Abstract
Background:
Autosomal dominant polycystic kidney disease (ADPKD), a multisystem disorder, is the most prevalent type of hereditary kidney disease. Here, we aimed to evaluate methylation of the PKD1 gene (PKD1) promoter and its correlation with PKD1 expression in peripheral blood.
Methods:
In this case-control study methylation of the PKD1 promoter was evaluated using methylation-sensitive high-resolution melt (MS-HRM) analysis. PKD1 expression was assessed by quantitative real-time PCR. The correlation was evaluated using the Pearson correlation test.
Results:
Twenty subjects from both the patient and control groups (n= 40 for each) were methylated at the PKD1 promoter to various levels (18.9% in patients and 62.5% in controls). This difference was statistically significant (p< 0.0001). PKD1 expression in blood samples was significantly greater in ADPKD patients than in controls (p= 0.0081). Significant correlation was seen between PKD1 expression and its promoter methylation status in peripheral blood (r case= -0.5300, p= 0.0162, and r control = -0.6265, p= 0.0031).
Conclusion:
Methylation of the PKD1 promoter in ADPKD patients was inversely correlated with PKD1 expression.
Key Words: Autosomal Dominant Polycystic Kidney Disease (ADPKD), Epigenetic, Methylation, methylation-sensitive high-resolution melt (MS-HRM) analysis, PKD1
Introduction
Autosomal dominant polycystic kidney disease (ADPKD), a ciliopathy with high morbidity and mortality, is the most prevalent type of hereditary kidney disease, affecting one per 400-1000 individuals. Overall, about 12.5 million individuals around the world struggle with this life-threatening genetic disease (1-3). Phenotypically, ADPKD is considered as the development of several fluid-filled cysts inside both kidneys with renal dysfunction that eventually, in the fourth or fifth decade of life, results in end-stage renal disease (ESRD) (4). Other extra-renal manifestations comprise the formation of cysts in the liver, hypertension, flank pain, and an increased risk of cardiovascular diseases and intracranial aneurysms. Several indices, including the nature of the mutated gene, and patient age, gender, kidney function, and kidney volume can be used to predict disease progression (5, 6). Formation of cysts in the liver may lead to polycystic liver disease (PLD) and congenital hepatic fibrosis (1).
Around 85% of ADPKD families were found to harbor a deleterious mutation in one copy of the PKD1 gene (PKD1), and about 15% of cases had mutations in the PKD2 gene (PKD2). Polycystins-1 and -2, are the protein products of PKD1 and PKD2, respectively, and have been shown to bind to each other through their C-terminal domains in renal tubule epithelium (7, 8). The extracellular N-terminal domain of polycystin-1 protrudes on the primary cilia, which are assumed to be activated in response to renal flow as a mechanical sensor, and consequently control the Ca2+ ion flow through polycystin-2 (9).
PKD1 is a large gene with 46 exons that spans a 52 kb region of chromosome 16p. It encodes a GC-rich transcript of 14 kb (10). Most of PKD1, including exons 1 to 33, is located in a six-fold duplicated genomic region, resulting in six highly homologous pseudogenes with 97.7% sequence similarity to the wild-type gene (11). To date, over 2300 disease-causing mutations in PKD1 have been recognized (12).
The "Two-Hit Model," which has been described as the underlying mechanism for familial cancers in which two different mutations affect proper genetic/cellular interactions, likely explains the clinical features observed in ADPKD patients. Based on this model, although the affected individual has inherited a germline mutation known as the first hit, cyst formation occurs only after the second hit occurs as mutations in PKD1 or PKD2 (7).
In some cases of ADPKD, no mutations have been found in either PKD1 or PKD2 (13). One possible explanation for this finding is epigenetic alteration. The two main mechanisms for epigenetic alterations include DNA methylation and histone post-translational modifications, by which gene expression is regulated through chromatin remodeling (14). DNA methylation is a mechanism by which the addition of methyl groups to cytosine in a CpG island occurs via methyltransferase function. Although promoter methylation often downregulates gene expression, gene body methylation, particularly at the 3′ end, correlates with increased transcription (15).
Altered methylation of gene promoters has a role in numerous diseases (16). To date, two studies evaluating the methylation status of ADPKD tissues using genome-wide profiling found that significant hypermethylation of the PKD1 gene body, but not the promoter, correlated with decreased PKD1 expression (17, 18). Here, we aimed to evaluate the methylation status of PKD1 using methylation-sensitive high-resolution melt (MS-HRM), and subsequently, its correlation with PKD1 expression. In contrast to the two previous studies, we used a larger sample size of 40 ADPKD patients and 40 patient first-relatives as controls, and a wider age range.
Materials and Methods
Participants
A total of 40 patients referred to the Labbafinezhad Hospital and diagnosed with ADPKD based on renal function tests and familial pedigree records, negative for PKD1 and PKD2 mutations, and forty healthy controls, each a first-relative of each patient, were included in this study. All patient and control conditions were confirmed by a nephrologist. Inclusion criteria included: age greater than 15 years, one cyst in each kidney in individuals aged 15-30 years, two cysts in each kidney in individuals aged 31-60 years, and two cysts in each kidney in individuals aged greater than 60 years. Written informed consent was obtained from all participants. Patients with known genetic diseases, including congenital blindness and tuberous sclerosis, were excluded from the study.
DNA extraction and bisulfite modification
DNA was isolated from 200 μL of peripheral blood samples using the Invisorb® Spin Blood Mini Kit (STRATEC Molecular GmbH, Germany) according to the manufacturer’s instructions. A NanoDrop™ 2000 (Eppendorf AG, Germany) was used to determine the DNA concentrations of all samples before and after bisulfite modification. To evaluate the methylation status of the PKD1 promoter, about one µg of extracted DNA was subjected to bisulfite modification using the EZ DNA Methylation Kit (Zymo Research, CA). Treatment of DNA sequences with sodium bisulfite leads to preferential deamination of unmethylated cytosines to uracils after PCR amplification, and leaves methylcytosines unmodified. Therefore, differentiation between methylated and unmethylated DNA sequences is possible using this selective modification. The final eluted DNA obtained from the above procedure was analyzed by MS-HRM.
MS-HRM analysis
MS-HRM was performed on the LightCycler® 96 Real-Time PCR System (Roche, Germany). Briefly, the designed primers have one or two CpG sites at or adjacent to the 5′ end of both the forward and reverse primers. The primer sets for PKD1 were designed according to the previously described protocol (19) and by MethPrimer software. The sequences were as follows: unmethylated forward, 5′TGGGAGTGTAGTGGTATAATTATGG3′ and unmethylated reverse, 5′AAAAACCAACCTAACCAACATAACA3′ for the unmethylated fragment and methylated forward, 5′GCGGGAGTGTAGTGGTATAATTAC3′, and methylated reverse, 5′ACCAACCTAACCAACATAACGAA3′ for the methylated fragment. HRM was performed in a 20 μL volume containing 4 µL of Master Mix Eva Green No Rox, 1 pmol/µL of each primer, 12 µL of ultra-pure water, and 2 µL of bisulfite-treated DNA template. The cycling conditions were as follows: 1 cycle of 95 °C for 15 min, 45 cycles of 95 °C for 15 sec, 60 °C for 20 sec, and 72 °C for 20 sec, followed by an HRM step of 95 °C for 1 min, 40 °C for 1 min, 65°C for 1 sec, 97 °C for 1 sec, and continuous acquisition to 95 °C at 1 acquisition per 0.1 °C. A standard curve with known methylation ratios was incorporated in each assay and used to infer the methylation ratio of each sample. To confirm and quantify methylation of the positive samples, bisulfite-modified DNA was sequenced using Sanger sequencing.
RNA extraction and reverse transcription PCR (RT-PCR)
RNA was isolated from the samples using the GeneAll Hybrid-R™ RNA purification kit (GeneAll Biotechnology Co. Ltd, Seoul, Korea). About 1 μg of DNase I-treated RNA was used to synthesize cDNA using the RevertAid First Strand cDNA Synthesis kit (Thermo Scientific). The qPCR was performed using the RealQ Plus Master Mix Green kit (Ampliqon, Denmark). To quantify PKD1 expression changes due to promoter hypermethylation, RT-PCR was performed with specific primers including 5′CTGCAGGAAGCACTCTACCC3′ as the forward primer and 5′CTCCCAGCCAACGTCGTAAT3′ as the reverse primer using the LightCycler 96 Real-Time PCR System (Roche, Germany). To normalize PKD1 expression, β-actin was used as an internal control.
Statistical analysis
RNA expression changes were evaluated using REST 2009 software. To evaluate the distribution normality, the Shapiro-Wilk test was used. The paired student’s t-test was used to assess PKD1 expression and promoter methylation differences between cases and controls. P values less than 0.05 were considered significant for all statistical tests. All statistics were analyzed using GraphPad Prism 8 (GraphPad Software).
Results
In this study we evaluated 40 ADPKD patients of both genders with deleterious mutations in PKD1 or PKD2 with mean ages of 27.8 and 27 years for males and females, respectively, and 40 controls with mean ages of 42.1 and 57.1 years, also for males and females, respectively. First, the PKD1 methylation status of all samples was evaluated using MS-HRM. Positive samples were confirmed and quantified using Sanger sequencing. Finally, PKD1 expression and its correlation with methylation status were evaluated.
PKD1 methylation
To evaluate the MS-HRM assay sensitivity to detect PKD1 promoter methylation, standard samples with 0, 25, 50, 75, and 100% methylation ware used (Fig. 1). Twenty subjects from each group were shown to be methylated at the PKD1 promoter. Positive samples were sequenced and the percentage of methylation was determined. The positive patient samples were 18.9% methylated, and the positive control samples were 62.5% methylated. This difference was statistically significant (p< 0.0001). Also, the mean percentage of methylation of the PKD1 promoter was significantly greater in patients
Fig. 1.
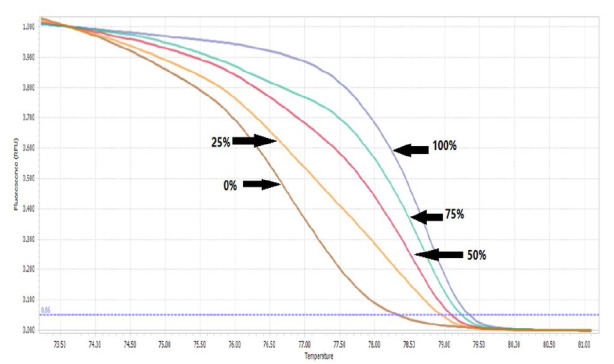
The melting curve for standard samples with 0, 25, 50, 75, and 100% methylation.
aged 15-30 years and 31-60 years than in controls of the same age groups (p= 0.01 and 0.001, respectively). Finally, the mean percentage of methylation of the PKD1 promoter was significantly greater in patients aged 15-30 than in patients aged 31-60 years (p= 0.455).
PKD1 expression
PKD1 expression was significantly greater in ADPKD patients than in controls (p= 0.0081, Fig. 2).
Fig. 2.
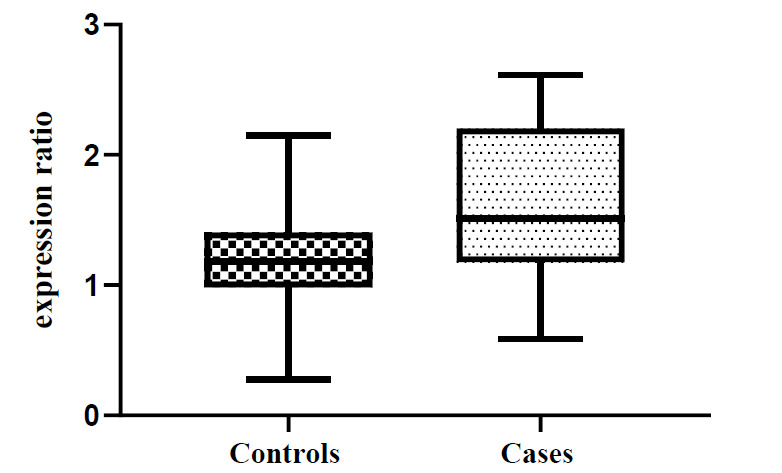
Relative expression of PKD1 in ADPKD patients and controls.
Correlation between PKD1 expression and promoter methylation status
Figure 3 is a scatter plot that shows PKD1 mRNA expression and promotor methylation status. In our study a significant correlation was found between PKD1 mRNA expression and its promoter methylation status in peripheral blood (r patients = -0.5300, p= 0.0162, and r controls = -0.6265, p= 0.0031).
Fig. 3.
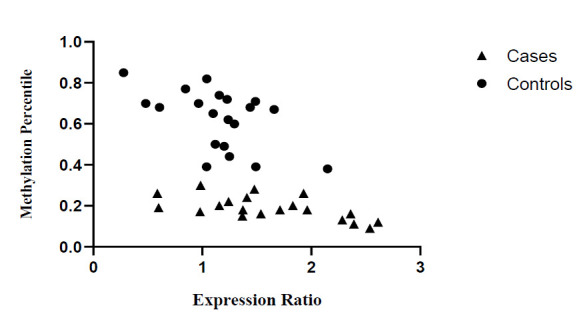
Scatter plot of PKD1 mRNA expression and promoter region methylation level in ADPKD patients and controls. Triangles represent patients and circles represent controls.
Discussion
ADPKD is an autosomal dominant disorder caused mainly by mutations in PKD1 and PKD2. These genes encode for polycystin-1 and -2 proteins, which function together as a transmembrane channel that plays a role in calcium influx. Over 85% of ADPKD cases are caused by mutations in PKD1 (20). In some cases of ADPKD, no deleterious mutations have been found in PKD1 or PKD2. In such cases, the disease etiology could be due to other modifiers of genetic factors including epigenetic modifications (13). Principally, DNA methylation in the gene promoter inversely correlates, while methylation in the gene body, particularly at its 3′ end, positively correlates, with gene expression (15, 21).
Therefore, due to the importance of PKD1 and epigenetic modifications, here we evaluated the methylation status of PKD1 in 40 ADPKD patients and 40 healthy controls using MS-HRM and determined the correlation between promoter methylation and PKD1 expression. Twenty subjects from each group were methylated at the PKD1 promoter to various levels.
Woo et al., using unbiased methylation profiling, assessed ADPKD patient genomes and showed that PKD1 gene-body regions were hypermethylated and PKD1 expression was reduced in ADPKD. Their results revealed a critical role for PKD1 hypermethylation in cystogenesis. However, they also reported that no methylation alterations were found in the promoter regions of either ADPKD or non-ADPKD and samples (18). However, in our study half of both case and control groups showed promoter methylation, but at significantly different levels.
Bowden and et al, recently showed that compared with DNA from non-ADPKD patient kidneys, the genomic DNA of ADPKD kidneys displays universal hypomethylation in all genomic elements, but more so in exons and to a lesser extent in fragments overlapping promoters. They showed increased PKD1 body methylation at the 3′ end but found no association between methylation and expression (17). Differences between our results and those of previous studies could be due to the different sample sizes. The Woo and Bowden studies were performed on seven and three ADPKD patients respectively. Our sample size was greater and our patients comprised a larger age range and included younger patients. Also, because we evaluated epigenetic alterations, we could not ignore the role of environmental factors.
Acknowledgements
We sincerely acknowledge the patients and their relatives for their cooperation in this project. There are no conflicts of interest.
References
- 1.nossen WR, Drenth JP. Polycystic liver disease: an overview of pathogenesis, clinical manifestations and management. Orphanet J Rare Dis. 2014;9:69. doi: 10.1186/1750-1172-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torres VE, Harris PC, Pirson Y . Autosomal dominant polycystic kidney disease. Lancet. . 2007;369(9569):1287–1301. doi: 10.1016/S0140-6736(07)60601-1. [DOI] [PubMed] [Google Scholar]
- 3.Bagheri M, Makhdoomi K, Taghizadeh Afshari A, Nikibakhsh AA, Abdi Rad I. Examining the Role of Polymorphisms in Exon 25 of the PKD1 Gene in the Pathogenesis of Autosomal Dominant Polycystic Kidney Disease in ranian Patients. . Rep Biochem Mol Biol. . 2019;8(2):102–110. [PMC free article] [PubMed] [Google Scholar]
- 4.Grantham JJ. Autosomal dominant polycystic kidney disease. N Engl J Med. 2008;359(14):1477–85. doi: 10.1056/NEJMcp0804458. [DOI] [PubMed] [Google Scholar]
- 5.Hopp K, Ward CJ, Hommerding CJ, Nasr SH, Tuan HF, Gainullin VG, et al. Functional polycystin-1 dosage governs autosomal dominant polycystic kidney disease severity. . J Clin Invest. 2012;122(11):4257–73. doi: 10.1172/JCI64313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ong AC, Devuyst O, Knebelmann B, Walz G, Diseases E-EWGfIK. Autosomal dominant polycystic kidney disease: the changing face of clinical management. Lancet. 2015;385(9981):1993–2002. doi: 10.1016/S0140-6736(15)60907-2. [DOI] [PubMed] [Google Scholar]
- 7.Cordido A, Besada-Cerecedo L, Garcia-Gonzalez MA. The Genetic and Cellular Basis of Autosomal Dominant Polycystic Kidney Disease-A Primer for Clinicians. Front Pediatr. 2017;5:279. doi: 10.3389/fped.2017.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian F, Germino FJ, Cai Y, Zhang X, Somlo S, Germino GG. PKD1 interacts with PKD2 through a probable coiled-coil domain. Nat Genet. 1997;16(2):179–83. doi: 10.1038/ng0697-179. [DOI] [PubMed] [Google Scholar]
- 9.Nauli SM, Zhou J. Polycystins and mechanosensation in renal and nodal cilia. . Bioessays. 2004;26(8):844–56. doi: 10.1002/bies.20069. [DOI] [PubMed] [Google Scholar]
- 10.Germino GG, Weinstat-Saslow D, Himmelbauer H, Gillespie GA, Somlo S, Wirth B, et al. The gene for autosomal dominant polycystic kidney disease lies in a 750-kb CpG-rich region. . Genomics. 1992;13(1):144–51. doi: 10.1016/0888-7543(92)90214-d. [DOI] [PubMed] [Google Scholar]
- 11.Cornec-Le Gall E, Audrezet MP, Le Meur Y, Chen JM, Ferec C. Genetics and pathogenesis of autosomal dominant polycystic kidney disease: 20 years on. Hum Mutat. 2014;35(12):1393–406. doi: 10.1002/humu.22708. [DOI] [PubMed] [Google Scholar]
- 12.Ranjzad F, Aghdami N, Tara A, Mohseni M, Moghadasali R, Basiri A. Identification of Three Novel Frameshift Mutations in the PKD1 Gene in Iranian Families with Autosomal Dominant Polycystic Kidney Disease Using Efficient Targeted Next-Generation Sequencing. Kidney Blood Press Res. . 2018;43(2):471–478. doi: 10.1159/000488471. [DOI] [PubMed] [Google Scholar]
- 13.Dasar N, Ghaderian SM, Azargashb E. Human Evaluation of the Glu298Asp Polymorphism in NOS3 Gene and its Relationship with Onset age of ESRD in Iranian Patients Suffering from ADPKD. Int J Mol Cell Med. 2012;1(2):105–12. [PMC free article] [PubMed] [Google Scholar]
- 14.Liyanage VR, Jarmasz JS, Murugeshan N, Del Bigio MR, Rastegar M, Davie JR. DNA modifications: function and applications in normal and disease States. Biology. 2014;3(4):670–723. doi: 10.3390/biology3040670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 16.Jin Z, Liu Y. DNA methylation in human diseases. Genes Dis. 2018;5(1):1–8. doi: 10.1016/j.gendis.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowden SA, Rodger EJ, Bates M, Chatterjee A, Eccles MR, Stayner C. Genome-Scale Single Nucleotide Resolution Analysis of DNA Methylation in Human Autosomal Dominant Polycystic Kidney Disease. . Am J Nephrol. 2018;48(6):415–424. doi: 10.1159/000494739. [DOI] [PubMed] [Google Scholar]
- 18.Woo YM, Bae JB, Oh YH, Lee YG, Lee MJ, Park EY, et al. Genome-wide methylation profiling of ADPKD identified epigenetically regulated genes associated with renal cyst development. Hum Genet. 2014;133(3):281–97. doi: 10.1007/s00439-013-1378-0. [DOI] [PubMed] [Google Scholar]
- 19.Wojdacz TK, Hansen LL, Dobrovic A. A new approach to primer design for the control of PCR bias in methylation studies. BMC Res Notes. 2008;1:54. doi: 10.1186/1756-0500-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barnawi RA, Attar RZ, Alfaer SS, Safdar OY. Is the light at the end of the tunnel nigh? A review of ADPKD focusing on the burden of disease and tolvaptan as a new treatment. . Int J Nephrol Renovasc Dis. 2018;11:53–67. doi: 10.2147/IJNRD.S136359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4(2):143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]


