Abstract
Background
The WRKY gene family, one of the major transcription factor families in plants, plays crucial regulatory roles in physiological and biological developmental processes, and the adaptation of plants to the environment. However, the systematic study of WRKY structure, expression profiling, and regulatory functions has not been extensively reported in Lycium ruthenicum, although these aspects have been comprehensively studied in most plant species.
Methods
In this study, the WRKY genes were identified from a L. ruthenicum transcriptome database by using bioinformatics. The identification, phylogenetic analysis, zinc-finger structures, and conserved motif prediction were extensively explored. Moreover, the expression levels of 23 selected genes with fragments per kilobase of exons per million mapped reads (FPKM) >5 were assayed during different fruit developmental stages with real-time quantitative polymerase chain reaction (RT-qPCR).
Results
A total of 73 putative WRKY proteins in the L. ruthenicum transcriptome database were identified and examined. Forty-four proteins with the WRKY domain were identified and divided into three major groups with several subgroups, in accordance with those in other plant species. All 44 LrWRKY proteins contained one or two conserved WRKY domains and a zinc-finger structure. Conserved motif prediction revealed conservation of the WRKY DNA-binding domain in L. ruthenicum proteins. The selected LrWRKY genes exhibited discrete expression patterns during different fruit developmental stages. Interestingly, five LrWRKYs (-20, -21, -28, -30, and -31) were expressed remarkably throughout the fruit developmental stages.
Discussion
Our results reveal the characteristics of the LrWRKY gene family, thus laying a foundation for further functional analysis of the WRKY family in L. ruthenicum.
Keywords: Lycium ruthenicum, WRKY gene, Fruit developmental stages, Gene expression, Conserved motif, Transcriptome data
Introduction
WRKY transcription factors (TFs) are the 7th largest family of regulatory genes screened from higher plants (Marchive et al., 2007; Chen et al., 2018a; Chen et al., 2018b). The first cDNA encoding a WRKY protein, SPF1, was cloned from Ipomoea batatas (Ulker & Somssich, 2004; Ling et al., 2011). Since then, several members of the WRKY proteins have been identified from several plant species (Liu & Ekramoddoullah, 2009; Liu et al., 2012). To date, only two WRKY homologs in non-plant species have been described, in Giardia lamblia and Dictyostelium discoideum (Wu et al., 2005). Notably, 72 WRKY members in Arabidopsis thaliana (Dong, Chen & Chen, 2003), 182 in Glycine max (Bencke-Malato et al., 2014), 59 in Fragaria vesca (Zhou et al., 2016), 127 in Malus domestica (Meng et al., 2016), 55 in Cucumis sativus (Ling et al., 2011), 80 in Vitis vinifera (Zhang & Feng, 2014), 86 in Brachypodium distachyon (Wen et al., 2014), 85 in Manihot esculenta (Wei et al., 2016), and 95 in Daucus carota (Li et al., 2016) have been identified. The name of the WRKY family is derived from the most common characteristic of these proteins, the WRKY domain, which is characterized by approximately 60 amino acid residues that often exhibit sequence-specific DNA-binding (Chen et al., 2018a; Chen et al., 2018b; Zhou et al., 2016). In the WRKY domain, the highly conserved heptapeptide sequence WRKYGQK is followed by a CX4−5 X4-CX22−23-HXH (C2H2) or CX7-CX23X23-HXC (C2HC-) type zinc-finger motif (Eulgem et al., 2000). The conserved WRKY domain may have a longer sequence, for example, WRKYGKK or WEKYGQK. It may also be replaced by WRRY, WKKY, WIKY, WSKY, WKRY, WRIC, WVKY, or WRMC. The conserved WRKY domain helps in binding of the protein to W-box elements or sugar-responsive cis-elements (SURE) in the promoter regions of target genes (Sun et al., 2003; Rushton et al., 2010). Depending on the number of WRKY domains and the zinc-finger motif, the WRKY proteins can be categorized into three main groups (1, 2, and 3) (Eulgem et al., 2000). Group 1 proteins have two WRKY domains: an N-terminal WRKY domain and a C-terminal WRKY domain, followed by a C2H2 zinc-finger motif (CX4 X4-CX22−23HXH). Group 2 proteins have only one WRKY domain and a C2H2 zinc-finger motif (CX4−5 X4-CX23-HXH); this group is further divided into five subgroups (2a to 2e) according to the phylogeny of the WRKY domain. Group 3 proteins also have a single WRKY domain and a C2HC zinc-finger motif (CX7-CX23 X23-HXC) (Eulgem et al., 2000; Rushton et al., 2010).
The WRKY gene family has recently gained attention for its involvement in a wide range of biological and physiological developmental processes in plants, including diverse biotic/abiotic stress responses (Li et al., 2016; Jiang et al., 2017; Yang et al., 2016). The WRKY gene family also plays a crucial role in the developmental and ripening processes of fruits; for example, FvWRKY4, -46, and -48 in group 2c are involved in fruit development and ripening in F. vesca (Zhou et al., 2016). In Ziziphus jujube ZjWRKY8, -26, -47, and -48 are involved in developmental process in younger fruits (Xue et al., 2019). In addition to the involvement in seed or fruit development, the WRKY genes also control seed pigmentation. AtWRKY44 (transparent testa glabra, TTG2) controls the epidermal color of Arabidopsis seeds by contributing to transcriptional regulation. It directly binds the upstream regulatory region of TT12. TTG1, TT2, and TT8 are involved in the biosynthesis of proanthocyanidins and pigmentation in Arabidopsis (Gonzalez et al., 2016). Moreover, OsWRKY11 in O. sativa is involved in controlling flower development (Cai et al., 2014). The roles of WRKYs in plant responses to abiotic stresses such as drought, cold, salinity, heat, ultraviolet B (UV-B), osmotic stress, and ABA have been extensively documented (Luo et al., 2013; Jiang, Liang & Yu, 2012; Ren et al., 2010; Tao et al., 2011; Li et al., 2010; Liu & Guan, 2012).
Lycium ruthenicum Murr. is a typical native halophyte and perennial shrub distributed across arid and semi-arid regions of Asia, America, and Africa (Liu et al., 2012; Dai, Wang & Zhang, 2016). Its excellent physiological features conferring drought resistance, coupled with its salt tolerance, make it a relevant plant for land desertification control and soil salinity improvement (alkalinity) (Zheng et al., 2011). Ripe L. ruthenicum fruits are considered the highest and richest wild source of anthocyanins and are unique from a pharmaceutical point of view (Chen et al., 2013). For instance, the fruits are used to treat health conditions, including early-onset diabetes, anemia, vision problems, impotency, and lung disorders. Recently evidence indicates that these fruits can improve liver and kidney function, boost the immune system, and promote longevity (Zheng et al., 2011; Dhar et al., 2011). Consequently, worldwide economic interest in the production of L. ruthenicum has recently grown. L. ruthenicum is not only an economically and ecologically important plant but could also be a model organism for research. The WRKY gene family has been comprehensively studied in many plant species (Jiang, Liang & Yu, 2012; Ren et al., 2010; Tao et al., 2011; Liu & Guan, 2012), but no previous studies have described the WRKY genes from L. ruthenicum. Because of the importance of the WRKY genes in various physiological and biological plant processes, systematic study of the identification, classification, expression, and functions of WRKY family in the fruit development of L. ruthenicum is of interest.
In the present study, we identified a total of 73 potential LrWRKY members in L. ruthenicum according to a transcriptome database. Forty-four LrWRKYs identified with WRKY domains were selected, and the identification, classification, phylogenetic analysis, conserved motif determination, and gene expression profiling were extensively examined. The expression levels of WRKY genes in different fruit developmental stages were further validated using RT-qPCR. This study reveals clues to aid in the functional characterization of LrWRKY genes and provides a basis for further genomic studies in L. ruthenicum.
Materials & Methods
Sequence database search and identification of the LrWRKY family in L. ruthenicum
A. thaliana and O. sativa WRKY protein sequences were obtained from The Arabidopsis Information Resource (TAIR) database (http://www.arabidopsis.org/) and the Joint Genome Institute (JGI) database (https://phytozome.jgi.doe.gov) separately and were used for comparative analysis. To obtain a comprehensive list of LrWRKY proteins, we obtained transcriptome data from the public Sequence Read Achieve database of NCBI (https://www.ncbi.nlm.nih.gov/sra) under accession number SRR7700825 (Ma et al., 2018). To obtain the transcriptome sequence encoding WRKY proteins, we performed a Blastx search in the NCBI database. A. thaliana WRKY domain sequences were used as query sequences, and Blast default settings were used. The nucleotide sequences of the possible LrWRKYs were translated into amino acid sequences in DNAMAN 8.0 (LynnonBioSoft, Quebec, Canada) software, to examine WRKY proteins encoded by the obtained transcriptome sequences (Data S1). The Protein families database (Pfam; http://pfam.xfam.org/) was used to verify whether the putative proteins had the distinctive features of WRKY proteins. Additionally, manual examination of the conserved WRKYGQK amino acid structure at the N-terminus and the zinc-finger structure at the C-terminus of the putative WRKY domain was conducted for further confirmation (Zhang & Wang, 2005).
Multiple sequence alignment, phylogenetic analysis, and conserved motif construction
All 44 LrWRKY proteins with WRKY domains and several selected AtWRKY proteins (AtWRKY6, -14, -25, -26, -31, -40, and -51) were used to generate multiple protein sequence alignments by using the default settings in ClustalW (Larkin et al., 2007). The 44 LrWRKY proteins with WRKY proteins from two model plant species, A. thaliana (72) and O. sativa (73) (Data S2), were used for phylogenetic tree construction in MEGA 5.0 software (Tamura et al., 2011) with the maximum likelihood method and 1000 bootstrap replications. The conserved motifs in LrWRKY proteins were determined with Multiple Expectation Maximization for Motif Elicitation (MEME) suite version 5.1.1 (http://meme-suite.org/doc/cite.html), and five motifs were searched.
Plant materials
L. ruthenicum fruit samples were collected from Shizuishan in Ningxia, China (38°56.799′ N, 106°24.711′ E, 1088 H) in three developmental stages, Rs1 (green ripeness stage), Rs2 (veraison ripeness stage), and Rs3 (complete ripeness stage), on the basis of the descriptions of Ma et al. (2018) and Guillaumie et al. (2011). Biological replicates of the material comprised three shrubs in each developmental stage. The fruit samples from the three shrubs were collected, immediately frozen in liquid nitrogen, and stored at −80 °C until total RNA isolation.
RNA extraction, cDNA synthesis, and RT-qPCR analysis
We extracted total RNA from nine samples (three biological replicates per treatment) by using a TransZol Up Plus RNA Kit (Lot# M31018) conferring to the manufacturer’s instructions. The RNA quantity and quality were determined with a TGen Spectrophotometer (TianGen), on the basis of the A260 nm/A280 nm and A260 nm/A230 nm ratios. An Evo M-MLV RT Kit AG11705 (Accurate Biotechnology) was used to reverse transcribe the total RNA into cDNA and remove any genomic DNA from the cDNA before RT-qPCR analysis, following the manufacturer’s instructions.
For the RT-qPCR analysis, primers were designed on the basis of the RNA sequences obtained from the NCBI SRA (Ma et al., 2018) in Primer 5 software and synthesized by TsingKe Biological Technology Co., Ltd. (Xi’an, China) (Data S3). To better understand the LrWRKY gene expression during different fruit developmental stages, we selected 23 genes with FPKM values >5 from the 44 genes with WRKY domains for RT-qPCR analysis. The LrEF1-a gene (JX427553) was used as a reference gene. We examined three biological replicates and performed triplicate quantitative analyses; each replicate was analyzed on the basis of 0.5 µl of each cDNA dilution with a Heiff® q-PCR SYBR® Green Master Mix kit (Yeasen Biotech Co., Ltd) according to the manufacturer’s protocol. The RT-qPCR analysis was performed with a QuantStudio™ 5 Real-Time PCR Instrument (ABI) (Data S4). The LrWRKY gene relative expression was calculated according to Livak & Schmittgen (2001).
A heatmap of all 44 LrWRKY gene expression patterns was constructed during fruit development and ripening stages, on the basis of FPKM values (Data S5) in TBtools software (https://github.com/CJ-Chen/TBtools) (Chen et al., 2018a; Chen et al., 2018b).
Data analysis
All reported gene expression levels are presented as means ± SE (n = 3). The significance level among means was analyzed with Duncan’s multiple range tests (p ≤ 0.05) after one-way ANOVA analysis in SPSS statistical software (Ver. 22.0, SPSS Inc., Chicago, IL, USA).
Results
Identification of LrWRKY genes
We searched 43,573 unigenes for all possible LrWRKY genes from the transcriptome data of L. ruthenicum and obtained a total of 73 best hits, which were validated by BLAST via the NCBI database. We use the prefix “Lr” for L. ruthenicum and assigned the WRKY numbering according to the order of the annotated sequences from the database, as CL10207Contig1 (LrWRKY1) to CL8621Contig1 (LrWRKY73), respectively (Data S6). The lengths of members of the WRKY family in L. ruthenicum ranged from 5207 aa (LrWRKY3) to 485 aa (LrWRKY67). The open reading frames (ORF) of all members could not be predicted, due to the limitations of the sequencing technique.
Phylogenetic analysis of LrWRKY genes
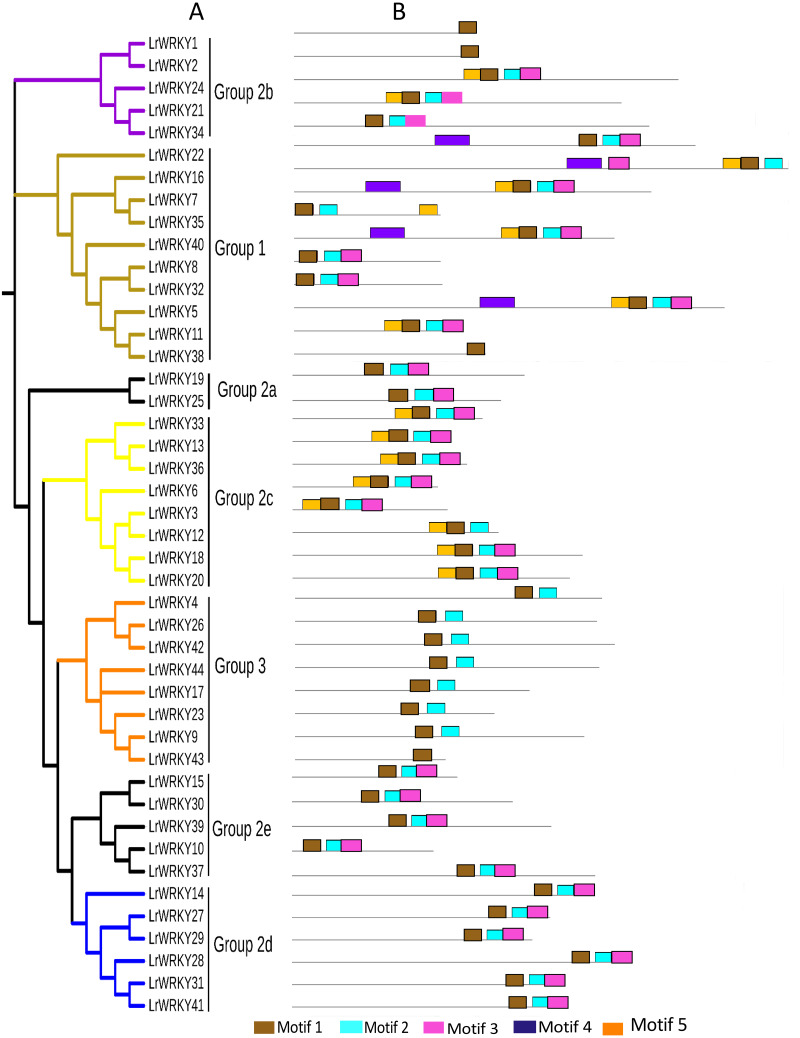
Phylogenetic analysis of plant WRKY genes is an effective way to understand the functions of uncharacterized WRKY members according to their evolutionary history and sequence similarity. Therefore, the 44 LrWRKYs identified to have WRKY domains were used to construct a phylogenetic tree. The phylogenetic analysis revealed that the LrWRKY proteins could be categorized into three main groups corresponding to groups 1, 2, and 3 in A. thaliana (Eulgem et al., 2000). Among the 44 LrWRKY proteins, 10 were classified into group 1 (representing 23%) consisting of N-terminal and C-terminal subgroups, 26 LrWRKY proteins were assigned to group 2 (representing 59%), and the remaining 8 LrWRKY proteins were classified in group 3 (representing 18%) (Fig. 1). Group 1 WRKY family members usually have two WRKY domains and a C2H2-type zinc-finger motif (CX4X4-CX22−23-HXH), but few LrWRKYs from group 1 contained only one WRKY domain (LrWRKY8, -11, -32, -35, and -38). LrWRKY proteins in group 2 had a single WRKY domain and a C2H2-type zinc-finger motif (CX4−5 X4-CX23-HXH), which was further subdivided into five distinct subgroups (2a to 2e): two LrWRKYs belonged to 2a, four LrWRKYs belonged to group 2b, eight belonged to group 2c, six belonged to group 2d, and five belonged to group 2e, according to their phylogeny. LrWRKYs from group 3 were characterized by a C2HC-type zinc-finger motif (CX7-CX23 X23-HXC) (Table 1).
Figure 1. Phylogenetic tree according to WRKY domain sequences from L. ruthenicum, O. sativa and A. thaliana.
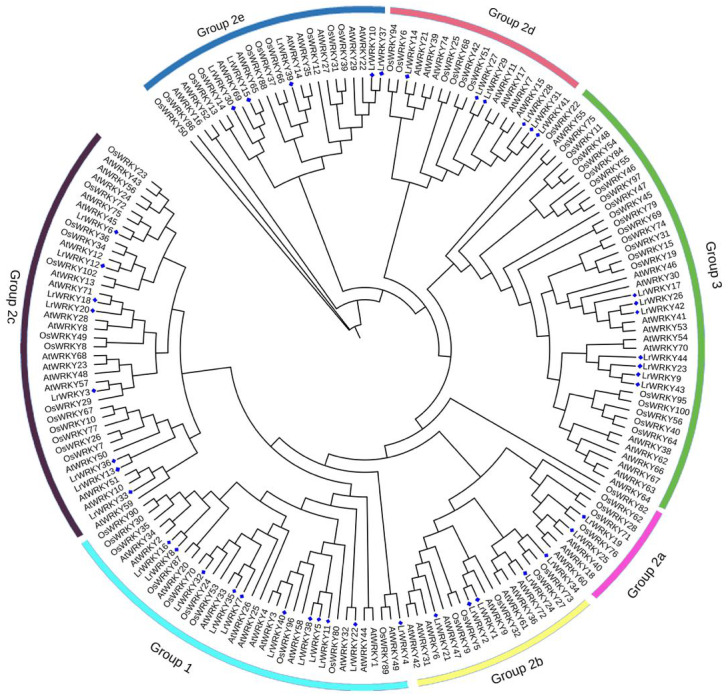
Consistency of the predicted tree was tested using the Maximum Likelihood method and bootstrapping with 1,000 replicates in MEGA 5. The different colored arcs indicate different groups or subgroups and the blue diamonds represent LrWRKYs.
Table 1. LrWRKYs grouping, zinc-finger structure and domain pattern.
| Gene name | Group classification | Zinc finger | Domain pattern | Gene name | Group classification | Zinc finger | Domain pattern |
|---|---|---|---|---|---|---|---|
| LrWRKY1 | 2b | C2H2 | N | LrWRKY23 | 3 | C2HC | CX7- CX23X23- HXC |
| LrWRKY2 | 2b | C2H2 | N | LrWRKY24 | 2b | C2H2 | CX5- CX23- HXH |
| LrWRKY3 | 2c | C2H2 | CX4- CX23- HXH | LrWRKY25 | 2a | C2H2 | CX5- CX23- HXH |
| LrWRKY4 | 3 | C2H2 | CX7- CX23X23- HXC | LrWRKY26 | 3 | C2HC | CX7- CX23X23- HXC |
| LrWRKY5 | 1 | C2H2 | CX4X4- CX22−23- HXH | LrWRKY27 | 2d | C2H2 | CX5- CX23- HXH |
| LrWRKY6 | 2c | C2H2 | CX4- CX23-HXH | LrWRKY28 | 2d | C2H2 | CX5- CX23- HXH |
| LrWRKY7 | 1 | C2H2 | CX4X4- CX22−23- HXH | LrWRKY29 | 2d | C2H2 | CX5- CX23- HXH |
| LrWRKY8 | 1 | C2H2 | CX4X4- CX22−23- HXH | LrWRKY30 | 2e | C2H2 | CX5- CX23- HXH |
| LrWRKY9 | 3 | C2HC | CX7- CX23X23- HXC | LrWRKY31 | 2d | C2H2 | CX5- CX23- HXH |
| LrWRKY10 | 2e | C2H2 | CX5X4- CX23- HXH | LrWRKY32 | 1 | C2H2 | CX4- CX22−23- HXH |
| LrWRKY11 | 1 | C2H2 | CX4X4- CX22−23- HXH | LrWRKY33 | 2c | C2H2 | CX4- CX23- HXH |
| LrWRKY12 | 2c | C2H2 | CX4- CX23- HXH | LrWRKY34 | 2b | C2H2 | CX5- CX23- HXH |
| LrWRKY13 | 2c | C2H2 | CX4- CX23- HXH | LrWRKY35 | 1 | C2H2 | CX4- CX22−23HXH |
| LrWRKY14 | 2d | C2H2 | CX5X4- CX23- HXH | LrWRKY36 | 2c | C2H2 | CX4- CX23-HXH |
| LrWRKY15 | 2e | C2H2 | CX5X4- CX23- HXH | LrWRKY37 | 2e | C2H2 | CX5- CX23- HXH |
| LrWRKY16 | 1 | C2H2 | CX4X4- CX22−23- HXH X22−23 | LrWRKY38 | 1 | C2H2 | N |
| LrWRKY17 | 3 | C2HC | CX7- CX23X23- HXC | LrWRKY39 | 2e | C2H2 | CX5- CX23-HXH |
| LrWRKY18 | 2c | C2H2 | CX4- CX23- HXH | LrWRKY40 | 1 | C2H2 | CX4- CX22−23- HXH |
| LrWRKY19 | 2a | C2H2 | CX5X4- CX23- HXH | LrWRKY41 | 2d | C2H2 | CX5- CX23- HXH |
| LrWRKY20 | 2c | C2H2 | CX4- CX23- HXH | LrWRKY42 | 3 | C2HC | CX7- CX23X23- HXC |
| LrWRKY21 | 2b | C2H2 | CX5X4- CX23- HXH | LrWRKY43 | 3 | C2HC | CX7- CX23X23- HXC |
| LrWRKY22 | 1 | C2H2 | CX4X4- CX22−23- HXH | LrWRKY44 | 3 | C2HC | CX7- CX23X23- HXC |
Notes.
N, missen domain pattern.
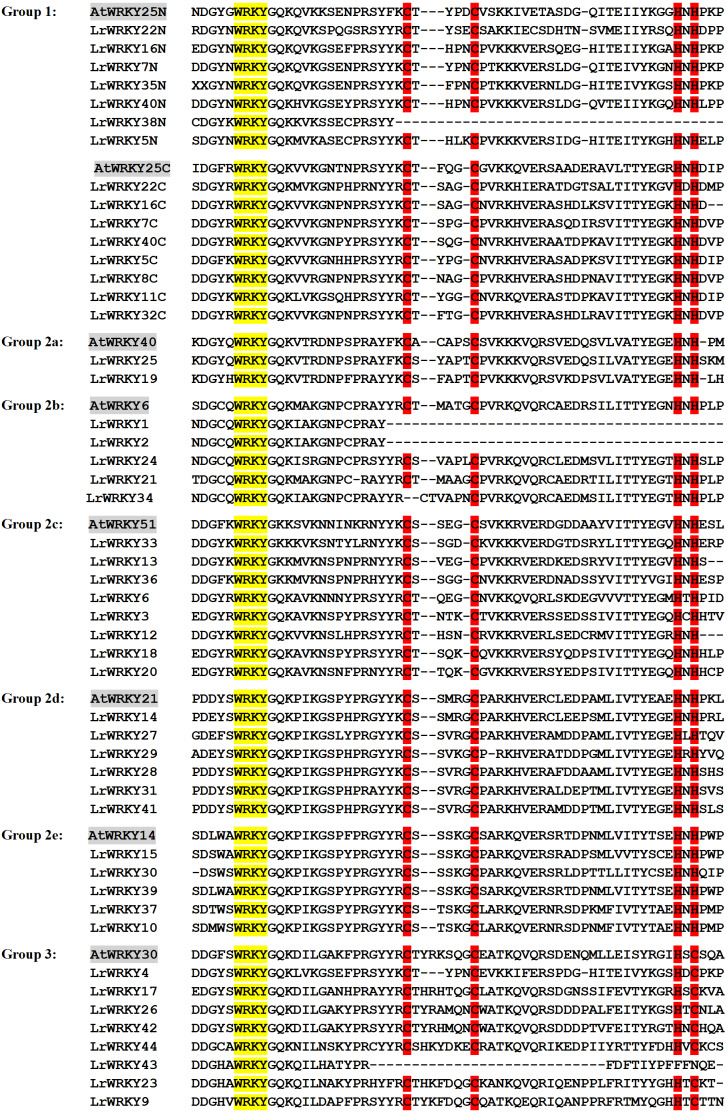
Multiple sequence alignment of LrWRKY genes
The evolutionary relationship of the LrWRKY genes was analyzed through multiple sequence alignment of their conserved WRKY domains. For examination of the core domains, WRKY domains of AtWRKY proteins were selected (AtWRKY6, −14, −21, −25, −30, −40, and −51) from each group or subgroup as representatives for further comparison. The 44 LrWRKY proteins contained the highly conserved WRKY domains, and 41 were discovered to have the conserved heptapeptide sequence of WRKYGQK, whereas three members of group 2c (LrWRKY13, −33 and −36) differed by one amino acid, with the glutamine (Q) substituted by a lysine (K), thus forming WRKYGKK (Fig. 2). For group 1 members, the two WRKY domains, which were denoted group 1 N-terminal and group 1 C-terminal, clustered into two separate groups because of divergence in their sequences. We discovered a CX4-CX22-HXH zinc-finger structure in the N-terminal subgroup and a CX4-CX23-HXH structure in the C-terminal subgroup.
Figure 2. Multiple sequence alignment of the 44 LrWRKY proteins and selected A. thaliana proteins. ‘N’ and ‘C’ indicate the N-terminal and C-terminal WRKY domains.
The conserved WRKY amino acid signature and the amino acids forming the zinc-finger motifs are highlighted in yellow and red, missing amino acids are represented by dashes.
Conserved motif analysis of LrWRKYs proteins
The conserved motifs of L. ruthenicum were examined with MEME, as described in the methods, to better understand the relationship and diversity of LrWRKY protein motif compositions. The results revealed that motifs 1 and 4 were WRKY DNA-binding motifs that encompassed the conserved heptapeptide WRKYGQK domain. The motif 4 WRKY domain belonged to the N-terminal domain, whereas motif 1 belonged to the C-terminal domain (Fig. 3, Table 2). Interestingly, motif 5 had the largest number of amino acid sequences and was present in groups 1, 2b, and 2c, whereas motif 3 had the smallest number of protein sequences but was present in all groups/subgroups except group 3. Most of the LrWRKY proteins contained at least two motifs. LrWRKY members belonging to the same group were discovered to share analogous motif compositions; for instance, motif 4 was exceptional to group 1 members. Groups 2a, 2e, and 2d contained three motifs (-1, -2, and -3) whereas group 2b and 2c contained four motifs (-1, -2, -3, and -5), except for LrWRKY1 and -2 which contained only motif 1. In addition, LrWRKY12 and -34 belonged to group 2b and 2c but did not contain motif 5. Group 3 contained motifs 1 and -2, except for LrWRKY43, which contained only motif 1. Comparisons between randomly selected AtWRKY members with their LrWRKY orthologs revealed that AtWRKY6, -14, -21, -25, -30, -40, and -51 shared the same motif compositions with their prospective LrWRKY group members (Data S7). Only LrWRKY1, -2, -38, and -43 motif compositions differed from the selected AtWRKY members.
Figure 3. Phylogenetic relations and conserved protein motifs in L. ruthenicum.
(A) The unrooted phylogenetic tree was constructed according to WRKY proteins sequences of L. ruthenicum with 1,000 bootstrap replicates. Details of groups are shown in different colors. (B) The motif composition of LrWRKY protein were identified using MEME. The different colored boxes represent different motifs and their position in each WRKY sequence. Each motif is indicated by a colored box indicated in the key at the bottom.
Table 2. Analysis and conserved motifs determination in LrWRKY proteins.
| Motif | E value | Width | Motif sequence |
|---|---|---|---|
| 1 | 8.9e−762 | 21 | GYSWRKYGQKPVKGSPYPRSY |
| 2 | 2.9e−500 | 21 | CPVRKQVERSSEDPSMLITTY |
| 3 | 5.8e−114 | 11 | EGEHNHPVPAA |
| 4 | 2.0e−112 | 21 | KKVRKPRVAVRTRSEVDILDD |
| 5 | 8.4e−104 | 41 | DGYNWRKYGQKQVKGSEYPRSYYKCTHPNCP VKKKVERSLD |
Expression analysis of LrWRKYs in different developmental stages of fruits
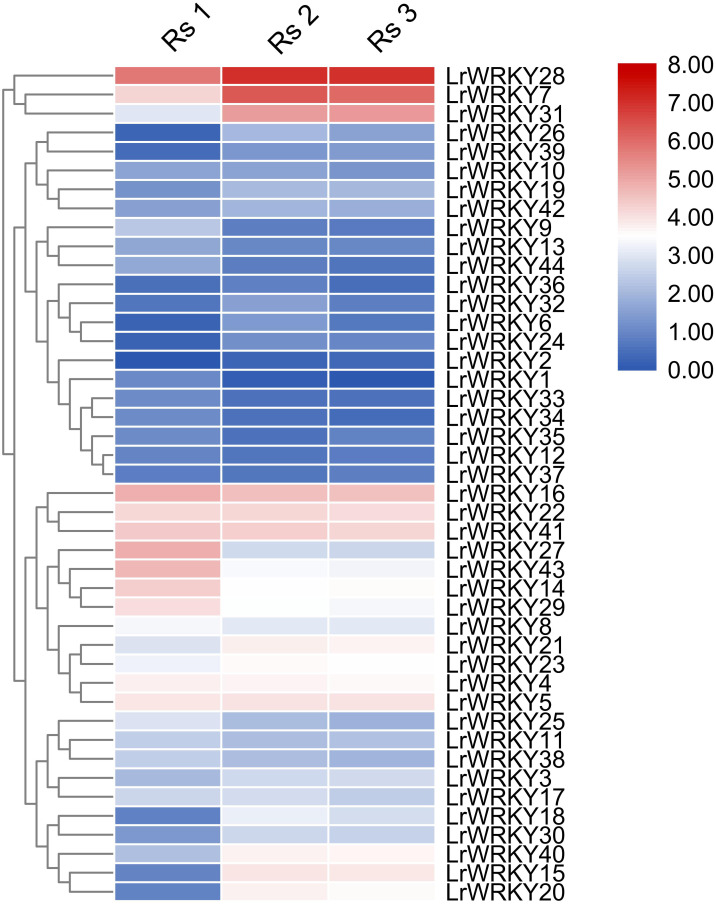
Expression profiles of the 44 LrWRKY genes were evaluated in three different fruit developmental stages (Rs 1, Rs 2, and Rs 3). The heatmap illustration of expression profiles of LrWRKY genes is shown in Fig. 4. The expression profiles revealed that all 44 LrWRKYs exhibited discrete expression during fruit development. Twenty-one LrWRKYs showed insignificant expression in all developmental stages of the fruit, and the FPKM values were <5 in all three stages. The remaining 23 members of LrWRKYs showed relatively higher expression, with FPKM >5 in at least one of the fruit developmental stages. Six LrWRKY genes exhibited markedly high expression patterns across all fruit developmental stages, with FPKM values >20. Interestingly, we also discovered that some WRKY genes classified in the same groups/subgroups had differential expression. For example, group 1, group 2d, LrWRKY3, and -20 from group 2c exhibited substantial expression with high FPKM values. However, group 2b members were weakly expressed in almost all developmental stages. The lowest FPKM value recorded in group 2d was 5.3, whereas some genes had FPKM values as high as 130.1 (Fig. 4, Data S4).
Figure 4. Heatmap representation of the LrWRKY genes during fruit developmental stages of L. ruthenicum.
The expression profiles of the 44 LrWRKY genes were based on FPKM-values. Red and blue boxes indicate high and low expression levels for each gene.
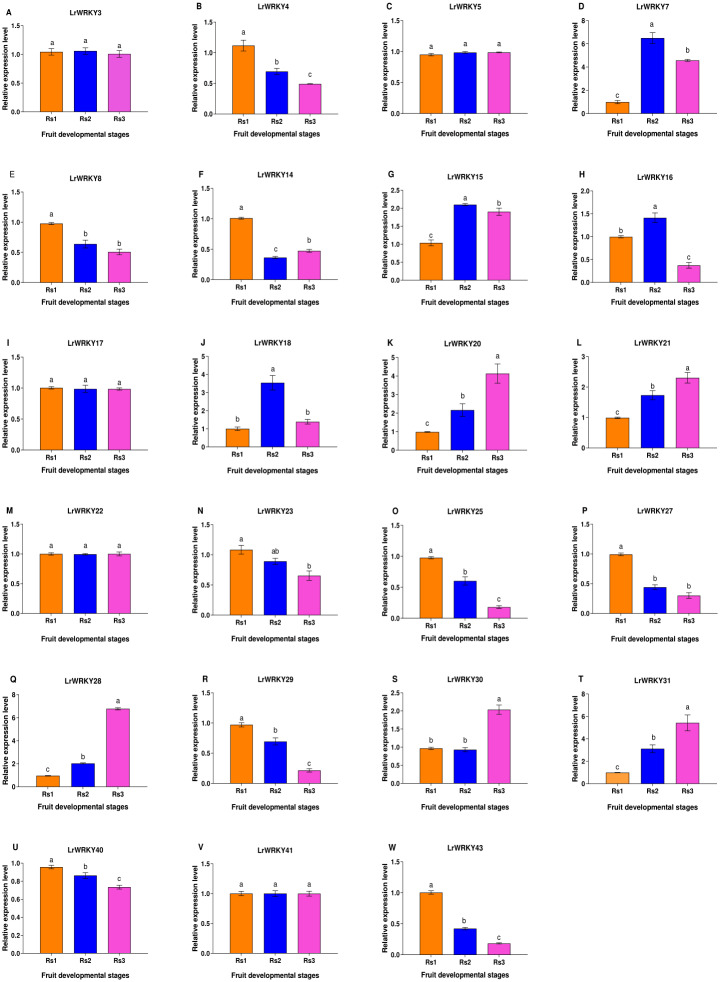
To gain a deeper knowledge of the gene expression patterns and to validate the LrWRKY genes, we performed RT-qPCR analysis of 23 selected genes. LrWRKYs exhibited different expression patterns in Rs 1, Rs 2, and Rs 3 (Fig. 5). Some LrWRKYs members maintained stable expression under different ripening stages, for example, LrWRKY3, -5, -17, -22, and -41. A total of nine LrWRKYs (-4, -8, -14, -23, -25, -27, -29, -40, and -43) decreased significantly from Rs 1 to Rs 3, among which LrWRKY 8, -23, and -27 maintained stable expression in Rs 2 and Rs 3. However, five LrWRKYs (-20, -21, -28, -30, and -31) continuously increased from Rs 1 to Rs 3, except for LrWRKY 30, which maintained stable expression in RS 1 and RS 2. In addition, three LrWRKYs increased first and peaked in RS 2, as compared with RS 1, and then decreased in RS 3.
Figure 5. RT-qPCR validation of twenty-three selected LrWRKYs expression levels during fruit developmental stages.
The Rs1 represent green ripeness stage, Rs2 represent veraison ripeness stage, and Rs3 represent complete ripeness stage. The name of the gene is written on top of each bar diagram; (A) LrWRKY3, (B) LrWRKY4, (C) LrWRKY5, (D) LrWRKY7, (E) LrWRKY8, (F) LrWRKY14, (G) LrWRKY15, (H) LrWRKY16, (I) LrWRKY17, (J) LrWRKY18, (K) LrWRKY20, (L) LrWRKY21, (M) LrWRKY22, (N) LrWRKY23, (O) LrWRKY25, (P) LrWRKY27, (Q) LrWRKY28, (R) LrWRKY29, (S) LrWRKY30, (T) LrWRKY31, (U) LrWRKY40, (V) LrWRKY41, (W) LrWRKY43. The different letters indicate significant differences among developmental stages (p < 0.05).
Discussion
The WRKY gene family is one of the major families of TFs with crucial roles in various plant physiological processes (Huang et al., 2012; Huang et al., 2015; He et al., 2012). Genome-wide analysis, including identification and expression profiling of WRKY genes in V. vinifera (Wang et al., 2014), F. vesca (Zhou et al., 2016), and A. thaliana (Miao et al., 2004), has been broadly investigated. Because of technical limitations, the reference genome database of L. ruthenicum is not available. In this present study, the WRKY family analysis in L. ruthenicum was mainly based on transcriptome data, and a total of 73 possible WRKY gene members were identified. Forty-four LrWRKY genes with the conserved WRKY domain were identified. Our results provided the first systematically identified set of WRKY family members in L. ruthenicum.
After construction of the evolutionary tree and multiple sequence alignment based on L. ruthenicum WRKY conserved domains, we discovered that the 44 LrWRKYs could be categorized into three major groups, as described earlier in A. thaliana by Eulgem et al. (2000). The N-terminal and C-terminal WRKY domains in group 1 were assembled into different groups, thus potentially revealing separate evolution of the two domains. Previous studies have described group 2 WRKY genes as the group with the most members, for example, A. thaliana (Eulgem et al., 2000), B. distachyon (Wen et al., 2014), Caragana intermedia (Wan et al., 2018), and M. esculenta (Wei et al., 2016). Our study also revealed that group 2 LrWRKY genes had the highest proportion of members (59%), thus suggesting the activity of group 2 during the evolutionary processes and indicating that this group might have regulatory functions in fruit development and ripening. Group 3 showed the smallest number of LrWRKY genes (18%) among the three groups, possibly because of transcriptome data limitations or underestimation during our analysis. The WRKY TFs have multiple diverse functions in plant growth and various stress responses, and WRKY genes within the same group or subgroup usually retain similar regulatory functions (Chen et al., 2018a; Chen et al., 2018b). Group 1 WRKY genes have been reported to participate in a wide range of plant growth and development processes. For instance, AtWRKY2, -26, -34, and -44 in group 1 are involved in seed germination, post-germination growth, leaf senescence, and root hair growth in Arabidopsis (Guan et al., 2014; Li et al., 2017; Gonzalez et al., 2016), and they share a similar homologous relationship with their L. ruthenicum counterparts (LrWRKY4, -5, -7, -8, -11 -16, -22, -32, -35, -38, and -40), thus suggesting related developmental roles among the genes. Most of the group 2a subfamily genes are involved in stress signaling, and group 2a genes in L. ruthenicum may share similar functions. Some members of group 2c (AtWRKY10, -28, -57, -75, and OsWRKY72) in A. thaliana and O. sativa have been reported to be involved in salt stress, flowering, seed development, and coloration (Chen et al., 2018a; Chen et al., 2018b); these members have homologous relationships with LrWRKY (-3, -6, -12, -13, -18, -20, -33, and -36) in group 2c. In addition, ZjWRKY26; VvWRKY14, -19, -52; and FvWRKY4, -46, and -48, belonging to group 2c, have been reported to be mainly involved in the developmental process of fruits in Z. jujube (Xue et al., 2019), V. vinifera (Wang et al., 2014), and F. vesca (Zhou et al., 2016) respectively. Similarly, the group 2d subfamily genes are involved in the regulation of growth and development. Among the WRKY groups, group 3 genes are considered to have evolved the most, and they appear to play a crucial role in plant adaptation and development (Tao et al., 2018). In A. thaliana, group 3 members (AtWRKY46, -54, and -70) are involved in brassinosteroid signaling and regulate both osmotic stress and plant development (Chen et al., 2017). Thus, the specific functions of WRKYs might provide clues regarding the same homologs in LrWRKYs for further functional identification.
Nevertheless, the conserved heptapeptide WRKY domain WRKYGQK is the most common characteristic of WRKY proteins, although this sequence is sometimes substituted by WRKYGKK through mutation (Rushton et al., 2010). Our analysis uncovered substitution of some WRKYGQK sequences (LrWRKY13, -33, and -36) in group 2c with a WRKYGKK sequence, a result similar to findings describing the substitution of glutamine (Q) by lysine (K) in A. thaliana (AtWRKY51, -52, and -59) (Eulgem et al., 2000), C. intermedia (CiWRKY2, -41, -50, and -51) (Wan et al., 2018), and Panicum miliaceum (PmWRKY2, -15, -23, -24, and -28) (Yue et al., 2016). According to Van Verk et al. (2008), the deviation of the WRKYGQK motif might affect DNA binding activity; therefore, LrWRKY13 -33, and -36 genes are recommended for further investigation in studies of their functions and binding specificities. This study also established that the C2HC zinc-finger was present in only group 3 WRKY domains, whereas C2H2 was found in group 1 and group 2 domains; this result confirmed similar reports in A. comosus (Tao et al., 2011), C. sativus (Ling et al., 2011), and A. thaliana (Eulgem et al., 2000). However, this finding differs from those from an earlier report in O. sativa (Wu et al., 2005), indicating that the zinc-finger structure C2HC and C2H2 concomitantly present in the N-terminus in group 1N. Group 1 proteins usually contain two conserved WRKY domains, but LrWRKY8, -11, -32, -35, and -38 group 1 proteins with a single conserved WRKY domain. This finding may be a result of the limitations of the transcriptome data.
The analysis of motif composition revealed that LrWRKYs share a similar motif composition within the same group or subgroup. Of note, AtWRKY6 -14 -21 -25 -30 -40, and -51 share similar motifs (Data S7) with their L. ruthenicum WRKY orthologs, thus presenting clues regarding their regulatory functions. Although the functions of the major motifs in LrWRKYs remain undefined, LrWRKYs with the same conserved motifs may have similar functions. The presence of all five motifs identified in LrWRKYs confirms their conserved nature in this study.
Many studies have shown that WRKY function in regulating plant growth and development, while also playing essential roles in plant adaptability to stress (Chen et al., 2018a; Chen et al., 2018b). L. ruthenicum is widely useful as an economically important shrub with fruits rich in anthocyanins. Because of the potential roles of WRKY genes in the developmental stages of fruit in L. ruthenicum, we analyzed the expression patterns of LrWRKY genes, on the basis of the available transcriptome data, and performed RT-qPCR validation. We identified 44 LrWRKYs showing different expression patterns in the heatmap and inferred their different roles in controlling fruit development. Some LrWRKY genes, for instance, LrWRKYs 3 -5 -17 -22, and -41, maintained stable expression under different developmental stages. Additionally, the expression of LrWRKYs -4 -8 -14 -23 -25 -27 -29 -40, and -43 decreased significantly during fruit development, similarly to observations in RcWRKY21 and -32 in Ricinus communis (Li et al., 2012). Moreover, LrWRKYs -20 -21 -28 -30, and -31 showed continuous significant increases in expression from green ripening to complete ripening during fruit development. Comparatively, AtWRKY10 and -44, and OsWRKY7 (in A. thaliana and O. sativa, respectively) are also highly expressed during seed/fruit development, biosynthesis of proanthocyanidins, and seed pigmentation (Zhang & Feng, 2014; Gonzalez et al., 2016; Luo et al., 2005), thus implicating similar functions in L. ruthenicum. In addition, Z. jujube WRKY8 -26 -47, and -48 and F. vesca WRKY4 -46, and -48 are expressed significantly in fruits, and their expression increases continuously from younger fruits to mature fruits (Zhou et al., 2016; Wang et al., 2014). Finally, four LrWRKYs (-7 -15 -16, and -18) increased first in Rs2 and then decreased in Rs3; likewise, in seed development in Jatropha curcas, three genes (JcWRKY14 -22, and -30) are also expressed at higher levels in the maturation stage (S2) than in the early development stage (Xiong et al., 2013), thus suggesting a putative role in regulating fruit development in L. ruthenicum. Nevertheless, more in-depth study is necessary to comprehensively verify the functions of all the LrWRKY genes.
Conclusions
In the present study, a total of 73 WRKY members were identified in L. ruthenicum according to the transcriptome data. Forty-four proteins were identified with the WRKY domain and, on the basis of their amino acid sequences, were divided into three major groups with several subgroups, in accordance with those in other plant species. All 44 LrWRKY proteins contained one or two conserved WRKY domains and a zinc-finger structure. The conserved motif prediction revealed that the WRKY DNA-binding domain has been conserved in L. ruthenicum proteins. Phylogenetic and gene expression analysis provided in-depth knowledge of the important functions of WRKYs during fruit development and ripening. These results may serve as a foundation enabling further genomic and functional analysis of the WRKY family in L. ruthenicum.
Supplemental Information
Funding Statement
This research was funded by the National Natural Science Foundation of China (No. 31760244, 31700338), the Fundamental Research Funds for the Central Public-interest Scientific Institution (No.1610322019012), the Innovation Project of Chinese Academy of Agricultural Sciences (No. CAAS-XTCX2016011-02), the Science and technology innovation project of Gansu Agricultural University (No. GAU-XKJS-2018-112), and Gansu Provincial Science and Technology Major Projects (No. 19ZD2NA002). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Huirong Duan, Email: duanhuirong@caas.cn.
Yanjun Ma, Email: mayanjun@gsau.edu.cn.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Richard John Tiika and Huirong Duan conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Jia Wei conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Rui Ma performed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Hongshan Yang performed the experiments, prepared figures and/or tables, and approved the final draft.
Guangxin Cui conceived and designed the experiments, analyzed the data, prepared figures and/or tables, and approved the final draft.
Yanjun Ma conceived and designed the experiments, analyzed the data, authored or reviewed drafts of the paper, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
Raw data is available in the Supplemental Files.
References
- Bencke-Malato et al. (2014).Bencke-Malato M, Cabreira C, Wiebke-Strohm B, Bucker-Neto L, Mancini E, Osorio MB, Homrich MS, Turchetto-Zolet AC, Carvalho MCCGD, Stolf R, Weber RLM, Westergaard G, Castagnaro AP, Abdelnoor RV, Marcelino-Guimarães FC, Margis-Pinheiro M, Bodanese-Zanettini MH. Genome-wide annotation of the soybean WRKY family and functional characterization of genes involved in response to Phakopsora pachyrhizi infection. BMC Plant Biology. 2014;14:236. doi: 10.1186/s12870-014-0236-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai et al. (2014).Cai Y, Chen X, Xie K, Xing Q, Wu Y, Li J, Du C, Sun Z, Guo Z. Dlf1, a WRKY transcription factor, is involved in the control of flowering time and plant height in rice. PLOS ONE. 2014;9:e102529. doi: 10.1371/journal.pone.0102529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen et al. (2013).Chen C, Shao Y, Tao YD, Mei LJ, Shu QY, Wang LS. Main anthocyanins compositions and corresponding H-ORAC assay for wild Lycium ruthenicum Murr. fruits from the Qaidam Basin. Journal of Pharmaceutical Technology & Drug Research. 2013;2:1–5. doi: 10.1080/15287394.2018.1451180. [DOI] [Google Scholar]
- Chen et al. (2018b).Chen C, Xia R, Chen H, He Y. TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv. 2018b:289660. doi: 10.1101/289660. [DOI] [Google Scholar]
- Chen et al. (2018a).Chen F, Hu Y, Vannozzi A, Wu K, Cai H, Qin Y, Mullis A, Lin Z, Zhang L. The WRKY transcription factor family in model plants and crops. Critical Reviews in Plant Sciences. 2018a;36:311–335. doi: 10.1080/07352689.2018.1441103. [DOI] [Google Scholar]
- Chen et al. (2017).Chen J, Nolan T, Ye H, Zhang M, Tong H, Xin P, Chu J, Chu C, Li Z, Yin Y. Arabidopsis WRKY46, WRKY54 and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought response. The Plant Cell. 2017;29:1425–1439. doi: 10.1105/tpc.17.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, Wang & Zhang (2016).Dai X, Wang Y, Zhang WH. OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. Journal of Experimental Botany. 2016;67:947–960. doi: 10.1093/jxb/erv515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar et al. (2011).Dhar P, Tayade1 A, Ballabh B, Chaurasia1 OP, Bhatt RP, Srivastava1 RB. Lycium ruthenicum murray: a less-explored but high value medicinal plant from trans-himalayan cold deserts of ladakh, India: a review. Plant Archives. 2011;11:583–586. [Google Scholar]
- Dong, Chen & Chen (2003).Dong J, Chen C, Chen Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Molecular Biology. 2003;51:21–37. doi: 10.1023/A:1020780022549. [DOI] [PubMed] [Google Scholar]
- Eulgem et al. (2000).Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends in Plant Science. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- Gonzalez et al. (2016).Gonzalez A, Brown M, Hatlestad G, Akhavan N, Smith T, Hembd A, Moore J, Montes D, Mosley T, Resendez J, Nguyen H, Wilson L, Campbell A, Sudarshan D, Lloyd A. TTG2 controls the developmental regulation of seed coat tannins in Arabidopsis by regulating vacuolar transport steps in the proanthocyanidin pathway. The Plant Cell. 2016;419(1):54–63. doi: 10.1016/j.ydbio.2016.03.031. [DOI] [PubMed] [Google Scholar]
- Guan et al. (2014).Guan Y, Meng X, Khanna R, LaMontagne E, Liu Y, Zhang S. Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in Arabidopsis. PLOS Genetics. 2014;109(5):e1004384. doi: 10.1371/journal.pgen.1004384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaumie et al. (2011).Guillaumie S, Fouquet R, Kappel C, Camps C, Terrier N, Moncomble D, Dunlevy JD, Davies C, Boss PK, Delrot S. Transcriptional analysis of late ripening stages of grapevine berry. BMC Plant Biology. 2011;11:165. doi: 10.1186/1471-2229-11-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He et al. (2012).He H, Dong Q, Shao Y, Jiang H, Zhu S, Cheng B, Xiang Y. Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Reports. 2012;31:1199–1217. doi: 10.1007/s00299-012-1241-0. [DOI] [PubMed] [Google Scholar]
- Huang et al. (2012).Huang S, Gao Y, Liu J, Peng X, Niu X, Fei Z, Cao S, Liu Y. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Molecular Genetics and Genomics. 2012;287:495–513. doi: 10.1007/s00438-012-0696-6. [DOI] [PubMed] [Google Scholar]
- Huang et al. (2015).Huang X, Li K, Xu X, Yao Z, Jin C, Zhang S. Genome-wide analysis of WRKY transcription factors in white pear (Pyrus bretschneideri) reveals evolution and patterns under drought stress. BMC Genomics. 2015;16:1104. doi: 10.1186/s12864-015-2233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang et al. (2017).Jiang J, Ma S, Ye N, Jiang M, Cao J, Zhang J. WRKY transcription factors in plant responses to stresses. Journal of Integrative Plant Biology. 2017;59:86–101. doi: 10.1111/jipb.12513. [DOI] [PubMed] [Google Scholar]
- Jiang, Liang & Yu (2012).Jiang Y, Liang G, Yu D. Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Molecular Plant. 2012;5(6):1375–1388. doi: 10.1093/mp/sss080. [DOI] [PubMed] [Google Scholar]
- Larkin et al. (2007).Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace MI, Wilm A, Lopez R, Thompson JD, Gibson TB, Higgins DG. ClustalW and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li et al. (2017).Li D, Liu P, Yu J, Wang L, Dossa K, Zhang Y, Zhou R, Wei X. Genome-wide analysis of WRKY gene family in the sesame genome and identification of the WRKY genes involved in responses to abiotic stresses. BMC Plant Biology. 2017;17:152. doi: 10.1186/s12870-017-1099-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2012).Li HL, Zhang LB, Guo D, Li CZ, Peng SQ. Identification and expression profiles of the WRKY transcription factor family in Ricinus communis. Gene. 2012;503:248–253. doi: 10.1016/j.gene.2012.04.069. [DOI] [PubMed] [Google Scholar]
- Li et al. (2016).Li MY, Xu ZS, Tian C, Huang Y, Wang F, Xiong AS. Genomic identification of WRKY transcription factors in carrot (Daucus Carota) and analysis of evolution and homologous groups for plants. Science Report. 2016;6:23101. doi: 10.1038/srep23101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2010).Li S, Zhou X, Chen L, Huang W, Yu D. Functional characterization of Arabidopsis Thaliana WRKY39 in heat stress. Molecules and Cells. 2010;29:475–483. doi: 10.1007/s10059-010-0059-2. [DOI] [PubMed] [Google Scholar]
- Ling et al. (2011).Ling J, Jiang W, Zhang Y, Yu H, Mao Z, Gu X, Huang S, Xie B. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genomics. 2011;12:471. doi: 10.1186/1471-2164-12-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu & Ekramoddoullah (2009).Liu JJ, Ekramoddoullah AK. Identification and characterization of the WRKY transcription factor family in Pinus monticola. Genome. 2009;52:77–88. doi: 10.1139/G08-106. [DOI] [PubMed] [Google Scholar]
- Liu & Guan (2012).Liu N, Guan L. Linkages between woody proliferation dynamics and plant physiological traits in Southwestern North America. Journal of Plant Ecology. 2012;5:407–416. doi: 10.1093/jpe/rts002. [DOI] [Google Scholar]
- Liu et al. (2012).Liu Z, Shu Q, Wang L, Yu M, Hu Y, Zhang H, Tao Y, Shao Y. Genetic diversity of the endangered and medically important Lycium ruthenicum Murr. revealed by sequence-related amplified polymorphism (SRAP) markers. Biochemical Systematics and Ecology. 2012;45:86–97. doi: 10.1016/j.bse.2012.07.017. [DOI] [Google Scholar]
- Livak & Schmittgen (2001).Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luo et al. (2005).Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2005;1029(48):17531–17536. doi: 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo et al. (2013).Luo X, Bai X, Sun X, Zhu D, Liu B, Ji W, Cai H, Cao L, Wu J, Hu M, Liu X, Tang L, Zhu Y. Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. Journal of Experimental Botany. 2013;64:2155–2169. doi: 10.1093/jxb/ert073. [DOI] [PubMed] [Google Scholar]
- Ma et al. (2018).Ma YJ, Duan HR, Zhang F, Li Y, Yang HS, Tian FP, Zhou XH, Wang CM, Ma R. Transcriptomic analysis of Lycium ruthenicum Murr. during fruit ripening provides insight into structural and regulatory genes in the anthocyanin biosynthetic pathway. PLOS ONE. 2018;13:e0208627. doi: 10.1371/journal.pone.0208627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchive et al. (2007).Marchive C, Mzid R, Deluc L, Barrieu F, Pirrello J, Gauthier A, Corio-Costet A, Regad F, Cailleteau B, Hamdi S, Lauvergeat V. Isolation and characterization of a Vitis vinifera transcription factor, VvWRKY1, and its effect on responses to fungal pathogens in transgenic tobacco plants. Journal of Experimental Botany. 2007;58:1999–2010. doi: 10.1093/jxb/erm062. [DOI] [PubMed] [Google Scholar]
- Meng et al. (2016).Meng D, Li Y, Bai Y, Li M, Cheng L. Genome-wide identification and characterization of WRKY transcriptional factor family in apple and analysis of their responses to waterlogging and drought stress. Plant Physiology and Biochemistry. 2016;103:71–83. doi: 10.1016/j.plaphy.2016.02.006. [DOI] [PubMed] [Google Scholar]
- Miao et al. (2004).Miao Y, Laun T, Zimmermann P, Zentgraf U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Molecular Biology. 2004;55:853–867. doi: 10.1007/s11103-004-2142-6. [DOI] [PubMed] [Google Scholar]
- Ren et al. (2010).Ren X, Chen Z, Liu Y, Zhang H, Zhang M, Liu Q, Hong X, Zhu JK, Gong Z. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant Journal. 2010;639(3):417–429. doi: 10.1111/j.1365-313X.2010.04248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton et al. (2010).Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends in Plant Science. 2010;15:247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Sun et al. (2003).Sun C, Palmqvist S, Olsson H, Boren M, Ahlandsberg S, Jansson CA. Novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar responsive elements of the iso1 promoter. The Plant Cell. 2003;15:2076–2092. doi: 10.1105/tpc.014597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura et al. (2011).Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao et al. (2018).Tao X, Chengjie C, Chuhao L, Jiarou L, Chaoyang L, Yehua H. Genome-wide investigation of WRKY gene family in pineapple: evolution and expression profiles during development and stress. BMC Genomics. 2018;19:490. doi: 10.1186/s12864-018-4880-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao et al. (2011).Tao Z, Kou Y, Liu H, Li X, Xiao J, Wang S. OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. Journal of Experimental Botany. 2011;62:4863–4874. doi: 10.1093/jxb/err144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulker & Somssich (2004).Ulker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function. Current Opinion in Plant Biology. 2004;7:491–498. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Van Verk et al. (2008).Van Verk MC, Pappaioannou D, Neeleman L, Bol JF, Linthorst HJ. A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiology. 2008;146:1983–1995. doi: 10.1104/pp.107.112789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan et al. (2018).Wan Y, Mao M, Wan D, Yang Q, Yang F, Mandlaa LG, Wang R. Identification of the WRKY gene family and functional analysis of two genes in Caragana intermedia. BMC Plant Biology. 2018;18:31. doi: 10.1186/s12870-018-1235-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2014).Wang M, Vannozzi A, Wang G, Liang YH, Tornielli GB, Zenoni S, Cavallini E, Pezzotti M, Cheng ZM. Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Horticulture Research. 2014;1:14016. doi: 10.1038/hortres.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei et al. (2016).Wei Y, Shi H, Xia Z, Tie W, Ding Z, Yan Y, Wang W, Hu W, Li K. Genome-wide identification and expression analysis of the WRKY gene family in cassava. Frontiers in Plant Science. 2016;7:25. doi: 10.3389/fpls.2016.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen et al. (2014).Wen F, Zhu H, Li P, Jiang M, Mao W, Ong C, Chu Z. Genome-wide evolutionary characterization and expression analyses of WRKY family genes in Brachypodium Distachyon. DNA Resources. 2014;21:327–339. doi: 10.1093/dnares/dst060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu et al. (2005).Wu KL, Guo ZJ, Wang HH, Li J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Research. 2005;12:9–26. doi: 10.1093/dnares/12.1.9. [DOI] [PubMed] [Google Scholar]
- Xiong et al. (2013).Xiong W, Xu X, Zhang L, Wu P, Chen Y, Li M, Jiang H, Wu G. Genome-wide analysis of the WRKY gene family in physic nut (Jatropha curcas L.) Gene. 2013;524:124–132. doi: 10.1016/j.gene.2013.04.047. [DOI] [PubMed] [Google Scholar]
- Xue et al. (2019).Xue C, Li H, Liu Z, Wang L, Zhao Y, Wei X, Fang H, Liu M, Zhao J. Genome-wide analysis of the WRKY gene family and their positive responses to phytoplasma invasion in Chinese jujube. BMC Genomics. 2019;20:464. doi: 10.1186/s12864-019-5789-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang et al. (2016).Yang Y, Chi Y, Wang Z, Zhou Y, Fan B, Chen Z. Functional analysis of structurally related soybean GmWRKY58 and GmWRKY76 in plant growth and development. Journal of Experimental Botany. 2016;67:4727–4742. doi: 10.1093/jxb/erw252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue et al. (2016).Yue H, Wang M, Liu S, Du X, Song W, Nie X. Transcriptome-wide identification and expression profiles of the WRKY transcription factor family in broomcorn millet (Panicum Miliaceum L.) BMC Genomics. 2016;17:343. doi: 10.1186/s12864-016-2677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang & Feng (2014).Zhang Y, Feng JC. Identification and characterization of the grape WRKY family. BioMed Research International. 2014;78:76–80. doi: 10.1155/2014/787680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang & Wang (2005).Zhang YJ, Wang LJ. The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evolutionary Biology. 2005;5:1. doi: 10.1186/1471-2148-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng et al. (2011).Zheng J, Ding CX, Wang LS, Li GL, Shi JY, Li H, Wang HL, Suo YR. Anthocyanins composition and antioxidant activity of wild Lycium ruthenicum Murr. from Qinghai-Tiber plateau. Food Chemistry. 2011;126:859–865. doi: 10.1016/j.foodchem.2010.11.052. [DOI] [Google Scholar]
- Zhou et al. (2016).Zhou H, Li Y, Zhang Q, Ren S, Shen Y, Qin L, Xing Y. Genome-wide analysis of the expression of wrky family genes in different developmental stages of wild strawberry (Fragaria vesca) fruit. PLOS ONE. 2016;11:e0154312. doi: 10.1371/journal.pone.0154312. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
Raw data is available in the Supplemental Files.