Abstract
Foot infections are the main disabling complication in patients with diabetes mellitus. These infections can lead to lower-limb amputation, increasing mortality and decreasing the quality of life. Biofilm formation is an important pathophysiology step in diabetic foot ulcers (DFU)—it plays a main role in the disease progression and chronicity of the lesion, the development of antibiotic resistance, and makes wound healing difficult to treat. The main problem is the difficulty in distinguishing between infection and colonization in DFU. The bacteria present in DFU are organized into functionally equivalent pathogroups that allow for close interactions between the bacteria within the biofilm. Consequently, some bacterial species that alone would be considered non-pathogenic, or incapable of maintaining a chronic infection, could co-aggregate symbiotically in a pathogenic biofilm and act synergistically to cause a chronic infection. In this review, we discuss current knowledge on biofilm formation, its presence in DFU, how the diabetic environment affects biofilm formation and its regulation, and the clinical implications.
Keywords: biofilm, commensal bacteria, diabetic foot infection, diabetic foot ulcer, pathogenic bacteria, pathogroups
1. Introduction
People suffering from diabetes mellitus have a 15–25% lifetime incidence of developing a diabetic foot ulcer (DFU) [1]. Infection is the most common, severe, and costly complication of diabetes mellitus [2], with high risk of mortality and morbidity due to lower limb amputation [3]. Wound infection, faulty wound healing, and ischemia are the most common precursors to diabetes-related amputations. Indeed, 80% of lower-limb amputations in diabetic patients are preceded by biofilm infected foot ulceration [4,5]. Infected wounds result in an increased risk of death within 18 months [6]. The host–microorganism interface plays a major role in DFU development. In DFU, bacteria are classically organized into functionally equivalent pathogroups (FEP), where pathogenic and commensal bacteria co-aggregate symbiotically in a pathogenic biofilm to maintain a chronic infection [7]. This polymicrobial biofilm has been observed both in pre-clinical studies using animal models and in clinical research on DFU. It represents the main cause of delayed healing.
2. Pathophysiology of Diabetic Foot Ulcers
2.1. Main Host-Related Factors
The triopathy induced by diabetes mellitus plays a role in the origin and chronicity of the DFU.
-
▪
Diabetic immunopathy: Diabetic patients have an altered function of polymorphonuclear cells and impaired phagocytosis, chemotaxis, and bactericidal activity (related to both non oxidative and oxidative mechanisms), which are more evident in the presence of high hyperglycemia [8]. A study on diabetic mice showed that persistent hyperglycemia had a deleterious effect on the innate immunity and could lead to skin and soft tissue infections by Staphylococcus aureus [9].
-
▪
Diabetic neuropathy: Neuropathy by C-fiber and autonomic nerve fiber dysfunction is a common and frequent complication of diabetes mellitus. An evolution of the deregulation of glycemic balance is the inhibition of nociception and the perception of pain, a process called loss of protective sensation [10]. Thus, patients may not initially notice small wounds in the legs and feet, and may fail to prevent infection. Studies have observed a reduction in foot skin innervation and the expression of neurogenic factors in DFU, correlated with low inflammatory cell accumulation and therefore in the chronicity of DFU. This contributes to enhancing susceptibility to infection of diabetic neuropathic foot ulcers [11].
-
▪
Diabetic angiopathy: Peripheral arterial disease (PAD) and microangiopathy are the main risk factors for DFU. The decrease in the oxygenation of tissues by thickening the capillary basement membrane is a hallmark of diabetic angiopathy [12]. Disease of arteries in the lower limb is a well-known risk factor for DFU. Indeed, studies have shown that PAD presents a 5.5-fold increased risk for DFU [13]. The ischemia caused by the angiopathy also enhances the severity of the infection as a result of a poor delivery of oxygen and nutrients in the infected wound and because of poor antibiotic tissue penetration [14].
Finally, the anatomical characteristics of the foot, with its division into compartments, participates in the pathophysiology by increasing the severity of the infectious process by promoting the spread of infection and aggravating tissue damage.
2.2. DFU Microbiota
The host–microbiota interface is often the key point in the development of wound infections. Defining the diabetic foot microbiota implies the possibility of distinguishing it from skin microbiota associated with other clinical statuses. Compared with the feet of non-diabetic men, those of diabetic men had decreased populations of Staphylococcus spp., increased populations of S. aureus, and increased bacterial diversity [15]. When compared with contralateral healthy skin, the DFU microbiota harbored less bacterial diversity with greater levels of opportunistic pathogens [16]. However, neither patient demographics nor wound type influenced the bacterial composition of the chronic wound microbiome [17]. Different studies have described this DFU microbiota [17,18,19,20,21,22,23,24,25]. Although they have produced interesting results and confirmed that the microbiota is a highly dynamic microbial community that maintains a relationship with the host, understanding the complex competitive or synergistic interaction between commensal and pathogenic microorganisms is necessary as it could play an important role in the severity and evolution of the wound.
2.3. Disturbances in the Host–Microorganism Interplay
-
▪
Bacterial virulence: The virulence of pathogens is a key element in the pathophysiology of DFU. The ability of a bacterium to be virulent is key to the precarious balance between colonization and infection [26]. Bacterial virulence has been characterized using DNA microarray-based genotyping, multiplex polymerase chain reaction (PCR), and in vivo assays [26,27]. Among the large panel of virulence factors, bacterial proteases (serine-, cysteine-, and metallo-proteases), produced by a wide range of pathogenic bacteria, could play a major role in the pathogenesis of wound healing [28]. However, these wounds, and especially DFU, are highly polymicrobial, and bacterial interactions should also be studied in order to better understand the mechanisms of infection and the role of each of the pathogens involved in DFU.
-
▪
Biofilm organization: In a 2008 study assessing wound tissue biopsies using electron microscopy, James et al. suggested that 60% of chronic wounds present biofilms versus 6% for acute wounds [29]. In the following sections of this review, we focus on the formation of biofilms, evidence of biofilms in DFU, influence of the diabetic environment, and finally the clinical implications of biofilms in DFU.
3. Overview of Biofilms in DFU
3.1. Biofilm Formation in DFU and Tools for Detection
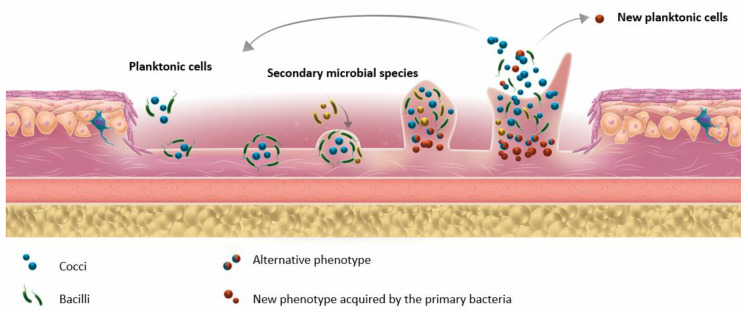
In the environment, microorganisms can exist in two main states, namely: planktonic and sessile. In the planktonic state, bacteria move freely in their environment. In the sessile state, microorganisms are attached either to solid surfaces (e.g., urinary catheter or prosthesis), or more frequently, to each other, constituting multicellular aggregates that can lead to biofilm formation. Biofilm formation is a multistep process (for review, see Percival et al., 2015 [30]; Figure 1) whereby heterogeneous communities of microorganisms (bacteria and/or fungi) [30] are embedded into a self-produced matrix of extracellular polymeric substance (EPS). EPS contains proteins, glycoproteins, and polysaccharides and confers the ability to adhere to biotic or abiotic surfaces [31]. Clinically, biopsy tissues are the most reliable samples for revealing biofilms in deep tissues. However, the use of swabs to collect biofilm samples from the wound surface is considered an improper technic because of contamination from the skin microbiota, the difficulty in detaching the biofilm from the host epithelium, and the growth of anaerobes in the deep tissues. If a moderate to severe soft tissue infection is suspected and a wound is present, a tissue sample from the base of the debrided wound should be examined. Biofilms in tissue samples are commonly quantified through microscopy. Techniques such as confocal laser scanning microscopy and scanning electron microscopy or fluorescence in situ hybridization (FISH) are the most appropriate for revealing biofilms in biopsies [32].
Figure 1.
Different bacterial steps of biofilm formation.
Some important features of chronic wounds, and notably DFU, could be noted as follows:
-
▪
Cells included in the biofilm can develop an intracellular communication mechanism called quorum sensing (QS) [33], which controls bacterial pathogenicity and biofilm formation. The bacterial density influences the biofilm production [34].
-
▪
Microbial cells within a biofilm can detach and disseminate in the wound environment. The behavior of the released bacteria may differ from that of the pioneering colonizing bacteria because of adaptation within the biofilm [30,35].
-
▪
The concept of FEP was proposed by Dowd et al. after observing that different bacterial species can collaborate and interact with each other. FEPs are responsible for the chronicity of infection and for the maintenance of the pathogenic biofilm [7].
3.2. Biofilm Studies in Animal Models of DFU
Several studies have described the presence of biofilms in animal wounds since the early 2000s, and experimental diabetic models were developed in 2010 (Table 1). Pioneering groups in this field have shown that in db/db mice (a model of diabetic dyslipidemia), Pseudomonas aeruginosa or S. aureus biofilms delayed wound healing, and that the diabetic condition slowed down healing and increased the biofilm thickness [36,37]. Hsu et al. also reported that high glucose levels encourage the formation of vancomycin-resistant S. aureus biofilms [38]. Other studies have shown that the host response and neutrophil oxidative burst activity were decreased in the wound, and that oxidative stress and reactive oxygen species promoted biofilm appearance [39,40]. James et al. suggested that biofilms in wounds induced oxygen-limiting conditions (and thus stress) by the following two mechanisms: (i) bacterial metabolic activities and (ii) oxygen-deprivation by the host defenses [41]. These findings were recently confirmed by Hunt et al., who showed delayed healing in diabetic mice concomitantly with increased pus production [42]. They also suggested that in db/db mice, the deleterious impact of P. aeruginosa on wound healing cannot be explained solely by its ability to form biofilms, and that the type-3 secretion system virulence structure was also involved in the wound damage caused by this pathogen [43] (Table 1).
Table 1.
Examples of biofilm studies in animal models of diabetes.
| Animal Model | Strain Used | Findings | Reference |
|---|---|---|---|
| db/db mice | P. aeruginosa (PAO1) | Biofilm evidence after a 6-mm punch biopsy wound on the dorsal skin | [36] |
| db/db mice | P. aeruginosa (PAO1) | Biofilm delays wound healing | [37] |
| TallyHo mice (Type 2 diabetes mellitus) | P. aeruginosa | Biofilm decreases TLR 2, TLR 4, IL-1α, and TNF-α expression and neutrophil oxidative burst activity | [39] |
| BALB/c mice with injection of STZ | Vancomycin-resistant S. aureus | Correlation between glucose concentration and biofilm formation | [38] |
| db/db mice | Wound microbiome | Oxidative stress and ROS favor biofilm formation and establish a chronic wound | [40] |
| db/db mice | P. aeruginosa | Bacteria in biofilm induce oxygen stress by producing metabolites and recruiting defense cells that reduce oxygen | [41] |
| Mice with injection of STZ | P. aeruginosa | Biofilm increases wound depth, mortality rate, and pus production | [42] |
| db/db mice | P. aeruginosa | P. aeruginosa infection is independent of its ability to form biofilm and primarily depends on T3SS | [43] |
db/db mice—diabetic mice; TLR—toll-like receptor; IL—interleukin; TNF—tumor necrosis factor; ROS—reactive oxygen species; STZ—streptozocin (a pancreatic β-cell toxin); T3SS—type-3 secretion system.
3.3. Biofilm Studies in Human Clinical DFU
Many clinical studies emerged in the 2010s demonstrating the impact of biofilms in chronic wounds (Table 2). In 2011, Neut et al. published two case studies of diabetic patients with non-healing ulcers. Using the confocal laser scanning microscope technique, they showed evidence of biofilms in diabetic wounds [44]. Subsequently, several studies have shown the presence and the impact of biofilms in clinical DFU. Malik et al. showed that on 162 diabetic foot infections (DFI), biofilms were present in 67.9% of the cases [45]. Other studies supported this and, in particular, the implication of S. aureus within the biofilms [46,47]. Oates et al. confirmed the importance of biofilms, using 26 human samples after debridement, employing FISH and scanning electron microscopy [48]. Recent research has shown that, during infection, in particular at the wound level, a single bacteria species is not responsible for biofilm formation [49]. Instead, microbes represent a complex polymicrobial biofilm community communicating with each other [50]. Interactions between microbes are complex and play an important role in the pathogenesis of the infection. These interactions range from competition for nutrients to evolving cooperative mechanisms that support their mutual growth in a specific environment [51]. Proximity and contact between bacteria in the biofilm promote communication and exchanges. To adapt their behavior, bacteria communicate through diffusible molecules like homoserines lactones or quinolones for Gram-negative bacteria, whereas Gram-positive cocci use short peptides [52]. Moreover, this proximity contributes to horizontal gene transfer, providing tolerance to antimicrobial agents and enhancing survival. Mottola et al. studied 53 staphylococci clinical isolates from DFU [53]. They discovered that biofilms cells were 10 to 1000 more tolerant to antibiotics than planktonic cells. In their work, of the 10 antibiotics tested, only gentamicin and ceftaroline were able to eradicate the biofilms. It has been reported that bacterial biofilms are also highly resistant to ultraviolet and heavy metals [54]. In addition to bacteria, fungi, especially Candida, are present in DFU biofilm-associated wound samples [55].
Table 2.
Examples of biofilm studies in clinical human DFU.
| Model | N° of Patients | Biofilm Visualization | Findings | Reference |
|---|---|---|---|---|
| DFU | 2 | CLSM | Evidence of biofilms | [44] |
| DFU | 162 | Microtiter plate assay | Biofilms in 67.9% of infected DFUs | [45] |
| DFU | 26 | FISH and ESEM | Observation of the formed biofilms and their bacterial constitution | [48] |
| DFU | 357 | Crystal violet | Observation of the formed biofilms | [46] |
| DFU | 100 | Congo Red dye, tissue culture plates, and crystal violet staining | Biofilm formation in 46.3% of isolates, predominantly by S. aureus (38.8% of isolates) and MDR bacteria (46.3%) | [47] |
| DFU | 49 | Calgary biofilm pin lid device with resazurin and PCR of genes associated with biofilm formation | Biofilms are resistant to antibiotics at concentrations 10–1000 times higher than those required to kill planktonic cells | [53] |
| DFU | 155 | Microtiter plate assay and ELISA, XTT formazan, and SEM | Presence and importance of non-Candida albicans species in biofilms | [55] |
| DFU | 95 | Microtiter plate assay and FISH | Polymicrobial biofilms are thicker | [56] |
DFU—diabetic foot ulcer; CLSM—confocal laser scanning microscopy; ELISA—enzyme-linked immunosorbent assay; ESEM—environmental scanning electron microscopy; FISH—fluorescent in situ hybridization; MDR—multidrug resistant; PIA—polysaccharide intercellular adhesin; SEM—scanning electron microscopy; XTT—2H-tetrazolium-5-carboxanilide.
3.4. Factors Influencing Biofilm Formation in DFU
DFUs are mainly colonized by commensal bacteria. Numerous papers have analyzed the DFU microbiome, showing that the wounds contain commensal microorganisms from different niches [57,58]. All of these studies highlight the high bacterial complexity of wounds. This complexity is one of the major characteristics of DFU, and the lack of knowledge regarding the interactions of these microorganisms in the wound renders these infections as being complicated to manage [59]. The microorganisms appear to be organized as multi-layered communities surrounded by a self-produced protective extracellular matrix, and are organized into different FEPs [7]. Biofilm formation is a multistep process, including random settlement of early bacterial colonizers, with increased competition among species and niche differentiation, resulting in highly heterogeneous biofilms [30]. The biofilms detected in patients with foot ulcers may be responsible for the delayed healing of these chronic wounds [18]. Moreover, the presence of some bacterial communities in the initial stages of the wounds has been associated with delayed healing [24].
Several microbial and host factors specific to DFU may interfere in the development and feature of the biofilms:
-
-
High bacterial diversity [7,15,60,61], including opportunistic pathogens [16] and anaerobic bacteria [57,62].
-
-
Increased S. aureus population [15], particularly in neuropathic DFUs [61]. However, their microbiota present a similar level of richness (number of different species in the wound community), abundance, and diversity compared to other chronic wounds [63], suggesting that the microbiota is not influenced by the wound type.
-
-
The wound depth with a more diverse and complex microbiota in the deep part of the wound [64] where pathogenic, particularly anaerobic, bacteria are sheltered.
-
-
Environmental factors (e.g., demographic characteristics, personal hygiene, geographical location of the patient, high glycemic level, and previous exposure to antimicrobial therapy) [65].
-
-
Patient immune status that modifies the role of low-virulence bacteria (e.g., Staphylococcus sp. and corynebacteria) towards a higher pathogenicity [66], and where excessive secretion of pro-inflammatory cytokines, pH, temperature, or antimicrobial treatment (topic or systemic administration) [67] can increase tissue destruction [68].
-
-
DFU duration is positively correlated with the ecological diversity of the bacteria present in the wounds, species richness, and relative abundance of Proteobacteria. It is also negatively correlated with the relative abundance of staphylococci [69].
-
-
Local tissue hypoxemia is often observed as a result of obstructive arteriopathy. This hypoxic environment influences bacterial diversity, with a higher prevalence of proteobacteria and strict anaerobic bacteria in deeper ulcers [61,68].
-
-
The development of a “unique microbiota” in each DFU (new or recurrent) [17].
3.5. Bacterial Organization Inside DFU
The main characteristic of DFU is the polymicrobial content that modulates bacterial virulence. Within DFU, microorganisms form a complex polymicrobial biofilm community and intercommunicate [7]. As described above, bacterial interactions play an important role in pathogenesis, competing and cooperating in order to support their mutual growth in a specific environment [51] via interactions through diffusible molecules [52].
The most studied bacterial interaction in DFU is the cooperation between S. aureus and P. aeruginosa, despite the location of P. aeruginosa being deeper in the wound bed than S. aureus [70]. Many substances produced by P. aeruginosa may play a protective role for S. aureus [17,70,71,72,73,74]. In a rat model of orthopedic wounds, even a low presence of both P. aeruginosa and S. aureus increased their infection rates in the wound [75]. A similar synergistic cooperation between P. aeruginosa and S. aureus also increased their tolerance to antibiotics, ability to form biofilms, and the secretion of virulence factors (hydrogen cyanide, exoenzyme S, exotoxin A, and pyocyanin for P. aeruginosa, and Panton-Valentine leukocidin and α hemolysin for S. aureus) [76]. These interactions can also be competitive, as exemplified by the competition for iron or the one-way growth inhibition of S. aureus [37,77]. Indeed, P. aeruginosa can simultaneously suppress S. aureus growth and enhance its resistance to aminoglycosides [71].
Other bacterial interactions have also been described. For instance, the combined inoculation of different pathogenic bacteria (Escherichia coli, Bacteroides fragilis, and Clostridium perfringens) increased the mortality rate in type-2 diabetic mice compared with those receiving inoculation of single strains [78]. Competition between commensal and pathogenic bacteria has been observed during cutaneous colonization [79]. In contrast, Helcococcus kunzii (a commensal Gram-positive coccus) and S. aureus cooperation led to a decrease of S. aureus virulence in Caenorhabditis elegans [80]. S. aureus shifts toward commensalism in response to Corynebacterium sp. [81]. Moreover, S. epidermidis, a commensal bacterium, produces a serine protease (Esp) that inhibits S. aureus biofilm formation [56,82]. Finally, the co-culture of Fusobacterium nucleatum (ATCC 25586) with Prevotella intermedia/Prevotella nigrescens promotes biofilm formation compared with single cultures [83].
Another pertinent aspect of polymicrobial biofilms in DFU is their ability to adapt under various circumstances via enhanced metabolic cooperation and gene regulation between sessile cells. Biofilm diversity promotes its survival by creating a thicker biofilm, resulting in more severe infections. In this context, Mottola et al. reported that the biofilms formed by P. aeruginosa and Enterococcus faecalis and Acinetobacter baumannii, and S. aureus resulted in a thicker biofilm than the bacteria alone, which were difficult to eradicate [84]. Furthermore, these microbial communities are heterogeneous. Interestingly, fungi can also form biofilms. Both yeasts and filamentous fungi can adhere to biotic and abiotic surfaces, and form highly organized communities that are resistant to antimicrobials and environmental conditions. Many fungi have been correlated with biofilm formation, however, Candida biofilms remain the most widely studied. The biofilms formed by yeast and filamentous fungi present differences, and studies of polymicrobial communities have become increasingly important. Interactions have been observed between bacterial and fungal species in chronic wounds [55]. Infections that are thought to involve polymicrobial biofilms are most frequently associated with the abiotic surfaces of indwelling medical devices. In a review written by Lynch and Robertson [85], they highlighted the indwelling medical devices commonly associated with biofilm formation. In all of the devices tested, the principal pathogen responsible for the biofilms was a bacterium, however in 70% of cases, fungi was found as a secondary species. Among fungal pathogens, Candida albicans, a commensal mucosal organism and opportunistic pathogen of the immunocompromised, was most commonly associated with biofilms. Numerous studies have described co-infections of fungi and bacteria in different diseases. For example, cystic fibrosis lungs are a major site of polymicrobial infection, with bacteria such as P. aeruginosa, S. aureus, Burkholderia cepacia, A. baumannii, and Haemophilus influenzae mixed with C. albicans, A. fumigatus, and Scedosporium sp. [86]. In a DFI context, Kalan et al. showed that the presence of fungal communities in the polymicrobial biofilms of chronic wounds is associated with a poor prognosis and delayed healing [87]. Further studies are needed in order to fully elaborate on the role of each microorganism in the polymicrobial biofilms of DFU.
4. Clinical Impact of Biofilms in DFU
As biofilms are implicated in 60 to 80% of chronic wounds [29,88], the clinical impact of biofilms is particularly relevant. For clinicians, the main difficulty is in distinguishing between infecting and colonizing bacteria. Misidentification can lead to inappropriate antibiotic prescriptions that may contribute to the emergence of multidrug resistant (MDR) bacteria, a major DFU health issue [30].
4.1. Antibiotics Resistance
Sessile cells involved in biofilm formation display different characteristics compared with non-biofilm-associated cells (i.e., planktonic cells) [89]. In particular, sessile cells show a higher tolerance towards antimicrobial agents, one of the main causes of treatment failure [90,91]. Antimicrobial agent tolerance arises by several mechanisms, namely: (i) inability of drugs to penetrate through the polymeric matrix; (ii) the lack of intracellular accumulation of antibiotics due to impermeability (e.g., excessive production of glucans by P. aeruginosa) or active efflux (e.g., increased expression of efflux pump genes in Gram-negative bacilli); (iii) the presence of sessile bacteria, whereby cells are metabolically inactive and thus tolerate the antibiotic action better; and (iv) the importance of horizontal gene transfer between bacteria for the diffusion of resistant traits [92,93]. Biofilms increase the opportunity of gene transfer of virulence factors and antibiotic-resistant genes to susceptible bacterial species. The rate of mutation occurring in biofilms is markedly higher compared with planktonic cells [94]. In addition, (v) stress response to hostile environmental conditions (e.g., leading to an overexpression of antimicrobial agent-destroying enzymes) can result in an altered microenvironment inside the biofilm matrix (pH and oxygen content) and may contribute to enhanced degradation of antimicrobial agents in the biofilm matrix [95]. Finally, the hypoxic environment present in DFU also modulates the tolerance of bacteria to some antibiotics. For instance, the in vitro bactericidal effect of vancomycin on S. aureus isolates is lower in anaerobic conditions [96].
4.2. Host Immune Response
Pioneering colonizing bacteria released from the biofilm can adapt to their environment and form a new biofilm. To our knowledge, the only study conducted in this field focused on Klebsiella pneumoniae [35]. In addition, EPS is a mechanical barrier to antimicrobials, as well as to immune system cells [97]. Bacteria within biofilms evade the host’s natural defenses and are resistant to the host immune defense by different mechanisms, including the following: (i) limited penetration of leukocytes and their products into the biofilm [98]; (ii) global response regulators and quorum sensing, which protect the biofilm bacteria [99]; (iii) decreased phagocytic capacity of host cells against biofilm bacteria [100]; (iv) genetic switches that increase the resistance of biofilm bacteria [101]; and (v) suppression of the leukocyte effector function, including softening the magnitude of the respiratory burst [102]. Indeed, stimulation of the immune system without effectively eradicating the infection causes collateral damage to surrounding tissue and causes chronic inflammation [103]. This persistent chronic inflammation, added to the diabetic immune context, leads to the production of auto-inflammatory cytokines that aggravate the wound and slow the healing process.
5. Therapeutic Perspectives
Biofilms have a crucial role in DFU and DFIs and contribute to delayed healing. They are especially difficult to treat using classical antibiotics because of EPS, which prevents diffusion into the biofilm. They also support gene transfer, the selection of strains with beneficial characteristics, and the development of new bacterial characteristics. This difficulty in treating DFU/DFI could be enhanced in the context of the diabetic environment.
Biofilms encountered in chronic wounds, such as DFU, are highly polymicrobial, which can enhance bacterial interactions. Bacterial cooperation is key to understanding the formation and regulation of biofilms at a wound level, but also for highlighting new therapeutic targets. The available approaches against biofilms are quite limited, and new prevention, diagnosis, and treatment methods are crucially needed, particularly because of the extent of the MDR bacteria in this pathology.
Targeting biofilm formation could be an interesting strategy to prevent or at least reduce this problem. Classically, clinicians reduce the bacteria load (constituting commensal and pathogen species) resulting from the biofilm organization and FEP. The best method involves physical removal, also called debridement, of the infected tissue in order to improve healing [104,105]. It is often performed using surgical instruments or by irrigation [105], and is the initial and essential stage in the management of infected wounds. This strategy is still the preferred method used to prepare the wound bed and to promote moist wound healing, but it might not completely remove the biofilms immediately. Therefore, it must be repeated at regular intervals [104]. The results obtained with ultrasound debridement could represent a promising approach [106]. Other approaches could be proposed with the aim to inhibit bacterial adhesion or biofilm metabolism, such as (i) blocking bacterial adhesins (using ions chelators such as ethylenediaminetetraacetic acid (EDTA) and citrate, the most promising compounds of this class [107]), (ii) inhibiting the adhesion structure biogenesis (e.g., plant-derived natural compounds [108]), (iii) modulating QS (e.g., furanone [109], savarine [110], or deferiprone [111]), and (iv) enhancing bacterial dispersion (such as the α-amylase enzyme [112], 2-aminoimidazole [113], or Cis-2-decenoic acid [114]). Physical inhibition could also represent an interesting method, such as photodynamic therapy-induced pathogen cell death by killing sessile bacteria [115].
Some antimicrobial strategies as alternatives to antibiotics have also be developed, such as phagotherapy [116,117], nanotechnologies [118], antimicrobial peptides (AMP) [119,120,121], or agent mimicking AMPs [122], as well as natural compounds (such as honey [123]). These approaches have an interesting potential, but further studies are required to really understand the mechanism of action of each of these solutions and to improve their role in DFI management.
Researchers are now aware of and consider the polymicrobial characteristics of DFI and biofilms. Further studies on bacterial interactions are required in order to really understand the pathophysiology and to help with the development of new therapeutic tools that will target polymicrobial biofilms. This needs to be done through the development of (i) validated, consistent, and robust animal wound models reproducing the clinical situation and biofilm constitution; (ii) ex vivo and in vivo imaging technologies to visualize bacterial biofilms and to confirm their eradication; and (iii) “omics” tools to detect biofilm formation at the bedside and to evaluate the best course of action for the debridement.
6. Conclusions
Biofilms have a crucial role in DFIs and contribute to delayed healing. These wounds are characterized by a complex microbiome and a polymicrobial organization, especially within the biofilm. Even if most experimental biofilm studies provide descriptive and interesting information, they are derived from in vitro studies or non-adapted in vivo models. The development of processes and methodologies to study biofilms is needed. This represents the next step to validating new antibiofilm molecules with a promising therapeutic potential.
Acknowledgments
We thank Sarah Kabani for her editing assistance. We thank the Nîmes University hospital for its structural, human, and financial support through the award obtained by our team during the internal call for tenders “Thématiques phares”. The authors belong to the FHU InCh (Federation Hospitalo Universitaire Infections Chroniques, Aviesan).
Author Contributions
Conceptualization, C.P., A.S., and J.-P.L.; validation, C.D.-R., A.S., and J.-P.L.; formal analysis, C.P. and A.P.; investigation, C.P. and S.S.; writing—original draft preparation, C.P. and J.-P.L.; writing—review and editing, C.D.-R., A.P., S.S., and A.S.; supervision, A.S. and J.-P.L. All of the authors have read and agreed to the published version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
Cassandra Pouget’s PhD is supported by a CIFRE grant (Biofilm Pharma).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Armstrong D.G., Boulton A.J.M., Bus S.A. Diabetic foot ulcers and their recurrence. N. Engl. J. Med. 2017;376:2367–2375. doi: 10.1056/NEJMra1615439. [DOI] [PubMed] [Google Scholar]
- 2.Prompers L., Schaper N., Apelqvist J., Edmonds M., Jude E., Mauricio D., Uccioli L., Urbancic V., Bakker K., Holstein P., et al. Prediction of outcome in individuals with diabetic foot ulcers: Focus on the differences between individuals with and without peripheral arterial disease. The EURODIALE Study. Diabetologia. 2008;51:747–755. doi: 10.1007/s00125-008-0940-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakker K., Apelqvist J., Lipsky B.A., Van Netten J.J., International Working Group on the Diabetic Foot The 2015 IWGDF guidance documents on prevention and management of foot problems in diabetes: Development of an evidence-based global consensus. Diabetes Metab. Res. Rev. 2016;32:2–6. doi: 10.1002/dmrr.2694. [DOI] [PubMed] [Google Scholar]
- 4.Palumbo P.J., Melton L.J.I. Diabetes in America: Diabetes Data Compiled 1984. Government Printing Office; Washington, DC, USA: 1985. Peripheral vascular disease and diabetes. [Google Scholar]
- 5.Adler A.I., Boyko E.J., Ahroni J.H., Smith D.G. Lower-extremity amputation in diabetes. The independent effects of peripheral vascular disease, sensory neuropathy, and foot ulcers. Diabetes Care. 1999;22:1029–1035. doi: 10.2337/diacare.22.7.1029. [DOI] [PubMed] [Google Scholar]
- 6.Ndosi M., Wright-Hughes A., Brown S., Backhouse M., Lipsky B.A., Bhogal M., Reynolds C., Vowden P., Jude E.B., Nixon J., et al. Prognosis of the infected diabetic foot ulcer: A 12-month prospective observational study. Diabet. Med. 2018;35:78–88. doi: 10.1111/dme.13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dowd S.E., Wolcott R.D., Sun Y., McKeehan T., Smith E., Rhoads D. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP) PLoS ONE. 2008;3:e3326. doi: 10.1371/journal.pone.0003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lecube A., Pachón G., Petriz J., Hernández C., Simó R. Phagocytic activity is impaired in type 2 diabetes mellitus and increases after metabolic improvement. PLoS ONE. 2011;6:e23366. doi: 10.1371/journal.pone.0023366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park S., Rich J., Hanses F., Lee J.C. Defects in innate immunity predispose C57BL/6J-Leprdb/Leprdb mice to infection by Staphylococcus aureus. Infect. Immun. 2009;77:1008–1014. doi: 10.1128/IAI.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yagihashi S., Mizukami H., Sugimoto K. Mechanism of diabetic neuropathy: Where are we now and where to go? J. Diabetes Investig. 2011;2:18–32. doi: 10.1111/j.2040-1124.2010.00070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galkowska H., Olszewski W.L., Wojewodzka U., Rosinski G., Karnafel W. Neurogenic factors in the impaired healing of diabetic foot ulcers. J. Surg. Res. 2006;134:252–258. doi: 10.1016/j.jss.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Chawla A., Chawla R., Jaggi S. Microvasular and macrovascular complications in diabetes mellitus: Distinct or continuum? Indian J. Endocrinol. Metab. 2016;20:546–551. doi: 10.4103/2230-8210.183480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters E.J., Lipsky B.A., Berendt A.R., Embil J.M., Lavery L.A., Senneville E., Urbančič-Rovan V., Bakker K., Jeffcoate W.J. A systematic review of the effectiveness of interventions in the management of infection in the diabetic foot. Diabetes Metab. Res. Rev. 2012;28:142–162. doi: 10.1002/dmrr.2247. [DOI] [PubMed] [Google Scholar]
- 14.Raymakers J.T., Houben A.J., van der Heyden J.J., Tordoir J.H., Kitslaar P.J., Schaper N.C. The effect of diabetes and severe ischaemia on the penetration of ceftazidime into tissues of the limb. Diabet. Med. 2001;18:229–234. doi: 10.1046/j.1464-5491.2001.00460.x. [DOI] [PubMed] [Google Scholar]
- 15.Redel H., Gao Z., Li H., Alekseyenko A.V., Zhou Y., Perez-Perez G.I., Weinstock G., Sodergren E., Blaser M.J. Quantitation and composition of cutaneous microbiota in diabetic and nondiabetic men. J. Infect. Dis. 2013;207:1105–1114. doi: 10.1093/infdis/jit005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gontcharova V., Youn E., Sun Y., Wolcott R.D., Dowd S.E. A comparison of bacterial composition in diabetic ulcers and contralateral intact skin. Open Microbiol. J. 2010;4:8–19. doi: 10.2174/1874285801004010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jneid J., Lavigne J.P., La Scola B., Cassir N. The diabetic foot microbiota: A review. Hum. Microbiome J. 2017;5–6:1–6. doi: 10.1016/j.humic.2017.09.002. [DOI] [Google Scholar]
- 18.Pereira S.G., Moura J., Carvalho E., Empadinhas N. Microbiota of Chronic Diabetic Wounds: Ecology, Impact, and Potential for Innovative Treatment Strategies. Front. Microbiol. 2017;8:1791. doi: 10.3389/fmicb.2017.01791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardiner M., Vicaretti M., Sparks J., Bansal S., Bush S., Liu M., Darling A., Harry E., Burke C.M. A longitudinal study of the diabetic skin and wound microbiome. PeerJ. 2017;5:e3543. doi: 10.7717/peerj.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malone M., Johani K., Jensen S.O., Gosbell I.B., Dickson H.G., Hu H., Vickery K. Next Generation DNA Sequencing of Tissues from Infected Diabetic Foot Ulcers. EBioMedicine. 2017;21:142–149. doi: 10.1016/j.ebiom.2017.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacDonald A., Brodell J.D., Daiss J.L., Schwarz E.M., Oh I. Evidence of differential microbiomes in healing versus non-healing diabetic foot ulcers prior to and following foot salvage therapy. J. Orthop. Res. 2019;37:1596–1603. doi: 10.1002/jor.24279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verbanic S., Shen Y., Lee J., Deacon J.M., Chen I.A. Microbial predictors of healing and short- term effect of debridement on the microbiome of chronic wounds. NPJ Biofilms Microbiomes. 2020;6:21. doi: 10.1038/s41522-020-0130-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sloan T.J., Turton J.C., Tyson J., Musgrove A., Fleming V.M., Lister M.M., Loose M.W., Sockett R.E., Diggle M., Game F.L., et al. Examining diabetic heel ulcers through an ecological lens: Microbial community dynamics associated with healing and infection. J. Med. Microbiol. 2019;68:230–240. doi: 10.1099/jmm.0.000907. [DOI] [PubMed] [Google Scholar]
- 24.Loesche M., Gardner S.E., Kalan L., Horwinski J., Zheng Q., Hodkinson B.P., Tyldsley A.S., Franciscus C.L., Hillis S.I., Mehta S., et al. Temporal Stability in Chronic Wound Microbiota is Associated with Poor Healing. J. Investig. Dermatol. 2017;137:237–244. doi: 10.1016/j.jid.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalan L.R., Meisel J.S., Loesche M.A., Horwinski J., Soaita I., Chen X., Uberoi A., Gardner S.E., Grice E.A. Strain- and Species- Level Variation in the Microbiome of Diabetic Wounds Is Associated with Clinical Outcomes and Therapeutic Efficacy. Cell Host Microbe. 2019;25:641–655. doi: 10.1016/j.chom.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spichler A., Hurwitz B.L., Armstrong D.G., Lipsky B.A. Microbiology of diabetic foot infections: From Louis Pasteur to ‘crime scene investigation’. BMC Med. 2015;13:2. doi: 10.1186/s12916-014-0232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavigne J.P., Sotto A., Dunyach-Remy C., Lipsky B.A. New molecular techniques to study the skin microbiota of diabetic foot ulcers. Adv. Wound Care. 2015;4:38–49. doi: 10.1089/wound.2014.0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarty S.M., Cochrane C.A., Clegg P.D., Percival S.L. The Role of Endogenous and Exogenous Enzymes in Chronic Wounds: A Focus on the Implications of Aberrant Levels of Both Host and Bacterial Proteases in Wound Healing. Wound Repair Regen. 2012;20:125–136. doi: 10.1111/j.1524-475X.2012.00763.x. [DOI] [PubMed] [Google Scholar]
- 29.James G.A., Swogger E., Wolcott R., Pulcini E.D., Secor P., Sestrich J., Costerton J.W., Stewart P.S. Biofilms in chronic wounds. Wound Repair Regen. 2008;16:37–44. doi: 10.1111/j.1524-475X.2007.00321.x. [DOI] [PubMed] [Google Scholar]
- 30.Percival S.L., McCarty S.M., Lipsky B. Biofilms and Wounds: An Overview of the Evidence. Adv. Wound Care. 2015;4:373–381. doi: 10.1089/wound.2014.0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bjarnsholt T. The role of bacterial biofilms in chronic infections. APMIS. 2013;121:1–58. doi: 10.1111/apm.12099. [DOI] [PubMed] [Google Scholar]
- 32.Høiby N., Bjarnsholt T., Moser C., Bassi G.L., Coenye T., Donelli G., Hall-Stoodley L., Holá V., Imbert C., Kirketerp-Møller K., et al. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin. Microbiol. Infect. 2015;21:S1–S25. doi: 10.1016/j.cmi.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 33.Solano C., Echeverz M., Lasa I. Biofilm dispersion and quorum sensing. Curr. Opin. Microbiol. 2014;18:96–104. doi: 10.1016/j.mib.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 34.Asfour H. Anti-quorum sensing natural compounds. J. Microsc. Ultrastruct. 2018;6:1. doi: 10.4103/JMAU.JMAU_10_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guilhen C., Forestier C., Balestrino D. Biofilm dispersal: Multiple elaborate strategies for dissemination of bacteria with unique properties. Mol. Microbiol. 2017;105:188–210. doi: 10.1111/mmi.13698. [DOI] [PubMed] [Google Scholar]
- 36.Zhao G., Usui M.L., Underwood R.A., Singh P.K., James G.A., Stewart P.S., Fleckman P., Olerud J.E. Time course study of delayed wound healing in a biofilm challenged diabetic mouse model. Wound Repair Regen. 2012;20:342–352. doi: 10.1111/j.1524-475X.2012.00793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen A.T., Oglesby-Sherrouse A.G. Interactions between Pseudomonas aeruginosa and Staphylococcus aureus during co-cultivations and polymicrobial infections. Appl. Microbiol. Biotechnol. 2016;100:6141–6148. doi: 10.1007/s00253-016-7596-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu C.Y., Shu J.C., Lin M.H., Chong K.Y., Chen C.C., Wen S.M., Hsieh Y.T., Lia W.T. High Glucose concentration promotes vancomycin-enhanced biofilm formation of vancomycin-non-susceptible Staphylococcus aureus in diabetic mice. PLoS ONE. 2015;10:e0134852. doi: 10.1371/journal.pone.0134852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nouvong A., Ambrus A.M., Zhang E.R., Hultman L., Coller H.A. Reactive oxygen species and bacterial biofilms in diabetic wound healing. Physiol. Genom. 2016;48:889–896. doi: 10.1152/physiolgenomics.00066.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dhall S., Do D.C., Garcia M., Kim J., Mirebrahim S.H., Lyubovitsky J., Lonardi S., Nothnagel E.A., Schiller N., Martins-Green M. Generating and reversing chronic wounds in diabetic mice by manipulating wound redox parameters. J. Diabetes Res. 2014;2014:1–18. doi: 10.1155/2014/562625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.James G.A., Ge Zhao A., Usui M., Underwood R.A., Nguyen H., Beyenal H., Pulcini E.D., Agostino Hunt A., Bernstein H.C., Fleckman P., et al. Microsensor and transcriptomic signatures of oxygen depletion in biofilms associated with chronic wounds. Wound Repair Regen. 2016;24:373–383. doi: 10.1111/wrr.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agostinho Hunt A.M., Gibson J.A., Larrivee C.L., O’Reilly S., Navitskaya S., Needle D.B., Abramowitch R.B., Busik J.V., Waters C.M. A bioluminescent Pseudomonas aeruginosa wound model reveals increased mortality of type 1 diabetic mice to biofilm infection. J. Wound Care. 2017;26:S24–S33. doi: 10.12968/jowc.2017.26.Sup7.S24. [DOI] [PubMed] [Google Scholar]
- 43.Goldufsky J., Wood S.J., Jayaraman V., Majdobeh O., Chen L., Qin S., Zhang C., DiPietro L.A., Shafikhani S.H. Pseudomonas aeruginosa uses T3SS to inhibit diabetic wound healing. Wound Repair Regen. 2015;23:557–564. doi: 10.1111/wrr.12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neut D., Tijdens-Creusen E.J., Bulstra S.K., van der Mei H.C., Busscher H.J. Biofilms in chronic diabetic foot ulcers—A study of 2 cases. Acta Orthop. 2011;82:383–385. doi: 10.3109/17453674.2011.581265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malik A., Mohammad Z., Ahmad J. The diabetic foot infections: Biofilms and antimicrobial resistance. Diabetes Metab. Syndr. 2013;7:101–107. doi: 10.1016/j.dsx.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 46.Murali T.S., Kavitha S., Spoorthi J., Bhat D.V., Prasat A.S.B., Upton Z., Ramachandra L., Acharya R.V., Satyamoorthy K. Characteristics of microbial drug resistance and its correlates in chronic diabetic foot ulcer infections. J. Med. Microbiol. 2014;63:1377–1385. doi: 10.1099/jmm.0.076034-0. [DOI] [PubMed] [Google Scholar]
- 47.Banu A., Noorul Hassan M.M., Rajkumar J., Srinivasa S. Spectrum of bacteria associated with diabetic foot ulcer and biofilm formation: A prospective study. Australas. Med. J. 2015;8:280–285. doi: 10.4066/AMJ.2015.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oates A., Bowling F.L., Boulton A.J., Bowler P.G., Metcalf D.G., McBain A.J. The visualization of biofilms in chronic diabetic foot wounds using routine diagnostic microscopy methods. J. Diabetes Res. 2014;2014:153586. doi: 10.1155/2014/153586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao G., Usui M.L., Lippman S.I., James G.A., Stewart P.S., Fleckman P., Olerud J.E. Biofilms and Inflammation in Chronic Wounds. Adv. Wound Care. 2013;2:389–399. doi: 10.1089/wound.2012.0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peters B.M., Jabra-Rizk M.A., O’May G.A., Costerton J.W., Shirtliff M.E. Polymicrobial interactions: Impact on pathogenesis and human disease. Clin. Microbiol. Rev. 2012;25:193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hibbing M.E., Fuqua C., Parsek M.R., Peterson S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rutherford S.T., Bassler B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012;2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mottola C., Semedo-Lemsaddek T., Mendes J.J., Melo-Cristino J., Tavares L., Cavaco-Silva P., Oliveira M. Molecular typing, virulence traits and antimicrobial resistance of diabetic foot staphylococci. J. Biomed. Sci. 2016;23:33. doi: 10.1186/s12929-016-0250-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yin W., Wang Y., Liu L., He J. Biofilms: The microbial “protective clothing’ in extreme environments. Int. J. Mol. Sci. 2019;20:3423. doi: 10.3390/ijms20143423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar D., Banerjee T., Chakravarty J., Singh S.K., Dwivedi A., Tilak R. Identification, antifungal resistance profile, in vitro biofilm formation and ultrastructural characteristics of Candida species isolated from diabetic foot patients in Northern India. Indian J. Med. Microbiol. 2016;34:308–314. doi: 10.4103/0255-0857.188320. [DOI] [PubMed] [Google Scholar]
- 56.Sugimoto S., Iwamoto T., Takada K., Okuda K.I., Tajima A., Iwase T., Mizunoe Y. Staphylococcus epidermidis Esp degrades specific proteins associated with Staphylococcus aureus biofilm formation and host-pathogen interaction. J. Bacteriol. 2013;195:1645–1655. doi: 10.1128/JB.01672-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolcott R.D., Hanson J.D., Rees E.J., Koenig L.D., Philips C.D., Wolcott R.A., Cox S.B., White J.S. Analysis of the chronic wound microbiota of 2,963 patients by 16S rDNA pyrosequencing. Wound Repair Regen. 2016;24:163–174. doi: 10.1111/wrr.12370. [DOI] [PubMed] [Google Scholar]
- 58.Jneid J., Cassir N., Schuldiner S., Jourdan N., Sotto A., Lavigne J.P., La Scola B. Exploring the microbiota of diabetic foot infections with culturomics. Front. Cell Infect. Microbiol. 2018;8:282. doi: 10.3389/fcimb.2018.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bowler P.G., Duerden B.I., Armstrong D.G. Wound Microbiology and Associated Approaches to Wound Management. Clin. Microbiol. Rev. 2001;14:244–269. doi: 10.1128/CMR.14.2.244-269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oates A., Bowling F.L., Boulton A.J.M., McBain A.J. Molecular and culture-based assessment of the microbial diversity of diabetic chronic foot wounds and contralateral skin sites. J. Clin. Microbiol. 2012;50:2263–2271. doi: 10.1128/JCM.06599-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gardner S.E., Hillis S.L., Heilmann K., Segre J.A., Grice E.A. The neuropathic diabetic foot ulcer microbiome is associated with clinical factors. Diabetes. 2013;62:923–930. doi: 10.2337/db12-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Percival S.L., Malone M., Mayer D., Salisbury A.M., Schultz G. Role of anaerobes in polymicrobial communities and biofilms complicating diabetic foot ulcers. Int. Wound J. 2018;15:776–782. doi: 10.1111/iwj.12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johnson T.R., Gómez B.I., McIntyre M.K., Dubick M.A., Christy R.J., Nicholson S.E., Burmeister D.M. The Cutaneous Microbiome and Wounds: New Molecular Targets to Promote Wound Healing. Int. J. Mol. Sci. 2018;19:2699. doi: 10.3390/ijms19092699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lipsky B.A., Richard J.L., Lavigne J.P. Diabetic foot ulcer microbiome: One small step for molecular microbiology... One giant leap for understanding diabetic foot ulcers? Diabetes. 2013;62:679–681. doi: 10.2337/db12-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Percival S.L., Thomas J.G., Williams D.W. Biofilms and bacterial imbalances in chronic wounds: Anti-Koch. Int. Wound J. 2010;7:169–175. doi: 10.1111/j.1742-481X.2010.00668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith K., Collier A., Townsend E.M., O’Donnell L.E., Bal A.M., Butcher J., MacKay W.G., Ramage G., Williams C. One step closer to understanding the role of bacteria in diabetic foot ulcers: Characterising the microbiome of ulcers. BMC Microbiol. 2016;16:54. doi: 10.1186/s12866-016-0665-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rahim K., Saleha S., Zhu X., Huo L., Basit A., Franco O.L. Bacterial contribution in chronicity of wounds. Microb. Ecol. 2017;73:710–721. doi: 10.1007/s00248-016-0867-9. [DOI] [PubMed] [Google Scholar]
- 68.Patel S., Srivastava S., Singh M.R., Singh D. Mechanistic insight into diabetic wounds: Pathogenesis, molecular targets and treatment strategies to pace wound healing. Biomed. Pharm. 2019;112:108615. doi: 10.1016/j.biopha.2019.108615. [DOI] [PubMed] [Google Scholar]
- 69.Cogen A.L., Nizet V., Gallo R.L. Skin microbiota: A source of disease or defence? Br. J. Dermatol. 2008;158:442–455. doi: 10.1111/j.1365-2133.2008.08437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fazli M., Bjarnsholt T., Kirketerp-Møller K., Jørgensen B., Andersen A.S., Krogfelt K.A., Givskov M., Tolker-Nielsen T. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J. Clin. Microbiol. 2009;47:4084–4089. doi: 10.1128/JCM.01395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hoffman L.R., Deziel E., D’Argenio D.A., Lépine F., Emerson J., McNamara S., Gibson R.L., Ramsey B.W., Miller S.I. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA. 2006;103:19890–19895. doi: 10.1073/pnas.0606756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chan K.G., Liu Y.C., Chang C.Y. Inhibiting N-acyl-homoserine lactone synthesis and quenching Pseudomonas quinolone quorum sensing to attenuate virulence. Front. Microbiol. 2015;6:1173. doi: 10.3389/fmicb.2015.01173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schurr M.J. Pseudomonas aeruginosa alginate benefits Staphylococcus aureus? J. Bacteriol. 2020;202 doi: 10.1128/JB.00040-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Price C.E., Brown D.G., Limoli D.H., Phelan V.V., O’Toole G.A. Exogenous alginate protects Staphylococcus aureus from killing by Pseudomonas aeruginosa. J. Bacteriol. 2020;202 doi: 10.1128/JB.00559-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hendricks K.J., Burd T.A., Anglen J.O., Simpson A.W., Christensen G.D., Gainor B.J. Synergy between Staphylococcus aureus and Pseudomonas aeruginosa in a rat model of complex orthopaedic wounds. J. Bone Joint Surg. 2001;83:855–861. doi: 10.2106/00004623-200106000-00006. [DOI] [PubMed] [Google Scholar]
- 76.Hotterbeekx A., Kumar-Singh S., Goossens H., Malhotra-Kumar S. In vivo and in vitro interactions between Pseudomonas aeruginosa and Staphylococcus spp. Front. Cell Infect. Microbiol. 2017;7:106. doi: 10.3389/fcimb.2017.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.DeLeon S., Clinton A., Fowler H., Everett J., Horswill A.R., Rumbaugh K.P. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect. Immun. 2014;82:4718–4728. doi: 10.1128/IAI.02198-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mastropaolo M.D., Evans N.P., Byrnes M.K., Stevens A.M., Robertson J.L., Melville S.B. Synergy in polymicrobial infections in a mouse model of Type 2 diabetes. Infect. Immun. 2005;73:6055–6063. doi: 10.1128/IAI.73.9.6055-6063.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nair N., Biswas R., Gotz F., Biswas L. Impact of Staphylococcus aureus on pathogenesis in polymicrobial infections. Infect. Immun. 2014;82:2162–2169. doi: 10.1128/IAI.00059-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ngba Essebe C., Visvikis O., Fines-Guyon M., Vergne A., Cattoir V., Lecoustumier A., Lemichez E., Sotto A., Lavigne J.P., Dunyach-Remy C. Decrease of Staphylococcus aureus virulence by Helcococcus kunzii in a Caenorhabditis elegans model. Front. Cell Infect. Microbiol. 2017;7:77. doi: 10.3389/fcimb.2017.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ramsey M.W., Freire M.O., Gabrilska R.A., Rumbaugh K.P., Lemon K.P. Staphylococcus aureus shifts toward commensalism in response to Corynebacterium Species. Front. Microbiol. 2016;7:1230. doi: 10.3389/fmicb.2016.01230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vandecandelaere I., Depuydt P., Nelis H.J., Coenye T. Protease production by Staphylococcus epidermidis and its effect on Staphylococcus aureus biofilms. Pathog. Dis. 2014;70:321–331. doi: 10.1111/2049-632X.12133. [DOI] [PubMed] [Google Scholar]
- 83.Okuda T., Kokubu E., Kawana T., Saito A., Okuda K., Ishihara K. Synergy in biofilm formation between Fusobacterium nucleatum and Prevotella species. Anaerobe. 2012;18:110–116. doi: 10.1016/j.anaerobe.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 84.Mottola C., Mendes J.J., Cristino J.M., Cavasco-Silva P., Tavares L., Oliveira M. Polymicrobial biofilms by diabetic foot clinical isolates. Folia Microbiol. 2016;61:35–43. doi: 10.1007/s12223-015-0401-3. [DOI] [PubMed] [Google Scholar]
- 85.Lynch A.S., Robertson G.T. Bacterial and fungal biofilm infections. Annu. Rev. Med. 2008;59:415–428. doi: 10.1146/annurev.med.59.110106.132000. [DOI] [PubMed] [Google Scholar]
- 86.Costa-Orlandi C.B., Sardi J.C.O., Pitangui N.S., de Oliveira H.C., Scorzoni L., Galeane M.C., Medina-Alarćon K.P., Melo W.C.M.A., Marcelino M.Y., Braz J.D., et al. Fungal Biofilms and Polymicrobial Diseases. J. Fungi. 2017;3:22. doi: 10.3390/jof3020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kalan L., Loesche M., Hodkinson B.P., Heilmann K., Ruthel G., Gardner S.E., Grice E.A. Redefining the chronic-wound microbiome: Fungal communities are prevalent, dynamic, and associated with delayed healing. mBio. 2016;7 doi: 10.1128/mBio.01058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Malone M., Bjarnsholt T., McBain A.J., James G.A., Stoodley P., Leaper D., Tachi M., Schultz G., Swanson T., Wolcott R.D. The prevalence of biofilms in chronic wounds: A systematic review and meta-analysis of published data. J. Wound Care. 2017;26:20–25. doi: 10.12968/jowc.2017.26.1.20. [DOI] [PubMed] [Google Scholar]
- 89.Donlan R.M., Costerton J.W. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hall-Stoodley L., Costerton J.W., Stoodley P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 91.Mah T.F., O’Toole G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 92.Alav I., Sutton J.M., Rahman K.M. Role of bacterial efflux pumps in biofilm formation. J. Antimicrob. Chemother. 2018;73:2003–2020. doi: 10.1093/jac/dky042. [DOI] [PubMed] [Google Scholar]
- 93.Ghigo J.M. Natural conjugative plasmids induce bacterial biofilm development. Nature. 2001;412:442–445. doi: 10.1038/35086581. [DOI] [PubMed] [Google Scholar]
- 94.Berlanga M., Guerrero R. Living together in biofilms: The microbial cell factory and its biotechnological implications. Microb. Cell Fact. 2016;15:165. doi: 10.1186/s12934-016-0569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gebreyohannes G., Nyerere C., Bii C., Sbhatu D.B. Challenges of intervention, treatment, and antibiotic resistance of biofilm-forming microorganism. Heliyon. 2019;5:e02192. doi: 10.1016/j.heliyon.2019.e02192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Suller M.T.E., Lloyd D. The antibacterial activity of vancomycin towards Staphylococcus aureus under aerobic and anaerobic conditions. J. Appl. Microbiol. 2002;92:866–872. doi: 10.1046/j.1365-2672.2002.01594.x. [DOI] [PubMed] [Google Scholar]
- 97.González J.F., Hahn M.M., Gunn J.S. Chronic biofilm-based infections: Skewing of the immune response. Pathog. Dis. 2018;76:fty023. doi: 10.1093/femspd/fty023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jefferson K.K. What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 2004;236:163–173. doi: 10.1111/j.1574-6968.2004.tb09643.x. [DOI] [PubMed] [Google Scholar]
- 99.Castillo-Juárez I., Maeda T., Mandujano-Tinoco E.A., Tomás M., Pérez-Eretza B., García-Contreras S.J., Wood T.K., García-Contreras R. Role of quorum sensing in bacterial infections. World J. Clin. Cases. 2015;3:575–598. doi: 10.12998/wjcc.v3.i7.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hirschfeld J. Dynamic interactions of neutrophils and biofilms. J. Oral Microbiol. 2014;6:26102. doi: 10.3402/jom.v6.26102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wolska K.I., Grudniak A.M., Rudnicka Z., Markowska K. Genetic control of bacterial biofilms. J. Appl. Genet. 2016;57:225–238. doi: 10.1007/s13353-015-0309-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Moser C., Pedersen H.T., Lerche C.J., Kolpen M., Line L., Thomsen K., Høiby N., Jensen P.O. Biofilms and host response—Helpful or harmful. APMIS. 2017;125:320–338. doi: 10.1111/apm.12674. [DOI] [PubMed] [Google Scholar]
- 103.Clinton A., Carter T. Chronic Wound Biofilms: Pathogenesis and Potential Therapies. Lab. Med. 2015;46:277–284. doi: 10.1309/LMBNSWKUI4JPN7SO. [DOI] [PubMed] [Google Scholar]
- 104.Lipsky B.A., Senneville E., Abbas Z.G., Aragón-Sánchez J., Diggle M., Embil J.M., Kono S., Lavery L.A., Malone M., van Asten S.A., et al. Guideline on the diagnostic and treatment of foot infection in persons with diabetes (IWGDF 2019 update) Diabetes Metab. Res. Rev. 2020;36:e3280. doi: 10.1002/dmrr.3280. [DOI] [PubMed] [Google Scholar]
- 105.Wolcott R.D., Kennedy J.P., Dowd S.E. Regular debridement is the main tool for maintaining a healthy wound bed in most chronic wounds. J. Wound Care. 2009;18:54–56. doi: 10.12968/jowc.2009.18.2.38743. [DOI] [PubMed] [Google Scholar]
- 106.Lázaro-Martinez J.L., Alvaro-Afonso F.J., Garcia-Alvarez Y., Molines-Barroso R.J., García-Morales E., Sevillano-Fernández D. Ultrasound-assisted debridement of neuroischaemic diabetic foot ulcers, clinical and microbiological effects: A case series. J. Wound Care. 2018;27:278–286. doi: 10.12968/jowc.2018.27.5.278. [DOI] [PubMed] [Google Scholar]
- 107.Raad I.I., Fang X., Keutgen X.M., Jiang Y., Sherertz R., Hachem R. The role of chelators in preventing biofilm formation and catheter-related bloodstream infections. Curr. Opin. Infect. Dis. 2008;21:385–392. doi: 10.1097/QCO.0b013e32830634d8. [DOI] [PubMed] [Google Scholar]
- 108.Cui H., Li W., Li C., Vittayapadung S., Lin L. Liposome containing cinnamon oil with antibacterial activity against methicillin-resistant Staphylococcus aureus biofilm. Biofouling. 2016;32:215–225. doi: 10.1080/08927014.2015.1134516. [DOI] [PubMed] [Google Scholar]
- 109.Kim S.G., Yoon Y.H., Choi J.W., Rha K.S., Park Y.H. Effect of furanone on experimentally induced Pseudomonas aeruginosa biofilm formation: In vitro study. Int. J. Pediatr. Otorhinolaryngol. 2012;76:1575–1578. doi: 10.1016/j.ijporl.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 110.Sully E.K., Malachowa N., Elmore B.O., Alexander S.M., Femling J.K., Gray B.M., DeLeo F.R., Otto M., Cheung A.L., Edwards B.S., et al. Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog. 2014;10:e1004174. doi: 10.1371/journal.ppat.1004174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Coraça-Huber D.C., Dichtl S., Steixner S., Nogler M., Weiss G. Iron chelation destabilizes bacterial biofilms and potentiates the antimicrobial activity of antibiotics against coagulase-negative Staphylococci. Pathog. Dis. 2018;76:fty052. doi: 10.1093/femspd/fty052. [DOI] [PubMed] [Google Scholar]
- 112.Kalpana B.J., Aarthy S., Pandian S.K. Antibiofilm activity of α-amylase from Bacillus subtilis S8–18 against biofilm forming human bacterial pathogens. Appl. Biochem. Biotechnol. 2012;167:1778–1794. doi: 10.1007/s12010-011-9526-2. [DOI] [PubMed] [Google Scholar]
- 113.Rogers S.A., Huigens R.W., III, Cavanagh J., Melander C. Synergistic effects between conventional antibiotics and 2-aminoimidazole-derived antibiofilm agents. Antimicrob. Agents Chemother. 2010;54:2112–2118. doi: 10.1128/AAC.01418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chung P.Y., Toh Y.S. Anti-biofilm agents: Recent breakthrough against multi-drug resistant Staphylococcus aureus. Pathog. Dis. 2014;70:231–239. doi: 10.1111/2049-632X.12141. [DOI] [PubMed] [Google Scholar]
- 115.Tardivo J.P., Adami F., Correa J.A., Pinhal M.A.S., Baptista M.S. A clinical trial testing the efficacy of PDT in preventing amputation in diabetic patients. Photodiagnosis Photodyn. Ther. 2014;11:342–350. doi: 10.1016/j.pdpdt.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 116.Taha O.A., Connerton P.L., Connerton I.F., El-Shibiny A. Bacteriophage ZCKP1: A potential treatment for Klebsiella pneumoniae isolated from diabetic foot patients. Front. Microbiol. 2018;9:2127. doi: 10.3389/fmicb.2018.02127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fish R., Kutter E., Bryan D., Wheat G., Kuhl S. Resolving digital staphylococcal osteomyelitis using bacteriophage—A case report. Antibiotics. 2018;7:87. doi: 10.3390/antibiotics7040087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ahmadi M., Adibhesami M. The effect of silver nanoparticles on wounds contaminated with Pseudomonas aeruginosa in mice: An experimental study. Iran. J. Pharm. Res. 2017;16:661–669. [PMC free article] [PubMed] [Google Scholar]
- 119.Thombare N., Jha U., Mishra S., Siddiqui M.Z. Guar gum as a promising starting material for diverse applications: A review. Int. J. Biol. Macromol. 2016;88:361–372. doi: 10.1016/j.ijbiomac.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 120.Cirioni O., Giacometti A., Ghiselli R., Kamysz W., Orlando F., Mocchegiani F., Silvestri C., Licci A., Chiodi L., Lukasiak J., et al. Citropin 1.1-treated central venous catheters improve the efficacy of hydrophobic antibiotics in the treatment of experimental staphylococcal catheter-related infection. Peptides. 2006;27:1210–1216. doi: 10.1016/j.peptides.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 121.Dutta P., Das S. Mammalian antimicrobial peptides: Promising therapeutic targets against infection and chronic inflammation. Curr. Top. Med. Chem. 2016;16:99–129. doi: 10.2174/1568026615666150703121819. [DOI] [PubMed] [Google Scholar]
- 122.Bilyayeva O.O., Neshta V.V., Golub A.A., Sams-Dodd F. Comparative clinical study of the wound healing effects of a novel micropore particle technology: Effects on wounds, venous leg ulcers, and diabetic foot ulcers. Wounds. 2017;29:1–9. [PubMed] [Google Scholar]
- 123.Minden-Bjrkenmajer B., Bowlin G. Honey-based templates in wound healing and tissue engineering. Bioengineering. 2018;5:46. doi: 10.3390/bioengineering5020046. [DOI] [PMC free article] [PubMed] [Google Scholar]