Abstract
Plasmopara viticola is one of the most important pathogens infecting Vitis vinifera plants. The interactions among P. viticola and both susceptible and resistant grapevine plants have been extensively characterised, at transcriptomic, proteomic and metabolomic levels. However, the involvement of plants ionome in the response against the pathogen has been completely neglected so far. Therefore, this study was aimed at investigating the possible role of leaf ionomic modulation during compatible and incompatible interactions between P. viticola and grapevine plants. In susceptible cultivars, a dramatic redistribution of mineral elements has been observed, thus uncovering a possible role for mineral nutrients in the response against pathogens. On the contrary, the resistant cultivars did not present substantial rearrangement of mineral elements at leaf level, except for manganese (Mn) and iron (Fe). This might demonstrate that, resistant cultivars, albeit expressing the resistance gene, still exploit a pathogen response mechanism based on the local increase in the concentration of microelements, which are involved in the synthesis of secondary metabolites and reactive oxygen species. Moreover, these data also highlight the link between the mineral nutrition and plants’ response to pathogens, further stressing that appropriate fertilization strategies can be fundamental for the expression of response mechanisms against pathogens.
Subject terms: Biotic, Plant physiology
Introduction
Grapevine is one of the most important crops worldwide, for production of fresh fruits, raisins, juices and wine1 and, within the genus Vitis, V. vinifera L. is the most relevant species for the wine industry. In 2018, the total surface cultivated with V. vinifera and the overall wine production was approximatively 7.5 million Ha and 30,000 million litres, respectively (https://www.oiv.int/). However, grapevine can be affected by a wide number diseases caused by various pathogenic organisms, which can cause production losses both in terms of quantity and quality with considerable negative impacts on wine industry2. Plasmopara viticola is one of the most important phytopathogens of V. vinifera, together with Botrytis cinerea and Erysiphe necator3. Downey mildew symptoms are characterised by initial yellow and oily spots on leaves and other green parts of the plant that evolve in necrosis and death of infected tissues, including bunches, possibly resulting in a total crop loss4. In order to control the disease, several fungicide treatments, including the copper (Cu)-based ones, are commonly applied in vineyards3,5,6. However, there are growing concerns about the possible negative impact of synthetic chemical fungicides and Cu on human health and the environment7–9. In particular, Cu can accumulate in the surface horizon of the soil and have adverse effects on soil biota10–12, as well as, on plants7,13–18. Therefore, with the aim of reducing the impact of synthetic chemical and Cu-based fungicides, the implementation of more sustainable practices aimed at controlling phytopathogen in viticulture are highly encouraged7.
Despite the high importance of downy mildew on grapevine production, the molecular bases of P. viticola pathogenesis are still largely unknown2. Nonetheless, several studies have highlighted that, during the early stages of infection (i.e. hyphal structures and haustoria development), P. viticola expresses genes involved in the ATP synthesis and in the active acquisition of mineral elements from the host19,20. On the other hand, grapevine plant responses to P. viticola have been thoroughly characterised at transcriptomic, proteomic and metabolomic levels21–26. In compatible interactions between the host and the pathogen, grapevine plants up-regulate genes related to the secondary metabolism, defence and response to external stimuli, whilst genes involved in the photosynthesis and the carbon metabolism are mostly down-regulated25. Transcriptional modulations were detected in micro-dissected stomata (i.e. the primary site of P. viticola infection) and surroundings cells, suggesting that grapevine plants can react to the pathogen by eliciting both a site-specific response and a short distance signal(s) from the stomata to neighbouring cells27. In contrast, incompatible interactions are characterised by the up-regulation of stress and defence-related genes, as for instance the Pathogenesis Related (PR) proteins3,28. Several Quantitative Trait Loci (QTLs), named Resistance to P. viticola (Rpv) genes, have been identified in different grapevine species and they have been associated with major phenotypic traits of resistance to downy mildew disease. In particular, Rpv1 and Rpv2 were found to be responsible for the resistance in Muscadinia rotundifolia29, Rpv3 was associated with the resistance in ‘Villard blanc’30, whilst the resistance traits in V. amurensis was ascribed to Rpv8, Rpv10 and Rpv1231–33. Since the discovery of the resistance traits to P. viticola, breeding programs have been undertaken in order to introduce the features related to downy mildew resistance in susceptible species34–38. Thus, the use of grapevine genotypes resistant to P. viticola infection might represent an important tool to improve the sustainability of viticulture39, in particular considering the current restriction on the use of Cu in the European Union40. Depending on the specific QTL and the grapevine genotype, the resistance mechanisms elicited by the Rpv genes can involve different responses, as for instance the accumulation of callose and lignin41–43, the hypersensitive response33,44, synthesis of phytoalexins45,46, the accumulation of phenolics in the infected tissues47,48, the induction of either cell necrosis38,44,49 or peroxidase activity50,51.
Recent studies have highlighted that also the ionome profile of plants might represent an important factor determining the success of the infection process by a pathogenic organism52,53. Mineral elements, both macro- and microelements, play a fundamental role in plants, being essential for the life cycles completion. They are involved in a plethora of cell functions, including primary and secondary metabolisms, energy production, defence, signals transduction, genes regulation, hormones perception, reproduction, enzyme functioning, as well as in maintaining cell structure54. In addition, an adequate intracellular concentration of essential metal ions was recognised as a necessary prerequisite for both pathogen virulence and plant defence responses55,56. In particular, considering the role played by metal ions in living organisms, it is conceivable that intracellular mineral concentrations of essential mineral elements can have a strong impact on a wide range of pathogens and on their ability to establish an interaction with host plants57. A recent research carried out on olive trees infected with the bacterial pathogen Xylella fastidiosa demonstrated that the resistant variety “Leccino”, which usually shows low or no symptoms, was characterised by a different ionomic signature as compared to the symptomatic susceptible variety (e.g. “Ogliarola salentina”)53. In particular, increases in calcium (Ca) and manganese (Mn) leaf concentration were detected, suggesting that these mineral elements might be related to the enhanced resistance of the “Leccino” variety against X. fastidiosa53. Similarly, it was reported that the ionomic balance in Lactuca sativa L. infected with the bacterium Xanthomonas campestris pv. vitians correlated with the degree of resistance showed by different lettuce varieties52.
Starting from this knowledge background, we investigated whether the resistance mechanisms showed by resistant grapevine cultivars, beside the specific activities of Rpv genes, might be also associated to the modulation of the nutrient balance in the plants. The ionomic signature of a plant tissue is the result of the regulation of transporter-based fluxes of mineral elements within the plant and among the different tissues, playing the leaf a relevant role58. The present study was undertaken on two Plasmopara-sensitive grapevine cultivars, Cabernet Sauvignon and Sauvignon Blanc, widely cultivated in wine-producing Countries (approx. 340 thousand and 120 thousand Ha, respectively) (https://www.oiv.int/public/medias/5888/en-distribution-of-the-worlds-grapevine-varieties.pdf), and the respective resistant cultivars (Sauvignon Kretos and Cabernet Volos), bearing the Rpv12 gene. After the inoculation with P. viticola, the ionomic signature of leaves was assessed using standard (e.g. Inductively Coupled Plasma-Optical Emission Spectroscopy) and chemical imaging (e.g. micro X-Ray Fluorescence spectroscopy) techniques, to unravel whether the concentration and/or the distribution of mineral elements could play a role in the response to the pathogen. To elucidate the mechanisms controlling the mineral element allocation and re-distribution upon P. viticola infection, the expression of selected genes, encoding mineral element transporters, was also assessed in leaf tissues.
Results
Mineral element distribution in Plasmopara viticola inoculated leaves
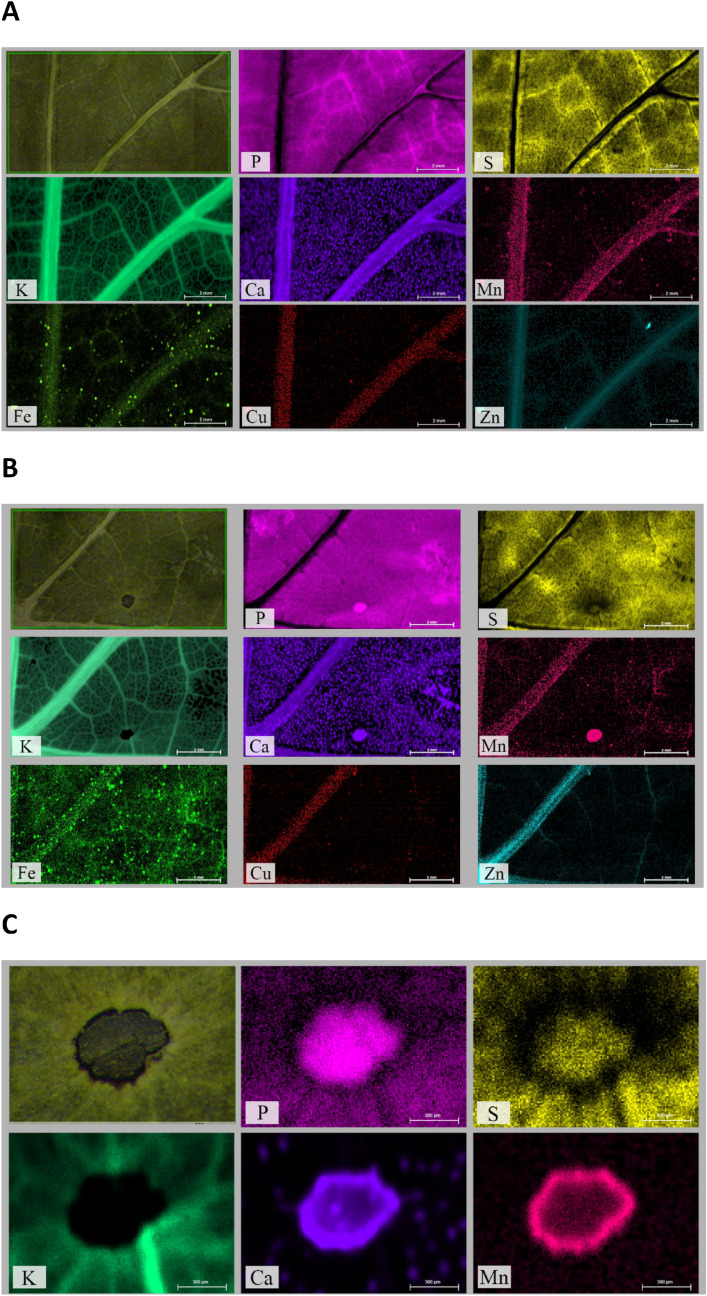
Six days after P. viticola inoculation, leaves of grapevine plants belonging to susceptible cultivars (Sauvignon Blanc and Cabernet Sauvignon) presented the typical symptoms of downy mildew, whilst resistant cultivars (Sauvignon Kretos and Cabernet Volos) did not (data not shown). Leaves of susceptible cultivars were subjected to µXRF analysis in order to unravel mineral element distribution as affected by the host–pathogen interaction. Figure 1A presents the typical mineral element distribution in a representative portion of grapevine leaves (approximately located near the petiole sinus including the midrib and nearby primary vein) belonging to the non-inoculated Sauvignon Blanc. Figure 1B reports the distribution maps of mineral elements in an inoculated Sauvignon Blanc leaf, recorded approximately in the same region as for the leaf of Fig. 1A. Although the spatial allocation of mineral elements in the grapevine mesophyll is comparable between inoculated and non-inoculated samples, several mineral elements revealed an abnormal distribution at the level of infected spots (Fig. 1B). Infected spots were completely devoid of potassium (K), whilst phosphorus (P), Ca and Mn seemed to be accumulated within the spots (Fig. 1B). When examined at higher magnification (Fig. 1C), it was possible to confirm the almost complete depletion of K and an accumulation of both P and Ca in the infected spot. In addition, Mn and Ca showed an accumulation in correspondence of the spot borders, whereas sulphur (S) displayed a preferential allocation at the centre of the infected spot (Fig. 1C). Iron (Fe) homogeneously increased in the tissue of inoculated leaves (Fig. 1B) compared to non-inoculated leaves (Fig. 1A). Zinc (Zn) appeared more concentrated in the primary veins and copper (Cu) did not show any variation in the allocation in inoculated as compared to non-inoculated tissues (Fig. 1A,B). The distribution maps of mineral elements obtained for leaves of Cabernet Sauvignon were substantially congruent to those described above, whilst no alteration in the distribution maps of mineral elements was highlighted in the inoculated Sauvignon Kretos and Cabernet Volos as compared to the non-inoculated samples (data not shown).
Figure 1.
Micro-focused X-ray fluorescence (μXRF) analyses of Sauvignon Blanc leaves. (A) Representative μ-XRF distribution maps for Sauvignon Blanc leaves. (B) Representative μ-XRF distribution maps for Sauvignon Blanc leaves inoculated with Plasmopara viticola. (C) Close-up of μ-XRF distribution maps for Sauvignon Blanc leaves inoculated with P. viticola at level of the infection spot shown in (B). For each mineral element specified by the abbreviation at the bottom of each panel, brighter colours correspond to higher mineral element concentrations.
Ionomic signatures of Plasmopara viticola inoculated leaves
To further investigate whether the alterations in the distribution maps of mineral elements were due either to a differential allocation of mineral elements or to an increase in the uptake and/or translocation of minerals, leaves ionomic profiling was undertaken using the ICP-OES technique. The Principal Component Analysis (PCA) carried out on the whole dataset (i.e. white and red cultivars, susceptible and resistant, inoculated and non-inoculated with P. viticola) generated a two components model explaining 86.88% of the total variance with no clustering of samples (Supplementary Figure 1). On the other hand, the PCA of white cultivars generated a two components model accounting for 89.60% of the total variance, showing a clear separation between inoculated and non-inoculated samples along the firs principal component 1 (PC1) (Fig. 2A). In addition, within the non-inoculated samples also the separation between Sauvignon Blanc and Sauvignon Kretos was highlighted (Fig. 2A). The mineral elements determining the separation along PC1 were mainly Fe and Mn (Fig. 2A), as found in the mineral element distribution maps (Fig. 1). Conversely, the PCA of the red varieties explained 90.44% of the total variance, with no clear clustering of samples (Fig. 2B).
Figure 2.
Principal Component Analyses of ionomic datasets. (A) Scatterplot representing the modification of leaves ionome in Sauvignon Blanc and Sauvignon Kretos both non-inoculated and inoculated with Plasmopara viticola. (B) Scatterplot representing the modification of leaves ionome in. Cabernet Sauvignon and Cabernet Volos both non-inoculated and inoculated with Plasmopara viticola. and by the infection with P. viticola.
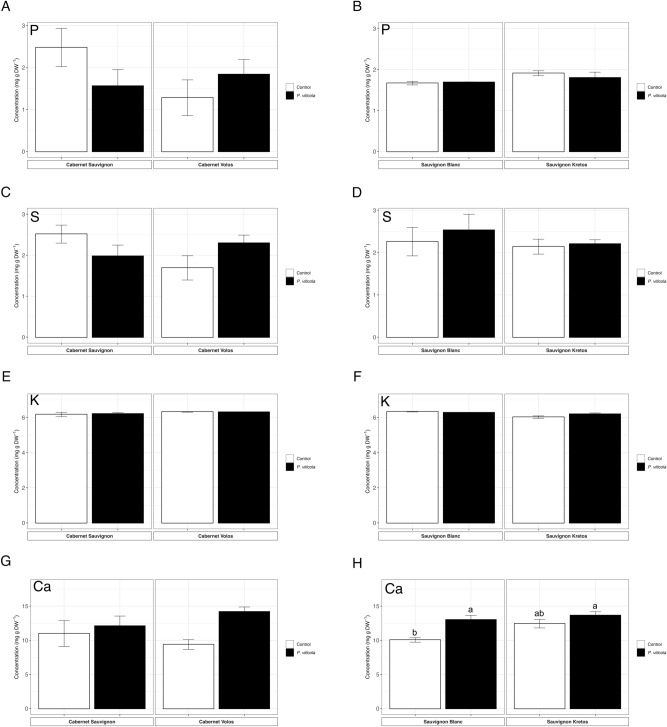
In spite of the visible alteration observed in the distribution maps of P, the ionomic analyses revealed no differences in the total concentration of P in the leaves of both cultivars, independently from the genotype and P. viticola inoculation (Fig. 3A,B). The same also applied for S (Fig. 3C,D), K (Fig. 3E,F) and Ca, at least in the red cultivars (Fig. 3G). Interestingly, the concentration of Ca in the leaves of Sauvignon Blanc was increased by the inoculation with P. viticola (Fig. 3H).
Figure 3.
Macroelement concentration. (A) Phosphorus concentration in Cabernet Sauvignon and Cabernet Volos, both non-inoculated and inoculated with Plasmopara viticola. (B) Phosphorus concentration in Sauvignon Blanc and Sauvignon Kretos, both non-inoculated and inoculated with P. viticola. (C) Sulphur concentration in Cabernet Sauvignon and Cabernet Volos, both non-inoculated and inoculated with P. viticola. (D) Sulphur concentration in Sauvignon Blanc and Sauvignon Kretos, both non-inoculated and inoculated with P. viticola. (E) Potassium concentration in Cabernet Sauvignon and Cabernet Volos, both non-inoculated and inoculated with P. viticola. (F) Potassium concentration in Sauvignon Blanc and Sauvignon Kretos, both non-inoculated and inoculated with P. viticola. (G) Calcium concentration in Cabernet Sauvignon and Cabernet Volos, both non-inoculated and inoculated with P. viticola. (H) Calcium concentration in Sauvignon Blanc and Sauvignon Kretos, both non-inoculated and inoculated with P. viticola. The data are reported as means ± SE of three biological replicates. Different letters indicate significant differences according to one-way ANOVA with Tukey post hoc tests (p < 0.05). Letters were omitted when no significant differences were found.
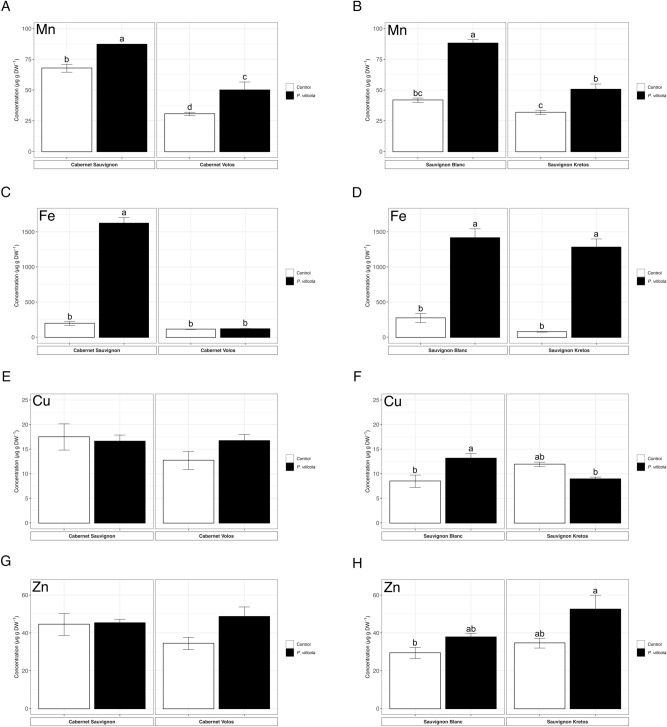
The Mn concentration increased in Cabernet Sauvignon and Sauvignon Blanc leaves inoculated with P. viticola as compared to non-inoculated plants (Fig. 4A,B), in agreement with the Mn distribution maps. Similarly, a higher concentration of Mn was also found in P. viticola-inoculated Cabernet Volos and Sauvignon Kretos by about 60% as compared to non-inoculated leaves (Fig. 4A,B). The Fe concentration was increased by about 8 and 5 times in P. viticola-inoculated Cabernet Sauvignon and Sauvignon Blanc as compared to non-inoculated samples, respectively (Fig. 4C,D). Consistently, a 16-fold increase in the Fe concentration was also detected in the P. viticola-inoculated leaves of Sauvignon Kretos with respect to non-inoculated Sauvignon Kretos leaves (Fig. 4D), whilst no alterations were detected in Cabernet Volos leaves (Fig. 4C).
Figure 4.
Microelement concentration. (A) Manganese concentration in Cabernet Sauvignon and Cabernet Volos, both non-inoculated and inoculated with Plasmopara viticola. (B) Manganese concentration in Sauvignon Blanc and Sauvignon Kretos, both non-inoculated and inoculated with P. viticola. (C) Iron concentration in Cabernet Sauvignon and Cabernet Volos, both non-inoculated and inoculated with P. viticola. (D) Iron concentration in Sauvignon Blanc and Sauvignon Kretos, both non-inoculated and inoculated with P. viticola. (E) Copper concentration in Cabernet Sauvignon and Cabernet Volos, both non-inoculated and inoculated with P. viticola. (F) Copper concentration in Sauvignon Blanc and Sauvignon Kretos, both non-inoculated and inoculated with P. viticola. (G) Zinc concentration in Cabernet Sauvignon and Cabernet Volos, both non-inoculated and inoculated with P. viticola. (H) Zinc concentration in Sauvignon Blanc and Sauvignon Kretos, both non-inoculated and inoculated with P. viticola. The data are reported as means ± SE of three biological replicates. Different letters indicate significant differences according to one-way ANOVA with Tukey post hoc tests (p < 0.05). Letters were omitted when no significant differences were found.
Concerning Cu, red and white cultivars showed a discrepancy in the behaviour (Fig. 4E,F). In both the Cabernet Sauvignon and Cabernet Volos, the P. viticola inoculation did not cause any variation in the Cu concentration as compared to the respective non-inoculates leaves (Fig. 4E). The Cu concentration was enhanced by about 70% in the inoculated Sauvignon Blanc leaves and reduced by about 30% in the inoculated Sauvignon Kretos leaves as compared to the respective non-inoculated leaves (Fig. 4F). Similarly, Zn concentration was not affected by P. viticola inoculation in Cabernet Sauvignon and in Cabernet Volos (Fig. 4G), whereas it was increased by 30% and 50% in Sauvignon Blanc and Sauvignon Kretos as compared to the non-inoculated leaves, respectively (Fig. 4H).
Gene expression analyses in Plasmopara viticola inoculated leaves
Sulphate transporters
On the bases of the phylogenetic analysis, five sulphate transporters belonging to the group 3 (i.e. VvSultr3.1, VvSultr3.2, VvSultr3.3, VvSultr3.4, VvSultr3.5) were identified in V. vinifera genome. However, the relative mRNA levels of VvSultr3.4 resulted not detectable in our experimental conditions.
The expression of VvSULTR3.1 was down-regulated in inoculated as compared to non-inoculated leaves of the susceptible cultivars and it showed a down-regulation trend in the inoculated resistant cultivars, albeit not significantly (Fig. 5A,B). VvSULTR3.2 was up-regulated in inoculated Cabernet Volos leaves and it was not modulated in the other samples analysed (Fig. 5C,D). The expression of VvSULTR3.3 had a different behaviour, depending on the cultivar (Fig. 5E,F). In the red cultivars, P. viticola inoculation caused a down-regulation of VvSULTR3.3 only in the susceptible one, whilst it was not modulated in Cabernet Volos (Fig. 5E). In the white cultivars, P. viticola inoculation decreased the expression of VvSULTR3.3 in Sauvignon Kretos leaves and not in Sauvignon Blanc compared to the respective non-inoculated leaves (Fig. 5F).
Figure 5.
Quantitative RT-PCR analyses of selected sulphate transport genes (VvSULTR) in grapevine leaves. (A) Assessment of VvSULTR3.1 gene expression in Cabernet Sauvignon and Cabernet Volos, both non-inoculated and inoculated with Plasmopara viticola. (B) Assessment of VvSULTR3.1 gene expression in Sauvignon Blanc and Sauvignon Kretos, both non-inoculated and inoculated with P. viticola. (C) Assessment of VvSULTR3.2 gene expression in Cabernet Sauvignon and Cabernet Volos, both non-inoculated and inoculated with P. viticola. (D) Assessment of VvSULTR3.2 gene expression in Sauvignon Blanc and Sauvignon Kretos, both non-inoculated and inoculated with P. viticola. (E) Assessment of VvSULTR3.3 gene expression in Cabernet Sauvignon and Cabernet Volos, both non-inoculated and inoculated with P. viticola. (F) Assessment of VvSULTR3.3 gene expression in Sauvignon Blanc and Sauvignon Kretos, both non-inoculated and inoculated with P. viticola. (G) Assessment of VvSULTR3.5 gene expression in Cabernet Sauvignon and Cabernet Volos, both non-inoculated and inoculated with P. viticola. (H) Assessment of VvSULTR3.5 gene expression in Sauvignon Blanc and Sauvignon Kretos, both non-inoculated and inoculated with P. viticola. The data were normalised to the tubulin and EF1α housekeeping genes. The relative expression ratios were calculated using untreated parental genotypes as a calibrator sample. The values reported are means ± SE of three biological replicates. Different letters indicate significant differences according to one-way ANOVA with Tukey post hoc tests (p < 0.05).
The expression of the VvSULTR3.5 was strongly up-regulated in inoculated as compared to non-inoculated susceptible cultivars, by about 1500 and 100-fold in the red and white cultivar, respectively (Fig. 5G,H). The pathogen inoculation up-regulated VvSULTR3.5 in Cabernet Volos leaves as compared to the non-inoculated plants (Fig. 5G).
Iron transporters
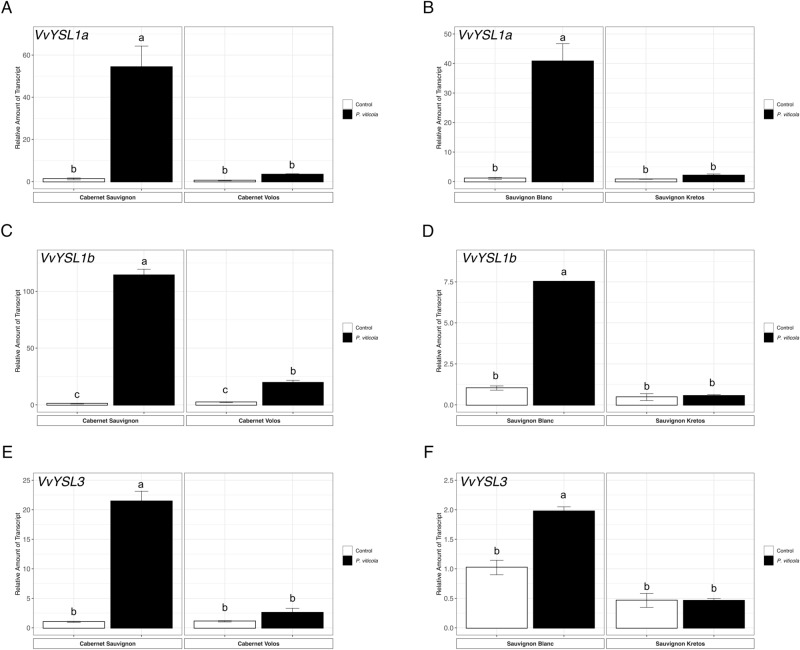
The expression of VvYSL transporters genes is generally higher in inoculated in comparison to the non-inoculated leaves of susceptible cultivars, in both Cabernet Sauvignon and Sauvignon Blanc (Fig. 6). In particular, VvYSL1a, VvYSL1b and VvYSL3 were 50-, 114- and 20-fold up-regulated in inoculated as compared to the non-inoculated Cabernet Sauvignon leaves, respectively (Fig. 6A,C,E). Similarly, VvYSL1a, VvYSL1b and VvYSL3 were up-regulated by 40-, 7- and 7-folds in inoculated as compared to non-inoculated Sauvignon Blanc leaves, respectively (Fig. 6B,D,F). Concerning the resistant cultivars, only the Cabernet Volos inoculated leaves showed an up-regulation of VvYSL1b as compared to the respective non-inoculated sample (Fig. 6C). Consistently, VvOPT3 and VvVIT1 were up-regulated by P. viticola inoculation in Cabernet Sauvignon and Sauvignon Blanc leaves (Fig. 7). In addition, VvOPT3 and VvVIT1 were up-regulated in inoculated as compared to non-inoculated Cabernet Volos leaves (Fig. 7A,C), whilst no modulations were detected in inoculated leaves of Sauvignon Kretos as compared to the respective non-inoculated samples (Fig. 7B,D).
Figure 6.
Quantitative RT-PCR analyses of genes possibly involved the Fe partitioning (VvYSL1a, VvYSL1b and VvYSL3) in grapevine leaves. (A) Assessment of VvYSL1a gene expression in Cabernet Sauvignon and Cabernet Volos, both non-inoculated and inoculated with P. viticola. (B) Assessment of VvYSL1a gene expression in Sauvignon Blanc and Sauvignon Kretos, both non-inoculated and inoculated with P. viticola. (C) Assessment of VvYSL1b gene expression in Cabernet Sauvignon and Cabernet Volos, both non-inoculated and inoculated with P. viticola. (D )Assessment of VvYSL1b gene expression in Sauvignon Blanc and Sauvignon Kretos, both non-inoculated and inoculated with P. viticola. (E) Assessment of VvYSL3 gene expression in Cabernet Sauvignon and Cabernet Volos, both non-inoculated and inoculated with P. viticola. (F) Assessment of VvYSL3 gene expression in Sauvignon Blanc and Sauvignon Kretos, both non-inoculated and inoculated with P. viticola. The data were normalised to the tubulin and EF1α housekeeping gene. The relative expression ratios were calculated using untreated parental genotypes as a calibrator sample. The values reported are means ± SE of three biological replicates. Different letters indicate significant differences according to one-way ANOVA with Tukey post hoc tests (p < 0.05).
Figure 7.
Quantitative RT-PCR analyses of VvOPT3 and VvVIT1 genes in grapevine leaves. (A) Assessment of VvOPT3 gene expression in Cabernet Sauvignon and Cabernet Volos, both non-inoculated and inoculated with Plasmopara viticola. (B) Assessment of VvOPT3 gene expression in Sauvignon Blanc and Sauvignon Kretos, both non-inoculated and inoculated with P. viticola. (C) Assessment of VvVIT1 gene expression in Cabernet Sauvignon and Cabernet Volos, both non-inoculated and inoculated with P. viticola. (D) Assessment of VvVIT1 gene expression in Sauvignon Blanc and Sauvignon Kretos, both non-inoculated and inoculated with P. viticola. The data were normalised to the tubulin and EF1α housekeeping gene. The relative expression ratios were calculated using untreated parental genotypes as a calibrator sample. The values reported are means ± SE of three biological replicates. Different letters indicate significant differences according to one-way ANOVA with Tukey post hoc tests (p < 0.05).
Manganese transporters
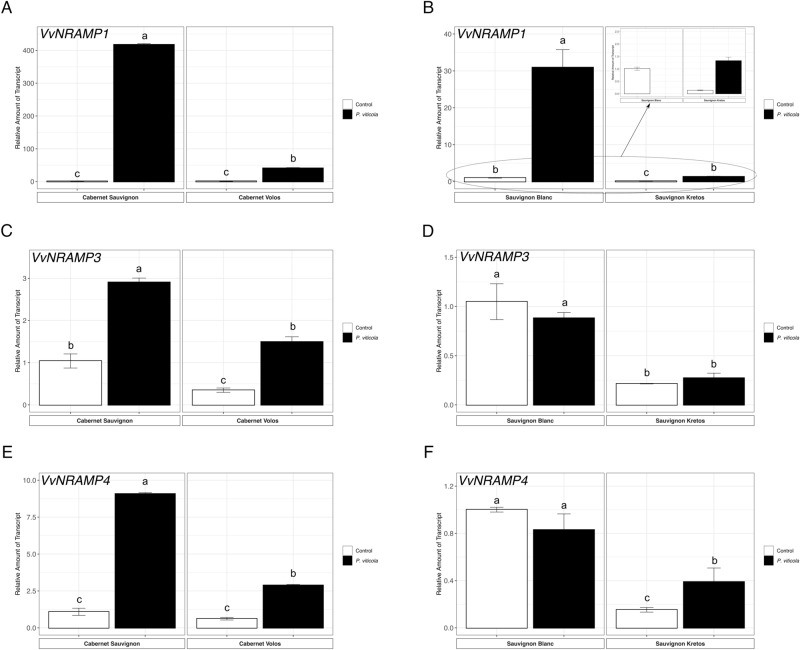
The NRAMP (Natural Resistance-Associated Macrophage Protein) genes encode primarily for manganese (Mn) transporters, albeit they can also transport other divalent cations (i.e. Fe, Cu and Zn) with a lower specificity14,59. According to a previous work, V. vinifera genome encodes three putative NRAMP transporters, i.e. VvNRAMP1, VvNRAMP3 and VvNRAMP414. The expression of VvNRAMP genes were up-regulted in inoculated as compared to non-inoculated leaves of both Cabernet Sauvignon and Cabernet Volos (Fig. 8A,C,E). Concerning the white cultivars, VvNRAMP1 was up-regulated in both Sauvignon Blanc and Sauvignon Kretos by P. viticola inoculation as compared to the respective non-inoculated samples (Fig. 8B). In addition, the inoculated leaves of Sauvignon Kretos showed the up-regulation of VvNRAPM4 as compared to the non-inoculated leaves (Fig. 8F). On the other hand, VvNRAMP3 was not modulated in both Sauvignon Blanc and Sauvignon Kretos (Fig. 8D).
Figure 8.
Quantitative RT-PCR analyses of NRAMP (natural resistance-associated macrophage protein) metal-ion transporter genes (VvNRAMP1, VvNRAMP3 and VvNRAMP4) in grapevine leaves. (A) Assessment of VvNRAMP1 gene expression in Cabernet Sauvignon and Cabernet Volos, both non-inoculated and inoculated with P. viticola. (B) Assessment of VvNRAMP1 gene expression in Sauvignon Blanc and Sauvignon Kretos, both non-inoculated and inoculated with P. viticola. (C) Assessment of VvNRAMP3 gene expression in Cabernet Sauvignon and Cabernet Volos, both non-inoculated and inoculated with P. viticola. (D) Assessment of VvNRAMP3 gene expression in Sauvignon Blanc and Sauvignon Kretos, both non-inoculated and inoculated with P. viticola. (E) Assessment of VvNRAMP4 gene expression in Cabernet Sauvignon and Cabernet Volos, both non-inoculated and inoculated with P. viticola. (F) Assessment of VvNRAMP4 gene expression in Sauvignon Blanc and Sauvignon Kretos, both non-inoculated and inoculated with P. viticola. The data were normalised to the tubulin and EF1α housekeeping gene. The relative expression ratios were calculated using untreated parental genotypes as a calibrator sample. The values reported are means ± SE of three biological replicates. Different letters indicate significant differences according to one-way ANOVA with Tukey post hoc tests (p < 0.05).
Discussion
The need to find sustainable alternative approaches to control P. viticola on grapevine requires to deepen our knowledge on the Rpv genes. Since the identification of Rpv genes, which have been associated with different phenotypic traits of downy mildew resistance in several grapevine species, breeding programs were undertaken in order to introduce resistant traits in susceptible grapevine species29,31–33,44,60,61. Because very little is known on the interaction between elements in leaf tissues and resistance to diseases, the present research aims at clarifying the role played by the ionomic signature of the leaf tissue in the response to the inoculation with P. viticola in susceptible cultivars, namely Cabernet Sauvignon and Sauvignon Blanc, in comparison to the respective resistant genotypes carrying the Rpv12 gene (Cabernet Volos and Sauvignon Kretos). With respect to this resistance trait, Rpv12 can activate an effective hypersensitive response within 24–48 h post-inoculation with the pathogen, thus preventing the sporulation of P. viticola33. However, recent pieces of evidence obtained in other species (e.g. Lactuca sativa and Olea europea) highlighted that also the mineral element balance of plants might play an important role in the resistance mechanisms against pathogens52,53. Although the compatible and the incompatible interaction between V. vinifera plants and P. viticola have been thoroughly characterized at transcriptomic, proteomic and metabolomic levels21–26, to the best of our knowledge, no information on the modulation of the ionomic profile in infected leaves of both susceptible and resistant cultivars is available. The analysis of the ionomic signature of V. vinifera leaves non-inoculated and inoculated with P. viticola highlighted a different concentration and/or distribution of several mineral nutrients in the leaf. In particular, in susceptible cultivars, in addition to their differential distribution in the leaves upon the infection with P. viticola, there is also an increase in specific microelement concentrations (i.e. Mn, Fe, Zn). Such differential allocation in the leaves has been also detected for several macronutrients (i.e. P, S, K and Ca), even though the total leaf concentration of these elements was not affected by the infection. Yet, in resistant cultivars, despite the infection with P. viticola could enhance the total leaf concentration of microelements (i.e. Mn and Fe), the spatial distribution in the leaf is not altered.
In susceptible cultivars, the ionomic profiling revealed that P. viticola infection caused an increase in Fe concentration, whilst the application of µXRF revealed that such increase was uniformly distributed all over the whole leaf. In Cabernet Sauvignon and Sauvignon Blanc, genes putatively related to Fe partitioning and translocation towards the sink tissues62–67, such as VvYSL1a, VvYSL1b, VvYSL3 and VvOPT3, were significantly up-regulated after P. viticola inoculation. Consistently, the up-regulation of the VvVIT1 gene, whose orthologous in other plants are involved in Fe, Mn and Zn trafficking across the tonoplast membrane68,69, suggested a re-arrangement in Fe distribution in the inoculated leaves. It has been postulated that the inoculation of plants with specific pathogens might induce a “Fe deficiency-like” signal70. Consistently, the silencing of AtYSL3 was related with an enhanced susceptibility of A. thaliana plants toward the pathogen Pseudomonas syringae pv. tomato DC300071. Although the µXRF did not highlight alterations in Fe distribution maps, the leaf micronutrient concentration was increased by P. viticola infection only in Sauvignon Kretos plants. Unexpectedly, genes putatively involved in Fe partitioning (i.e. VvYSL1a, VvYSL1b, VvYSL3, VvOPT3 and VvVTI1) were not modulated in inoculated Sauvignon Kretos leaves, whilst transporters belonging to the NRAMP family, (VvNRAMP1 and VvNRAMP4), were up-regulated by P. viticola infection. Interestingly, NRAMP transporters have been shown to be both involved in the Fe starvation response, in the uptake and translocation of divalent cations, like Mn, Fe, Cu and Zn14,59,72–74 and to be induced by infections in A. thaliana plants75. Inoculated Cabernet Volos leaves showed an up-regulation VvYSL1b, VvOPT3 and VvVTI1 that suggested a redistribution of Fe in the leaf, even though the total concentration was not affected. A possible involvement of Fe in the resistance mechanisms against pathogens has been for long time postulated; for instance, at rhizosphere level, the competition for Fe between pathogens and beneficial microbes (i.e. plant growth-promoting rhizobacteria) can limit the growth of dangerous microorganisms, thus indirectly protecting plants from diseases76,77. However, there is also an increasing body of evidence suggesting that the inoculation with pathogens can lead to the disturbance of Fe homeostasis78–81. The local increase in Fe concentration has been hypothesised to be associated with the production of Reactive Oxygen Species (ROS), which are well known to be involved in the hypersensitive response82. Moreover, this resistance trait has been identified as distinctive for the grapevine resistant cultivars carrying the Rpv12 gene33. Even though free Fe can catalyse ROS formation83, up to date its role in the ROS production in response to pathogen inoculation is still under investigation70. Considering the possible role of this micronutrient in the hypersensitive response82, the difference in Fe concentration and the differential modulation of genes putatively involved in Fe translocation and partitioning in the leaves of resistant cultivars inoculated with the pathogen might be ascribable to a different timing in the response of plants toward the pathogen. In fact, at the sampling time, Sauvignon Kretos seemed to have already translocated Fe to the leaves (i.e. high Fe concentration in the leaves and reduced expression of genes involved in partitioning) suggesting a faster reaction to the Plasmopara. On the other hand, inoculated leaves of Cabernet Volos featured a Fe concentration similar to non-inoculated ones, whilst the genes involved in Fe partitioning were up-regulated, thus possibly suggesting an ongoing Fe translocation process at the time of sampling.
In Cabernet Sauvignon and Sauvignon Blanc cultivars, the inoculation with P. viticola increases Mn concentration in the leaf. In particular, the micronutrient was strongly accumulated on the necrotic lesion induced by the pathogen, with the mineral element mainly localised at the borders of infected spots. Such increase in Mn concentration in the infected leaves of susceptible cultivars might be associated to the enhanced expression of VvNRAMP genes, which could be putatively involved in the uptake and translocation of this cation84,85. Manganese is indeed accounted as one of the most effective micro-elements in the suppression of plant diseases86, being able to affect several biochemical reactions involved in the synthesis of phenolic compounds (e.g. flavonoids) and of lignin and suberin, with some of them showing antimicrobial activities56,87. In cucumber plants infected with pathogenic fungi, Mn was shown to reduce the sensitivity by regulating the synthesis of the cell wall polysaccharides and the cell water status88. These pieces of evidence might therefore suggest an active process of the cell wall strengthening in the lesions, in particular at the boundaries of the P. viticola inoculation spots. Similar dynamics for Mn were also observed in resistant cultivars, which showed both a higher Mn concentration and an increased expression of VvNRAMP genes in the infected leaves as compared to non-infected ones. Noteworthy, both the concentration of Mn and the expression levels achieved in the infected leaves of resistant cultivars were significantly lower as compared to the infected leaves of the respective susceptible cultivars.
In spite of being primarily characterised as Mn transporters in plants84,85, members of the NRAMP gene family have been also shown to mediate the uptake and translocation of other divalent cations, like Fe, Cu and Zn, albeit less specifically14,59. Therefore, the up-regulation VvNRAMP genes in the susceptible cultivars infected with P. viticola might be also related to the modification in the distribution of Zn, which has been suggested to have a role in plant cell protection against the oxidative stress and in the production of important disease resistance signalling proteins56,89. Consistently with the dynamics observed in the susceptible cultivars, the up-regulation of VvNRAMP genes in the inoculated resistant cultivars did not cause a differential accumulation of Zn in the leaves as compared to the respective non-inoculated controls.
The inoculation of susceptible cultivars with P. viticola also modified the distribution of S, even though the total leaf concentration did not change as compared to the non-inoculated plants. Indeed, the modulation of transporter genes might suggest a redistribution of S at cellular/subcellular levels, whereas the steady S concentration in the tissues might be accounted to the presence of S in the organic form. On the other hand, the S distribution was not affected by P. viticola inoculation in resistant cultivars (i.e. Cabernet Volos and Sauvignon Kretos). However, the inoculation up-regulated VvSULTR3.5 in leaves of Cabernet Sauvignon, Cabernet Volos and Sauvignon Blanc, albeit to significantly lower levels in the resistant cultivar. Interestingly, the ortholog gene PtSULTR3.5 was up-regulated in poplar leaves inoculated with the fungus Melampsora larici-populina90, further strengthening the connection between S dynamics in leaves and plants response to biotic stresses. Indeed, S is involved in different metabolic pathways in plant cells, such as the synthesis of hydrogen sulphide, cysteine, methionine, glutathione, glucosinolates and phytoalexins, to tackle with biotic stressors91,92.
The distribution of other macroelements, like K, Ca and P, resulted modified only in the inoculated leaves of susceptible varieties, even though their possible roles in the response to pathogen are still under investigation. An adequate K nutrition in plants can contribute in reducing the susceptibility towards bacterial and fungal infections56,93. At cell level, K fluxes are involved in the control on the plasma membrane polarization93; whose alterations are often observed in the presence of fugal effectors94. Changes in the membrane polarity quickly occur after the infection and they trigger intracellular signalling cascades, involving, for instance, Ca as second messenger95. Besides being involved in transient intracellular oscillation activating signalling cascades, Ca can also play a structural role in cells, stabilizing and strengthening the cell wall56; this would explain the increased Ca concentration in the P. viticola-induced lesion. In addition, P also showed an accumulation at the infection lesions; although P plays a paramount role in plants life cycle, its involvement in the resistance mechanisms against pathogens is still controversial, due to contrasting observations86. Nonetheless, it has been observed that the foliar application of orthophosphoric acid, or its salts, can inhibit the diseases caused by Oomycota belonging to the Phytophthora and Pythium species96.
In conclusion, by investigating the compatible and the incompatible interaction between V. vinifera plant and P. viticola, we demonstrated that in susceptible cultivars, the inoculation with P. viticola caused a dramatic redistribution of several micro- and macronutrients (e.g. Fe, Mn, Zn, S, K, Ca and P), in terms of concentration and localization at leaf level. This evidence might therefore suggest a role of Fe, Mn, Zn, S, K, Ca and P in the response to the pathogen, which could be exerted according with the specific functions of single elements in the plant cell during the compatible interaction. Such differential accumulation and allocation of elements was associated to the modulation of specific transporters by P. viticola inoculation (i.e. VvYSL1a, VvYSL1b, VvYSL3, VvOPT3, VvVIT1, VvNRAMP1, VvNRAMP3, VvNRAMP4 and VvSULTR3.5). On the contrary, the resistant cultivars did not display a substantial alteration in the element distribution in the leaves infected with P. viticola. However, Mn and Fe showed an increased concentration in inoculated leaves that is also mirrored by the transcriptional regulation of putative transporters, suggesting that Cabernet Volos and Sauvignon Kretos included some pathogen defence mechanisms based on the increase of Mn ad Fe concentration. These two elements are involved in the synthesis of secondary metabolites (e.g. phenolic compounds)56,87 and ROS70,82,83, which play a key role in the hypersensitive response observed in incompatible interactions. Our results clearly indicate the link between the mineral nutrition (in terms of nutrient contents and localisation) and plants’ response to pathogens. This evidence suggests that an appropriate management of the mineral element availability for grapevine plants at field scale could play a pivotal role in guaranteeing the expression of the response mechanisms against pathogens. In fact, a mineral element shortage, even though latent or induced by toxicity/excess of other mineral elements, as already demonstrated for Cu14, can seriously limit the ability of either susceptible or resistant grapevine plants to cope with P. viticola infections.
Materials and methods
Plant material, growing conditions and Plasmopara viticola inoculation
Tissues were sampled from one-year old grapevine (V. vinifera) cuttings grafted to SO4 rootstock plants, belonging to four different cultivars, two resistant (Sauvignon Kretos and Cabernet Volos) and two susceptible (Sauvignon Blanc and Cabernet Sauvignon). The resistant cultivars were obtained through two crosses between V. vinifera cv Sauvignon Blanc or Cabernet Sauvignon with the Vitis ‘breeding line 20/3’, obtained by Pal Kozma in Ungary and kindly offered to the University of Udine. The crosses were performed at the University of Udine (Udine, Italy). The Vitis 20/3 has been obtained from a crossing between Vitis ‘Bianca’ x ‘SK77-4/5’ (bred by crossing ‘Kumbarát’, originated from hybridisation of V. amurensis x V. vinifera, and V. vinifera ‘Traminer’)97–100. The two susceptible cultivars (Sauvignon Blanc and Cabernet Sauvignon) were used as control. All the plants were obtained from Vivai Cooperativi Rauscedo (Rauscedo, Italy).
Plants were grown in 4.2 L pots (12 × 12 × 30 cm) where at the bottom was placed a layer of expanded clay. Before the planting, the peat soil of each pot was supplemented with the urea fertilizer (to a concentration of 20 kg N Ha−1) and manually mixed. The roots of each grapevine were cut to approximately 10 cm length. The pots were manually irrigated, every 2–3 days, in order to maintain constant soil humidity during the whole experiment. Plants were grown in an experimental greenhouse under controlled relative humidity (RH) and temperature conditions (65% ± 5% RH, 24 °C ± 1 °C).
Grapevine plants were inoculated by spraying the abaxial side of the leaves with a solution containing 5 × 105 spores of P. viticola as previously described101. Inoculated and non-inoculated, were kept at 100% RH overnight to allow pathogen infection and then incubated under greenhouse conditions. Six days after the inoculation the RH of the chambers was again increased up to 100%, in order to allow the pathogen sporulation. The day after, leaves were sampled, immediately frozen in liquid N2 and stored at -80 °C until further processing. For qPCR and mineral analysis, each sample (biological replicate) comprised three leaves taken from the same plant and only leaves of the 4th–5th node from the top of the shoot were collected to avoid ontogenic resistance effects101. Three independent biological replicates were analysed; each biological replicate was analysed three times, as for technical replicates.
Ionomic analysis
The determination of leaves ionomic signature was carried out as previously described102. Briefly, samples of leaves tissue were digested with concentrated HNO3 [65% (v/v), Carlo Erba] using a single reaction chamber (SRC, UltraWAVE, Milestone Inc, Shelton, USA). The mineral elements concentration was subsequently determined by Inductively Coupled Plasma-Optical Emission Spectroscopy (ICP-OES; Arcos Ametek, Spectro, Germany), using tomato leaves (SRM 1573a) and spinach leaves (SRM 1547) as external certified reference material.
Micro-focused X-ray fluorescence (μ-XRF) imaging
Micro X-ray fluorescence maps were collected with a laboratory benchtop μ-XRF spectrometer (M4 Tornado, Bruker Nano GmbH, Berlin, Germany) at the “Micro X-ray Lab” of the University of Bari, Italy, following the procedure previously reported103. This instrument is equipped with a micro-focus Rh X-ray source (50 kV, 600 μA), a polycapillary X-ray optics with a spotsize of 25 μm and two XFlash energy dispersive silicon drift detectors with 30 mm2 sensitive area and an energy resolution of 140 eV @ Mn Kα. The two detectors, placed at opposite sites compared to the X-ray optics, allow reducing shadowing effects in the mineral element distribution maps and obtaining a better signal-to-noise ratio (S/N). Immediately after sampling, grapevine leaves were inserted between two circles of filter paper and kept tightly pressed within a Petri dish to preserve the sample flat during the freeze-drying process; the Petri dishes containing the flat leaves were quickly frozen in liquid nitrogen and then freeze-dried under vacuum. All the µ-XRF analyses were performed under reduced pressure (20 mbar) by using 25 μm stepsize and an acquisition time of 10 ms per step. In order to increase the S/N, each scan was repeated 30 times. For each of the leaves, a rectangular area of 220–240 pixels (ca. 5.5–6 mm) height and 400–440 pixels (ca. 10–11 mm) width (depending on leaf dimension), was selected approximately near the petiole sinus (including the midrib and nearby primary vein) of the leaf for the analysis. Smaller areas (approximately 1 mm height and 1.5 mm width) containing infection lesions were also analysed keeping the acquisition conditions unchanged, except for the use of a smaller step size (10 µm) to improve map details through oversampling. µ-XRF distribution maps were obtained with the instrument ESPRIT software (Bruker Nano GmbH, Berlin, Germany) version 1.3.0.3273. All the mineral element distribution maps were collected using the same analytical conditions. The same intensity scale was adopted to visualise the distributions of the same mineral element in all the maps. Therefore, the mineral element distribution maps of the same mineral element can be directly compared. Brighter colours in the map correspond to a higher concentration of the mineral element. Several areas on many different leaves were investigated and the reported data can be considered representative of the main and more relevant features observed in all the studied samples.
Bioinformatics
The identification of transporter gene sequences, namely VIT1, OPT3, YSL1, SULTR3, involved in the translocation and allocation of selected mineral elements was carried out in the 12X release of Vitis vinifera genome in the Grape Genome Browser (https://www.cns.fr/externe/GenomeBrowser/Vitis/). The isolation of putative gene sequences from the grapevine genome has been primarily based on amino acid sequence similarity between already characterised transporters (Supplementary Table 1), retrieved from public databases like https://www.ncbi.nlm.nih.gov/, https://www.uniprot.org/uniprot/. The predicted transporter sequences were identified in grapevine genome through a BLASTP104 search. BLASTP analysis was performed using each known protein, selecting the putative proteins encoded by the predicted coding sequences on the basis of the highest sequence homology value (the threshold value for sequence homology was set at 80%); afterwards a phylogenetic analysis was performed. The amino acid sequences were aligned by the ClustalW ver. 2.1 algorithm (https://clustalw.ddbj.nig.ac.jp/). Phylogenetic tree was built using the Phylogenetic Interference Package program (PHYLIP; University of Washington, USA, https://evolution.genetics.washington.edu/phylip.html) and visualised by the Figtree software (https://tree.bio.ed.ac.uk/software/figtree/).
Sulphate transporters
On the basis of the predicted functions, the bioinformatic analysis was aimed at identifying specifically members of the group 3, by using orthologous sequences from A. thaliana and Oryza sativa (Supplementary Table 1). The BLASTP algorithm identified eight putative transporters in the genome of V. vinifera; one sequence (GSVIVT01022159001) resulted more similar to group 2 transporters (Supplementary Figure 2), whereas seven sequences (GSVIVT01001198001, GSVIVT01015413001, GSVIVT01018027001, GSVIVT01015414001, GSVIVT01018028001, GSVIVT01011744001, GSVIVT01015412001) clustered with other transporter sequences belonging to the group 3. Among the putative group 3 transporters, five sequences (GSVIVT01018027001, GSVIVT01018028001, GSVIVT01011744001, GSVIVT01015412001 and GSVIVT01015413001, hereafter referred to as VvSultr3.1, VvSultr3.2, VvSultr3.3, VvSultr3.4, VvSultr3.5) were kept for the gene expression analyses, whereas GSVIVT01015414001and GSVIVT01001198001 were discarded since a duplication and a truncated form of VvSultr3.5 and VvSultr3.1, respectively (Supplementary Figure 2).
Iron transporters
Members of two distinct clades of the Oligopeptide Transporter family, the Yellow Stripe-like (YSL) proteins and the Oligopetide Transporters (OPTs), have been shown to be involved in Fe partitioning in plants62–64,105. Known YSL protein sequences from different plants, as for instance A. thaliana, Oryza sativa, Hordeum vulgare, Zea mays and Arachis hypogea (Supplementary Table 1), were used to retrieve six sequences (i.e. GSVIVT01019645001, GSVIVT01019646001, GSVIVT01038611001, GSVIVT01029329001, GSVIVT01029331001, GSVIVT01012033001) in the V. vinifera genome. According to the phylogenetic analysis (Supplementary Figure 3), the six sequences isolated in the grapevine genome clustered in two different branches of the phylogenetic tree; GSVIVT01029329001, GSVIVT01029331001, GSVIVT01012033001 clustered with AtYSL5, AtYSL7 and AtYSL8, whereas GSVIVT01019645001 (hereafter referred to as VvYSL1a), GSVIVT01019646001 (hereafter referred to as VvYSL1b), GSVIVT01038611001 (hereafter referred to as VvYSL3) showed a closer relationship with the A. thaliana sequences (AtYSL1, AtYSL2 and AtYSL3), known to be involved in the Fe partitioning in plants65–67.
The already characterised OPT3 sequences from A. thaliana and Populus trichocarpa (Supplementary Table 1) allowed the identification of five putative OPT sequences in grapevine genome, GSVIVT01014721001, GSVIVT01014724001, GSVIVT01009222001, GSVIVT01005133001, GSVIVT01038599001, already annotated as VvOPT1, VvOPT1a, VvOPT2, VvOPT3 and VvOPT7 respectively. The phylogenetic analysis confirmed that VvOPT3 had the highest homology with the OPT3 sequences from A. thaliana and P. trichocarpa (Supplementary Figure 4), thus suggesting a similar function in plants.
Two putative V. vinifera sequences (i.e. GSVIVT01011629001 and GSVIVT01011628001) showed a high homology with AtVIT1, OsVIT1, OsVIT2 and PtVIT1 from poplar (Supplementary Table 1). According to the phylogenetic analysis (Supplementary Figure 5), GSVIVT01011629001, hereafter referred to as VvVIT1, was the sequence most closely related to AtVIT1; on the bases of this similarity, VvVIT1 was selected for further molecular analyses.
RNA extraction and cDNA synthesis
RNA samples were extracted from leaf tissues sampled as described above. Total RNA was prepared using Spectrum Plant Total RNA Kit (Sigma-Aldrich Co. LLC) according to the users’ guide, as previously described106. Afterwards, 1 μg of total RNA was subjected to DNAse digestion with 10 U of DNAse RQ1 and cDNA was synthesised using the ImProm-II Reverse Transcription System (Promega, Madison, WI, USA). The quality of total RNA and cDNA was checked through a PCR using couples of primers specific for housekeeping genes.
Real-time reverse transcription–PCR
Specific primers were designed for the target genes as well as for the housekeeping genes (Supplemental Table 2). Real-time reverse transcription–PCR (RT-PCR) experiments were carried out in biological triplicates and the reaction was performed by using the SsoFast EvaGreen Supermix (Bio-Rad, Segrate, Italy) as previously described106. Nevertheless, the identity of each amplicon was confirmed by sequencing. The amplification efficiency was calculated from raw data using LinRegPCR software107. The expression data were normalized to the tubulin and EF1α housekeeping genes, whereas the relative expression ratios were calculated using non-inoculated susceptible cultivar as a calibrator sample according to the Pfaffl equation108. Standard error values were calculated according to Pfaffl et al. (2002).
Statistical analyses
The results are reported as mean ± standard error (SE). The significance of differences among means was calculated by One-way ANOVA with post-hoc Tukey HSD with α = 0.05 using R software (version 3.6.0). The following R packages were used for data visualization and for statistical analyses: ggplot2 v.3.2.0110, Agricolae v.1.3-1111 and ggfortify112.
Supplementary information
Acknowledgements
The research was supported by grants from the Free University of Bolzano (TN2071 and TN2081) and by the “Micro X-ray Lab” of the University of Bari. Ignazio Allegretta was supported by a research grant on the project PON R&I "Studio del sistema suolo-pianta mediante tecniche analitiche innovative che impiegano raggi X"—Progetto AIM1809249—attività 1, linea 1.
Author contributions
Experimental Design: GC, IP, TM, SC, YP. Experiments execution: OG, MP, SN, SR, FV, YP. Data collection: IA, SN, SR, CP, RT, AT, FV, YP. Data analyses and visualization: IA, CP, RT, YP. Data interpretation: SC, GC, TM, MP, IP, RT, YP. Manuscript writing and critical revision: SC, GC, OG, MP, TM, IP, RT, YP. Financial support: TM, RT, YP.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-75990-x.
References
- 1.Vivier MA, Pretorius IS. Genetically tailored grapevines for the wine industry. Trends Biotechnol. 2002;20:472–478. doi: 10.1016/S0167-7799(02)02058-9. [DOI] [PubMed] [Google Scholar]
- 2.Armijo G, et al. Grapevine pathogenic microorganisms: Understanding infection strategies and host response scenarios. Front. Plant Sci. 2016;7:382. doi: 10.3389/fpls.2016.00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gessler C, Pertot I, Perazzolli M. Plasmopara viticola: a review of knowledge on downy mildew of grapevine and effective disease management. Phytopathol. Mediterr. 2011;50:3–44. [Google Scholar]
- 4.Yin L, et al. Genome sequence of Plasmopara viticola and insight into the pathogenic mechanism. Sci. Rep. 2017;7:1–12. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong FP, Burr HN, Wilcox WF. Heterothallism in Plasmopara viticola. Plant Pathol. 2001;50:427–432. doi: 10.1046/j.1365-3059.2001.00573.x. [DOI] [Google Scholar]
- 6.Fontaine S, Remuson F, Caddoux L, Barrès B. Investigation of the sensitivity of Plasmopara viticola to amisulbrom and ametoctradin in French vineyards using bioassays and molecular tools. Pest Manag. Sci. 2019;75:2115–2123. doi: 10.1002/ps.5461. [DOI] [PubMed] [Google Scholar]
- 7.Brunetto G, et al. Copper accumulation in vineyard soils: Rhizosphere processes and agronomic practices to limit its toxicity. Chemosphere. 2016;162:293–307. doi: 10.1016/j.chemosphere.2016.07.104. [DOI] [PubMed] [Google Scholar]
- 8.Komárek M, Čadková E, Chrastný V, Bordas F, Bollinger JC. Contamination of vineyard soils with fungicides: A review of environmental and toxicological aspects. Environ. Int. 2010;36:138–151. doi: 10.1016/j.envint.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Wightwick AM, Mollah MR, Partington DL, Allinson G. Copper fungicide residues in Australian vineyard soils. J. Agric. Food Chem. 2008;56:2457–2464. doi: 10.1021/jf0727950. [DOI] [PubMed] [Google Scholar]
- 10.Kandeler E, Kampichler C, Horak O. Influence of heavy metals on the functional diversity of soil microbial communities. Biol. Fertil. Soils. 1996;23:299–306. doi: 10.1007/BF00335958. [DOI] [Google Scholar]
- 11.Merrington G, Rogers SL, Van Zwieten L. The potential impact of long-term copper fungicide usage on soil microbial biomass and microbial activity in an avocado orchard. Aust. J. Soil Res. 2002;40:749–759. doi: 10.1071/SR01084. [DOI] [Google Scholar]
- 12.Keiblinger KM, et al. Assessment of Cu applications in two contrasting soils—Effects on soil microbial activity and the fungal community structure. Ecotoxicology. 2018;27:217–233. doi: 10.1007/s10646-017-1888-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marastoni L, et al. Synergism and antagonisms between nutrients induced by copper toxicity in grapevine rootstocks: Monocropping vs. intercropping. Chemosphere. 2019;214:563–578. doi: 10.1016/j.chemosphere.2018.09.127. [DOI] [PubMed] [Google Scholar]
- 14.Marastoni L, et al. Morphological root responses and molecular regulation of cation transporters are differently affected by copper toxicity and cropping system depending on the grapevine rootstock genotype. Front. Plant Sci. 2019;10:946. doi: 10.3389/fpls.2019.00946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwalbert R, et al. Physiological responses of soybean (Glycine max (L.) Merrill) cultivars to copper excess. An. Acad. Bras. Cienc. 2019;91:e20190121. doi: 10.1590/0001-3795201920190121. [DOI] [PubMed] [Google Scholar]
- 16.De Conti L, et al. Photosynthesis and growth of young grapevines intercropped with native grasses in soils contaminated with copper. Acta Hortic. 2018;1217:179–184. doi: 10.17660/ActaHortic.2018.1217.23. [DOI] [Google Scholar]
- 17.Ambrosini VG, et al. High copper content in vineyard soils promotes modifications in photosynthetic parameters and morphological changes in the root system of ‘Red Niagara’ plantlets. Plant Physiol. Biochem. 2018;128:89–98. doi: 10.1016/j.plaphy.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 18.Hammerschmitt RK, et al. Copper and zinc distribution and toxicity in ‘Jade’/‘Genovesa’ young peach tree. Sci. Hortic. (Amsterdam) 2020;259:108763. doi: 10.1016/j.scienta.2019.108763. [DOI] [Google Scholar]
- 19.Mestre P, Piron MC, Merdinoglu D. Identification of effector genes from the phytopathogenic Oomycete Plasmopara viticola through the analysis of gene expression in germinated zoospores. Fungal Biol. 2012;116:825–835. doi: 10.1016/j.funbio.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 20.Luis P, Gauthier A, Trouvelot S, Poinssot B, Frettinger P. Identification of Plasmopara viticola genes potentially involved in pathogenesis on grapevine suggests new similarities between oomycetes and true fungi. Phytopathology. 2013;103:1035–1044. doi: 10.1094/PHYTO-06-12-0121-R. [DOI] [PubMed] [Google Scholar]
- 21.Nascimento R, et al. Early stage metabolic events associated with the establishment of Vitis vinifera–Plasmopara viticola compatible interaction. Plant Physiol. Biochem. 2019;137:1–13. doi: 10.1016/j.plaphy.2019.01.026. [DOI] [PubMed] [Google Scholar]
- 22.Chitarrini G, et al. Identification of biomarkers for defense response to Plasmopara viticola in a resistant grape variety. Front. Plant Sci. 2017;8:1524. doi: 10.3389/fpls.2017.01524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milli A, et al. Proteomic analysis of the compatible interaction between Vitis vinifera and Plasmopara viticola. J. Proteomics. 2012;75:1284–1302. doi: 10.1016/j.jprot.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 24.Nascimento-Gavioli MCA, et al. Proteome of Plasmopara viticola-infected Vitis vinifera provides insights into grapevine Rpv1/Rpv3 pyramided resistance to downy mildew. J. Proteomics. 2017;151:264–274. doi: 10.1016/j.jprot.2016.05.024. [DOI] [PubMed] [Google Scholar]
- 25.Polesani M, et al. cDNA-AFLP analysis of plant and pathogen genes expressed in grapevine infected with Plasmopara viticola. BMC Genomics. 2008;9:1–14. doi: 10.1186/1471-2164-9-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polesani M, et al. General and species-specific transcriptional responses to downy mildew infection in a susceptible (Vitis vinifera) and a resistant (V. riparia) grapevine species. BMC Genomics. 2010;11:117. doi: 10.1186/1471-2164-11-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lenzi L, Caruso C, Bianchedi PL, Pertot I, Perazzolli M. Laser microdissection of grapevine leaves reveals site-specific regulation of transcriptional response to Plasmopara viticola. Plant Cell Physiol. 2016;57:69–81. doi: 10.1093/pcp/pcv166. [DOI] [PubMed] [Google Scholar]
- 28.Malacarne G, et al. Resistance to Plasmopara viticola in a grapevine segregating population is associated with stilbenoid accumulation and with specific host transcriptional responses. BMC Plant Biol. 2011;11:1–13. doi: 10.1186/1471-2229-11-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peressotti E, et al. Breakdown of resistance to grapevine downy mildew upon limited deployment of a resistant variety. BMC Plant Biol. 2010;10:147. doi: 10.1186/1471-2229-10-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foria S, et al. Gene duplication and transposition of mobile elements drive evolution of the Rpv3 resistance locus in grapevine. Plant J. 2020;101:529–542. doi: 10.1111/tpj.14551. [DOI] [PubMed] [Google Scholar]
- 31.Blasi P, et al. Construction of a reference linkage map of Vitis amurensis and genetic mapping of Rpv8, a locus conferring resistance to grapevine downy mildew. Theor. Appl. Genet. 2011;123:43–53. doi: 10.1007/s00122-011-1565-0. [DOI] [PubMed] [Google Scholar]
- 32.Schwander F, et al. Rpv10: A new locus from the Asian Vitis gene pool for pyramiding downy mildew resistance loci in grapevine. Theor. Appl. Genet. 2012;124:163–176. doi: 10.1007/s00122-011-1695-4. [DOI] [PubMed] [Google Scholar]
- 33.Venuti S, et al. Historical introgression of the downy mildew resistance gene Rpv12 from the Asian species Vitis amurensis into grapevine varieties. PLoS ONE. 2013;8:e61228. doi: 10.1371/journal.pone.0061228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fuller KB, Alston JM, Sambucci OS. The value of powdery mildew resistance in grapes: Evidence from California. Wine Econ. Policy. 2014;3:90–107. doi: 10.1016/j.wep.2014.09.001. [DOI] [Google Scholar]
- 35.Pedneault K, Provost C. Fungus resistant grape varieties as a suitable alternative for organic wine production: Benefits, limits, and challenges. Sci. Hortic. 2016;208:57–77. doi: 10.1016/j.scienta.2016.03.016. [DOI] [Google Scholar]
- 36.Di Gaspero G, Foria S. Molecular grapevine breeding techniques. In: Reynolds A, editor. Grapevine Breeding Programs for the Wine Industry. Amsterdam: Elsevier; 2015. pp. 23–37. [Google Scholar]
- 37.Emanuelli F, Battilana J, Costantini L, Grando MS. Molecular breeding of grapevine for aromatic quality and other traits relevant to viticulture. In: Jenks MA, Bebeli PJ, editors. Breeding for Fruit Quality. New York: Wiley; 2011. pp. 247–260. [Google Scholar]
- 38.Zini E, et al. Applying a defined set of molecular markers to improve selection of resistant grapevine accessions. Acta Hortic. 2015;1082:73–78. doi: 10.17660/ActaHortic.2015.1082.9. [DOI] [Google Scholar]
- 39.Bove F, Bavaresco L, Caffi T, Rossi V. Assessment of resistance components for improved phenotyping of grapevine varieties resistant to downy mildew. Front. Plant Sci. 2019;10:1559. doi: 10.3389/fpls.2019.01559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.COMMISSION IMPLEMENTING REGULATION (EU) 2018/1981. (European Commission, 2018).
- 41.Dai GH, Andary C, Mondolot-Cosson L, Boubals D. Involvement of phenolic compounds in the resistance of grapevine callus to downy mildew (Plasmopara viticola) Eur. J. Plant Pathol. 1995;101:541–547. doi: 10.1007/BF01874479. [DOI] [Google Scholar]
- 42.Kortekamp A, Wind R, Zyprian E. The role of callose deposits during infection of two downy mildew-tolerant and two -susceptible Vitis cuitivars. Vitis. 1997;36:103–104. [Google Scholar]
- 43.Gindro K, Pezet R, Viret O. Histological study of the responses of two Vitis vinifera cultivars (resistant and susceptible) to Plasmopara viticola infections. Plant Physiol. Biochem. 2003;41:846–853. doi: 10.1016/S0981-9428(03)00124-4. [DOI] [Google Scholar]
- 44.Bellin D, et al. Resistance to Plasmopara viticola in grapevine ‘Bianca’ is controlled by a major dominant gene causing localised necrosis at the infection site. Theor. Appl. Genet. 2009;120:163–176. doi: 10.1007/s00122-009-1167-2. [DOI] [PubMed] [Google Scholar]
- 45.Pezet R, Gindro K, Viret O, Richter H. Effects of resveratrol, viniferins and pterostilbene on Plasmopara viticola zoospore mobility and disease development. Vitis - J. Grapevine Res. 2004;43:145–148. [Google Scholar]
- 46.Gindro K, Spring JL, Pezet R, Richter H, Viret O. Histological and biochemical criteria for objective and early selection of grapevine cultivars resistant to Plasmopara viticola. Vitis - J. Grapevine Res. 2006;45:191–196. [Google Scholar]
- 47.Langcake P. Disease resistance of Vitis spp. and the production of the stress metabolites resveratrol, ε-viniferin, α-viniferin and pterostilbene. Physiol. Plant Pathol. 1981;18:213–226. doi: 10.1016/S0048-4059(81)80043-4. [DOI] [Google Scholar]
- 48.Alonso-Villaverde V, Voinesco F, Viret O, Spring JL, Gindro K. The effectiveness of stilbenes in resistant Vitaceae: ultrastructural and biochemical events during Plasmopara viticola infection process. Plant Physiol. Biochem. 2011;49:265–274. doi: 10.1016/j.plaphy.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 49.Boso S, Kassemeyer HH. Different susceptibility of European grapevine cultivars for downy mildew. Vitis - J. Grapevine Res. 2008;47:39–49. [Google Scholar]
- 50.Kortekamp A, Wind R, Zyprian E. Investigation of the interaction of Plasmopara viticola with susceptible and resistant grapevine cultivars/Untersuchungen zur Interaktion von Plasmopara viticola mit anfälligen und resistenten Rebsorten. Zeitschrift für Pflanzenkrankheiten und Pflanzenschutz/J. Plant Dis. Prot. 1998;105:475–488. [Google Scholar]
- 51.Toffolatti SL, Venturini G, Maffi D, Vercesi A. Phenotypic and histochemical traits of the interaction between Plasmopara viticola and resistant or susceptible grapevine varieties. BMC Plant Biol. 2012;12:124. doi: 10.1186/1471-2229-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nicolas O, et al. The ionomics of lettuce infected by Xanthomonas campestris pv. vitians. Front. Plant Sci. 2019;10:351. doi: 10.3389/fpls.2019.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.D’attoma G, et al. Ionomic differences between susceptible and resistant olive cultivars infected by Xylella fastidiosa in the outbreak area of Salento, Italy. Pathogens. 2019;8:272. doi: 10.3390/pathogens8040272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marschner, P. Marschner’s mineral nutrition of higher plants (2012).
- 55.Poschenrieder C, Tolrà R, Barceló J. Can metals defend plants against biotic stress? Trends Plant Sci. 2006;11:288–295. doi: 10.1016/j.tplants.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 56.Elmer, W. H. & Datnoff, L. E. Mineral nutrition and suppression of plant disease. in Encyclopedia of Agriculture and Food Systems 231–244 (Elsevier, 2014). 10.1016/B978-0-444-52512-3.00251-5
- 57.Fones H, Preston GM. The impact of transition metals on bacterial plant disease. FEMS Microbiol. Rev. 2013;37:495–519. doi: 10.1111/1574-6976.12004. [DOI] [PubMed] [Google Scholar]
- 58.Zanin L, et al. Iron allocation in leaves of Fe-deficient cucumber plants fed with natural Fe complexes. Physiol. Plant. 2015;154:82–94. doi: 10.1111/ppl.12296. [DOI] [PubMed] [Google Scholar]
- 59.Pittman JK. Managing the manganese: Molecular mechanisms of manganese transport and homeostasis. New Phytol. 2005;167:733–742. doi: 10.1111/j.1469-8137.2005.01453.x. [DOI] [PubMed] [Google Scholar]
- 60.Casagrande K, Falginella L, Castellarin SD, Testolin R, Di Gaspero G. Defence responses in Rpv3-dependent resistance to grapevine downy mildew. Planta. 2011;234:1097–1109. doi: 10.1007/s00425-011-1461-5. [DOI] [PubMed] [Google Scholar]
- 61.Di Gaspero G, et al. Selective sweep at the Rpv3 locus during grapevine breeding for downy mildew resistance. Theor. Appl. Genet. 2012;124:277–286. doi: 10.1007/s00122-011-1703-8. [DOI] [PubMed] [Google Scholar]
- 62.Zhai Z, et al. OPT3 is a phloem-specific iron transporter that is essential for systemic iron signaling and redistribution of iron and cadmium in Arabidopsis. Plant Cell. 2014;26:2249–2264. doi: 10.1105/tpc.114.123737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stacey MG, Koh S, Becker J, Stacey G. AtOPT3, a member of the oligopeptide transporter family, is essential for embryo development in Arabidopsis. Plant Cell. 2002;14:2799–2811. doi: 10.1105/tpc.005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stacey MG, et al. The arabidopsis AtOPT3 protein functions in metal homeostasis and movement of iron to developing seeds. Plant Physiol. 2008;146:589–601. doi: 10.1104/pp.107.108183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chu HH, et al. Successful reproduction requires the function of Arabidopsis YELLOW STRIPE-LIKE1 and YELLOW STRIPE-LIKE3 metal-nicotianamine transporters in both vegetative and reproductive structures. Plant Physiol. 2010;154:197–210. doi: 10.1104/pp.110.159103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ishimaru Y, et al. Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J. 2010;62:379–390. doi: 10.1111/j.1365-313X.2010.04158.x. [DOI] [PubMed] [Google Scholar]
- 67.Waters BM, et al. Mutations in Arabidopsis Yellow Stripe-Like1 and Yellow Stripe-Like3 reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiol. 2006;141:1446–1458. doi: 10.1104/pp.106.082586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim SA, et al. Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science (80-.) 2006;314:1295–1298. doi: 10.1126/science.1132563. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Y, Xu Y-H, Yi H-Y, Gong J-M. Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J. 2012;72:400–410. doi: 10.1111/j.1365-313X.2012.05088.x. [DOI] [PubMed] [Google Scholar]
- 70.Aznar A, Chen NWG, Thomine S, Dellagi A. Immunity to plant pathogens and iron homeostasis. Plant Sci. 2015;240:90–97. doi: 10.1016/j.plantsci.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 71.Chen CC, Chien WF, Lin NC, Yeh KC. Alternative functions of Arabidopsis YELLOW STRIPELIKE3: From metal translocation to pathogen defense. PLoS ONE. 2014;9:e98008. doi: 10.1371/journal.pone.0098008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kobayashi T, Nozoye T, Nishizawa NK. Iron transport and its regulation in plants. Free Radical Biol. Med. 2019;133:11–20. doi: 10.1016/j.freeradbiomed.2018.10.439. [DOI] [PubMed] [Google Scholar]
- 73.Thomine S, Wang R, Ward JM, Crawford NM, Schroeder JI. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc. Natl. Acad. Sci. 2000;97:4991–4996. doi: 10.1073/pnas.97.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Curie C, Alonso JM, Le Jean M, Ecker JR, Briat JF. Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochem. J. 2000;347:749–755. doi: 10.1042/bj3470749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Segond D, et al. NRAMP genes function in Arabidopsis thaliana resistance to Erwinia chrysanthemi infection. Plant J. 2009;58:195–207. doi: 10.1111/j.1365-313X.2008.03775.x. [DOI] [PubMed] [Google Scholar]
- 76.Pii Y, et al. Microbial interactions in the rhizosphere: Beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. A review. Biol. Fertil. Soils. 2015;51:403–415. doi: 10.1007/s00374-015-0996-1. [DOI] [Google Scholar]
- 77.Crecchio C, et al. Beneficial soil microbiome for sustainable agriculture production. In: Lichtfouse E, et al., editors. Sustainable Agriculture Reviews. Berlin: Springer; 2018. pp. 443–481. [Google Scholar]
- 78.Dellagi A, et al. Siderophore-mediated upregulation of Arabidopsis ferritin expression in response to Erwinia chrysanthemi infection. Plant J. 2005;43:262–272. doi: 10.1111/j.1365-313X.2005.02451.x. [DOI] [PubMed] [Google Scholar]
- 79.Liu G, et al. Targeted alterations in iron homeostasis underlie plant defense responses. J. Cell Sci. 2007;120:596–605. doi: 10.1242/jcs.001362. [DOI] [PubMed] [Google Scholar]
- 80.García Mata C, Lamattina L, Cassia RO. Involvement of iron and ferritin in the potato-Phytophthora infestans interaction. Eur. J. Plant Pathol. 2001;107:557–562. doi: 10.1023/A:1011228317709. [DOI] [Google Scholar]
- 81.Calla B, Blahut-Beatty L, Koziol L, Simmonds DH, Clough SJ. Transcriptome analyses suggest a disturbance of iron homeostasis in soybean leaves during white mould disease establishment. Mol. Plant Pathol. 2014;15:576–588. doi: 10.1111/mpp.12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Torres MA. ROS in biotic interactions. Physiol. Plant. 2010;138:414–429. doi: 10.1111/j.1399-3054.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- 83.Pierre JL, Fontecave M. Iron and activated oxygen species in biology: The basic chemistry. Biometals. 1999;12:195–199. doi: 10.1023/A:1009252919854. [DOI] [PubMed] [Google Scholar]
- 84.Nevo Y, Nelson N. The NRAMP family of metal-ion transporters. Biochim Biophys Acta Mol Cell Res. 2006;1763:609–620. doi: 10.1016/j.bbamcr.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 85.Gao H, et al. NRAMP2, a trans-Golgi network-localized transporter, is required for Arabidopsis root growth under manganese deficiency. New Phytol. 2018;217:179–193. doi: 10.1111/nph.14783. [DOI] [PubMed] [Google Scholar]
- 86.Dordas C. Role of nutrients in controlling plant diseases in sustainable agriculture. A review. Agron. Sustain. Dev. 2008;28:33–46. doi: 10.1051/agro:2007051. [DOI] [Google Scholar]
- 87.Burnell JN. The biochemistry of manganese in plants. In: Graham RD, Hannam RJ, Uren NC, editors. Manganese in Soils and Plants. Dordrecht: Springer; 1988. pp. 125–137. [Google Scholar]
- 88.Eskandari S, Sharifnabi B. The modifications of cell wall composition and water status of cucumber leaves induced by powdery mildew and manganese nutrition. Plant Physiol. Biochem. 2019;145:132–141. doi: 10.1016/j.plaphy.2019.10.037. [DOI] [PubMed] [Google Scholar]
- 89.Cabot C, et al. A role for Zinc in plant defense against pathogens and herbivores. Front. Plant Sci. 2019;10:1171. doi: 10.3389/fpls.2019.01171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Petre B, et al. RNA-Seq of early-infected poplar leaves by the rust pathogen Melampsora larici-populina uncovers PtSultr3;5, a fungal-induced host sulfate transporter. PLoS ONE. 2012;7:e44408. doi: 10.1371/journal.pone.0044408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ali Abdalla M, Mühling KH. Plant-derived sulfur containing natural products produced as a response to biotic and abiotic stresses: A review of their structural diversity and medicinal importance. J. Appl. Bot. Food Qual. 2019;92:204–215. [Google Scholar]
- 92.Kopriva S, Malagoli M, Takahashi H. Sulfur nutrition: impacts on plant development, metabolism, and stress responses. J. Exp. Bot. 2019;70:4069–4073. doi: 10.1093/jxb/erz319. [DOI] [PubMed] [Google Scholar]
- 93.Amtmann A, Troufflard S, Armengaud P. The effect of potassium nutrition on pest and disease resistance in plants. Physiol. Plant. 2008;133:682–691. doi: 10.1111/j.1399-3054.2008.01075.x. [DOI] [PubMed] [Google Scholar]
- 94.Rossard S, Luini E, Pérault JM, Bonmort J, Roblin G. Early changes in membrane permeability, production of oxidative burst and modification of PAL activity induced by ergosterol in cotyledons of Mimosa pudica. J. Exp. Bot. 2006;57:1245–1252. doi: 10.1093/jxb/erj090. [DOI] [PubMed] [Google Scholar]
- 95.Yang Y, Shah J, Klessig DF. Signal perception and transduction in plant defense responses. Genes Dev. 1997;11:1621–1639. doi: 10.1101/gad.11.13.1621. [DOI] [PubMed] [Google Scholar]
- 96.Brunings, A. M., Datnoff, L. E. & Simonne, E. H. Phosphorous acid and phosphoric acid : When all P sources are not equal. University of Florida IFAS Extension 8 (2005).
- 97.Koleda I. Ergebnisse von Kreuzungen zwischen Vitis amurensis und Vitis vinifera in der Züchtung frostwiderstandsfähiger Reben. Vitis. 1975;14:1–5. [Google Scholar]
- 98.Kozma P., Jr Winegrape breeding for fungus disease resistance. Acta Hort. 2000;528:505–510. [Google Scholar]
- 99.Cindric P, Korac N, Kovac V. Grape breeding in the vojvodina province. Acta Hort. 2000;528:499–504. [Google Scholar]
- 100.Di Gaspero G, Cipriani G, Adam-Blondon AF, Testolin R. Linkage maps of grapevine displaying the chromosomal locations of 420 microsatellite markers and 82 markers for R-gene candidates. Theor. Appl. Genet. 2007;114:1249–1263. doi: 10.1007/s00122-007-0516-2. [DOI] [PubMed] [Google Scholar]
- 101.Perazzolli M, et al. Downy mildew resistance induced by Trichoderma harzianum T39 in susceptible grapevines partially mimics transcriptional changes of resistant genotypes. BMC Genomics. 2012;13:660. doi: 10.1186/1471-2164-13-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pii Y, Cesco S, Mimmo T. Shoot ionome to predict the synergism and antagonism between nutrients as affected by substrate and physiological status. Plant Physiol. Biochem. 2015;94:48–56. doi: 10.1016/j.plaphy.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 103.Valentinuzzi F, et al. Root-shoot-root Fe translocation in cucumber plants grown in a heterogeneous Fe provision. Plant Sci. 2020;293:110431. doi: 10.1016/j.plantsci.2020.110431. [DOI] [PubMed] [Google Scholar]
- 104.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stacey MG, Osawa H, Patel A, Gassmann W, Stacey G. Expression analyses of Arabidopsis oligopeptide transporters during seed germination, vegetative growth and reproduction. Planta. 2006;223:291–305. doi: 10.1007/s00425-005-0087-x. [DOI] [PubMed] [Google Scholar]
- 106.Pii Y, et al. Time-resolved investigation of molecular components involved in the induction of NO3– high affinity transport system in maize roots. Front. Plant Sci. 2016;7:1657. doi: 10.3389/fpls.2016.01657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003;339:62–66. doi: 10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 108.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36–e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wickham H. Ggplot2: Elegant Graphics for Data Analysis. Berlin: Springer; 2016. [Google Scholar]
- 111.de Mendiburu, F. & de Mendiburu, M. F. Package ‘agricolae’. R Packag. Version 1–2 (2019).
- 112.Tang Y, Horikoshi M, Li W. Ggfortify: Unified interface to visualize statistical results of popular r packages. R J. 2016;8:478–489. doi: 10.32614/RJ-2016-060. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.