Abstract
Background:
The Ccr4-Not protein complex (CNOT complex) is a key regulator of gene expression in eukaryotic cells. Ccr4-Not Complex is composed of at least nine conserved subunits in mammalian cells with two main enzymatic activities. CNOT8 is a subunit of the complex with deadenylase activity that interacts transiently with the CNOT6 or CNOT6L subunits. Here, we focused on the role of the human CNOT8 subunit in the DNA damage response (DDR).
Methods:
Cell viability was assessed to measure ATP level using a Cell Titer-Glo Luminescence reagent up to 4 days’ post CNOT8 siRNA transfection. In addition, expression level of phosphorylated proteins in signalling pathways were detected by western blotting and immunofluorescence microscopy. CNOT8- depleted Hela cells post- 3 Gy ionizing radiation (IR) treatment were considered as a control.
Results:
Our results from cell viability assays indicated a significant reduction at 72-hour post CNOT8 siRNA transfection (p= 0.04). Western blot analysis showed slightly alteration in the phosphorylation of DNA damage response (DDR) proteins in CNOT8-depleted HeLa cells following treatment with ionizing radiation (IR). Increased foci formation of γH2AX, RPA, 53BP1, and RAD51 foci was observed after IR in CNOT8-depleted cells compared to the control cells.
Conclusion:
We conclude that CNOT8 deadenylase subunit is involved in the cellular response to DNA damage.
Key Words: CNOT complex, CNOT8, DNA damage response, SiRNA
Introduction
The Ccr4-Not (CNOT) complex is a unique and multi-subunit complex that is highly conserved. Its components are involved at the different levels of gene expression including transcription, elongation, translation, mRNA degradation, ubiquitination, and protein modification (1-3). Two main enzymatic activities associated with the CNOT complex are mRNA deadenylation and ubiquitination. The CNOT complex contains nine subunits in yeast while there are more human orthologues, CNOT1 to CNOT10, and the novel interactor Tab 182 (Tankyrase 1-binding protein 1), and CNOT11 (C2ORF29) (2,4,5). CNOT1 is identified as a scaffold subunit which its depletion prevents assembly of the CNOT complex and eliminates deadenylase activity (6-7). CNOT4 is a RING E3 ligase subunit involved in post-translational modification by ubiquitination and targeting for degradation by the proteasomal pathway; it is not consistently associated with the whole CNOT complex (8). Ccr4 and Caf1 subunits in yeast and their human orthologues CNOT6/CNOT6L subunits and CNOT7/CNOT8 subunits, respectively, are responsible for deadenylase activity (4,9). The human CNOT complex exists in four variable structures consisting of CNOT7-CNOT6, CNOT7-CNOT6L, CNOT8-CNOT6, and CNOT8-CNOT6L heterodimers that may provide alterations in mRNA decay mechanism. Furthermore, the purification of deadenylase subunits suggested that CNOT6 and CNOT6L are in more constant interaction with CNOT7 than that CNOT8 (4). Deadenylation has an important role in splicing, transport, localization, and mRNA degradation. Deadenylase activity has been classified into two types based on nuclease domains that contain DEDD (CNOT7 and CNOT8) and Exonuclease-Endonuclease-Phosphatase (EEP) family (CNOT6 and CNOT6L) which constantly shorten the poly(A) tails of mRNA (4,10). CNOT7 and CNOT8 with high similarity in amino acid sequences have overlapping activity with roles in cell proliferation (11).
Several studies have revealed an important role of Ccr4 and Caf1 subunits in the resistance to DNA damaging agents in yeast (12-14). Deletion of each deadenylase subunits behaved in the similar manner with hypersensitivity effects, following DNA damage induced by IR, hydroxyurea (HU) and methyl methane sulphate (MMS) (12-14). Diverse cellular signalling pathways are initiated in response to different forms of DNA damage (15-16).
Here, we report the role of the human CNOT8 subunit in the cellular response to DNA damage in mammalian cells. We show that cell viability is reduced in CNOT8 siRNA transfected HeLa cells and, in addition, the depletion of CNOT8 in HeLa cells effects on the level of protein phosphorylation in the DDR pathway.
Materials and Methods
Cell Culture and Transfection
HeLa cells (ATCC, Manassas, VA, USA) were cultured in Dulbecco's Modified Eagle Medium (DMEM) (Sigma, St. Louis, USA) supplemented with L-glutamine and 10% Fetal calf serum (FCS)(Sigma, St. Louis, USA) at 37 °C in a humidified atmosphere of 5% CO2. For 70-80% confluency, they were plated at a density of 5×10 6 cells per 10 cm 2 dish 24 h before use. They were then transfected with 0.5 nM control siRNA (GE Dharmacon) and 25µl Oligofectamine (Invitrogen, Carlsbad, CA) in 5 ml OPTI-MEM (Gibco, Grand Island, NY, USA) or with a mixture of 0.5 nM CNOT8 siRNAs (GE Dharmacon), 25 µl Oligofectamine (Invitrogen, Carlsbad, CA), and 5 ml Opti-MEM (Gibco, Grand Island, NY, USA) following the manufacture’s protocol and were incubated for 5-6 h.
Cell viability assay
Transfected HeLa cells were rinsed with phosphate-buffered saline (PBS) and detached with 0.05% Trypsin. For cell viability, 2×10 4 cells in DMEM suspension per well were seeded in 96- well microplates in triplicate. Control wells just containing DMEM were also set up to assess luminescence background. Fifty μl Cell Titer-Glo Reagent was added to each well at the appropriate time and incubated for 30 mins. Relative Luminescence Units (RLU) were read over four days using a Pheraster Microplate Reader (BMG Labtech, Germany).
Cell irradiation
CNOT8 siRNA-transfected and control siRNAtransfected HeLa cells were split 1 to 6 into 6 cm2-plates. After 24 h, plates were exposed to 3 Gy ionizing radiation (IR). Each plate was harvested at the indicated time-point (1, 2, 4, 8, and 24 h) after DNA damage. A plate of control and CNOT8 siRNA-transfected HeLa cells were left as a negative control for DNA damage.
Western blots analysis
Treated HeLa cells were harvested and lysed by sonicating (5 watts, 15 sec) in 300-400 μl UTB lysis buffer (9 M urea, 40 mM Tris [pH7.4 an] and 150 mM beta;-mercaptoethanol). The protein concentration of the lysates was determined by Bradford protein assay (Bio-Rad, Hercules, CA, USA). Equal amounts of each protein sample were fractionated on Sodium Dodecyl Sulphate- Polyacrylamide Gel Electrophoresis (SDSPAGE) using standard procedures. Proteins were transferred to nitrocellulose membranes which were incubated with appropriate antibodies overnight at 4 °C. Primary antibodies used in the study are as follows: CNOT8, NSB1, pNSB1343s (all from Gene Tex), Chk1 (Santa Cruz Biotechnology), pChk1 317S, pRPA S4/S8, KAP1, pKAP1 824S, (all from Bethyl), pChk1 345S, (Cell signalling), RPA (Calbiochem), H2AX, and γH2AX 139S (Merck Millipore). Then, membranes were incubated with anti-mouse and anti-rabbit Horseradishperoxidase (HRP) conjugated secondary antibodies (Dako Laboratories) as appropriate. Enhanced Chemical Luminescence (ECL) reagent (GE Healthcare) was used to visualize proteins. Chemiluminescent signals were visualized using autoradiography film (Kodak).
Immunofluorescence microscopy
CNOT8 siRNA-transfected and control siRNA-transfected HeLa cells were grown in 24-well plates containing poly-L-lysine coated circular coverslips (Dixon Glass Ltd Marketplace); 5×104 cells were seeded in each well. On the following day, HeLa cells were exposed to 3 Gy ionizing radiation and were fixed at the appropriate time-points (0, 8, and 24 h). Cells were fixed in 200 μl icecold 3.6% paraformaldehyde (PFA) in PBS for 10 mins and permeabilized in 200 μl 0.5% Triton-X100 in PBS for 5 mins followed by a PBS wash. Cells were then blocked in 10% FCS diluted in PBS for 30 mins before being stained with primary antibodies. Two hundred μl of the diluted primary antibody was added to each well for an hour at room temperature. Cells were washed with blocking solution three times and incubated with 200 μl of secondary antibodies containing goat antimouse or anti-rabbit IgG (1:1000 diluted, Alexa-Fluor, Invitrogen) and maintained in the dark for an hour. Cells were washed in PBS three times before the addition of DAPI (4´,6-Diamidino-2-Phenylindole) for 5 mins and then coverslips were mounted on glass slides for microscopy. Fluorescence images were observed using a Nikon Eclipse E600 microscope with Velocity software 334 v4.1 (Improvisation). At least 100 cells in each group were counted three times by eye and cells categorized into two groups with less or more than 10 foci. Primary antibodies used for immunofluorescence microscopy were: γH2AX (1:1000 diluted, Millipore), RPA (1:1000 diluted, Calbiochem), RAD51 (1:200 diluted, Santa Cruz), and 53BP1 (1:500 diluted, Novus).
Statistical analysis
Student’s t-test and correlation analysis were performed by Excel 2013 to evaluate differences between groups. Also, the statistically significant p-values were considered at < 0.05.
Results
CNOT8 reduction affected on cell viability
Cell Titer-Glo luminescent cell viability assay was used to determine the ATP present in CNOT8 siRNA transfected HeLa cells at several time points. Results showed that the CNOT8 depleted cells presented a significant reduction of the ATP level at 72 h (p= 0.04) in comparison with control cells. There were no statistically significant changes at 24 h (p= 0.69), 48 h (p= 0.51), and 96 h (p= 0.06) (Fig.1). The reduction was also observed at 96-h incubation treatment.
Fig. 1.
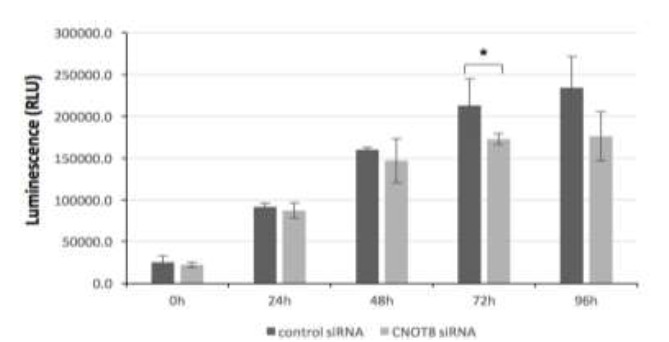
The CNOT8 depletion affects the viability of HeLa cells at 72 h after siRNA transfection. Significant differences were calculated using the student t-test p< 0.05 (*)
Phosphorylation of DDR pathway proteins is altered after CNOT8 Depletion following IR
CNOT8 siRNA transfected and control HeLa cells were exposed to IR for DDR activation. Western blot analysis shows CNOT8 depletion after CNOT8 siRNA transfection. Phosphorylation of Chk1 at Ser345, H2AX at Ser139, and RPA at Ser4/8 was initiated within one hour in both groups while the level of pChk1, γH2AX, and pRPA was greater in control cells compared to CNOT8 depleted cells. Slight phosphorylation of NBS1 at ser343 was observed 2 to 4 h post-IR in the control group. There were no notable changes in phosphorylation of KAP1 at Ser824 in CNOT8 depleted cells compared to controls after IR treatment (Fig.2).
Fig. 2.
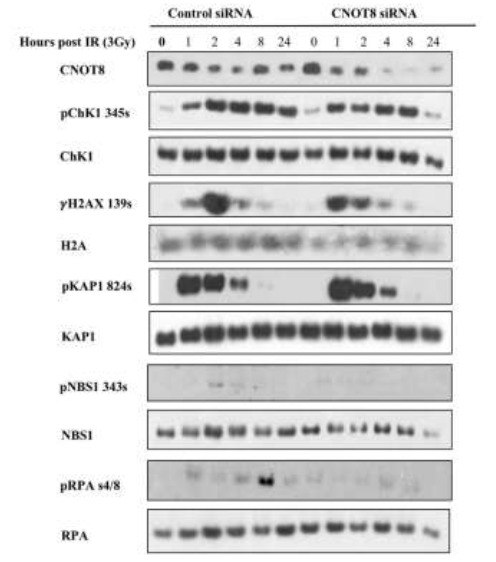
The immunoblotting analysis of control and CNOT8-depleted HeLa cells following 3Gy IR. High- affinity antibodies as shown identify the level of phosphorylated and total proteins at different time points.
Irradiation-induced-foci formation in CNOT8 depleted cells in response to IR
To further analyse CNOT8’s role in the CNOT complex following DDR, we determined the level of discrete foci formation in CNOT8 depleted cells in comparison to control cells after IR by immunofluorescence microscopy. RPA, γH2AX, 53BP1, and RAD51 proteins are known to locate to nuclear foci at the sites of doublestrand breaks (DSBs). Antibodies with high specificity were used to detect foci in both control HeLa cells and after siRNA transfection (Fig.3). After exposure to 3 Gy IR, RPA foci were significantly increased in CNOT8-depleted compared to control cells at both 8 h and 24 h time points (p-values 0.003 and 0.011,respectively) (Fig.4A). The peak value of γH2AX foci was obvious at 8 h with significant differences between both groups and was reduced by 24 h when differences between the groups were much less (p-values 0.001 and 0.065, respectively) (Fig.4B). Similarly, much increased 53BP1 foci were present at 8 h and these were significantly higher in CNOT8 depleted cells; they disappeared by 24 h with no significant differences between both groups (pvalues 0.016 and 0.220, respectively) (Fig.4C). RAD51 significantly peaked at 8 h and was stable by 24 h in both groups while the level of RAD51 foci formation was significantly higher in CNOT8 depleted cells at 8 h and 24 h (Pvalues 0.021 and 0.009, respectively) (Fig.4D).
Fig. 3.
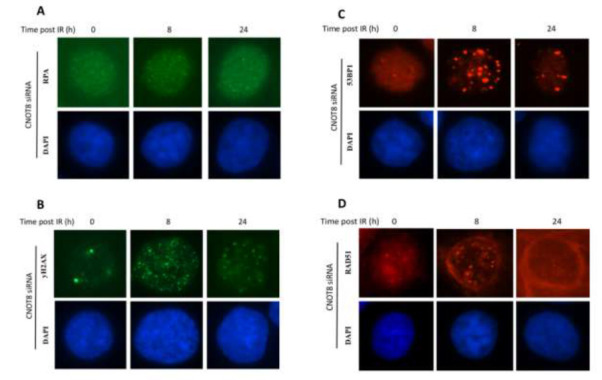
The immunoblotting analysis of control and CNOT8-depleted HeLa cells following 3Gy IR. High- affinity antibodies as shown identify the level of phosphorylated and total proteins at different time points.
Fig. 4.
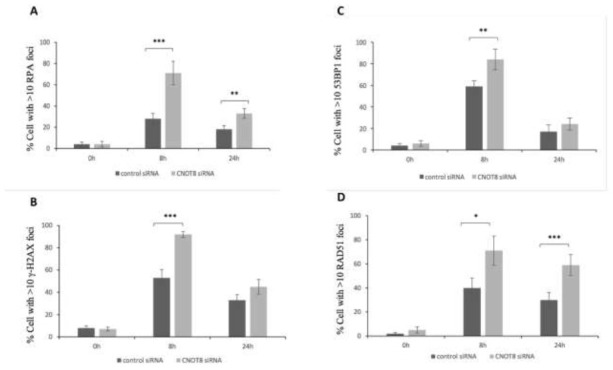
The level of foci number in control and CNOT8-depleted HeLa cells quantified following 3Gy IR. Significant differences of RPA (A), γH2AX (B), 53BP1(C) and RAD51 (D) foci/cell between two groups were calculated using the student t-test p< 0.05 (*), p<0.01(**) or p< 0.001 (***).
Discussion
The CNOT complex is an evolutionarily conserved complex with multiple cellular functions at different levels of gene regulation. Although most studies of the versatility of the CNOT complex have been in yeast, other reports are expanding to reveal complicated functions and networks in eukaryotes, including humans (1 - 3),17). The CNOT complex is equipped with deadenylase subunits that have a major role in cytoplasmic mRNA degradation by shortening and removing the poly-A tails from mRNAs (2, 4,11,18). CNOT7 /CNOT8 were identified in a distinct complex that binds to the MIF4G domain of CNOT1; they are required with CNOT6/CNOT6L for deadenylase activity (5, 19). In Drosophila , Caf1 is considered the dominant deadenylase subunit while, in yeast, catalytic activity is present in Ccr4 (20, 21). Gene Ontology (GO) analysis by Lau and colleagues indicated that the CNOT7 subunit is involved in mRNA splicing, transport and localization, whereas CNOT8 is involved in splicing only (4. Similarly, microarray analysis showed that CNOT7 and CNOT8, despite high homology, have specific substrates for their enzymatic activity (10, 17). Some other studies have suggested, combined depletion of CNOT7 and CNOT8 subunits cause mRNA decay in human HTGM5 cells, reduce cell proliferation in MCF7 breast cancer cells and result in cell viability reduction and cell death in mouse embryonic fibroblasts (MEFs) (11, 22,23). Accordingly, it has been suggested that alterations of CNOT7 and CNOT8 expression are relevant to the development and progression of cancer (17, 23,24).
Here, in agreement with previous reports, we show that CNOT8 depletion can reduce cell activity and viability at 72 h post siRNA transfection, suggesting that CNOT8 is essential for cell viability and growth. Limited data are available on the role of the human CNOT8 deadenylase subunit in the resistance to DNA damage. Ccr4 and Caf1 depletion following HU treatment results in ribonucleotide reductase (RNR) inhibition and a replication block that leads to cell hypersensitivity (12, 13). Caf1 contributes in the localization of RNR by releasing regulatory subunits (13, 25).
The most dangerous form of DNA damage are DSBs that can be induced by IR. ATR-Chk1 and ATM-Chk2 signalling is mediated early upon DNA DSBs and leads to the phosphorylation of many other DDR proteins (15, 26). The phosphorylation of KAP-1 and NBS1 are dependent on the kinase activity of ATM and the phosphorylation of H2AX, ChK1, and RPA32 are dependent on ATR (15, 16),27), although H2AX can also be phosphorylated by ATM and DNAPK. In the present study, greater phosphorylation of DDR proteins H2AX, Chk1, NBS1, and RPA were observed in control cells compared to CNOT8-depleted cells that suggest a possible involvement of the CNOT complex in both ATR and ATM-dependent signalling cascades post-IR. Also, it has been suggested CNOT8 depletion reduces CNOT complex activity in response to DNA damage.
In further experiments to assess the potential role of the CNOT8 subunit in DDR, we quantified RPA32, 53BP1, γ2AX, and RAD51 foci in CNOT8 depleted HeLa cells compared to control cells. γH2AX and p53 binding protein1 (53BP1) foci co-localized to unrepaired DSBs rapidly within the cellular nucleus after DNA Damage (28, 29). The level and stability of γH2AX and 53BP1 are relevant to dose and irradiation time (30). Results from our analysis indicated that the level of 53BP1 foci and γH2AX foci focus formation was significantly higher at 8 h in CNOT8 depleted cells compared to control cells that suggested increased response/sensitisation to DNA damage. RPA foci accumulate in response to single-strand breaks and also following a break within ribosomal DNA (31 , 32). Here, we saw that RPA foci formation in CNOT8 depleted cells was significantly greater compared to control cells while on the contrary, very weak phosphorylation was observed in CNOT8 depleted cells in immunoblot analysis compared to control cells. This may suggest a different DDR pathway is mediated or perhaps RPA is replaced by RAD51 to facilitate the formation of a presynaptic complex and homologous recombination (HR) (33) . The localization of RAD51 foci acts as a biomarker in HR repair (31 , 34). CNOT8 siRNA-transfected cells showed a significantly higher rate of RAD51 focus formation compared to control cells showing cellular hypersensitivity to IR.
Finally, the present study provides evidence that the CNOT8 deadenylase subunit plays an essential role in cellular response following DNA damage in HeLa cells; its loss probably sensitising cells to damage Here we identified the role of CNOT8 in cell viability and hypersensitivity of CNOT8 depleted cells after ionizing radiation by evaluating of DDR pathway proteins and foci focus formation. Each subunit of CNOT complex has much broader roles in cellular programming and gene expression and can be identified as a target of cancer therapeutic agents that are still being debated.
Acknowledgement
The project was supported through a joint grant provided by the University Of Birmingham, United Kingdom and the Islamic Azad University, Science and Research Branch, Tehran, Iran. We are most grateful to Nafiseh Chalabi Hagkarim for invaluable advice during this project. The authors declare that they have no conflicts of interest.
We appreciate the help of the Animal Facility workers of Sohag Faculty of Medicine in taking care of the rats.
References
- 1.Collart MA, Panasenko OO. The Ccr4–Not complex. Gene. 2012;492(1):42–53. doi: 10.1016/j.gene.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 2.Collart MA. The Ccr4-Not complex is a key regulator of eukaryotic gene expression . Wiley Interdiscip Rev RNA. 2016;7(4):438–454. doi: 10.1002/wrna.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller JE, Reese JC. Ccr4-Not complex: the control freak of eukaryotic cells. Crit Rev Biochem Mol Biol. 2012;47(4):315–33. doi: 10.3109/10409238.2012.667214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lau NC, Kolkman A, van Schaik FM, Mulder KW, Pijnappel WP, Heck AJ, Timmers HT. Human Ccr4–Not complexes contain variable deadenylase subunits. Biochem J. 2009;442(3):443–53. doi: 10.1042/BJ20090500. [DOI] [PubMed] [Google Scholar]
- 5.Boland A, Chen Y, Raisch T, Jonas S, Kuzuoğlu-Öztürk D, Wohlbold L, et al. Structure and assembly of the NOT module of the human CCR4-NOT complex . Nat Struct Mol Biol. 2013;20(11):1298–97. doi: 10.1038/nsmb.2681. [DOI] [PubMed] [Google Scholar]
- 6.Temme C, Zaessinger S, Meyer S, Simonelig M, Wahle E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila . EMBO J. 2004;23(14):2862–2871. doi: 10.1038/sj.emboj.7600273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito K, Takahashi A, Morita M, Suzuki T, Yamamoto The role of the CNOT1 subunit of the CCR4-NOT complex in mRNA deadenylation and cell viability. Protein Cell. 2011;2(9):755–763. doi: 10.1007/s13238-011-1092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panasenko OO. The role of the E3 ligase Not4 in cotranslational quality control. Front genet. 2014;19(5):141. doi: 10.3389/fgene.2014.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maryati M, Airhihen B, Winkler GS. The enzyme activities of Caf1 and Ccr4 are both required for deadenylation by the human Ccr4–Not nuclease module. Biochem J. 2015;469(PT 1 ):169–176. doi: 10.1042/BJ20150304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mark Bartlam TY. The structural basis for deadenylation by the CCR4-NOT complex. Protein Cell. 2010;1(5):443–453. doi: 10.1007/s13238-010-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aslam A, Mittal S, Koch F, Andrau JC, Winkler GS. The Ccr4–Not Deadenylase Subunits CNOT7 and CNOT8 Have Overlapping Roles and Modulate Cell Proliferation. Mol Biol Cell. 2009;20(17):3840–3850. doi: 10.1091/mbc.E09-02-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Traven A, Hammet A, Tenis N, Denis CL, Heierhorst J. Ccr4-Not complex mRNA deadenylase activity contributes to DNA damage responses in Saccharomyces cerevisiae. Genetics. 2005;169(1):65–75. doi: 10.1534/genetics.104.030940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulder KW, Winkler GS, Timmers HT. DNA damage and replication stress induced transcription of RNR genes is dependent on the Ccr4-Not complex. . Nucleic Acids Res. 2005;33(19):6384–6392. doi: 10.1093/nar/gki938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woolstencroft RN, Beilharz TH, Cook MA, Preiss T, Durocher D, Tyers M. Ccr4 contributes to tolerance of replication stress through control of CRT1 mRNA poly(A) tail length. J Cell Sci. 2006;119(Pt 24):5178–92. doi: 10.1242/jcs.03221. [DOI] [PubMed] [Google Scholar]
- 15.Yajima H, Lee KJ, Zhang S, Kobayashi J, Chen BP. DNA Double Strand Break Formation upon UV-Induced Replication Stress Activates ATM and DNA-PKcs Kinases. J Mol Biol. 2009;385(3):800–810. doi: 10.1016/j.jmb.2008.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol Cell. 2017;66(6):801–817. doi: 10.1016/j.molcel.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 17.Shirai YT, Suzuki T, Morita M, Takahashi A, Yamamoto T. Multifunctional roles of the mammalian CCR4–NOT complex in physiological phenomena. Front Genet. 2014;5(286) doi: 10.3389/fgene.2014.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104(3):377–86. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 19.Petit AP, Wohlbold L, Bawankar P, Huntzinger E, Schmidt S, Izaurralde E, et al. The structural basis for the interaction between the CAF1 nuclease and the NOT1 scaffold of the human CCR4-NOT deadenylase complex. Nucleic Acids Res. 2012;40(21):11058–72. doi: 10.1093/nar/gks883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. . EMBO J. 2002;21(6):1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Temme C, Zhang L, Kremmer E, Ihling C, Chartier A, Sinz A, et al. Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA. 2010;16(7):1356–1370. doi: 10.1261/rna.2145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwede A, Ellis L, Luther J, Carrington M, Stoecklin G, Clayton C. A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res. 2008;36(10):3374–3388. doi: 10.1093/nar/gkn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mostafa D, Takahashi A, Yanagiya A, Yamaguchi T, Abe T, Kureha T, et al. Essential functions of the CNOT7/8 catalytic subunits of the CCR4-NOT complex in mRNA regulation and cell viability. RNA Biol. 2020;17(3):403–416. doi: 10.1080/15476286.2019.1709747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maragozidis P, Karangeli M, Labrou M, Dimoulou G, Papaspyrou K, Salataj E, et al. Alterations of deadenylase expression in acute leukemias: evidence for poly(a)-specific ribonuclease as a potential biomarker. Acta Haematol. 2012;128(1):39–46. doi: 10.1159/000337418. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi s, Kontani k, Araki Y, Katada T. Caf1 regulates translocation of ribonucleotide reductase by releasing nucleoplasmic Spd1– Suc22 assembly. Nucleic Acids Res. 2007;35(4):1157–1197. doi: 10.1093/nar/gkm015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halazonetis TD, Shiloh Y. Many faces of ATM: eighth interna- tional workshop on ataxiatelangiectasia. Biochim Biophys Acta. 1999;1424(2-3):R45–55. doi: 10.1016/s0304-419x(99)00023-2. [DOI] [PubMed] [Google Scholar]
- 27.Giunta S, Belotserkovskaya R, Jackson SP. DNA damage signaling in response to doublestrand breaks during mitosis. J Cell Biol. 2010;190(2):197–207. doi: 10.1083/jcb.200911156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. 2000;10(15):886–95. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 29.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol. 2000;151(7):1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DiTullio RA, Mochan TA, Venere M, Bartkova J, Sehested M, Bartek J, et al. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. . Nat Cell Biol. . 2002;4(12):998–1002. doi: 10.1038/ncb892. [DOI] [PubMed] [Google Scholar]
- 31.Glover L, Marques CA, Suska O, Horna D. Persistent DNA Damage Foci and DNA Replication with a Broken Chromosome in the African Trypanosome. mBio. 2019;10(4):e01252–19. doi: 10.1128/mBio.01252-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saldivar JC, Cortez D, Cimprich KA. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat Rev Mol Cell Biol. 2017;18(10):622–636. doi: 10.1038/nrm.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hustedt N, Durocher D. The control of DNA repair by the cell cycle. Nat Cell Biol. 2016;19(1):1–9. doi: 10.1038/ncb3452. [DOI] [PubMed] [Google Scholar]
- 34.Cruz C, Castroviejo-Bermejo M, Gutie ´rrez-Enr ´ıquez M, Llop-Guevara A, Ibrahim YH, Gris-Oliver A, et al. RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCAmutated breast cancer. Ann Oncol. 2018;29(5):1203–1210. doi: 10.1093/annonc/mdy099. [DOI] [PMC free article] [PubMed] [Google Scholar]


