Abstract
Background:
We aimed to evaluate the effectiveness of Highly Upregulated in Liver Cancer (HULC) and microRNA-372 (miR-372) as biochemical markers in Hepatocellular carcinoma (HCC) and HCV-infected patients.
Methods:
The present study was conducted on 100 Egyptian individuals divided into 3 groups, 40 patients with HCC and HCV infection, 40 patients only HCV-infected, and 20 individuals as normal controls. They were subject to full history taking, full clinical and laboratory examination, and assessment of HULC and miR-372 levels by real-time PCR.
Results:
A statistically significant difference was found with p< 0.05 between HCC and each of HCV and control groups as regards HULC level with high mean among HCC followed by HCV patients. Our results also show a statistically significant difference with p< 0.05 between each of HCC and HCV compared to control as regards miR-372 level with low mean among HCC patients.
Conclusion:
HULC could be considered as a potential non-invasive marker for detection and early diagnosis of HCC. Also, it may play an important role in the early prophylaxis and control measures to reduce the incidence of HCC. However, miR-372 cannot be considered as a reliable marker as HULC for early detection of HCC especially in HCV patients.
Key Words: HCC, HCV, HULC, MiRNA-372
Introduction
Hepatocellular carcinoma (HCC) represents a significant clinical problem, being the second leading cause of cancer deaths in the world (1). The risk of developing HCC is 17-fold higher in HCV-infected patients (2).
LncRNAs are non-coding RNAs with more than 200 nucleotides in length, which play an important role in the regulation of gene expression. These are involved in translational regulation, alternative splicing, epigenetics, and other biological processes (3). Highly Upregulated in Liver Cancer (HULC) is considered as the most over-expressed LncRNAs in human HCC (4). MicroRNAs are endogenous, small, and non-coding RNAs with 22 nucleotides in length that their role in sequence-specific negative regulation of the translation and stability of target mRNAs is very essential (5).
It has been documented that HULC has a conserved binding site for miR-372, thus suggesting that it can exert an antisense effect on miR-372. LncRNA HULC may cause downregulation of miR-372 and the tumor suppressor gene P18 leading to promote HCC cell proliferation (6).
Materials and Methods
Subjects
The present study was conducted on 100 Egyptian individuals. They were classified into 3 groups:
Group I: 40 patients with HCC on top of HCV, divided into 29 males (72.5%) and 11 females (27.5%).
Group II: 40 patients with HCV, divided into 29 males (72.5%) and 11 females (27.5%).
Group III: 20 age-and-sex-matched healthy individuals, divided into 11 males (55%) and 9 females (45%) as normal controls.
All the patients were selected from Tropical Medicine Department, Kasr Alainy Hospital, Faculty of Medicine, Cairo University. The study was performed with the approval of Faculty of Medicine, Cairo University local ethics committee and carried out in compliance with Helsinki Declaration, 2008. Informed consent was obtained from all the subjects enrolled in this study. Inclusion criteria included naive patients of both sexes with either HCV or HCC on top of HCV infection. Exclusion criteria included liver diseases other than Hepatitis C infection such as Hepatitis B, evidence of diseases such as cardiovascular disease, nervous system, pulmonary, renal, and endocrine or gastrointestinal disorders, other types of cancer (e.g. cholangiocarcinoma, gastric, colorectal cancer).
All participants were subjected to full history taking, full clinical examination, and full basal laboratory investigations. Samples were collected for quantitative determination of serum non-coding RNAs "HULC and miRNA -372" using quantitative Real-time Polymerase Chain Reaction technique (qRT-PCR).
Blood sampling
Five mL venous blood samples were withdrawn from all subjects using BD Vacutainer system. The samples were collected in serum separator tubes, allowed to clot for 15 minutes, then centrifuged at 4000 xg for 10 minutes. Sera were stored at -80 °C until the time of analysis.
RNA extraction
RNA extraction was performed using miRNeasy Mini Kit, a protocol for purification of serum total RNA, including miRNA (Qiagen, Germany. Cat. No. 217004), according to the manufacturer's instructions. Extracted RNA was subjected to RNA quantitation and purity assessment using NanoDrop® (ND)-1000 spectrophotometer (NanoDrop Technologies, Inc. Wilmington, USA).
Reverse transcription into complementary DNA (cDNA)
Extracted total RNA in a volume of 20 μL RT reactions was incubated for 60 min at 37 °C, followed by 5 min at 95 °C using miScript® II RT Kit (Qiagen, Germany. Cat. No. 218161), according to the manufacturer's instructions. The cDNA of each sample was stored at -80 °C until the next step.
Detection of non-coding RNAs using qRT-PCR
The miScript SYBR® Green PCR Kit (Qiagen, Germany. Cat. No. 218073) as a part of miScript PCR system and target-specific miScript primers assay for miR-372 and HULC were used. The qRT-PCR cycler was programmed for the following cycling conditions: Initial activation step for 15 min at 95 °C then 3 steps cycling denaturation for 15 sec at 94 °C, annealing for 30 sec at 55 °C and extension for 30 sec at 70 °C and these steps were repeated for 40 cycles in Rotor-gene qRT-PCR system thermocycler (Qiagen, USA).
After PCR cycles, melting curves were analyzed to validate the specific expression of the targeted RNAs. Due to the lack of an endogenous reference housekeeping gene or miRNA in the serum, a small nucleolar RNA C/D box 68 (SNORD-68) was used as an endogenous housekeeping gene because its expression is not affected and doesn’t vary under different experimental conditions or in different states of the same sample. It was used to normalize the expression pattern and for relative quantification of the target miRNAs. Also, Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control in calculating the long noncoding HULC. Then, the fold changes (FC) of expression or relative quantitation (Rq) for the target miRNA and long noncoding HULC were calculated using 2-ΔΔCt method.
The mathematical relationship between Ct, ΔCt, ΔΔCt, and FC (Rq) was calculated as following:
ΔCt (patients) = Ct (non-coding RNA) - Ct (endogenous control)
ΔCt (control) = Ct (non-coding RNA) - Ct (endogenous control)
ΔΔCt (patients) = ΔCt (patients) - ΔCt (control) FC (Rq) = 2 - ΔΔCt
The fold change is the expression ratio: if the fold change is positive, it means that the gene is upregulated; if the fold change is negative, it means it is downregulated. So, the results were expressed as fold change compared to the control sample, and not expressed as an absolute value. Control value, considered the normal value, was assumed equaled 1 because –ΔΔCt for control subjects equals zero and 20 equals one.
Statistical Analysis
All statistics were analyzed using SPSS software version 18, running on Windows 7.
The nominal and qualitative data were reported as percent and numbers. If the data had a normal distribution, we applied parametric tests, such as the independent t-test, to compare the normal data between the two groups, and ANOVA tests to compare the normal data among three groups or more. But if the related data did not have a normal distribution, nonparametric tests, such as Mann-Whitney and Kruskal Wallis tests, were performed to compare the non-normal data between two and among three groups or more, respectively. To remove the confounding effects of confounding variables such as age and sex, multiple regression analyses were performed (7).
For qualitative data, Chi-Square test was used to compare two of more than two qualitative groups (8). Bivariate (Pearson) correlation test was used to test the association between variables (9), while sensitivity and specificity test was used for testing the performance of a new test, using the Receiver Operating Characteristic (ROC) curve.
The p≤ 0.05 was considered the cut-off value for significance.
Results
Comparisons of demographic characters in the different studied groups
As shown in Table 1, regarding age, there is a statistically significant difference between the HCC group and each of HCV and control groups with high mean age among HCC patients. Regarding the sex, there is no statistically significant difference between the three study groups
Table 1.
Comparisons of Demographic characters in the different studied groups.
| HCC (n= 40) | HCV (n= 40) | Control (n= 20) | p | Sig. | ||
|---|---|---|---|---|---|---|
| Age (years) | Mean ± SD | 60.3 ± 7.6 | 44.4 ± 11.1 | 49 ± 8.6 | < 0.001a, b | HS |
| 0.2c | NS | |||||
| Sex | Male | 29 (72.5%) | 29 (72.5%) | 11 (55%) | 0.2 | NS |
| Female | 11 (27.5%) | 11 (27.5%) | 9 (45%) | |||
significance between HCC and HCV,
significance between HCC and control,
significance between HCV and control
Comparisons of routine investigations in the different studied groups
Table 2 shows a statistically significant difference between HCC and HCV groups regarding hemoglobin, platelets, ALT and AST levels with higher levels of ALT and AST and lower levels of hemoglobin and platelets among HCC patients. Comparing HCC and control groups, our results show statistically significant difference as regards hemoglobin, PLT, TLC, AST, ALT, ALP, total bilirubin and albumin level with low mean hemoglobin, PLT, and albumin level and high mean of AST, ALT, ALP, and total bilirubin among HCC patients. Comparing HCV to control groups, there is a statistically significant difference with low mean PLT and albumin levels and high mean of AST, ALT, ALP, total and direct bilirubin levels among HCV patients.
Table 2.
Comparisons of routine investigations in different study groups.
| HCC (n= 40) | HCV (n= 40) | Control (n= 20) | p | Sig. | ||
|---|---|---|---|---|---|---|
| Mean ± SD | Mean ± SD | Mean ± SD | ||||
| CBC | Hb (gm/dl) | 11.3 ± 2.6 | 13.4 ± 1.8 | 14.6 ± 0.89 | < 0.001a, b | HS |
| 0.08c | NS | |||||
| TLC * 1000 | 5.6 ± 2.2 | 6.8 ± 2.5 | 7.5 ± 1.5 | 0.9a | NS | |
| 0.01b | S | |||||
| 0.8c | NS | |||||
| PLT | 126.2 ± 77.8 | 213.3 ± 75.8 | 312.3 ± 45.9 | < 0.001a,b,c | HS | |
| Liver function tests | AST (U/L) | 112.1 ± 64.4 | 46.4 ± 22.2 | 19.4 ± 6.8 | < 0.001a,b,c | HS |
| ALT (U/L) | 80 ± 53.3 | 55.8 ± 27.1 | 20.8 ± 6.1 | 0.01a | S | |
| 0.001b | HS | |||||
| 0.003c | S | |||||
| ALP (U/L) | 176.3 ± 147.6 | 122.7 ± 63.9 | 50.3 ± 14.6 | 0.06a | NS | |
| 0.001b | HS | |||||
| 0.03c | S | |||||
| Albumin | 3.7 ± 1.3 | 3.8 ± 0.67 | 4.5 ± 0.29 | 0.9a | NS | |
| 0.01b | S | |||||
| 0.04c | S | |||||
| Median / IQR | Median / IQR | Median / IQR | ||||
| Total bilirubin (mg/dl) | 1.5 / 1.1 | 0.9 / 0.28 | 0.7 / 0.18 | 0.001a | S | |
| 0.001b | S | |||||
| 0.008c | HS | |||||
| Direct bilirubin (mg/dl) | 0.5 / 0.93 | 0.6 / 0.42 | 0.3 / 0.1 | 0.3a | NS | |
| < 0.001b,c | HS | |||||
significance between HCC and HCV,
significance between HCC and control,
significance between HCV and control.
AFP levels among the different studied groups
There is a statistically significant increase in AFP levels in the HCC group in comparison with HCV and control groups as shown in Figure 1.
Fig. 1.
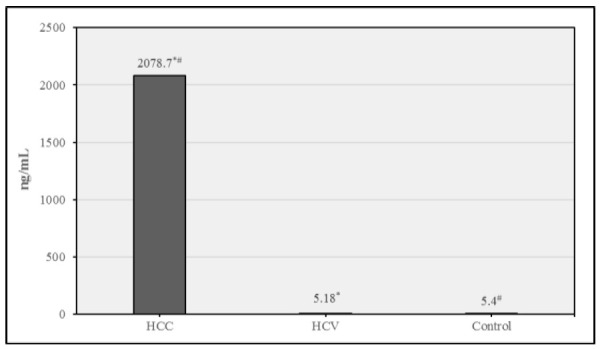
Mean AFP in different study groups. Data were expressed as Mean ± SD, p< 0.05 was significant. *denotes significant difference between HCC & HCV. #denotes significant difference between HCC & control.
lncRNA HULC is upregulated in HCC patients compared to normal healthy controls
Here, lncRNA HULC shows a highly significant fold increase in its expression level in the HCC group compared to both HCV and control groups confirming that it can be used for early detection of HCC as shown in Figure 2.
Fig. 2.
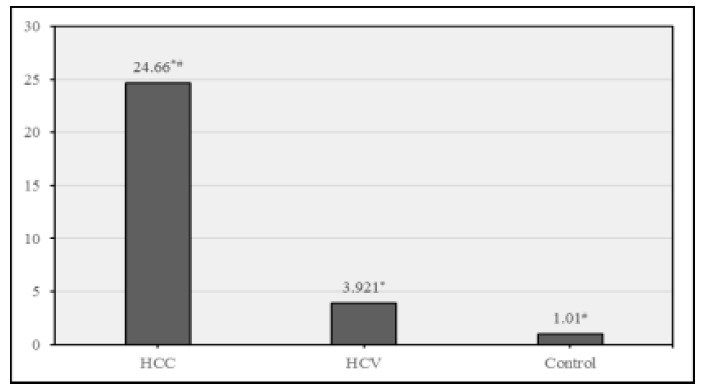
. Fold expression of HULC in different study groups. Data were expressed as Mean ± SD, p< 0.05 was significant. * denotes significant difference between HCC & HCV. # denotes significant difference between HCC & control.
miR-372 is downregulated in HCV and HCC compared to normal healthy controls
This study shows a statistically significant fold decrease in miR-372 levels in HCC and HCV groups compared to control group and non-significant fold decrease in its levels in HCC group, compared to HCV group, suggesting that it cannot be utilized for early detection of HCC in HCV patients, as shown in Fig 3.
Fig. 3.
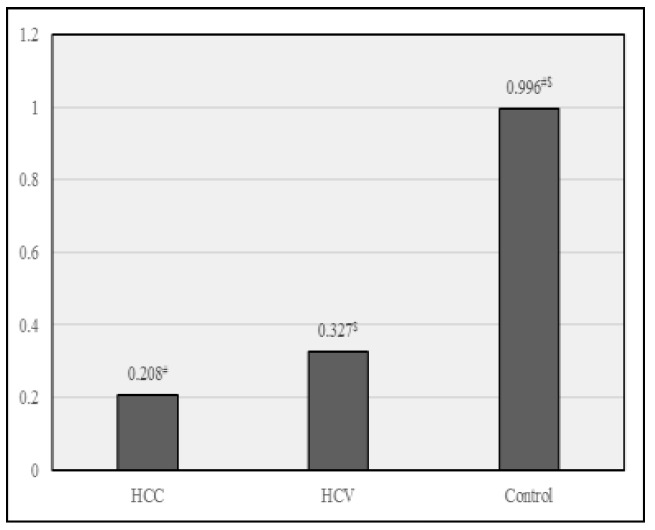
Fold expression miRNA-372 in different study groups. Data were expressed as Mean ± SD, p< 0.05 was significant, * denotes a significant difference between HCC & HCV, # denotes a significant difference between HCC & control., $ denotes a significant difference between HCV & control.
Correlation between miR-372 and HULC with the standard routine investigations done in all studied groups
As shown in Table 3, there is a statistically significant positive correlation with p< 0.05 between the levels of miR-372 and PLT, and a statistically significant negative correlation between miR-372 and AST levels, which indicates that a decrease in PLT and an increase in AST level will be associated with a decrease in miR-372 level.
Also, there is a statistically significant negative correlation with p< 0.05 between the level of HULC and each of hemoglobin, PLT, and albumin levels, and a statistically significant positive correlation between the level of HULC with AST, and ALT levels, which indicates that a decrease in hemoglobin, PLT, and albumin level, and an increase in ALT and AST levels will be associated with an increase in HULC level.
Sensitivity and specificity of miR-372 and HULC in the diagnosis of HCC
Fig 4 illustrates that the probability of being true positive is (92.3%) more than being false positive when repeating the test 100 times with sensitivity (90.6%) and specificity (100%) at a cut-off point of (0.665).
Fig. 4.
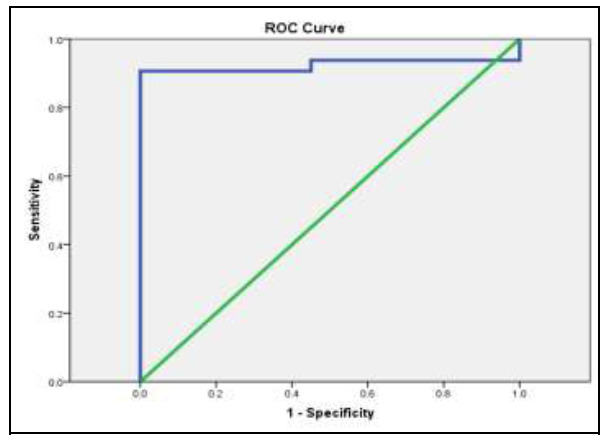
ROC curve for miR-372: for sensitivity and specificity.
The sensitivity and specificity test for HULC in the diagnosis of HCC cases is shown in Figure 5. It illustrates that the probability of being true positive is (94.4%) more than being false positive when repeating the test 100 times with sensitivity (94.4%) and specificity (95%) at a cut-off point of (1.25).
Fig. 5.
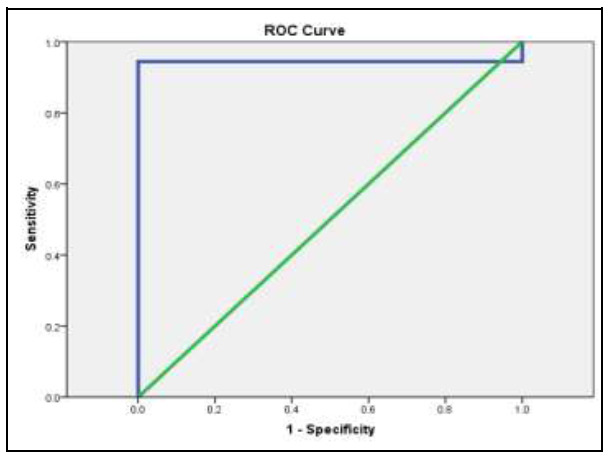
ROC curve for HULC: for sensitivity and specificity.
Measuring the MIC in all positive samples for M. pneumoniae using broth micro-dilution method, showed that all specimens were sensitive to erythromycin. Moreover, no ML resistance was reported (Table 3). Therefore, mutations in domain V of 23S rRNA gene at A2431G and G2491A were not associated with resistance of M. pneumoniae against.
Table 3.
Correlation between miR-372 and HULC with study variables among the whole study group.
| miR-372 | HULC | ||||
|---|---|---|---|---|---|
| r | p | r | p | ||
| CBC | H b | 0.09 | 0.4 | -0.31 | 0.002* |
| TLC * 1000 | -0.04 | 0.7 | -0.08 | 0.4 | |
| PLT | 0.31 | 0.003* | -0.23 | 0.02* | |
| Liver function tests | AST (U/L) | -0.26 | 0.01* | 0.34 | 0.001* |
| ALT (U/L) | -0.18 | 0.09 | 0.22 | 0.03* | |
| ALP (U/L) | -0.15 | 0.2 | 0.18 | 0.08 | |
| Total bilirubin | -0.18 | .08 | 0.15 | 0.1 | |
| Direct bilirubin | -0.15 | 0.2 | 0.001 | 0.9 | |
| Albumin | 0.19 | 0.07 | -0.31 | 0.002* | |
| Other investigations | AFP | -0.09 | 0.3 | 0.04 | 0.7 |
statistical significance with p< 0.05.
Discussion
Targeting to refine HCC risk, scientists started to study the role of molecular biomarkers which allow objective stratification of risk and help in individualized therapy and surveillance for HCV individuals. Moreover, molecular biomarker-based stratification plays an important role in HCC chemoprevention clinical trials, saves us time and costs and allows a smaller sample size (10).
Some of these molecular biomarkers are lncRNAs, which play a vital role in the regulation of gene expression (3). Evidences show that lncRNAs are involved in many types of cancer, such as colon, lung, gastric, and pancreatic cancers in addition to melanoma, glioma, and HCC (11).
In this work, one of these lncRNAs was studied, which is known as HULC. HULC is aberrantly upregulated in many cancer types (12). It has been proved that HULC can promote different pro-tumorigenic phenotypes, like cell proliferation and survival in vitro as well as tumor growth (13).
Another type of possible biomarkers is microRNAs (miRNAs), which were found to act as post-transcriptional regulators of gene expression (5) and to have an important role in tumorigenesis (14).
One of these miRNAs is miR-372, which was found to play a role in many types of cancer, including HCC, where it may regulate the cell cycle, apoptosis, growth, and invasion (15).
The aim was to find the association between lnc-HULC and miR-372 gene expressions in HCC and HCV patients compared to healthy controls and to correlate these expression levels with the levels of different biochemical markers in HCC and HCV patients.
Concerning the age of patients, it was statistically significantly higher in HCC patients which could be explained by the natural history of HCC on top of HCV related cirrhosis. A similar finding was observed before by Gamal et al. and Gopal et al. (13,14).
Concerning liver function tests, ALT and AST were statistically significantly higher in HCC patients compared to HCV and control groups, which might be explained by the continued hepatocellular damage in HCV-related HCC. This observation is in accordance with several studies as those done by Ataseven et al. and Axley et al. (15,16).
Also ALP and bilirubin levels were statistically significantly higher in HCC and HCV groups as compared with the control group. Similar findings were detected by Axley et al. (16). As for albumin, it was statistically significantly lower in HCC and HCV patients as compared to the control. This observation is in accordance with the study of Jing et al. and Taverna et al. (17,18) who found that during systemic inflammation, albumin level is decreased, and it works as a protective agent that scavenges the reactive nitrogen and oxygen species.
In this work, concerning CBC, platelet levels were found to be statistically significantly decreased in the HCC group compared to both HCV and control groups, which agrees with Gopal et al. (14). This can be explained by the crucial role of platelets in predicting liver fibrosis and cirrhosis; being lower with further progression of cirrhosis, so lower platelet count is found in HCC cases (16). As regards Hb levels, there was a statistically significant decrease in the HCC group compared to both HCV and control groups. This was consistent with the study done by Knight et al. (19) who stated that cancer-related anemia is a common complication in nearly all types of malignant diseases including HCC. Moreover, anemia is considered as a cause of death in HCC patients (23).
Regarding serum AFP, there were significant differences in its level between the different studied groups. It was significantly higher in the HCC group. Our findings are consistent with Axley et al. and Montaser et al. (16,21) who found a highly significant increase in the serum AFP in the HCC cases.
In this work, HULC expression levels were highly significantly increased in the HCC group compared to HCV and control groups (p< 0.05) and non-significantly increased in the HCV group compared to the control group.
To some extent, these results are similar to the results of Panzitt et al. (4) who found that HULC is specifically expressed in hepatocytes and is highly upregulated in HCC than controls. In agreement with us, Wang et al. (12) proved that HULC is highly expressed in HCC patients and also revealed that HULC plays an important role in tumorigenesis through inhibiting miR-372. Also, Huang et al. (8) confirmed our results and demonstrated the functional involvement of HULC in the pathogenesis of HCC with high expression in the HCC group compared to normal control. Also, our results are supported by Li et al. (22), which found that HULC was aberrantly upregulated in HCC tissues and associated with the tumor node and metastases (TNM) stage, HCC recurrence, intra-hepatic metastases, and postoperative survival.
Concerning the results regarding miR-372, its expression levels were statistically significantly decreased in both HCC and HCV groups compared to the control group, but there was a non-significant decrease in its expression levels in the HCC group compared to HCV group.
Wu et al. (23) agreed with us and demonstrated that miR-372 was expressed in HCC at low levels and stated that it plays an anti-oncogenic role by negatively controlling its target gene ATAD2. These results also came in agreement with Wu et al. (24) who tested miR-372 expression not only by RT-PCR but also by in-situ hybridization and the results from both approaches showed that miR-372 was expressed at low levels in HCC than the control group. However, on the contrary, this study found a significant difference between HCC and HCV groups as regards miR-372 level which was not detected in our study.
Jalali et al. (6) also supported these results and stated that there are evidences that suggest an important interaction between lncRNAs and miRNAs. It has been illustrated that HULC has a conserved binding site for miR-372.Thus, suggesting that it can mediate an antisense effect on miR-372. LncRNA HULC may downregulate miR-372 and the tumor suppressor gene P18, leading to the promotion of HCC cell proliferation.
Shrivastava et al. (25) also confirmed the results as regards the expression of miR-372 and noted that its level was higher in HCV than HCC patients and illustrated that there is complex crosstalk between HCV and miR-372 in liver steatosis, fibrosis, and HCC. However, the exact role of miR-372 in HCV-mediated hepatocyte injury and pathogenesis of liver disease remains to be elucidated.
On the contrary, Ramachandran et al. (26) showed that changes in miRNAs induced by HCV infection affect progression of liver fibrosis. After comparing chronic HCV patients with normal controls, they identified that miR-372, which is highly expressed in chronic HCV patients, downregulates FAS-associated protein (FAP)-1 and subsequently activates Src kinase-mediated profibrotic signaling pathway, suggesting that it can be considered as a factor, which promotes hepatic fibrosis, This is against our results that indicated a low level of miR-372 among HCV patients than controls.
In addition, Liu et al. (27) demonstrated that miR-372 expression levels changed by more than 2-fold in response to HCV infection and that it was more in HCC than chronic HCV infected patients and control. On the other hand, our results were against the results of Gu et al. (28) who reported that miR-372 was expressed at high levels in HCC and may exert a proto-oncogenic effect in hepatic carcinogenesis.
As for the correlation between HULC expression levels and the laboratory data of all the studied groups, a significant negative correlation between the level of HULC and each of hemoglobin, PLT, and albumin levels was found and a significant positive correlation with AST and ALT levels confirming that HULC can be used as a marker for diagnosing HCC. These findings are in accordance with those of Zhu et al. (29), who showed that downregulation of HULC reduced the ALT and AST in the rats serum and inhibited lesions and fibrosis of the hepatic tissue in rats.
As for the correlation between miR-372 expression levels and the laboratory data of all the studied groups, a significant positive correlation between the level of miR-372 and PLT and a significant negative correlation with AST level were found confirming the promising role of miR-372 to detect liver affection.
In this study, using the ROC curve to predict HCC using the fold change of HULC, it showed that the probability of being true positive is (94.4%) more than being false positive, on repeating the test 100 times with sensitivity (94.4%) and specificity (95%) at a cut-off point of (1.25). So, we conclude that HULC fold changes below this cut-off value are mostly predictive of HCC. This coincides with the results of Yang et al. [30] where ROC curves demonstrated that HULC expression level in tumor tissues could significantly predict HCC overall survival (area under ROC = 0.608; p= 0.004).
When the ROC curve was used to predict HCC using the fold change of miR-372, it showed that the probability of being true positive is (92.3%) more than being a false positive. Repeating the test 100 times with sensitivity (90.6%) and specificity (100%) at a cut-off point of (0.665) detected that miR-372 fold changes below this cut-off value are mostly predictive of HCC.
According to our results, we conclude that HULC could be considered as a potential non-invasive marker for detection and early diagnosis of HCC, and it may play an important role in the early prophylaxis and control measures to reduce the incidence of HCC. However, miR-372 cannot be considered as a reliable marker as HULC for early detection of HCC especially in HCV patients, as there was a non-significant decrease in its expression levels in HCC group compared to HCV group in our study.
Acknowledgements
We would like to thank the supervisors of the medical biochemistry laboratories at Kasr Alainy hospital of Cairo University for their helpful cooperation. We are grateful to all of our patients who participated in this study.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. . A Cancer Journal for Clinicians. . 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Donato F, Tagger A, Gelatti U, Parrinello G, Boffetta P, Albertini A, et al. Alcohol and hepatocellular carcinoma: The effect of lifetime intake and hepatitis virus infections in men and women. . Am J Epidemiol. 2002;155(4):323–31. doi: 10.1093/aje/155.4.323. [DOI] [PubMed] [Google Scholar]
- 3.Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491(7424):454–7. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 4.Panzitt K, Tschernatsch MMO, Guelly C, Moustafa T, Stradner M, Strohmaier HM, et al. Characterization of HULC, a Novel Gene with Striking Up-Regulation in Hepatocellular Carcinoma, as Noncoding RNA. Gastroenterology. 2007;132(1):330–42. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Jalali S, Bhartiya D, Lalwani MK, Sivasubbu S, Scaria V. Systematic Transcriptome Wide Analysis of lncRNA-miRNA Interactions. PLoS One. 2013;8(2):e53823. doi: 10.1371/journal.pone.0053823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan YH. Biostatistics 102: Quantitative Data - Parametric & Non-parametric Tests. Singapore Med J. . 2003;44(8):391–396. [PubMed] [Google Scholar]
- 8.Chan YH. Biostatistics 103: Qualitative Data - Tests of Independence. . Singapore Med J. 2003;44(10):498–503. [PubMed] [Google Scholar]
- 9.Chan YH. Biostatistics104: Correlational Analysis. Singapore Med J. 2003;44(12):614–9. [PubMed] [Google Scholar]
- 10.Goossens N, Hoshida Y. Hepatitis C virus-induced hepatocellular carcinoma. Clin Mol Hepatol. 2015;21(2):105–114. doi: 10.3350/cmh.2015.21.2.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang M De, Chen WM, Qi FZ, Xia R, Sun M, Xu TP, et al. Long non-coding RNA ANRIL is upregulated in hepatocellular carcinoma and regulates cell apoptosis by epigenetic silencing of KLF2. J Hematol Oncol. 2015;8:50. doi: 10.1186/s13045-015-0146-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun XH, Yang L Bin, Geng XL, Wang R, Zhang ZC. Increased expression of lncRNA HULC indicates a poor prognosis and promotes cell metastasis in osteosarcoma. Int J Clin Exp. Pathol. 2015;8(3):2994–3000. [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Z, Xiao Z, Liu F, Cui M, Li W, Yang Z, et al. Long non-coding RNA HULC promotes tumor angiogenesis in liver cancer by up-regulating sphingosine kinase 1 (SPHK1). Oncotarget. 2016;7(1):241–54. doi: 10.18632/oncotarget.6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garofalo M, Croce CM. microRNAs: Master Regulators as Potential Therapeutics in Cancer. Annu Rev Pharmacol Toxicol. 2011;51:25–43. doi: 10.1146/annurev-pharmtox-010510-100517. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38(16):5366–83. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamal E-SS, Osama EE agroudy, Seham MS, Hazem HE, Ibrahim AE-M. MicroRNA 122 as adiagnostic biomarker for hepatitis C- related hepatocellular carcinoma. Medical Journal of Viral Hepatitis. 2019;1(1):35–43. [Google Scholar]
- 17.Gopal P, Yopp AC, Waljee AK, Chiang J, Nehra M, Kandunoori P, et al. Factors that affect accuracy of α-fetoprotein test in detection of hepatocellular carcinoma in patients with cirrhosis. . Clin Gastroenterol Hepatol. . 2014;12(5):870–7. doi: 10.1016/j.cgh.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ataseven H, Bahcecioglu IH, Kuzu N, Yalniz M, Celebi S, Erensoy A, et al. The levels of ghrelin, leptin, TNF-alpha, and IL-6 in liver cirrhosis and hepatocellular carcinoma due to HBV and HDV infection. Mediators Inflamm. . 2006;2006(4):78380. doi: 10.1155/MI/2006/78380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Axley P, Ahmed Z, Ravi S, Singal AK. Hepatitis C Virus and Hepatocellular Carcinoma: A Narrative Review. J Clin Transl Hepatol. . 2018;6(1):79–84. doi: 10.14218/JCTH.2017.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jing CY, Fu YP, Shen HJ, Zheng SS, Lin JJ, Yi Y, et al. Albumin to gamma-glutamyltransferase ratio as a prognostic indicator in intrahepatic cholangiocarcinoma after curative resection. . Oncotarget. 2017;8(8):13293–13303. doi: 10.18632/oncotarget.14530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taverna M, Marie AL, Mira JP, Guidet B. Specific antioxidant properties of human serum albumin. Ann Intensive Care. 2013;3(1):4. doi: 10.1186/2110-5820-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knight K, Wade S, Balducci L. Prevalence and outcomes of anemia in cancer: A systematic review of the literature. . Am J Med. 2004;116 Suppl 7A:11S–26S. doi: 10.1016/j.amjmed.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 23.Finkelmeier F, Bettinger D, Köberle V, Schultheiß M, Zeuzem S, Kronenberger B, et al. Single measurement of hemoglobin predicts outcome of HCC patients. Med Oncol. . 2014;31(1):806. doi: 10.1007/s12032-013-0806-2. [DOI] [PubMed] [Google Scholar]
- 24.Fawzy Montaser M, Amin Sakr M, Omar Khalifa M. Alpha-L-fucosidase as a tumour marker of hepatocellular carcinoma. Arab J Gastroenterol. 2012;13(1):9–13. doi: 10.1016/j.ajg.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Li SP, Xu HX, Yu Y, He JD, Wang Z, Xu YJ, et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget. 2016;7(27):42431–42446. doi: 10.18632/oncotarget.9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu G, Liu H, He H, Wang Y, Lu X, Yu Y, et al. miR-372 down-regulates the oncogene ATAD2 to influence hepatocellular carcinoma proliferation and metastasis. BMC Cancer. 2014;14:107. doi: 10.1186/1471-2407-14-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu G, Wang Y, Lu X, He H, Liu H, Meng X, et al. Low miR-372 expression correlates with poor prognosis and tumor metastasis in hepatocellular carcinoma. BMC Cancer. 2015;15(1):182. doi: 10.1186/s12885-015-1214-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shrivastava S, Mukherjee A, Ray RB. Hepatitis C virus infection, microRNA and liver disease progression. . World J Hepatol. 2013;5(9):479–486. doi: 10.4254/wjh.v5.i9.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramachandran S, Ilias Basha H, Sarma NJ, Lin Y, Crippin JS, Chapman WC, et al. Hepatitis C Virus Induced miR200c Down Modulates FAP-1, a Negative Regulator of Src Signaling and Promotes Hepatic Fibrosis. PLoS One. . 2013;8(8):e70744. doi: 10.1371/journal.pone.0070744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X, Wang T, Wakita T, Yang W. Systematic identification of microRNA and messenger RNA profiles in hepatitis C virus-infected human hepatoma cells. Virology. 2010;398(1):57–67. doi: 10.1016/j.virol.2009.11.036. [DOI] [PubMed] [Google Scholar]
- 31.Gu H, Guo X, Zou L, Zhu H, Zhang J. Upregulation of microRNA-372 associates with tumor progression and prognosis in hepatocellular carcinoma. Mol Cell Biochem. . 2013;375(1-2):23–30. doi: 10.1007/s11010-012-1521-6. [DOI] [PubMed] [Google Scholar]
- 32.Zhu Y, Chen X, Zheng C, Rao X, Peng X. Down-regulation of LncRNA UCA1 alleviates liver injury in rats with liver cirrhosis. Int J Clin Exp Pathol. 2019;12(2):455–465. [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Z, Lu Y, Xu Q, Tang B, Park C-K, Chen X. HULC and H19 Played Different Roles in Overall and Disease-Free Survival from Hepatocellular Carcinoma after Curative Hepatectomy: A Preliminary Analysis from Gene Expression Omnibus. . Dis Markers. 2015;2015:191029. doi: 10.1155/2015/191029. [DOI] [PMC free article] [PubMed] [Google Scholar]


