SUMMARY
RNA interference (RNAi) is an essential regulatory mechanism in all animals. In Caenorhabditis elegans, several classes of small RNAs act to silence or license expression of mRNA targets. ERI-6/7 is required for the production of some endogenous small interfering RNAs (siRNAs) and acts as a negative regulator of the exogenous RNAi pathway. We find that the genomic locus encoding eri-6/7 contains two distinct regions that are targeted by endogenous siRNAs. Loss of these siRNAs disrupts eri-6/7 mRNA expression, resulting in increased production of siRNAs from other small RNA pathways because these pathways compete with eri-6/7-dependent transcripts for access to the downstream siRNA amplification machinery. Thus, the pathway acts like a small-RNA-mediated feedback loop to ensure homeostasis of gene expression by small RNA pathways. Similar feedback loops that maintain chromatin homeostasis have been identified in yeast and Drosophila melanogaster, suggesting an evolutionary conservation of feedback mechanisms in gene regulatory pathways.
Graphical Abstract

In Brief
Rogers and Phillips identify a small-RNA-mediated feedback mechanism that regulates the expression of endogenous siRNAs. By modulating the expression of the endogenous siRNA biogenesis factor ERI-6/7, this feedback mechanism ensures homeostasis of small RNA production and ultimately mRNA expression.
INTRODUCTION
The evolutionarily conserved RNA interference (RNAi) pathways in metazoans are essential for the proper regulation of endogenous and exogenous gene expression. Argonaute proteins and their associated small RNAs are integral components of RNA-induced silencing complexes (RISCs), which transcriptionally and post-transcriptionally regulate target transcripts (Buckley et al., 2012; Burkhart et al., 2011; Claycomb, 2014; Gu et al., 2012; Guang et al., 2008, 2010; Hutvagner and Simard, 2008). Several mechanisms exist to generate the small RNA guides within RISC, each with distinct features. For instance, the RNase-III-like enzyme Dicer generates small interfering RNAs (siRNAs) by cleaving exogenous and endogenous long double-stranded RNAs (dsRNAs) (Bernstein et al., 2001; Ketting et al., 2001). In Caenorhabditis elegans, these siRNAs initiate amplification of additional secondary siRNAs within a perinuclear granule referred to as the Mutator focus (Gent et al., 2010; Gu et al., 2009; Pak and Fire, 2007; Phillips et al., 2012; Sijen et al., 2007; Vasale et al., 2010). MUT-16 is required to assemble the mutator complex, which includes an RNA-dependent RNA polymerase (RdRP) that is responsible for the amplification of 22G-RNAs (Phillips et al., 2012; Zhang et al., 2011). Secondary siRNAs dependent on the mutator complex are essential for robust RISC-mediated silencing (Gent et al., 2010; Ghildiyal and Zamore, 2009; Gu et al., 2009; Lee and Collins, 2007; Pak and Fire, 2007; Sijen et al., 2007; Vasale et al., 2010). RISC-meditated regulation is responsible for maintaining homeostasis, appropriate gene expression, and silencing of foreign genetic elements, by either post-transcriptionally regulating targets in the cytoplasm or transcriptionally regulating mRNAs at the chromatin level by directing the establishment of the repressive chromatin mark H3K9me3 (Buckley et al., 2012; Burkhart et al., 2011; Castel and Martienssen, 2013; Claycomb, 2014; Guang et al., 2008, 2010; Ketting, 2011).
ERI-6/7 is an RNA helicase that is required for the biogenesis of ERGO-1-class 26G-RNAs in embryos and thus is necessary to target complementary transcripts to the mutator complex for the production of high levels of 22G-RNAs in adults (Figure S1; Fischer et al., 2011). In C. elegans, the eri-6/7 transcript is produced by a trans-splicing event between the eri-6 and eri-7 pre-messenger RNAs (pre-mRNAs) (Fischer et al., 2008). If the function of ERI-6/7 is disrupted, animals display an enhanced RNAi (Eri) phenotype (Fischer et al., 2008, 2011). It has been proposed that this Eri phenotype stems from a requirement by both the ERGO-1 26G-RNA pathway and the RDE-1 exogenous siRNA pathway for the shared downstream use of the mutator complex for siRNA amplification (Duchaine et al., 2006; Lee et al., 2006).
The genomic locus of eri-6 contains several annotated isoforms. Four of the isoforms (eri-6[a–d]) encode the eri-6 sequence that corresponds to the functional eri-6/7 protein, whereas the remaining annotated isoforms (eri-6[e–f], formerly annotated as T01A4.3) do not contain a sequence that encodes the ERI-6/7 protein. Nested within the eri-6 genomic locus is the intronless gene C41D11.6, whose function has not previously been described. Based on our findings described in this work, we have named C41D11.6 sosi-1 (sensor of siRNAs-1). Transcriptional silencing of sosi-1 is mediated by the nuclear Argonaute protein HRDE-1 in the C. elegans germline by deposition of the repressive chromatin mark H3K9 trimethylation (H3K9me3) (Ni et al., 2014). In hrde-1 mutants, which are incapable of mediating H3K9me3 deposition at germline nuclear RNAi target loci, H3K9me3 and small RNAs are lost at the sosi-1 locus, which corresponds to increased sosi-1 mRNA expression (Ni et al., 2014).
It has been observed that in mut-16 mutant embryos, which are incapable of synthesizing 22G-RNAs complementary to mRNA targets of multiple small RNA pathways, ERGO-1-class 26G-RNAs are strongly depleted relative to wild-type embryos (Zhang et al., 2011). This finding is surprising, as it is generally thought that MUT-16 acts only in the secondary siRNA pathway, placing it functionally downstream of ERI-6/7 and ERGO-1 (Figure S1). Here, we show that the eri-6 genomic locus contains two regions that are targeted in a MUT-16-dependent manner by 22G-RNAs—the eri-6 isoforms, eri-6[e–f], and sosi-1. Loss of 22G-RNAs targeting these regions results in reduced expression of the eri-6/7 trans-spliced mRNA and loss of ERGO-1-class 26G-RNAs. We propose that these regions act independently of one another as sensors for HRDE-1-loaded MUT-16-dependent 22G-RNA levels to regulate ERI-6/7 function, which ultimately feeds back to regulate the biogenesis of 22G-RNAs associated with other small RNA pathways. Thus, these results reveal the existence of a regulatory network that modulates the expression of the exogenous and endogenous RNAi pathways by targeting sensors contained within the eri-6 gene locus. Furthermore, this regulatory network explains the depletion of 26G-RNAs in mut-16 mutants, despite the well-documented role of MUT-16 in siRNA amplification downstream of 26G-RNA biogenesis.
RESULTS
eri-6/7 Expression Is Reduced in mut-16 Mutants
MUT-16 is a core component of the small RNA amplification complex that is responsible for synthesis of 22G-RNAs within the Mutator focus (Phillips et al., 2012; Zhang et al., 2011). Previously, we generated mRNA sequencing (mRNA-seq) and small RNA-seq libraries from synchronized day 1 adult wild-type and mut-16 mutant animals grown at 20°C (Rogers and Phillips, 2020). Interestingly, we noticed significant differential expression of the eri-6/7 locus in mut-16 mutants compared with wild-type animals (Figures 1A and 1B). The eri-6/7 transcript is produced by a trans-splicing event of separate eri-6 and eri-7 pre-mRNAs (Fischer et al., 2008). The eri-6 locus contains the protein-coding isoforms (eri-6[a–d]) and other annotated isoforms (eri-6[e–f]) that do not encode an eri-6 pre-mRNA that can be trans-spliced to form eri-6/7. In wild-type animals, eri-6d, which is the primary protein-coding isoform, is the most highly expressed eri-6 isoform; however, in mut-16 mutants the expression of the eri-6d isoform is reduced significantly, and instead, the eri-6e and eri-6f isoforms are predominantly expressed (Figures 1A and 1B). The eri-7 locus also exhibited significantly reduced transcript levels in mut-16 mutants compared with those of wild-type animals (Figures 1A and 1B). Furthermore, in mut-16 mutants, we observed a significant increase in the expression of the sosi-1 gene, which is found in an intron of eri-6 (Figures 1A and 1B). These results indicate that in mut-16 mutants, the expression of both sosi-1 and eri-7 are altered, and there is a change in the selection of isoforms from the eri-6 genomic locus.
Figure 1. eri-6/7 Expression Is Altered in mut-16 Mutants.

(A) mRNA and small RNA reads per million (RPMs) across the eri-7 and eri-6 genomic locus in wild-type and mut-16 mutant animals. The sosi-1 and eri-6[e–f] regions are boxed.
(B) Shown are transcripts per kilobase million (TPMs) for eri-6 isoforms, sosi-1, and eri-7 in wild-type and mut-16 mutant mRNA-seq libraries. Error bars indicate standard deviation. n = 3 biological replicates.
(C) Shown are 22G-RNA levels in RPMs mapping to the sosi-1 and eri-6[e–f] regions in small RNA libraries from wild-type animals and mut-16 mutants. Error bars indicate standard deviation. n = 3 biological replicates.
(D) Shown are size profiles of all reads mapping to the sosi-1 region in wild-type small RNA libraries. Error bars indicate standard deviation between replicates. n = 3 biological replicates.
(E) Shown is the nucleotide represented in the first position of 22-nucleotide (nt) reads mapping to the sosi-1 region in wild-type small RNA libraries.
(F) Shown are size profiles of all reads mapping to the eri-6[e–f] region in wild-type small RNA libraries. Error bars indicate standard deviation between replicates. n = 3 biological replicates.
(G) Shown is the nucleotide represented in the first position of 22-nt reads mapping to the eri-6[e–f] region in wild-type small RNA libraries. See also Figures S1 and S2.
To determine whether the observed differential expression of the eri-6 and eri-7 pre-mRNAs correlated with changes in small RNAs, we compared small RNA-seq libraries generated from wild-type and mut-16 mutants. In wild-type animals, we observed high levels of small RNA reads mapping to the genomic locus of sosi-1 (genomic coordinates I:4459750–4462935, which we call the sosi-1 region) and to the eri-6[e–f] isoforms of eri-6 (genomic coordinates I:4463370–4469204, which we call the eri-6[e–f] region) (Figures 1A and 1C). In contrast, small RNAs mapping to the sosi-1 region and eri-6[e–f] region are depleted in mut-16 mutants (Figures 1A and 1C). Further analysis revealed the small RNAs mapping to the sosi-1 region and eri-6[e–f] region are predominantly 22G-RNAs, which is not surprising due to their dependence on MUT-16 (Figures 1D–1G). It should be noted that the eri-6f portion of the eri-6[e–f] region is 97.3% identical to the uncharacterized intronless gene K09B11.4, which is a 22G-RNA target situated near a Piwi-interacting RNA (piRNA) cluster (Figure S2). The proximity of K09B11.4 to a piRNA cluster (Ruby cluster genomic coordinates IV:13.5M-17.2M) is intriguing; however, further experiments will need to be performed to determine whether the eri-6[e–f] region or K09B11.4 are the source of the 22G-RNAs. Therefore, our data reveal that mutator-complex-dependent 22G-RNAs targeting sosi-1 and the eri-6 isoforms eri-6[e–f] are lost in mut-16 mutants, which correlates with the upregulation of sosi-1 and eri-6[e–f] mRNA and downregulation of the protein-coding regions of eri-6 (eri-6d) and eri-7 pre-mRNAs.
The sosi-1 and eri-6[e–f] Regions Are Targeted by HRDE-1-Loaded MUT-16-Dependent 22G-RNAs
Secondary WAGO-class 22G-RNAs, which require the mutator complex for their biogenesis, can be dependent on upstream primary small RNA pathways including piRNAs, also referred to as 21U-RNAs in C. elegans, and ERGO-1-class 26G-RNAs (Phillips et al., 2012; Zhang et al., 2011). To determine whether the 22G-RNAs targeting the sosi-1 region and eri-6[e–f] region are dependent on piRNAs or ERGO-1-class 26G-RNAs, we generated small RNA libraries from prg-1, ergo-1, and prg-1; ergo-1 double mutants, respectively. Previously, it was shown that piRNA-initiated silencing can be maintained independent of PRG-1 through the activity of the mutator complex and the Argonaute protein HRDE-1 (Ashe et al., 2012; Luteijn et al., 2012; Shirayama et al., 2012). To ensure we effectively disrupted all piRNA-initiated gene silencing, we also generated small RNA libraries from prg-1; hrde-1 double mutants. We assessed 22G-RNA levels mapping to the sosi-1 region and eri-6[e–f] region in mut-16, prg-1, and ergo-1 single mutants and in prg-1; ergo-1 and prg-1; hrde-1 double mutants compared with those of wild-type animals. We found that 22G-RNA levels mapping to the sosi-1 region are not significantly changed in ergo-1 mutants and are slightly upregulated in prg-1 and prg-1; ergo-1 mutants (Figure 2A). However, 22G-RNA levels mapping to the sosi-1 region are significantly reduced in prg-1; hrde-1 mutants (Figures 2A and 2B). Similar results were observed previously in hrde-1 single mutants (Ni et al., 2014). Using piRTarBase, a database that curates both predicted and experimentally identified piRNA target sites, we found that the mRNA sequence of sosi-1 harbors 12 sequence-predicted piRNA-binding sites (Wu et al., 2019; Zhang et al., 2018), suggesting that the MUT-16-dependent and HRDE-1-loaded 22G-RNAs mapping to the sosi-1 region may be initiated by piRNAs.
Figure 2. The sosi-1 and eri-6[e–f] Regions Are Targeted by HRDE-1-Loaded MUT-16-Dependent 22G-RNAs.
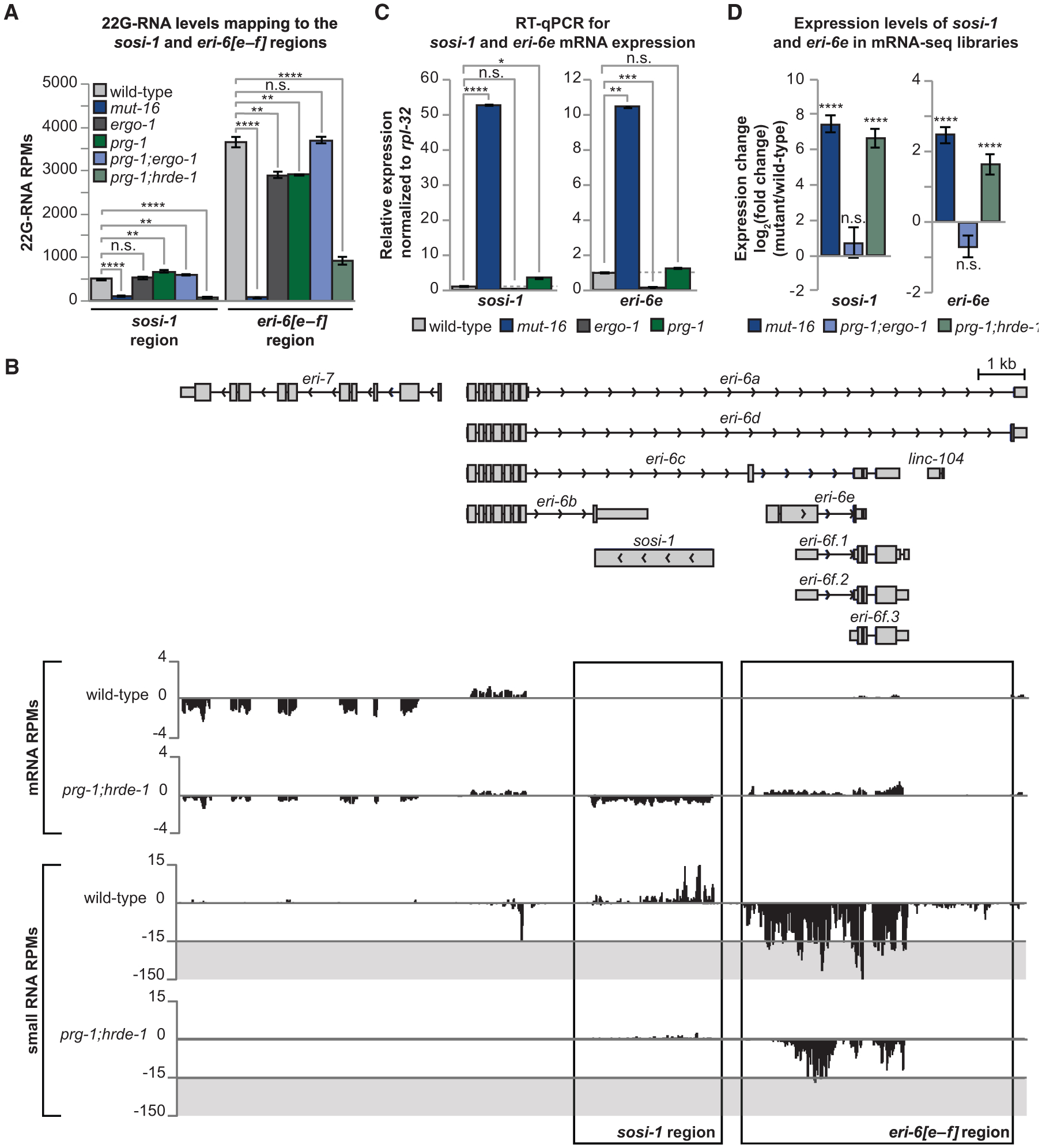
(A) Shown are 22G-RNA levels in RPMs mapping to the sosi-1 and eri-6[e–f] regions in small RNA libraries from wild-type animals, mut-16 mutants, ergo-1 mutants, prg-1 mutants, prg-1; ergo-1 mutants, and prg-1; hrde-1 mutants. Error bars represent standard deviation. n = 3 biological replicates.
(B) mRNA and small RNA RPMs across the eri-7 and eri-6 genomic locus in wild-type and prg-1; hrde-1 mutant animals. The sosi-1 and eri-6[e–f] regions are boxed.
(C) qRT-PCR assay of sosi-1 and eri-6e expression in mut-16 mutants, ergo-1 mutants, and prg-1 mutants normalized to expression in wild-type animals. Error bars represent standard deviation. Dashed line represents 1. n = 3 biological replicates.
(D) Shown is the the log2(fold change) of sosi-1 and eri-6e transcript levels in mRNA-seq libraries of mut-16 mutants, prg-1; ergo-1 mutants, and prg-1; hrde-1 mutants compared to those of wild-type animals. Error bars represent log2(standard error). n = 3 biological replicates. n.s., not significant and indicates a p > 0.05; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001.
Levels of 22G-RNAs mapping to the eri-6[e–f] region are moderately, but significantly, reduced in both ergo-1 and prg-1 single mutants and were more strongly reduced in prg-1; hrde-1 double mutants (Figures 2A and 2B). Surprisingly, 22G-RNA levels targeting the eri-6[e–f] region were not affected in prg-1; ergo-1 double mutants (Figure 2A); however, these libraries are depleted for two major classes of 22G-RNAs that may result in increased sampling of other 22G-RNA classes. Thus, another class of 22G-RNAs targeting the eri-6[e–f] region may be overrepresented in the prg-1; ergo-1 mutant libraries. Because hrde-1 mutants were previously shown to exhibit reduced small RNA levels mapping to the eri-6[e–f] region (Ni et al., 2014), we were unable to definitively conclude whether the HRDE-1-loaded 22G-RNAs are piRNA initiated. However, using piRTarBase, we found both sequence-predicted and cross-linking, ligation, and sequencing of hybrids (CLASH)-identified piRNA-binding sites within the mRNA sequences of eri-6 [e–f] (25 sequence predicted, 14 CLASH identified) and its paralog K09B11.4 (6 sequence predicted, 6 CLASH identified) (Shen et al., 2018; Wu et al., 2019; Zhang et al., 2018), suggesting the HRDE-1-loaded 22G-RNAs may be piRNA initiated. The piRNAs identified as binding to K09B11.4 were also found to bind eri-6f transcripts (Shen et al., 2018). Overall, these data suggest that the MUT-16-dependent 22G-RNAs that map to the eri-6[e–f] region may include ERGO-1-dependent 22G-RNAs, piRNA-dependent 22G-RNAs, HRDE-1-loaded 22G-RNAs, and potentially other 22G-RNA classes.
Next, to determine whether ERGO-1-dependent 22G-RNAs or piRNA-dependent 22G-RNAs affect the expression of sosi-1 or eri-6[e–f] transcripts, we performed qRT-PCR using cDNA generated from wild-type animals, mut-16 mutants, ergo-1 mutants, and prg-1 mutants with primers targeting sosi-1 and eri-6e transcripts. We designed our qPCR primers to detect the expression of the eri-6e transcript to prevent detecting the expression of the K09B11.4 transcript, which is highly similar to eri-6f. It should be noted that sosi-1 and eri-6e are expressed at very low levels in wild-type animals, and therefore, only increased expression can be accurately assessed. We found that, compared to wild-type animals, expression of sosi-1 is significantly upregulated in mut-16 mutants and is slightly upregulated in prg-1 mutants but is not upregulated in ergo-1 mutants (Figure 2C). Expression of eri-6e is significantly upregulated in mut-16 mutants but is not upregulated in ergo-1 or prg-1 mutants (Figure 2C). Thus, our qRT-PCR results corroborate our mRNA-seq results demonstrating that sosi-1 and eri-6e expression is significantly upregulated in mut-16 mutants (Figures 1A, 2B, and 2C). Next, we generated mRNA-seq libraries generated from prg-1; hrde-1 and prg-1; ergo-1 libraries and assessed the expression levels of sosi-1 and eri-6e. We observed that, compared to wild-type animals, prg-1; ergo-1 mutants did not exhibit changes in the expression levels of sosi-1 or eri-6e (Figure 2D). In contrast, prg-1; hrde-1 mutants had increased levels of expression of both sosi-1 and eri-6e, compared to wild-type animals (Figures 2B and 2D). Similar results were observed previously in hrde-1 single mutants (Ni et al., 2014), indicating that HRDE-1 is essential for maintaining the repression of both sosi-1 and eri-6e in wild-type animals. Taken together, our small RNA-seq and mRNA-seq data suggest that sosi-1 and eri-6 [e–f] are targeted predominately by HRDE-1-loaded 22G-RNAs.
sosi-1 and eri-6[e–f] Regions Are Independently Regulated by MUT-16-Dependent 22G-RNAs
To address whether the small RNAs mapping to the sosi-1 region, eri-6[e–f] region, or both regions combined were directly responsible for the regulation of eri-6/7, we generated C. elegans strains that lacked either the sosi-1 region (sosi-1Δ(cmp262)), the eri-6[e–f] region (eri-6[e–f]Δ(cmp261)), or both regions (sosi-1 eri-6[e–f]Δ(cmp263)) in wild-type and mut-16 mutants (Figure 3A). First, we assessed how expression of the sosi-1 and eri-6e transcript levels are affected in our mutants using qRT-PCR and mRNA-seq. Compared to wild-type animals, only eri-6[e–f]Δ mut-16 double mutants exhibited upregulated sosi-1 expression, which was similar to the mut-16 mutant alone (Figures 3B and S3). Both sosi-1Δ and sosi-1 eri-6[e–f]Δ, with and without the mut-16 mutant, did not exhibit upregulated sosi-1 expression, as expected, because the sosi-1 locus is deleted in these strains (Figures 3B and S3). Similarly, eri-6e expression is significantly upregulated only in sosi-1Δ mut-16 double mutants, similar to the mut-16 mutant alone (Figures 3B and S3). sosi-1Δ mutants expressed eri-6e transcripts at levels similar to those of wild-type animals, and both eri-6[e–f]Δ and sosi-1 eri-6[e–f]Δ, lacking the eri-6e locus, either in the presence or absence of the mut-16 mutant, displayed severely reduced expression of eri-6e transcripts (Figures 3B and S3). In addition, we generated small RNA-seq libraries from sosi-1Δ, eri-6[e–f]Δ, and sosi-1 eri-6[e–f]Δ mutants in wild-type and mut-16 backgrounds and found that the 22G-RNAs mapping to the sosi-1 region were not dependent on the existence of the eri-6[e–f] region and vice versa (Figure 3C). These data indicate that the sosi-1 and eri-6 [e–f] regions are regulated independently of one another by MUT-16-dependent 22G-RNAs and that deletion of either region is not sufficient to prevent upregulation of the other region in a mut-16 mutant.
Figure 3. sosi-1 and eri-6[e–f] Regions Are Independently Regulated.

(A) Schema of transgenic C. elegans in which the sosi-1 region (sosi-1Δ), eri-6[e–f] region (eri-6[e–f]Δ), or both regions (sosi-1 eri-6[e–f]Δ) are removed.
(B) The log2(fold change) of sosi-1 and eri-6e transcript levels in mRNA-seq libraries of mut-16 mutants, eri-6[e–f]Δ mutants, eri-6[e–f]Δ mut-16 mutants, sosi-1Δ mutants, sosi-1Δ mut-16 mutants, sosi-1 eri-6[e–f]Δ mutants, and sosi-1 eri-6[e–f]Δ mut-16 mutants compared to wild-type animals. Error bars represent log2(standard error). n = 3 biological replicates. Black triangles denote mutants in which the region assayed is deleted.
(C) The log2(fold change) of 22G-RNAs mapping to the sosi-1 region and eri-6[e–f] region in small RNA libraries generated from wild-type animals, mut-16 mutants, eri-6[e–f]Δ mutants, eri-6[e–f]Δ mut-16 mutants, sosi-1Δ mutants, sosi-1Δ mut-16 mutants, sosi-1 eri-6[e–f]Δ mutants, and sosi-1 eri-6[e–f]Δ mut-16 mutants compared to wild-type animals. Error bars represent log2(standard error). n = 3 biological replicates. Black triangles denote mutants in which the region assayed is deleted. n.s., not significant and indicates a p > 0.05; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001. See also Figure S3.
sosi-1 and eri-6[e–f] Regions Regulate ERI-6/7 Function and Expression
ERI-6/7 is required for the production of ERGO-1-class 26G-RNAs (Fischer et al., 2011). When ERI-6/7 function is disrupted, animals display an enhanced RNAi (Eri) phenotype (Fischer et al., 2008). To assess whether the sosi-1Δ, eri-6[e–f]Δ, or sosi-1 eri-6 [e–f]Δ deletions affect ERI-6/7 function, we performed an RNAi feeding assay to determine whether the mutants exhibited wild-type, enhanced, or deficient RNAi capabilities. Enhanced RNAi capabilities can be assessed using lir-1 and dpy-13 RNAi. The first generation of Eri mutants exposed to lir-1 RNAi exhibit lethality as a consequence of enhanced nuclear silencing of the lir-1/lin-26 polycistronic pre-mRNA, whereas the first generation of wild-type animals exposed to lir-1 RNAi do not exhibit a phenotype (Bosher et al., 1999; Guang et al., 2008; Figure 4A). Similarly, RNAi of the collagen gene dpy-13 in wild-type animals results in only a modestly shorter animal, whereas RNAi of the same gene in Eri mutant animals causes a severe Dumpy (Dpy) phenotype, possibly due to knockdown of multiple paralogous collagen genes (Kennedy et al., 2004; Zhuang et al., 2013; Figure 4A). Therefore, we assayed the RNAi competency of each deletion, with and without the mut-16 mutation, compared to our control strains (wild-type [RNAi competent], mut-16 [RNAi deficient], and ergo-1 [enhanced RNAi]) following exposure to dpy-13, lir-1, or a control RNAi clone (L4440). The effects of the RNAi clones were scored based on whether the strains exhibited no phenotype (−), a weak phenotype (+), or a strong phenotype (+++) (Figure 4A). We observed that the sosi-1Δ, eri-6[e–f]Δ, and sosi-1 eri-6[e–f]Δ deletion mutants exhibited wild-type RNAi competency for both dpy-13 and lir-1 RNAi (Figure 4A). As expected, all strains carrying the mut-16 mutant, which is required for response to exogenous dsRNA, were RNAi defective for dpy-13 RNAi (Figure 4A). RNAi of lir-1 does not elicit a phenotype in wild-type animals until the second generation of exposure; therefore, after a single generation of lir-1 RNAi, we did not observe a phenotypic difference between wild-type and mut-16 mutants (Figure 4A). These data indicate that the three deletions do not affect the function of the ERI-6/7 protein.
Figure 4. sosi-1 and eri-6[e–f] Regions Regulate ERI-6/7 Function and Expression.

(A) Sensitivity of wild-type animals, ergo-1 mutants, mut-16 mutants, eri-6[e–f]Δ mutants, eri-6[e–f]Δ mut-16 mutants, sosi-1Δ mutants, sosi-1Δ mut-16 mutants, sosi-1 eri-6[e–f]Δ mutants, and sosi-1 eri-6[e–f]Δ mut-16 mutants to dsRNA clones—L4440 (control RNAi), dpy-13, or lir-1. Animals were scored as follows—for dpy-13 RNAi: −, no phenotype; +, weak phenotype (slightly Dumpy); and +++, strong phenotype (severely Dumpy); and for lir-1 RNAi: −, no phenotype; and +++, strong phenotype (lethal). For each strain, n = 120 individuals for each RNAi clone. Shown are representative images for the phenotypes scored in the Eri RNAi Assay of plated L1 animals after exposure for 3 days. *, dead larva.
(B) TaqMan qPCR assay for 26GsiR-O1 and 26GsiR-O2 levels in embryos of ergo-1 mutants, mut-16 mutants, eri-6[e–f]Δ mutants, eri-6[e–f]Δ mut-16 mutants, sosi-1Δ mutants, sosi-1Δ mut-16 mutants, sosi-1 eri-6[e–f]Δ mutants, and sosi-1 eri-6[e–f]Δ mut-16 mutants normalized to mir-35 expression and graphed relative to wild-type. Error bars represent standard deviation. Dashed line represents 1. n = 3 biological replicates.
(C) TaqMan qPCR assay for 22GsiR-O1 levels in embryos of ergo-1 mutants, mut-16 mutants, eri-6[e–f]Δ mutants, eri-6[e–f]Δ mut-16 mutants, sosi-1Δ mutants, sosi-1Δ mut-16 mutants, sosi-1 eri-6[e–f]Δ mutants, and sosi-1 eri-6[e–f]Δ mut-16 mutants normalized to mir-35 expression and graphed relative to wild-type. Error bars represent standard deviation. Dashed line represents 1. n = 3 biological replicates.
(D) The log2(fold change) of 26G-RNAs mapping to ERGO-1 targets in mut-16 mutants, eri-6[e–f]Δ mutants, eri-6[e–f]Δ mut-16 mutants, sosi-1Δ mutants, sosi-1Δ mut-16 mutants, sosi-1 eri-6[e–f]Δ mutants, and sosi-1 eri-6[e–f]Δ mut-16 mutants compared to wild-type animals. 26G-RNAs mapping to ERGO-1 targets are normalized to all reads mapping to mir-35 family members (mir-35–42). Error bars represent log2(standard error). n = 3 biological replicates.
(E) The log2(fold change) of 22G-RNAs mapping to ERGO-1 targets in mut-16 mutants, eri-6[e-f]Δ mutants, eri-6[e–f]Δ mut-16 mutants, sosi-1Δ mutants, sosi-1Δ mut-16 mutants, sosi-1 eri-6[e–f]Δ mutants, and sosi-1 eri-6[e–f]Δ mut-16 mutants compared to wild-type animals. 22G-RNAs mapping to ERGO-1 targets are normalized to all reads mapping to mir-35 family members (mir-35–42). Error bars represent log2(standard error). n = 3 biological replicates.
(F) The log2(fold change) of the eri-6/7 transcript levels in mRNA-seq libraries of prg-1; hrde-1 mutants, mut-16 mutants, eri-6[e–f]Δ mutants, eri-6[e–f]Δ mut-16 mutants, sosi-1Δ mutants, sosi-1Δ mut-16 mutants, sosi-1 eri-6[e–f]Δ mutants, and sosi-1 eri-6[e–f]Δ mut-16 mutants compared to wild-type animals. Error bars represent log2(standard error). n = 3 biological replicates. n.s., not significant and indicates a p > 0.05; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001.
Because we could not assess the function of ERI-6/7 in the mut-16 mutants using the Eri RNAi assay, in parallel, we directly assessed the ability of each strain to generate ERGO-1-class 26G-RNAs using TaqMan qRT-PCR from embryos. We quantified the levels of two ERGO-1-class 26G-RNAs, 26G-siR-O1 and 26G-siR-O2, derived from the C40A11.10 and E01G4.7 loci, respectively. In addition, we quantified the levels of the 22G-RNAs generated downstream of 26G-siR-O1 (22G-siR-O1). As previously observed, 26G-siR-O1 and 26G-siR-O2 levels, as well as 22G-siR-O1 levels, were strongly depleted in embryos of ergo-1 and mut-16 mutants compared to those in wild-type embryos (Figures 4B and 4C; Fischer et al., 2011; Zhang et al., 2011). Consistent with the results of our RNAi assay, sosi-1Δ, eri-6[e–f]Δ, and sosi-1 eri-6[e–f]Δ mutants maintained the ability to generate ERGO-1-class 26G-RNAs and their downstream 22G-RNAs for 26G-siR-O1 and 26G-siR-O2, at levels comparable to or higher than in wild-type embryos (Figures 4B and 4C).These data further support that these deletions do not affect ERI-6/7 function. 26G-siR-O1, 26GsiR-O2, and 22G-siR-O1 levels were strongly depleted in sosi-1Δ mut-16 mutants and eri-6[e–f]Δ mut-16 mutants (Figures 4B and 4C). In contrast, the sosi-1 eri-6[e–f]Δ mut-16 mutant was able to produce 26G-siR-O1 and 26G-siR-O2, but not 22G-siR-O1, at levels comparable to wild-type embryos and significantly higher than mut-16 mutants alone (Figures 4B and 4C), which indicates that ERI-6/7 function is restored in this strain. Furthermore, an analysis of the levels of all 26G-RNAs and 22G-RNAs mapping to ERGO-1 class targets relative to the small RNAs mapping to members of the germline-specific mir-35 family (mir-35–42), which are not amplified by the mutator complex, indicated that, compared to wild-type animals, the sosi-1Δ, eri-6[e–f]Δ, and sosi-1 eri-6[e–f]Δ mutants did not have reduced levels of 26G-RNAs or 22G-RNAs mapping to ERGO-1 class targets (Figures 4D and 4E). In fact, sosi-1Δ and eri-6[e–f]Δ single mutants produce significantly more ERGO-1-class 26G-RNAs than wild-type animals, although this increase in 26G-RNAs did not lead to a corresponding increase in ERGO-1-dependent 22G-RNAs, and the explanation for this increase in 26G-RNA levels is unknown (Figures 4D and 4E). In contrast, sosi-1Δ mut-16, eri-6[e–f]Δ mut-16, and sosi-1 eri-6[e–f]Δ mut-16 mutants had reduced levels of 22G-RNAs mapping to ERGO-1 class targets due to the loss of the MUT-16 function (Figures 4D and 4E). Similar to our Taqman qPCR results, sosi-1Δ mut-16 and eri-6 [e–f]Δ mut-16 mutants had reduced levels of 26G-RNAs mapping to ERGO-1 class targets, whereas the sosi-1 eri-6[e–f]Δ mut-16 mutant was able to produce 26G-RNAs, but not 22G-RNAs, mapping to ERGO-1 class targets at levels comparable to wild-type embryos and significantly higher than mut-16 mutants alone (Figures 4D and 4E). Taken together, with the RNAi assay, these data further indicate that the three deletions do not affect the function of the ERI-6/7 protein.
Next, we sought to determine whether the sosi-1Δ, eri-6[e–f]Δ, or sosi-1 eri-6[e–f]Δ deletions affect eri6/7 transcript levels. We assessed the expression of the eri-6/7 transcript by measuring the expression levels of eri6[a–d] and eri-7 in our mRNA-seq libraries generated from wild-type animals, prg-1; hrde-1 mutants, mut-16 mutants, and sosi-1Δ, eri-6[e–f]Δ, and sosi-1 eri-6[e–f]Δ mutants in both the wild-type and mut-16 backgrounds. We found that eri-6/7 transcript levels are strongly reduced (log2(fold change) ≤ −2) in the prg-1; hrde-1 mutants, mut-16 mutants, sosi-1Δ mut-16 mutants, and eri-6[e–f]Δ mut-16 mutants, further supporting that a loss of sosi-1 and eri-6[e–f] siRNAs in the mut-16 and prg-1; hrde-1 mutants causes a reduced expression of the eri-6/7 transcript and that a loss of either the sosi-1 or eri-6[e–f] locus alone is not sufficient to rescue eri-6/7 expression (Figure 4F). We also found that the sosi-1Δ, eri-6[e–f]Δ, and sosi-1 eri-6[e–f]Δ mutants exhibited modestly reduced levels of eri-6/7 transcripts compared to those of wild-type animals (log2(fold change) ≥ −1.5) (Figure 4F); however, this reduction in eri-6/7 expression is not sufficient to affect the function of ERI-6/7 (Figures 4A–4E). In the sosi-1 eri-6[e–f]Δ mut-16 mutants, which lack both sosi-1 and eri-6[e–f] regions, the expression levels of eri-6/7 were restored nearly to the level of wild-type animals (Figure 4F). Taken together, these data demonstrate that MUT-16 and the mutator complex are required for the production of HRDE-1-loaded 22G-RNAs that target the eri-6[e–f] and sosi-1 regions, which, in turn, regulate the expression of the eri-6/7 coding transcript and ultimately the production of ERGO-1-class 26G-RNAs. The reduction in ERGO-1-class 26G-RNAs in the sosi-1Δ mut-16 and eri-6[e–f]Δ mut-16 mutants indicates that mRNA expression from either eri-6[e–f] or sosi-1 is sufficient to disrupt the expression of the eri-6 and eri-7 coding regions and production of the ERI-6/7 protein.
Modulation of the ERGO-1 26G-RNA Pathway Leads to Fine Tuning of the Production of MUT-16-Dependent 22G-RNA Classes
Previously, it was proposed that ERI proteins inhibit the exogenous RNAi pathway by competing for factors shared between the ERGO-1-class 26G-RNA pathway and the exogenous RNAi pathway (Duchaine et al., 2006; Lee et al., 2006). ERI-6/7 is required exclusively for the production of 26G-RNAs bound by the Argonaute ERGO-1, and both eri-6/7 and ergo-1 mutants exhibit an enhanced RNAi (Eri) phenotype (Fischer et al., 2008; Yigit et al., 2006). If mRNAs targeted by ERGO-1-class 26G-RNAs compete with mRNAs targeted by other primary siRNA pathways for siRNA amplification by the mutator complex, depletion of ERGO-1-class 26G-RNAs could lead to an increased production of other classes of endogenous mutator-dependent 22G-RNAs (Figure 5A). Based on our observation that MUT-16-dependent 22G-RNAs targeting the sosi-1 and eri-6[e–f] regions is required for proper ERI-6/7 function, we hypothesized that these regions act as sensors for the production of non-ERGO-1-class 22G-RNAs and modulate expression of eri-6/7 to maintain homeostasis and proper functioning of the exogenous and endogenous RNAi pathways. To determine whether we could observe changes in mutator-dependent small RNA populations as a result of a loss of ERGO-1-class 26G-RNAs, we assessed changes in the mutator-dependent small RNA classes in ergo-1 mutants compared to wild-type animals. As expected, we found that loci annotated as ERGO-1 targets had significantly reduced levels of mapping small RNAs compared to miRNAs, which are not amplified by the mutator complex (Figure 5B). In contrast, we found that annotated piRNA target genes, mutator target genes, and RDE-1 target genes had significantly increased levels of corresponding small RNAs compared to miRNAs (Figure 5B). It should be noted that levels of small RNAs mapping to miRNAs in ergo-1 mutants appear reduced compared to those in wild-type animals; however, because this analysis depends on normalizing to total library depth, an increase in the production of many MUT-16-dependent 22G-RNAs could result in an apparent decrease in other classes of small RNAs, including miRNAs. As an alternative analysis, we performed an enrichment analysis on genes with increased levels of small RNAs in ergo-1 mutants compared to wild-type animals. We found that genes with significantly increased levels of mapped small RNAs in ergo-1 mutants were enriched for piRNA targets, mutator targets, and RDE-1 targets and depleted for ERGO-1 target genes. CSR-1 target genes, whose 22G-RNAs are not amplified by the mutator complex, were neither significantly enriched nor significantly depleted (Figure 5C). These bioinformatic analyses confirm that when ERGO-1-class 26G-RNAs, and their downstream 22G-RNAs, are depleted, there is a corresponding increase in the other major classes of MUT-16-dependent small RNAs.
Figure 5. Modulation of ERGO-1-Class 26G-RNAs Leads to Fine Tuning of the Production of MUT-16-Dependent 22G-RNA Classes.

(A) Schema of competition for resources during amplification of MUT-16-dependent 22G-RNA classes.
(B) Comparison of total small RNA levels in wild-type animals compared to ergo-1 mutants for known small RNA pathway targets. Notches indicate the 95% confidence interval of the median; black line indicates median. Significance between the log2(fold change) for miRNAs compared to each class is indicated.
(C) Enrichment analysis for piRNA target genes, ERGO-1 target genes, mutator target genes, RDE-1 target genes, and CSR-1 target genes among the genes with increased mapped small RNAs in ergo-1 mutants. **p ≤ 0.01; **** p ≤ 0.0001.
Taken together, our results reveal a regulatory feedback mechanism contained within the small RNA pathway in which the levels of ERGO-1 class 26G-RNAs and their downstream 22G-RNAs can be modulated to allow for increased production of other MUT-16-dependent 22G-RNA classes by HRDE-1-loaded 22G-RNAs, which ultimately may help maintain the robustness of the RNAi pathways.
DISCUSSION
Typically, when a locus is regulated by a small RNA pathway, its transcript is directly targeted by complementary Argonaute-loaded small RNAs that conduct transcriptional or post-transcriptional silencing of the target locus (Buckley et al., 2012; Burkhart et al., 2011; Claycomb, 2014; Gu et al., 2012; Guang et al., 2008, 2010; Hutvagner and Simard, 2008). Here, we show that HRDE-1-loaded 22G-RNAs targeting the regions of sosi-1 and eri-6[e–f] downregulate sosi-1 and eri-6[e–f], which ultimately promotes the expression of the trans-spliced eri-6/7 mRNA. The loss of the sosi-1 region, the eri-6[e–f] region, or both regions does not affect ERI-6/7 function in wild-type animals. Furthermore, deletion of both regions eliminates the dependency of the eri-6/7 trans-spliced transcript on mut-16 expression. These results indicate that, in mut-16 mutants, the loss of 22G-RNAs targeting the sosi-1 and eri-6[e–f] regions, and subsequent reduced expression of trans-spliced eri-6/7, is the underlying cause of the failure to produce ERGO-1-class 26G-RNAs. This small-RNA-mediated feedback loop explains the long-standing question of why mut-16 mutants are defective in the synthesis of ERGO-1-class 26G-RNAs, which are produced upstream of MUT-16 in the small RNA pathway. Furthermore, when ERGO-1 class 26G-RNAs are lost, we observe increased levels of 22G-RNAs at other MUT-16-dependent loci (piRNA targets, mutator targets, and RDE-1 targets). Because the machinery for the production of other mutator-dependent 22G-RNAs seems to be a limiting resource (Duchaine et al., 2006; Lee et al., 2006), we propose that the 22G-RNAs targeting sosi-1 and eri-6[e–f], within the eri-6/7 locus, may act as a sensor for functioning of the mutator 22G-RNA biogenesis pathway. Thus, when small RNA levels at sosi-1 and eri-6[e–f] regions are reduced, this change allows for reallocation of resources away from the ERGO-1 class 26G-RNA pathway and toward the exogenous RNAi, piRNA, and other small RNA pathways more critical for fertility and maintaining appropriate germline gene expression (Figure 6A).
Figure 6. A Small-RNA-Mediated Feedback Mechanism Regulates the Production of Distinct 22G-RNA Classes.

(A and B) Model of the feedback loop regulating eri-6/7 expression and function through sensing of MUT-16-dependent 22G-RNAs.
Furthermore, it was previously reported that hrde-1 mutants, which cannot perform germline nuclear RNAi and therefore fail to promote H3K9me3 deposition at RNAi target loci, exhibited a 50% reduction in eri-6/7 expression (Ni et al., 2014). Based on our discovery of 22G-RNA sensors nested within the eri-6 genomic locus, we propose that the reduction of eri-6/7 expression in hrde-1 mutants is a direct result of this feedback mechanism. De-repression of sosi-1, and the resulting significant reduction of the eri-6/7 mRNA, can occur whether there is a loss of secondary siRNAs or loss of H3K9me3 deposition by the germline nuclear RNAi pathway targeting the locus.
Biological circuits use feedback mechanisms to provide homeostatic regulation by maintaining appropriate levels of proteins. A recent study in Drosophila melanogaster that focused on the SUMO ligase Su(var)2–10, which links the piRNA-loaded Piwi complex to the silencing effector complex that induces H3K9me3 deposition at target loci, identified an autoregulatory feedback loop in which several factors involved in heterochromatin formation and maintenance were marked by H3K9me3 in a Su(var)2–10-dependent manner to maintain the proper ratio and boundaries of heterochromatin versus euchromatin (Ninova et al., 2020). In addition, the loss of Su(var)2–10-dependent H3K9me3 at target gene loci resulted in spurious differential expression of isoforms or internal genes (Ninova et al., 2020), similar to the spurious expression of sosi-1 and eri-6[e–f] we observed in mut-16 mutants. In yeast, a similar feedback mechanism was identified in which the H3K9 methyltransferase clr4 is suppressed by H3K9me3 to ensure there is not inappropriate spreading of heterochromatin (Wang et al., 2015). Our work revealed a small-RNA-mediated feedback mechanism in which a factor involved in the RNAi pathways contains a sensor repressed by H3K9me3 deposition guided by 22G-RNAs. Future studies to determine the existence of other such feedback mechanisms in the evolutionarily conserved small RNA pathways will provide invaluable insights into how these regulatory mechanisms maintain homeostatic regulation in order to maintain proper levels of each class of siRNAs.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to, and will be fulfilled by the Lead Contact, Carolyn M. Phillips (cphil@usc.edu).
Materials Availability
C. elegans strains generated in this study are deposited and maintained in the Phillips Lab strain collection (USC1332 – eri-6[e–f]Δ(cmp261) I, USC1333 – eri-6[e–f]Δ(cmp261) mut-16(pk710) I, USC1335 – sosi-1Δ(cmp262) I, USC1337 – sosi-1Δ(cmp262) mut-16(pk710) I, USC1338 – sosi-1Δ eri-6[e–f]Δ(cmp263) I, USC1355 – sosi-1 eri-6[e–f]Δ(cmp263) mut-16(pk710) I, USC1387 – prg-1(n4357) I; hrde-1(tm1200) III, and USC1388 – prg-1(n4357); ergo-1(tm1860) V).
Data and Code Availability
The accession number for the de-multiplexed raw sequencing data, in fastq format, for prg-1, ergo-1, eri-6[e–f]Δ, eri-6[e–f]Δ mut-16, sosi-1Δ, sosi-1Δ mut-16, sosi-1 eri-6[e–f]Δ, sosi-1 eri-6[e–f]Δ mut-16, prg-1; hrde-1, and prg-1; ergo-1 mutants small RNA-seq and mRNA-seq libraries reported in this paper is NCBI’s Gene Expression Omnibus GEO: GSE145217.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans Strains
Unless otherwise stated, synchronized hermaphroditic C. elegans worms were grown to adulthood at 20°C according to standard conditions (Brenner, 1974).
METHOD DETAILS
Strain Construction
For the generation of the eri-6[e–f] deletion mutant, the sosi-1 deletion mutant, and the sosi-1 eri-6[e–f] double deletion mutant, CRISPR injections were performed according to published protocols using an oligo repair template and RNA guides (Table S1; Dokshin et al., 2018; Paix et al., 2015). The injection mixes included 0.25 μg/μl Cas9 protein (IDT), 100 ng/μl tracrRNA (IDT), 14 ng/μl dpy- 10 crRNA, 21 ng/μl each gene-specific crRNA, 110 ng/μl dpy-10 repair template and 110 ng/μl gene-specific repair template, and were injected into the wild-type (N2) strain (Dokshin et al., 2018; Paix et al., 2015). Post-injection, F1 animals with the Roller (Rol) or Dumpy (Dpy) phenotypes were plated individually and their progeny were PCR genotyped for the presence of the eri-6[e–f], sosi-1, or sosi-1 eri-6[e–f] deletion mutants (Arribere et al., 2014).
RNAi Assay
Feeding RNAi assays were done at 20°C. For L4440 (control), dpy-13, and lir-1 RNAi, 120 synchronized L1s of each genotype were placed on E. coli expressing the dsRNA. P0 animals were scored after ~3 days on RNAi for the following phenotypes – dpy-13 RNAi: - indicates no phenotype, + indicates a weak phenotype (slightly Dumpy), and +++ indicates a strong phenotype (severely Dumpy) and for lir-1 RNAi: - indicates no phenotype and +++ indicates a strong phenotype (Lethal). Representative images of scored phenotypes are shown in Figure 4A. All images were taken with 5x zoom on a Nikon SMZ645 stereomicroscope using an iPhone camera.
RNA Extraction
Synchronized L1s of each strain were cultured for ~68hrs at 20°C and harvested as adults for RNA extraction. Worms were washed off plates using water and then settled on ice to form a pellet. For embryo RNA samples, embryos were extracted from adult animals by bleaching (14% bleach + 10% 5M NaOH) until adult animals degraded and embryos released, and then washed in water. Adults or embryos were resuspended in 1mL TRIzol reagent (Life Technologies) and freeze-thawed on dry ice followed by vortexing. Debris was pelleted using centrifugation and the supernatant containing RNA was collected. 0.2 volume chloroform was added to the supernatant, vortexed, centrifuged, and then the aqueous phase was transferred to a new tube. Samples were precipitated using isopropanol and rehydrated in 50 μL nuclease-free H2O.
cDNA Preparation and qPCR Reactions
Adult RNA samples were DNase treated using DNase I Amplification Grade (ThermoFisher 18068015) and reverse transcribed with SuperScript III Reverse Transcriptase (ThermoFisher 18080093), following manufacturer’s protocols. All real time PCR reactions were performed using the 2x iTaq Universal SYBER Green Supermix (Biorad 1725121). Quantitative RT-PCR for small RNA (TaqMan, Life Technologies) levels in embryo RNA samples were done according to Life Technologies recommendations using the TaqMan MicroRNA Reverse Transcription kit (ThermoFisher 4366597). All real time TaqMan PCR reactions were performed using the 2x TaqMan Fast Advanced Master Mix (ThermoFisher 4444557). All qRT-PCR reactions were run in a CFX96 Touch Real-Time PCR System (Biorad 1855196). Primers and small RNA sequences used are listed in Table S1.
Small RNA Library Preparation
Small RNAs (18- to 30- nt) were size selected on denaturing 15% polyacrylamide gels (Criterion 3450091) from total RNA samples. Libraries were prepared as previously described (Montgomery et al., 2012). Library quality was assessed (Agilent BioAnalyzer Chip) and concentration was determined using the Qubit 1X dsDNA HS Assay kit. Libraries were sequenced on the Illumina NextSeq500 (SE 75-bp reads) platform. Three biological replicates were generated for prg-1, ergo-1, eri-6[e–f]Δ, eri-6[e–f]Δ mut-16, sosi-1Δ, sosi-1Δ mut-16, sosi-1 eri-6[e–f]Δ, sosi-1 eri-6[e–f]Δ mut-16, prg-1; hrde-1, and prg-1; ergo-1 mutants.
mRNA-seq library preparation
Nuclease-free H2O was added to 7.5 μg of each RNA sample, extracted from whole animals, to a final volume of 100 μL. Samples were incubated at 65°C for 2 minutes then incubated on ice. The Dynabeads mRNA Purification Kit (ThermoFisher 61006) was used according to the manufacturer’s protocol. 20 μL of Dynabeads was used for each sample. 100ng of each mRNA sample was used to prepare libraries with the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB E7760S) according to the manual, using NEBNext multiplex oligos for Illumina (NEB E7335S). Library quality was assessed (Agilent BioAnalyzer Chip) and concentration was determined using the Qubit 1X dsDNA HS Assay kit (ThermoFisher Q33231). Libraries were sequenced on the Illumina NextSeq500 (SE 75-bp reads) platform. Three biological replicates were generated for eri-6[e–f]Δ, eri-6[e–f]Δ mut-16, sosi-1Δ, sosi-1Δ mut-16, sosi-1 eri-6[e–f]Δ, sosi-1 eri-6[e–f]Δ mut-16, prg-1; hrde-1, and prg-1; ergo-1 mutants.
QUANTIFICATION AND STATISTICAL ANALYSIS
Bioinformatic Analysis of mRNA-seq and Small RNA-seq Libraries
Sequences were parsed from adapters using Cutadapt (Martin, 2011) and mapped to the C. elegans genome, WS258, using HISAT2 and Bowtie2 (Langmead and Salzberg, 2012; Kim et al., 2015) and the transcriptome using Salmon (Patro et al., 2017). Data analysis was done using samtools (Li et al., 2009), R, Excel, and custom Python scripts. Reads per million were plotted along the WS258 genome using Integrative Genomics Viewer 2.3.68 (Robinson et al., 2011). ERGO-1 target genes were defined using our ergo-1 libraries, and mutator target genes, piRNA target genes, RDE-1 target genes, and CSR-1 target genes were previously described (Gu et al., 2009; Lee et al., 2012; Phillips et al., 2014; Svendsen et al., 2019; Tsai et al., 2015; Zhang et al., 2011). Sequencing data is summarized in Table S2. For all sequencing experiments, n = 3 biological replicates for each condition examined. Statistical parameters, including log2(fold change), standard deviation, and statistical significance are reported in the figures.
Statistical Analysis of qPCR Reactions
For each qPCR experiment, n = 3 biological replicates, with 3 technical replicates, for each condition were examined. Statistical parameters, including log2(fold change), normalized as indicated, standard deviation, and statistical significance are reported in the figures.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| TRIzol Reagent | Life Technologies | Cat# 15596026 |
| RNaseOUT | ThermoFisher Scientific | Cat# 10777019 |
| TURBO DNase | ThermoFisher Scientific | Cat# AM2238 |
| SuperScript III Reverse Transcriptase | ThermoFisher Scientific | Cat# 18080093 |
| 2× iTaq Universal SYBER Green Supermix | Biorad | Cat# 1725121 |
| Multiscribe Reverse Transcriptase | ThermoFisher Scientific | Cat# 4311235 |
| 2× TaqMan Fast Advanced Master Mix | ThermoFisher Scientific | Cat# 4444557 |
| Alt-R S.p. Cas9 Nuclease V3 | Integrated DNA Technologies | Cat# 1081058 |
| Critical Commercial Assays | ||
| Qubit 1X dsDNA HS Assay Kit | ThermoFisher Scientific | Cat# Q33230 |
| Dynabeads mRNA Purification Kit | ThermoFisher Scientific | Cat# 61006 |
| NEBNext Ultra II Directional RNA Library Prep Kit for Illumina | New England Biolabs | Cat# E7760S |
| NEBNext multiplex oligos for Illumina | New England Biolabs | Cat# E7335S |
| Deposited Data | ||
| Raw and analyzed data | This paper. | GEO: GSE145217 |
| Raw and analyzed data for wild-type and mut-16 mutant small RNA-seq and mRNA-seq | Rogers and Phillips, 2020 | GEO: GSE134573 |
| Experimental Models: Organisms/Strains | ||
| C. elegans: Strain N2 (wild-type) | Caenorhabditis Genetics Center | RRID:WB-STRAIN:N2_(ancestral) |
| C. elegans: Strain NL1810 - mut-16(pk710) I. | Caenorhabditis Genetics Center | RRID:WB-STRAIN:NL1810; Wormbase: WBVar00239329(pk710) |
| C. elegans: Strain WM158 - ergo-1(tm1860) V. | Caenorhabditis Genetics Center | RRID:WB-STRAIN:WBStrain00040450 |
| C. elegans: Strain SX922 - prg-1(n4357) I. | Caenorhabditis Genetics Center | RRID:WB-STRAIN:WBStrain00034642 |
| C. elegans: Strain USC1332 - eri-6[e-f]Δ(cmp261) I. | The Phillips Laboratory | USC1332 |
| C. elegans: Strain USC1333 - eri-6[e-f]Δ(cmp261) mut-16(pk710) I. | The Phillips Laboratory | USC1333 |
| C. elegans: Strain USC1335 - sosi-1 Δ(cmp262) I. | The Phillips Laboratory | USC1335 |
| C. elegans: Strain USC1337 - sosi-1 Δ(cmp262) mut-16(pk710) I. | The Phillips Laboratory | USC1337 |
| C. elegans: Strain USC1338 - sosi-1Δ eri-6[e-f]Δ(cmp263) I. | The Phillips Laboratory | USC1338 |
| C. elegans: Strain USC1355 - sosi-1 eri-6[e-f]Δ(cmp263) mut-16(pk710) I. | The Phillips Laboratory | USC1355 |
| C. elegans: Strain USC1387 -prg-1(n4357) I; hrde-1(tm1200) III. | The Phillips Laboratory | USC1387 |
| C. elegans: Strain USC1388 -prg-1(n4357) I; ergo-1(tm1860) V. | The Phillips Laboratory | USC1388 |
| Oligonucleotides | ||
| Primers used for experiments, See Table S1. | This paper. | N/A |
| Software and Algorithms | ||
| Cutadapt | Martin, 2011 | https://cutadapt.readthedocs.io/en/stable/ |
| HISAT2 | Kim et al., 2015 | https://ccb.jhu.edu/software/hisat2/index.shtml |
| Salmon | Patro et al., 2017 | https://combine-lab.github.io/salmon/ |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Integrative Genomics Viewer 2.3.68 | Robinson et al., 2011 | https://software.broadinstitute.org/software/igv/ |
| Other | ||
| 15% Criterion TBE-Urea Polyacrylamide Gel, 12+2 well | Criterion | Cat# 3450091 |
| 4–12% Bis-Tris polyacrylamide Gels | ThermoFisher Scientific | Cat# NP0322BOX |
Highlights.
The eri-6/7 genomic locus harbors two regions targeted by MUT-16-dependent 22G-RNAs
Loss of these 22G-RNAs disrupts expression of eri-6/7 mRNA and thus ERGO-1 26G-RNAs
Modulation of ERGO-1 26G-RNA biogenesis tunes MUT-16-dependent 22G-RNAs production
This pathway acts like a feedback loop to ensure homeostasis of 22G-RNA production
ACKNOWLEDGMENTS
We thank the members of the Phillips lab for helpful discussions and feedback on the manuscript. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). Next-generation sequencing was performed by the USC Molecular Genomics Core, which is supported by award number P30 CA014089 from the National Cancer Institute. This work was supported in part by a Basil O’Connor Starter Scholar Research Award from the March of Dimes Foundation grant no. 5-FY17-38 (to C.M.P.) and the National Institutes of Health grant R35 GM119656 (to C.M.P.). C.M.P. is a Pew Scholar in the Biomedical Sciences supported by the Pew Charitable Trusts. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108279.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, and Fire AZ (2014). Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, et al. (2012). piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150, 88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, and Hannon GJ (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366. [DOI] [PubMed] [Google Scholar]
- Bosher JM, Dufourcq P, Sookhareea S, and Labouesse M (1999). RNA interference can target pre-mRNA: consequences for gene expression in a Caenorhabditis elegans operon. Genetics 153, 1245–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, and Kennedy S (2012). A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489, 447–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart KB, Guang S, Buckley BA, Wong L, Bochner AF, and Kennedy S (2011). A pre-mRNA-associating factor links endogenous siRNAs to chromatin regulation. PLoS Genet. 7, e1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel SE, and Martienssen RA (2013). RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet 14, 100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb JM (2014). Ancient endo-siRNA pathways reveal new tricks. Curr. Biol 24, R703–R715. [DOI] [PubMed] [Google Scholar]
- Dokshin GA, Ghanta KS, Piscopo KM, and Mello CC (2018). Robust Genome Editing with Short Single-Stranded and Long, Partially Single-Stranded DNA Donors in Caenorhabditis elegans. Genetics 210, 781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D Jr., Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, et al. (2006). Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124, 343–354. [DOI] [PubMed] [Google Scholar]
- Fischer SE, Butler MD, Pan Q, and Ruvkun G (2008). Trans-splicing in C. elegans generates the negative RNAi regulator ERI-6/7. Nature 455, 491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer SE, Montgomery TA, Zhang C, Fahlgren N, Breen PC, Hwang A, Sullivan CM, Carrington JC, and Ruvkun G (2011). The ERI-6/7 helicase acts at the first stage of an siRNA amplification pathway that targets recent gene duplications. PLoS Genet. 7, e1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, Kennedy S, and Fire AZ (2010). Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol. Cell 37, 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, and Zamore PD (2009). Small silencing RNAs: an expanding universe. Nat. Rev. Genet 10, 94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D Jr., Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. (2009). Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell 36, 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu SG, Pak J, Guang S, Maniar JM, Kennedy S, and Fire A (2012). Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat. Genet 44, 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lacho-wiec J, and Kennedy S (2008). An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science 321, 537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, and Kennedy S (2010). Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature 465, 1097–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, and Simard MJ (2008). Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol 9, 22–32. [DOI] [PubMed] [Google Scholar]
- Kennedy S, Wang D, and Ruvkun G (2004). A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427, 645–649. [DOI] [PubMed] [Google Scholar]
- Ketting RF (2011). The many faces of RNAi. Dev. Cell 20, 148–161. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, and Plasterk RH (2001). Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, and Collins K (2007). Physical and functional coupling of RNA-dependent RNA polymerase and Dicer in the biogenesis of endogenous siRNAs. Nat. Struct. Mol. Biol 14, 604–610. [DOI] [PubMed] [Google Scholar]
- Lee RC, Hammell CM, and Ambros V (2006). Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA 12, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Gu W, Shirayama M, Youngman E, Conte D Jr., and Mello CC (2012). C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 150, 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Sub-group (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn MJ, van Bergeijk P, Kaaij LJ, Almeida MV, Roovers EF, Berezikov E, and Ketting RF (2012). Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 31, 3422–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17, 3. [Google Scholar]
- Montgomery TA, Rim YS, Zhang C, Dowen RH, Phillips CM, Fischer SE, and Ruvkun G (2012). PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS Genet. 8, e1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JZ, Chen E, and Gu SG (2014). Complex coding of endogenous siRNA, transcriptional silencing and H3K9 methylation on native targets of germline nuclear RNAi in C. elegans. BMC Genomics 15, 1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninova M, Godneeva B, Chen YA, Luo Y, Prakash SJ, Jankovics F, Erdélyi M, Aravin AA, and Fejes Tóth K (2020). The SUMO Ligase Su(var)2–10 Controls Hetero- and Euchromatic Gene Expression via Establishing H3K9 Trimethylation and Negative Feedback Regulation. Mol. Cell 77, 571–585.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Rasoloson D, and Seydoux G (2015). High Efficiency, Homology-Directed Genome Editing in Caenorhabditis elegans Using CRISPR-Cas9 Ribonucleoprotein Complexes. Genetics 201, 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J, and Fire A (2007). Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315, 241–244. [DOI] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, and Kingsford C (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Montgomery TA, Breen PC, and Ruvkun G (2012). MUT-16 promotes formation of perinuclear mutator foci required for RNA silencing in the C. elegans germline. Genes Dev. 26, 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Montgomery BE, Breen PC, Roovers EF, Rim YS, Ohsumi TK, Newman MA, van Wolfswinkel JC, Ketting RF, Ruvkun G, and Montgomery TA (2014). MUT-14 and SMUT-1 DEAD box RNA helicases have overlapping roles in germline RNAi and endogenous siRNA formation. Curr. Biol 24, 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Biotechnol 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers AK, and Phillips CM (2020). RNAi pathways repress reprogramming of C. elegans germ cells during heat stress. Nucleic Acids Res. 48, 4256–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen EZ, Chen H, Ozturk AR, Tu S, Shirayama M, Tang W, Ding YH, Dai SY, Weng Z, and Mello CC (2018). Identification of piRNA Binding Sites Reveals the Argonaute Regulatory Landscape of the C. elegans Germline. Cell 172, 937–951.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D Jr., and Mello CC (2012). piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150, 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, and Plasterk RH (2007). Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315, 244–247. [DOI] [PubMed] [Google Scholar]
- Svendsen JM, Reed KJ, Vijayasarathy T, Montgomery BE, Tucci RM, Brown KC, Marks TN, Nguyen DAH, Phillips CM, and Montgomery TA (2019). henn-1/HEN1 Promotes Germline Immortality in Caenorhabditis elegans. Cell Rep. 29, 3187–3199.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HY, Chen CC, Conte D Jr., Moresco JJ, Chaves DA, Mitani S, Yates JR 3rd, Tsai MD, and Mello CC (2015). A ribonuclease coordinates siRNA amplification and mRNA cleavage during RNAi. Cell 160, 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC, and Conte D Jr. (2010). Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc. Natl. Acad. Sci. USA 107, 3582–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Reddy BD, and Jia S (2015). Rapid epigenetic adaptation to un-controlled heterochromatin spreading. Elife 4, e06179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WS, Brown JS, Chen TT, Chu YH, Huang WC, Tu S, and Lee HC (2019). piRTarBase: a database of piRNA targeting sites and their roles in gene regulation. Nucleic Acids Res. 47, D181–D187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, and Mello CC (2006). Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127, 747–757. [DOI] [PubMed] [Google Scholar]
- Zhang C, Montgomery TA, Gabel HW, Fischer SE, Phillips CM, Fahlgren N, Sullivan CM, Carrington JC, and Ruvkun G (2011). mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 108, 1201–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Tu S, Stubna M, Wu WS, Huang WC, Weng Z, and Lee HC (2018). The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science 359, 587–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang JJ, Banse SA, and Hunter CP (2013). The nuclear argonaute NRDE-3 contributes to transitive RNAi in Caenorhabditis elegans. Genetics 194, 117–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the de-multiplexed raw sequencing data, in fastq format, for prg-1, ergo-1, eri-6[e–f]Δ, eri-6[e–f]Δ mut-16, sosi-1Δ, sosi-1Δ mut-16, sosi-1 eri-6[e–f]Δ, sosi-1 eri-6[e–f]Δ mut-16, prg-1; hrde-1, and prg-1; ergo-1 mutants small RNA-seq and mRNA-seq libraries reported in this paper is NCBI’s Gene Expression Omnibus GEO: GSE145217.


