Abstract
Simple Summary
Cetuximab is an antibody that blocks EGFR signaling and stimulates an immune response against cancer cells. For patients with advanced colorectal cancer, tumor sidedness and RAS mutation status are the primary factors used to select systemic therapy. Additional biomarkers are needed to better predict which patients will benefit from cetuximab-based regimens. The aim of our retrospective study was to assess the predictive and prognostic value of 12 germline single nucleotide polymorphisms in five immune related genes in 924 patients with advanced colorectal cancer undergoing therapy with cetuximab. We identified a CD24 germline genetic variant which independently predicted survival in a discovery cohort and confirmed these findings in a validation cohort. If confirmed in prospective studies, CD24 and other immune related polymorphisms may guide the use of cetuximab in patients with advanced colorectal cancer.
Abstract
Cetuximab, an IgG1 EGFR-directed antibody, promotes antibody-dependent cell-mediated cytotoxicity. We hypothesized that single-nucleotide polymorphisms (SNPs) in immune regulatory pathways may predict outcomes in patients with metastatic colorectal cancer treated with cetuximab-based regimens. A total of 924 patients were included: 105 received cetuximab in IMCL-0144 and cetuximab/irinotecan in GONO-ASL608LIOM01 (training cohort), 225 FOLFIRI/cetuximab in FIRE-3 (validation cohort 1), 74 oxaliplatin/cetuximab regimens in JACCRO CC-05/06 (validation cohort 2), and 520 FOLFIRI/bevacizumab in FIRE-3 and TRIBE (control cohorts). Twelve SNPs in five genes (IDO1; PD-L1; PD-1; CTLA-4; CD24) were evaluated by PCR-based direct sequencing. We analyzed associations between genotype and clinical outcomes. In the training cohort; patients with the CD24 rs52812045 A/A genotype had a significantly shorter median PFS and OS than those with the G/G genotype (PFS 1.3 vs. 3.6 months; OS 2.3 vs. 7.8 months) in univariate (PFS HR 3.62; p = 0.001; OS HR 3.27; p = 0.0004) and multivariate (PFS HR 3.18; p = 0.009; OS HR 4.93; p = 0.001) analyses. Similarly; any A allele carriers in the JACCRO validation cohort had a significantly shorter PFS than G/G carriers (9.2 vs. 11.8 months; univariate HR 1.90; p = 0.011; multivariate HR 2.12; p = 0.018). These associations were not demonstrated in the control cohorts. CD24 genetic variants may help select patients with metastatic colorectal cancer most likely to benefit from cetuximab-based therapy.
Keywords: CD24, biomarker, cetuximab, colorectal cancer, single nucleotide polymorphism
1. Introduction
Cetuximab monotherapy and combinations with FOLFIRI (leucovorin, fluorouracil, irinotecan) or FOLFOX (leucovorin, fluorouracil, oxaliplatin) have prolonged the survival of metastatic colorectal cancer (mCRC) patients with RAS wild-type tumors [1,2]. Nonetheless, one-third of RAS-selected patients do not benefit from cetuximab-based regimens [3], and treatment resistance develops independent of RAS/RAF/MEK signaling.
Cetuximab, a chimeric IgG1 monoclonal antibody against the epidermal growth factor receptor (EGFR), promotes antibody-dependent cell-mediated cytotoxicity (ADCC) [4,5]. Upon binding to the FCγ receptor on natural killer (NK) and other immune cells, cetuximab facilitates Fas-FasL-induced tumor cell apoptosis. In response to cetuximab-mediated ADCC, compensatory immunosuppressive pathways (e.g., PDL-1, CTLA-4) may be activated by cancer cells [6]. While extended RAS testing accounts for compensatory pathways that renders tumors resistant to cetuximab-mediated EGFR inhibition [3], there are no validated markers to predict the benefit from its ADCC-dependent mode of action. Identifying such markers could refine patient selection and reveal novel actionable alterations.
We previously demonstrated the prognostic value of FCγ-receptor polymorphisms in mCRC patients receiving cetuximab [7]. With the advent of immune checkpoint inhibitors in mCRC, identifying relevant biomarkers is an area of active investigation. We hypothesized that single nucleotide polymorphisms (SNPs) within critical immune regulatory pathways may serve as clinically meaningful markers in mCRC patients receiving cetuximab-based therapy. We examined 12 SNPs in five genes (CD24, IDO1, PD-L1, PD-1, CTLA-4) to identify predictive and prognostic genetic variants in a training cohort of mCRC patients treated with cetuximab with or without irinotecan in two phase 2 studies [8]. Then, we tested SNPs with significant prognostic associations in two validation cohorts of mCRC patients, one treated with FOLFIRI-cetuximab in FIRE-3 [2] and the other treated with oxaliplatin-cetuximab regimens in JACCRO CC-05/06. Lastly, we examined these associations in two control cohorts who received FOLFIRI-bevacizumab in FIRE-3 and TRIBE [2,9].
2. Results
Patient baseline characteristics and primary outcome data are summarized in Table 1. Patients in the training cohort had relatively refractory disease, with lower overall response rate (RR) (20.0%; p < 0.001) and disease control rate (61.9%; p < 0.001), and an expectedly shorter median PFS (3.7 months, 95% CI: 3.2, 4.9 months; p < 0.001) and OS (10.6 months, 95% CI: 7.8, 13.2 months; p < 0.001) than those in the validation or control cohorts who underwent first-line therapy.
Table 1.
Baseline patient and tumor characteristics.
| Characteristic | Training Cohort (N = 105) |
FIRE-3 Validation Cohort 1 (N = 225) |
Japanese Validation Cohort 2 (N = 74) |
FIRE-3 Control Cohort 1 (N = 292) |
TRIBE Control Cohort 2 (N = 228) |
p-Value * |
|---|---|---|---|---|---|---|
| Age, years | ||||||
| Median (range) | 63 (35–83) | 64 (38–79) | 63 (40-79) | 65 (31-76) | 60 (29–75) | <0.001 |
| Sex | ||||||
| Male | 62 (59%) | 159 (71%) | 42 (57%) | 194 (66%) | 138 (61%) | 0.059 |
| Female | 43 (41%) | 66 (29%) | 32 (43%) | 98 (34%) | 90 (39%) | |
| Overall Response Rate (ORR) | 20.0% | 61.3% | 71.6% | 57.2% | 56.6% | <0.001 |
| Disease Control Rate (DCR) | 61.9% | 80.0% | 91.9% | 83.2% | 89.0% | <0.001 |
| Median PFS, months ** (95% CI) |
3.7 (3.2–4.9) |
9.8 (8·7–10.8) |
10.0 (8.9–13.7) |
10.1 (9.6–11.2) |
9·7 (9.2–10.8) |
<0.001 |
| Median OS, months ** (95% CI) |
10.6 (7.8–13.2) |
28.6 (23.7–36.3) |
33.9 (26.5+) |
23.7 (21.4–26.4) |
26.1 (22.7-30.9) |
<0.001 |
* p-value was based on the Kruskal-Wallis test for age, chi-square test for categorical variables, and log-rank test for PFS and OS. ** Median follow-up was 18.2 months (range: 1.2–42.4 months), 35.3 months (range: 0.0–70.7 months), 31.0 months (range: 5.4–42.9 months), 40.7 months (range: 0.0–69.7 months), and 33.5 months (range: 2.8–53.2 months) in the training, validation 1, validation 2, FIRE-3 control, and TRIBE control cohorts, respectively.
2.1. Gene Variants and OS
Within the training cohort, three polymorphisms (CD24 rs52812045, IDO1 rs3739319, IDO1 rs9657182) were significantly associated with OS.
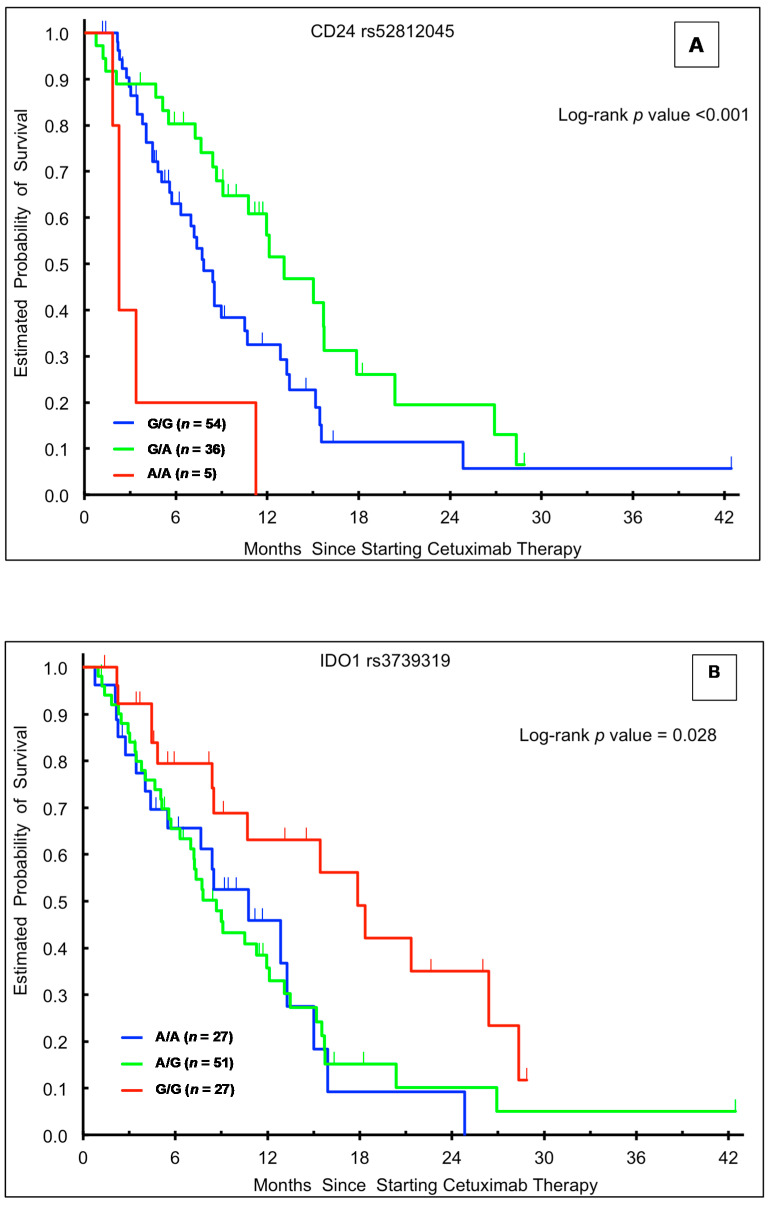
In the univariable analysis, patients in the training cohort with the CD24 rs52812045 G/A genotype had a superior median OS relative to those with either homozygous genotype (G/A 13.1 vs. G/G 7.8 vs. A/A 2.3 months, HR 0.58, p < 0.001; Table 2, Figure 1A). This association held significance in a multivariable model accounting for sex, age, ethnicity, and presence of rash (HR 0.50, 95% CI: 0.28–0.88, p = 0.001; Table 2). There were no significant associations between CD24 rs52812045 and OS in the validation (Table 3 and Table 4) or control (Table 5 and Table 6) cohorts.
Table 2.
Immune regulatory single nucleotide polymorphisms (SNPs) and outcomes in patients with advanced colorectal cancer (CRC) treated with cetuximab-based therapy (training cohort, Italian + USC).
| SNP | Tumor Response | Progression-Free Survival | Overall Survival | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N | PR | SD + PD | Median, ms (95% CI) | Univariable HR (95% CI) † |
Multivariable HR (95% CI) ‡ |
Median, ms (95% CI) |
Univariable HR (95% CI) † |
Multivariable HR (95% CI) ‡ |
|
| IDO1 rs9657182 | |||||||||
| C/C | 25 | 3 (12%) | 22 (88%) | 3.3 (2.2, 4.4) | 1 (reference) | 1 (reference) | 8.5 (5.1, 10.8) | 1 (reference) | 1 (reference) |
| C/T | 55 | 11 (20%) | 43 (80%) | 4.6 (3.4, 5.7) | 0.77 (0.47, 1.26) | 0.71 (0.42, 1.19) | 15.0 (7.8, 15.9) | 0.49 (0.27, 0.89) | 0.39 (0.20, 0.74) |
| T/T | 24 | 7 (30%) | 16 (70%) | 4.1 (2.4, 7.4) | 0.56 (0.30, 1.04) | 0.47 (0.25, 0.89) | 12.0 (5.7, 20.4) | 0.54 (0.27, 1.07) | 0.38 (0.18, 0.79) |
| p-value * | 0.33 | 0.13 | 0.068 | 0.021 | 0.008 | ||||
| IDO1 rs3739319 | |||||||||
| G/G | 27 | 8 (31%) | 18 (69%) | 5.2 (4.0, 8.0) | 1 (reference) | 1 (reference) | 17.9 (8.5, 26.4) | 1 (reference) | 1 (reference) |
| A/G | 51 | 9 (18%) | 41 (82%) | 3.6 (2.5, 5.2) | 1.35 (0.82, 2.23) | 1.55 (0.92, 2.60) | 8.7 (6.3, 12.0) | 2.12 (1.15, 3.93) | 2.32 (1.20, 4.50) |
| A/A | 27 | 4 (15%) | 23 (85%) | 2.8 (2.4, 4.7) | 1.72 (0.97, 3.08) | 1.55 (0.83, 2.88) | 10.8 (4.4, 15.0) | 2.13 (1.04, 4.36) | 2.06 (0.95, 4.47) |
| p-value * | 0.32 | 0.15 | 0.22 | 0.028 | 0.043 | ||||
| CD24 rs52812045 | |||||||||
| G/G | 54 | 8 (15%) | 45 (85%) | 3.6 (2.5, 4.6) | 1 (reference) | 1 (reference) | 7.8 (5.7, 10.5) | 1 (reference) | 1 (reference) |
| G/A | 36 | 10 (29%) | 25 (71%) | 5.0 (3.3, 6.6) | 0.74 (0.47, 1.16) | 0.62 (0.39, 1.00) | 13.1 (8.7, 17.9) | 0.58 (0.34, 0.99) | 0.50 (0.28, 0.88) |
| A/A | 5 | 0 (%) | 5 (100%) | 1.3 (1.0, 3.3) | 3.62 (1.35, 9.70) | 3.18 (1.10, 9.20) | 2.3 (1.8, 11.3) | 3.27 (1.23, 8.68) | 4.93 (1.65, 14.77) |
| p-value * | 0.20 | 0.001 | 0.009 | <0.001 | 0.001 | ||||
* p-value was based on Fisher’s exact test for response, log-rank test for PFS and OS in the univariate analysis (†), and Wald test for PFS and OS in the multivariable Cox regression model (‡) that was adjusted for sex, age, rash, and racial background.
Figure 1.
Kaplan-Meier plots according to genotype and outcome in the training cohort. (A) Overall survival (OS) stratified by CD24 rs52812045 genotype; (B) OS stratified by IDO1 rs3739319 genotype; (C) OS stratified by IDO1 rs9657182 genotype; (D) progression-free survival stratified by CD24 rs52812045 genotype.
Table 3.
Immune regulatory SNPs and outcomes in patients receiving first-line FOLFIRI and cetuximab in FIRE-3 (validation cohort 1).
| SNP | Tumor Response | Progression-Free Survival | Overall Survival | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N | PR | PD + SD | Median, ms (95% CI) |
Univariable HR (95% CI) |
Multivariable HR (95% CI) |
Median, ms (95% CI) |
Univariable HR (95% CI) |
Multivariable HR (95% CI) |
|
| IDO1 rs9657182 | |||||||||
| C/C | 63 | 39 (70%) | 17 (30%) | 9.6 (8.2, 13.0) | 1 (reference) | 1 (reference) | 28.7 (16.8, 38.3) | 1 (reference) | 1 (reference) |
| C/T | 86 | 48 (70%) | 21 (30%) | 9.7 (6.9, 11.3) | 1.18 (0.83, 1.69) | 1.09 (0.74, 1.61) | 25.2 (22.6, 33.6) | 1.15 (0.75, 1.76) | 0.90 (0.56, 1.43) |
| T/T | 72 | 49 (73%) | 18 (27%) | 10.0 (7.9, 11.5) | 1.06 (0.74, 1.54) | 1.13 (0.76, 1.68) | 36.6 (21.3, 52.0) | 0.84 (0.53, 1.32) | 0.73 (0.45, 1.17) |
| p-value * | 0.88 | 0.64 | 0.83 | 0.34 | 0.41 | ||||
| C/T or T/T | 158 | 97 (71%) | 39 (29%) | 10.0 (8.2, 10.9) | 1.12 (0.81, 1.55) | 1.11 (0.78, 1.57) | 28.0 (23.7, 37.5) | 0.99 (0.67, 1.45) | 0.81 (0.54, 1.22) |
| p-value * | 0.86 | 0.48 | 0.56 | 0.96 | 0.32 | ||||
| IDO1 rs3739319 | |||||||||
| G/G | 94 | 54 (69%) | 24 (31%) | 9.2 (7.9, 10.6) | 1 (reference) | 1 (reference) | 23.9 (20.6, 36.6) | 1 (reference) | 1 (reference) |
| G/A | 78 | 50 (69%) | 22 (31%) | 10.3 (7.8, 12.6) | 0.81 (0.59, 1.12) | 0.68 (0.48, 0.97) | 30.6 (22.6, 49.8) | 0.83 (0.56, 1.23) | 0.75 (0.49, 1.16) |
| A/A | 46 | 29 (74%) | 10 (26%) | 12.1 (8.7, 14.1) | 0.68 (0.46, 1.00) | 0.67 (0.45, 0.99) | 38.7 (27.9, 68.7) | 0.57 (0.35, 0.94) | 0.61 (0.36, 1.04) |
| p-value * | 0.88 | 0.12 | 0.040 | 0.079 | 0.14 | ||||
| G/A or A/A | 124 | 79 (71%) | 32 (29%) | 10.5 (8.8, 12.6) | 0.76 (0.57, 1.02) | 0.67 (0.50, 0.91) | 33.1 (27.6, 41.2) | 0.72 (0.50, 1.03) | 0.70 (0.48, 1.02) |
| p-value * | 0.87 | 0.059 | 0.011 | 0.068 | 0.063 | ||||
| CD24 rs52812045 | |||||||||
| G/G | 137 | 84 (72%) | 33 (28%) | 9.3 (7.9, 10.6) | 1 (reference) | 1 (reference) | 25.2 (20.6, 33.4) | 1 (reference) | 1 (reference) |
| G/A | 66 | 43 (68%) | 20 (32%) | 10.4 (8.0, 12.3) | 0.92 (0.67, 1.27) | 1.01 (0.72, 1.44) | 33.8 (24.5, 40.0) | 0.79 (0.53, 1.18) | 0.61 (0.38, 0.97) |
| A/A | 11 | 4 (57%) | 3 (43%) | 14.1 (5.7, 19.9) | 0.61 (0.32, 1.17) | 0.54 (0.26, 1.11) | 41.2 (19.9, 56.2) | 0.72 (0.33, 1.56) | 1.15 (0.50, 2.65) |
| p-value * | 0.56 | 0.31 | 0.24 | 0.40 | 0.093 | ||||
| G/A or A/A | 77 | 47 (67%) | 23 (33%) | 11.3 (9.0, 12.8) | 0.86 (0.64, 1.16) | 0.90 (0.65, 1.25) | 37.5 (26.5, 41.2) | 0.77 (0.53, 1.13) | 0.68 (0.44, 1.04) |
| p-value * | 0.51 | 0.32 | 0.55 | 0.18 | 0.074 | ||||
* p-value was based on Fisher’s exact test for tumor response, log-rank test for PFS and OS in the univariable analysis, and Wald test in the multivariable Cox proportional hazards regression model adjusting for sex, primary tumor site, liver metastases, number of metastatic sites, LDH, Kohne score, RAS and BRAF mutation status.
Table 4.
Immune regulatory SNPs and outcomes in Japanese patients receiving first-line cetuximab plus oxaliplatin-based therapy (validation cohort 2).
| SNP | Tumor Response | Progression-Free Survival | Overall Survival | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N | PR | SD + PD | Median, ms (95% CI) | Univariable HR (95% CI) † |
Multivariable HR (95% CI) ‡ |
Median, ms (95% CI) |
Univariable HR (95% CI) † |
Multivariable HR (95% CI) ‡ |
|
| IDO1 rs9657182 | |||||||||
| C/C | 26 | 19 (79%) | 5 (21%) | 11.6 (9.2, 15.2) | 1 (reference) | 1 (reference) | 41.1+ (17.9, 41.1+) | 1 (reference) | 1 (reference) |
| C/T | 30 | 19 (68%) | 9 (32%) | 9.5 (5.6, 13.8) | 1.14 (0.63, 2.06) | 1.11 (0.61, 2.04) | 26.5 (18.4, 36.2) | 2.18 (0.98, 4.85) | 1.98 (0.86, 4.56) |
| T/T | 16 | 13 (81%) | 3 (19%) | 9.1 (5.1, 16.7) | 1.35 (0.66, 2.76) | 1.21 (0.57, 2.55) | 42.9+ (27.1, 42.9+) | 0.92 (0.33, 2.58) | 0.87 (0.31, 2.47) |
| p-value * | 0.63 | 0.68 | 0.88 | 0.049 | 0.12 | ||||
| C/T or T/T | 46 | 32 (73%) | 12 (27%) | 9.5 (8.0, 13.6) | 1.21 (0.70, 2.07) | 1.14 (0.65, 2.00) | 27.5 (23.2, 42.8) | 1.64 (0.76, 3.52) | 1.47 (0.66, 3.25) |
| p-value * | 0.77 | 0.48 | 0.65 | 0.19 | 0.34 | ||||
| IDO1 rs3739319 | |||||||||
| G/G | 22 | 17 (81%) | 4 (19%) | 10.0 (8.9, 15.2) | 1 (reference) | 1 (reference) | 27.1 (16.0, 42.8) | 1 (reference) | 1 (reference) |
| G/A | 34 | 21 (66%) | 11 (34%) | 9.1 (6.1, 11.8) | 0.96 (0.52, 1.75) | 0.98 (0.53, 1.81) | 26.7 (18.4, 42.9) | 0.98 (0.47, 2.05) | 0.94 (0.45, 1.98) |
| A/A | 16 | 13 (87%) | 2 (13%) | 13.6 (5.6, 15.2) | 0.79 (0.37, 1.69) | 0.75 (0.35, 1.64) | 41.4+ (23.5, 41.4+) | 0.53 (0.20, 1.41) | 0.51 (0.19, 1.39) |
| p-value * | 0.27 | 0.80 | 0.73 | 0.35 | 0.38 | ||||
| G/A or A/A | 50 | 34 (72%) | 13 (28%) | 9.7 (8.0, 13.8) | 0.90 (0.51, 1.59) | 0.91 (0.51, 1.62) | 36.2 (21.5, 42.9) | 0.80 (0.40, 1.61) | 0.78 (0.39, 1.58) |
| p-value * | 0.55 | 0.71 | 0.74 | 0.52 | 0.49 | ||||
| CD24 rs52812045 | |||||||||
| G/G | 34 | 23 (74%) | 8 (26%) | 11.8 (9.4, 18.9) | 1 (reference) | 1 (reference) | 33.9 (19.0, 39.6) | 1 (reference) | 1 (reference) |
| G/A a | 28 | 21 (78%) | 6 (22%) | 9.2 (6.7, 13.8) | 1.90 (1.05, 3.42) | 2.12 (1.14, 3.95) | 36.2 (23.5, 42.9) | 0.88 (0.44, 1.76) | 0.85 (0.41, 1.76) |
| A/A a | 6 | 5 (83%) | 1 (17%) | ||||||
| p-value * | 1.00 | 0.011 | 0.018 | 0.71 | 0.66 | ||||
* p-value was based on Fisher’s exact test for response, log-rank test for PFS and OS in the univariable analysis (†), and Wald test for PFS and OS in the multivariable Cox regression model (‡) adjusting for ECOG performance status (0 vs. 1), and chemo backbone (FOLFOX vs. SOX). a Grouped together to estimate hazard ratios.
Table 5.
Immune regulatory SNPs and outcomes in patients treated with first-line FOLFIRI and bevacizumab in FIRE-3 (control cohort 1).
| SNP | Tumor Response | Progression-Free Survival | Overall Survival | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N | PR | SD + PD | Median, ms (95% CI) |
Univariable HR (95% CI) † |
Multivariable HR (95% CI) ‡ |
Median, ms (95% CI) |
Univariable HR (95% CI) † |
Multivariable HR (95% CI) ‡ |
|
| IDO1 rs9657182 | |||||||||
| C/C | 76 | 47 (66%) | 24 (34%) | 13.2 (10.4, 13.9) | 1 (reference) | 1 (reference) | 26.4 (21.3, 32.8) | 1 (reference) | 1 (reference) |
| C/T | 112 | 65 (62%) | 40 (38%) | 9.2 (8.6, 10.5) | 1.61 (1.17, 2.22) | 1.58 (1.12, 2.21) | 22.1 (18.4, 26.3) | 1.28 (0.89, 1.83) | 1.28 (0.88, 1.87) |
| T/T | 74 | 39 (57%) | 29 (43%) | 10.1 (8.8, 11.8) | 1.26 (0.89, 1.79) | 1.29 (0.90, 1.85) | 24.8 (20.6, 29.1) | 0.98 (0.66, 1.46) | 1.21 (0.81, 1.82) |
| p-value * | 0.58 | 0.010 | 0.031 | 0.22 | 0.42 | ||||
| C/T or T/T | 186 | 104 (60%) | 69 (40%) | 9.8 (8.9, 10.5) | 1.45 (1.08, 1.95) | 1.44 (1.06, 1.96) | 23.1 (20.1, 26.7) | 1.14 (0.82, 1.59) | 1.25 (0.89, 1.76) |
| p-value * | 0.39 | 0.010 | 0.019 | 0.43 | 0.20 | ||||
| IDO1 rs3739319 | |||||||||
| G/G | 94 | 51 (61%) | 33 (39%) | 9.9 (8.6, 11.5) | 1 (reference) | 1 (reference) | 21.9 (17.5, 25.6) | 1 (reference) | 1 (reference) |
| G/A | 129 | 75 (62%) | 46 (38%) | 9.7 (8.8, 11.3) | 0.87 (0.65, 1.16) | 0.85 (0.63, 1.15) | 25.0 (21.5, 28.4) | 0.89 (0.64, 1.22) | 0.87 (0.62, 1.22) |
| A/A | 47 | 30 (68%) | 14 (32%) | 11.9 (9.8, 14.1) | 0.75 (0.52, 1.10) | 0.80 (0.54, 1.18) | 27.6 (18.5, 35.0) | 0.82 (0.53, 1.25) | 0.89 (0.57, 1.38) |
| p-value * | 0.72 | 0.30 | 0.43 | 0.59 | 0.71 | ||||
| G/A or A/A | 176 | 105 (64%) | 60 (36%) | 10.3 (9.2, 11.8) | 0.83 (0.63, 1.09) | 0.83 (0.63, 1.10) | 25.1 (22.3, 28.6) | 0.87 (0.64, 1.17) | 0.88 (0.64, 1.20) |
| p-value * | 0.68 | 0.18 | 0.21 | 0.34 | 0.41 | ||||
| CD24 rs52812045 | |||||||||
| G/G | 130 | 68 (56%) | 53 (44%) | 9.6 (8.8, 11.7) | 1 (reference) | 1 (reference) | 23.7 (19.4, 27.4) | 1 (reference) | 1 (reference) |
| G/A a | 129 | 79 (66%) | 40 (34%) | 10.5 (9.8, 12.4) | 0.86 (0.66, 1.12) | 0.77 (0.59, 1.02) | 26.1 (21.5, 28.8) | 1.01 (0.76, 1.35) | 1.00 (0.74, 1.36) |
| A/A a | 7 | 4 (57%) | 3 (43%) | ||||||
| p-value * | 0.25 | 0.25 | 0.068 | 0.93 | 0.98 | ||||
* p-value was based on Fisher’s exact test for tumor response, log-rank test for PFS and OS in the univariable analysis (†), and Wald test in the multivariable Cox regression model (‡) adjusting for sex, ECOG performance status, primary tumor resection, number of metastatic sites, adjuvant chemotherapy, LDH, RAS and BRAF mutation status. a Grouped together to estimate hazard ratios.
Table 6.
Immune regulatory SNPs and outcomes in patients receiving first-line FOLFIRI and bevacizumab in TRIBE (control cohort 2).
| SNP | Tumor Response | Progression-Free Survival | Overall Survival | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N | PR | SD + PD | Median, ms (95% CI) |
Univariable HR (95% CI) † | Multivariable HR (95% CI) ‡ | Median, ms (95% CI) |
Univariable HR (95% CI) † | Multivariable HR (95% CI) ‡ | |
| IDO1 rs9657182 | |||||||||
| C/C | 44 | 20 (49%) | 21 (51%) | 8.8 (7.8, 10.5) | 1 (reference) | 1 (reference) | 23.6 (14.7, 33.9) | 1 (reference) | 1 (reference) |
| C/T | 112 | 66 (61%) | 42 (39%) | 9.4 (8.6, 10.5) | 0.82 (0.55, 1.22) | 0.87 (0.57, 1.32) | 25.6 (21.1, 28.6) | 0.94 (0.64, 1.40) | 0.99 (0.65, 1.49) |
| T/T | 65 | 38(58%) | 27(42%) | 11.3 (9.6, 12.7) | 0.67 (0.44, 1.04) | 0.70 (0.45, 1.11) | 30.9 (22.7, 39.7) | 0.74 (0.48, 1.14) | 0.74 (0.47, 1.16) |
| p-value * | 0.40 | 0.18 | 0.29 | 0.28 | 0.24 | ||||
| A/G or G/G | 177 | 104 (60%) | 69 (40%) | 10.3 (9.3, 11.1) | 0.76 (0.52, 1.10) | 0.80 (0.54, 1.19) | 26.2 (22.7, 30.8) | 0.86 (0.59, 1.25) | 0.88 (0.60, 1.30) |
| p-value * | 0.22 | 0.14 | 0.27 | 0.42 | 0.52 | ||||
| IDO1 rs3739319 | |||||||||
| G/G | 78 | 45 (60%) | 30 (40%) | 11.1 (8.3, 12.3) | 1 (reference) | 1 (reference) | 23.9 (19.4, 28.7) | 1 (reference) | 1 (reference) |
| G/A | 90 | 44 (50%) | 44 (50%) | 9.4 (8.7, 10.4) | 1.19 (0.84, 1.69) | 1.25 (0.87, 1.80) | 26.5 (22.0, 33.0) | 0.82 (0.59, 1.16) | 0.84 (0.59, 1.20) |
| A/A | 48 | 33 (70%) | 14 (30%) | 9.7 (8.6, 10.8) | 1.01 (0.67, 1.55) | 1.12 (0.71, 1.76) | 29.2 (19.6, 39.2) | 0.71 (0.47, 1.08) | 0.80 (0.52, 1.23) |
| p-value * | 0.070 | 0.55 | 0.48 | 0.25 | 0.50 | ||||
| G/A or A/A | 138 | 77 (57%) | 58 (43%) | 9.5 (9.0, 10.4) | 1.13 (0.82, 1.55) | 1.20 (0.87, 1.67) | 27.4 (23.9, 33.6) | 0.78 (0.57, 1.07) | 0.83 (0.60, 1.14) |
| p-value * | 0.77 | 0.45 | 0.27 | 0.12 | 0.24 | ||||
| CD24 rs52812045 | |||||||||
| G/G | 123 | 71 (59%) | 49 (41%) | 9.5 (8.8, 10.8) | 1 (reference) | 1 (reference) | 28.7 (23.9, 35.1) | 1 (reference) | 1 (reference) |
| G/A | 79 | 42 (55%) | 34 (45%) | 9.9 (9.3, 12.6) | 0.98 (0.71, 1.35) | 0.78 (0.55, 1.10) | 24.0 (20.3, 27.3) | 1.30 (0.94, 1.79) | 1.38 (0.99, 1.94) |
| A/A | 21 | 13 (62%) | 8 (38%) | 9.2 (6.7, 11.3) | 1.22 (0.71, 2.11) | 1.00 (0.56, 1.79) | 22.3 (15.6, 48.7) | 1.17 (0.70, 1.97) | 1.34 (0.78, 2.30) |
| p-value * | 0.82 | 0.73 | 0.35 | 0.27 | 0.14 | ||||
| G/A or A/A | 100 | 55 (57%) | 42 (43%) | 9.7 (9.2, 11.6) | 1.02 (0.75, 1.38) | 0.82 (0.59, 1.13) | 23.5 (20.3, 27.3) | 1.27 (0.94, 1.71) | 1.38 (1.00, 1.89) |
| p-value * | 0.78 | 0.91 | 0.22 | 0.11 | 0.049 | ||||
* p-value was based on Fisher’s exact test for tumor response, log-rank test for PFS and OS in the univariable analysis (†), and Wald test in the multivariable Cox proportional hazards regression model (‡) adjusting for sex, age, ECOG performance status, primary tumor site, number of metastatic sites, BRAF mutation status, resection of the primary tumors, and adjuvant therapy.
Patients in the training cohort carrying the IDO1 rs3739319 G/G variant had a significantly longer median OS (17.9 months) compared to those with the A/A (10.8 months, HR 2.13, 95% CI: 1.04–4.36) or A/G (8.7 months, HR 2.12, 95% CI: 1.15–3.93) genotypes (p = 0.028; Table 2, Figure 1B). This association held significance in multivariable analysis (p = 0.043). There were no significant associations between IDO1 rs3739319 genotype and OS in the other cohorts. In the FIRE-3 validation cohort, patients with an IDO1 rs3739319 A allele had a numerically longer OS (33.1 months) in univariate (HR 0.72, 95% CI: 0.50–1.03, p = 0.068) and multivariate (HR 0.70, 95% CI: 0.48–1.02, p = 0.063) analyses, though this association did not reach statistical significance (Table 3).
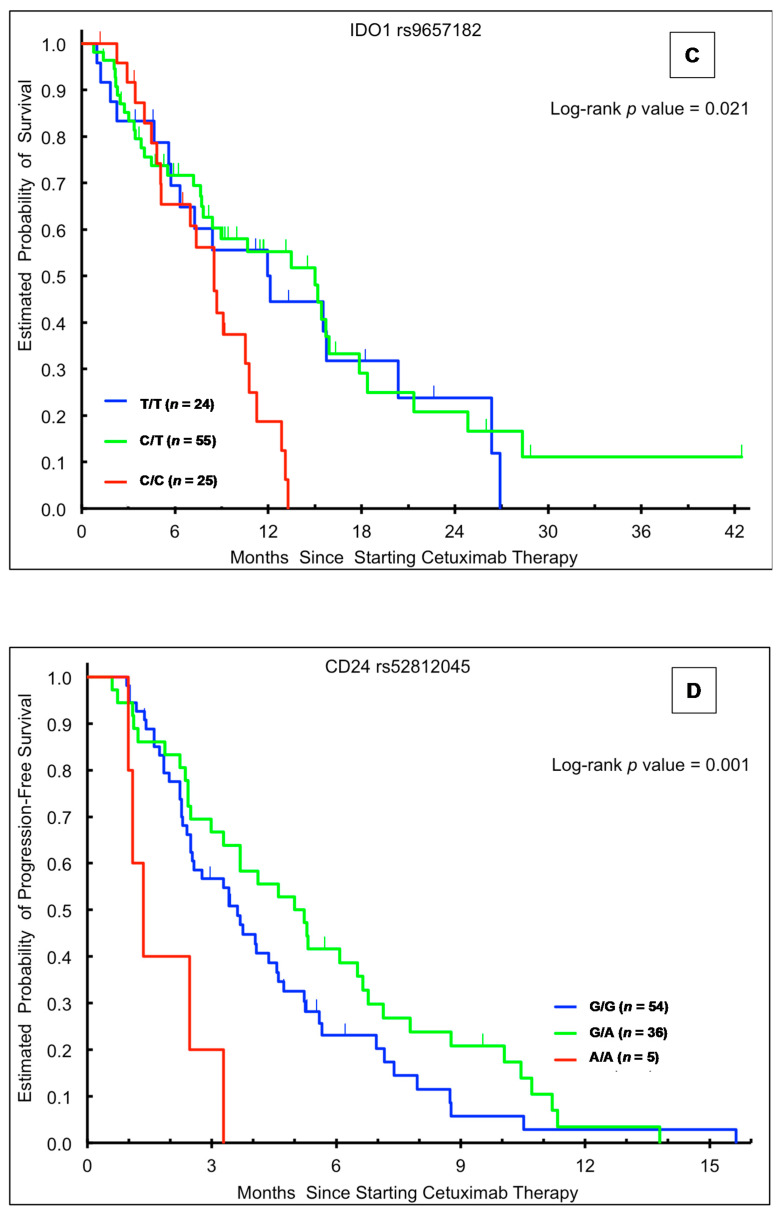
Patients in the training cohort with the IDO1 rs9657182 C/T (15.0 months, HR 0.49, 95% CI: 0.27–0.89) genotype had a significantly longer OS than those harboring the C/C genotype (8.5 months) (p = 0.021; Table 2, Figure 1C). This relationship remained significant in multivariable analysis (p = 0.008; Table 2). Conversely, in the Japanese validation cohort, patients with the heterozygote C/T genotype had the shortest median OS (26.5 vs. 41.1+ months for C/C genotype; univariate HR 2.18, 95% CI 0.98–4.85), though with borderline significance (p = 0.049). There was a similar but non-significant trend in multivariate analysis (p = 0.12) (Table 4). IDO1 rs9657182 genotype was not significantly associated with OS in the other cohorts (Table 3,Table 5 and Table 6).
Patients in the TRIBE control cohort with any PD-L1 rs2297137 A allele had a significantly prolonged OS (33.9 months) compared to those with the G/G genotype (23.4 months) in univariable (HR 0.63, 95% CI 0.45-0.89, p = 0.007) and multivariable analyses (HR 0.65, 95% CI 0.45–0.94, p = 0.021) (Table S6). This association was not seen in the other cohorts.
2.2. Gene Variants and PFS
One polymorphism (CD24 rs52812045) was significantly associated with PFS in the training cohort.
In the training cohort, the CD24 rs52812045 A/A genotype was associated with shorter median PFS (1.3 months) in both univariable (HR 3.62, 95% CI: 1.35–9.70, p = 0.001) and multivariable analyses (HR 3.18, 95% CI: 1.10–9.20, p = 0.009) (Table 2, Figure 1D). Similarly, in the Japanese validation cohort, the A allele was associated with significantly shorter PFS in univariable (HR 1.90, 95% CI: 1.05–3.42, p = 0.011) and multivariable (HR 2.12, 95% CI: 1.14–3.95, p = 0.018) analyses (Table 4). There were no significant associations between CD24 rs52812045 and PFS in the other cohorts (Table 3,Table 5 and Table 6).
In the FIRE-3 control cohort, patients harboring a T allele of IDO1 rs9657182 had a significantly shorter PFS (9.8 months) compared to those with the C/C genotype (13.2 months) in univariable (HR 1.45, 95% CI: 1.08–1.95, p = 0.010) and multivariable analyses (HR 1.44, 95% CI: 1.06–1.96, p = 0.019) (Table 5). This association was not demonstrated in the other cohorts.
Patients in the FIRE-3 validation cohort with any A allele of IDO1 rs3739319 achieved a prolonged PFS compared to those with the G/G genotype, but only in multivariable analysis (10.5 vs. 9.2 months; univariable HR 0.76, 95% CI: 0.57–1.02, p = 0.059; multivariable HR 0.67, 95% CI: 0.50–0.91, p = 0.011) (Table 3). This trend was not evident in the other cohorts.
Patients in the training cohort with the CTLA4 rs231777 C/T genotype had a significantly shorter PFS (2.6 months) compared to those with the C/C variant (4.1 months), but only in multivariable analysis (HR 1.76, 95% CI: 1.10–2.81, p = 0.019) (Table S2). This association was not seen in the other cohorts.
2.3. Gene Variants and Tumor Response
In the training cohort, patients with the PD-L1 rs2297137 A/A genotype demonstrated a greater RR (56%) than patients with G/A (19%) or G/G (16%) genotypes (two-sided Fisher’s exact test p = 0.029) (Table S2). There was no association between PD-L1 genotype and RR in the validation or control cohorts.
3. Discussion
Cetuximab affects EGFR inhibition as well as innate and adaptive immunity [10,11,12,13]. While RAS mutation status is a validated predictive marker for cetuximab in mCRC patients, it may not completely account for its immunomodulatory effects.
The wide-ranging immunogenic effects of cetuximab have been demonstrated in multiple studies [6,10]. Cetuximab binds the Fc portion of the activating receptor on NK cells, which may lead to direct cancer cell lysis and the release of tumor antigens which can be presented by dendritic cells to cytotoxic T-cells [6]. Moreover, activated NK cells may use cytokines and interferon to activate tumor suppressing macrophages [6]. In CRC cell lines and murine xenograft models, cetuximab has been shown to induce immunogenic cell death, marked by increased phagocytosis by dendritic cells [10], as well as stimulating and enhancing NK cell activity [14]. The cytotoxic agents, 5-FU and irinotecan, have also been shown to improve the ADCC activity of cetuximab across different CRC cell lines, in part by increasing cell surface EGFR expression [15]. The interaction between KRAS mutations and the ability of cetuximab to mediate ADCC is not completely understood. Some studies have shown ADCC to be dependent on KRAS-wildtype status [16], while others show the effects to be independent of KRAS status [6]. In mCRC patients with liver-limited metastases undergoing pre-operative chemotherapy with or without cetuximab, cetuximab was shown to increase the degree of tumor infiltration with CD3+ and CD8+ T-cells in addition to CD56+ NK cells [11]. Clinically, the degree of ADCC activity (as measured in peripheral blood mononuclear cells and by NK-dependent LDH release) has been positively associated with improved survival [17], as well as degree of tumor shrinkage [18] in mCRC patients receiving cetuximab.
Much of the literature regarding the predictive and prognostic impact of immune-related germline polymorphisms on cetuximab efficacy has focused on the FCγ-receptor family [7,19,20,21]. As immunotherapeutics become integrated into the management of advanced CRC, discovering markers of efficacy within immune mediated pathways will become increasingly important. To our knowledge, this is the first study to evaluate the predictive and prognostic utility of common genetic variants within checkpoint and other immune regulatory pathways in mCRC patients receiving cetuximab-based therapy.
CD24, a membrane surface glycosyl-phosphatidyl-inositol-(GPI)-anchored protein, is overexpressed in 90% of colorectal tumors [22] and has been implicated in inflammatory bowel disease (IBD) [23,24]. As a putative marker of EGF and Wnt-expressing metaplastic Paneth cells [25,26], CD24 provides a crucial link between host mucosal immunity, colonic crypt homeostasis [27], and the intestinal stem cell niche [28,29]. Indeed, CD24 knockout mice models exhibit reduced T-cell proliferation [30], blunted polyp formation, and suppressed colorectal tumorigenesis [22,31]. Furthermore, the effects of CD24 on ADCC and chronic inflammation are thought to be partly mediated by autophagy [27,32], another purported mechanism of cetuximab efficacy [33].
CD24 maps to chromosome 6q21 and rs52812045 is located in the GPI-anchor cleavage site [34]. Clinically, this polymorphism has been associated with pathologic complete response and concomitant intra-tumoral lymphocyte infiltration in breast cancer patients [35,36], and poor prognosis in esophageal cancer patients [37]. However, a recent meta-analysis of CD24 SNPs and cancer risk was non-confirmatory [38]. In our study, carriers of the CD24 rs52812045 A/A variant in the training cohort had the shortest PFS and OS compared to those with the G/G and G/A genotypes, with heterozygotes appearing to have an OS advantage (albeit with the 95% CI boundary near 1 in univariable analysis). Similarly, in the Japanese validation cohort, the A allele was associated with shorter PFS. It is important to note that very few patients carried the A/A genotype which limits the power of the analysis. In addition, patients in the training cohort received cetuximab monotherapy or cetuximab combined with irinotecan, while patients in the Japanese cohort received cetuximab combined with a fluoropyrimidine (5-FU or S-1) and oxaliplatin. Each cytotoxic agent has its own immunomodulatory effects or may impact those seen with cetuximab, and this may have influenced our results. Nonetheless, our findings warrant further evaluation. If confirmed in larger, prospective studies, CD24 rs52812045 genotype may be used to select mCRC patients most likely to benefit from cetuximab-based regimens in clinical practice.
Another critical modulator of intestinal immunity and stem cell signaling is IDO1, an enzyme which catalyzes the rate-limiting step of tryptophan catabolism within the kynurenine pathway [39]. Released by tumor and myeloid derived suppressor cells, IDO1 depletes T-cells of tryptophan, thereby attenuating their activity [40]. Plasmacytoid dendritic cells express IDO1 to convert naïve T-cells into Treg-cells and increase PD-L1/PD-L2 expression, leading to T-cell suppression [41,42]. Independent of its immunoregulatory effects, IDO1 has been shown to activate canonical Wnt-β-catenin signaling and promote tumor progression by decreasing the tryptophan/kynurenine ratio [43].
Clinically, IDO1 mRNA expression has been correlated with colitis severity [44], peri-tumoral immune tolerance, and worse survival in CRC patients [45,46]. IDO1 genetic variants have been associated with IBD severity [47], and we previously demonstrated the prognostic value of IDO1 rs3739319 in patients with liver-limited mCRC [48]. Consistent with prior findings, the IDO1 rs3739319 A allele was a negative prognosticator in our training cohort, though an opposite relationship was observed in the FIRE-3 validation cohort. Notably, the IDO1 rs9657182 T allele conferred a positive prognostic effect in patients receiving irinotecan-cetuximab in the training cohort, but a negative effect in patients receiving oxaliplatin-cetuximab regimens in the Japanese validation cohort, and bevacizumab-treated patients of the FIRE-3 control cohort. Similarly, PD-L1 rs2297137 predicted outcomes in both cetuximab and bevacizumab-treated patients. This may reflect the immunomodulatory activity of bevacizumab [49] and suggests distinct roles for IDO1 and PD-L1 in mediating these effects, which may vary by chemotherapy regimen.
Our study has its limitations, the first being its retrospective nature. Prospective studies are needed to further validate our findings in patients with mCRC receiving cetuximab-based therapy. Clinical trials combining cetuximab with immune therapy and chemotherapy in patients with mCRC are ongoing and incorporating the CD24 or IDO1 genotype as a stratification factor may guide the use of these SNPs as predictive markers. Notably, there were certain genotypes that were carried by a small number of patients, and this limited our ability to test combinations of alleles and limits the strength of our conclusions. The heterogeneity of concomitant chemotherapy received between the cohorts may have also impacted our results, and future studies controlling for this variable will be important to translate the utility of these SNPs in the clinic. In addition, the RAS mutation status was unknown for a proportion of patients. As the role of RAS signaling on cetuximab-mediated ADCC is unclear [16,50,51,52], deciphering differential effects of these SNPs based on the RAS status could guide future study design.
Studies examining the effect of these SNPs on gene function as well as associations between genotype/haplotype and immune cell subsets, serum cytokine levels, PDL-1 status, tumor mutational burden, and pathologic features (e.g., poor differentiation, tumor infiltrating lymphocytes, mucinous histology) may provide mechanistic explanations for the observed findings and better characterize the link between genotype and clinical outcomes. The presence of microsatellite instability (MSI) and BRAF V600E mutations also affect the tumor immune milieu and may have influenced the relationship between germline variants and outcomes; this information was not available in the present study. Therefore, further investigations should evaluate the effect of MSI and BRAF mutation status (V600E and perhaps non-V600E) on the predictive and prognostic utility of CD24 or IDO1 genotype, especially as it pertains to the efficacy of immune therapy combinations [53]. For instance, avelumab is currently being tested in combination with FOLFOXIRI plus cetuximab in the mCRC frontline setting in the phase 2 AVETRIC trial (NCT04513951). Others are examining the efficacy of first-line FOLFIRI plus cetuximab in mCRC patients based on FCγ-receptor genotype in the phase 2 CIFRA trial (NCT03874026). Specific germline polymorphisms or composite haplotypes, in conjunction with tumor features (e.g., sidedness) and molecular profile (e.g., RAS, BRAF, HER2, MSI status, CMS subtype) may serve to inform the selection of mCRC patients to treat with cetuximab and other targeted or immune therapies.
4. Materials and Methods
4.1. Study Design and Patient Population
A total of 924 mCRC patients with sufficient tissue for analysis were included in this study. The training cohort consisted of 105 patients enrolled in two phase II clinical trials. Thirty-two patients with EGFR-expressing mCRC refractory to irinotecan, oxaliplatin, and fluoropyrimidines were recruited October 2002-March 2003, at the University of Southern California/Norris Comprehensive Cancer Center (USC/NCCC) to the phase 2, open-label, multicenter ImClone-0144 (IMCL-0144) study [8], and treated with single-agent cetuximab. The remaining 73 patients were recruited to a phase 2 trial (protocol ASL608LIOM01) conducted by the Gruppo Oncologico Nord Ovest (EudraCT 2008-003160-19) and received irinotecan-cetuximab.
Among the twelve SNPs tested in the training cohort, five (CD24 rs52812045, IDO1 rs9657182 and rs3739319, PDL1 rs2297137, CTLA4 rs231777) showed significant associations with response rate (RR), progression-free survival (PFS), and/or overall survival (OS). These five SNPs were tested in two independent validation and two independent control cohorts. The first validation cohort consisted of 225 patients with KRAS exon 2 wildtype mCRC treated with FOLFIRI-cetuximab in FIRE-3 (NCT00433927). FIRE-3 was a phase 3 trial randomizing patients with mCRC to first-line FOLFIRI-cetuximab versus FOLFIRI-bevacizumab. The second validation cohort consisted of 74 patients enrolled in two phase 2 trials of first-line therapy with cetuximab-oxaliplatin-containing chemotherapy, either mFOLFOX6 (JACCRO-CC-05; n = 57, UMIN000004197) or SOX (JACCRO-CC-06; n = 67, UMIN000007022). Two control cohorts consisted of patients treated with FOLFIRI-bevacizumab in FIRE-3 (n = 292) and TRIBE (n = 228) (NCT00719797) [2,9]. TRIBE was a phase 3 trial randomizing patients with mCRC to first-line FOLFOXIRI-bevacizumab versus FOLFIRI-bevacizumab. The KRAS status was analyzed in 407 (80.1%) patients from TRIBE, of which 38.7% were wildtype in the FOLFIRI-bevacizumab arm [9].
Eligible patients had stage 4 histologically confirmed colorectal adenocarcinoma, with measurable disease per response evaluation criteria in solid tumors (RECIST) 1.0 criteria, and life expectancy of ≥12 weeks. Standard inclusion and exclusion criteria were applied. Treatment was administered until disease progression, intolerable toxicities, or patient withdrawal. The current study was conducted at the USC/NCCC and approved by the USC Institutional Review Board (HS-997018). All patients signed informed consent for molecular correlates analysis. This study adheres to the reporting recommendations for tumor MARKer prognostic studies (REMARK).
4.2. Candidate Single-Nucleotide Polymorphisms
Polymorphisms were selected based on a minimum allele frequency of 10% in Caucasians, functional or predicted functional relevance of the respective gene or protein, TagSNP, and location. Functional significance was predicted based on information derived from the National Institute of Environmental Health Sciences SNP Function Prediction [54], and the Queen’s University F-SNP [55]. Selected polymorphisms are presented in Table S1.
4.3. Genotyping
Genotyping was performed on DNA extracted from whole blood or tissue samples using the QIAamp Kit (Qiagen, Germantown, MD, USA), followed by PCR-based direct sequencing as previously published [56,57]. All primers (Table S1) were validated on known DNA sequences. Sequencing results were analyzed using the ABI Sequencing Scanner v1.0 (Applied Biosystems, Foster, CA, USA) by one investigator (N.B.V.) blinded to the patients’ identifying data and outcomes.
4.4. Statistical Analyses
The primary endpoint of this retrospective correlative analysis was OS, defined as the period from the randomization start date to death from any cause, or the date of last follow-up if patients were still alive. The secondary endpoints included RR per RECIST 1.0 and PFS. PFS was defined as the period from the randomization start date to first observation of tumor progression or death, whichever came first. PFS was censored at the date of last follow-up if patients remained alive and progression-free.
Patients were compared between cohorts using the Kruskal-Wallis test for continuous variables, chi-square test for categorical variables, and log-rank test for PFS and OS. Associations between SNPs and clinical outcomes were tested in univariable and multivariable analyses. The log-rank test and Kaplan-Meier curves were performed to investigate associations between SNPs and OS or PFS in univariable analysis. Multivariable Cox proportional hazard regression models were used to re-examine associations between SNPs and OS or PFS when adjusting for patient baseline characteristics. Fisher’s exact test was used to test associations between SNPs and RR. SNPs without significant associations with clinical outcomes are outlined in Tables S2–S6.
SAS 9.3 (SAS Institute, Cary, NC, USA) was used to perform all analyses. Case-wise deletion was applied when patients with missing SNPs were excluded in the analyses. All tests were two-sided at a significance level of 0.05.
5. Conclusions
Cetuximab is an established therapy for mCRC, which affects both EGFR signaling and ADCC. As the role of immune checkpoint inhibition in mCRC is being actively defined, identifying biomarkers of the immunomodulatory effects of cetuximab may refine its clinical use and inform the development of chemo-immunotherapy combinations. We examined common genetic variants within immune regulatory pathways and identified CD24 and IDO1 polymorphisms, which independently predicted outcomes. Further studies are needed to evaluate the functional relevance and clinical utility of these genetic variants in patients with mCRC receiving cetuximab-based therapy.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/10/2947/s1, Table S1: Candidate single-nucleotide polymorphisms (SNPs); Table S2: Immune regulatory SNPs and outcomes in patients with advanced CRC treated with cetuximab-based therapy (training cohort, Italian + USC); Table S3: Immune regulatory SNPs and outcomes in patients receiving first-line FOLFIRI and cetuximab in FIRE-3 (validation cohort 1); Table S4: Immune regulatory SNPs and outcomes in Japanese patients receiving first-line cetuximab plus oxaliplatin-based therapy (validation cohort 2); Table S5: Immune regulatory SNPs and outcomes in patients treated with first-line FOLFIRI and bevacizumab in FIRE-3 (control cohort 1); Table S6: Immune regulatory SNPs and outcomes in patients receiving first-line FOLFIRI and bevacizumab in TRIBE (control cohort 2).
Author Contributions
Conceptualization, N.B.V., D.L.H., S.S., F.L., A.F., and H.-J.L.; methodology, N.B.V., W.Z., Y.N., and H.-J.L.; software, D.Y. and S.C.; validation, D.Y. and S.C.; formal analysis, D.Y. and S.C.; investigation, N.B.V., Y.N., S.S., S.M., Y.S., M.D.B., C.C., F.L., A.F., and H.-J.L.; resources, S.S., W.Z., S.M., Y.S., M.D.B., C.C., F.L., A.F., and H.-J.L.; data curation, D.Y. and S.C.; writing—original draft preparation, N.B.V. and D.L.H.; writing—review and editing, N.B.V., D.L.H., and H.-J.L.; visualization, N.B.V., D.L.H., D.Y., and S.C.; supervision, H.-J.L.; project administration, S.S., W.Z., F.L., A.F., and H.-J.L.; funding acquisition, H.-J.L. All authors have read and agreed to the published version of the manuscript.
Funding
This project was supported in part by the award number P30CA014089-27S1 grant from the National Cancer Institute. This project was also supported in part by the Dhont and Wunderglo foundations, and the Daniel Butler Research Fund. Sebastian Stintzing is supported by a postdoctoral fellowship from the German Cancer Aid (Mildred-Scheel Foundation).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Venook A.P., Niedzwiecki D., Lenz H.J., Innocenti F., Mahoney M.R., O’Neil B.H., Goldberg R.M. CALGB/SWOG 80405: Phase III trial of irinotecan/5-FU/leucovorin (FOLFIRI) or oxaliplatin/5-FU/leucovorin (mFOLFOX6) with bevacizumab (BV) or cetuximab (CET) for patients (pts) with KRAS wild-type (wt) untreated metastatic adenocarcinoma of the colon or rectum (MCRC) J. Clin. Oncol. 2014;32:5. [Google Scholar]
- 2.Heinemann V., von Weikersthal L.F., Decker T., Kiani A., Vehling-Kaiser U., Al-Batran S.E., Kullmann F. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): A randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15:1065–1075. doi: 10.1016/S1470-2045(14)70330-4. [DOI] [PubMed] [Google Scholar]
- 3.Van Cutsem E., Lenz H.-J., Köhne C.-H., Heinemann V., Tejpar S., Melezínek I., Beier F., Stroh C., Rougier P., Van Krieken J.H., et al. Fluorouracil, Leucovorin, and Irinotecan Plus Cetuximab Treatment and RAS Mutations in Colorectal Cancer. J. Clin. Oncol. 2015;33:692–700. doi: 10.1200/JCO.2014.59.4812. [DOI] [PubMed] [Google Scholar]
- 4.Kurai J., Chikumi H., Hashimoto K., Yamaguchi K., Yamasaki A., Sako T., Touge H., Makino H., Takata M., Miyata M., et al. Antibody-Dependent Cellular Cytotoxicity Mediated by Cetuximab against Lung Cancer Cell Lines. Clin. Cancer Res. 2007;13:1552–1561. doi: 10.1158/1078-0432.CCR-06-1726. [DOI] [PubMed] [Google Scholar]
- 5.Kimura H., Sakai K., Arao T., Shimoyama T., Tamura T., Nishio K. Antibody-dependent cellular cytotoxicity of cetuximab against tumor cells with wild-type or mutant epidermal growth factor receptor. Cancer Sci. 2007;98:1275–1280. doi: 10.1111/j.1349-7006.2007.00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferris R.L., Lenz H.-J., Trotta A.M., García-Foncillas J., Schulten J., Audhuy F., Merlano M., Milano G. Rationale for combination of therapeutic antibodies targeting tumor cells and immune checkpoint receptors: Harnessing innate and adaptive immunity through IgG1 isotype immune effector stimulation. Cancer Treat. Rev. 2018;63:48–60. doi: 10.1016/j.ctrv.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W., Gordon M., Schultheis A.M., Yang D., Nagashima F., Azuma M., Chang H.-M., Borucka E., Lurje G., Sherrod A.E., et al. FCGR2A and FCGR3A Polymorphisms Associated With Clinical Outcome of Epidermal Growth Factor Receptor–Expressing Metastatic Colorectal Cancer Patients Treated With Single-Agent Cetuximab. J. Clin. Oncol. 2007;25:3712–3718. doi: 10.1200/JCO.2006.08.8021. [DOI] [PubMed] [Google Scholar]
- 8.Lenz H.-J., Van Cutsem E., Khambata-Ford S., Mayer R.J., Gold P., Stella P., Mirtsching B., Cohn A.L., Pippas A.W., Azarnia N., et al. Multicenter Phase II and Translational Study of Cetuximab in Metastatic Colorectal Carcinoma Refractory to Irinotecan, Oxaliplatin, and Fluoropyrimidines. J. Clin. Oncol. 2006;24:4914–4921. doi: 10.1200/JCO.2006.06.7595. [DOI] [PubMed] [Google Scholar]
- 9.Loupakis F., Cremolini C., Masi G., Lonardi S., Zagonel V., Salvatore L., Cortesi E., Tomasello G., Ronzoni M., Spadi R., et al. Initial Therapy with FOLFOXIRI and Bevacizumab for Metastatic Colorectal Cancer. N. Engl. J. Med. 2014;371:1609–1618. doi: 10.1056/NEJMoa1403108. [DOI] [PubMed] [Google Scholar]
- 10.Pozzi C., Cuomo A., Spadoni I., Magni E., Silvola A., Conte A., Sigismund S., Ravenda P.S., Bonaldi T., Zampino M.G., et al. The EGFR-specific antibody cetuximab combined with chemotherapy triggers immunogenic cell death. Nat. Med. 2016;22:624–631. doi: 10.1038/nm.4078. [DOI] [PubMed] [Google Scholar]
- 11.Inoue Y., Hazama S., Suzuki N., Tokumitsu Y., Kanekiyo S., Tomochika S., Tsunedomi R., Tokuhisa Y., Iida M., Sakamoto K., et al. Cetuximab strongly enhances immune cell infiltration into liver metastatic sites in colorectal cancer. Cancer Sci. 2017;108:455–460. doi: 10.1111/cas.13162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang X., Zhang X., Mortenson E.D., Radkevich-Brown O., Wang Y., Fu Y. Cetuximab-mediated Tumor Regression Depends on Innate and Adaptive Immune Responses. Mol. Ther. 2012;21:91–100. doi: 10.1038/mt.2012.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L., Wei Y., Fang W., Lu C., Chen J., Cui G., Diao H. Cetuximab Enhanced the Cytotoxic Activity of Immune Cells during Treatment of Colorectal Cancer. Cell. Physiol. Biochem. 2017;44:1038–1050. doi: 10.1159/000485404. [DOI] [PubMed] [Google Scholar]
- 14.Chen S., Li X., Chen R., Yin M., Zheng Q. Cetuximab intensifies the ADCC activity of adoptive NK cells in a nude mouse colorectal cancer xenograft model. Oncol. Lett. 2016;12:1868–1876. doi: 10.3892/ol.2016.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Correale P., Marra M., Remondo C., Migali C., Misso G., Arcuri F.P., Del Vecchio M.T., Carducci A., LoIacono L., Tassone P., et al. Cytotoxic drugs up-regulate epidermal growth factor receptor (EGFR) expression in colon cancer cells and enhance their susceptibility to EGFR-targeted antibody-dependent cell-mediated-cytotoxicity (ADCC) Eur. J. Cancer. 2010;46:1703–1711. doi: 10.1016/j.ejca.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 16.Nakadate Y., Kodera Y., Kitamura Y., Shirasawa S., Tachibana T., Tamura T., Koizumi F. KRAS mutation confers resistance to antibody-dependent cellular cytotoxicity of cetuximab against human colorectal cancer cells. Int. J. Cancer. 2013;134:2146–2155. doi: 10.1002/ijc.28550. [DOI] [PubMed] [Google Scholar]
- 17.Nigro C.L., Ricci V., Vivenza D., Monteverde M., Strola G., Lucio F., Tonissi F., Miraglio E., Granetto C., Fortunato M., et al. Evaluation of antibody-dependent cell-mediated cytotoxicity activity and cetuximab response inKRASwild-type metastatic colorectal cancer patients. World J. Gastrointest. Oncol. 2016;8:222–230. doi: 10.4251/wjgo.v8.i2.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trotta A.M., Ottaiano A., Romano C., Nasti G., Nappi A., De Divitiis C., Napolitano M., Zanotta S., Casaretti R., D’Alterio C., et al. Prospective Evaluation of Cetuximab-Mediated Antibody-Dependent Cell Cytotoxicity in Metastatic Colorectal Cancer Patients Predicts Treatment Efficacy. Cancer Immunol. Res. 2016;4:366–374. doi: 10.1158/2326-6066.CIR-15-0184. [DOI] [PubMed] [Google Scholar]
- 19.Bibeau F., Lopez-Crapez E., Di Fiore F., Thezenas S., Ychou M., Blanchard F., Sabourin J.-C., Michel P., Lamy A., Frebourg T., et al. Impact of Fc{gamma}RIIa-Fc{gamma}RIIIa polymorphisms and KRAS mutations on the clinical outcome of patients with metastatic colorectal cancer treated with cetuximab plus irinotecan. J. Clin. Oncol. 2009;27:1122–1129. doi: 10.1200/JCO.2008.18.0463. [DOI] [PubMed] [Google Scholar]
- 20.Etienne-Grimaldi M.-C., Bennouna J., Formento J.-L., Douillard J.-Y., Francoual M., Hennebelle I., Chatelut E., Francois E., Faroux R., El Hannani C., et al. Multifactorial pharmacogenetic analysis in colorectal cancer patients receiving 5-fluorouracil-based therapy together with cetuximab-irinotecan. Br. J. Clin. Pharmacol. 2012;73:776–785. doi: 10.1111/j.1365-2125.2011.04141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodríguez J., Zarate R., Bandres E., Boni V., Hernández A., Sola J.J., Honorato B., Bitarte N., García-Foncillas J. Fc gamma receptor polymorphisms as predictive markers of Cetuximab efficacy in epidermal growth factor receptor downstream-mutated metastatic colorectal cancer. Eur. J. Cancer. 2012;48:1774–1780. doi: 10.1016/j.ejca.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 22.Naumov I., Zilberberg A., Rosin-Arbesfeld R., Ilan A., Kazanov D., Arber N., Kraus S.R. Abstract 1338: Knockdown of CD24 prevents colorectal polyp formation in APCmin/CD24 double knockout transgenic mice. Tumor Biol. 2012;72:1338. doi: 10.1158/1538-7445.am2012-1338. [DOI] [Google Scholar]
- 23.Huang X.-L., Xu D.-H., Wang G.-P., Zhang S., Yu C. Associations betweenCD24gene polymorphisms and inflammatory bowel disease: A meta-analysis. World J. Gastroenterol. 2015;21:6052–6059. doi: 10.3748/wjg.v21.i19.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Limbergen J., Geddes K., Henderson P., Russell R.K., Drummond H.E., Satsangi J., Griffiths A.M., Philpott D.J., Wilson D.C. Paneth cell marker CD24 in NOD2 knockout organoids and in inflammatory bowel disease (IBD) Gut. 2013;64:353–354. doi: 10.1136/gutjnl-2013-305077. [DOI] [PubMed] [Google Scholar]
- 25.Sato T., Van Es J.H., Snippert H.J., Stange D.E., Vries R.G., Van Den Born M., Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cunliffe R.N., Rose F.R.A.J., Keyte J., Abberley L., Chan W.C., Mahida Y.R. Human defensin 5 is stored in precursor form in normal Paneth cells and is expressed by some villous epithelial cells and by metaplastic Paneth cells in the colon in inflammatory bowel disease. Gut. 2001;48:176–185. doi: 10.1136/gut.48.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bevins C.L., Salzman N. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Genet. 2011;9:356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 28.Gracz A.D., Fuller M.K., Wang F., Li L., Stelzner M., Dunn J.C., Martin M.G., Magness S.T. Brief report: CD24 and CD44 mark human intestinal epithelial cell populations with characteristics of active and facultative stem cells. Stem Cells. 2013;31:2024–2030. doi: 10.1002/stem.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vermeulen L., Todaro M., Melo F.D.S.E., Sprick M.R., Kemper K., Alea M.P., Richel D.J., Stassi G., Medema J.P. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc. Natl. Acad. Sci. USA. 2008;105:13427–13432. doi: 10.1073/pnas.0805706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li O., Zheng P., Liu Y. CD24 Expression on T Cells Is Required for Optimal T Cell Proliferation in Lymphopenic Host. J. Exp. Med. 2004;200:1083–1089. doi: 10.1084/jem.20040779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahmed M.A., Jackson D., Seth R., Robins A., Lobo D.N., Tomlinson I.P., Ilyas M. CD24 is upregulated in inflammatory bowel disease and stimulates cell motility and colony formation. Inflamm. Bowel Dis. 2010;16:795–803. doi: 10.1002/ibd.21134. [DOI] [PubMed] [Google Scholar]
- 32.Diessner J., Bruttel V., Stein R.G., Horn E., Häusler S.F.M., Dietl J., Honig A., Wischhusen J. Targeting of preexisting and induced breast cancer stem cells with trastuzumab and trastuzumab emtansine (T-DM1) Cell Death Dis. 2014;5:e1149. doi: 10.1038/cddis.2014.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X., Fan Z. The epidermal growth factor receptor antibody cetuximab induces autophagy in cancer cells by downregulating HIF-1alpha and Bcl-2 and activating the beclin 1/hVps34 complex. Cancer Res. 2010;70:5942–5952. doi: 10.1158/0008-5472.CAN-10-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zarn J., Jackson D., Bell M., Jones T.A., Weber É., Sheer D., Waibel R., Stahel R. The small cell lung cancer antigen cluster-4 and the leukocyte antigen CD24 are allelic isoforms of the same gene (CD24) on chromosome band 6q21. Cytogenet. Genome Res. 1995;70:119–125. doi: 10.1159/000134075. [DOI] [PubMed] [Google Scholar]
- 35.Marmé F., Werft W., Walter A., Keller S., Wang X., Benner A., Burwinkel B., Sinn P., Hug S., Sohn C., et al. CD24 Ala57Val polymorphism predicts pathologic complete response to sequential anthracycline- and taxane-based neoadjuvant chemotherapy for primary breast cancer. Breast Cancer Res. Treat. 2011;132:819–831. doi: 10.1007/s10549-011-1759-9. [DOI] [PubMed] [Google Scholar]
- 36.Buck K., Hug S., Seibold P., Ferschke I., Altevogt P., Sohn C., Schneeweiss A., Burwinkel B., Jäger D., Flesch-Janys D., et al. CD24 polymorphisms in breast cancer: Impact on prognosis and risk. Breast Cancer Res. Treat. 2013;137:927–937. doi: 10.1007/s10549-012-2325-9. [DOI] [PubMed] [Google Scholar]
- 37.Sadot E., Kraus S., Stein M., Naboishchikov I., Toledano O., Kazanov D., Arber N., Kashtan H. CD24 Gene Polymorphism—A Novel Prognostic Factor in Esophageal Cancer. Int. J. Biol. Markers. 2014;29:e49–e54. doi: 10.5301/jbm.5000071. [DOI] [PubMed] [Google Scholar]
- 38.Yan S., Xu D., Jiang T., Wang P., Yin Y., Wang X., Hua C., Zhang B., Li Z., Lu L., et al. CD24 single nucleotide polymorphisms and cancer risk. Tumor Biol. 2014;35:8927–8932. doi: 10.1007/s13277-014-2127-2. [DOI] [PubMed] [Google Scholar]
- 39.Forouhar F., Anderson J.L.R., Mowat C.G., Vorobiev S.M., Hussain A., Abashidze M., Bruckmann C., Thackray S.J., Seetharaman J., Tucker T., et al. Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase. Proc. Natl. Acad. Sci. USA. 2006;104:473–478. doi: 10.1073/pnas.0610007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pardoll D. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma M.D., Baban B., Chandler P., Hou D.-Y., Singh N., Yagita H., Azuma M., Blazar B.R., Mellor A.L., Munn D.H. Plasmacytoid dendritic cells from mouse tumor-draining lymph nodes directly activate mature Tregs via indoleamine 2,3-dioxygenase. J. Clin. Investig. 2007;117:2570–2582. doi: 10.1172/JCI31911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muller A.J., Sharma M.D., Chandler P.R., DuHadaway J.B., Everhart M.E., Johnson B.A., Kahler D.J., Pihkala J., Soler A.P., Munn D.H., et al. Chronic inflammation that facilitates tumor progression creates local immune suppression by inducing indoleamine 2,3 dioxygenase. Proc. Natl. Acad. Sci. USA. 2008;105:17073–17078. doi: 10.1073/pnas.0806173105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thaker A.I., Rao M.S., Bishnupuri K.S., Kerr T.A., Foster L., Marinshaw J.M., Newberry R.D., Stenson W.F., Ciorba M.A. IDO1 metabolites activate beta-catenin signaling to promote cancer cell proliferation and colon tumorigenesis in mice. Gastroenterology. 2013;145:416–425. doi: 10.1053/j.gastro.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolf A. Overexpression of indoleamine 2,3-dioxygenase in human inflammatory bowel disease. Clin. Immunol. 2004;113:47–55. doi: 10.1016/j.clim.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 45.Ferdinande L., Decaestecker C., Verset L., Mathieu A., Lopez X.M., Negulescu A.-M., Van Maerken T., Salmon I., Cuvelier C.A., Demetter P. Clinicopathological significance of indoleamine 2,3-dioxygenase 1 expression in colorectal cancer. Br. J. Cancer. 2011;106:141–147. doi: 10.1038/bjc.2011.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brandacher G., Perathoner A., Ladurner R., Schneeberger S., Obrist P., Winkler C., Werner E.R., Werner-Felmayer G., Weiss H.G., Gobel G., et al. Prognostic Value of Indoleamine 2,3-Dioxygenase Expression in Colorectal Cancer: Effect on Tumor-Infiltrating T Cells. Clin. Cancer Res. 2006;12:1144–1151. doi: 10.1158/1078-0432.CCR-05-1966. [DOI] [PubMed] [Google Scholar]
- 47.Lee A., Kanuri N., Zhang Y., Sayuk G.S., Li E., Ciorba M.A. IDO1 and IDO2 Non-Synonymous Gene Variants: Correlation with Crohn’s Disease Risk and Clinical Phenotype. PLoS ONE. 2014;9:e115848. doi: 10.1371/journal.pone.0115848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stremitzer S., Sunakawa Y., Zhang W., Yang D., Ning Y., Stintzing S., Sebio A., Yamauchi S., Matsusaka S., El-Khoueiry R., et al. Variations in genes involved in immune response checkpoints and association with outcomes in patients with resected colorectal liver metastases. Pharmacogenomics J. 2015;15:521–529. doi: 10.1038/tpj.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Michielsen A.J., Noonan S., Martin P., Tosetto M., Marry J., Biniecka M., Maguire A.A., Hyland J.M., Sheahan K.D., O’Donoghue D.P., et al. Inhibition of Dendritic Cell Maturation by the Tumor Microenvironment Correlates with the Survival of Colorectal Cancer Patients following Bevacizumab Treatment. Mol. Cancer Ther. 2012;11:1829–1837. doi: 10.1158/1535-7163.MCT-12-0162. [DOI] [PubMed] [Google Scholar]
- 50.Schlaeth M., Berger S., Derer S., Klausz K., Lohse S., DeChant M., Lazar G.A., Schneider-Merck T., Peipp M., Valerius T. Fc-engineered EGF-R antibodies mediate improved antibody-dependent cellular cytotoxicity (ADCC) againstKRAS-mutated tumor cells. Cancer Sci. 2010;101:1080–1088. doi: 10.1111/j.1349-7006.2010.01505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gerdes C.A., Nicolini V.G., Herter S., Van Puijenbroek E., Lang S., Roemmele M., Moessner E., Freytag O., Friess T., Ries C.H., et al. GA201 (RG7160): A Novel, Humanized, Glycoengineered Anti-EGFR Antibody with Enhanced ADCC and Superior In Vivo Efficacy Compared with Cetuximab. Clin. Cancer Res. 2012;19:1126–1138. doi: 10.1158/1078-0432.CCR-12-0989. [DOI] [PubMed] [Google Scholar]
- 52.Kasper S., Breitenbuecher F., Reis H., Brandau S., Worm K., Kohler J., Paul A., Trarbach T., Schmid K.W., Schuler M. Oncogenic RAS simultaneously protects against anti-EGFR antibody-dependent cellular cytotoxicity and EGFR signaling blockade. Oncogene. 2012;32:2873–2881. doi: 10.1038/onc.2012.302. [DOI] [PubMed] [Google Scholar]
- 53.Le D.T., Uram J.N., Wang H., Bartlett B.R., Kemberling H., Eyring A.D., Skora A.D., Luber B.S., Azad N.S., Laheru D., et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu Z., Taylor J.A. SNPinfo: Integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37:W600–W605. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee P.H., Shatkay H. F-SNP: Computationally predicted functional SNPs for disease association studies. Nucleic Acids Res. 2007;36:D820–D824. doi: 10.1093/nar/gkm904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paez D., Gerger A., Zhang W., Yang D., Labonte M.J., Benhanim L., Wakatsuki T. Association of common gene variants in the WNT/beta-catenin pathway with colon cancer recurrence. Pharm. J. 2013;14:142–150. doi: 10.1038/tpj.2013.20. [DOI] [PubMed] [Google Scholar]
- 57.Lurje G., Nagashima F., Zhang W., Yang D., Chang H.M., Gordon M.A., El-Khoueiry A., Husain H., Wilson P.M., Ladner R.D., et al. Polymorphisms in Cyclooxygenase-2 and Epidermal Growth Factor Receptor Are Associated with Progression-Free Survival Independent of K-ras in Metastatic Colorectal Cancer Patients Treated with Single-Agent Cetuximab. Clin. Cancer Res. 2008;14:7884–7895. doi: 10.1158/1078-0432.CCR-07-5165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.