Abstract
Background & objectives:
The presence of Cat Que virus (CQV) in Culex mosquitoes and pigs has been reported in China and Vietnam. Due to the spread of similar species of the Culex mosquitoes in India, there is a need to understand the replication kinetics of this virus in mosquito models. As a part of preparedness and to identify the presence of this CQV in humans and swine, this study was carried out to develop diagnostic tests.
Methods:
Serological and molecular diagnostic assays were developed for testing the mosquito population, human and swine serum samples. In this line, RNA-dependent RNA polymerase (L), glycoprotein (M) and nucleocapsid (S) genes-based reverse transcription-polymerase chain reaction (RT-PCR) assays were developed for CQV. Real-time RT-PCR was used for screening of retrospectively collected human serum samples (n=1020) with acute febrile illness during 2014-2017. Simultaneously, an in-house anti-CQV swine and human IgG ELISAs were also developed to detect anti-CQV IgG antibody. Human serum samples (n=883) with post-onset of disease (POD) >4 days and swine serum samples (n=459) were tested for the presence of anti-CQV IgG antibodies. CQV NIV 612,045 isolate was used for susceptibility and replication kinetics experiment using three different species of mosquitoes to understand its behaviour in Indian mosquitoes.
Results:
All human serum samples (n=1020) screened for the presence of CQV using real-time RT-PCR were found to be negative. Anti-CQV IgG antibody positivity was recorded in two of 883 human serum samples tested. Virus susceptibility experiments indicated that three species of mosquito, namely Aedes aegypti, Culex quinquefasciatus and Cx. tritaeniorhynchus supported multiplication of CQV by intrathoracic as well as artificial membrane/oral feeding routes.
Interpretation & conclusions:
Anti-CQV IgG antibody positivity in human serum samples tested and the replication capability of CQV in mosquitoes indicated a possible disease causing potential of CQV in Indian scenario. Screening of more human and swine serum samples using these assays is required as a proactive measure for understanding the prevalence of this neglected tropical virus.
Keywords: Anti-CQV IgG, Cat Que virus, ELISA, mosquito, real-time RT-PCR
Genus Orthobunyavirus is the largest genus of the Peribunyaviridae family consisting of more than 170 viruses. Orthobunyaviruses are classified into 49 species and 19 serogroups complexes1,2,3,4, which consist of highly diverse arboviruses that infect both humans as well as economically important livestock species5,6,7,8,9. Simbu serogroup is the largest of these with 25 viruses isolated so far5,10 and with seven species complexes namely Akabane, Manzanilla, Oropouche, Sathuperi, Simbu, Shamonda and Shuni. Cat Que virus (CQV) is a member of the Manzanilla (MAN) species complex belonging to Simbu serogroup. Isolation of CQV from mosquitoes was first reported in 2004 during surveillance of arboviruses in cases of acute paediatric encephalitis in Vietnam11 and later in Uganda12.
Earlier in 1961, a virus from Jungle Myna (Acridotheres fuscus) serum sample was isolated from Sagar district of Karnataka State, India13. Efforts for its characterization failed for many years. In 2016, using the next-generation sequencing technology, it was identified as CQV [JM1 isolate-National Institute of Virology (NIV) 612,045)14. CQV NIV 612,045 was found to be closely related to Oya virus (OYAV) and Ingwavuma virus (INGV) which are also reported from India and characterized in 2016. OYAV, INGV and MANV form distinct core MAN clade with an amino acid identity of 96.2 to 99.6 per cent while comparing with CQV15. Viruses belonging to MAN clade have a high degree of genetic similarity and are difficult to differentiate at the molecular level. Confirmation of the specific virus can only be done using conventional reverse transcription-polymerase chain reaction (RT-PCR) followed by sequencing13.
CQV has been isolated and reported from its natural host, mosquito (Culex tritaeniorhynchus)16, but the role of birds as a host or vector for CQV transmission and report of human infection with CQV are not documented. Domestic pigs are the primary mammalian host of CQV and anti-CQV IgM and IgG antibodies have been reported in swine reared locally in China, indicating that CQV has formed a natural cycle in the local area16. Considering the isolations of CQV from neighbouring countries such as China and Vietnam in mosquitoes and assuming the spread of the vector mosquitoes (Culex) in India, an attempt was made to develop diagnostic assays for rapid detection of CQV.
Material & Methods
The study was conducted during 2017 to 2018 in the ICMR-National Institute of Virology, Pune, India, after obtaining prior approval from the Institutional Ethics Committee. For standardization of molecular and serological assays and replication kinetics in mosquitoes, the CQV NIV 612,045 isolate was grown in Vero CCL-81 (ATCC, USA) with a titre of 105.5/mlTCID50 (Median tissue culture infectious dose).
Human serum samples [n=1020, Karnataka (806), Maharashtra (116), Kerala (51), Madhya Pradesh (20) and Gujarat (27) States] obtained from patients with acute febrile illness who were negative for Crimean congo haemorrhagic fever (CCHF) and Kyasanur forest disease (KFD), Dengue (DENV) and Chikungunya viruses, received during 2014-2017 and available at the ICMR-NIV were used for molecular screening and samples with post-onset of disease (POD) more than four days [(n=883, Karnataka (669), Maharashtra (116), Kerala (51), Madhya Pradesh (20) and Gujarat (27)] were used for anti-CQV human IgG ELISA. Swine serum samples [(n=459, Patna (30), Meghalaya (4), Uttar Pradesh (24), Gorakhpur (10), Karnataka (5), Kerala (10), Nagaland (19), Mizoram (334) Odisha (13) and Indian Veterinary Research Institute (IVRI), Izatnagar (10)] received during 2015 to 2017 were screened for the presence of anti-CQV IgG antibodies. Mosquito samples from virus susceptibility experiments study were used for CQV screening by real-time RT PCR.
Development of RT-PCR for detection of CQV: Using the CQV NIV 612,045 isolate (Accession No. MH507152, MH507153, MH507154) primers were designed for nucleocapsid (S), glycoprotein (M) and RNA-dependent RNA polymerase (L) genes, using online tool Primer 3 v0.4.017 and were synthesized by Sigma-Aldrich, USA. RNA extraction was performed using QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) as per the manufacturer's instruction. For RT-PCR, 5 μl of extracted RNA was used. The reactions were carried out using Superscript III single step RT-PCR system with Platinum Taq High Fidelity (Invitrogen, USA), according to the manufacturer's instructions. The thermal cycling conditions optimized for RT-PCR for S gene (primers: CQV S_101forward 5'-TCCGGAGGCACAATATGTGGC-3'; CQV_ S_549 reverse 5'-AGTACGCGGTGCATCTCAATCAC-3')and M gene (primers: CQV M_1901forward 5'-TACTGTCAGAGTGCTGATATTGATGCC-3'; CQV_M_2487reverse 5'-CTTGATAGCAGTATCCGCATCTAGCCTA-3') were 50°C for 45 min, 94°C for 5 min, 94°C for 45 sec, 55°C for 45 sec and 68°C for 45 sec and repeat 40 cycles and the final extension of 68°C for 5 minutes. The thermal cycling conditions for L gene (primers: CQV_L_358forward 5'-GGAATAGATTATTGAGATTCGTGATTATA-3'; CQV_L_653 reverse 5'-CGTTCTTCTTCAGGCAT AGATTCTA-3') were 50°C for 30 min, 94°C for 5 min, 94°C for 30 sec, 46°C for 30 sec and 68°C for 30 sec and repeat 40 cycles and the final extension of 68°C for 5 minutes. Primers were checked for specificity with Nairovirus (Dugbe orthonairovirus), Phlebovirus (Malsoor), flaviviruses including DENV, Japanese encephalitis virus (JEV) and other orthobunyaviruses such as Umbre virus (UMBV) and Batai virus (BATV).
DNA sequencing of the amplified products was carried out using an ABI 3100 Automated DNA Sequencer (ABI PRISM® 3100 Genetic Analyzer, Applied Biosystems, USA) and the Big Dye Terminator® Kit (Thermo Fisher Scientific, USA). The sequences generated were analyzed using the nucleotide BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi?LINK_LOC=blasthome&PAGE_TYPE= BlastSearch&PROGRAM=blastn).
Screening of human serum samples for CQV using RT-PCR: Real-time RT-PCR primers and probe as described by Yadav et al14 were used for screening of human samples. These primers and probes were also able to detect CQV because of similarity at nucleotide sequences level among CQV and OYAV. Using SuperScript III One-Step qRT-PCR kit with Platinum Taq (Invitrogen, USA), these primers/probes were checked for sensitivity for detection of CQV NIV 612,045 isolate. A total of 1020 human serum samples collected earlier were screened by real-time RT-PCR assay.
Susceptibility of Culex and Aedes mosquito species to CQV NIV 612,045 isolate
Intra-thoracic (ITI) inoculation: Indian Aedes aegypti, Culex quinquefasciatus and Cx. tritaeniorhynchus mosquito species used in this study were from laboratory colonies maintained at the ICMR-NIV, Pune. To study the susceptibility to the CQV NIV 612,045 isolate, female mosquitoes of the three different species were inoculated with CQV (105.5/mlTCID50) via intrathoracic (ITI) route. The inoculation technique was essentially similar to that employed by Mourya et al18. The inoculated mosquitoes were placed in three separate plastic jars (3 cm in diameter and 7 cm in height) covered with mosquito netting. These jars were kept within the wire mesh cages (30 × 30 × 38 cm), which were covered with a wet lint cloth to provide the required temperature 28±2°C and relative humidity 85±5 per cent inside a Biochemical Oxygen Demand (BOD) incubator (CI-10S, Remi, Mumbai). During the entire period of incubation (10-14) days, these were maintained on 10 per cent glucose solution soaked in cotton wool pledget16. A total of 50 mosquitoes from each of the species were infected with the virus from which five infected mosquitoes from each species were stored at alternate post-inoculation days (PIDs) from 0 to 14th PID. These were tested for the presence CQV real-time RT-PCR.
The artificial membrane feeding system: Ten-fold dilution of the virus was made in defibrinated chicken (white leghorn) blood, 0.5 ml of the neat CQV NIV 612,045 isolate (105.5/mlTCID50) was added to 1.5 ml of the defibrinated blood and pulse vortexed for 30 seconds. Four to five day old female mosquitoes Ae. aegypti, Cx. quinquefasciatus and Cx. tritaeniorhynchus were fed on the blood virus mixture through an artificial membrane (Parafilm, American National Can, USA)18. This membrane feeding method mimics the natural route of virus infection in mosquitoes through a blood meal on any infected patient. The membrane fed mosquitoes were also incubated in the same manner as described for the ITI inoculated mosquitoes. The remaining pre- and post-fed virus-blood mixture was then taken for RNA extraction (Qiagen, Hilden, Germany), and 5 μl of the extracted RNA was used for CQV real-time RT-PCR.
Development of anti-CQV IgG ELISA for the screening of human and swine serum samples: Gamma-inactivated CQV infected Vero CCL-81 cell lysate antigen was coated on the maxisorp microwell plates (Merck, Darmstadt, Germany). Microwell plates were coated from rows A-D with NIV 612,045 (positive antigen) and uninfected Vero CCL-81 cell lysate (control antigen) from rows E-H at 1:10 dilution in carbonate buffer (pH 9.2, 0.025 M) and incubated overnight at 4°C (100 μl/well). Plates were blocked with two per cent bovine serum albumin (BSA) (100 μl/well) and kept at room temperature for two hours. The plates were stored at 4°C after drying until further use. One in hundred diluted human serum samples and one in 500 diluted swine serum samples [in 1% BSA in 10 mM phosphate-buffered saline (PBS) containing 0.1% Tween-20] were added in positive as well as control antigen-coated wells and incubated at 37°C for one hour in respective human and swine ELISA tests. Anti-human IgG horseradish peroxidase (HRP) (Thermo Fisher Scientific, USA) (1:30,000; 100 μl/well) and anti-swine IgG HRP (1:20,000; 1:100 μl/well) were added to the wells in human/swine IgG ELISA, respectively, and incubated at 37°C for one hour. TMB (3,3', 5,5'-tetramethylbenzidine) (Clinical Science Product incorporation NeA blue, USA) substrate (100 μl/well) was further added to the wells and incubated at 37°C for one hour, the reaction was terminated with 1 N H2 SO4 and absorbance was measured at 450 nm. At the end of each step of this assay except the addition of TMB, all the wells were washed five times using 10 mM PBS, pH 6.8 with 0.1 per cent Tween-20 (Sigma-Aldrich, USA). For an unknown sample, OD value more than or equal to 0.2 and the P/N ratio more than or equal to 1.5 were considered as positive or else the sample was considered as negative for the presence of anti-CQV IgG antibodies. The assay was checked for cross-reactivity using anti-CCHF virus (CCHFV), KFD virus (KFDV), DENV and JEV IgG-positive human samples. Human serum samples having POD >4 days (n=883) and all swine serum samples (n=459) were used for screening of anti-CQV IgG antibodies.
Results
Development of RT-PCR for CQV: Using CQV isolate (105.5/mlTCID50) RNA and negative controls the primers were standardized for S, M and L genes. The expected size of PCR products (S gene - 448 bp, M gene - 586 bp, L gene - 295 bp) were generated using these primers (Fig. 1). The assay was found to detect CQV as well as OYAV and no cross-reactivity was observed for Dugbe orthonairovirus, Malsoor virus, DENV, JEV, UMBV and BATV. To determine and confirm the sequences, the amplified product of the all three (L, M and S) genes were purified and sequenced.
Fig. 1.
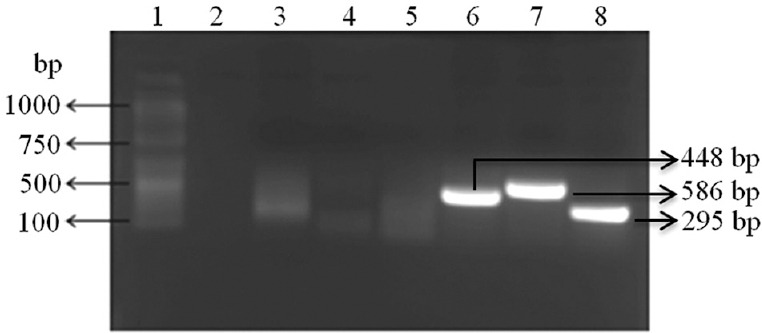
Cat Que virus reverse transcription-polymerase chain reaction for L, M and S genes. Lane 1, 100 bp DNA ladder; lane 2, no template control; lane 3, negative extraction control for S gene; lane 4, negative extraction control for M gene; lane 5, negative extraction control for L gene; lane 6, S gene PCR product; lane 7, M gene PCR product; lane 8, L gene PCR product.
BLAST analysis of the sequences generated showed 100 per cent homology with CQV sequences available in NCBI database.
Screening of human serum samples and screening for CQV in mosquitoes using real-time RT-PCR: To determine the sensitivity of real-time RT-PCR specific to CQV NIV 612,045 (Fig. 2), the CQV RNA was serially diluted (10-fold) and tested, and the assay was found to be sensitive to detect CQV RNA copies up to 10−6 dilutions (Fig. 3). All the 1020 human samples were found to be negative for the presence of MANV clade-specific viruses. CQV RNA was detected from 2nd PID onwards in the ITI infected Indian strain of Ae. aegypti, Cx. Quinquefasciatus and Cx. tritaeniorhynchus mosquitoes by CQV-specific real-time RT-PCR (Table I). The real-time data depicted that the CQV multiplied in all three species of mosquitoes over a period of 12 days post-infection via ITI inoculation as well as membrane feeding (natural mode of infection) routes (Table II).
Fig. 2.
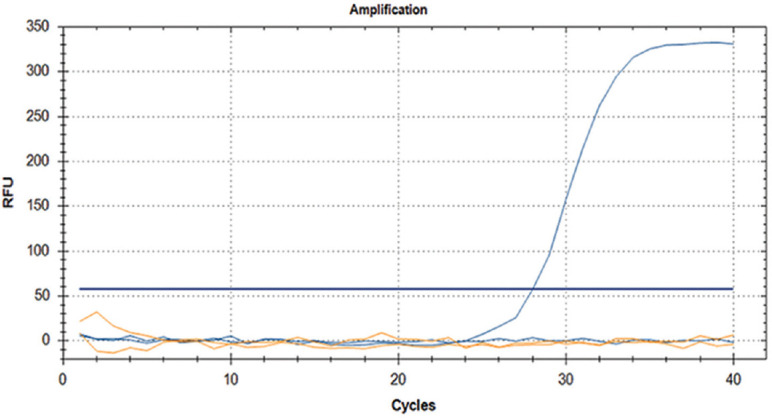
Cat Que virus real-time reverse transcription-polymerase chain reaction using CQV NIV 612,045 isolate. RFU, relative fluorescence units.
Fig. 3.
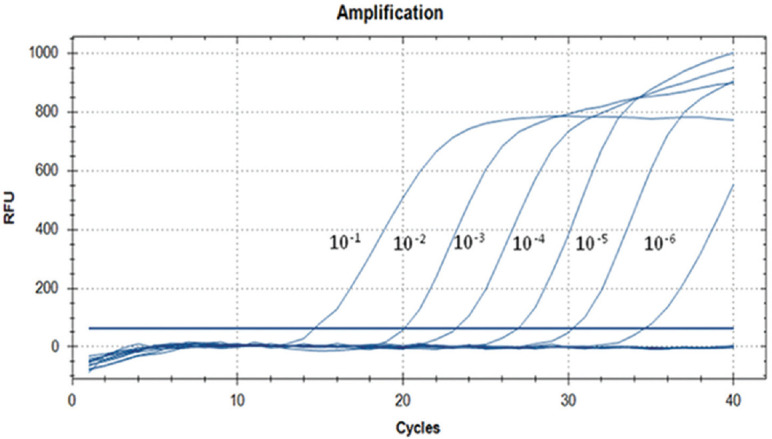
Sensitivity of Cat Que virus real-time reverse transcription-polymerase chain reaction.
Table I.
Comparative results of Cat Que virus (CQV) replication in different species of Indian mosquitoes infected with CQV NIV 612,045 via intrathoracic inoculation route
| Post-infection day | Results of CQV real-time RT-PCR (Ct value) | ||
|---|---|---|---|
| Aedes aegypti | Culex quinquefasciatus | Culex tritaeniorhynchus | |
| 0 | 27.5 | 25.35 | 25.7 |
| 2 | 17.2 | 20.5 | 22.8 |
| 4 | 16.7 | 21.4 | 22.5 |
| 6 | 17.1 | 19.2 | 23.9 |
| 8 | 15.5 | 20.1 | 23.5 |
| 10 | 17.2 | 18.5 | 20.6 |
| 12 | 17.1 | 18.8 | All dead |
| 14 | 18.4 | 18.6 | All dead |
Real-time RT-PCR, real-time reverse transcription-polymerase chain reaction; Ct, threshold cycle
Table II.
Comparative results of Cat Que virus (CQV) replication kinetics in different species of Indian mosquitoes infected with CQV NIV 612,045 via membrane fed (oral feeding) route
| Post-infection day | Results of CQV real-time RT-PCR vi (Ct value) | ||
|---|---|---|---|
| Aedes aegypti | Culex quinquefasciatus | Culex tritaeniorhynchus | |
| 0 | 25.2 | 22 | 22.2 |
| 2 | 34 | No Ct | 35 |
| 4 | No Ct | Since number of fed mosquitoes was less, so each day, mosquitoes was not stored | No Ct |
| 6 | Since number of fed mosquitoes was less, so each day, mosquitoes were not stored | Since number of fed mosquitoes was less, so each day, mosquitoes were not stored | |
| 8 | |||
| 10 | |||
| 12 | 31.3 | 30.1 | |
Screening of human and swine serum samples for anti-CQV IgG: Human serum samples (n=833) were screened for the presence of anti-CQV antibodies using indigenously developed anti-CQV IgG ELISA. Two samples collected from Karnataka in 2014 and 2017 respectively were found to be positive for the presence of anti-CQV IgG antibodies (OD: 0.289, P/N: 3.43; OD: 0.242, P/N: 1.77); all the remaining samples were found to be negative (OD: range: 0.05-0.199 and P/N 1.0-1.5). No cross-reactivity was recorded using anti-CCHFV, KFDV, DENV and JEV IgG-positive samples. All swine serum samples (n=459) were found to be negative for the presence of anti-CQV IgG antibodies (OD: range: 0.05-0.199 and P/N 1.0-1.5).
Discussion
Arthropod-borne viruses (arboviruses) have become a significant public health concern, with the emergence and re-emergence of arboviral diseases worldwide. Humans get affected with viruses belonging to Orthobunyavirus genus, transmitted by mosquitoes like Cache valley virus causing meningitis, La Crosse virus causing paediatric encephalitis, Jamestown canyon encephalitis, Guaroa virus causing febrile illness7,8,19,20,21. Presence of CQV in Culex mosquitoes in China16 and pigs in Vietnam11 suggested susceptibility of Asian countries to CQV. Availability of vector, primary mammalian host (swine) and confirmation of CQV from jungle myna signifies the potential of this Orthobunyavirus as a public health pathogen in India. This led us to develop molecular and serological tests for CQV, screening of host population (human and swine) and its replication kinetics in mosquitoes as a part of preparedness against the likely emergence of CQV. Data showed that Indian mosquitoes (Ae. aegypti, Cx. quinquefasciatus and Cx. tritaeniorhynchus) were susceptible to CQV when the virus was inoculated by both the ITI and oral routes. Thus, mosquitoes were found to be a potential vector for CQV transmission to mammalian hosts. The diagnostic assays developed during this study could be used for rapid detection of CQV as part of vigilance for future CQV outbreaks. Mosquito susceptibility data revealed involvement of mosquitoes as potential vectors for CQV transmission.
This study had limitations: (i) Due to the unavailability of sufficient positive serum sample, virus neutralization test could not be performed to confirm its specificity; (ii) More human and swine samples need to be screened for further strengthening the outcome of the study; and (iii) To confirm the presence of CQV from OYAV, sequencing of the amplified PCR product for L, M and S genes is suggested as RT-PCR can amplify both CAQV as well as OYAV as they have high degree of genetic similarity.
In conclusion, the study reports the development of molecular and serological assays for detection of the CQV. Replication kinetics in Indian species of mosquitoes supported the multiplication of CQV. Detection of antibodies against CQV in human serum samples indicates the need for the cross-sectional surveillance to understand the circulation of this virus in India. The diagnostic tools developed in this study may be useful in the preparedness for detection of CQV in human and swine populations.
Footnotes
Financial support & sponsorship: This study was funded by an intramural research grant from ICMR-National Institute of Virology, Pune.
Conflicts of Interest: None.
References
- 1.Maes P, Alkhovsky SV, Bào Y, Beer M, Birkhead M, Briese T, et al. Taxonomy of the family Arenaviridae and the order Bunyavirales: Update 2018. Arch Virol. 2018;163:2295–310. doi: 10.1007/s00705-018-3843-5. [DOI] [PubMed] [Google Scholar]
- 2.de Brito Magalhães CL, Drumond BP, Novaes RFV, Quinan BR, de Magalhães JC, dos Santos JR, et al. Identification of a phylogenetically distinct Orthobunyavirus from group C. Arch Virol. 2011;156:1173–84. doi: 10.1007/s00705-011-0976-1. [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann B, Scheuch M, Höper D, Jungblut R, Holsteg M, Schirrmeier H, et al. Novel Orthobunyavirus in cattle, Europe, 2011. Emerg Infect Dis. 2012;18:469–72. doi: 10.3201/eid1803.111905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briese T, Calisher CH, Higgs S. Viruses of the family Bunyaviridae: Are all available isolates reassortants? Virology. 2013;446:207–16. doi: 10.1016/j.virol.2013.07.030. [DOI] [PubMed] [Google Scholar]
- 5.Ladner JT, Savji N, Lofts L, Travassos da Rosa A, Wiley MR, Gestole MC, et al. Genomic and phylogenetic characterization of viruses included in the manzanilla and oropouche species complexes of the genus Orthobunyavirus, family Bunyaviridae. J Gen Virol. 2014;95:1055–66. doi: 10.1099/vir.0.061309-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saeed MF, Li L, Wang H, Weaver SC, Barrett AD. Phylogeny of the Simbu serogroup of the genus Bunyavirus. J Gen Virol. 2001;82:2173–81. doi: 10.1099/0022-1317-82-9-2173. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen NL, Zhao G, Hull R, Shelly MA, Wong SJ, Wu G, et al. Cache valley virus in a patient diagnosed with aseptic meningitis. J Clin Microbiol. 2013;51:1966–9. doi: 10.1128/JCM.00252-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bewick S, Agusto F, Calabrese JM, Muturi EJ, Fagan WF. Epidemiology of La Crosse virus emergence, Appalachia region, United States. Emerg Infect Dis. 2016;22:1921–9. doi: 10.3201/eid2211.160308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oluwayelu D, Wernike K, Adebiyi A, Cadmus S, Beer M. Neutralizing antibodies against Simbu serogroup viruses in cattle and sheep, Nigeria, 2012-2014. BMC Vet Res. 2018;14:277. doi: 10.1186/s12917-018-1605-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodrigues DS, Medeiros DB, Rodrigues SG, Martins LC, de Lima CP, de Oliveira LF, et al. Pacui virus, Rio Preto Da Eva virus, and tapirape virus, three distinct viruses within the family. Bunyaviridae Genome Announc. 2014;2 doi: 10.1128/genomeA.00923-14. pii: e00923-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryant JE, Crabtree MB, Nam VS, Yen NT, Duc HM, Miller BR, et al. Isolation of arboviruses from mosquitoes collected in Northern Vietnam. Am J Trop Med Hyg. 2005;73:470–3. [PubMed] [Google Scholar]
- 12.Mossel EC, Crabtree MB, Mutebi JP, Lutwama JJ, Borland EM, Powers AM, et al. Arboviruses isolated from mosquitoes collected in Uganda, 2008-2012. J Med Entomol. 2017;54:1403–9. doi: 10.1093/jme/tjx120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitmer SLM, Yadav PD, Sarkale P, Chaubal GY, Francis A, Klena J, et al. Characterization of unknown Orthobunya-like viruses from India. Viruses. 2018;10:451. doi: 10.3390/v10090451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yadav P, Shete A, Bondre V, Patil D, Kokate P, Chaudhari S, et al. Isolation and characterization of Oya virus a member of Simbu serogroup, family Bunyaviridae, isolated from Karnataka, India. Infect Genet Evol. 2016;44:122–6. doi: 10.1016/j.meegid.2016.06.049. [DOI] [PubMed] [Google Scholar]
- 15.Whitmer SLM, Yadav PD, Sarkale P, Chaubal GY, Francis A, Klena J, et al. Characterization of unknown orthobunya-like viruses from India. Viruses. 2018;10 doi: 10.3390/v10090451. pii: E451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Wang J, Wang L, Fu S, Li M, Zhao G, et al. Molecular characterization and seroprevalence in swines of SC0806, a Cat Que virus isolated from mosquitoes in Sichuan Province, China. Vector Borne Zoonotic Dis. 2015;15:424–31. doi: 10.1089/vbz.2014.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abd-Elsalam KA. Bioinformatics tools and guideline for PCR primer design. Afr J Biotechnol. 2003;2:91–5. [Google Scholar]
- 18.Mourya DT, Gokhale MD, Majumdar TD, Yadav PD, Kumar V, Mavale MS. Experimental Zika virus infection in Aedes aegypti: Susceptibility, transmission & co-infection with dengue & chikungunya viruses. Indian J Med Res. 2018;147:88–96. doi: 10.4103/ijmr.IJMR_1142_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Treangen TJ, Schoeler G, Phillippy AM, Bergman NH, Turell MJ. Identification and genomic analysis of a novel group C Orthobunyavirus isolated from a mosquito captured near Iquitos, Peru. PLoS Negl Trop Dis. 2016;10:e0004440. doi: 10.1371/journal.pntd.0004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pastula DM, Hoang Johnson DK, White JL, Dupuis AP, 2nd, Fischer M, Staples JE. Jamestown canyon virus disease in the United States-2000-2013. Am J Trop Med Hyg. 2015;93:384–9. doi: 10.4269/ajtmh.15-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguilar PV, Morrison AC, Rocha C, Watts DM, Beingolea L, Suarez V, et al. Guaroa virus infection among humans in Bolivia and Peru. Am J Trop Med Hyg. 2010;83:714–21. doi: 10.4269/ajtmh.2010.10-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]


