdiagnosis. Studies have observed that the X-rays used for mammography could be a contributor to the onset of cancer. Overdiagnosis through breast cancer mammogram screening has been recognized as the most important adverse event and the estimates of overdiagnosis range from 0% to 40–50% depending on invitational age and methods [10,11]. In addition, mammograms cannot detect small tumors and are less accurate in cancer detection in women with dense breasts. Therefore, it is imperative to have alternative tools for early detection of breast cancer [12].
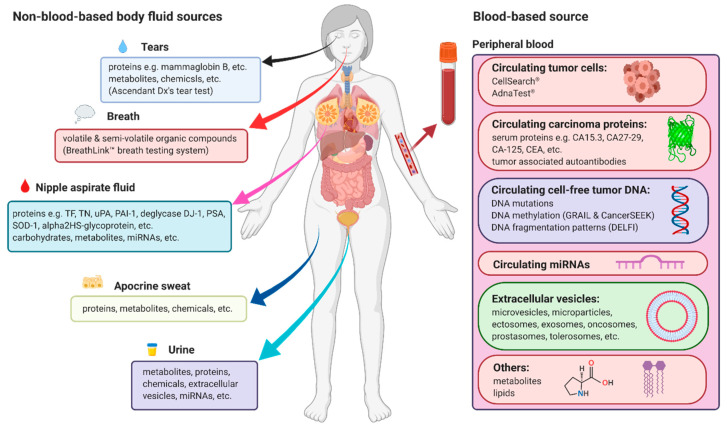
Breast cancer diagnosis is usually confirmed by using needle or surgical biopsy, which is not only invasive but also not necessary in most cases when tumors are benign. Therefore, much attention and many research efforts have been focused on the development of non-invasive and more convenient biomarkers that allow earlier detection of breast cancer. It has been reported that non-invasive body fluid-based tests, including circulating carcinoma antigens (CAs), circulating tumor cells (CTCs), circulating cell-free tumor nucleic acids (DNA or RNA), circulating microRNAs (miRNAs), circulating extracellular vesicles (EVs) in the peripheral blood, nipple aspirate fluid (NAF), sweat, urine, and tears, as well as volatile organic compounds (VOCs) in exhaled breath, have the potential to supplement current clinical approaches to earlier detection of breast cancer (Figure 1) [13,14]. In this review, we summarize the current progress of research in these areas (Table 1).
Figure 1.
Overview of current sources and main measurement types of non-invasive biomarkers for early detection of breast cancer. Figures were created with BioRender.com (https://biorender.com/).
Table 1.
Summary of potential non-invasive biomarkers for early detection of breast cancer.
| Sources | Types | Biomarkers Measured | Detection Types (Sample Size) | Sensitivity | Specificity | Notes | References |
|---|---|---|---|---|---|---|---|
| Peripheral blood | Circulating carcinoma proteins | serum proteins: CEA, FASL, OPN, VEGFC, VEGFD, HGF. | breast cancer (100) vs. non-breast cancer subjects (110). | 74.7%. | 77.0%. | AUC*: 0.79. | [15] |
| tumor-associated autoantibodies: FRS3, RAC3, HOXD1, GPR157, ZMYM6, EIF3E, CSNK1E, ZNF510, BMX, SF3A1, SOX2 | breast cancer (100) vs. non-breast cancer subjects (110) | 72.2% | 70.8% | AUC: 0.77 | |||
| serum proteins and tumor-associated autoantibodies: FASL, IL6, IL8, OPN, VEGFD, HGF, FRS3, MYOZ2, RAC3GPR157, ZMYM6, EIF3E, CSNK1E, ZNF510, BMXSF3A1, SOX2 | breast cancer (100) vs. non-breast cancer subjects (110) | 81.0% | 78.8% | AUC: 0.89 | |||
| Videssa Breast consisting of serum proteins: AFP, CA19-9, CEA, TNF-α, VEGF-C, ErbB2 (HER2); and tumor-associated autoantibodies: ANXA1, ATF3, ATP6AP1, BAT4 (GPANK1), BDNF, CTBP1, DBT, HOXD1, IGF2BP1, IGFBP2, ErbB2 (HER2) | women aged 25–75 (1145) | 93.0% | 64.0% | negative predictive value: 98.0% | [16] | ||
| Videssa Breast consisting of 11 serum protein biomarkers and 33 tumor-associated autoantibodies | women aged under 50 years (545): Breast cancer (32) vs. benign breast tumor (513) | 87.5% | 83.8% | negative predictive value: 99.1% | [17] | ||
| Circulating tumor cells | CellSearch® system | metastatic breast cancer (55) | 47% | N/A | overall positive agreement for both assays: 73% (CTC ≥ 2) and 69% (CTC ≥ 5) | [18] | |
| AdnaTest® assay | metastatic breast cancer (55) | 53% | N/A | ||||
| Circulating cell-free tumor DNA | ctDNA concentrations (≥0.75%) | colorectal and breast cancer patients (10) vs. healthy subjects (10) | >90.0% | >99.0% | [19] | ||
| ctDNA levels (amplifiable per ml of plasma) with CA15-3 expression level | breast cancer (27) | 96.0% | N/A | [20] | |||
| ctDNA levels with CTC numbers | breast cancer (30) | 97.0% | N/A | ||||
| ctDNA levels with PIK3CA E545K and H1047R mutations | early-stage breast cancer (29) | 93.3% | 100.0% | accuracy: 96.7% | [21] | ||
| SCGB3A1 DNA methylation in cfDNA | breast cancer (108) vs. female asymptomatic controls (103) | 16.8% | 80.0% | accuracy: 53.0% | [22,23] | ||
| methylation patterns of six genes in cfDNA: SFN, P16, hMLH1, HOXD13, PCDHGB7, and RASSF1a | breast cancer (125) vs. healthy subjects (104) | 79.6% | 72.4% | AUC: 0.727 | [24] | ||
| methylation analysis of a panel of 16 genes: 12 novel epigenetic markers: JAK3, RASGRF1, CPXM1, SHF, DNM3, CAV2, HOXA10, B3GNT5, ST3GAL6, DACH1, P2RX3, and chr8:23572595; and four internal control markers: CREM, GLYATL3, ELMOD3, and KLF9 |
breast cancer (87) vs. healthy subjects (80) | 86.2% | 82.7% | [25] | |||
| GRAIL DNA methylation patterns of cfDNA for detecting various types of cancers including breast cancer | training set: 844 | N/A | 99.8% | false-positive rate: <1% | [26] | ||
| validation set: 359 | N/A | 99.3% | false-positive rate: <1% | ||||
| GRAIL DNA methylation patterns of cfDNA for detecting breast cancer | training set: Breast cancer (247) | N/A | N/A | precision: 96% (82/85) | |||
| validation set: Breast cancer (104) | N/A | N/A | precision: 93% (40/43) | ||||
| DELFI fragmentation patterns of cfDNA | breast cancer (54) vs. healthy subjects (245) | 57.0% | 98.0% | [27] | |||
| DELFI fragmentation patterns of cfDNA combined with mutations of cfDNA | breast cancer (54) vs. healthy subjects (245) | 65.0% | 98.0% | ||||
| Circulating miRNAs | a panel of five miRNAs: miR-1246, miR-1307-3p, miR-4634, miR-6861-5p, and miR-6875-5p | breast cancer (1206), non-cancer controls (1343), benign breast diseases (54) | 97.3% | 82.9% | accuracy: 89.7% | [28] | |
| a panel of seven miRNAs: Hsa-miR-126-5p, hsa-miR-144-5p, hsa-miR-144-3p, hsa-miR-301a-3p, hsa-miR- 126-3p, hsa-miR-101-3p, and hsa-miR-664b-5p | triple-negative breast cancer (21) vs. healthy subjects (21) | 83.8% | 74.2% | accuracy: 79.0% AUC: 0.814 |
[29] | ||
| Extracellular vesicles | fibronectin | breast cancer (240) vs. non-cancer individuals (205) | 65.1% | 83.2% | AUC: 0.81 | [30] | |
| developmental endothelial locus-1 protein (Del-1) | breast cancer (100) vs. benign breast tumor (38), noncancerous diseases (58), and healthy subjects (46) | 92.31% | 86.62% | [31] | |||
| Metabolites | seven metabolites in the plasma: Glu, Orn, Thr, Trp, Met-SO, C2, and C3. | training cohort: breast cancer (80) vs. healthy subjects (100) validation cohort: breast cancer (109) vs. healthy subjects (50) |
N/A | N/A | AUC in training cohort: 0.87; AUC in validation cohort: 0.80 |
[32] | |
| three metabolites (8-hydroxy-2ʹ-deoxyguanosine, 1-methylguanosine, 1-methyl adenosine) combined with CA15-3 | malignant breast cancer (120) vs. benign breast disease (47) and healthy subjects (55) | 88.8% | 86.8% | AUC: 0.94 | [33] | ||
| seven metabolites in serum: Dimethyldodecane, galactose, α-glyceryl stearate, methyl stearate, 1(1-methoxycarbonyethyl)-4-(2-methyl-2-trimethylsilyl-oxypropyl) benzene, tetradecane, glucopyranoside | pre-operative breast cancer patients (152) vs. healthy subjects (155) | 96% | 100% | [34] | |||
| four plasma metabolites: L-octanoylcarnitine, 5-oxoproline, hypoxanthine, docosahexaenoic acid | discovery set: Breast cancer (40) vs. healthy subjects (30); validation set: Breast cancer (30) vs. healthy subjects (16) |
N/A | N/A | positive predictive value: 100.0% | [35] | ||
| metabolomics signature in the plasma | breast cancer (91) vs. healthy subjects (20) | up to 100.0% | up to 100.0% | [36] | |||
| Lipids | a panel of five serum free fatty acids: C16:1, C18:3, C18:2, C20:4, C22:6. | breast cancer (140) vs. healthy subjects (202) | 83.3% | 87.1% | AUC: 0.953 | [37] | |
| Multi-analyte tests | CancerSEEK testing of eight circulating proteins: CA-125, CA19-9, CEA, HGF, myeloperoxidase, OPN, prolactin, and TIMP-1; and mutations in 16 genes in cfDNA in the blood: NRAS, HRAS, KRAS, CTNNB1, PIK3CA, FBXW7, APC, EGFR, BRAF, CDKN2A, PTEN, FGFR2, AKT1, TP53, PPP2R1A, and GNAS | patients with one of the eight cancer types including breast cancer (1005) vs. individuals without known cancers (812) | 70.0% | >99% | [38] | ||
| breast cancer (209) vs. healthy subjects (812) | 33.0% | >99.0% | |||||
| CancerSEEK tests for different types of cancers | patients with cancers including breast cancer (96) vs. subjects without cancers (9,815) | 27.1% | 98.9% | positive predictive value: 19.4% | [39] | ||
| CancerSEEK tests for breast cancer | breast cancer (27) | 3.7% | N/A | ||||
| CancerSEEK remodeling with CancerA1DE method | first dataset: 1817 patient blood test records second dataset: 626 patient blood test records |
70.0% | 99.0% | remodeling of the CancerSEEK dataset to improve the sensitivity | [40] | ||
| Other body fluids | Urine | miRNAs (miR-21, miR-34a, miR-125b, miR-155, miR-195, miR-200b, miR-200c, miR-375, miR-451) | breast cancer (24) vs. healthy subjects (24) | 91.7% | 91.7% | AUC: 0.932 | [41] |
| four urinary microRNA types (miR-424, miR-423, miR-660, and let7-i) | breast cancer (69) vs. healthy subjects (40) | 98.6% | 100.0% | [42] | |||
| miRNA-21 and MMP-1 | breast cancer (22) vs. healthy subjects (26) | 95.0% | 79.0% | [43] | |||
| succinic acid and dimethyl-heptanoylcarnitine | breast cancer (31) vs. healthy subjects (29) | 93.5% | 86.2% | [44] | |||
| Breath | a set of VOCs of oxidative stress | breast cancer (51) vs. healthy subjects (42) | 94.1% | 73.8% | [45] | ||
| 3-methylhexane | breast cancer (10) vs. healthy subjects (10) | 100% | 40% | [46] | |||
| decene | 100% | 40% | |||||
| caryophyllene | 100% | 60% | |||||
| naphthalene | 90% | 70% | |||||
| trichlorethylene | 80% | 70% | |||||
| five breath VOCs: 2-propanol, 2,3-dihydro-1-phenyl-4(1H)-quinazolinone,1-phenyl-ethanone, heptanal and isopropyl myristate | breast cancer (51) vs. healthy subjects (42) | 93.8% | 84.6% | [47] | |||
| two commercial electronic noses | breast cancer (48) vs. healthy subjects (45) | N/A | N/A | an average of 95% accuracy | [48] | ||
| BreathLink™ point-of-care breath testing systems | breast cancer (50) vs. non-breast cancer subjects (543) | 82.0% | 77.1% | accuracy: 83% | [49] | ||
| VOCs collected by ultra-clean breath collection balloons | breast cancer (54) vs. non-breast cancer subjects (124) | N/A | N/A | low DF values: Negative predictive value > 99.9%; high DF values: Positive predictive value rising to 100% |
[50] | ||
| NAF | Thomsen–Freidenreich (TF) antigen and its biosynthetic precursor Tn antigen | breast cancer (25) vs. healthy subjects (25) | N/A | N/A | detected in 92% of the cancerous breast NAF samples | [51] | |
| TF, Tn, and age information | breast cancer (83) vs. benign disease (41) | N/A | N/A | AUC: 0.83 | [52] | ||
| combination of TF and uPA | breast cancer (83) vs. benign disease (41) | N/A | N/A | accuracy: 84-92% | [53] | ||
| combination of TF, uPA and PAI-1 | breast cancer (83) vs. benign disease (41) | N/A | N/A | predictive ability reached 100% | |||
| deglycase DJ-1 protein | 136 patients with nipple discharge (benign: 63; malignant: 73) | 75.0% | 85.9% | [54] | |||
| proteomic profiles of NAFs | breast cancer (18) vs. healthy subjects (4) | N/A | N/A | 39 proteins differentially expressed in tumor-bearing vs. disease-free breasts | [55] | ||
| dehydroepiandrosterone (DHEA) concentration | breast cancer (160) vs. healthy subjects (157) | N/A | N/A | higher DHEA concentrations in NAFs were associated with breast cancer | [56] | ||
| Tears | proteomic profiles of tears | breast cancer (10) vs. healthy subjects (10) | ~90.0% | ~90.0% | [57] | ||
| a panel of 20 proteins in tears | breast cancer (50) vs. healthy subjects (50) | ~70.0% | ~70.0% | [58] | |||
| MALDI-TOF-TOF-MS-driven semi-quantitative comparison of tear protein levels | breast cancer (25) vs. healthy subjects (25) | N/A | N/A | more than 20 proteins were differentially expressed in the tears | [59] | ||
| surface-enhanced Raman scattering (SERS) spectra of tear fluid | breast cancer (5) vs. healthy subjects (5) | 92% | 100% | accuracy: 96% | [60] | ||
| Apocrine sweat | 20 sweat markers | breast cancer (70) vs. healthy subjects (53) | 97% | 72% | [61] |
* AUC, area under the receiver operating characteristic (ROC) curve; CTC, circulating tumor cell; cfDNA, cell-free DNA; ctDNA, circulating tumor DNA; DELFI, DNA evaluation of fragments for early interception; MALDI-TOF-TOF-MS, matrix-assisted laser desorption/ionization time-of-flight/time-of-flight mass spectrometry; NAF, nipple aspirate fluid; uPA, urinary plasminogen activator; PAI-1, plasminogen activator inhibitor-1; VOC, volatile organic compound.
2. Blood-Based Biomarkers
Compared with imaging and biopsy cancer detection approaches, blood tests are not only convenient and non-invasive (or minimally invasive), but also widely acceptable, readily reproducible and cost effective. Cancer cells often produce specific proteins, nucleic acids or other cellular vesicles, and shed live cells or dead cell debris into the blood. Analyzing for the existence of those components in the blood may provide a method of detection of the cancer.
2.1. Circulating Carcinoma Proteins
Circulating carcinoma proteins are associated with proliferation, invasion, metastasis, aggressiveness, angiogenesis, oncogenic signaling, and immune regulation of tumor cells. Thus, they have the potential to serve as markers for cancer detection [62]. A number of serum carcinoma protein markers in breast cancer have been identified, including CA15-3, CA27-29, CA-125, carcinoembryonic antigen (CEA), tissue polypeptide antigen (TPA), circulating extracellular domain of human epidermal growth factor receptor 2 (HER2), and tissue polypeptide-specific antigen (TPS) [63,64]. These markers are mainly used in monitoring response to therapy in patients with advanced disease and none of them has been used alone for screening because of low diagnostic sensitivity for early disease or lack of specificity [65,66]. Although a combination of several such biomarkers may increase the sensitivity and specificity, a study demonstrated that no combination of the selected ten breast cancer serum protein markers, including CA15-3, CA125, and CEA, could accurately predict early-stage breast cancer [67]. However, when combined with tumor-associated autoantibodies, serum protein biomarkers were able to achieve 81.0% sensitivity and 78.8% specificity for detection of breast cancer with an area under the receiver operating characteristic (ROC) curve (AUC) of 0.89 [15]. Combined with clinical patient characteristics, a combinatorial serum biomarker panel, Videssa Breast (Table 1), could achieve a comprehensive 93% sensitivity and 98% negative predictive value for women aged 25–75 [16], and a 99.1% negative predictive value, 87.5% sensitivity, and 83.8% specificity for women under age 50 [17]. However, in one study, the negative predictive value of the Videssa Breast liquid biopsy was not high enough to defer tissue biopsy, and its specificity was also too low to help predict results in high-risk solid lesions [68]. Thus, although Videssa Breast tests, in combination with breast imaging results, could improve the accuracy and reduce the false-positive rate of breast cancer detection in women aged over 50 compared with mammography alone, further improvements of detection sensitivity by using new technologies are still needed [69,70].
Of note, other secreted oncoproteins, such as TFF1 [71], TFF3 [72], ARTN [73], and SHON [74,75], have been described in breast cancer. Whether these proteins could be used for early detection of breast cancer remains to be investigated.
2.2. Circulating Tumor Cells
Tumor cells may enter into the peripheral blood of cancer patients either through active intravasation or passive shedding from the primary or metastatic tumors. The presence of CTCs in early-stage breast cancer increases the risk of recurrence and death [76,77]. However, the measurement of CTCs has not been recommended for breast cancer diagnosis because of low sensitivity and reproducibility [64]. CTCs are rare and there is as few as one CTC per billion normal blood cells [78]. Many technologies have been assessed for enumeration and analyses of CTCs based on physical characteristics, immunomagnetic separation, and immunofluorescence/enzyme-linked immunosorbent assays, as well as reverse transcription polymerase chain reaction (RT-PCR) assays [78,79,80,81]. Among the many CTC technologies, CellSearch® (Janssen Diagnostics, Raritan, NJ, USA) and AdnaTest® (AdnaGen AG, Langenhagen, Germany) have been widely studied in clinical trials. The CellSearch® system is a semiautomated antibody-based assay based on immunofluorescence and flow cytometry. CTCs are enriched using epithelial cell adhesion molecule (EpCAM) antibodies, and counted by cytokeratin positivity, positive nuclear staining, and CD45 negativity. The AdnaTest® assay is an RT-PCR based test for detecting cancer-specific mRNA markers, e.g., GA733-2, MUC1, and HER2 for breast cancer, after immunomagnetic enrichment of tumor cells with MUC1 and EpCAM antibodies. Both assays have a similar detection sensitivity in detecting two or more CTCs, but their overall positive agreement was only 73% for CTC ≥ 2 and 69% for CTC ≥ 5, respectively [18]. However, the CellSearch® system was shown to be superior to the AdnaTest® assay in predicting clinical outcome in advanced breast cancer [82], while the AdnaTest® assay was superior to CellSearch® system in terms of overall detection rates [83]. Nevertheless, the CellSearch® system is currently the only FDA-approved CTC assessment for detecting and enumerating CTCs of metastatic breast cancer for prognosis. The presence of CTCs above a cut-off level—five cells per 7.5 mL blood—is associated with shorter survival, which makes CTCs a useful prognostic biomarker [84,85]. However, the low frequency of CTCs, together with the heterogeneity of antigens expressed on the surface of CTCs, makes the detection very difficult and limits their value as a diagnostic tool for patients with early-stage breast cancer. Therefore, more sensitive technologies to target a broad range and variety of CTCs are still needed [86].
2.3. Circulating Cell-Free Tumor DNA
Circulating tumor DNA (ctDNA) in the bloodstream has emerged as a promising biomarker of disease status for breast cancer [20,87,88,89,90]. An elevated level of ctDNA has been found to be associated with advanced-stage breast cancer and metastasis [91,92]. Breast cancer patients had significantly higher levels of ctDNA than healthy controls [91]. The ctDNA concentrations at levels ≥0.75% could be detected in breast cancer patients with a sensitivity of >90% and a specificity of >99% [19]. Some ctDNA was also detected in 97% (29/30) women with metastatic breast cancer [20]. It has also been demonstrated that analyses of mutations in ctDNA could detect early-stage tumors [93]. For example, ctDNA assays targeting known tumor mutations using droplet digital PCR (ddPCR) assays demonstrated a sensitivity of 93.3% and a specificity of 100% in detection of early-stage breast cancer [21]. Similarly, TP53 mutations detected in ctDNA may be useful for breast cancer screening for those with BRCA1 mutations [94]. Chromosome instability is one of the hallmarks of tumors. We have shown that chromosome instability analysis of cell-free DNA (cfDNA) using low-pass whole-genome sequencing can detect breast cancer recurrence more accurately than traditional serum CA15-3 and CEA biomarkers [90]. Another study also shows that amplification of chromosome 1q21.3 detected in ctDNA from blood is significantly associated with breast cancer early relapse [95].
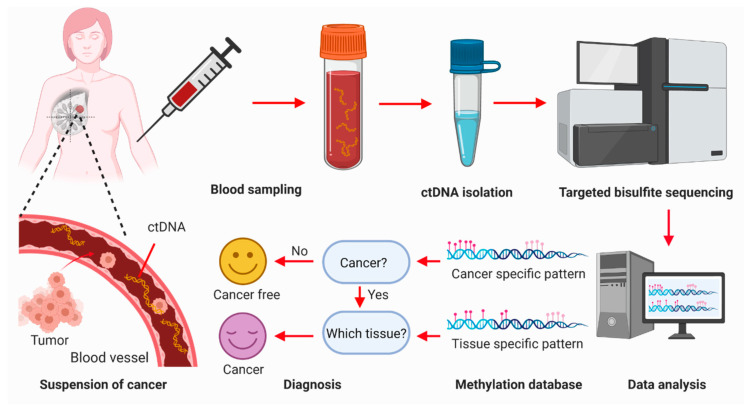
DNA methylation alteration has been frequently observed in cancer. A number of genes, such as p16 [96], BRCA1 [97,98], RASSF1A [22,23], APC [99], and GSTP1 [100], are reported to be hypermethylated in breast cancer. Technological advances in epigenetics have opened an era to detect breast cancer using methylation signatures of cfDNA [101,102,103]. Although the sensitivity of a single gene methylation is too low to be used for early screening of breast cancer [22,99,104], a panel of epigenetic markers can greatly increase the sensitivity required for breast cancer detection [105,106]. For example, the methylation patterns of six genes in cfDNA achieved 79.6% sensitivity and 72.4% specificity in the diagnosis of breast cancer (n = 749) (Table 1) [24]. The methylation analysis of a panel of 16 genes, including 12 novel epigenetic markers and four internal control markers, could discriminate cancer patients from normal healthy controls with a sensitivity of 86.2% and a specificity of 82.7% (Table 1) [25]. Of note, a targeted methylation panel covering 103,456 distinct DNA regions (17.2 Mb) and 1,116,720 CpGs was developed by GRAIL Inc (Menlo Park, CA, USA) (Figure 2). In a prospective case–control sub-study of 6689 participants (2482 cancer of > 50 cancer types and 4207 non-cancer), the GRAIL DNA methylation patterns of cfDNA can detect various types of cancers at 99.8% specificity (training set: N = 844) or 99.3% specificity (validation set: N = 359) with a <1% false-positive rate [26]. The precision for breast cancer was 96% (82/85) for training and 93% (40/43) for validation [26].
Figure 2.
Diagram of the procedures of breast cancer detection through methylation patterns of circulating cell-free DNA in the blood. Breast tumor and normal tissue shed DNA fragments into the blood. Peripheral blood is withdrawn from human subjects and circulating tumor DNA (ctDNA) is isolated from the blood sample. The methylation status of the ctDNA is determined by targeted bisulfite methylation sequencing. The methylation patterns of the ctDNA fragments are analyzed. The presence or absence of cancer cells is thus determined by comparing the methylation patterns with those in the methylation database, which is constructed from those individuals with and without cancer. The origin of cancer is determined using unique tissue-specific methylation patterns. Figures were created with BioRender.com (https://biorender.com/).
Except methylations, fragmentation patterns of cfDNA could also be used for early detection of breast cancer. An approach called “DNA evaluation of fragments for early interception” (DELFI) was developed to detect a large number of abnormalities in cfDNA by genome-wide analysis of fragmentation patterns [27]. DELFI profiles of cfDNA could detect breast cancer at a sensitivity of 57% (31/54) and a specificity of 98%. When DELFI fragmentation patterns of cfDNA are combined with mutations detected in cfDNA [93], the detection sensitivity increased to 65% (35/54 patients) with a specificity of 98% [27].
Compared with carcinoma proteins and CTCs, ctDNA has several advantages, being a better and more sensitive marker for monitoring tumor burden [107,108]. It has a greater dynamic range representing the heterogenetic features of tumors, a shorter half-life, and is more specific to malignancy [109]. Moreover, ctDNA mutation and methylation assays are able to identify the presence of relatively early cancers as well as localize the organ of origin of cancers [38]. However, the application of ctDNA as a non-invasive diagnostic clinical biomarker still faces several technical issues. Most cfDNA is released by normal cells into the circulation as a result of cell death. The amount of ctDNA is very low, highly variable and comprises <0.1% of the total cfDNA, particularly in patients with early-stage cancers [110], making detection of the ctDNA challenging [107]. The lower copy numbers of ctDNA compared with those of wild-type cfDNA and the limited accuracy of current sequencing technologies contribute to the limitation in ctDNA detection [111]. Therefore, a highly sensitive technique to detect ctDNA is still required. For example, the CRISPR-based diagnostic platform may be used to identify mutations in cell-free tumor DNA with high sensitivity and specificity [112].
2.4. Circulating miRNAs
The miRNAs are small regulatory RNA molecules that mediate target mRNA expression by base pairing to complementary sequences in the 3′ untranslated region [113]. It was shown that circulation miRNAs in the serum and plasma of patients could distinguish patients with prostate cancer from healthy controls [114]. In breast cancer, a large number of miRNAs have been observed to be significantly upregulated in the plasma of patients, although a small number of miRNAs were found to be downregulated when compared to healthy controls [115,116]. Studies have demonstrated that a combination of certain circulating miRNAs were able to distinguish breast cancer from normal and healthy controls, as well as differentiate breast cancer from benign lesions [28,29,117,118,119,120,121,122]. For example, a panel of five miRNAs could detect breast cancer with a sensitivity of 97.3%, a specificity of 82.9%, and an accuracy of 89.7% (Table 1) [28]. Another panel of seven miRNAs could differentiate triple-negative breast cancer from healthy women with an accuracy of 79%, a specificity of 74.2%, and a sensitivity of 83.8% (Table 1) [29]. Therefore, differentially expressed circulating miRNAs are potential diagnostic biomarkers for breast cancer detection [14,122,123]. However, there is little consistency among the circulating miRNA panels identified in different studies. So far, there are no panels of circulating miRNAs that are ready for breast cancer diagnosis in a clinical setting [122].
2.5. Extracellular Vesicles
The term EV is a generic term used to refer to all types of vesicles that exist in the extracellular space, including microvesicles, microparticles, ectosomes, exosomes, oncosomes, prostasomes, and tolerosomes [124,125]. EVs, secreted from normal and cancer cells, are a complex of lipids, proteins, DNAs, various RNAs, and other biomolecules enclosed by a lipid bilayer with transmembrane proteins [126]. Because EVs can mirror the features of the origin and the state of the tumor, they have gained increasing attention as cancer biomarkers. An elevated number of EVs has been found in the peripheral blood of breast cancer patients compared with healthy controls [127,128,129,130,131]. However, the number of EVs alone is not specific enough for cancer diagnosis [132].
As an alternative to EV count, their molecular cargos may be a better cancer biomarker, especially if the enclosed contents are cancer related, such as amplified oncogenes and oncoproteins [133,134]. For example, the levels of cancer-associated fibronectin and developmental endothelial locus-1 (Del-1) proteins detected in circulating EVs were significantly elevated at all stages of breast cancer, and returned to normal after tumor removal [30,31]. Similarly, focal adhesion kinase (FAK), epidermal growth factor receptor proteins [130], survivin [128], EMMPRIN [135], and various miRNAs [136] were also significantly enriched in EVs isolated from the plasma of breast cancer patients. Moreover, compared with healthy controls, specific miRNAs that were overexpressed in breast cancer sera were enriched in EVs [136]. Thus, analyzing cancer-related contents enclosed in EVs could help early-stage breast cancer diagnosis and distinguish breast cancer from benign and noncancerous diseases. Such analysis could provide a diagnostic tool with higher sensitivity and specificity compared with whole-blood analysis, as cancer-derived EVs preserve molecules that are relevant to diagnosis [126,132].
EVs possess great potential as novel non-invasive diagnostic molecular biomarkers of breast cancer. However, there are still issues in how to identify and isolate EVs, such as contamination with cells and platelet remnants [137]. There are many technologies that have been used to isolate and characterize EVs [138,139,140,141,142]. Currently, there are no standardized methods of isolation and quantification of EVs. It has been observed that the profiles of EVs captured from the same source of materials are dependent on isolation methods used [142]. This has made the use of EVs as a diagnostic biomarker more challenging. Optimization and standardization of the methods and protocols of EV isolation and purification are urgently required [141,142]. Moreover, our knowledge of EVs is still limited. The precise molecular mechanisms of biogenesis, release, and functions of EVs also remain to be investigated [143].
2.6. Other Emerging Blood-Based Biomarkers
Metabolite profiles in the blood are also potential biomarkers to differentiate primary breast cancer patients from healthy controls. A panel of seven metabolites (Glu, Orn, Thr, Trp, Met-SO, C2, and C3) in the plasma could detect breast cancer with an AUC of 0.87 in the training cohort (80 cancer vs. 100 healthy controls) and an AUC of 0.80 in the validation cohort (109 cancer vs. 50 healthy controls) [32]. The expression levels of three metabolites (8-hydroxy-2ʹ-deoxyguanosine, 1-methylguanosine, and 1-methyl adenosine), together with CA15-3, could achieve 88.8% sensitivity and 86.8% specificity in the detection of early-stage breast cancer based on a serum metabolome score [33]. In a prospective case–control study, the signature of seven metabolites (dimethyldodecane, galactose, α-glyceryl stearate, methyl stearate, 1(1-methoxycarbonyethyl)-4-(2-methyl-2-trimethylsilyl-oxypropyl) benzene, tetradecane, and glucopyranoside) in serum revealed a distinct separation between healthy controls and breast cancer patients at a sensitivity of 96% and a specificity of 100% [34]. Four plasma metabolites (L-octanoylcarnitine, 5-oxoproline, hypoxanthine, and docosahexaenoic acid) were also identified as potential biomarkers for diagnosis of breast cancer, with L-octanoylcarnitine showing a 100.0% positive predictive value [35]. Interestingly, one study identified metabolites, which included four knowns (caproic acid, taurine, stearamide and linoleic acid) and five unknowns (C26H43ClN4S3, C26H51N5O4, C9H16O3S, C23H30N2S, and 278.1552@9.641), in the plasma that could distinguish between breast cancer patients and healthy controls, irrespective of the breast cancer type, at a high sensitivity and specificity up to 100% with high accuracy [36]. However, these need to be validated.
One study identified 18 serum free fatty acids from serum samples of 16 breast cancer patients and 18 breast adenosis patients. Five candidates (palmitic acid, oleic acid, cis-8,11,14-eicosatrienoic acid, docosanoic acid, and the ratio of oleic acid to stearic acid) were shown to be potential serum biomarkers for differential diagnosis of breast cancer [144]. A panel of five serum free fatty acids (C16:1, C18:3, C18:2, C20:4, and C22:6) was shown to possess the ability to differentiate early-stage breast cancer patients (n = 140) from healthy controls (n = 202), with an AUC of 0.953, a sensitivity of 83.3%, and a specificity of 87.1% [37].
2.7. Multi-Analyte Blood Tests
Multi-analyte blood tests combine the detection advantages of several blood marker assays for cancer detection and localization, thus improving the accuracy. The CancerSEEK test is such a multi-analyte test that evaluates the levels of eight circulating protein markers and the presence of mutations in 16 genes in cfDNA in the blood (Table 1) [38]. CancerSEEK has a median sensitivity of 70% and a specificity of >99% in identifying patients with one of the eight cancer types evaluated [38]. However, the cancer detection effectiveness of CancerSEEK ranges from 98% for ovarian cancer to only 33% for breast cancer. A feasibility study of CancerSEEK tests has been conducted on a general population of 10,000 women with no history of cancer [39]. The blood test detected 26 previously unknown cancers of different types with a sensitivity (26/96) of 27.1%, a specificity of 98.9% (9707/9815), and a positive predictive value of 19.4% (26/134) [39]. Among the participants, 27 breast cancers were identified. Among them, only one breast cancer was first detected by the blood test, 20 by standard-of-care screening and six by other imaging means [39]. Therefore, CancerSEEK tests only have a low sensitivity of 3.7% (1/27) in breast cancer detection.
The CancerSEEK test was developed from a case–control study, which introduced a possible bias in the utility of the assay [145,146]. Although a remolding of the CancerSEEK test, e.g., the CancerA1DE method, increased the sensitivity to 70% for breast cancer detection at a 99% specificity [40], it is unlikely that the CancerSEEK test will be effective in breast cancer detection [39].
3. Non-Blood-Based Biomarkers
Besides blood, other body liquids, including urine, NAF, tears, and sweat, as well as the breath of patients, have also been investigated for breast cancer diagnosis.
3.1. Biomarkers in Urine
Human urine is one of the most useful body biofluids for routine testing. A number of studies have suggested that urine could contain potential biomarkers for breast cancer screening, ranging from metabolome profiling and proteomic profiling to exosomic analysis [147,148,149,150].
Phospholipids are building blocks for cellular membranes. Phosphatidylcholine (PC), phosphatidylethanolamine (PE), and sphingomyelin are the most abundant phospholipids and comprise up to 80% of the membrane. Increased phospholipid metabolism, particularly PC and PE or their precursor molecules, has been observed in breast cancer tissues [151,152]. Loss of the wild-type BRCA1 allele has been shown to increase lipid production in breast cancer cells [153]. Using nanoflow liquid chromatography/electrospray ionization tandem mass spectrometry, the urine samples of patients with breast cancer, both before and after surgery, were qualitatively and quantitatively analyzed [154]. In that analysis, twenty-one PCs and 12 PEs were identified in the patient samples, and two PCs (16:0/16:0 and 14:1/20:4) and one PE (18:3/18:0) were not detectable after surgery. Moreover, compared with the controls, the total concentrations of PCs and PEs of patient urine samples increased by 44% and 71%, respectively, but significantly decreased after surgery [154].
One study has demonstrated that 59 urinary proteins are differentially detected (>3-fold change) in breast cancer patients compared with healthy control individuals, and of them, 36 urinary proteins are stage specific [155]. A panel of 13 upregulated proteins could be used for breast cancer detection. The panel consists of leucine LRC36, MAST4, DYH8, HBA, PEPA, filaggrin, MMRN2, AGRIN, NEGR1, FIBA, keratin KIC10, and two uncharacterized proteins, C4orf14 (CD014) and CI131 [155]. These stage-specific markers are associated with pre-invasive breast cancer in ductal carcinoma in situ (DCIS), early invasive breast cancer, and metastatic breast cancer [155].
It was demonstrated that the urinary miRNA profile of primary breast cancer patients was different from that of healthy controls. The urinary miRNA profiles could separate the patients from healthy controls with a sensitivity and specificity of 91.7% and an AUC of 0.932 [41]. A study explored the diagnostic potential of urinary exosomal miRNAs in a case–control study of 69 breast cancer patients and 40 healthy controls [42]. They identified a specific panel of four urinary miRNA types (miR-424, miR-423, miR-660, and let7-i) that could discriminate breast cancer patients from healthy controls with a sensitivity of 98.6% and a specificity of 100% (Table 1) [42].
Urine is also a source of exosomes. One study isolated urine exosomes from patients with breast cancer and 26 healthy females and determined the expression of miRNA-21 and matrix metalloproteinase-1 (MMP-1) in the isolated exosomes by quantitative RT-PCR. They discovered that the expression of urine exosome-derived miRNA-21 and MMP-1 could be diagnostic markers for cancer detection with a combined sensitivity of 95% and a specificity of 79% [43].
Many studies have analyzed the metabolomic and proteomic profiles using technologies such as NMR spectroscopy [150,156], gas chromatography/mass spectrometry [48], liquid chromatography coupled to mass spectrometry [157,158,159,160], and capillary electrophoresis coupled to mass spectrometry [161]. Based on these profiles, they identified cancer-specific patterns and used calculated models for early diagnosis of breast cancer. For example, the combination of succinic acid and dimethyl-heptanoylcarnitine were able to separate breast cancer patients (n = 31) from healthy controls (n = 29) with a sensitivity of 93.5% and a specificity of 86.2% [44].
Because urine collection is non-invasive and convenient, it has been used for the discovery of cancer biomarkers. Changes in proteins, metabolites, miRNAs, or other cellular components in urine could potentially indicate the presence of breast cancer. However, current reported urinary breast cancer biomarkers are still in the biomarker discovery phase, and their specificity and sensitivity have to be validated in cohort studies [162].
3.2. Volatile Biomarkers in the Breath
Human breath contains VOCs and semi-volatile compounds that are released by the body as a result of normal metabolic activity or due to pathological disorders. The presence of cancer cells can affect both the identity and abundance of these compounds in the exhaled breath of cancer patients. Chemical analyses of exhaled breath from patients have been exploited for diagnosing many different types of cancers [163,164].
In the 1990s, the use of breath testing for breast cancer was demonstrated with the finding that women with breast cancer had an increased level of pentane in their breath [165]. It was soon found that a set of VOC markers of oxidative stress in the breath could distinguish breast cancer patients from healthy volunteers with a sensitivity of 94.1% (48/51) and a specificity of 73.8% (31/42) [45]. More importantly, this breath test accurately identified women with breast cancer with a negative predictive value superior to a screening mammogram test, although the positive predictive value was lower than that of a screening mammogram test [45]. This clearly suggested that breath tests could be employed as a primary screen for breast cancer.
The breath samples of patients with breast cancer and pair-matched healthy controls were analyzed in a prospective study by gas chromatography/mass spectrometry [46]. A total of 109 different VOCs was identified in the breath of breast cancer patients and controls. Of them, five specific VOCs, 3-methylhexane, decene, caryophyllene, naphthalene, and trichlorethylene in the breath, could distinguish those with breast cancer from healthy controls [46]. In another study, a fuzzy logic model was constructed with five breath VOC biomarkers, 2-propanol, 2,3-dihydro-1-phenyl-4(1H)-quinazolinone, 1-phenyl-ethanone, heptanal, and isopropyl myristate in the training set (n = 64); and the model could predict breast cancer with 93.8% sensitivity and 84.6% specificity in the test set (n = 29) [47]. More importantly, the same model showed a negative prediction of breast cancer for 16 out of 50 (32.0%) women with abnormal mammograms and their cancer-free prediction was confirmed by biopsy [47].
The ability of a breath test for breast cancer detection has been further demonstrated in a number of other studies [166,167,168,169]. With improvements in breath collection methods, analytic techniques and data modeling, a clinical breath test for breast cancer is being actively developed. One study used inexpensive commercial electronic noses to identify unique breath patterns in women with breast cancer and an average of 95% accuracy in the classification of breast cancer patients was achieved through an artificial neural network model based on the data obtained from the electronic noses [48]. Menssana Research Inc. (Fort Lee, NJ, USA) has developed BreathLink™ point-of-care breath testing systems for the collection, concentration, and analysis of VOCs in human breath to detect breast cancer in women. A pilot study showed that this system could accurately identify women with breast cancer and abnormal mammograms [169]. The results of this prospective clinical validation study were updated recently [49]. To facilitate the collection of breath and analysis of VOCs, ultra-clean breath collection balloons have been developed by Menssana Research Inc. The balloons were used to collect breath samples from 54 female breast cancer patients and 124 cancer-free controls. Breath VOC biomarkers of breast cancer identified by a Monte Carlo analysis in the training set were incorporated into a multivariate algorithm to predict disease in the validation set to generate a discriminant function (DF) value from the predictive algorithms [50]. Breast cancer risks can be accurately predicted based on the DF scores, with a low DF value indicating a low risk of breast cancer at a negative predictive value of > 99.9% and a high DF value indicating a high risk of breast cancer at a positive predictive value rising to 100% [50].
Breath VOC tests are certainly a very promising approach to screening women for breast cancer. Breath tests are non-invasive, painless, completely safe, and cost-effective. They could potentially reduce the use of mammograms in clinics for the screening and monitoring of breast cancer. However, a number of factors, including the breath collection methods, patient’s physiologic conditions, test environments, and methods of analysis, will affect the accuracy of VOC breath test results, and standardized procedures are still required [164].
3.3. Biomarkers in NAF
NAF is a natural secretion produced in the breasts by breast epithelial duct cells. It can be collected by nipple aspiration or by other methods in healthy nonlactating women. The color of NAF and the presence of specific biomarkers in NAF could be used to diagnose breast cancer. It has been observed that women with bloody or brown nipple discharge suffer a higher risk of breast cancer than those with white, cream, green, or yellow nipple discharge [170]. NAF contains high concentrations of proteins, carbohydrates and metabolites, such as amino acids, organic acids, and fatty acids. Several studies have suggested that the levels of drug, protein, and hormone levels in NAF more closely reflect the products of breast tissue metabolism and exposures than those in plasma or serum [171]. A study analyzed NAFs collected from healthy women using both nuclear magnetic resonance and gas chromatography/mass spectrometry [171]. In that study, a total of 38 metabolites, including amino acids, organic acids, fatty acids, and carbohydrates, were identified by the two analytical techniques. Eight metabolites are unique to NAF, 19 unique to plasma, and 24 are shared metabolites, indicating that the metabolic profile of NAF is distinct from that of matched plasma samples [171]. In addition, NAF also contains exfoliated breast epithelial cells, from which breast cancer originates [172]. Therefore, NAF is arguably a better source than other body fluids for the discovery of biomarkers for breast cancer.
The Thomsen–Freidenreich (TF) antigen and its biosynthetic precursor, Tn antigen, are often aberrantly glycosylated in cancer cells. A study examined the expression of TF and Tn in NAF samples from 25 breasts with cancer and 25 normal breasts [51]. Of the 25 NAF samples from breasts with cancer, 19 samples had TF and 20 samples had Tn, whereas only one of the 25 NAF samples from breasts without cancer contained Tn and neither TF nor Tn was detected in the rest, indicating that TF and Tn can be used as detection biomarkers for breast cancer [51]. A direct immunoassay of TF expression levels in NAFs of 124 women requiring biopsy because of a suspicious breast lesion demonstrated that TF concentrations in NAFs could distinguish women with precancer and cancer from those with benign disease [52]. Urinary plasminogen activator (uPA) and its inhibitor PAI-1 have been shown to promote tumor invasion and metastases by regulating the degradation of the extracellular matrix. Compared with women with benign disease, women with atypia and cancer were found to have higher concentrations of uPA and PAI-1 in NAFs [53]. In combination with TF, uPA could predict breast cancer in both pre- and post-menopausal women with 84-92% accuracy [53]. When TF, uPA, and PAI-1 were all combined, the predictive ability reached 100% in the cohort studied [53]. With a cut-off level of 3.0 ng/mL, the protein deglycase DJ-1 in NAFs could detect the presence of breast cancer with 85.9% specificity and 75% sensitivity [54].
High prostate-specific antigen (PSA) levels have been widely used as a diagnostic biomarker for prostate cancer. However, high levels of PSA in NAFs were found in all women with no risk factors or 90% of those with a family history of breast cancer, whereas women with precancerous or invasive cancer had reduced levels of PSA [173,174]. Such an inverse correlation was also found with the superoxide dismutase SOD-1. Breast cancer patients had a lower concentration of SOD-1 in their NAFs compared with healthy individuals [175].
Proteomic analyses of NAFs have been conducted to screen for diagnostic markers for breast cancer. One study analyzed paired NAFs of 18 women with low invasive breast carcinoma (stage I or II) and four healthy controls and identified 39 differentially expressed proteins between tumor-bearing and disease-free breasts [55]. Tumor-bearing breasts overexpressed lipophilin B, beta-globin, hemopexin, and vitamin D-binding protein precursor, but underexpressed alpha2HS-glycoprotein [55]. Higher dehydroepiandrosterone (DHEA) concentrations in NAFs were associated with breast cancer, especially estrogen receptor-positive cases [56].
NAF is also a source of miRNAs and contains more miRNA species than serum [176]. It was found that miR-3646 and miR-4484 were upregulated while miR-4732-5p was downregulated in the NAFs of patients with breast cancer, compared with those with benign breast lesions [177]. Therefore, miRNA analyses of NAFs are potentially very useful for the detection of breast cancer [178,179]. More studies comparing miRNA profiles of cancerous and healthy NAFs are underway [176].
Despite the low volume of samples, NAF collection is simple, quick, reliable, and reproducible [55]. Moreover, analytical reproducibility of NAF samples was high across different extraction and analysis days [171]. Therefore, NAF is an ideal source of biomarkers for early diagnosis of breast cancer [55]. Furthermore, because NAF is derived directly from the breast ductal system, future characterization of NAF may provide valuable, more specific, and sensitive information on breast cancer diagnosis and treatment [176]. However, in order to provide an accurate screening approach for breast cancer-specific biomarkers, increased success in NAF sample collection methods, normalization of the volume of the sample, and standardization of the analysis will be required.
3.4. Biomarkers in Tears
Tears are mainly produced by the lacrimal glands underneath the skin of the upper eyelids through filtration from blood plasma, which circulates in the organs and tissues of our body. Therefore, tears have been used as a source for external drug screening and pharmacokinetic studies, as well as for biomarker discovery of many diseases, including cancer [180].
Mammaglobin B (also called SCGB2A1 or lacryglobin) is overexpressed in breast cancer and serves as a biomarker for axillary lymph node micrometastases in breast cancer patients [181,182]. Mammaglobin B was first discovered in normal human reflex tears [183]. However, it is also detected in the tears of 88% of breast cancer patients [184]. A study analyzed the proteomic profiles of tears (and blood) of 10 breast cancer patients and 10 healthy controls using surface-enhanced laser desorption/ionization-time-of-flight mass spectroscopy and distinct protein expression profiles were identified [57]. This study demonstrated that breast cancer patients could be distinguished from controls based on a proteomic pattern in tear (and serum) fluid with a specificity and sensitivity of approximately 90% [57]. A further analysis of a larger cohort of breast cancer patients (n = 50) and healthy controls (n = 50) identified a panel of 20 differentiating protein biomarkers in tears, which could separate breast cancer patients from healthy controls with a specificity and sensitivity of approximately 70% [58]. Using a semi-quantitative method, more than 20 proteins were found to be differentially expressed in the tears of 25 patients with primary invasive breast carcinoma and 25 age-matched healthy controls [59]. Taken together, these studies have clearly indicated that tears are a source for potential non-invasive biomarkers for breast cancer diagnosis.
A tear protein-based breast cancer screening test is currently under development by Ascendant Dx, a diagnostic company. It was claimed to have up to 90% accuracy, implying a higher sensitivity than a mammogram [185].
Compared with blood, tears as a source of biomarkers have advantages. Tear sample collection is less invasive. Tears can be easily obtained from the surface of the eyes inside or outside the clinic for rapid and continuous monitoring of health. No prior protein filtration before analysis is needed as tears contain few solid proteins such as albumin. However, one particular analytical challenge is the much lower concentration of some molecules in tears compared with blood. Therefore, a sensitive and reliable screening technique is required for reliable and reproducible detection and quantification. More recently, a label-free surface-enhanced Raman spectroscopy biosensor has been designed for detecting or predicting breast cancer from human tears. It is a highly sensitive nanoplasmonic biosensor and allows for a rapid quantitative and qualitative analysis of tears on-site using a portable Raman spectrometer. Based on the preprocessed surface-enhanced Raman scattering (SERS) spectra of tear fluid obtained from five patients with breast cancer and five healthy controls, breast cancer can be predicted with a sensitivity of 92% and a specificity of 100% [60].
3.5. Biomarkers in Apocrine Sweat
Sweat is one of the less used non-invasively collectable biofluids for the discovery of biomarkers. Interests in screening for sweat biomarkers have been increasing with the advance in proteomic and metabolomic technologies. The composition of sweat is highly dynamic and alters significantly in various skin and other disorders. An in-depth profiling of sweat has identified 861 unique proteins and 32,818 endogenous peptides [186]. Sweat chloride tests have long been used clinically to diagnose cystic fibrosis [187]. There are reports that use sweat testing for drugs of abuse [188] and tuberculosis diagnosis [189]. A metabolite analysis of the sweat of patients with lung cancer versus smokers as control individuals has identified a panel of five metabolites that can detect lung cancer with 80% specificity and 79% sensitivity [190]. A similar sweat test for breast cancer has demonstrated that a mathematical–statistical model of 20 sweat markers was able to detect breast cancer with 97% sensitivity and 72% specificity [61]. However, these sweat-based cancer tests have not been clinically validated.
4. Conclusions
Over the last few decades, a lot of non-invasive or minimally invasive methods for early detection of breast cancer have been described to overcome the limitations of the current mammogram screening. Blood, other body fluids, and breath all contain potential biomarkers for breast cancer diagnosis. Even though blood has been the main source for biomarker discovery, a number of very promising biomarkers have been identified from other body fluids including urine, sweat, tears, breath, and NAF. In theory, NAF may be the best liquid source for developing a screening diagnostic tool for breast cancer. This is because NAF is in close proximity to the disease area [176], and breast cancer-specific molecules should exist at a much higher concentration in NAFs than in other circulating body fluids. Among many types of biomarkers, circulating cell-free tumor DNA and circulating miRNAs, especially EV-enclosed and tumor-associated miRNAs, hold great promise for early breast cancer detection [12]. With technological advances in epigenetics to detect DNA methylation at a large scale, cancers including breast cancer can be accurately detected by a simple analysis of methylation patterns of ctDNA through a blood test [26,27,38,39]. Although the current ctDNA methylation signatures do not work well for breast cancer detection, a tailored algorithm specifically developed for breast cancer using breast-derived ctDNA may increase the sensitivity for breast cancer detection [40,191,192]. Of note, breath tests have demonstrated the highest potential to be used in clinics given that a rapid breath test has already been shown to accurately predict the risk of breast cancer and abnormal mammograms in women with breast-related symptoms [156,157,158]. We envision that the breath test, combined with mammography, will significantly reduce the false-positive and false-negative diagnosis of mammograms. Breath tests are non-invasive, easy to use, and could potentially detect cancer at a relatively early stage, and they are also suitable for large-scale population screening [163].
It takes time to develop a clinical biomarker. The process of development of a biomarker for early cancer detection is normally divided into five phases [193]. Although many potential non-invasive biomarkers have been reported, most of them remain in phase 1 (preclinical exploratory) or 2 (clinical assay and validation), a few in phase 3 (retrospective longitudinal) or 4 (prospective screening), and none in phase 5 (cancer control). A phase 2 clinical trial to evaluate the use of protein signature in tears for the detection of breast and other cancers was completed (www.clinicaltrials.gov NCT00574678, https://clinicaltrials.gov/ct2/show/NCT00574678) [194]. Another prospective clinical trial to validate the breath test for breast cancer detection was completed (NCT02888366, https://clinicaltrials.gov/ct2/show/NCT02888366) [195] and the results are very promising [49]. A prospective clinical trial, the ASCEND study (NCT04213326, https://clinicaltrials.gov/ct2/show/NCT04213326), is under way to evaluate the CancerSEEK test for multiple cancer detection [196]. GRAIL Inc. has been very active in developing a ctDNA methylation-based blood test for early detection of multiple cancers, including breast cancer. They have already completed two clinical trials, the CCGA case–control study (NCT02889978, https://clinicaltrials.gov/ct2/show/NCT02889978) [197,198] and the STRIVE cohort study (NCT03085888, https://clinicaltrials.gov/ct2/show/NCT03085888) [26,199]. The results obtained from these two studies are being used to conduct two real-world prospective cohort studies, the UK SUMMIT study (NCT03934866, https://clinicaltrials.gov/ct2/show/NCT03934866) [200] and the US PATHFINDER study (NCT04241796, https://clinicaltrials.gov/ct2/show/NCT04241796) [201].
A series of white papers on bioanalysis have been published by the US FDA to guide analytical validation of potential biomarkers. Analytical validity of any biomarkers has to be evaluated for accuracy, reproducibility, and reliability before they become a clinical utility [202,203]. For the clinical application of biomarkers, the standardization of specimen collection, handling, storage, and analysis is key for quality assurance, reliability and reproducibility. With the significant advances in analytical techniques and tumor biology [204], there is a great possibility of a more accurate, sensitive, and cost-effective non-invasive test for early detection of breast cancer being developed in the near future.
Author Contributions
Conceptualization, J.L., W.-M.C. and D.-X.L.; Writing–original draft preparation, J.L., X.G., L.-M.C. and D.-X.L.; Writing–review and editing, J.L., X.G., Z.F., L.-M.C., Y.L., X.W., W.-M.C. and D.-X.L; Supervision, D.-X.L.; Funding acquisition, Z.F., X.W., W.-M.C. and D.-X.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by the key research–development program of Zhejiang Province (grant numbers: 2020C04012 and 2019C04001) (to X.W., W.-M.C., and D.-X.L.), the Breast Cancer Foundation New Zealand (to D.-X.L., no grant number), the New Zealand Breast Cancer Cure (to D.-X.L., no grant number), and the Auckland Medical Research Foundation (grant number: 5117014) (to D.-X.L., L.-M.C., and Y.L.).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L., Jemal A. GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Galizia D., Milani A., Geuna E., Martinello R., Cagnazzo C., Foresto M., Longo V., Berchialla P., Solinas G., Calori A. Self-evaluation of duration of adjuvant chemotherapy side effects in breast cancer patients: A prospective study. Cancer Med. 2018;7:4339–4344. doi: 10.1002/cam4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Relative Survival by Stage at Diagnosis (Female Breast Cancer) [(accessed on 3 September 2020)]; Available online: https://ncci.canceraustralia.gov.au/relative-survival-stage-diagnosis-female-breast-cancer.
- 4.Marmot M.G., Altman D.G., Cameron D.A., Dewar J.A., Thompson S.G., Wilcox M. The benefits and harms of breast cancer screening: An independent review. Lancet. 2012;380:1778–1786. doi: 10.1038/bjc.2013.177. [DOI] [PubMed] [Google Scholar]
- 5.Marmot M.G., Altman D.G., Cameron D.A., Dewar J.A., Thompson S.G., Wilcox M. The benefits and harms of breast cancer screening: An independent review. A report jointly commissioned by Cancer Research UK and the Department of Health (England) October 2012. Br. J. Cancer. 2013;108:2205–2240. doi: 10.1038/bjc.2013.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stout N.K., Lee S.J., Schechter C.B., Kerlikowske K., Alagoz O., Berry D., Buist D.S., Cevik M., Chisholm G., de Koning H.J., et al. Benefits, harms, and costs for breast cancer screening after US implementation of digital mammography. J. Natl. Cancer Inst. 2014;106:dju092. doi: 10.1093/jnci/dju092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loberg M., Lousdal M.L., Bretthauer M., Kalager M. Benefits and harms of mammography screening. Breast Cancer Res. 2015;17:63. doi: 10.1186/s13058-015-0525-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Ravesteyn N.T., Stout N.K., Schechter C.B., Heijnsdijk E.A., Alagoz O., Trentham-Dietz A., Mandelblatt J.S., de Koning H.J. Benefits and harms of mammography screening after age 74 years: Model estimates of overdiagnosis. J. Natl. Cancer Inst. 2015;107 doi: 10.1093/jnci/djv103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van den Ende C., Oordt-Speets A.M., Vroling H., van Agt H.M.E. Benefits and harms of breast cancer screening with mammography in women aged 40–49 years: A systematic review. Int. J. Cancer. 2017;141:1295–1306. doi: 10.1002/ijc.30794. [DOI] [PubMed] [Google Scholar]
- 10.Jacklyn G., McGeechan K., Houssami N., Bell K., Glasziou P.P., Barratt A. Overdiagnosis due to screening mammography for women aged 40 years and over. Cochrane Database Syst. Rev. 2018;2018:CD013076. doi: 10.1002/14651858.CD013076. [DOI] [Google Scholar]
- 11.Heinavaara S., Sarkeala T., Anttila A. Overdiagnosis due to breast cancer screening: Updated estimates of the Helsinki service study in Finland. Br. J. Cancer. 2014;111:1463–1468. doi: 10.1038/bjc.2014.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zubor P., Kubatka P., Kajo K., Dankova Z., Polacek H., Bielik T., Kudela E., Samec M., Liskova A., Vlcakova D., et al. Why the gold standard approach by mammography demands extension by multiomics? application of liquid biopsy miRNA profiles to breast cancer disease management. Int. J. Mol. Sci. 2019;20:2878. doi: 10.3390/ijms20122878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alimirzaie S., Bagherzadeh M., Akbari M.R. Liquid biopsy in breast cancer: A comprehensive review. Clin. Genet. 2019;95:643–660. doi: 10.1111/cge.13514. [DOI] [PubMed] [Google Scholar]
- 14.Loke S.Y., Lee A.S.G. The future of blood-based biomarkers for the early detection of breast cancer. Eur. J. Cancer. 2018;92:54–68. doi: 10.1016/j.ejca.2017.12.025. [DOI] [PubMed] [Google Scholar]
- 15.Henderson M.C., Hollingsworth A.B., Gordon K., Silver M., Mulpuri R., Letsios E., Reese D.E. Integration of serum protein biomarker and tumor associated autoantibody expression data increases the ability of a blood-based proteomic assay to identify breast cancer. PLoS ONE. 2016;11:e0157692. doi: 10.1371/journal.pone.0157692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson M.C., Silver M., Tran Q., Letsios E.E., Mulpuri R., Reese D.E., Lourenco A.P., LaBaer J., Anderson K.S., Alpers J. A noninvasive blood-based combinatorial proteomic biomarker assay to detect breast cancer in women over age 50 with BI-RADS 3, 4, or 5 Assessment. Clin. Cancer Res. 2019;25:142–149. doi: 10.1158/1078-0432.CCR-18-0843. [DOI] [PubMed] [Google Scholar]
- 17.Lourenco A.P., Benson K.L., Henderson M.C., Silver M., Letsios E., Tran Q., Gordon K.J., Borman S., Corn C., Mulpuri R., et al. A Noninvasive Blood-based Combinatorial Proteomic Biomarker Assay to Detect Breast Cancer in Women under the Age of 50 Years. Clin. Breast Cancer. 2017;17:516–525 e516. doi: 10.1016/j.clbc.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Andreopoulou E., Yang L.Y., Rangel K.M., Reuben J.M., Hsu L., Krishnamurthy S., Valero V., Fritsche H.A., Cristofanilli M. Comparison of assay methods for detection of circulating tumor cells in metastatic breast cancer: AdnaGen AdnaTest BreastCancer Select/Detect versus Veridex CellSearch system. Int. J. Cancer. 2012;130:1590–1597. doi: 10.1002/ijc.26111. [DOI] [PubMed] [Google Scholar]
- 19.Leary R.J., Sausen M., Kinde I., Papadopoulos N., Carpten J.D., Craig D., O’Shaughnessy J., Kinzler K.W., Parmigiani G., Vogelstein B., et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci. Transl. Med. 2012;4:162ra154. doi: 10.1126/scitranslmed.3004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dawson S.-J., Tsui D.W., Murtaza M., Biggs H., Rueda O.M., Chin S.-F., Dunning M.J., Gale D., Forshew T., Mahler-Araujo B. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 21.Beaver J.A., Jelovac D., Balukrishna S., Cochran R.L., Croessmann S., Zabransky D.J., Wong H.Y., Toro P.V., Cidado J., Blair B.G. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin. Cancer Res. 2014;20:2643–2650. doi: 10.1158/1078-0432.CCR-13-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M., Wang C., Yu B., Zhang X., Shi F., Liu X. Diagnostic value of RASSF1A methylation for breast cancer: A meta-analysis. Biosci. Rep. 2019;39 doi: 10.1042/BSR20190923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nunes S.P., Moreira-Barbosa C., Salta S., Palma de Sousa S., Pousa I., Oliveira J., Soares M., Rego L., Dias T., Rodrigues J., et al. Cell-Free DNA Methylation of Selected Genes Allows for Early Detection of the Major Cancers in Women. Cancers. 2018;10:357. doi: 10.3390/cancers10100357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shan M., Yin H., Li J., Li X., Wang D., Su Y., Niu M., Zhong Z., Wang J., Zhang X., et al. Detection of aberrant methylation of a six-gene panel in serum DNA for diagnosis of breast cancer. Oncotarget. 2016;7:18485–18494. doi: 10.18632/oncotarget.7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uehiro N., Sato F., Pu F., Tanaka S., Kawashima M., Kawaguchi K., Sugimoto M., Saji S., Toi M. Circulating cell-free DNA-based epigenetic assay can detect early breast cancer. Breast Cancer Res. Earch BCR. 2016;18:129. doi: 10.1186/s13058-016-0788-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu M.C., Oxnard G.R., Klein E.A., Swanton C., Seiden M.V., Consortium C. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020;31:745–759. doi: 10.1016/j.annonc.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cristiano S., Leal A., Phallen J., Fiksel J., Adleff V., Bruhm D.C., Jensen S.O., Medina J.E., Hruban C., White J.R., et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature. 2019;570:385–389. doi: 10.1038/s41586-019-1272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimomura A., Shiino S., Kawauchi J., Takizawa S., Sakamoto H., Matsuzaki J., Ono M., Takeshita F., Niida S., Shimizu C., et al. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Sci. 2016;107:326–334. doi: 10.1111/cas.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kahraman M., Roske A., Laufer T., Fehlmann T., Backes C., Kern F., Kohlhaas J., Schrors H., Saiz A., Zabler C., et al. MicroRNA in diagnosis and therapy monitoring of early-stage triple-negative breast cancer. Sci. Rep. 2018;8:11584. doi: 10.1038/s41598-018-29917-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moon P.-G., Lee J.-E., Cho Y.-E., Lee S.J., Chae Y.S., Jung J.H., Kim I.-S., Park H.Y., Baek M.-C. Fibronectin on circulating extracellular vesicles as a liquid biopsy to detect breast cancer. Oncotarget. 2016;7:40189. doi: 10.18632/oncotarget.9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moon P.-G., Lee J.-E., Cho Y.-E., Lee S.J., Jung J.H., Chae Y.S., Bae H.-I., Kim Y.-B., Kim I.-S., Park H.Y. Identification of developmental endothelial locus-1 on circulating extracellular vesicles as a novel biomarker for early breast cancer detection. Clin. Cancer Res. 2016;22:1757–1766. doi: 10.1158/1078-0432.CCR-15-0654. [DOI] [PubMed] [Google Scholar]
- 32.Yuan B., Schafferer S., Tang Q., Scheffler M., Nees J., Heil J., Schott S., Golatta M., Wallwiener M., Sohn C., et al. A plasma metabolite panel as biomarkers for early primary breast cancer detection. Int. J. Cancer. 2019;144:2833–2842. doi: 10.1002/ijc.31996. [DOI] [PubMed] [Google Scholar]
- 33.Rashed R., Darwish H., Omran M., Belal A., Zahran F. A novel serum metabolome score for breast cancer diagnosis. Br. J. Biomed. Sci. 2020:1–6. doi: 10.1080/09674845.2020.1784568. [DOI] [PubMed] [Google Scholar]
- 34.Hadi N.I., Jamal Q., Iqbal A., Shaikh F., Somroo S., Musharraf S.G. Serum metabolomic profiles for breast cancer diagnosis, grading and staging by gas chromatography-mass spectrometry. Sci. Rep. 2017;7:1715. doi: 10.1038/s41598-017-01924-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park J., Shin Y., Kim T.H., Kim D.H., Lee A. Plasma metabolites as possible biomarkers for diagnosis of breast cancer. PLoS ONE. 2019;14:e0225129. doi: 10.1371/journal.pone.0225129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jove M., Collado R., Quiles J.L., Ramirez-Tortosa M.C., Sol J., Ruiz-Sanjuan M., Fernandez M., de la Torre Cabrera C., Ramirez-Tortosa C., Granados-Principal S., et al. A plasma metabolomic signature discloses human breast cancer. Oncotarget. 2017;8:19522–19533. doi: 10.18632/oncotarget.14521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Song L., Liu N., He C., Li Z. Decreased serum levels of free fatty acids are associated with breast cancer. Clin. Chim. Acta. 2014;437:31–37. doi: 10.1016/j.cca.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Cohen J.D., Li L., Wang Y., Thoburn C., Afsari B., Danilova L., Douville C., Javed A.A., Wong F., Mattox A., et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lennon A.M., Buchanan A.H., Kinde I., Warren A., Honushefsky A., Cohain A.T., Ledbetter D.H., Sanfilippo F., Sheridan K., Rosica D., et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science. 2020;369 doi: 10.1126/science.abb9601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong K.C., Chen J., Zhang J., Lin J., Yan S., Zhang S., Li X., Liang C., Peng C., Lin Q., et al. Early cancer detection from multianalyte blood test results. iScience. 2019;15:332–341. doi: 10.1016/j.isci.2019.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erbes T., Hirschfeld M., Rucker G., Jaeger M., Boas J., Iborra S., Mayer S., Gitsch G., Stickeler E. Feasibility of urinary microRNA detection in breast cancer patients and its potential as an innovative non-invasive biomarker. BMC Cancer. 2015;15:193. doi: 10.1186/s12885-015-1190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirschfeld M., Rücker G., Weiß D., Berner K., Ritter A., Jäger M., Erbes T. Urinary Exosomal MicroRNAs as Potential Non-invasive Biomarkers in Breast Cancer Detection. Mol. Diagn. Ther. 2020;28:1–18. doi: 10.1007/s40291-020-00453-y. [DOI] [PubMed] [Google Scholar]
- 43.Ando W., Kikuchi K., Uematsu T., Yokomori H., Takaki T., Sogabe M., Kohgo Y., Otori K., Ishikawa S., Okazaki I. Novel breast cancer screening: Combined expression of miR-21 and MMP-1 in urinary exosomes detects 95% of breast cancer without metastasis. Sci. Rep. 2019;9:1–10. doi: 10.1038/s41598-019-50084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cala M., Aldana J., Sanchez J., Guio J., Meesters R.J.W. Urinary metabolite and lipid alterations in Colombian Hispanic women with breast cancer: A pilot study. J. Pharm. Biomed. Anal. 2018;152:234–241. doi: 10.1016/j.jpba.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 45.Phillips M., Cataneo R.N., Ditkoff B.A., Fisher P., Greenberg J., Gunawardena R., Kwon C.S., Rahbari-Oskoui F., Wong C. Volatile markers of breast cancer in the breath. Breast J. 2003;9:184–191. doi: 10.1046/j.1524-4741.2003.09309.x. [DOI] [PubMed] [Google Scholar]
- 46.Mangler M., Freitag C., Lanowska M., Staeck O., Schneider A., Speiser D. Volatile organic compounds (VOCs) in exhaled breath of patients with breast cancer in a clinical setting. Ginekol. Pol. 2012;83:730–736. [PubMed] [Google Scholar]
- 47.Phillips M., Cataneo R.N., Ditkoff B.A., Fisher P., Greenberg J., Gunawardena R., Kwon C.S., Tietje O., Wong C. Prediction of breast cancer using volatile biomarkers in the breath. Breast Cancer Res. Treat. 2006;99:19–21. doi: 10.1007/s10549-006-9176-1. [DOI] [PubMed] [Google Scholar]
- 48.Herman-Saffar O., Boger Z., Libson S., Lieberman D., Gonen R., Zeiri Y. Early non-invasive detection of breast cancer using exhaled breath and urine analysis. Comput. Biol. Med. 2018;96:227–232. doi: 10.1016/j.compbiomed.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 49.Phillips M., Bevers T., Larsen L.H., Wilkes N.P., Pathak S. Rapid point-of-care breath test predicts breast cancer and abnormal mammograms in symptomatic women. medRxiv. 2020 doi: 10.1101/2020.04.07.20042895. [DOI] [Google Scholar]
- 50.Phillips M., Cataneo R.N., Cruz-Ramos J.A., Huston J., Ornelas O., Pappas N., Pathak S. Prediction of breast cancer risk with volatile biomarkers in breath. Breast Cancer Res. Treat. 2018;170:343–350. doi: 10.1007/s10549-018-4764-4. [DOI] [PubMed] [Google Scholar]
- 51.Kumar S.R., Sauter E.R., Quinn T.P., Deutscher S.L. Thomsen-Friedenreich and Tn antigens in nipple fluid: Carbohydrate biomarkers for breast cancer detection. Clin. Cancer Res. 2005;11:6868–6871. doi: 10.1158/1078-0432.CCR-05-0146. [DOI] [PubMed] [Google Scholar]
- 52.Deutscher S.L., Dickerson M., Gui G., Newton J., Holm J.E., Vogeltanz-Holm N., Kliethermes B., Hewett J.E., Kumar S.R., Quinn T.P., et al. Carbohydrate antigens in nipple aspirate fluid predict the presence of atypia and cancer in women requiring diagnostic breast biopsy. BMC Cancer. 2010;10:519. doi: 10.1186/1471-2407-10-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qin W., Gui G., Zhang K., Twelves D., Kliethermes B., Sauter E.R. Proteins and carbohydrates in nipple aspirate fluid predict the presence of atypia and cancer in women requiring diagnostic breast biopsy. BMC Cancer. 2012;12:52. doi: 10.1186/1471-2407-12-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oda M., Makita M., Iwaya K., Akiyama F., Kohno N., Tsuchiya B., Iwase T., Matsubara O. High levels of DJ-1 protein in nipple fluid of patients with breast cancer. Cancer Sci. 2012;103:1172–1176. doi: 10.1111/j.1349-7006.2012.02267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pawlik T.M., Hawke D.H., Liu Y., Krishnamurthy S., Fritsche H., Hunt K.K., Kuerer H.M. Proteomic analysis of nipple aspirate fluid from women with early-stage breast cancer using isotope-coded affinity tags and tandem mass spectrometry reveals differential expression of vitamin D binding protein. BMC Cancer. 2006;6:68. doi: 10.1186/1471-2407-6-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chatterton R.T., Heinz R.E., Fought A.J., Ivancic D., Shappell C., Allu S., Gapstur S., Scholtens D.M., Gann P.H., Khan S.A. Nipple Aspirate Fluid Hormone Concentrations and Breast Cancer Risk. Horm. Cancer. 2016;7:127–136. doi: 10.1007/s12672-016-0252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lebrecht A., Boehm D., Schmidt M., Koelbl H., Grus F.H. Surface-enhanced laser desorption/ionisation time-of-flight mass spectrometry to detect breast cancer markers in tears and serum. Cancer Genom. Proteom. 2009;6:75–83. [PubMed] [Google Scholar]
- 58.Lebrecht A., Boehm D., Schmidt M., Koelbl H., Schwirz R.L., Grus F.H. Diagnosis of breast cancer by tear proteomic pattern. Cancer Genom. Proteom. 2009;6:177–182. [PubMed] [Google Scholar]
- 59.Bohm D., Keller K., Pieter J., Boehm N., Wolters D., Siggelkow W., Lebrecht A., Schmidt M., Kolbl H., Pfeiffer N., et al. Comparison of tear protein levels in breast cancer patients and healthy controls using a de novo proteomic approach. Oncol. Rep. 2012;28:429–438. doi: 10.3892/or.2012.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim S., Kim T.G., Lee S.H., Kim W., Bang A., Moon S.W., Song J., Shin J.H., Yu J.S., Choi S. Label-free surface-enhanced Raman spectroscopy biosensor for on-site breast cancer detection using human tears. ACS Appl. Mater. Interfaces. 2020;12:7897–7904. doi: 10.1021/acsami.9b19421. [DOI] [PubMed] [Google Scholar]
- 61.Zadák Z., Klemera P., Hyšpler R., Tichá A., Adam T., Friedecký D., Janeč;ková H., Gardlo A., Karlíková; R. A Method of Diagnosing Breast Sancer from a Sample of Apocrine Sweat. CZ307724B6 and PCT/CZ2018/050045. U.S. Patent. 2019
- 62.Gebrehiwot A.G. Ph.D. Thesis. Hokkaido University; Sapporo, Japan: 2019. Human Serum N-glycans as Highly Sensitive Cancer Biomarkers: Potential Benefits and the Risks [an Abstract of Dissertation and a Summary of Dissertation Review] [Google Scholar]
- 63.Duffy M.J. Serum tumor markers in breast cancer: Are they of clinical value? Clin. Chem. 2006;52:345–351. doi: 10.1373/clinchem.2005.059832. [DOI] [PubMed] [Google Scholar]
- 64.Harris L., Fritsche H., Mennel R., Norton L., Ravdin P., Taube S., Somerfield M.R., Hayes D.F., Bast R.C., Jr. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J. Clin. Oncol. 2007;25:5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 65.Kazarian A., Blyuss O., Metodieva G., Gentry-Maharaj A., Ryan A., Kiseleva E.M., Prytomanova O.M., Jacobs I.J., Widschwendter M., Menon U. Testing breast cancer serum biomarkers for early detection and prognosis in pre-diagnosis samples. Br. J. Cancer. 2017;116:501–508. doi: 10.1038/bjc.2016.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hollingsworth A.B., Reese D.E. Potential use of biomarkers to augment clinical decisions for the early detection of breast cancer. Oncol. Hematol. Rev. 2014;10:103–109. doi: 10.17925/OHR.2014.10.2.103. [DOI] [Google Scholar]
- 67.Opstal-van Winden A.W., Rodenburg W., Pennings J.L., Van Oostrom C., Beijnen J.H., Peeters P.H., Van Gils C.H., De Vries A. A bead-based multiplexed immunoassay to evaluate breast cancer biomarkers for early detection in pre-diagnostic serum. Int. J. Mol. Sci. 2012;13:13587–13604. doi: 10.3390/ijms131013587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen C., Poole L., Sizer L., Carter W.B., Frazier T.G. Diagnostic accuracy of the Videssa protein-based liquid biopsy for breast cancer in suspicious mammography. Am. Soc. Clin. Oncol. 2019;37 doi: 10.1200/JCO.2019.37.15_suppl.e13034. [DOI] [Google Scholar]
- 69.Choi J.W., Moon B.I., Lee J.W., Kim H.J., Jin Y., Kim H.J. Use of CA153 for screening breast cancer: An antibodylectin sandwich assay for detecting glycosylation of CA153 in sera. Oncol. Rep. 2018;40:145–154. doi: 10.3892/or.2018.6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reese D.E., Henderson M.C., Silver M., Mulpuri R., Letsios E., Tran Q., Wolf J.K. Breast density does not impact the ability of Videssa(R) Breast to detect breast cancer in women under age 50. PLoS ONE. 2017;12:e0186198. doi: 10.1371/journal.pone.0186198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Amiry N., Kong X., Muniraj N., Kannan N., Grandison P.M., Lin J., Yang Y., Vouyovitch C.M., Borges S., Perry J.K., et al. Trefoil factor-1 (TFF1) enhances oncogenicity of mammary carcinoma cells. Endocrinology. 2009;150:4473–4483. doi: 10.1210/en.2009-0066. [DOI] [PubMed] [Google Scholar]
- 72.Kannan N., Kang J., Kong X., Tang J., Perry J.K., Mohankumar K.M., Miller L.D., Liu E.T., Mertani H.C., Zhu T., et al. Trefoil factor 3 is oncogenic and mediates anti-estrogen resistance in human mammary carcinoma. Neoplasia. 2010;12:1041–1053. doi: 10.1593/neo.10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kang J., Perry J.K., Pandey V., Fielder G.C., Mei B., Qian P.X., Wu Z.S., Zhu T., Liu D.X., Lobie P.E. Artemin is oncogenic for human mammary carcinoma cells. Oncogene. 2009;28:2034–2045. doi: 10.1038/onc.2009.66. [DOI] [PubMed] [Google Scholar]
- 74.Abdel-Fatah T.M.A., Broom R.J., Lu J., Moseley P.M., Huang B., Li L., Liu S., Chen L., Ma R.Z., Cao W., et al. SHON expression predicts response and relapse risk of breast cancer patients after anthracycline-based combination chemotherapy or tamoxifen treatment. Br. J. Cancer. 2019;120:728–745. doi: 10.1038/s41416-019-0405-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jung Y., Abdel-Fatah T.M., Chan S.Y., Nolan C.C., Green A.R., Ellis I.O., Li L., Huang B., Lu J., Xu B., et al. SHON is a novel estrogen-regulated oncogene in mammary carcinoma that predicts patient response to endocrine therapy. Cancer Res. 2013;73:6951–6962. doi: 10.1158/0008-5472.CAN-13-0982. [DOI] [PubMed] [Google Scholar]
- 76.Zhang L., Riethdorf S., Wu G., Wang T., Yang K., Peng G., Liu J., Pantel K. Meta-analysis of the prognostic value of circulating tumor cells in breast cancer. Clin. Cancer Res. 2012;18:5701–5710. doi: 10.1158/1078-0432.CCR-12-1587. [DOI] [PubMed] [Google Scholar]
- 77.Mansouri S., Mokhtari-Hesari P., Naghavi-Al-Hosseini F., Majidzadeh A.K., Farahmand L. The Prognostic Value of Circulating Tumor Cells in Primary Breast Cancer Prior to any Systematic Therapy: A Systematic Review. Curr. Stem. Cell Res. Ther. 2019;14:519–529. doi: 10.2174/1574888X14666190306103759. [DOI] [PubMed] [Google Scholar]
- 78.Eroglu Z., Fielder O., Somlo G. Analysis of circulating tumor cells in breast cancer. J. Natl. Compr. Cancer Netw. 2013;11:977–985. doi: 10.6004/jnccn.2013.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alix-Panabières C., Schwarzenbach H., Pantel K. Circulating tumor cells and circulating tumor DNA. Annu. Rev. Med. 2012;63:199–215. doi: 10.1146/annurev-med-062310-094219. [DOI] [PubMed] [Google Scholar]
- 80.Ferreira M.M., Ramani V.C., Jeffrey S.S. Circulating tumor cell technologies. Mol. Oncol. 2016;10:374–394. doi: 10.1016/j.molonc.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Banko P., Lee S.Y., Nagygyorgy V., Zrinyi M., Chae C.H., Cho D.H., Telekes A. Technologies for circulating tumor cell separation from whole blood. J. Hematol. Oncol. 2019;12:48. doi: 10.1186/s13045-019-0735-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Muller V., Riethdorf S., Rack B., Janni W., Fasching P.A., Solomayer E., Aktas B., Kasimir-Bauer S., Pantel K., Fehm T., et al. Prognostic impact of circulating tumor cells assessed with the CellSearch System and AdnaTest Breast in metastatic breast cancer patients: The DETECT study. Breast Cancer Res. Earch BCR. 2012;14:R118. doi: 10.1186/bcr3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Danila D.C., Samoila A., Patel C., Schreiber N., Herkal A., Anand A., Bastos D., Heller G., Fleisher M., Scher H.I. Clinical validity of detecting circulating tumor cells by AdnaTest assay compared with direct detection of tumor mRNA in stabilized whole blood, as a biomarker predicting overall survival for metastatic castration-resistant prostate cancer patients. Cancer J. 2016;22:315–320. doi: 10.1097/PPO.0000000000000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cohen S.J., Punt C., Iannotti N., Saidman B.H., Sabbath K.D., Gabrail N.Y., Picus J., Morse M., Mitchell E., Miller M.C. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. Clin. Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 85.De Bono J.S., Scher H.I., Montgomery R.B., Parker C., Miller M.C., Tissing H., Doyle G.V., Terstappen L.W., Pienta K.J., Raghavan D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 86.Andree K.C., van Dalum G., Terstappen L.W. Challenges in circulating tumor cell detection by the CellSearch system. Mol. Oncol. 2016;10:395–407. doi: 10.1016/j.molonc.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wan J.C.M., Massie C., Garcia-Corbacho J., Mouliere F., Brenton J.D., Caldas C., Pacey S., Baird R., Rosenfeld N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer. 2017;17:223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 88.De Mattos-Arruda L., Caldas C. Cell-free circulating tumour DNA as a liquid biopsy in breast cancer. Mol. Oncol. 2016;10:464–474. doi: 10.1016/j.molonc.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Buono G., Gerratana L., Bulfoni M., Provinciali N., Basile D., Giuliano M., Corvaja C., Arpino G., Del Mastro L., De Placido S., et al. Circulating tumor DNA analysis in breast cancer: Is it ready for prime-time? Cancer Treat. Rev. 2019;73:73–83. doi: 10.1016/j.ctrv.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 90.Zhou H., Wang X.J., Jiang X., Qian Z., Chen T., Hu Y., Chen Z.H., Gao Y., Wang R., Ye W.W., et al. Plasma cell-free DNA chromosomal instability analysis by low-pass whole-genome sequencing to monitor breast cancer relapse. Breast Cancer Res. Treat. 2019;178:63–73. doi: 10.1007/s10549-019-05375-w. [DOI] [PubMed] [Google Scholar]
- 91.Nicolini C., Ens C., Cerutti T., Roehe A.V., Agnes G., Damin A.P., Alexandre C.O. Elevated level of cell-free plasma DNA is associated with advanced-stage breast cancer and metastasis. Clin. Chem. Lab. Med. 2013;51:e277–e278. doi: 10.1515/cclm-2013-0120. [DOI] [PubMed] [Google Scholar]
- 92.Adalsteinsson V.A., Ha G., Freeman S.S., Choudhury A.D., Stover D.G., Parsons H.A., Gydush G., Reed S.C., Rotem D., Rhoades J., et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 2017;8:1324. doi: 10.1038/s41467-017-00965-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Phallen J., Sausen M., Adleff V., Leal A., Hruban C., White J., Anagnostou V., Fiksel J., Cristiano S., Papp E., et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bidard F.C., Pierga J.Y. Clinical utility of circulating tumor cells in metastatic breast cancer. J. Clin. Oncol. 2015;33:1622. doi: 10.1200/JCO.2014.57.9714. [DOI] [PubMed] [Google Scholar]
- 95.Goh J.Y., Feng M., Wang W., Oguz G., Yatim S., Lee P.L., Bao Y., Lim T.H., Wang P., Tam W.L., et al. Chromosome 1q21.3 amplification is a trackable biomarker and actionable target for breast cancer recurrence. Nat. Med. 2017;23:1319–1330. doi: 10.1038/nm.4405. [DOI] [PubMed] [Google Scholar]
- 96.Berman H., Zhang J., Crawford Y.G., Gauthier M.L., Fordyce C.A., McDermott K.M., Sigaroudinia M., Kozakiewicz K., Tlsty T.D. Cold Spring Harbor Symposia on Quantitative Biology. Volume 70. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 2005. Genetic and epigenetic changes in mammary epithelial cells identify a subpopulation of cells involved in early carcinogenesis; pp. 317–327. [DOI] [PubMed] [Google Scholar]
- 97.Azzollini J., Pesenti C., Pizzamiglio S., Fontana L., Guarino C., Peissel B., Plebani M., Tabano S., Sirchia S.M., Colapietro P., et al. Constitutive BRCA1 Promoter Hypermethylation Can Be a Predisposing Event in Isolated Early-Onset Breast Cancer. Cancers. 2019;11:58. doi: 10.3390/cancers11010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tang Q., Cheng J., Cao X., Surowy H., Burwinkel B. Blood-based DNA methylation as biomarker for breast cancer: A systematic review. Clin. Epigenetics. 2016;8:115. doi: 10.1186/s13148-016-0282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Qian X., Ruan L. APC gene promoter aberrant methylation in serum as a biomarker for breast cancer diagnosis: A meta-analysis. Thorac. Cancer. 2018;9:284–290. doi: 10.1111/1759-7714.12580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gurioli G., Martignano F., Salvi S., Costantini M., Gunelli R., Casadio V. GSTP1 methylation in cancer: A liquid biopsy biomarker? Clin. Chem. Lab. Med. 2018;56:702–717. doi: 10.1515/cclm-2017-0703. [DOI] [PubMed] [Google Scholar]
- 101.Bailleux C., Lacroix L., Barranger E., Delaloge S. Using methylation signatures on cell-free DNA for early cancer detection: A new era in liquid biopsy? Ann. Oncol. 2020 doi: 10.1016/j.annonc.2020.03.307. [DOI] [PubMed] [Google Scholar]
- 102.Van De Voorde L., Speeckaert R., Van Gestel D., Bracke M., De Neve W., Delanghe J., Speeckaert M. DNA methylation-based biomarkers in serum of patients with breast cancer. Mutat. Res. 2012;751:304–325. doi: 10.1016/j.mrrev.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 103.Fece de la Cruz F., Corcoran R.B. Methylation in cell-free DNA for early cancer detection. Ann. Oncol. 2018;29:1351–1353. doi: 10.1093/annonc/mdy134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Widschwendter M., Evans I., Jones A., Ghazali S., Reisel D., Ryan A., Gentry-Maharaj A., Zikan M., Cibula D., Eichner J., et al. Methylation patterns in serum DNA for early identification of disseminated breast cancer. Genome Med. 2017;9:115. doi: 10.1186/s13073-017-0499-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Panagopoulou M., Karaglani M., Balgkouranidou I., Biziota E., Koukaki T., Karamitrousis E., Nena E., Tsamardinos I., Kolios G., Lianidou E., et al. Circulating cell-free DNA in breast cancer: Size profiling, levels, and methylation patterns lead to prognostic and predictive classifiers. Oncogene. 2019;38:3387–3401. doi: 10.1038/s41388-018-0660-y. [DOI] [PubMed] [Google Scholar]
- 106.Shan M., Zhang L., Liu Y., Gao C., Kang W., Yang W., He Y., Zhang G. DNA Methylation Profiles and Their Diagnostic Utility in BC. Dis. Markers. 2019;2019:6328503. doi: 10.1155/2019/6328503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chang Y., Tolani B., Nie X., Zhi X., Hu M., He B. Review of the clinical applications and technological advances of circulating tumor DNA in cancer monitoring. Ther. Clin. Risk Manag. 2017;13:1363–1374. doi: 10.2147/TCRM.S141991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Eigeliene N., Saarenheimo J., Jekunen A. Potential of Liquid Biopsies for Breast Cancer Screening, Diagnosis, and Response to Treatment. Oncology. 2019;96:115–124. doi: 10.1159/000495615. [DOI] [PubMed] [Google Scholar]
- 109.Duffy M.J., McDermott E.W., Crown J. Blood-based biomarkers in breast cancer: From proteins to circulating tumor cells to circulating tumor DNA. Tumour. Biol. 2018;40:1010428318776169. doi: 10.1177/1010428318776169. [DOI] [PubMed] [Google Scholar]
- 110.Bettegowda C., Sausen M., Leary R.J., Kinde I., Wang Y., Agrawal N., Bartlett B.R., Wang H., Luber B., Alani R.M., et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014;6:224ra224. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ossandon M.R., Agrawal L., Bernhard E.J., Conley B.A., Dey S.M., Divi R.L., Guan P., Lively T.G., McKee T.C., Sorg B.S. Circulating tumor DNA assays in Clinical Cancer Research. JNCI J. Natl. Cancer Inst. 2018;110:929–934. doi: 10.1093/jnci/djy105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A., et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schwarzenbach H., Nishida N., Calin G.A., Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014;11:145–156. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- 114.Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L., Peterson A., Noteboom J., O’Briant K.C., Allen A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Guo J., Liu C., Wang W., Liu Y., He H., Chen C., Xiang R., Luo Y. Identification of serum miR-1915-3p and miR-455-3p as biomarkers for breast cancer. PLoS ONE. 2018;13:e0200716. doi: 10.1371/journal.pone.0200716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li G., Hu J., Hu G. Translational Research in Breast Cancer. Springer; Berlin/Heidelberg, Germany: 2017. Biomarker studies in early detection and prognosis of breast cancer; pp. 27–39. [DOI] [PubMed] [Google Scholar]
- 117.Fang R., Zhu Y., Hu L., Khadka V.S., Ai J., Zou H., Ju D., Jiang B., Deng Y., Hu X. Plasma MicroRNA Pair Panels as Novel Biomarkers for Detection of Early Stage Breast Cancer. Front. Physiol. 2018;9:1879. doi: 10.3389/fphys.2018.01879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.An X., Quan H., Lv J., Meng L., Wang C., Yu Z., Han J. Serum microRNA as potential biomarker to detect breast atypical hyperplasia and early-stage breast cancer. Future Oncol. 2018;14:3145–3161. doi: 10.2217/fon-2018-0334. [DOI] [PubMed] [Google Scholar]
- 119.Sochor M., Basova P., Pesta M., Dusilkova N., Bartos J., Burda P., Pospisil V., Stopka T. Oncogenic microRNAs: miR-155, miR-19a, miR-181b, and miR-24 enable monitoring of early breast cancer in serum. BMC Cancer. 2014;14:448. doi: 10.1186/1471-2407-14-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schwarzenbach H., Milde-Langosch K., Steinbach B., Muller V., Pantel K. Diagnostic potential of PTEN-targeting miR-214 in the blood of breast cancer patients. Breast Cancer Res. Treat. 2012;134:933–941. doi: 10.1007/s10549-012-1988-6. [DOI] [PubMed] [Google Scholar]
- 121.Heneghan H.M., Miller N., Kelly R., Newell J., Kerin M.J. Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease. Oncologist. 2010;15:673–682. doi: 10.1634/theoncologist.2010-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hamam R., Hamam D., Alsaleh K.A., Kassem M., Zaher W., Alfayez M., Aldahmash A., Alajez N.M. Circulating microRNAs in breast cancer: Novel diagnostic and prognostic biomarkers. Cell Death Dis. 2017;8:e3045. doi: 10.1038/cddis.2017.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang H., Peng R., Wang J., Qin Z., Xue L. Circulating microRNAs as potential cancer biomarkers: The advantage and disadvantage. Clin. Epigenetics. 2018;10:59. doi: 10.1186/s13148-018-0492-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gould S.J., Raposo G. As we wait: Coping with an imperfect nomenclature for extracellular vesicles. J. Extracell. Vesicles. 2013;2:20389. doi: 10.3402/jev.v2i0.20389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lötvall J., Hill A.F., Hochberg F., Buzás E.I., Di Vizio D., Gardiner C., Gho Y.S., Kurochkin I.V., Mathivanan S., Quesenberry P. Minimal Experimental Requirements for Definition of Extracellular Vesicles and their Functions: A Position Statement from the International Society for Extracellular Vesicles. Taylor & Francis; Oxfordshire, UK: 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kosaka N., Kogure A., Yamamoto T., Urabe F., Usuba W., Prieto-Vila M., Ochiya T. Exploiting the message from cancer: The diagnostic value of extracellular vesicles for clinical applications. Exp. Mol. Med. 2019;51:1–9. doi: 10.1038/s12276-019-0219-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rupp A.K., Rupp C., Keller S., Brase J.C., Ehehalt R., Fogel M., Moldenhauer G., Marme F., Sultmann H., Altevogt P. Loss of EpCAM expression in breast cancer derived serum exosomes: Role of proteolytic cleavage. Gynecol. Oncol. 2011;122:437–446. doi: 10.1016/j.ygyno.2011.04.035. [DOI] [PubMed] [Google Scholar]
- 128.Khan S., Bennit H.F., Turay D., Perez M., Mirshahidi S., Yuan Y., Wall N.R. Early diagnostic value of survivin and its alternative splice variants in breast cancer. BMC Cancer. 2014;14:176. doi: 10.1186/1471-2407-14-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Melo S.A., Sugimoto H., O’Connell J.T., Kato N., Villanueva A., Vidal A., Qiu L., Vitkin E., Perelman L.T., Melo C.A., et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Galindo-Hernandez O., Villegas-Comonfort S., Candanedo F., Gonzalez-Vazquez M.C., Chavez-Ocana S., Jimenez-Villanueva X., Sierra-Martinez M., Salazar E.P. Elevated concentration of microvesicles isolated from peripheral blood in breast cancer patients. Arch. Med. Res. 2013;44:208–214. doi: 10.1016/j.arcmed.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 131.Gonzalez-Villasana V., Rashed M.H., Gonzalez-Cantu Y., Bayraktar R., Menchaca-Arredondo J.L., Vazquez-Guillen J.M., Rodriguez-Padilla C., Lopez-Berestein G., Resendez-Perez D. Presence of Circulating miR-145, miR-155, and miR-382 in Exosomes Isolated from Serum of Breast Cancer Patients and Healthy Donors. Dis. Markers. 2019;2019:6852917. doi: 10.1155/2019/6852917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sadovska L., EGLĪTIS J., LINĒ A. Extracellular vesicles as biomarkers and therapeutic targets in breast cancer. AntiCancer Res. 2015;35:6379–6390. [PubMed] [Google Scholar]
- 133.Al-Nedawi K., Meehan B., Micallef J., Lhotak V., May L., Guha A., Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 134.Balaj L., Lessard R., Dai L., Cho Y.J., Pomeroy S.L., Breakefield X.O., Skog J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Menck K., Scharf C., Bleckmann A., Dyck L., Rost U., Wenzel D., Dhople V.M., Siam L., Pukrop T., Binder C., et al. Tumor-derived microvesicles mediate human breast cancer invasion through differentially glycosylated EMMPRIN. J. Mol. Cell Biol. 2015;7:143–153. doi: 10.1093/jmcb/mju047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Eichelser C., Stuckrath I., Muller V., Milde-Langosch K., Wikman H., Pantel K., Schwarzenbach H. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget. 2014;5:9650–9663. doi: 10.18632/oncotarget.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Aatonen M.T., Ohman T., Nyman T.A., Laitinen S., Gronholm M., Siljander P.R. Isolation and characterization of platelet-derived extracellular vesicles. J. Extracell. Vesicles. 2014;3 doi: 10.3402/jev.v3.24692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Momen-Heravi F., Balaj L., Alian S., Mantel P.Y., Halleck A.E., Trachtenberg A.J., Soria C.E., Oquin S., Bonebreak C.M., Saracoglu E., et al. Current methods for the isolation of extracellular vesicles. Biol. Chem. 2013;394:1253–1262. doi: 10.1515/hsz-2013-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Brenner A.W., Su G.H., Momen-Heravi F. Isolation of extracellular vesicles for cancer diagnosis and functional studies. Methods Mol. Biol. 2019;1882:229–237. doi: 10.1007/978-1-4939-8879-2_21. [DOI] [PubMed] [Google Scholar]
- 140.Doyle L.M., Wang M.Z. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells. 2019;8:727. doi: 10.3390/cells8070727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Charest A. Experimental and biological insights from proteomic analyses of extracellular vesicle cargos in normalcy and disease. Adv. Biosyst. 2020 doi: 10.1002/adbi.202000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Royo F., Thery C., Falcon-Perez J.M., Nieuwland R., Witwer K.W. Methods for separation and characterization of extracellular vesicles: Results of a worldwide survey performed by the ISEV rigor and standardization subcommittee. Cells. 2020;9:1955. doi: 10.3390/cells9091955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zha Q.B., Yao Y.F., Ren Z.J., Li X.J., Tang J.H. Extracellular vesicles: An overview of biogenesis, function, and role in breast cancer. Tumour. Biol. 2017;39:1010428317691182. doi: 10.1177/1010428317691182. [DOI] [PubMed] [Google Scholar]
- 144.Tan B., Zhang Y., Zhang T., He J., Luo X., Bian X., Wu J., Zou C., Wang Y., Fu L. Identifying potential serum biomarkers of breast cancer through targeted free fatty acid profiles screening based on a GC-MS platform. Biomed. Chromatogr. 2020 doi: 10.1002/bmc.4922. [DOI] [PubMed] [Google Scholar]
- 145.Young R.P., Christmas T., Hopkins R.J. Multi-analyte assays and early detection of common cancers. J. Thorac. Dis. 2018;10:S2165–S2167. doi: 10.21037/jtd.2018.06.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pencina M.J. Caution is needed in the interpretation of added value of biomarkers analyzed in matched case control studies. Clin. Chem. 2012;58:1176–1178. doi: 10.1373/clinchem.2012.188383. [DOI] [PubMed] [Google Scholar]
- 147.Beretov J., Wasinger V.C., Graham P.H., Millar E.K., Kearsley J.H., Li Y. Proteomics for breast cancer urine biomarkers. Adv. Clin. Chem. 2014;63:123–167. doi: 10.1016/b978-0-12-800094-6.00004-2. [DOI] [PubMed] [Google Scholar]
- 148.Hesari A., Golrokh Moghadam S.A., Siasi A., Rahmani M., Behboodi N., Rastgar-Moghadam A., Ferns G.A., Ghasemi F., Avan A. Tumor-derived exosomes: Potential biomarker or therapeutic target in breast cancer? J. Cell. Biochem. 2018;119:4236–4240. doi: 10.1002/jcb.26364. [DOI] [PubMed] [Google Scholar]
- 149.Gasparri M.L., Casorelli A., Bardhi E., Besharat A.R., Savone D., Ruscito I., Farooqi A.A., Papadia A., Mueller M.D., Ferretti E., et al. Beyond circulating microRNA biomarkers: Urinary microRNAs in ovarian and breast cancer. Tumour. Biol. 2017;39:1010428317695525. doi: 10.1177/1010428317695525. [DOI] [PubMed] [Google Scholar]
- 150.Slupsky C.M., Steed H., Wells T.H., Dabbs K., Schepansky A., Capstick V., Faught W., Sawyer M.B. Urine metabolite analysis offers potential early diagnosis of ovarian and breast cancers. Clin. Cancer Res. 2010;16:5835–5841. doi: 10.1158/1078-0432.CCR-10-1434. [DOI] [PubMed] [Google Scholar]
- 151.Punnonen K., Hietanen E., Auvinen O., Punnonen R. Phospholipids and fatty acids in breast cancer tissue. J. Cancer Res. Clin. Oncol. 1989;115:575–578. doi: 10.1007/BF00391361. [DOI] [PubMed] [Google Scholar]
- 152.Mistry D.A., French P.W. Circulating Phospholipids as Biomarkers of Breast Cancer: A Review. Breast Cancer (Auckl) 2016;10:191–196. doi: 10.4137/BCBCR.S40693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Moreau K., Dizin E., Ray H., Luquain C., Lefai E., Foufelle F., Billaud M., Lenoir G.M., Venezia N.D. BRCA1 affects lipid synthesis through its interaction with acetyl-CoA carboxylase. J. Biol. Chem. 2006;281:3172–3181. doi: 10.1074/jbc.M504652200. [DOI] [PubMed] [Google Scholar]
- 154.Kim H., Min H.K., Kong G., Moon M.H. Quantitative analysis of phosphatidylcholines and phosphatidylethanolamines in urine of patients with breast cancer by nanoflow liquid chromatography/tandem mass spectrometry. Anal. Bioanal. Chem. 2009;393:1649–1656. doi: 10.1007/s00216-009-2621-3. [DOI] [PubMed] [Google Scholar]
- 155.Beretov J., Wasinger V.C., Millar E.K., Schwartz P., Graham P.H., Li Y. Proteomic Analysis of Urine to Identify Breast Cancer Biomarker Candidates Using a Label-Free LC-MS/MS Approach. PLoS ONE. 2015;10:e0141876. doi: 10.1371/journal.pone.0141876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sreekumar A., Poisson L.M., Rajendiran T.M., Khan A.P., Cao Q., Yu J., Laxman B., Mehra R., Lonigro R.J., Li Y., et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 157.Woo H.M., Kim K.M., Choi M.H., Jung B.H., Lee J., Kong G., Nam S.J., Kim S., Bai S.W., Chung B.C. Mass spectrometry based metabolomic approaches in urinary biomarker study of women’s cancers. Clin. Chim. Acta. 2009;400:63–69. doi: 10.1016/j.cca.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 158.Frickenschmidt A., Frohlich H., Bullinger D., Zell A., Laufer S., Gleiter C.H., Liebich H., Kammerer B. Metabonomics in cancer diagnosis: Mass spectrometry-based profiling of urinary nucleosides from breast cancer patients. Biomarkers. 2008;13:435–449. doi: 10.1080/13547500802012858. [DOI] [PubMed] [Google Scholar]
- 159.Cho S.H., Jung B.H., Lee S.H., Lee W.Y., Kong G., Chung B.C. Direct determination of nucleosides in the urine of patients with breast cancer using column-switching liquid chromatography-tandem mass spectrometry. Biomed. Chromatogr. 2006;20:1229–1236. doi: 10.1002/bmc.689. [DOI] [PubMed] [Google Scholar]
- 160.Chen Y., Zhang R., Song Y., He J., Sun J., Bai J., An Z., Dong L., Zhan Q., Abliz Z. RRLC-MS/MS-based metabonomics combined with in-depth analysis of metabolic correlation network: Finding potential biomarkers for breast cancer. Analyst. 2009;134:2003–2011. doi: 10.1039/b907243h. [DOI] [PubMed] [Google Scholar]
- 161.Kim Y., Koo I., Jung B.H., Chung B.C., Lee D. Multivariate classification of urine metabolome profiles for breast cancer diagnosis. BMC Bioinform. 2010;11(Suppl. S2):S4. doi: 10.1186/1471-2105-11-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Dinges S.S., Hohm A., Vandergrift L.A., Nowak J., Habbel P., Kaltashov I.A., Cheng L.L. Cancer metabolomic markers in urine: Evidence, techniques and recommendations. Nat. Rev. Urol. 2019;16:339–362. doi: 10.1038/s41585-019-0185-3. [DOI] [PubMed] [Google Scholar]
- 163.Kabir K.M.M., Donald W.A. Cancer breath testing: A patent review. Expert Opin. Ther. Pat. 2018;28:227–239. doi: 10.1080/13543776.2018.1423680. [DOI] [PubMed] [Google Scholar]
- 164.Hanna G.B., Boshier P.R., Markar S.R., Romano A. Accuracy and methodologic challenges of volatile organic compound–based exhaled breath tests for cancer diagnosis: A systematic review and meta-analysis. JAMA Oncol. 2019;5:e182815. doi: 10.1001/jamaoncol.2018.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hietanen E., Bartsch H., Bereziat J.C., Camus A.M., McClinton S., Eremin O., Davidson L., Boyle P. Diet and oxidative stress in breast, colon and prostate cancer patients: A case-control study. Eur. J. Clin. Nutr. 1994;48:575–586. [PubMed] [Google Scholar]
- 166.Barash O., Zhang W., Halpern J.M., Hua Q.L., Pan Y.Y., Kayal H., Khoury K., Liu H., Davies M.P., Haick H. Differentiation between genetic mutations of breast cancer by breath volatolomics. Oncotarget. 2015;6:44864–44876. doi: 10.18632/oncotarget.6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Li J., Peng Y., Liu Y., Li W., Jin Y., Tang Z., Duan Y. Investigation of potential breath biomarkers for the early diagnosis of breast cancer using gas chromatography-mass spectrometry. Clin. Chim. Acta. 2014;436:59–67. doi: 10.1016/j.cca.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 168.Peng G., Hakim M., Broza Y.Y., Billan S., Abdah-Bortnyak R., Kuten A., Tisch U., Haick H. Detection of lung, breast, colorectal, and prostate cancers from exhaled breath using a single array of nanosensors. Br. J. Cancer. 2010;103:542–551. doi: 10.1038/sj.bjc.6605810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Phillips M., Beatty J.D., Cataneo R.N., Huston J., Kaplan P.D., Lalisang R.I., Lambin P., Lobbes M.B., Mundada M., Pappas N., et al. Rapid point-of-care breath test for biomarkers of breast cancer and abnormal mammograms. PLoS ONE. 2014;9:e90226. doi: 10.1371/journal.pone.0090226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sauter E.R., Winn J.N., Dale P.S., Wagner-Mann C. Nipple aspirate fluid color is associated with breast cancer. Cancer Detect. Prev. 2006;30:322–328. doi: 10.1016/j.cdp.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 171.Tredwell G.D., Miller J.A., Chow H.H., Thompson P.A., Keun H.C. Metabolomic characterization of nipple aspirate fluid by (1)H NMR spectroscopy and GC-MS. J. Proteome. Res. 2014;13:883–889. doi: 10.1021/pr400924k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sauter E.R., Ross E., Daly M., Klein-Szanto A., Engstrom P.F., Sorling A., Malick J., Ehya H. Nipple aspirate fluid: A promising non-invasive method to identify cellular markers of breast cancer risk. Br. J. Cancer. 1997;76:494–501. doi: 10.1038/bjc.1997.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mannello F., Malatesta M., Sebastiani M., Battistelli S., Gazzanelli G. Molecular forms and ultrastructural localization of prostate-specific antigen in nipple aspirate fluids. Clin. Chem. 1999;45:2263–2266. doi: 10.1093/clinchem/45.12.2263. [DOI] [PubMed] [Google Scholar]
- 174.Sauter E.R., Tichansky D.S., Chervoneva I., Diamandis E.P. Circulating testosterone and prostate-specific antigen in nipple aspirate fluid and tissue are associated with breast cancer. Environ. Health Perspect. 2002;110:241–246. doi: 10.1289/ehp.02110241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Shaheed S.U., Tait C., Kyriacou K., Linforth R., Salhab M., Sutton C. Evaluation of nipple aspirate fluid as a diagnostic tool for early detection of breast cancer. Clin. Proteom. 2018;15:3. doi: 10.1186/s12014-017-9179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Moelans C.B., Patuleia S.I.S., van Gils C.H., van der Wall E., van Diest P.J. Application of nipple aspirate fluid miRNA profiles for early breast cancer detection and management. Int. J. Mol. Sci. 2019;20:5814. doi: 10.3390/ijms20225814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang K., Zhao S., Wang Q., Yang H.S., Zhu J., Ma R. Identification of microRNAs in nipple discharge as potential diagnostic biomarkers for breast cancer. Ann. Surg. Oncol. 2015;22(Suppl. S3):536–544. doi: 10.1245/s10434-015-4586-0. [DOI] [PubMed] [Google Scholar]
- 178.Do Canto L.M., Marian C., Willey S., Sidawy M., Da Cunha P.A., Rone J.D., Li X., Gusev Y., Haddad B.R. MicroRNA analysis of breast ductal fluid in breast cancer patients. Int. J. Oncol. 2016;48:2071–2078. doi: 10.3892/ijo.2016.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zhang K., Wang Y.W., Ma R. Bioinformatics analysis of dysregulated microRNAs in the nipple discharge of patients with breast cancer. Oncol. Lett. 2017;13:3100–3108. doi: 10.3892/ol.2017.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Barmada A., Shippy S.A. Tear analysis as the next routine body fluid test. Eye. 2020;6:1–3. doi: 10.1038/s41433-020-0930-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Aihara T., Fujiwara Y., Ooka M., Sakita I., Tamaki Y., Monden M. Mammaglobin B as a novel marker for detection of breast cancer micrometastases in axillary lymph nodes by reverse transcription-polymerase chain reaction. Breast Cancer Res. Treat. 1999;58:137–140. doi: 10.1023/A:1006335817889. [DOI] [PubMed] [Google Scholar]
- 182.Al Joudi F.S. Human mammaglobin in breast cancer: A brief review of its clinical utility. Indian J. Med. Res. 2014;139:675–685. [PMC free article] [PubMed] [Google Scholar]
- 183.Molloy M.P., Bolis S., Herbert B.R., Ou K., Tyler M.I., van Dyk D.D., Willcox M.D., Gooley A.A., Williams K.L., Morris C.A., et al. Establishment of the human reflex tear two-dimensional polyacrylamide gel electrophoresis reference map: New proteins of potential diagnostic value. Electrophoresis. 1997;18:2811–2815. doi: 10.1002/elps.1150181516. [DOI] [PubMed] [Google Scholar]
- 184.Evans V., Vockler C., Friedlander M., Walsh B., Willcox M.D. Lacryglobin in human tears, a potential marker for cancer. Clin. Exp. Ophthalmol. 2001;29:161–163. doi: 10.1046/j.1442-9071.2001.00408.x. [DOI] [PubMed] [Google Scholar]
- 185.Morton S., Crucian B., Hagan S., Satyamitra M., Daily A. NASA Technical Reports Server. U.S. National Aeronautics and Space Administration; Washington, DC, USA: 2018. Pilot study on the investigation of tear fluid biomarkers as an indicator of ocular, neurological, and immunological health in astronauts. [Google Scholar]
- 186.Yu Y., Prassas I., Muytjens C.M., Diamandis E.P. Proteomic and peptidomic analysis of human sweat with emphasis on proteolysis. J. Proteom. 2017;155:40–48. doi: 10.1016/j.jprot.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 187.Wagener J.S., Zemanick E.T., Sontag M.K. Newborn screening for cystic fibrosis. Curr. Opin. Pediatr. 2012;24:329–335. doi: 10.1097/MOP.0b013e328353489a. [DOI] [PubMed] [Google Scholar]
- 188.De Giovanni N., Fucci N. The current status of sweat testing for drugs of abuse: A review. Curr. Med. Chem. 2013;20:545–561. doi: 10.2174/0929867311320040006. [DOI] [PubMed] [Google Scholar]
- 189.Adewole O.O., Erhabor G.E., Adewole T.O., Ojo A.O., Oshokoya H., Wolfe L.M., Prenni J.E. Proteomic profiling of eccrine sweat reveals its potential as a diagnostic biofluid for active tuberculosis. Proteom. Clin. Appl. 2016;10:547–553. doi: 10.1002/prca.201500071. [DOI] [PubMed] [Google Scholar]
- 190.Calderón-Santiago M., Priego-Capote F., Turck N., Robin X., Jurado-Gámez B., Sanchez J.C., Luque de Castro M.D. Human sweat metabolomics for lung cancer screening. Anal. Bioanal. Chem. 2015;407:5381–5392. doi: 10.1007/s00216-015-8700-8. [DOI] [PubMed] [Google Scholar]
- 191.Moss J., Zick A., Grinshpun A., Carmon E., Maoz M., Ochana B.L., Abraham O., Arieli O., Germansky L., Meir K., et al. Circulating breast-derived DNA allows universal detection and monitoring of localized breast cancer. Ann. Oncol. 2020;31:395–403. doi: 10.1016/j.annonc.2019.11.014. [DOI] [PubMed] [Google Scholar]
- 192.Szyf M. DNA methylation signatures for breast cancer classification and prognosis. Genome. Med. 2012;4:26. doi: 10.1186/gm325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Pepe M.S., Etzioni R., Feng Z., Potter J.D., Thompson M.L., Thornquist M., Winget M., Yasui Y. Phases of biomarker development for early detection of cancer. J. Natl. Cancer Inst. 2001;93:1054–1061. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
- 194.Analyzing the Composition of Tears to Identify Cancer (ACT) [(accessed on 23 September 2020)]; Available online: https://clinicaltrials.gov/ct2/show/NCT00574678.
- 195.A Prospective Validation Study of a Rapid Point-of-Care Breath Test for Breast Cancer. [(accessed on 23 September 2020)]; Available online: https://clinicaltrials.gov/ct2/show/NCT02888366.
- 196.Detecting Cancers Earlier through Elective Plasma-based CancerSeek Testing (ASCEND) [(accessed on 23 September 2020)]; Available online: https://clinicaltrials.gov/ct2/show/Nct04213326.
- 197.The Circulating Cell-Free Genome Atlas Study (CCGA) [(accessed on 23 September 2020)]; Available online: https://clinicaltrials.gov/ct2/show/NCT02889978.
- 198.Cohn A., Seiden M., Kurtzman K., Hubbell E., Gross S., Venn O., Fung E., Liu M., Klein E., Oxnard G., et al. The Circulating Cell-free Genome Atlas (CCGA) Study: Follow-up (F/U) on non-cancer participants with cancer-like cell-free DNA signals. J. Clin. Oncol. 2019;37:5574. doi: 10.1200/JCO.2019.37.15_suppl.5574. [DOI] [Google Scholar]
- 199.The STRIVE Study: Development of a Blood Test for Early Detection of Multiple Cancer Types. [(accessed on 23 September 2020)]; Available online: https://clinicaltrials.gov/ct2/show/NCT03085888.
- 200.The SUMMIT Study: A Cancer Screening Study (SUMMIT) [(accessed on 23 September 2020)]; Available online: https://clinicaltrials.gov/ct2/show/NCT03934866.
- 201.Assessment of the Implementation of an Investigational Multi-Cancer Early Detection Test into Clinical Practice. [(accessed on 23 September 2020)]; Available online: https://clinicaltrials.gov/ct2/show/NCT04241796.
- 202.Hayes D.F. Biomarker validation and testing. Mol. Oncol. 2015;9:960–966. doi: 10.1016/j.molonc.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Goossens N., Nakagawa S., Sun X., Hoshida Y. Cancer biomarker discovery and validation. Transl. Cancer Res. 2015;4:256–269. doi: 10.3978/j.issn.2218-676X.2015.06.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Heitzer E., Perakis S., Geigl J.B., Speicher M.R. The potential of liquid biopsies for the early detection of cancer. NPJ Precis. Oncol. 2017;1:36. doi: 10.1038/s41698-017-0039-5. [DOI] [PMC free article] [PubMed] [Google Scholar]