Abstract
Purpose
It is difficult to predict the prognosis of COVID-19 patients at the disease onset. This study was designed to add new biomarkers into conventional inflammatory panels to build an optimal combination panel, to better triage patients and predict their outcomes.
Patients and Methods
Biochemical parameters representing multi-organ functions, cytokines, acute-phase proteins, and other inflammatory markers were measured in COVID-19 patients on hospital admission. Receiver operating characteristic (ROC) curves, logistic regression, event-free survival (EFS), and Cox analyses were performed to screen and compare the predictive capabilities of the new panel in patients with different illness severity and outcome.
Results
This study included 120 patients with COVID-19, consisting of 32 critical, 28 severe, and 60 mild/moderate patients. Initial levels of the selected biomarkers showed a significant difference in the three groups, all of which influenced patient outcome and EFS to varying degrees. Cox proportional hazard model revealed that procalcitonin (PCT) and interleukin 10 (IL-10) were independent risk factors, while superoxide dismutase (SOD) was an independent protective factor influencing EFS. In discriminating the critical and mild patients, a panel combining PCT, IL-6, and neutrophil (NEUT) yielded the best diagnostic performance with an AUC of 0.99, the sensitivity of 90.60% and specificity of 100%. In distinguishing between severe and mild patients, SOD’s AUC of 0.89 was higher than any other single biomarker. In differentiating the critical and severe patients, the combination of white blood cell count (WBC), PCT, IL-6, IL-10, and SOD achieved the highest AUC of 0.95 with a sensitivity of 75.00% and specificity of 100%.
Conclusion
The optimal combination panel has a substantial potential to better triage COVID-19 patients on admission. Better triage of patients will benefit the rational use of medical resources.
Keywords: COVID-19, diagnostic panel, cytokine, acute-phase protein
Plain Language Summary
1. The pandemic of COVID-19 has brought unprecedented difficulty to hospitals. It is of utmost importance to establish a triage risk score for identifying patients who may need a different therapeutic regimen, and timely treatment of severely ill patients can potentially improve the prognosis.
2. Three optimal combination panels (panel of WBC, PCT, IL-6, IL-10, SOD, a panel of NEUT, IL-6, PCT, and a panel of SOD) offer substantial potentials in discriminating the critical, severe, and mild patients at disease onset. This could assist medical professionals in triaging patients appropriately when allocating limited healthcare resources.
3. Initial of selected biomarkers showed a significant difference in patients with different degree of disease severity or outcome. PCT and IL-10 were independent risk factors, while SOD was an independent protective indicator affecting event-free survival (EFS). Our study demonstrated that the addition of APP to conventional inflammatory board might improve the discriminative ability regarding patients’ prognosis.
Introduction
The novel coronavirus disease (COVID-19) pandemic is a rapidly developing global emergency. At the time of writing, more than twenty million people have been infected worldwide. The high pathogenicity, mortality, and rapid human-to-human transmission render COVID-19 a significant threat to global public health. Although many patients presented with mild symptoms at the onset of illness, organ dysfunction including shock, acute respiratory distress syndrome, acute cardiac and kidney injuries, and death often occur in severe cases.1,2 Some patients with mild symptoms may experience sudden deterioration. The median time from symptom onset to intensive care unit (ICU) admission has been reported to be ten days.3 Prognostic uncertainty is a unique feature of COVID-19. Risk models for progression to severe disease, mortality risk, or length of hospital stay could assist medical staff in triaging patients when allocating limited healthcare resources.4 Currently, the most reported predictors of severe prognosis in patients with COVID −19 included age, sex, image change derived from computed tomography (CT) scans, laboratory parameters including C-reactive protein (CRP), lactic dehydrogenase (LDH), and lymphocyte count. However, proposed models are poorly reported and at high risk of bias, raising concern that their predictions could be unreliable when applied in daily practice.4
In COVID-19, the difference in clinical characteristics between severe and non-severe cases was not reported.1 Secondary bacterial infections can significantly aggravate the viral disease. Septic shock was frequently found in seriously ill patients with COVID-19, especially those in ICU who often developed viral and bacterial co-infection. Most patients received antibacterial therapy or glucocorticoid therapy besides antiviral treatment.1 However, high doses of antibacterial or glucocorticoid therapy does not only cause severe sequelae, like femoral head necrosis and enhanced bacterial resistance, but also increases the hospital cost. Real-time monitoring of the infectious status helps physicians to choose optimal therapy. CRP and procalcitonin (PCT) are the conventional biomarkers most frequently used for monitoring the severity of inflammation. The higher CRP levels correlated with more severe symptoms5,6. In respiratory diseases, CRP guiding antibiotic therapy reduces prescription rates without compromising patient outcomes. PCT has been used as a biomarker to aid in differentiating bacterial pneumonia from viral pneumonia.7 PCT was suggested to have superior efficacy compared with CRP in predicting bacteremia in patients with community-acquired pneumonia8. In patients with acute dyspnea, PCT is an accurate diagnostic marker for bacterial pneumonia.9,10 Clinical observation from COVID-19 revealed that the proportion of elevated PCT in ICU patients was significantly higher than that in non-ICU patients.1 However, no cutoff point of PCT and CRP could divide the patients into high- or low-risk group. Several limitations also exist in conventional inflammatory markers regarding suboptimal sensitivity and specificity.11
COVID-19 patients in ICU developed cellular immunodeficiency, coagulation activation, myocardial, hepatic, and kidney injuries compared with non-ICU patients.1 Cytokine storms may play a leading role in COVID-19 victims. Amongst the severe patients, cytokine storm is involved in organ failure leading to death.2,3 Cytokine storm is also the primary cause of morbidity for some critical COVID-19 patients3,12. Biomarkers representing immunopathological mechanisms and distinct biological signals hold high potentials in determining patient outcomes. Another host defense mechanism to pathogen invasion is the activation of acute-phase response (APR) that includes the production of inflammatory cytokines and many proteins such as CRP, serum amyloid proteins (SAA, SAP) and pentraxins (PTX). Acute-phase proteins (APP) are vital components of the antimicrobial response, which very often are involved directly or indirectly in the inhibition of viral replication and spread within the host.13 Serum APP in viral respiratory illness can reach the hundred thousand-fold over healthy samples.14 We also found COVID-19 patients had significantly higher levels of inflammatory cytokines (especially IL-6 and IL-10), indicating that cytokines may be sensitive biomarkers to predict disease outcome. SAA is a vital component released into the circulation in correlation with antimicrobial and anti-inflammatory activity.15 Blood cystatin C (Cys-C) is a sensitive biomarker that is superior to serum creatinine (Cr) in the assessment of renal function, which is crucial in the early diagnose of renal injury16,17. Myoglobin (MYO) and hypersensitive troponin I (HSTNI), as well as aspartate aminotransferase (AST) and alanine aminotransferase (ALT), are the parameters reflecting heart and liver functions, respectively. Multiple organs were affected by the SARS-CoV-2 infection, and the differential modulation of those proteins upon microbial challenge may make them ideal markers for predicting disease prognosis and triage patients. For COVID-19 patients, timely treatment of severe cases is of paramount importance.18 Risk stratification is vital to reduce time to effective therapy in high-risk patients and prevent them from the development of organ impairment. In this study, we investigated the incremental usefulness of multiple biomarkers in risk stratification and the prognostic prediction.
Methods
Patients
The Ethics committee of Zhongnan Hospital of Wuhan University approved the study (No. 2020051K). Written consent was obtained from all enrolled patients. COVID-19 patients with different degrees of disease severity were included in this study. From January 20 to March 10, 2020, a total of 120 confirmed cases were enrolled in the study. The classification criteria were the guidelines based on the New Coronavirus Pneumonia Prevention and Control Program (7th edition) published by the National Health Commission of China. The patients’ main symptoms, vital signs, and comorbidities were recorded. All biomarkers were selected from the baseline measurements taken when patients were admitted to the hospital. The primary outcome was evaluated by vital signs, imaging, and laboratory tests. COVID-19 patients who had no complete medical record and test results were excluded.
Measurement of Cytokines, Inflammatory Parameters and Other Laboratory Data
All biochemical and hematologic parameters were obtained via standard automated laboratory methods and using commercially available kits following the manufacturers’ protocols. The serum cytokines, including IL2, IL4, IL6, IL8, IL10, tumor necrosis factor-α (TNF-α), and Interferon-Ƴ (IFN-Ƴ) were measured using BD FACSCalibur flow cytometry (Becton, Dickinson and Company, New Jersey, USA), according to the manufacturer’s instructions (Node Company, Jiangxi, China). In brief, 25uL of serum was added to 25uL fluorescence detection reagent to capture microsphere mixture. The mixture was then incubated at room temperature for 2.5 hours. Then, the microsphere was resuspended with 100ul sheath fluid after being washed, and then analyzed using flow cytometry. The serum levels of CRP, SAA, SOD were determined using Beckman Coulter AU5800 (Beckman Coulter, Inc., California, USA). Latex immunoturbidimetry technique was used to detect CRP (Sekisui, Medical Company, Tokyo, Japan) and SAA (Ningbo Purebio Biotechnology Company, Zhe Jiang, China). SOD was determined by pyrogallol autoxidation method (Iprocom Biotechnology Company, Anhui, China). The complete blood count (CBC) was performed by a Beckman Coulter UniCelDxH 800 Hematology Analyzer (Beckman Coulter, Incorporated, Miami, USA). Coagulation function was assessed by an ACL TOP1000 coagulation analyzer using matched reagent (Instrumentation Laboratory Company, Bedford, MA, USA), and D-D was measured by immunoturbidimetry methods. PCT was determined by the VIDAS analyzer using matched Enzyme-linked fluorescent assay reagent (BioMerieux, Chemin de, L’Orme Marcy L’Etoile, France).
Measurement of Nucleic Acid (NA) and Antibody of SARS-CoV-2
NA of SARS-CoV-2 was performed on throat swabs following a previously described method.1 Total RNA was extracted using the respiratory sample RNA isolation kit (Zhongzhi, Wuhan, China). Two target genes, Target 1 (ORF1ab) and Target 2 (N) were measured by reverse transcription-polymerase chain reaction (RT-PCR) (DAAN GENE Company, Guangzhou, China). Primer information followed the WHO guidelines for qRT-PCR.1,19 A cycle threshold value (Ct-value) less than 37 was defined as a positive test result, and a Ct-value of 40 or more was identified as a negative test. SARS-CoV-2 IgM and IgG antibody were tested with YHLO iFlash 3000-C chemiluminescence instrument using original reagent (YHLO Biotechnology Company, Shenzhen, China).
Statistical Analyses
A comparison of clinical and laboratory parameters among different groups was analyzed by Kruskal–Wallis H, Mann–Whitney U and chi-square tests due to the abnormal distribution. All studied groups were characterized by median values and the interquartile ranges (IQRs) (low quartile–high quartile) or mean ± standard error (SE) according to the type and distribution of data. The inflammatory biomarkers and APP were selected as candidates for evaluating disease severity and prognosis. Three models were used in further statistical analysis: the first model (critical vs mild), the second model (severe vs mild), and the third model (critical vs severe). Binary logistic regression of candidates was applied to determine the optimal panel mirroring disease severity independently.
Furthermore, the diagnostic performance of a single biomarker or optimal combination panel was analyzed by receiver-operating characteristic (ROC) curves. Event-free survival (EFS) was defined as the time from the start of illness to the first event (respiratory distress) or until the last follow up. Kaplan–Meier estimation was used to plot survival curves; the Log-rank was utilized to test the difference among groups. Multivariate regression analysis by the Cox proportional hazard model was applied to determine independent factors affecting survival. SPSS 20.0 was used for statistical analyses (SPSS Incorporated, Chicago, IL, USA). All statistical tests were two-sided, P <0.05 was considered statistically significant.
Results
Clinical Features, Treatment, and Outcome of the Patients Stratified by Disease Severity
Clinical Features
The final diagnosis of the 120 patients was classified as critical (group1: 32 cases), severe (group2: 28 cases), and mild/moderate (group3: 60 cases) COVID-19 cases (Table 1). The median age and the proportion of females in group 3 were significantly lower than the other two groups (P<0.05). The fever was highest in group 1, and lowest in group 3 (39.0°C vs 38.0°C) (P<0.001). There was no significant difference in clinical symptoms (including fever, myalgia, sore throat, coryza, cough, expectoration, anorexia, and vomiting) among three groups (P>0.05). Fatigue, dyspnea, and diarrhea were more likely to be seen in group 1 and group 2 than group 3 (P<0.05). There were significantly more patients with hypertension, diabetes, and cardiovascular disease in group 1 than those in group3 (P<0.05). In contrast, other comorbidities, including chronic lung disease, malignancy, digestive diseases, neurological disease, and autoimmune disorders, were not significantly different among the three groups (P>0.05).
Table 1.
Baseline Characteristics of Patients Infected by 2019-nCoV with Various Severity
| NO (%) | Group 1 | Group 2 | Group 3 | P value | |||
|---|---|---|---|---|---|---|---|
| Total N=120 | Critical N=32 | Severe N=28 | Mild N=60 | 1 vs 2 | 1 vs 3 | 2 vs 3 | |
| Characteristics | |||||||
| Age, median (IQR), years | 59 (47–68) | 65 (55–76) | 62 (48–72) | 55 (44–63) | 0.222 | <0.001 | 0.032 |
| Sex | |||||||
| Female | 66 (55) | 22 (68.75) | 18 (64.29) | 26 (43.33) | 0.714 | 0.020 | 0.067 |
| Male | 54 (45) | 10 (31.25) | 10 (35.71) | 34 (56.67) | |||
| BMI, median (IQR) | 22.86 (20.76–25.22) | 23.43 (21.91–25.22) | 22.60 (21.22–25.39) | 18.00 (8.74–25.29) | 0.988 | 0.455 | 0.444 |
| Signs and symptoms | |||||||
| T, median (IQR), °C | 38.20 (37.80–38.88) | 38.95 (38.13–39.38) | 38.35(37.58–39) | 38 (37.58–38.48) | 0.021 | <0.001 | 0.131 |
| HR, median (IQR) | 84 (78.50–90.75) | 87 (79–99.75) | 80(76–89.75) | 84.50 (80–90) | 0.098 | 0.226 | 0.660 |
| RR, median (IQR) | 20 (19.25–21) | 22(20–25) | 20 (20–21) | 20 (18–20) | 0.028 | 0.001 | 0.222 |
| BP1, median (IQR) | 130 (117.25–141) | 130(112–148) | 133.50 (117.75–140) | 126.50 (120–140.50) | 0.715 | 0.897 | 0.860 |
| BP2, median (IQR) | 75 (65.50–85) | 70.50 (62–80) | 77 (65.75–85.50) | 76 (70–86) | 0.152 | 0.005 | 0.279 |
| SpO2, median (IQR) | 93(99–95) | 81 (75–89) | 91 (86–93) | 95 (94–97) | 0.001 | <0.001 | <0.001 |
| Fever | 103 (85.83) | 30 (93.75) | 24 (85.71) | 49 (81.67) | 0.404 | 0.207 | 0.766 |
| Fatigue | 45 (37.50) | 16 (50) | 14 (50) | 15 (25) | 1.000 | 0.016 | 0.020 |
| Myalgia | 18 (15) | 7 (21.88) | 3 (10.71) | 8 (13.33) | 0.312 | 0.291 | 1.000 |
| Sore throat | 5 (4.17) | 1 (3.13) | 0 | 4 (6.67) | 1.000 | 0.655 | 0.302 |
| Coryza | 2 (1.67) | 1 (3.13) | 1 (3.57) | 0 | 1.000 | 0.348 | 0.318 |
| Cough | 71 (59.17) | 21 (65.63) | 16 (57.14) | 34 (56.67) | 0.500 | 0.404 | 0.966 |
| Expectoration | 18 (15) | 4 (12.50) | 4 (14.29) | 10 (16.67) | 1.000 | 0.764 | 1.000 |
| Dyspnea | 56 (46.67) | 23 (71.88) | 16 (57.14) | 17 (28.33) | 0.233 | <0.001 | 0.009 |
| Anorexia | 17 (14.17) | 7 (21.88) | 5 (17.86) | 5 (8.33) | 0.698 | 0.102 | 0.278 |
| Vomiting | 3 (2.50) | 1 (3.13) | 1 (3.57) | 1 (1.67) | 1.000 | 1.000 | 0.538 |
| Diarrhea | 11 (9.17) | 7 (21.88) | 2 (7.14) | 2 (3.33) | 0.155 | 0.008 | 0.589 |
| Oliguria | 1 (0.83) | 1 (3.13) | 0 | 0 | 1.000 | 0.348 | – |
| Hematuria | 2 (1.67) | 2 (6.25) | 0 | 0 | 0.494 | 1.000 | – |
| Dizziness | 4 (3.33) | 2 (6.25) | 1 (3.57) | 1 (1.67) | 1.000 | 0.276 | 0.538 |
| Headache | 4 (3.33) | 1 (3.13) | 2 (7.14) | 1 (1.67) | 0.594 | 1.000 | 0.237 |
| Syncope | 10 (8.33) | 6 (18.75) | 3 (10.71) | 1 (1.67) | 0.482 | 0.007 | 0.093 |
| Comorbidities | |||||||
| Hypertension | 41 (34.17) | 17 (53.13) | 10 (35.71) | 14 (23.33) | 0.176 | 0.004 | 0.224 |
| Diabetes | 21 (17.50) | 9 (28.13) | 5 (17.86) | 7 (11.67) | 0.348 | 0.047 | 0.509 |
| Cardiovascular disease | 24 (20) | 14 (43.75) | 5 (17.86) | 5 (8.33) | 0.031 | <0.001 | 0.278 |
| Chronic lung disease | 7 (5.83) | 2 (6.25) | 2 (7.14) | 3 (5) | 1.000 | 1.000 | 0.651 |
| Malignancy | 5 (4.17) | 1 (3.13) | 1 (3.57) | 3 (5) | 1.000 | 1.000 | 1.000 |
| Digestive diseases | 13 (10.83) | 5 (15.63) | 3 (10.71) | 5 (8.33) | 0.712 | 0.309 | 0.706 |
| Neurological disease | 6 (5) | 2 (6.25) | 3 (10.71) | 1 (1.67) | 0.657 | 0.276 | 0.093 |
| Autoimmune disorders | 2 (1.67) | 1 (3.13) | 0 | 1 (1.67) | 1.000 | 1.000 | 1.000 |
| Treatment | |||||||
| Antiviral therapy | 114 (95) | 30 (93.75) | 25 (89.29) | 59 (98.33) | 0.657 | 0.276 | 0.093 |
| Antibiotic treatment | 98 (81.67) | 32 (100.0) | 27 (96.43) | 39 (65) | 0.467 | <0.001 | 0.002 |
| Glucocorticoid therapy | 71 (59.17) | 30 (93.75) | 22 (78.57) | 19 (31.67) | 0.130 | <0.001 | <0.001 |
| Immunoglobulin therapy | 23 (19.17) | 10 (31.25) | 7 (25) | 6 (10) | 0.592 | 0.010 | 0.104 |
| Oxygen inhalation | 77 (64.17) | 25 (78.13) | 28 (100) | 24 (40) | 0.012 | <0.001 | <0.001 |
| NIV | 31 (25.83) | 25 (78.13) | 6 (21.43) | 0 | <0.001 | <0.001 | 0.001 |
| IMV | 26 (21.67) | 26 (81.25) | 0 | 0 | <0.001 | <0.001 | – |
| ECMO | 9 (7.50) | 9 (28.13) | 0 | 0 | 0.002 | <0.001 | – |
| CRRT | 13 (10.83) | 13 (40.63) | 0 | 0 | <0.001 | <0.001 | – |
| CPR | 17 (14.17) | 17 (53.13) | 0 | 0 | <0.001 | <0.001 | – |
| Blood transfusion | 19 (15.83) | 18 (56.25) | 1 (3.57) | 0 | <0.001 | <0.001 | 0.318 |
| Bronchial lavage | 9 (7.50) | 9 (28.13) | 0 | 0 | 0.002 | <0.001 | – |
| Clinical trial | 16 (13.33) | 4 (12.50) | 8 (28.57) | 4 (6.67) | 0.121 | 0.442 | 0.015 |
| Complications | |||||||
| ARDS | 50 (41.67) | 32 (100) | 18 (64.29) | 0 | <0.001 | <0.001 | <0.001 |
| Abnormal liver function | 47 (39.17) | 22 (68.75) | 16 (57.14) | 9 (15) | 0.352 | <0.001 | <0.001 |
| Acute cardiac injury | 29 (24.17) | 24 (75) | 4 (14.29) | 1 (1.67) | <0.001 | <0.001 | 0.034 |
| Acute kidney injury | 37 (30.83) | 25 (78.13) | 10 (35.71) | 2 (3.33) | 0.001 | <0.001 | <0.001 |
| Shock | 7 (5.83) | 7 (21.88) | 0 | 0 | 0.012 | <0.001 | – |
| Clinical outcome | |||||||
| Died | 16 (13.33) | 16 (50) | 0 | 0 | <0.001 | <0.001 | – |
| Remained in ICU | 10 (8.33) | 6 (18.75) | 4 (14.29) | 0 | 0.737 | 0.001 | 0.009 |
| Condition improve | 28 (23.33) | 5 (15.63) | 10 (35.71) | 13 (21.67) | 0.073 | 0.487 | 0.162 |
| Hospital discharge | 66 (55) | 5 (15.63) | 14 (50) | 47 (78.33) | 0.004 | <0.001 | 0.007 |
Notes: P values comparing Critical, Severe and Mild are from One-way analysis of variance, Pearson chi-square test, or Fisher’s exact test. P value <0.05 was considered statistically significant. - Not applicable. Data were available for 71 patients.
Abbreviations: IQR, interquartile range; T, temperature; HR, heart rate; RR, respiratory rate; BP, blood pressure (mmHg); NIV, non-invasive ventilation; IMV, intermittent mandatory ventilation; ECMO, extracorporeal membrane oxygenation; CRRT, continuous renal replacement therapy; CPR, cardiopulmonary resuscitation; ARDS, acute respiratory distress syndrome. Data are n (%), n/N (%), mean (SEM), and median (1QR).
Treatment
Group 1 had the highest number of patients receiving antibiotics (100%), glucocorticoid therapy (93.75%), and non-invasive ventilation (NIV) (78.13%), while group 3 had the lowest number receiving antibiotics (65%), glucocorticoid therapy (31.67%), and oxygen inhalation (40%), (P<0.001). Intermittent mandatory ventilation (IMV) (81.25%), extracorporeal membrane oxygenation (ECMO) (28.13%), continuous renal replacement therapy (CRRT) (40.63%), cardio-pulmonary resuscitation (CPR) (53.13%), and bronchial lavage (28.13%) were only instituted for patients in group 1 (Table 1).
Complications
Shock was observed only in group1 (21.88%), while ARDS was found in groups 1 (100%) and 2 (64.29%). Patients with abnormal liver function, acute cardiac injury, and acute kidney injury were significantly more in groups 1 and 2 than in group 3 (P<0.05) (Table 1).
Outcome
In group 1, 50% of patients died, 18.75% of patients remained in ICU at the time of writing, 15.6% of patients improved, and 15.63% of patients were discharged from hospital. In group 2, 14.29% of patients remained in ICU, 35.71% of patients improved, and 50% of patients were discharged. In group 3, 21.67% of patients improved, whereas 78.33% of patients were discharged (Table 1).
Laboratory Parameters
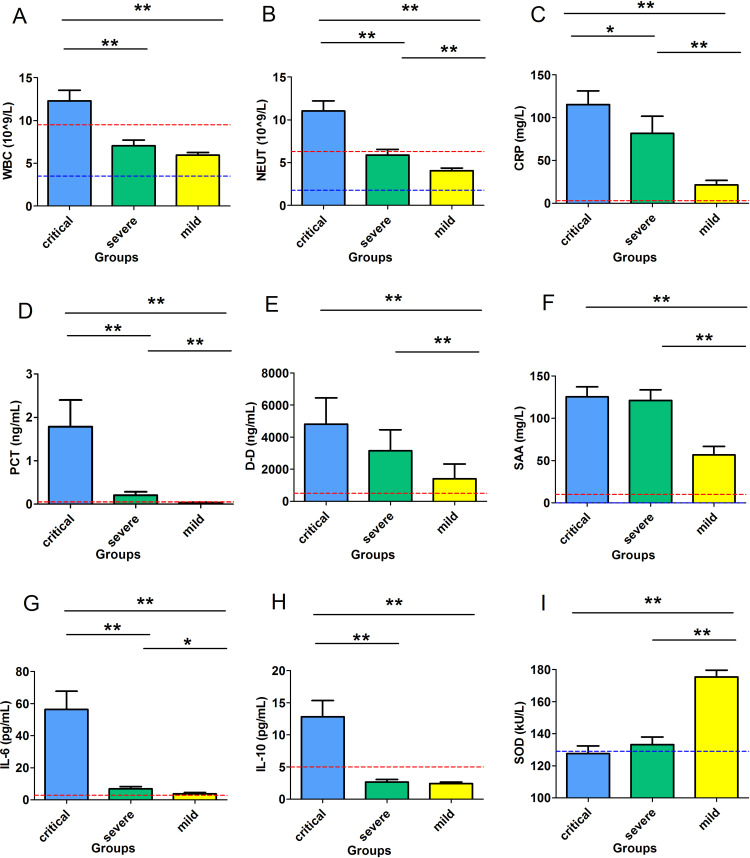
WBC (Figure 1A) was significantly higher in group 1 (12.29±1.23) than that in groups 2 (7.05±0.66) and 3 (5.97±0.31) (P<0.001), while there was no significant difference between groups 2 and 3(P>0.05). There was a substantial difference in neutrophil (NEUT) among the three groups. Group 1 vs group 2 vs group 3 were 11.04±1.19 vs 5.88±0.67 vs 4.07±0.30, respectively, (P<0.001) (Figure 1B). Hemoglobin (HGB), red blood cell (RBC), and hematocrit (HCT) levels all showed an increasing trend from group 1 to group 3. Platelet (PLT) was significantly higher in group 3 than the other two groups (P<0.05) (Table 2). For parameters reflecting coagulation activation, there was no significant difference in prothrombin time (PT), activated partial thromboplastin time (APTT), thrombin time (TT), fibrinogen content (FIB), and D-dimer (D-D) between groups 1 and 2 (P>0.05). In contrast, PT, FIB, and D-D in groups 1 and 2 were significantly higher than those in group3 (P<0.05) (Table 2). For liver function, total protein (TP) and albumin (ALB) in groups 1 and 2 were significantly lower than those in group 3 (P<0.01). Among the three groups, the highest level of AST was seen in group 1. However, there was no significant difference in ALT, Alpha-glutamyl transpeptidase (GGT), alkaline phosphatase (ALP), and total bile acid (TBA) among the three groups (P>0.05). For biomarkers of renal function, blood urea nitrogen (BUN) and CYS-C in group1 and 2 were significantly higher than those in group 3 (P<0.05) and Creatinine (CREA) in group 1 was significantly higher than those in group 3 (P<0.05). For heart function, MYO, LDH, creatine kinase-MB (CK-MB) and HSTNI in group 1 were significantly higher than those in group 3 (P<0.05) (Table 2).
Figure 1.
Nine inflammatory-related parameters in various clinical subgroups. (A) (H) The levels of WBC and IL-10 were significantly higher in group 1 than group 2 and 3 (P<0.001). (B) (C) (D) (G) NEUT, CRP, PCT, and IL-6 successively decreased from group 1 to group 3(P<0.001). (E) (F) Higher levels of D-D and SAA were found in groups 1 and 2 than in group3 (P<0.001) (I) Level of SOD successively increased from group 1 to group 3 (P<0.001). WBC: white blood cell; NEUT: absolute neutrophil count; CRP: C-reactive protein; PCT: procalcitonin; D-D: D-dimer; SAA: serum amyloid A; IL-6: interleukin-6; IL-10: interleukin-10; SOD: superoxide dismutase; The red and blue dashed lines represents the upper and lower limits of the reference range. Group1: critical type, Group 2: severe type, and Group 3: mild type; *P<0.05 and **P<0.01 were considered to be statistically significant.
Table 2.
Laboratory Characteristics of the NCIP Patients with Various Disease Severity
| Clinical Indicator | Normal Range | Group 1 | Group 2 | Group 3 | P value | ||
|---|---|---|---|---|---|---|---|
| Critical N=32 | Severe N=28 | Mild N=60 | 1 vs 2 | 1 vs 3 | 2 vs 3 | ||
| Blood RT | |||||||
| WBC,10^9/L | 3.50–9.50 | 12.29±1.23 | 7.05±0.66 | 5.97±0.31 | 0.001 | <0.001 | 0.370 |
| NEUT#,10^9/L | 1.80–6.30 | 11.04±1.19 | 5.88±0.67 | 4.07±0.30 | 0.001 | <0.001 | 0.052 |
| NEUT%,% | 40–75 | 89.15±1.30 | 79.85±2.27 | 65.60±1.67 | 0.002 | <0.001 | <0.001 |
| LYMPH#,10^9/L | 1.10–3.20 | 0.59±0.06 | 0.66±0.06 | 1.30±0.07 | 0.824 | <0.001 | <0.001 |
| LYMPH%,% | 20–50 | 6.28±0.93 | 11.58±1.56 | 24±1.35 | 0.022 | <0.001 | <0.001 |
| HGB, g/L | 115–150 | 121.65±3.97 | 123.57±4.57 | 133.29±1.65 | 0.984 | 0.029 | 0.149 |
| RBC,10^12/L | 3.80–5.10 | 3.87±0.13 | 4±0.16 | 4.18±0.06 | 0.905 | 0.107 | 0.650 |
| HCT,% | 35–45 | 35.31±1.14 | 36.13±1.32 | 38.15±0.49 | 0.952 | 0.079 | 0.404 |
| PLT,10^9/L | 125–350 | 181.28±16.15 | 186.18±15.09 | 226.57±10.93 | 0.825 | 0.017 | 0.041 |
| Coagulogram | |||||||
| PT, s | 9.40–12.50 | 14.18±0.40 | 13.12±0.27 | 12.04±0.18 | 0.092 | <0.001 | 0.002 |
| APTT, s | 25.10–36.50 | 38.82±6.06 | 28.28±0.79 | 30.70±0.40 | 0.252 | 0.463 | 0.027 |
| TT, s | 10.30–16.60 | 29.37±7.94 | 15.71±0.29 | 15.57±0.27 | 0.256 | 0.248 | 0.978 |
| FIB, mg/dL | 238–498 | 436.56±22.73 | 457.86±20.31 | 382.09±12.86 | 0.442 | 0.028 | 0.004 |
| D-D, ng/mL | 0–500 | 4812.06±1639.09 | 3152.86±1315.49 | 1405.19±919.54 | 0.445 | <0.001 | 0.002 |
| Liver function | |||||||
| ALT, U/L | 7–45 | 49.88±6.71 | 62.82±14.22 | 31.88±3.19 | 0.795 | 0.057 | 0.119 |
| AST, U/L | 13–35 | 68.25±10.65 | 44.82±6.78 | 27.48±2.07 | 0.192 | 0.002 | 0.058 |
| TP, g/L | 65–85 | 59.60±0.92 | 62.10±1.43 | 69.46±0.85 | 0.375 | <0.001 | <0.001 |
| ALB, g/L | 40–55 | 30.53±0.66 | 31.29±1.04 | 41.93±3.09 | 0.867 | 0.008 | 0.003 |
| GLB, g/L | 20–30 | 29.07±0.90 | 30.82±1.16 | 29.61±0.67 | 0.211 | 0.644 | 0.329 |
| ALB/GLB | 1.5–2.5 | 1.10±0.06 | 1.05±0.05 | 1.38±0.04 | 0.564 | <0.001 | <0.001 |
| GGT, U/L | 8–57 | 63.59±11.50 | 63.32±14.64 | 39.52±4.22 | 1.000 | 0.158 | 0.331 |
| ALP, U/L | 30–120 | 77.19±4.83 | 82.89±5.92 | 83.48±3.41 | 0.431 | 0.304 | 0.926 |
| TBA, μmol/L | 0–15 | 4.17±0.40 | 5.68±1.98 | 3.50±0.28 | 0.838 | 0.438 | 0.627 |
| TBIL, μmol/L | 5–21 | 16.75±1.33 | 13.91±1.68 | 12.35±0.58 | 0.464 | 0.012 | 0.763 |
| DBIL, μmol/L | 0–7 | 6.58±0.72 | 3.77±0.50 | 2.48±0.21 | 0.007 | <0.001 | 0.067 |
| GLU, mmol/L | 3.90–6.10 | 10.87±1.02 | 8.43±0.63 | 6.38±0.33 | 0.134 | <0.001 | 0.019 |
| Renal function | |||||||
| BUN, mmol/L | 2.80–7.60 | 8.25±0.84 | 6.91±0.63 | 4.41±0.18 | 0.496 | <0.001 | 0.002 |
| CREA, μmol/L | 49–90 | 102.02±21.42 | 72.88±5.24 | 62.13±1.79 | 0.347 | 0.001 | 0.348 |
| UA, μmol/L | 155–357 | 294.46±28.18 | 247.41±19.76 | 315.47±13.24 | 0.439 | 0.874 | 0.018 |
| CYSC, mg/L | 0–1.20 | 1.37±0.15 | 1.23±0.10 | 0.93±0.04 | 0.818 | 0.020 | 0.015 |
| CO2, mmol/L | 21–29 | 21.19±0.84 | 24.09±0.91 | 25.95±0.39 | 0.068 | <0.001 | 0.190 |
| Cardiac function | |||||||
| CK, U/L | <145 | 377.45±154.45 | 225.32±103.39 | 99.56±12.06 | 0.798 | 0.226 | 0.548 |
| LDH, U/L | 125–243 | 511.62±44.90 | 322.89±38.59 | 197.88±10.75 | 0.007 | <0.001 | 0.012 |
| CKMB, U/L | 0–25 | 48.21±14.92 | 34.48±12.25 | 10.29±0.62 | 0.857 | 0.049 | 0.164 |
| MYO, ng/mL | <140.10 | 308.94±72.56 | 175.37±58.44 | 76.25±24.30 | 0.398 | 0.013 | 0.334 |
| HSTNI, pg/mL | 0–26.20 | 794.11±645.40 | 41.21±20.42 | 6.83±4.39 | 0.041 | <0.001 | 0.142 |
| Electrolyte | |||||||
| K, mmol/L | 3.50–5.30 | 3.96±0.10 | 4.13±0.10 | 4.27±0.06 | 0.198 | 0.006 | 0.226 |
| Na, mmol/L | 137–147 | 135.96±1.20 | 138.38±0.55 | 139.48±0.42 | 0.201 | 0.025 | 0.307 |
| Cl, mmol/L | 99–110 | 101.27±1.04 | 103.01±0.74 | 102.87±0.56 | 0.441 | 0.450 | 0.998 |
| Ca, mmol/L | 2.11–2.52 | 1.97±0.02 | 2.04±0.03 | 2.24±0.02 | 0.081 | <0.001 | <0.001 |
| Mg, mmol/L | 0.85–1.15 | 1.27±0.31 | 1.34±0.36 | 1±0.01 | 0.998 | 0.765 | 0.709 |
| Phos, mmol/L | 0.85–1.51 | 1.01±0.07 | 0.98±0.04 | 1.16±0.03 | 0.977 | 0.160 | 0.003 |
Notes: P value <0.05 was considered statistically significant; #, absolute count; %, percentage.
Abbreviations: Blood RT: WBC, white blood cell; NEUT, neutrophils; LYMPH, lymphocyte; HGB, haemoglobin; RBC, red blood cell; HCT, hematocrit; PLT, platelet. Coagulogram: PT, prothrombin time; APTT, activated partial thromboplastin time; TT, thrombin time; FIB, fibrinogen content; D-D, D-dimer. Liver function: ALT, alanine aminotransferase; AST, aspartate aminotransferase; TP, total protein; ALB, albumin; GLB, globin; GGT, Alpha-glutamyl transpeptidase; ALP, alkaline phosphatase; TBA, total bile acid. Renal function: BUN, blood urea nitrogen; CREA, creatinine; UA, uric Acid; CO2, carbon dioxide; CYSC, cystatin C. Cardiac function: CK, creatine kinase; LDH, lactate dehydrogenase; CKMB, creatine kinase-MB; MYO, myoglobin; HSTNI, hypersensitive troponin I.
Inflammatory Parameters and Acute-Phase Proteins in Mild, Severe and Critical Patients
For cytokines, IL-10 was significantly higher in group 1 (3:12.80±2.56) than that in group 2 (2.65±0.40) and group 3 (2.41±0.26) (P<0.001) (Figure 1H). Similar trend was observed with IL-6 (group 1 vs group 2 vs group 3 were 56.45±11.26 vs 6.92±1.29 vs 3.83±0.74, respectively, P<0.001) (Figure 1G), CRP (group 1 vs group 2 vs group 3 were 111.58±14.85 vs 78.07±17.96 vs 21.88±5.31, P<0.001), and PCT (group 1 vs group 2 vs group 3 were 1.78±0.62 vs 0.21±0.08 vs 0.04±0.01, P<0.001) (Figure 1C and D). The levels of D-D in group 1 vs group 2 vs group 3 were 4812.06±1639.09 vs 3152.86±1315.49 vs 1405.19±919.54, respectively (P<0.001) (Figure 1E). SAA in group 1 vs group 2 vs group 3 were 125.57±11.77 vs 121.07±12.44 vs 55.93±9.98, respectively (P<0.001) (Figure 1F). Unlike above parameters, SOD levels were the lowest in group 1, highest in group 3, and levels of SOD in group 3 were markedly higher than those in groups 1 and 2 (Figure 1I) (group 1 vs group 2 vs group 3: 127.64±4.82 vs 133.82±4.81 vs 175.47±4.14, P<0.001).
Association of Possible Risk Factors and Disease Outcome
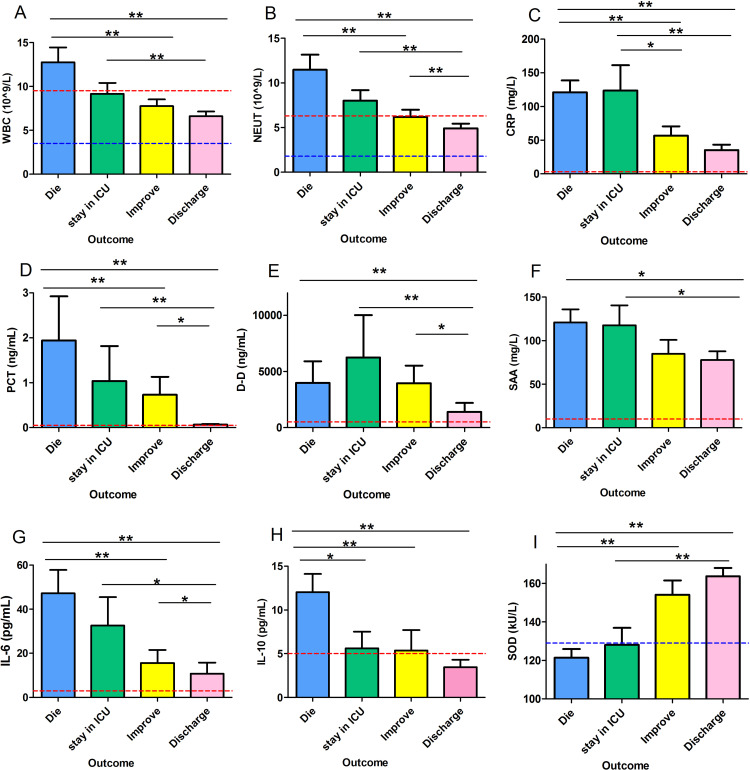
According to the consequences, the patients were divided into four categories (Category 1: died, Category 2: remained in ICU, Category 3: improved, Category 4: discharged) (Figure 2). A significantly different downward trend of WBC count was seen from categories 1 to 4 (from category 1 to category 4: 12.76±1.68 vs 9.16±1.23 vs 7.75±0.77 vs 6.61±0.54, respectively, P<0.001) (Figure 2A). Similar trends were also found in NEUT (from category 1 to category 4: 11.47±1.69 vs 8.01±1.18 vs 6.20±0.81vs. 4.92±0.53, respectively, P<0.001)(Figure 2B), PCT (from category 1 to category 4: 1.95±0.98 vs 1.04±0.78 vs 0.73±0.40 vs 0.07±0.02, respectively, P<0.001)(Figure 2D), and IL-6 (from category 1 to category 4: 47.20±10.60 vs 32.59±12.85 vs 15.60±5.91 vs 10.79±4.93, respectively, P<0.001)(Figure 2G). Level of CRP (from category 1 to category 4:120.89±17.81 vs 123.72±37.48 vs 56.87±13.52 vs 35.33±8.08, respectively, P<0.001)(Figure 2C) and D-D (from category 1 to category 4: 3994.63±1921.21 vs 6249.10±3762.01 vs 3953.74±1574.62 vs 1391.74±802.03, respectively, P<0.001) (Figure 2E) were higher in category 2 than other three categories. The level of IL-10 was significantly higher in category 1 than that in the other three categories (from category 1 to category 4:12.03±2.08 vs 5.61±1.90 vs 5.37±2.33 vs 3.47±0.83, respectively, P<0.001) (Figure 2H). In comparison, there was no significant difference among the other three categories (P>0.05). The levels of SAA were significantly higher in categories 1 and 2 than those in categories 3 and 4 (from group 1 to group 4:120.96±15.03 vs 117.68±22.85 vs 84.75±16.11 vs 77.87±9.94, respectively, P<0.05) (Figure 2F). On the contrary, lower levels of SOD were found in categories with more severe disease (from category 1 to category 4: 121.38±4.48 vs 128.03±8.87 vs 154.12±7.48 vs 163.71±4.29, respectively, P<0.001) (Figure 2I).
Figure 2.
The possible risk factors in COVID-19 patients with different outcomes. (A) (B) (D) (F) (G) (H) Higher levels of WBC, NEUT, PCT, SAA, IL-6, and IL-10 were found in patients with worse outcomes (P<0.05). (C) (E) The same trend was observed in CRP and D-D, except that the highest level was found in group 2, not group 1 (P<0.05). (I) SOD showed a reverse trend to that in other factors (P<0.001). Group1: patients died, Group 2: remained in ICU, Group 3: improved, Group 4: discharge; *P<0.05 and **P<0.01 were considered to be statistically significant.
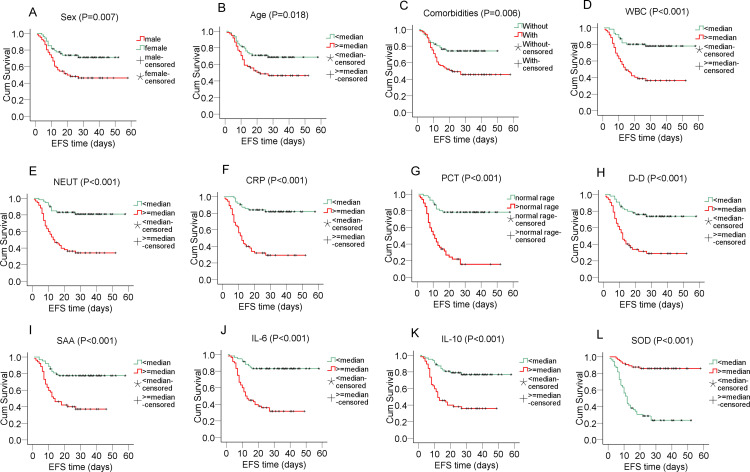
Prognostic Value of Biomarkers on Overall Survival and Event-Free Survival (EFS) in COVID-19 Patients
Event-free survival (EFS) was defined as the time from start of illness to the first event (respiratory distress) or until the last follow up (Figure 3). Male patients tended to have shorter EFS than their female counterparts (male vs female: 32.48±2.99 vs 40.19±2.60, P=0.007) (Figure 3A). Older patients had shorter EFS (old vs young: 29.91±2.72 vs 43.13±2.93, P=0.018) (Figure 3B). COVID-19 patients with comorbidities experienced shorter EFS (with vs without: 32.56±2.88 vs 39.31±2.57, P=0.006) (Figure 3C). Patients with higher WBC (high vs low: 25.51±2.70 vs 47.76±2.51, P<0.001), NEUT (high vs low: 24.48±2.65 vs 49.26±2.39, P<0.001), CRP (high vs low: 22.25±2.71vs.49.78±2.26, P<0.001), PCT (high vs low: 17.36±2.45 vs 47.70±2.64, P<0.001), D-D (high vs low: 22.16±2.78 vs 46.08±2.77, P<0.001), SAA (high vs low: 23.35±2.53 vs 47.25±2.54, P<0.001), IL-6 (high vs low: 22.85±2.43 vs.49.83±2.32, P<0.001), IL-10 (high vs low: 23.92±2.58 vs.47.51±2.48, P<0.001) had decreased EFS (Figure 3D-K). On the contrary, patients with lower levels of SOD had shorter EFS (low vs high: 20.42±2.52 vs.51.34±2.07, P<0.001) (Figure 3L). By taking the parameters in Figure 3 as co-variables, Cox proportional hazard model with forwarding stepwise selection revealed that PCT (hazard ratio: 1.439, 95% CI: 1.223–1.694, P<0.001), and IL-10 (hazard ratio: 1.043, 95% CI: 1.020–1.065, P<0.001) were the independent risk factors affecting EFS. While SOD (hazard ratio: 0.986, 95% CI: 0.979–0.993, P<0.001) was an independent favorable factor for EFS.
Figure 3.
The Kaplan-Meier survival curve and Log-rank test was used to evaluate the suitability of these molecules as prognostic factors. (A) (B) (C) (D) (E) (F) (G) (H) (I) (J) (K) Event-free survival (EFS) was shorter in patients with male gender, older age, with comorbidities, higher levels of WBC, NEUT, CRP, PCT, D-D, SAA, IL-6, and IL-10. These parameters were considered risk factors. (L) However, higher SOD appeared as a protective factor with longer EFS. P<0.05 was considered to be statistically significant.
Diagnostic Performance of Single and Combined Biomarkers in Prediction of COVID-19 Severity
To estimate the diagnostic value of single and combined biomarkers in discriminating between severe and mild patients, we used binary logistic regression models (Table 3) and receiver operating characteristic curve (ROC) analyses (Table 4). For single biomarkers, the AUC of SAA, D-D, WBC, SOD, CRP, NEUT were more than 0.80, AUC of PCT, IL-10, and IL-6 were more than 0.90 in discriminating the critical and mild patients (Model 1). In separating severe and mild patients (Model 2), only AUC of CRP and SOD was greater than 0.8. In Model 3 (critical vs severe), AUC of WBC, NEUT, IL-6, and IL-10 were more than 0.80. As the binary logistic results shown in Table 3, in the Model 1, IL-6 (OR: 1.21, P=0.012, 95% CI: 1.04–1.40), NEUT (OR: 1.62, P=0.014, 95% CI: 1.11–2.35), and PCT (OR: 1372.34, P=0.062, 95% CI: 0.71–2657795.58) were chosen as the optimal panel. In the Model 2, results revealed that only SOD (OR: 0.96, P<0.001, 95% CI: 0.94–0.98) entered the logistic regression equation. In the Model 3, the panel of WBC (OR: 1.44, P=0.055, 95% CI: 0.99–2.10), PCT (OR: 4.85, P=0.156, 95% CI: 0.55–42.87), IL-6 (OR: 1.29, P= 0.05, 95% CI: 1.00–1.67), SOD (OR: 1.08, P=0.031, 95% CI: 1.01–1.16) and IL-10 (OR: 1.81, P=0.043, 95% CI: 1.02–3.23) were selected as best combination. In the next step, when the risk prediction of the selected panel above was analyzed, better diagnostic efficiency was observed in the panels than in a single biomarker. In Model 1, the combined panel of PCT, IL-6, and NEUT showed an AUC of 0.99 with a sensitivity of 90.6% and specificity of 100%, which was better than any single biomarker. In Model 3, the combination of WBC, PCT, IL-6, IL-10, and SOD achieved the highest AUC of 0.95 (95% CI: 0.90,1.00) with a sensitivity of 75% and specificity of 100%.
Table 3.
Regression Analysis of Clinical Model with Critical, The Severe and the Mild
| Clinical Model | OR | P value | 95% CI |
|---|---|---|---|
| Model 1: Critical vs Mild | |||
| IL-6 | 1.21 | 0.012 | (1.04,1.40) |
| NEUT | 1.62 | 0.014 | (1.11,2.35) |
| PCT | 1372.34 | 0.062 | (0.71,2657795.58) |
| Model 2: Severe vs Mild | |||
| SOD | 0.96 | <0.001 | (0.94,0.98) |
| Model 3: Critical vs Severe | |||
| WBC | 1.44 | 0.055 | (0.99,2.10) |
| PCT | 4.85 | 0.156 | (0.55,42.87) |
| IL-6 | 1.29 | 0.050 | (1.00,1.67) |
| SOD | 1.08 | 0.031 | (1.01,1.16) |
| IL-10 | 1.81 | 0.043 | (1.02,3.23) |
Notes: P value <0.05 was considered statistically significant.
Abbreviation: OR, odd ratio.
Table 4.
Clinical Model and Biomarker Outcome Prediction of the Critical, The Severe and the Mild
| Variable(s) | AUC (95% CI) | P value | SD | Cut-off point | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | LR+ | LR- |
|---|---|---|---|---|---|---|---|---|---|---|
| Model 1: Critical vs Mild | ||||||||||
| SAA | 0.81(0.72,0.90) | <0.001 | 0.05 | 16.92 | 100 | 61.70 | 55.80 | 100 | 2.61 | 0 |
| D-D | 0.84(0.75,0.93) | <0.001 | 0.05 | 501.00 | 78.10 | 85.10 | 78.10 | 85.10 | 5.24 | 0.26 |
| WBC | 0.87(0.78,0.96) | <0.001 | 0.05 | 8.06 | 84.40 | 85 | 75.01 | 91.08 | 5.63 | 0.18 |
| SOD | 0.89(0.82,0.96) | <0.001 | 0.04 | 154.05 | 90.0 | 87.5 | 93.1 | 82.4 | 7.2 | 0.1 |
| CRP | 0.89(0.83,0.96) | <0.001 | 0.03 | 32.35 | 87.10 | 83.10 | 73.03 | 92.46 | 5.15 | 0.16 |
| NEUT | 0.89(0.81,0.97) | <0.001 | 0.04 | 5.40 | 90.60 | 86.70 | 78.42 | 94.53 | 6.81 | 0.11 |
| PCT | 0.91(0.83,0.98) | <0.001 | 0.04 | 0.07 | 87.50 | 90.70 | 87.50 | 90.70 | 9.41 | 0.14 |
| IL-10 | 0.91(0.85,0.96) | <0.001 | 0.03 | 3.69 | 87.50 | 78.30 | 68.26 | 92.15 | 4.03 | 0.16 |
| IL-6 | 0.95(0.91,0.99) | <0.001 | 0.02 | 8.42 | 87.50 | 88.30 | 79.95 | 92.98 | 7.48 | 0.14 |
| Panel 1 | 0.99(0.98,1.00) | <0.001 | 0.01 | N/A | 90.60 | 100 | 100 | 93.46 | N/A | 0.09 |
| Model 2: Severe vs Mild | ||||||||||
| IL-10 | 0.53(0.40,0.66) | 0.625 | 0.07 | 2.40 | 42.90 | 66.70 | 37.55 | 71.45 | 1.29 | 0.86 |
| WBC | 0.61(0.47,0.74) | 0.107 | 0.07 | 7.08 | 50 | 81.70 | 56.04 | 77.78 | 2.73 | 0.61 |
| IL6 | 0.67(0.55,0.79) | 0.012 | 0.06 | 0.64 | 89.30 | 40 | 40.99 | 88.90 | 1.49 | 0.27 |
| PCT | 0.68(0.54,0.81) | 0.011 | 0.07 | 0.08 | 46.40 | 90.70 | 97.22 | 19.47 | 4.99 | 0.59 |
| NEUT | 0.69(0.56,0.82) | 0.004 | 0.07 | 5.38 | 57.10 | 86.70 | 66.71 | 81.24 | 4.29 | 0.49 |
| D-D | 0.75(0.63,0.86) | <0.001 | 0.06 | 238.00 | 82.10 | 66 | 58.99 | 86.09 | 2.41 | 0.27 |
| SAA | 0.78(0.69,0.88) | <0.001 | 0.05 | 17.28 | 96.20 | 61.70 | 52.12 | 97.40 | 2.51 | 0.06 |
| CRP | 0.83(0.75,0.92) | <0.001 | 0.04 | 12.26 | 92.9 | 67.80 | 57.79 | 95.27 | 2.89 | 0.10 |
| SOD | 0.89(0.80,0.97) | <0.001 | 0.04 | 156.00 | 88.30 | 89.30 | 94.65 | 78.08 | 8.25 | 0.13 |
| Panel 2 | 0.89(0.80,0.97) | <0.001 | 0.04 | 156.00 | 89.30 | 88.30 | 78.08 | 94.65 | 7.63 | 0.12 |
| Model 3: Critical vs Severe | ||||||||||
| SAA | 0.52(0.37,0.68) | 0.787 | 0.08 | 111.96 | 65.50 | 46.20 | 57.59 | 54.56 | 1.22 | 0.75 |
| SOD | 0.57(0.43,0.72) | 0.328 | 0.08 | 113.45 | 92.90 | 37.50 | 56.53 | 85.79 | 1.49 | 0.19 |
| D-D | 0.60(0.46,0.75) | 0.181 | 0.08 | 477.00 | 78.10 | 48.10 | 64.07 | 64.95 | 1.50 | 0.46 |
| CRP | 0.67(0.52,0.81) | 0.030 | 0.07 | 81.88 | 64.50 | 75 | 74.07 | 65.61 | 2.58 | 0.47 |
| PCT | 0.76(0.64,0.88) | 0.001 | 0.06 | 0.10 | 81.30 | 60.70 | 70.28 | 73.96 | 2.07 | 0.31 |
| WBC | 0.81(0.70,0.93) | <0.001 | 0.06 | 8.05 | 84.40 | 78.60 | 81.84 | 81.51 | 3.94 | 0.20 |
| NEUT | 0.81(0.70,0.93) | <0.001 | 0.06 | 6.69 | 84.40 | 78.60 | 81.84 | 81.51 | 3.94 | 0.20 |
| IL-6 | 0.88(0.80,0.96) | <0.001 | 0.04 | 9.33 | 84.40 | 71.40 | 77.13 | 80.00 | 2.95 | 0.22 |
| IL-10 | 0.89(0.82,0.97) | <0.001 | 0.04 | 3.77 | 87.5 | 75 | 80 | 84 | 3.50 | 0.17 |
| Panel 3 | 0.95(0.90,1.00) | <0.001 | 0.02 | N/A | 75 | 100 | 100 | 77.78 | N/A | 0.25 |
Notes: Panel 1: IL-6, NEUT, PCT; Panel 2: SOD; Panel 3: WBC, PCT, IL-6, SOD, IL-10. AUC, areas under the receiver-operating characteristic curve; PPV, positive predictive value; NPV, negative predictive value; LR, likelihood ratio; P value <0.05 was considered statistically significant.
Abbreviations: WBC, white blood cell; NEUT, absolute neutrophil count; CRP, C-reactive protein; PCT, procalcitonin; D-D, D-dimer; SAA, serum amyloid A; IL-6, interleukin-6; IL-10, interleukin-10; SOD, superoxide dismutase.
Discussion
Due to the severity of the pandemic and patients with COVID-19 are far beyond the hospital’s capacity, it is of utmost importance to set a diagnostic standard in discriminating patients for hospital admission, so that every patient could have appropriate treatment in time, meanwhile saving medical resources. Generally, patients with COVID-19 are divided into four categories according to disease severity: no symptoms, mild/moderate, severe, and critical, their prognosis is significantly different, with deaths often occurring in critical patients. Asymptomatic or some mild cases do not require admission to the hospital; however, severe or critical patients should get treatment immediately. It is hard to triage patients at the disease onset, since the initial symptoms are generally similar. In our patients with COVID-19, there was no significant difference in initial symptoms between severe and mild illness. Fatigue, dyspnea, and diarrhea were more likely to be seen in critical patients, but they are non-specific. Currently, there are no ideal diagnostic biomarkers to classify patients effectively. The disease severity is supposed to be determined by initial viral titers, age, and comorbidity of the infected individual. Old age and comorbidity were more prevalent in our critical and severe patients; however, no marked difference was found between these two groups. Both older and young critical patients developed multi-organ failure. The objective metrics in risk stratification of patients is particularly important, as suboptimal triage causes delays in initial treatment for these patients.
A viral infection may provoke a cytokine storm, inducing immunopathological damage in multi-organs.20–27 Although it is unclear what determinants of the host response to infection are responsible for triggering the inflammatory sequence leading to the clinical syndrome, a cytokine storm is typical in COVID-19 patients with severe-to-critical symptoms.28 IL-6 may be the master cytokine that serves in both pro- and anti-inflammatory.29 Clinically, anti-IL-6 treatment, such as tocilizumab, may stabilize an advanced case from transitioning to a more critical state.30 Besides IL-6, a lack of negative feedback mechanism by IL-10 and IL-4 would be expected to increase the severity of cytokine responses toward a pathogenic cytokine storm.31 IL-2R/lymphocyte was a prominent biomarker for early identification of severe COVID-19, with a sensitivity of 90.9% and a specificity of 66.5% to differentiate between critical and severe patients. Still, the performance of IL-2R/lymphocytes to distinguish severe from mild patients was poor.32 Therefore, developing criteria to predict and diagnose a cytokine storm in COVID-19 patients with surrogate biomarkers is vital because the peak levels of circulating cytokines are not routinely monitored for a change in kinetics.28
In our study, most patients with extremely high levels of IL-6 and IL-10 at disease onset developed a critical and severe illness. We then included IL-6 and IL-10 into the new panel. The liver synthesizes APP in response to IL-6.33 SAA has a regulatory activity on innate immunity and inflammatory cytokines.34 Serum SAA was the best indicator of disease progression in swine influenza cases.35 Combined high levels of both CRP and SAA are indicative of bacterial infection in cases of viral infection, while single elevated SAA points to viral infection.36 In both critical and severe patients, there were significantly higher levels of SAA and CRP compared with the mild patients, indicating that SAA and CRP may be used to stratify disease severity.
Virus-induced oxidative stress could be mediated by an early phase of pro-inflammatory cytokine release. Leukocytes generate reactive oxygen species (ROS) during the oxidative burst to neutralize pathogens,37 the ROS induced by the viral infection has been linked with innate antiviral signaling pathways.38,39 ROS may damage lung parenchyma cells in viral infection40,41. The superoxide dismutase (SOD) is a class of enzymes that scavenges superoxide explicitly in biological systems.42 SODs are the only enzymes that interact with superoxide specifically and thus control the levels of ROS, loss of SOD activity is associated with increased levels of oxidative damage.43 The expression of SOD1 is downregulated by IFN-I signaling during viral infection and that loss of SOD1 results in oxidative damage in the liver2. In COVID-19, significantly lower levels of SOD were found in the critical patients compared with the mild patients, indicating that SOD levels could be biomarkers mirroring tissue damage.
The above biomarkers play a crucial role in the immunopathological mechanisms of viral disease. Our clinical observations revealed that they rose rapidly at the early phase of disease and significantly high levels were observed in critical patients. Therefore, these biomarkers may have the potential to form a panel for patients’ initial triage. Our results showed that they had an excellent performance in the initial screening, especially PCT, IL-10, and IL-6 (AUC>0.90). Among all biomarkers, SAA, SOD, and NEUT had very high sensitivity (>90%), while only PCT had high specificity (>90%). Their diagnostic performance of negative predictive value (NPV) was better than the positive predictive value (PPV). Combining PCT, IL-6, and NEUT (panel one) yielded a higher AUC (0.99), with the sensitivities 90.6% and specificities 100%, PPV was 100.0 NPV was 93.46. This panel has been shown to possess high discriminative power regarding disease severity in patients. For COVID-19, about 50.0% of critical patients died, 18.75% of patients remained in ICU, only 15.63% of patients were discharged from hospital in our study. On the contrary, in mild patients, 21.67% of patients improved, and 78.33% of patients were discharged. Therefore, the distinction between patients who would progress to critically severe illness and those who would remain mildly ill is particularly important. This distinction may help doctors select patients who need immediate care and hospitalization. This optimal panel’s ability to accurately identify high- or low-risk subjects helps to “rule out” the need for immediate medical measures. Therefore, it has a substantial potential to better triage unselected medical patients on admission and during hospitalization.
As opposed to distinguishing between critical and mild patients, it is even harder to separate critical and severe ones. The rate of admission to ICU was 18.75% vs.14.29%; while discharge rate was 15.63% vs 50.0%, for the critical vs severe patients, respectively. When WBC, IL-10, and SOD were added to panel one, AUCs were achieved 0.95, with the sensitivities 75.0% and specificities 100%. Panel three (WBC, PCT, IL-6, IL-10, and SOD) had much better performance in the initial triage model in risk prediction for the critical vs severe patients. When there is diagnostic uncertainty between critical and severe patients, panel three provided good discrimination for identifying those who require urgent treatment to prevent adverse medical outcomes, reducing the mortality in patients with critical disease.
Conclusion
Our study demonstrated that the addition of APP to the conventional inflammatory board might improve the discriminative ability regarding patients’ prognosis. The new panel provides optimal operating characteristics for the disease, which could assist physicians in more rational decisions regarding emergencies. Serum specimens are easily obtainable, quick measurement in the laboratory, and results could be quickly translatable to clinical decision-making44. Therefore, the panels have high practicability. However, the efficacy and safety of the new panels still need to be verified in a large population. Established triage risk score at the hospital admission has a substantial potential to improve triage of unselected medical patients, which may improve patient management and optimize hospital resources.
Funding Statement
This study was funded by the National Key Research and Development Program of China (grant number 2018YFE0204500), National Natural Science Foundation of China (Grant No. 81702273), the Key Project for Anti-2019 Novel Coronavirus Pneumonia from the National Key Research and Development Program of China (Grant No. 2020YFC0845500),Science and technology key project of Guangdong Province: Study on the source and epidemiology of COVID-19 (No.2020B111107001), and Scientific research project of COVID-19 epidemic prevention and control in Guangdong universities (No. 2020KZDZX1087).
Abbreviation
COVID-19, coronavirus disease 2019; ROC, receiver-operating characteristic; AUC, area under the curve; IQR, inter-quartile range; PCT, procalcitonin; IL-6, interleukin-6; IL-10, interleukin-10; NEUT, neutrophils; SOD, superoxide dismutase; WBC, white blood cell; CT, computed tomography; CPR, cardiopulmonary resuscitation; LDH, lactic dehydrogenase; ICU, intensive care unit; PCT, procalcitonin; APR, acute-phase response; APP, acute-phase proteins; SAA, serum amyloid A; PTX, pentraxins; Cr, creatinine; MYO, myoglobin; HSTNI, hypersensitive troponin I; AST, aspartateaminotransferase; ALT, alanineaminotransferase; TNF-α, tumor necrosis factor-α; IFN-Ƴ, interferon-Ƴ; C1q, complement C1q; CBC, complete blood count; ACL, automatic coagulation analyzer; IgG, immunoglobulin G; SE, standard error; EFS, event-free survival; IMV, intermittent mandatory ventilation; ECMO, extracorporeal membrane oxygenation; CRRT, continuous renal replacement therapy; ARDS, acute respiratory distress syndrome; HGB, hemoglobin; RBC, red blood cell; HCT, hematocrit; PLT, platelet; PT, prothrombin time; APTT, activated partial thromboplastin time; TT, thrombin time; FIB, fibrinogen content; D-D, D-dimer; TP, total protein; ALB, albumin; GGT, alpha-glutamyl transpeptidase; ALP, alkaline phosphatase; TBA, total bile acid; BUN, blood urea nitrogen; CYS-C, cystatin C; ROS, reactive oxygen species; PPV, positive predictive value; NPV, negative predictive value.
Ethical Approval
The study was reviewed and approved by Medical Ethics Committee Zhongnan Hospital of Wuhan University in the Scientific Research project No 2020051K. All patients provided informed consent, and that this study was conducted in accordance with the Declaration of Helsinki.
Author Contributions
TYT and XYJ designed and drafted the manuscript. YRL revised the final manuscript. HQP collected and summarized the clinical laboratory and clinical data. JYS, FY and TYT processed statistical data. YRL and HQP had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest for this work.
References
- 1.Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;7. doi: 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharya A, Hegazy AN, Deigendesch N, et al. Superoxide dismutase 1 protects hepatocytes from type I interferon-driven oxidative damage. Immunity. 2015;43(5):974–986. doi: 10.1016/j.immuni.2015.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakraborty C, Sharma AR, Sharma G, Bhattacharya M. SARS-CoV-2 causing pneumonia-associated respiratory disorder (COVID-19): diagnostic and proposed therapeutic options. Eur Rev Med Pharmacol Sci. 2020;24(7):4016–4026. doi: 10.26355/eurrev_202004_20871 [DOI] [PubMed] [Google Scholar]
- 4.Wynants L, Van Calster B, Collins GS, et al. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ. 2020;369:m1328. doi: 10.1136/bmj.m1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zakham F, Pillonel T, Brunel AS, et al. Molecular diagnosis and enrichment culture identified a septic pseudoarthrosis due to an infection with Erysipelatoclostridium ramosum. Int J Infect Dis. 2019;81:167–169. doi: 10.1016/j.ijid.2019.02.001 [DOI] [PubMed] [Google Scholar]
- 6.Gao R, Wang L, Bai T, Zhang Y, Bo H, C-Reactive Protein SY. Mediating immunopathological lesions: a potential treatment option for severe influenza A diseases. EBioMedicine. 2017;22:133–142. doi: 10.1016/j.ebiom.2017.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meynaar IA, Droog W, Batstra M, Vreede R, Herbrink P. In critically ill patients, serum procalcitonin is more useful in differentiating between sepsis and SIRS than CRP, Il-6, or LBP. Crit Care Res Pract. 2011;2011:594645. doi: 10.1155/2011/594645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller F, Christ-Crain M, Bregenzer T, et al. Procalcitonin levels predict bacteremia in patients with community-acquired pneumonia: a prospective cohort trial. Chest. 2010;138(1):121–129. doi: 10.1378/chest.09-2920 [DOI] [PubMed] [Google Scholar]
- 9.Sager R, Kutz A, Mueller B, Schuetz P. Procalcitonin-guided diagnosis and antibiotic stewardship revisited. BMC Med. 2017;15(1):15. doi: 10.1186/s12916-017-0795-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alba GA, Truong QA, Gaggin HK, et al. Diagnostic and prognostic utility of procalcitonin in patients presenting to the emergency department with dyspnea. Am J Med. 2016;129(1):96–104 e7. doi: 10.1016/j.amjmed.2015.06.037 [DOI] [PubMed] [Google Scholar]
- 11.Mitsuma SF, Mansour MK, Dekker JP, et al. Promising new assays and technologies for the diagnosis and management of infectious diseases. Clin Infect Dis. 2013;56(7):996–1002. doi: 10.1093/cid/cis1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oberfeld B, Achanta A, Carpenter K, et al. SnapShot: COVID-19. Cell. 2020;181(4):954–954e1. doi: 10.1016/j.cell.2020.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez L. Acute phase protein response to viral infection and vaccination. Arch Biochem Biophys. 2019;671:196–202. doi: 10.1016/j.abb.2019.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vollmer AH, Gebre MS, Barnard DL. Serum amyloid A (SAA) is an early biomarker of influenza virus disease in BALB/c, C57BL/2, Swiss-Webster, and DBA.2 mice. Antiviral Res. 2016;133:196–207. doi: 10.1016/j.antiviral.2016.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Buck M, Gouwy M, Wang JM, et al. Structure and expression of different serum amyloid A (SAA) variants and their concentration-dependent functions during host insults. Curr Med Chem. 2016;23(17):1725–1755. doi: 10.2174/0929867323666160418114600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haase-Fielitz A, Bellomo R, Devarajan P, et al. Novel and conventional serum biomarkers predicting acute kidney injury in adult cardiac surgery–a prospective cohort study. Crit Care Med. 2009;37(2):553–560. doi: 10.1097/CCM.0b013e318195846e [DOI] [PubMed] [Google Scholar]
- 17.Roos JF, Doust J, Tett SE, Kirkpatrick CM. Diagnostic accuracy of cystatin C compared to serum creatinine for the estimation of renal dysfunction in adults and children–a meta-analysis. Clin Biochem. 2007;40(5–6):383–391. doi: 10.1016/j.clinbiochem.2006.10.026 [DOI] [PubMed] [Google Scholar]
- 18.Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Organization WH. Laboratory testing for 2019 novel coronavirus (2019‐nCoV) in suspected human cases. https://apps.who.int/iris/bitstream/handle/10665/330676/9789240001275-chi.pdf. Accessed September21, 2020.
- 20.Povoa TF, Oliveira ER, Basilio-de-Oliveira CA, et al. Peripheral organs of dengue fatal cases present strong pro-inflammatory response with participation of IFN-gamma-, TNF-alpha- and RANTES-producing cells. PLoS One. 2016;11(12):e0168973. doi: 10.1371/journal.pone.0168973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J, Lau YF, Lamirande EW, et al. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection. J Virol. 2010;84(3):1289–1301. doi: 10.1128/JVI.01281-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dahlke C, Lunemann S, Kasonta R, et al. Comprehensive characterization of cellular immune responses following Ebola virus infection. J Infect Dis. 2017;215(2):287–292. doi: 10.1093/infdis/jiw508 [DOI] [PubMed] [Google Scholar]
- 23.Caballero IS, Honko AN, Gire SK, et al. In vivo Ebola virus infection leads to a strong innate response in circulating immune cells. BMC Genomics. 2016;17:707. doi: 10.1186/s12864-016-3060-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McElroy AK, Akondy RS, Davis CW, et al. Human Ebola virus infection results in substantial immune activation. Proc Natl Acad Sci U S A. 2015;112(15):4719–4724. doi: 10.1073/pnas.1502619112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feldmann H, Geisbert TW. Ebola haemorrhagic fever. Lancet. 2011;377(9768):849–862. doi: 10.1016/S0140-6736(10)60667-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam CW, Chan MH, Wong CK. Severe acute respiratory syndrome: clinical and laboratory manifestations. Clin Biochem Rev. 2004;25(2):121–132. [PMC free article] [PubMed] [Google Scholar]
- 27.Martina BE, Koraka P, Osterhaus AD. Dengue virus pathogenesis: an integrated view. Clin Microbiol Rev. 2009;22(4):564–581. doi: 10.1128/CMR.00035-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kahn R, Schmidt T, Golestani K, et al. Mismatch between circulating cytokines and spontaneous cytokine production by leukocytes in hyperinflammatory COVID-19. J Leukoc Biol. 2020. doi: 10.1002/JLB.5COVBCR0720-310RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunter CA, Jones SA. IL-6 as a keystone cytokine in health and disease. Nat Immunol. 2015;16(5):448–457. doi: 10.1038/ni.3153 [DOI] [PubMed] [Google Scholar]
- 30.Quartuccio L, Sonaglia A, McGonagle D, et al. Profiling COVID-19 pneumonia progressing into the cytokine storm syndrome: results from a single Italian Centre study on tocilizumab versus standard of care. J Clin Virol. 2020;129:104444. doi: 10.1016/j.jcv.2020.104444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, Katze MG. Into the eye of the cytokine storm. Microbiol Mol Biol Rev. 2012;76(1):16–32. doi: 10.1128/MMBR.05015-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou H, Zhang B, Huang H, et al. Using IL-2R/lymphocytes for predicting the clinical progression of patients with COVID-19. Clin Exp Immunol. 2020;201(1):76–84. doi: 10.1111/cei.13450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pepys MB, Hirschfield GM. C-reactive protein: a critical update. J Clin Invest. 2003;111(12):1805–1812. doi: 10.1172/JCI18921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cox N, Pilling D, Gomer RH. Serum amyloid P: a systemic regulator of the innate immune response. J Leukoc Biol. 2014;96(5):739–743. doi: 10.1189/jlb.1MR0114-068R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pomorska-Mol M, Markowska-Daniel I, Kwit K. Immune and acute phase response in pigs experimentally infected with H1N2 swine influenza virus. FEMS Immunol Med Microbiol. 2012;66(3):334–342. doi: 10.1111/j.1574-695X.2012.01026.x [DOI] [PubMed] [Google Scholar]
- 36.Todorov I, Gospodinova M, Bocheva Y, Popcheva G. Serum amyloid A protein in the course of infectious mononucleosis. Ther Adv Infect Dis. 2019;6:2049936118811208. doi: 10.1177/2049936118811208 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Tauber AI, Babior BM. Evidence for hydroxyl radical production by human neutrophils. J Clin Invest. 1977;60(2):374–379. doi: 10.1172/JCI108786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Indukuri H, Castro SM, Liao SM, et al. Ikkepsilon regulates viral-induced interferon regulatory factor-3 activation via a redox-sensitive pathway. Virology. 2006;353(1):155–165. doi: 10.1016/j.virol.2006.05.022 [DOI] [PubMed] [Google Scholar]
- 39.Gonzalez-Dosal R, Horan KA, Rahbek SH, et al. HSV infection induces production of ROS, which potentiate signaling from pattern recognition receptors: role for S-glutathionylation of TRAF3 and 6. PLoS Pathog. 2011;7(9):e1002250. doi: 10.1371/journal.ppat.1002250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suliman HB, Ryan LK, Bishop L, Folz RJ. Prevention of influenza-induced lung injury in mice overexpressing extracellular superoxide dismutase. Am J Physiol Lung Cell Mol Physiol. 2001;280(1):L69–L78. doi: 10.1152/ajplung.2001.280.1.L69 [DOI] [PubMed] [Google Scholar]
- 41.Geiler J, Michaelis M, Naczk P, et al. N-acetyl-L-cysteine (NAC) inhibits virus replication and expression of pro-inflammatory molecules in A549 cells infected with highly pathogenic H5N1 influenza A virus. Biochem Pharmacol. 2010;79(3):413–420. doi: 10.1016/j.bcp.2009.08.025 [DOI] [PubMed] [Google Scholar]
- 42.Fridovich I. The biology of oxygen radicals. Science. 1978;201(4359):875–880. doi: 10.1126/science.210504 [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Branicky R, Noe A, Hekimi S. Superoxide dismutases: dual roles in controlling ROS damage and regulating ROS signaling. J Cell Biol. 2018;217(6):1915–1928. doi: 10.1083/jcb.201708007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schultz M, Rasmussen LJH, Kallemose T, et al. Availability of suPAR in emergency departments may improve risk stratification: a secondary analysis of the TRIAGE III trial. Scand J Trauma Resusc Emerg Med. 2019;27(1):43. doi: 10.1186/s13049-019-0621-7 [DOI] [PMC free article] [PubMed] [Google Scholar]