Abstract
Pseudomonas aeruginosa MPAO1 is the parental strain of the widely utilized transposon mutant collection for this important clinical pathogen. Here, we validate a model system to identify genes involved in biofilm growth and biofilm-associated antibiotic resistance. Our model employs a genomics-driven workflow to assemble the complete MPAO1 genome, identify unique and conserved genes by comparative genomics with the PAO1 reference strain and genes missed within existing assemblies by proteogenomics. Among over 200 unique MPAO1 genes, we identified six general essential genes that were overlooked when mapping public Tn-seq data sets against PAO1, including an antitoxin. Genomic data were integrated with phenotypic data from an experimental workflow using a user-friendly, soft lithography-based microfluidic flow chamber for biofilm growth and a screen with the Tn-mutant library in microtiter plates. The screen identified hitherto unknown genes involved in biofilm growth and antibiotic resistance. Experiments conducted with the flow chamber across three laboratories delivered reproducible data on P. aeruginosa biofilms and validated the function of both known genes and genes identified in the Tn-mutant screens. Differential protein abundance data from planktonic cells versus biofilm confirmed the upregulation of candidates known to affect biofilm formation, of structural and secreted proteins of type VI secretion systems, and provided proteogenomic evidence for some missed MPAO1 genes. This integrated, broadly applicable model promises to improve the mechanistic understanding of biofilm formation, antimicrobial tolerance, and resistance evolution in biofilms.
Subject terms: Pathogens, Biofilms, Antimicrobials
Introduction
Pseudomonas aeruginosa is a Gram-negative bacterium ubiquitously present in the soil, water, and different animal hosts1. As an opportunistic human pathogen2, it can cause sepsis, and chronic wound and lung infections, especially in immunocompromised and cystic fibrosis patients. Two mechanisms complicate the treatment of P. aeruginosa infections. It forms recalcitrant biofilms in which the bacterial cells have an increased tolerance against antimicrobial compounds3,4. In addition, worldwide, multiple genetic variants have acquired antimicrobial resistance (AMR) traits5, either through the acquisition of resistance genes on mobile genetic elements such as plasmids6 or through de novo mutations of chromosomal genes7. Furthermore, mutations affecting outer membrane porins and multidrug efflux pumps can mediate resistance to almost all major antibiotic classes and several important biocides8,9. P. aeruginosa thus also belongs to the notorious group of ESKAPE pathogens, which represent the leading causes of worldwide nosocomial infections (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, P. aeruginosa, and Enterobacter species)10,11. Clinically most relevant are the resistances of P. aeruginosa strains against fluoroquinolones, aminoglycosides, and beta-lactams, and against the last-resort antibiotic colistin (polymyxin). In 2017, the World Health Organization (WHO) classified carbapenem-resistant P. aeruginosa strains in the highest priority group of “critical pathogens”. New treatment options informed by a more detailed molecular understanding of how and why resistance emerges during treatment, and how resistance is transmitted, are urgently needed for such critical pathogens.
Increased antimicrobial tolerance, a fundamental property of biofilms12 is well-studied13 and four mechanisms have a major role: (i) under nutrient-limited conditions in biofilms, P. aeruginosa expresses phenotypic variants, i.e., dormant cells that are less susceptible to antibiotics which target actively dividing cells14; (ii) P. aeruginosa form a protective extracellular matrix composed of polysaccharides, proteins, and DNA that limits the diffusion of antimicrobial substances or sequesters them, such that biofilm cells experience a decreased antimicrobial dosage15; (iii) anoxic conditions exist within the biofilm limiting the efficacy of antibiotics that require aerobic metabolic activity and the generation of reactive oxygen species16; (iv) sub-inhibitory concentrations of antibiotics induce increased rates of mutation, recombination, and lateral gene transfer. The mutation rate in biofilms has been reported to be up to 100 times higher than in planktonic cells17, significantly accelerating the development of antibiotic-resistant mutants. Together, these mechanisms lead to hard-to-treat, chronic infections during which P. aeruginosa can persist and further evolve within the host in the presence of antimicrobial substances. Evolution within biofilms is highly parallel and differs significantly from the evolution of planktonic cells18. However, the evolutionary drivers of within-biofilm AMR evolution remain poorly understood. Their study requires well-defined model systems and tools, including model strains with complete genomic background information, genetic tools, and flow chambers allowing representative and reproducible growth of P. aeruginosa biofilms and deep sequencing data18.
The canonical reference model strain for P. aeruginosa is PAO1, also referred to as PAO1-UW. Its complete genome sequence was published in 20002, which allowed many breakthrough discoveries. However, a number of closely related PAO1 strains exist that differ in their phenotypic appearances19. These include P. aeruginosa strain MPAO120, the parental strain of the widely utilized transposon insertion mutant library from the University of Washington (UW)21. Such mutant collections represent highly valuable resources to uncover new functions and condition-specific essential genes in genome-wide screens21, for example, genes relevant for resistance against certain antibiotics22,23. They have also been used to define so-called general essential genes, i.e., genes that were identified as essential under more than one relevant growth condition24,25. As a subset of the conserved core genes of P. aeruginosa PAO1 and PA14 were shown to exhibit differential essentiality25, the approach to focus on those general essential genes that are furthermore conserved among key pathogen strains of a species is particularly promising26. However, the utility of such libraries to identify gain of function mutations is limited and polar effects need to be controlled for27. Notably, no complete MPAO1 genome sequence was available. Improvements in next-generation sequencing (NGS) technologies28 and assembly algorithms nowadays allow researchers to readily generate complete de novo genome assemblies for most prokaryotes except a few percent of strains with highly complex repeat regions29. Such fully resolved genomes are advantageous compared to fragmented short read-based genome assemblies that can sometimes even miss conserved core genes30; they are an ideal basis for subsequent functional genomics and systems biology studies and allow researchers to identify so far missed genes in genome annotations by proteogenomics31.
Here, we set out to develop, validate, and make available a model system to study the biofilm-associated adaptation to antimicrobials and AMR evolution in P. aeruginosa MPAO1. Conceptually, the model was designed to integrate genotype information with phenotypic data and to leverage the valuable genetic tools and wealth of functional genomics data sets that exist for important bacterial model organisms. Important elements include the complete MPAO1 genome sequence and the design for a standardized flow chamber based on accessible soft lithography replication in poly(dimethylsiloxane) (PDMS) that can deliver laminar flow conditions relevant to typical biofilm niches. Comparative genomics with the PAO1-UW reference strain uncovered numerous MPAO1-unique genes. Strikingly, these included 39 essential genes that had been missed so far by performing reference-based mapping of public Tn-seq data sets. Proof of principle experiments highlighted reproducible biofilm growth using the microfluidic flow chamber and identified hitherto unknown genes important for biofilm growth and biofilm-associated AMR through microtiter plate screening of the mutant library. A differential (planktonic vs. biofilm) proteomic data set uncovered gene products known to have a role in biofilm formation. Finally, a publicly available, integrated proteogenomics search database enables the identification of unannotated genes in MPAO1.
Results
De novo genome assembly of MPAO1
The availability of a complete genome sequence is an important pre-requisite to study the phenotypic adaptation and evolution of resistance to antimicrobials in biofilms. An analysis of over 9300 completely sequenced, publicly available bacterial genomes29 (see “Methods” section) listed 106 P. aeruginosa strains overall, two of which were P. aeruginosa PAO1 strains, including the PAO1 type strain (Genbank AE004091), also called PAO1-UW2. In contrast, the only strain annotated as MPAO1, i.e., the founder strain of the transposon mutant library available from the UW21, had been sequenced with Illumina short reads, assembled into 140 contigs32 and deposited (http://www.pseudomonas.com/strain/show?id=659; Genbank ASM24743v2) in the Pseudomonas genome DB33. To provide an optimal basis for subsequent functional genomics and evolution studies for P. aeruginosa strain MPAO1, we thus first sequenced and de novo assembled its complete genome. Due to the genomic differences reported for MPAO1 and PAO120 and the fact that many of the 106 completely sequenced P. aeruginosa strains have difficult to assemble genomes with long repeat pairs in excess of 10 kilobases (kb) (38/106), so-called class III genomes29, we used third-generation long reads from Pacific Biosciences’ (PacBio) RSII platform. By relying on size-selected fragments (average length 9 kb; see “Methods” section), a single bacterial chromosome could be assembled. Additional genome polishing steps with Illumina MiSeq data (300 bp, PE reads) allowed the removal of remaining homopolymer errors in the PacBio assembly34. The final, high-quality MPAO1 genome consisted of one chromosome of 6,275,467 bp and coded for 5926 genes (Genbank CP027857; Table 1). An overview of selected predicted genome features (see “Methods” section) is shown in Supplementary Table 1. To facilitate data mining and comparison, we also provide an extensive annotation of all 5799 protein-coding genes. This includes information on conserved and MPAO1-unique genes compared to PAO1, the respective reciprocal best BLAST hits, protein domains, families, Gene Ontology (GO) classification, predictions of subcellular localization, lipoproteins, secreted and described membrane-localized proteins, as well as gene essentiality status and protein abundance data below (Supplementary Data 1).
Table 1.
Summary over the core and strain-specific CDS of strains MPAO1 and PAO1-UW.
| Category | P. aeruginosa MPAO1 | P. aeruginosa PAO1-UW |
|---|---|---|
| Total no. of genes | 5926 | 5697 |
| Total no. of CDS | 5799 | 5572 |
| No. of core CDS (clustersa) | 5548 (5534) | 5545 (5534) |
| No. of unique (strain-specific) CDS (clusters) | 234 (232) | 19 (21) |
| Unique ncRNA | – | 3 |
| CDS ≤ 120 bpb | 17 | 5 |
aAll individual CDS are shown including those that are grouped in gene clusters (paralogs) in Fig. 1c.
bCDS of 120 bp or below are not considered (see “Methods” section).
Comparative genomics of MPAO1 and PAO1 strains
An alignment of our de novo assembled MPAO1 genome with that of the MPAO1/P1 strain32 revealed that overall, 42,813 bp of our complete genome sequence were missed by the 140 contigs of the available fragmented Illumina assembly (Fig. 1a). This comprised 66 genes (52 protein-coding genes, (CDS)) either missed completely or partially, including eight of 12 rRNA genes (75%) and six of 63 tRNA genes (11%). Among the CDS, the essential gene ftsY encoding the signal recognition particle-docking protein FtsY was missing, four of eight (50%) non-ribosomal peptide synthetase (NRPS) genes, three of six (50%) filamentous hemagglutinin N-terminal domain protein-coding genes, and three of 10 (30%) type VI secretion system (T6SS) VgrG effector proteins (Supplementary Data 1). The analysis of the number of interrupted genes or pseudogenes also confirmed the fragmented nature of the MPAO1/P1 genome compared to the complete genomes of both our assembly and the PAO1-UW type strain (Supplementary Fig. 1). Importantly, a key study of the genotypic and phenotypic diversity of P. aeruginosa PAO1 strains recently reported 10 PAO1/MPAO1 laboratory isolates as complete genomes19. As all 10 genomes have been assembled using Illumina data into sets of contigs, strictly speaking, they are not fully assembled, closed genome sequences. Indeed, the genomes of the two MPAO1 strains in that list (PAO1-2017-E, 71 contigs, whole genome shotgun (WGS) QZGA00000000 and PAO1-2017-I, 70 contigs, WGS QZGE00000000) also lacked a similar amount of genomic sequence (56.5 and 59.4 kb) and number of genes (55, 62) or CDS (40, 47) respectively, compared to our complete genome (Supplementary Data 1).
Fig. 1. Genome map of P. aeruginosa MPAO1 and comparison to other strains.
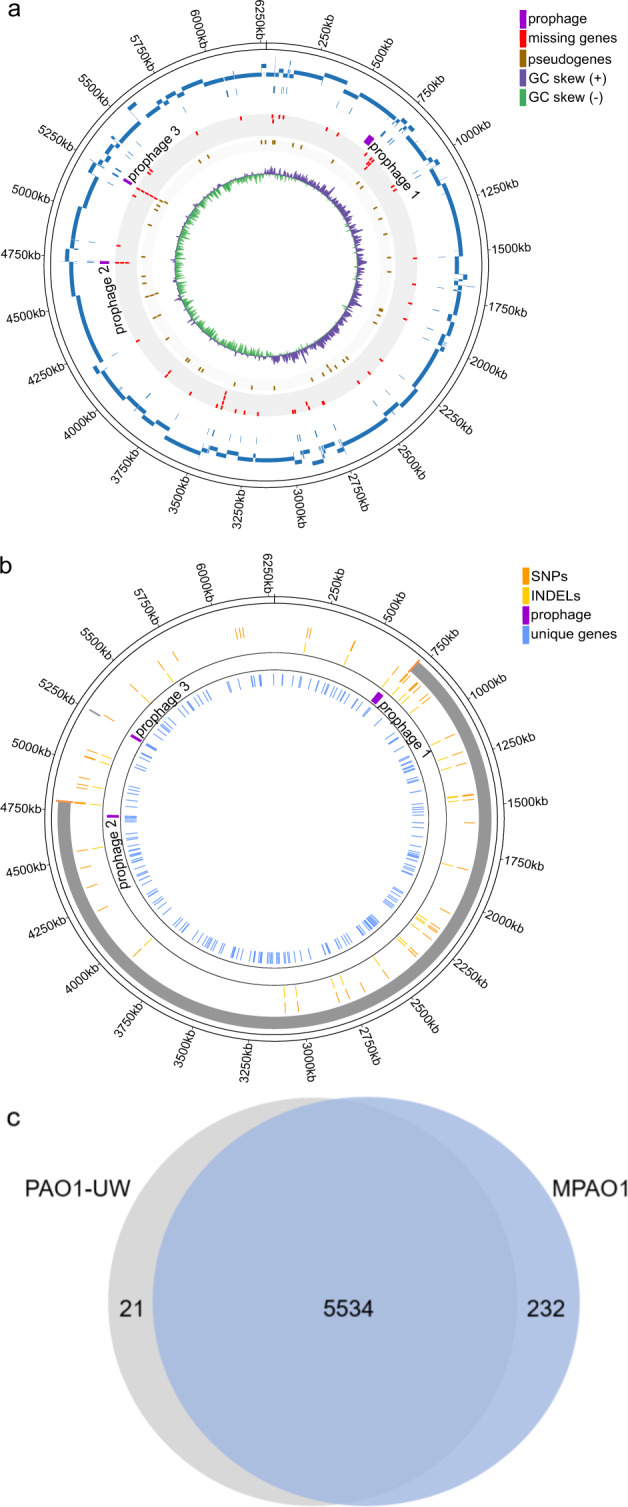
a The Circos plot visualizes the comparison of our complete MPAO1 genome (outer circle with genome coordinates) and that of strain MPAO1/P1 (second circle; blue), the respective gaps (third circle; blue) followed by annotated prophages (fourth circle; purple), missing genes (fifth circle, red), pseudogenes (sixth circle; brown), and GC skew (seventh circle; positive - purple; negative - green). b Differences of the MPAO1 genome compared to the PAO1 reference strain. Going from outer towards inner circles, the following genome features are shown: (1) a large inversion (gray) flanked by rRNAs (not shown), (2) SNPs (dark orange), (3) INDELs (light orange) (4) prophages (purple), (5) genes unique to MPAO1 (blue). c Comparative genomic analysis of P. aeruginosa strains MPAO1 and PAO1-UW. The Venn diagram shows the core gene clusters (paralogous genes are grouped into the same cluster provided they belong to a syntenic genomic region) and the respective number of strain-specific CDS clusters.
Next, to explore the extent of strain-specific genomic differences, we created an alignment of our de novo assembled MPAO1 genome with that of P. aeruginosa strain PAO1-UW. This analysis confirmed the major differences reported previously20, i.e., the presence of a third prophage region (12.8 kb, 20 genes; genome coordinates 5,241,813–5,254,613) in strain MPAO1 (Fig. 1b) and the absence of a ~1 kb genome fragment (leading to a pseudogene annotation for MPAO1_24940 in MPAO1). An analysis of smaller differences between the genomes confirmed the 16 SNPs reported previously20 and identified 176 additional SNPs and INDELs between MPAO1 and PAO1 that had not been reported by Klockgether and colleagues20 (Supplementary Data 2).
Notably, while the overall number of predicted genes was close for both strains (Table 1), we observed 232 gene clusters specific to strain MPAO1 and 21 clusters specific to strain PAO1-UW (Fig. 1c), suggestive of potentially relevant differences between the strains. The annotation of the shared (core) and strain-specific (unique) gene clusters is provided in Supplementary Data 3. This analysis indicated that a sizeable set of genes were specific to the MPAO1 genome and that mapping data sets obtained from this strain back to the PAO1-UW genome could overlook important genes (see below). A gene ontology (GO) enrichment analysis of the MPAO1-unique proteins against all CDS in its genome revealed that the biological process “protein phosphorylation” was significantly enriched (p value < 0.01) with 10 hits among all genes including three among the unique genes (including a DNA helicase and 2 serine/threonine protein kinases; Supplementary Table 2). Furthermore, for the biological process “Bacteriocin immunity” five hits were found among all genes, two of which were among the unique MPAO1 genes (Supplementary Table 2).
Tn-seq data mapping
The complete MPAO1 genome sequence allowed us to re-analyze public Tn-seq data sets without the limitation of any remaining “genomic blind spots” that otherwise might preclude identification of all essential genes26, and the drawbacks of mapping Tn-seq data to a closely related reference genome. A re-mapping of MPAO1 Tn-seq data sets obtained from several conditions (LB medium, minimal medium, sputum, and brain-heart infusion BHI medium)24 against both the PAO1-UW genome and our MPAO1 genome (see “Methods” section), confirmed our expectation. We indeed observed a higher percentage of mapped reads for MPAO1 (roughly 0.1–0.35% of all mapped reads per sample; Supplementary Table 3) and unique insertion sites (roughly 0.2% more in MPAO1, Supplementary Table 3). Genes with no insertion or genes whose p-value was less than 0.001 were considered essential (see “Methods” section). Overall, 577 genes were classified as condition-specific essential in one of the three primary growth conditions LB medium, minimal medium, sputum (Supplementary Data 4), and 312 genes represented general essential genes, i.e., were essential in all three growth conditions, respectively (Supplementary Fig. 2). Importantly, close to 40 MPAO1 unique genes were linked here with an essentiality status, as they were essential in one or more of the 16 Tn-seq libraries (Supplementary Data 4). By mapping data against the PAO1-UW genome, these genes had been previously overlooked in the analysis of essential P. aeruginosa genes.
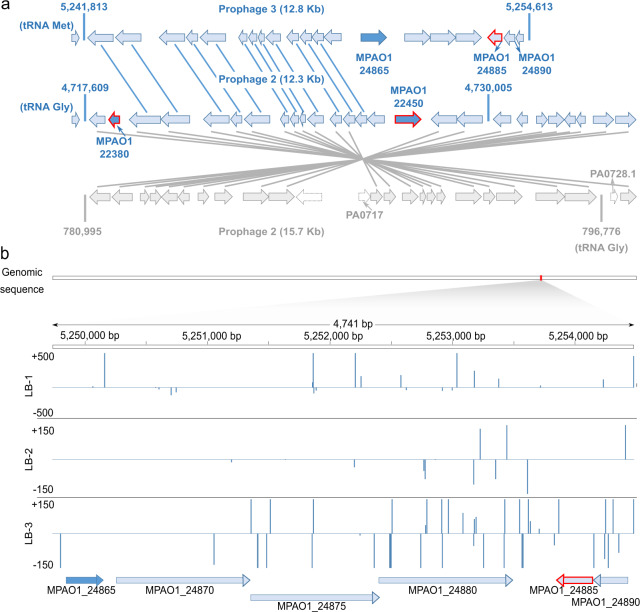
Among these MPAO1-unique genes, we identified 18 genes that were essential in 50% or more of the Tn-seq runs, six of which represented general essential genes (Table 2). The general essential genes included two genes located in the prophage 2 region, i.e., MPAO1_22380, a type II Phd/YefM family antitoxin gene located next to MPAO1_22375, coding for a RelE/ParE type toxin, and MPAO1_22450, a DNA-binding protein (Fig. 2a; arrows framed in red). A further general essential gene was MPAO1_00215 encoding for a hypothetical protein. MPAO1_00215 is located in a genomic region that harbors another essential gene (MPAO1_00230, Supplementary Data 1), that may represent an operon.
Table 2.
List of 18 selected MPAO1-unique genes along with their essentiality classification in all 16 Tn-seq samples24 and comments about their genomic location.
| Locus | Gene annotation | General essential | Essential in x/16 samples | Comment |
|---|---|---|---|---|
| MPAO1_22380 | Type II toxin-antitoxin system Phd/YefM family antitoxin | Yes | 16 | Prophage 2 |
| MPAO1_00215 | Hypothetical protein | Yes | 15 | *Operon? |
| MPAO1_10410 | Hypothetical protein | Yes | 14 | |
| MPAO1_22450 | DNA-binding protein | Yes | 14 | Prophage 2 |
| MPAO1_25260 | Cytidine deaminase | 12 | ||
| MPAO1_12950 | Hypothetical protein | Yes | 11 | |
| MPAO1_00230 | Hypothetical protein | 10 | *Operon? | |
| MPAO1_20095 | Hypothetical protein | 10 | ||
| MPAO1_02335 | Dihydropyrimidinase | 9 | ||
| MPAO1_15010 | 6-O-methylguanine DNA methyltransferase | 9 | ||
| MPAO1_15215 | Amino acid permease | 9 | ||
| MPAO1_18025 | Ferredoxin | 9 | ||
| MPAO1_02315 | Oxidoreductase | 8 | ||
| MPAO1_05695 | Hypothetical protein | Yes | 8 | Bacteriocin (GO) |
| MPAO1_08710 | DUF3304 domain-containing protein | 8 | ||
| MPAO1_10195 | Universal stress protein | 8 | ||
| MPAO1_14380 | Glycosyltransferase | 8 | ||
| MPAO1_24865 | Hypothetical protein | 8 | Prophage 3 |
Information about all MPAO1-unique essential genes is available in Supplementary Data 4.
Fig. 2. An overview of annotated genes in selected prophage regions and their essentiality classification.
MPAO1-unique essential genes are shown in dark blue, general essential MPAO1 genes with a red arrow outline. a Genes located in prophage region 2 of PAO1-UW (gray), the corresponding inverted region in strain MPAO1 (light blue arrows in middle), and the prophage region 3 (light blue arrows on top) unique to MPAO1 are shown (not drawn to scale), the genomic positions of their boundaries (5′–3′) and flanking tRNAs. Genes connected by lines are orthologous to each other based on comparative genomics combined with a Blast analysis. b Transposon insertions in selected genes of prophage region 3 of MPAO1. Insertion frequencies in six genes are shown using data mapped from the LB-1 (3 replicates), LB-2 (2 replicates), and LB-3 (1 sample) Tn-seq libraries. Non-essential genes (based on a data set of 577 genes essential in one of three primary growth conditions) are shown in light blue.
Furthermore, the prophage 3 region unique to strain MPAO1, harbored a gene encoding a hypothetical protein (MPAO1_24865; Fig. 2b) that was essential in eight of 16 samples (Table 2). Conversely, MPAO1_24885 (addiction module antidote protein from the HigA family toxin-antitoxin (TA) system) from this region was classified as general essential (Table 2; 14 of 16 samples). Due to its homology to PA4674 in PAO1-UW, which is listed among the 352 general essential genes reported by Lee and colleagues and encodes the HigA antitoxin24, it is not unique to MPAO1 (Fig. 2b). Together with the non-essential MPAO1_24890 (plasmid maintenance system killer protein; most similar to RelE-like toxins of the type II TA system HigB), MPAO1_24885 encodes for a TA system. However, there is no homolog annotated for MPAO1_24890 in PAO1-UW. Therefore, due to this missing gene, the TA system was not identified in PAO1-UW. This finding again underlines the importance of having the actual and complete genome sequence to map functional data.
Reproducible formation of MPAO1 biofilms
The second important objective of our integrated model system was to enable the reliable generation of phenotypic data under conditions relevant for biofilm growth. For this purpose, we focused on the development of a microfluidic flow chamber for reproducible biofilm formation that would allow us to subsequently identify genes relevant for biofilm growth and biofilm-associated AMR. The flow chamber was designed in such a way that we could assess the effects of hydrodynamic conditions35, such as shear stress and controlled flow conditions. Our flow chamber was replicated in PDMS, a simple to use, transparent, and breathable elastomer material that naturally adheres to the glass. A straight microfluidic channel design was used (30 mm length × 2 mm width × 0.200 mm depth) (Fig. 3a, see “Methods” section for further details). PDMS was selected due to its broad application in indwelling devices and implant materials36. The inlet and outlet of the microfluidic flow chamber comprised of sterile polytetrafluoroethylene (PTFE) tubing, a material that was chosen because it generally exhibits low bacterial adhesion. A syringe pump was used to deliver 5 μL/min (ū ≈ 208 µm/s) flow inside the chamber to provide laminar flow conditions for bacterial adhesion and biofilm growth (the calculated Reynolds Number corrected for the transport of water at 37 °C was 0.103; for details see Supplementary Table 4).
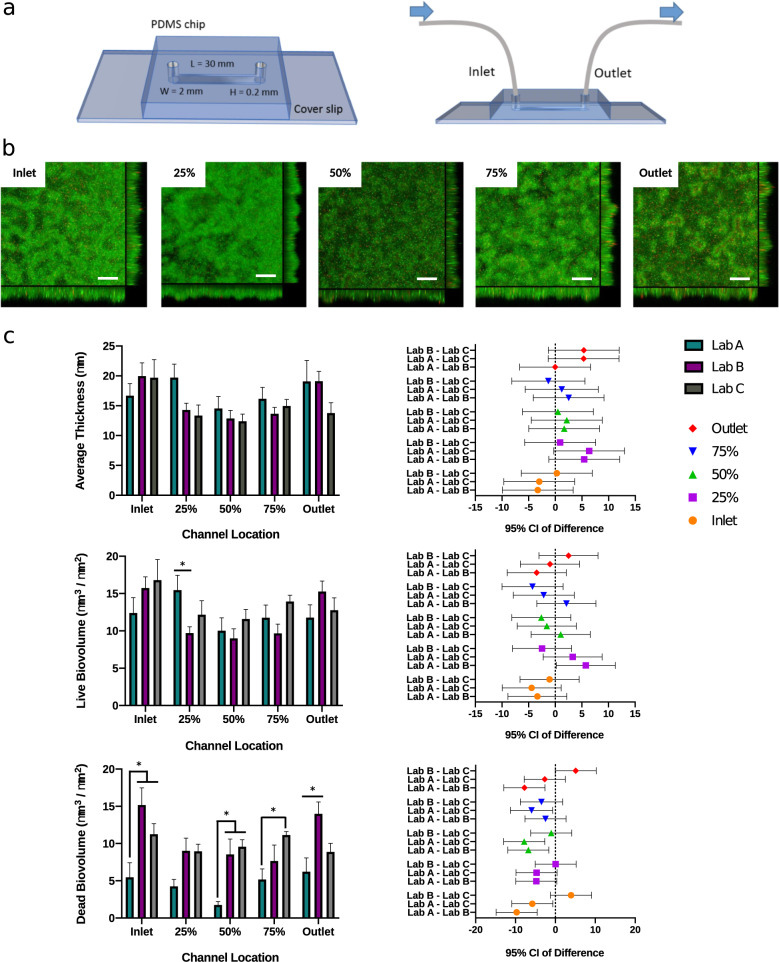
Fig. 3. The publicly available mold design for the microfluidic flow chamber allows reproducible biofilm formation as confirmed by an inter-laboratory comparison.
a Schematic and dimensions of the flow chamber. b Representative images of 72 h MPAO1 WT biofilms grown on the PDMS surface of the device under laminar flow conditions at five different locations along the channel. Biofilms were treated with live/dead staining (green – live cells stained with Syto9; red – dead cells stained with propidium iodide). Scale bar in confocal XY plane: 40 µm. Sagittal XZ section represents biofilm thickness. c COMSTAT data for average thickness, and live/dead biovolume of 72 h MPAO1 WT biofilms generated by three different laboratories, with 95% confidence interval comparisons (3 biological repeats comprising 3 technical repeats per site, i.e., n = 9 biological/n = 27 technical repeats overall; error bars - standard error of mean; 2-way ANOVA with lab and channel location as variables followed by multiple comparisons Tukey test). *p value < 0.05.
The reproducibility of a 72 h mature MPAO1 biofilm on the PDMS surface of the device was investigated by confocal laser scanning microscopy (CLSM) combined with live/dead staining using the dyes Syto9 and propidium iodide in three separate consortium laboratories all using the same microfluidic chamber mold (design publicly available; see Data availability) (Fig. 3b, c). The biofilms formed in the three laboratories were consistent with data falling within 95% confidence intervals, the only difference being the observation of a reduced dead biovolume in one laboratory’s model (Lab A; p value < 0.05). Biofilm formation was relatively uniform throughout the flow channel with an average thickness of 16 µm and a small reduction observed towards the center of the channel (Inlet—18.8 µm, 25%—15.8 µm, 50%—13.3 µm, 75%—14.9 µm, Outlet—17.3 µm). An average biovolume of 12.5 µm3/µm2 and dead biovolume of 8.4 µm3/μm2 was observed, again reducing towards the center of the device commensurate with the average biomass.
Screening experiments identify known and new genes relevant for biofilm formation and antibiotic resistance
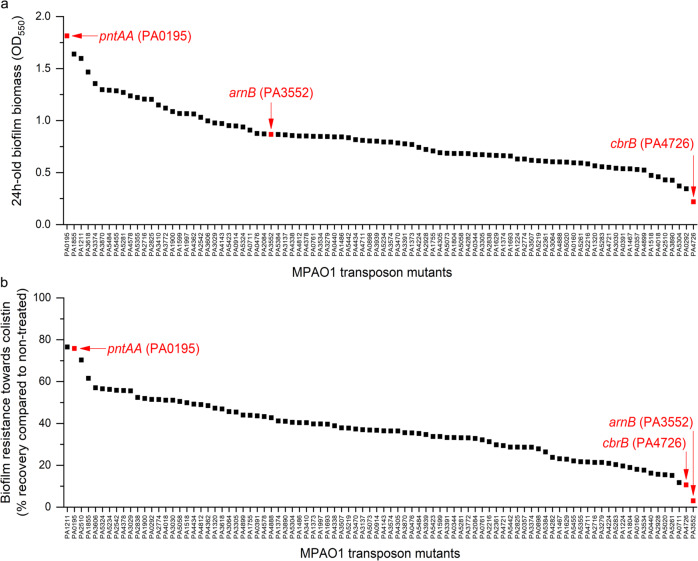
The MPAO1 transposon mutant library was tested with a 96-well plate screening system that was devised to enable the identification of genes that affect biofilm formation and/or have a role in the development of biofilm-associated AMR (see Supplementary Fig. 3). A batch of 95 selected mutants (Supplementary Table 5) was taken from the library to test the reliability of our protocol and to identify genes related to biofilm formation (in duplicate). Strain PW8965 harboring an insertion in cbrB (PAO1 identifier PA4726, MPAO1_25185), a transcriptional activator that forms part of the CbrA/CbrB two-component system important in catabolite repression37, was found to produce the least amount of biofilm (Fig. 4a). In contrast, strain PW9283 mutated in pntAA (PA0195; MPAO1_01040), an NADPH/NAD+ redox balance transhydrogenase38, exhibited the highest biofilm biomass.
Fig. 4. Proof of principle that biofilm growth-relevant and AMR-related genes can be identified in adequate screens using the MPAO1 transposon mutant library.
A diagram of the protocol is shown in Supplementary Fig. 3. a Biofilm formation of 90 MPAO1 mutant strains (X axis) after 24 h incubation in the M9 medium (average of two independent wells). Biofilm biomass was quantified by crystal violet. b Ability of biofilms formed by 90 MPAO1 mutant strains to recover after colistin treatment (see “Methods” section). The recovery of treated biofilm cells was normalized to the recovery of non-treated biofilm cells (defined as 100%). The arnB mutant (PA3552) is highlighted in red, as well as the highest biofilm former missing pntAA (PA0195) and the lowest biofilm former missing cbrB (PA4726).
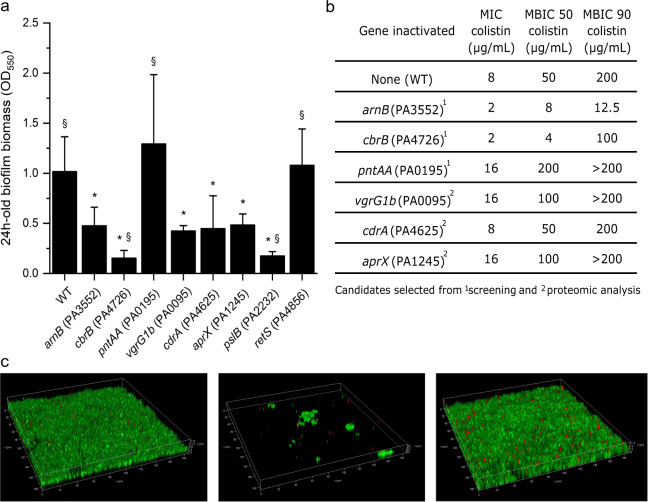
In a second step, selected mutants identified by the screening and the proteomic analysis (see below) were compared to positive and negative controls for biofilm formation (Fig. 5a). The pslB mutant (PA2232; MPAO1_14370), a gene whose product is involved in the synthesis and export of polysaccharides, was used as a reference point for low biofilm formation39, while a retS mutant (PA4856; MPAO1_25880), encoding a pleiotropic regulator of multiple virulence factors, was used as a reference point for high biofilm formation40. Overall, MPAO1 WT produced roughly twice the biofilm biomass of transposon mutants, suggesting that the transposon has an influence on biofilm formation and that it is more reliable to compare transposon mutants amongst each other. The transposon mutant PW7021 (an arnB mutant; PA3552; MPAO1_07345, see below) was chosen as an internal reference for biofilm formation as its biomass was found approximately midway through the 24 h biofilm readings in Fig. 4a. We confirmed that the cbrB mutant produced significantly less biofilm biomass (p value < 0.001) than the arnB mutant, similar to the low biofilm-forming pslB mutant. Biofilm growth of the cbrB mutant was also performed within the flow chamber to confirm the capacity of the device to assess differential biofilm formation. Similar to the 96-well plate screening assay, the cbrB mutant produced substantially less biofilm compared to the MPAO1 WT over 18 h in the flow chamber (Fig. 5c) and displayed a delayed exponential growth compared to WT and the other mutants tested (Supplementary Fig. 4). We also confirmed that the pntAA mutant produced higher biofilm biomass than other transposon mutants, similar to the high biofilm former retS mutant (Fig. 5a). However, compared to the WT, the retS mutant produced comparable biofilm biomass, which is likely caused by a decrease of strain fitness due to the transposon insertion. An alternative explanation is that the effect of RetS cannot be measured after 24 h because it has been shown previously that RetS turns non-functional in P. aeruginosa WT after 8 h following initial attachment41. Genes identified by the proteomic analysis (vgrG1b, cdrA, aprX; see “Result” section below) did not seem to affect the biofilm formation of MPAO1 in the conditions tested.
Fig. 5. Confirmation of the phenotypes identified in our screening.
a Biofilm formation was quantified after 24 h incubation in M9 medium by crystal violet staining (average of at least 18 wells from two independent cultures). The pslB and retS mutants were used as a reference for low and high biofilm formation, respectively. The cbrB and pntAA mutants demonstrated substantially reduced and increased biofilm formation, respectively. Symbols (* and §) indicate significant differences (Student’s tests with p value < 0.001) in comparison to MPAO1 WT and the arnB mutant, respectively. PAO1 genes are shown in brackets, the respective MPAO1 genes are mentioned in the text. b Resistance of planktonic and biofilm cells towards colistin was evaluated for a subset of mutant strains identified in the screening (1) or based on differential proteomics abundance (2). The MIC was determined as the lowest concentration resulting in a 90% reduction of bacterial growth after 24 h in M9 medium compared to the non-treated condition (average of four replicates from two independent cultures). The MBIC was determined as the lowest concentration resulting in a 50% or 90% reduction of the biofilm cell recovery after 24 h treatment compared to the non-treated condition (average of four replicates from two independent cultures). c Comparative confocal micrographs after live/dead staining (green – live cells stained with Syto9; red – dead cells stained with propidium iodide) of 18 h MPAO1 WT, cbrB and arnB biofilms grown under microfluidic conditions using the publicly available mold confirm reduced biofilm formation for the cbrB mutant and robust biofilm formation of the arnB mutant in the absence of treatment.
Next, we tested the strains for their biofilm resistance to colistin and included the arnB mutant strain PW7021 as a positive control (Supplementary Fig. 3). ArnB is a well-studied protein known to modify lipopolysaccharide (LPS) and have a key role in the resistance to colistin42,43. The recovery of biofilm cells after treatment with 25 µg/mL colistin was compared to the recovery of non-treated biofilm cells (Fig. 4b) (see “Methods” section), as described previously13. This concentration of colistin was much higher than the minimal inhibitory concentration (MIC) used for the planktonic P. aeruginosa MPAO1 (8 µg/mL) allowing us to focus specifically on biofilm cells. As expected, the arnB mutant exhibited a very low recovery after colistin treatment (97% less than the control without colistin) (Fig. 4b). In contrast, the arnB mutant produced robust biofilms in the biofilm screening assay (Fig. 4a), a phenotype that was confirmed using the microfluidic chamber (Fig. 4c). Notably, the cbrB mutant strain grown as a biofilm was also found to be sensitive to colistin (90% less recovery than the control without colistin; Fig. 4a), which might be related to the low amount of biofilm produced by this mutant. In contrast, the high biofilm former pntAA mutant exhibited high resistance towards colistin with recovery close to the non-treated biofilm.
In a second step, the resistance profile of the identified mutants was characterized in more detail by measuring the MIC of planktonic cells and the minimal biofilm inhibitory concentration (MBIC) towards colistin (Fig. 5b). Two independent experiments (with four replicates in total) confirmed the significantly higher sensitivity of planktonic cells of arnB and cbrB mutants compared to the WT (Fig. 5b). Additionally, inactivation of the genes arnB and cbrB reduced the biofilm recovery by 50% when 6 and 12 times less colistin was used, respectively, compared to the WT. Inactivating cbrB made MPAO1 biofilms more sensitive to low concentrations of colistin, but high concentrations seemed necessary to reach complete eradication (Fig. 5b). In contrast, inactivating pntAA increased P. aeruginosa resistance towards colistin both as planktonic and biofilm cells. Characterization of the genes identified in the proteomic study (see below) revealed that inactivating cdrA had no impact on MPAO1 resistance, but inactivation of vgrG1b44 and aprX45 increased MPAO1 resistance towards colistin both for planktonic and biofilm cells (Fig. 5b).
Protein abundance profiling of MPAO1 grown planktonically and in biofilms
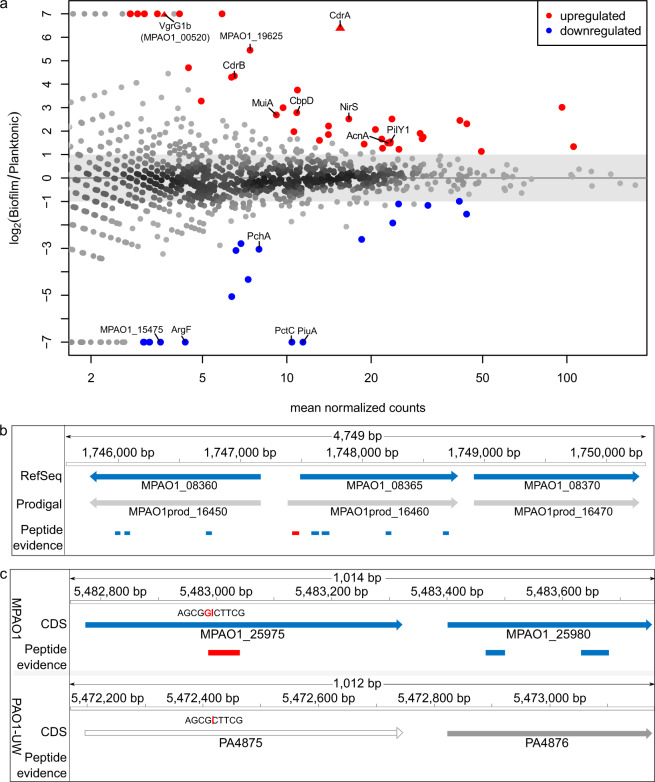
To assess if we could identify proteins known to have a role in biofilm formation with the microfluidic chamber, we next generated shotgun proteomics data for MPAO1 cells grown to mid-exponential planktonic phase or as 72 h biofilms (3 replicates each). 1530 and 1728 proteins were identified in planktonic cells and biofilm, respectively, resulting in a combined 1922 of the 5799 annotated proteins (33.1%). Among the most significantly differentially abundant proteins (log2 fold change (FC) of ≥ 1 or ≤−1 and adjusted p value ≤ 0.05; see “Methods” section and Supplementary Fig. 5) several candidates were identified that have previously been linked with a role in biofilm formation. These included MuiA (MPAO1_18330)46, CbpD (MPAO1_21730)47, AcnA (MPAO1_17965)48 and PilY1 (MPAO1_24155)49 (Fig. 6a, Table 3; see “Discussion” section). In addition, MPAO1_19625 was highly upregulated in biofilms (Fig. 6a). Notably, its PAO1 homolog AprX was reported to be secreted by a type I secretion system45, indicating that hypothetical proteins or proteins of unknown function can be linked to roles in biofilm formation and growth. We next looked for protein expression evidence for CDS missed in the fragmented short-read genome assemblies. We found that 21 of the 52 CDS missed in the MPAO1/P1 assembly were detected at the protein level (Supplementary Data 1). Notably, this included two proteins significantly upregulated in the biofilm, namely MPAO1_00520 (T6SS tip protein VgrG1b) located close to the H1-T6SS cluster44 and MPAO1_24535 (Fig. 6a), the homolog of PAO1 CdrA, a cyclic-di-GMP-regulated adhesin known to reinforce the biofilm matrix50, again underlining the importance of a complete genome sequence for downstream functional genomics analyses. Notably, nine of 14 structural genes of H1-T6SS, one of overall three T6SSs in P. aeruginosa that helps it to prevail under stressful conditions51, were upregulated around two-fold or more in biofilm (Supplementary Fig. 6). Similarly, all three VgrG1 proteins (1a-1c) that are co-regulated with the H1-TS6652 were upregulated in biofilm, while none of the other seven VgrG family members were expressed. Among the proteins downregulated in 72 h biofilms, three are associated with the iron acquisition; isochorismate synthase (MPAO1_03800), a rate-limiting enzyme involved in the production of salicylate (precursor of the siderophore pyochelin)53, the siderophore receptor MPAO1_23930 (PuiA), and the siderophore-interacting protein MPAO1_15475. Iron acquisition is deemed necessary for P. aeruginosa biofilm formation54 so their down-regulation was unexpected, however, this response is likely circumvented by the utilization of alternative iron acquisition strategies including the high-affinity siderophore pyoverdine.
Fig. 6. Proteomic experiments identify known biofilm-related proteins and new information.
a Differential protein abundance between MPAO1 mid-exponential planktonic cells and 72 h biofilms. Selected significantly upregulated proteins (red dots) known to have a role in biofilm formation/growth are labeled, proteins downregulated in planktonic growth are shown in blue. Red triangles denote proteins encoded by genes missed in the MPAO1/P1 genome. b Proteogenomic expression evidence for a longer protein than annotated by RefSeq: the Prodigal predicted protein MPAO1prod_16460 (gray arrow; 447 aa; amino acid) is 44 aa longer than the RefSeq annotated MPAO1_08365 and encodes a glutamine synthetase (blue arrow; 413 aa). The NH-terminal extension is supported by 1 peptide (red) with seven PSMs and harbors a 40 aa longer glutamine synthetase N-terminal domain compared to the RefSeq protein. c Proteogenomic expression evidence for a single nucleotide insertion (red) in the MPAO1_25975 gene (blue arrow) compared to its PAO1 homolog PA4875 (annotated as pseudogene; gray open arrow). The change is supported by peptide evidence (1 red bar).
Table 3.
List of 61 proteins with significant differential abundance (see text) or unique expression when comparing biofilm grown and planktonic cells.
| Locus tag | Gene | Product | log2 FC | padj | Comment, reference |
|---|---|---|---|---|---|
| Biofilm only | |||||
| MPAO1_19985 | napA | Nitrate reductase catalytic subunit NapA | 5.02 | 0.05 | |
| MPAO1_04195 | SH3 domain-containing protein | 5.02 | 0.05 | ||
| MPAO1_10705 | Methyl-accepting chemotaxis protein | 5.11 | 0.03 | ||
| MPAO1_17160 | EscC/YscC/HrcC family-type III secretion system outer membrane ring protein | 5.11 | 0.03 | ||
| MPAO1_21585 | Itaconyl-CoA hydratase | 5.19 | 0.04 | ||
| MPAO1_17195 | Translocator outer membrane protein PopD | 5.19 | 0.02 | ||
| MPAO1_17200 | Hypothetical protein | 5.34 | 0.01 | ||
| MPAO1_00520 | vgrG1b* | Type VI secretion system tip protein VgrG1b52 | 5.41 | 0.01 | H1-T6SS44 |
| MPAO1_20935 | Beta-keto-ACP synthase | 5.61 | 0.04 | ||
| MPAO1_24325 | Cytochrome c551 peroxidase | 6.11 | 0.00 | ||
| Diff. Abundant | |||||
| MPAO1_07815 | Osmoprotectant NAGGN system M42 family peptidase | 4.70 | 0.02 | ||
| MPAO1_19625 | aprX | Hypothetical protein | 5.45 | 0.00 | 45 |
| MPAO1_24535 | cdrA* | Filamentous hemagglutinin N-terminal domain-containing protein | 6.54 | 0.00 | 50 |
| MPAO1_02725 | nirF | Protein NirF | 4.30 | 0.01 | |
| MPAO1_24530 | cdrB* | ShlB/FhaC/HecB family hemolysin secretion/activation protein | 4.35 | 0.01 | 50 |
| MPAO1_25250 | BON domain-containing protein | 3.28 | 0.05 | ||
| MPAO1_19595 | Serralysin | 3.75 | 0.01 | ||
| MPAO1_22090 | putA | Bifunctional proline dehydrogenase/L-glutamate gamma-semialdehyde dehydrogenase PutA | 3.00 | 0.01 | |
| MPAO1_18330 | muiA* | Mucoidy inhibitor MuiA | 2.69 | 0.01 | 46 |
| MPAO1_21730 | cbpD* | Chitin-binding protein CbpD | 2.79 | 0.00 | 47 |
| MPAO1_06120 | Copper chaperone PCu(A)C | 1.98 | 0.03 | ||
| MPAO1_14990 | NAD(P)-dependent alcohol dehydrogenase | 2.22 | 0.01 | ||
| MPAO1_02740 | nirS | Nitrite reductase | 2.52 | 0.00 | |
| MPAO1_25230 | DUF748 domain-containing protein | 1.85 | 0.02 | ||
| MPAO1_18000 | ccoP | Cytochrome-c oxidase, cbb3-type subunit III | 1.60 | 0.05 | |
| MPAO1_28880 | adhP | Alcohol dehydrogenase AdhP | 2.52 | 0.00 | |
| MPAO1_07010 | Phosphoketolase | 2.07 | 0.00 | ||
| MPAO1_00100 | LysM peptidoglycan-binding domain-containing protein | 1.44 | 0.03 | ||
| MPAO1_02290 | TonB-dependent receptor | 1.66 | 0.01 | ||
| MPAO1_27435 | Amino acid ABC transporter substrate-binding protein | −3.09 | 0.03 | ||
| MPAO1_05385 | DUF1302 domain-containing protein | −2.80 | 0.03 | ||
| MPAO1_17965 | acnA* | Aconitate hydratase | 1.49 | 0.01 | 48 |
| MPAO1_24155 | pilY1* | Type 4a pilus biogenesis protein PilY1 | 1.54 | 0.01 | 49 |
| MPAO1_05375 | Fatty acid--CoA ligase | −5.06 | 0.01 | ||
| MPAO1_04650 | OmpW family protein | 1.49 | 0.01 | ||
| MPAO1_00495 | tssH | Type VI secretion system ATPase TssH | 1.27 | 0.03 | H1-T6SS44 |
| MPAO1_14010 | Non-ribosomal peptide synthetase (NRPS) | 1.90 | 0.02 | ||
| MPAO1_26210 | azu | Azurin | 2.45 | 0 | |
| MPAO1_13620 | Xanthine dehydrogenase family protein molybdopterin-binding subunit | −4.33 | 0.01 | ||
| MPAO1_03800 | pchA | Salicylate biosynthesis isochorismate synthase | −3.04 | 0.01 | |
| MPAO1_06095 | TonB-dependent copper receptor | 1.74 | 0.00 | ||
| MPAO1_03775 | Catalase | 1.67 | 0.00 | ||
| MPAO1_02430 | clpG | AAA family protein disaggregase ClpG | 2.31 | 0.00 | |
| MPAO1_26945 | Poly(3-hydroxyalkanoate) granule-associated protein PhaF | 1.22 | 0.03 | ||
| MPAO1_23990 | Prepilin-type cleavage/methylation domain-containing protein | 3.01 | 0.00 | ||
| MPAO1_02180 | Response regulator | 1.13 | 0.00 | ||
| MPAO1_05390 | DUF1329 domain-containing protein | −2.62 | 0.00 | ||
| MPAO1_13900 | NADP-dependent glyceraldehyde-3-phosphate dehydrogenase | −1.11 | 0.05 | ||
| MPAO1_13035 | Multidrug efflux RND transporter periplasmic adaptor subunit MexE | −1.92 | 0.00 | ||
| MPAO1_25100 | TonB-dependent hemoglobin/transferrin/lactoferrin family receptor | −1.17 | 0.02 | ||
| MPAO1_09260 | Carbohydrate ABC transporter substrate-binding protein | −0.99 | 0.02 | ||
| MPAO1_16835 | Porin | 1.34 | 0.00 | ||
| MPAO1_09280 | Porin | −1.54 | 0.00 | ||
| Planktonic only | |||||
| MPAO1_23930 | puiA** | TonB-dependent siderophore receptor | −6.91 | 0.00 | |
| MPAO1_22860 | pctC | Methyl-accepting chemotaxis protein PctC | −6.78 | 0.00 | |
| MPAO1_07425 | argF | Ornithine carbamoyltransferase | −5.51 | 0.01 | |
| MPAO1_21260 | Chain-length determining protein | −5.22 | 0.02 | ||
| MPAO1_15475 | Siderophore-interacting protein | −5.09 | 0.02 | ||
| MPAO1_29055 | Class I SAM-dependent methyltransferase | −5.08 | 0.03 | ||
| MPAO1_22680 | Biliverdin-producing heme oxygenase | −5.02 | 0.03 | ||
| MPAO1_09305 | pgl | 6-phosphogluconolactonase | −5.01 | 0.03 | |
Publications linking the genes/proteins with various roles in biofilms are listed for proteins highlighted in Fig. 6. Two genes missed in MPAO1/P1 are shown in bold. Gene names stem from the National Center for Biotechnology Information (NCBI) annotation or were deduced from the eggNOG annotation or the respective PAO1 homolog (*) or the Pseudomonas genome database (**); see also Supplementary Data 1.
Finally, to identify unannotated short ORFs that may carry out important functions or new start sites, we created an integrated proteogenomics search database (iPtgxDB) for strain MPAO1 and PAO1 (Supplementary Table 6), which cover their entire coding potential31. A search combined with stringent result filtering (see “Methods” section) allowed us to identify unambiguous peptide evidence55 for a 44 aa longer proteoform of MPAO1_08365 (predicted by Prodigal, an ab initio gene prediction algorithm; Fig. 6b). In addition, we obtained proteogenomic evidence supporting a single nucleotide insertion in MPAO1_25975 in strain MPAO1 as compared to PA4875 (annotated as pseudogene) in strain PAO1 (Fig. 6c). The peptide that supported this single nucleotide change at the amino acid level was identified with seven peptide spectrum matches (PSMs), illustrating the ability to identify SNP changes at the protein level, with implications for clinical proteomics.
Discussion
P. aeruginosa is a member of the ESKAPE pathogens, the lead cause of worldwide nosocomial infections10. Along with many other clinically relevant bacteria, it can form biofilms whose emergent properties56 include a much higher tolerance to antimicrobials. Together with the increased mutation rates in biofilm compared to planktonic cells17, this further complicates treatment and cure of biofilm-based infections12,13. The development of model systems allowing the study of antimicrobial tolerance mechanisms and the evolutionary dynamics that lead to AMR development in biofilms is thus of utmost priority.
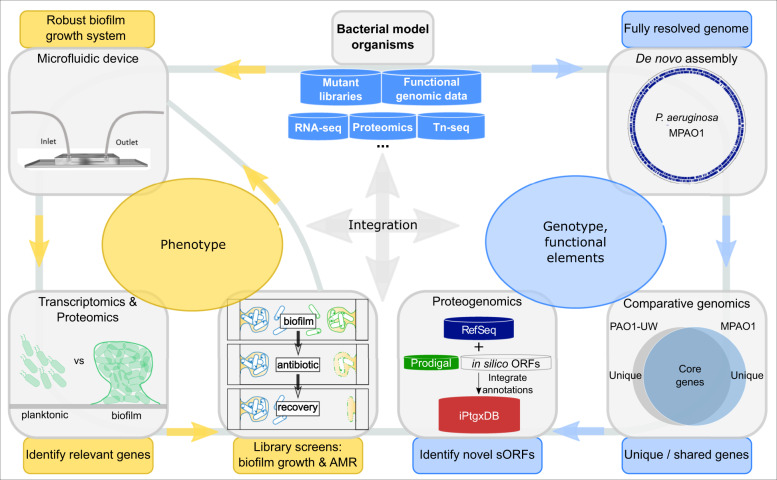
We here develop and validate such a model system for P. aeruginosa MPAO1 (Fig. 7). Conceptually, the model was designed to integrate genotype data with phenotypic information and to leverage the wealth of existing public genetic resources and functional genomics data sets. A complete, fully resolved genome sequence is one critical element31,57, which recently allowed linking of genotypic differences of nine Pseudomonas plant microbiome isolates with their varying biocontrol potential58. While a complete genome existed for P. aeruginosa PAO12, only three fragmented Illumina-based genome assemblies were available for MPAO1, the parental strain of the popular UW transposon mutant library21. These included strains MPAO1/P132 and the recently sequenced PAO1-2017-E and PAO1-2017-I19. On average, they lacked between 55 to 66 genes (40–52 CDS) compared to our complete MPAO1 genome (Supplementary Data 1). For MPAO1/P1, these included the essential ftsY, an adhesin, several T6SS effectors (see below), and four of the overall eight NRPSs. NRPSs are highly relevant for AMR as they often represent enzymes involved in the biosynthesis of antibiotics59. In fact, due to the multi-resistant phenotype of ESKAPE pathogens, concerted efforts aim to describe their NRPS gene clusters in search of new therapeutic approaches60, reinforcing the need for complete genome sequences.
Fig. 7. Integrated model system to identify and validate genes relevant for biofilm growth and AMR.
A sequential genomics-driven workflow (blue arrows) to de novo assemble the complete genome, identify unique and conserved genes among key reference strains by comparative genomics and missed genes by proteogenomics is integrated with an experimental workflow in the form of an iterative cycle that can be entered at various points (yellow arrows). This workflow allows the study of biofilm grown cells, to explore differentially abundant genes or proteins compared to planktonic cells, and to screen mutant libraries to identify functionally relevant genes. The model leverages the enormous value of genetic resources like gene knockout or transposon insertion mutant libraries and functional genomics data sets (RNA-seq, Tn-seq, etc.; blue containers). Additionally, it allows for phenotypic characterization of biofilms formed by mutant strains, thereby allowing us to determine the impact of specific genes on biofilm formation and assess their role in AMR (yellow arrows).
Comparative genomics with the PAO1 type strain uncovered an inventory of conserved and strain-specific genes, and a list of genome-wide SNPs, extending an earlier study that had compared a subset of genomic regions20. Among the 232 MPAO1-unique gene clusters, bacteriocins61 were enriched, which have a role in restricting the growth of closely related microbial competitors to gain an advantage in colonizing a variety of environments62. The complete MPAO1 genome enabled us to remap valuable existing Tn-seq data sets from relevant conditions24, thereby identifying 39 MPAO1-unique essential genes that had escaped detection so far due to reference-based PAO1 mapping. 18 of these genes were essential in at least 50% of the 16 Tn-seq samples, and six represented general essential genes, including a Phd/YefM family type antitoxin (MPAO1_22380), which was essential in all samples. This is worth noting given the relevance of toxin-antitoxin systems for bacterial growth arrest and persistence63. Importantly, our data do not conflict with results from previous studies; rather, they open the field to study the roles of additional MPAO1-unique essential genes. Furthermore, our results suggest that groups planning to construct inventories of core essential genes in other pathogens, following the elegant approach of Poulsen et al.26 who had considered both relevant media mimicking different infection types and nine strains from different lineages of a P. aeruginosa phylogenetic tree, should ideally select complete genomes without any genomic blind spots.
To leverage the experimental arm of our model (Fig. 7), the consortium developed a PDMS microfluidic flow chamber for biofilm growth, which offers several significant advantages. It provides laminar flow conditions inside the channels (Supplementary Table 4), allows gas exchange, decreases the amount of growth medium, facilitates heat transfer, is inexpensive to replicate, and permits imaging of the biofilm and easy harvesting for biochemical characterization. While the flow chamber can be used to monitor biofilm formation on both glass (oxygen impermeable) and PDMS, it is more relevant to investigate biofilm formation on PDMS as a widely applied biomaterial used in indwelling devices and implants36. We observed that biofilms on PDMS formed a more homogeneous layer (Fig. 3b) as compared to the commonly observed mushroom-like structures of P. aeruginosa biofilms on glass64. This effect is not related to hydrodynamics as a flow chamber that previously has been shown to produce mushroom-like structures65 has hydrodynamics (ū ≈ 208 µm/s and Re = 0.24) comparable to our microfluidic chip. We speculate that the effect is most likely explained by two differences: (i) PDMS is oxygen permeable and can transport oxygen to the base of the biofilm that then manifests in overall biofilm structure, or (ii) slight differences in media composition.
The microfluidic data from the inter-laboratory trial on strain MPAO1 validated the utility of the flow chamber and allowed us to compare the phenotypes of WT and mutant strains of the UW transposon library. Important genes were identified with a microtiter plate screening assay and subsequently validated with the flow chamber. Proof of principle experiments confirmed the role of arnB (PA3552), i.e., a gene relevant for colistin resistance42,43, both in biofilms grown in the 96-well plate screen and the flow chamber. In addition, a mutant lacking cbrB (PA4726) showed reduced resistance to colistin in biofilm and planktonic cells and formed very low amounts of biofilm in both the microtiter plate and flow chamber. In addition, inactivating cbrB was found to be as inhibitory for biofilm formation as inactivating the gene pslB, known to negatively influence biofilm matrix synthesis39. As part of the two-component system CbrAB, a mutation in the response regulator cbrB is known to negatively affect the use of several carbon and nitrogen sources37. Such a defect could explain the low growth rate, the low biofilm biomass and therefore the low resistance to colistin of this mutant. Using P. aeruginosa PA14, it was shown that a mutation in CbrA improved biofilm formation, while a mutation in CbrB did not66. However, these differences might be explained by strains (MPAO1 versus PA14) or growth media used (M9 versus BM2-biofilm medium). In contrast, our screening revealed that inactivating the transhydrogenase pntAA induced high biofilm formation, comparable to the known gene retS. While redox balance is known to correlate with biofilm morphology67, the precise role of pntAA remains to be investigated. Together, the combined data of the screen and flow chamber experiments demonstrated that genes previously not implicated in AMR and biofilm formation can be identified and that the function of known genes can be validated.
The differential proteomics data confirmed proteins known to have a role in biofilm formation and growth. These included MuiA, which inhibited swarming motility and enhanced biofilm formation (roles, that were validated in knockout strains)46, and CbpD, for which higher protein abundance had been observed in late phases of biofilm growth; accordingly, mutants displayed a lower amount of biofilm growth and exopolysaccharides (EPS)47. Similarly, for two other proteins with significantly higher abundance in biofilms, inactivation studies showed that the gene encoding AcnA impaired biofilm formation and was required for microcolony formation48, while an increased abundance of PilY1 repressed swarming and increased biofilm formation, as confirmed by knockout experiments49. Biofilm exclusive protein expression was observed for MPAO1_00520, the T6SS VgrG1b effector protein52, while the adhesin CdrA (MPAO1_24535)50 was highly upregulated in biofilms. Both genes were missed in the MPAO1/P1 genome. CdrA forms a two-partner secretion system with CdrB, and both were upregulated under elevated c-di-GMP levels50, in line with the upregulation we observed in biofilm. Moreover, an NRPS (MPAO1_14010) and the hypothetical protein MPAO1_19625 were significantly upregulated in biofilm (Table 3). The data provided insights beyond the top differentially abundant proteins. Notable examples included immunity protein TplEi68 (PA1509, MPAO1_18250), a bacteriocin of the H2-T6SS51, which was exclusively expressed in biofilm (Supplementary Data 1), and upregulation of nine of 14 structural members of H1-T6SS51 (Supplementary Fig. 6). Active T6SSs have been associated with chronic infections in cystic fibrosis patients52, and H1-T6SS has an important role in the dominance of P. aeruginosa in multi-species biofilms69. More sensitive and comprehensive proteomics studies are needed to overcome the limitation that only a third of the theoretical proteome was identified with our shotgun proteomics approach, e.g., by combining data-dependent and data-independent acquisition and the use of spectral libraries70, allowing a more comprehensive identification of lower abundant and small proteins, or by analyzing additional conditions or mutant strains under which tightly regulated proteins such as the Tse toxins (secreted substrates of the H1-T6SS) are expressed (Supplementary Fig. 6)71.
The public MPAO1 (and PAO1) iPtgxDBs allows the identification of missed genes by proteogenomics31, which often encode short proteins (sProteins) that can carry out important functions72,73. Interestingly, Tn-seq data from the Manoil group had implied an essential genomic region in the PF1 phage region of PAO1-UW24. Re-mapping their data, we identified a general essential gene (MPAO1_22380) annotated in our MPAO1 genome whose homolog had been missed in the PAO1 genome annotation, and which appeared to encode the antitoxin member of a ParDE-like TA system (PA0728.1, Fig. 2). However, we did not identify expression evidence for the antitoxin MPAO1_22380 (83 aa) with our iPtgxDB, most likely because our data set (33% of MPAO1 proteins) was not as extensive as that used in a comprehensive proteogenomic study (85% of Bartonella henselae proteins)31, whose complete membrane proteome coverage included expression evidence for all T4SS members74. Nevertheless, we observed proteogenomic evidence for gene products missed in the fragmented MPAO1/P1 genome, for new start sites, and for single amino acid variations, underlining the potential value of proteogenomics for application in clinical proteomics.
Our proof of principle experiments uncovered several candidates for follow-up studies and illustrated the benefit of the complete MPAO1 genome, which led to the discovery of six general essential genes not contained in the transposon library, and which will allow identifying evolutionary changes that lead to AMR in biofilm by deep sequencing in the future. Having been validated for the generation of reproducible inter-laboratory P. aeruginosa biofilm results, a milestone en route to a community standard (see Data availability), the microfluidic platform can be instrumental to investigate other biofilms, notably clinical pathogens and mixed-species biofilms69. The upregulation of the H1-T6SS highly relevant for dominance of P. aeruginosa69 implies that our microfluidic chamber should also be valuable for this extension. Our proposed workflow (Fig. 7) with feedback between genotypic and phenotypic assessment of biofilm characteristics can thus be leveraged across the field of biofilm research and helps bridge the gap between genome-wide and reductionist approaches to study phenomena related to biofilm-associated AMR.
Methods
Bacterial growth and genomic DNA extraction
P. aeruginosa strain MPAO1 (originating from the lab of Dr. Barbara Iglewski) was obtained from Prof. Colin Manoil, UW (Seattle, USA) together with the transposon insertion mutant collection of ~5000 mutated genes (9437 strains)21. For DNA extraction, the MPAO1 cryoculture was streaked out on 20% BHI solid medium (7.4 g in 1 L water) containing 1.5% agar (both Sigma, Switzerland). Shaken 20% BHI fluid cultures were inoculated from a single colony and grown at 30 °C until the mid-exponential phase (OD600 = 0.5). Genomic DNA (gDNA) was extracted with the GeneElute kit (Sigma, Switzerland), following the Gram-negative protocol, including RNase treatment. A study that had analyzed 9331 complete bacterial genomes29 (NCBI RefSeq, assembly category: ‘complete genome’; status Feb. 23, 2018; see their TableS4) reported that 106 P. aeruginosa strains have been sequenced completely, which included only two PAO1 strains (and no complete genome of strain MPAO1). 38/106 (36%) had difficulty assemble genomes with repeat pairs >10 kilo base pairs (bp).
Sequencing, de novo genome assembly, and annotation
PacBio SMRT sequencing was carried out on an RSII machine (1 SMRT cell, P6-C4 chemistry). A size selection step (BluePippin) was used to enrich for fragments longer than 10 kb. The PacBio run yielded 105,221 subreads (132 Gbp sequence data). Subreads were de novo assembled using the SMRT Analysis portal v5.1.0 and HGAP475, and polished with Arrow. In addition, a 2 × 300 bp paired-end library (Illumina Nextera XT DNA kit) was sequenced on a MiSeq. Polishing of the assembly with Illumina reads, circularization, start alignment using dnaA, and final verification of assembly completeness was performed as described previously76. The quality of the aligned reads and the final chromosome was assessed using Qualimap77. In addition, we checked for any potential large scale mis-assemblies using Sniffles v1.0.878 by mapping the PacBio subreads using NGMLR v0.2.678. SPAdes v3.7.179 was run on the Illumina data to detect smaller plasmids that might have been lost in the size selection step. The genome was annotated with the NCBI’s prokaryotic genome annotation pipeline (v3.3)80. Prophages were identified with Phaster81. Detailed annotations for all CDS were computed as described previously82; this included assignment to Cluster of Orthologous Groups (COG) categories using eggnog-mapper (v1.0.3) and EggNOG 4.5, an Interproscan analysis and prediction or /integration of subcellular localizations, lipoproteins, transmembrane helices and signal peptides (for details, see Supplementary Data 1).
Comparative genomics of selected PAO1 genomes
The genome of the P. aeruginosa PAO1-UW reference strain2 was compared to our complete MPAO1 genome using the software Roary (v3.8.0)83 to define core and strain-specific gene clusters as described before30,83. A BlastP analysis helped to correctly identify conserved genes with ribosomal slippage (prfB; peptide chain release factor B) or that encodes selenocysteine (MPAO1_25645), which otherwise can be misclassified as unique genes; genes of 120 bp or below (17 in MPAO1) were not considered. ProgressiveMauve84 was used to align the genomes globally and to identify larger genomic differences. Smaller differences (indels, single nucleotide polymorphisms (SNPs)), were identified and annotated against the PAO1 reference strain as described previously82. Furthermore, contigs from the MPAO1/P1 genome32 were aligned to our complete MPAO1 genome assembly using BWA mem85. Bedtools v2.16.1 “genomecov”86 was used to calculate a gene-wise coverage, allowing to identify genes that were missed in the 140 contigs.
Re-mapping of Transposon-sequencing data
MPAO1 Tn-seq data sets24 were downloaded from NCBI’s SRA (SRP052838) and mapped back both to the PAO1-UW reference strain genome2 and to our MPAO1 assembly following the scripts and notes provided in the Supplement. Insertion sites were computed as described by the authors, reads mapping to multiple genome positions were assigned randomly, and the number of insertion sites per gene was used to differentiate essential and non-essential genes as described24. Genes with zero insertions were considered essential; for the remaining genes, normalized read counts across all insertion sites per gene (considering insertions falling within 5–90% of the length of each gene) were log2 transformed and fitted to a normal distribution. Genes with a p value < 0.001 were added to the list of essential genes. Finally, essential and conditionally essential genes were identified among the three main growth conditions (LB medium, minimal medium, sputum) as described24. Data from each growth condition consisted of multiple mutant pools; for LB, two mutant pools additionally contained multiple replicates (LB-1: 3 replicates; LB-2: 2 replicates). For LB, genes were considered essential in the mutant pool LB-1 and LB-2 if at least two of three (LB-1) and one of two replicates (LB-2) agreed. Next, a consensus set of essential genes in LB and the minimal medium was derived from those genes that were essential in at least two of three mutant pools (LB-1, LB-2, and LB-3) in LB and minimal respectively. Similarly, essential genes in sputum (four mutant pools) were derived if data from at least three of four pools agreed. Finally, genes that were essential in all three growth conditions were called “general essential genes (312)” and genes essential to a specific growth condition were called “condition-specific essential genes”. Together, they comprise “all essential genes (577)”; for further details, see Supplement.
Microfluidic chamber used for biofilm growth
The standardized microfluidic flow chamber consisted of a PDMS chip with a straight microfluidic channel (30 mm length × 2 mm width × 0.200 mm depth) that naturally adhered to a glass coverslip (26 × 60 mm; thickness no.1). The wafer master was fabricated using SU-8 spin-coated to a thickness of 200 μm on a silicon wafer in advance of standard soft lithography replication into PDMS [84]. From this, polyurethane clones of the structures were prepared to upscale production and for sharing microfluidic molds between laboratories. A degassed 10:1 mixture of Sylgard 184 PDMS base and curing agent was cured in an oven at 60 °C for 2 h. Following cooling and retrieval from the SU-8 wafer the structured PDMS was attached, structures facing upwards, to a silicone baking mold using a transparent double-sided adhesive (3 M). The PDMS part was degassed, while the two-component polyurethane (Smooth-Cast™ 310) solutions were each thoroughly shaken for 10 min and then combined in a 1:1 ratio followed by thorough mixing (by repeat inversion and then shaking). The PDMS device was then submerged in the mixture, with degassing for 10 min, after which the mold was left overnight in a well-ventilated area followed by a hard bake at 60 °C for 4 h. Once cooled the PDMS device was retrieved leaving the polyurethane mold in readiness for replica molding fresh PDMS devices again at 60 °C for 2 h. Importantly, PDMS devices are only retrieved after the polyurethane mold has cooled to room temperature to allow the repeated replication (>100 times) of precision PDMS microfluidic chambers. Inlet and outlet ports were prepared using 1-mm-diameter biopsy punches (Miltex™) and then the device was enclosed using a coverslip that was cleaned with 2% RBS 35 detergent (prepared in demineralized water), rinsed with tap water, then immersed in 96% ethanol and sonicated for 5 min, followed by a final rinse with demineralized water and then autoclaved. The inlet and outlet of the microfluidic flow chamber were connected to a syringe pump with a 25 G needle and waste container, respectively, via sterile PTFE tubing (Smiths Medical, ID 0.38 mm, OD 1.09 mm). The chamber was disinfected by flowing 70% ethanol for 15 min at a rate of 20 μL/min, before rinsing with sterile PBS for 15 min and then flushing with M9 minimal medium (Formedium Ltd, Hunstanto, England) for another 15 min at the same flow rate.
Device inoculation, biofilm staining, and confocal laser microscopy
P. aeruginosa MPAO1 was inoculated with 500 µL of an M9-grown overnight pre-culture and grown for ~16–18 h in 10 mL M9 medium (1x M9 salts supplemented with 2 mM MgSO4, 100 μM CaCl2 and 5 mM glucose) at 37 °C with gentle rotation (150 rpm) until a cell concentration of 1.5 × 109 bacteria/mL was reached. One mL of the culture was then washed twice with PBS (pH 7.0) by centrifugation at 5000×g for 5 min at 10 °C. The bacterial pellet was resuspended and diluted in PBS + 2% M9 such that the final cell suspension contained 3 × 108 bacteria/mL. The microfluidic chamber was set on a hotplate at 37 °C with the glass coverslip in direct contact with the hotplate surface. The freshly prepared bacterial suspension was flown through at a rate of 5 μL/min for 1 h. After 1 h, the bacterial suspension was replaced by M9 medium and run through the system at 5 μL/min for 72 h. After 72 h, CLSM images were taken. The biofilm was stained by flowing 1 mL of Live/Dead (Life Technologies, Oregon, USA) staining solution (1.5 μL Syto9 + 1.5 μL propidium iodide in 1 mL of sterile demineralized water) through the flow chamber at 5 μL/min. Once the channel was filled, the flow was stopped and the biofilm kept in the dark for 30 min to allow dye penetration. Finally, PBS was flown through the system at 5 μL/min for 30 min to remove the staining agent. Confocal imaging was performed using a Leica SP8 with ×63 oil immersion lens (HC PL APO CS2 ×63/1.30, Southampton; LabA), a Leica SP8 with ×63 water immersion lens (HC PL APO ×63/1.20 W CORR CS2; BAM, LabB), and a Leica SP2 with x63 water lens (HCX APO L ×63/0.9 W; Groningen, LabC) for 3 biological repeats comprising 3 technical repeats per site (n = 9 biological/n = 27 technical). Z-stacks (1 μm) were taken of the biofilms formed on the PDMS surface of the device at five separate regions (besides the inlet, 25%, 50%, 75%, and beside the outlet). COMSTAT 2.1 (Image J) analysis of combined confocal data was performed to provide a quantification of average biofilm thickness and Live/Dead biovolume87. A 2-way ANOVA multiple comparisons was performed with Tukey’s post hoc test to determine 95% confidence intervals. Similar conditions were applied to strain PA4726 (cbrB) that had shown reduced biofilm growth during screening, and PA3552 (arnB) which demonstrated robust biofilm formation. Biofilm formation of both mutant strains was compared to the MPAO1 WT strain after 18 h growth in the flow chamber.
Screening the public MPAO1 transposon library for antibiotic resistance
The protocol to assess the antibiotic resistance of biofilm-forming MPAO1 cells was adapted from a previous study88. Frozen mutant stocks of 95 randomly selected mutants of the UW Genome Center’s P. aeruginosa PAO1 transposon mutant library21, each harboring a transposon insertion inactivating the function of the respective gene, were allowed to recover in 20% BHI overnight at 150 rpm and 37 °C. All subsequent incubations were done at 37 °C in 96-well plates (TPP tissue culture 96-well plates, Z707902, Sigma-Aldrich) covered with an air-permeable foil (Breathe-Easy sealing membrane, Z380059, Sigma-Aldrich) without further shaking. The overnight cultures were diluted 10 fold in M9 medium and 100 µL each was distributed in six plates (1 well/mutant/plate). After 24 h incubation, the biofilm formation from two plates was quantified by crystal violet staining, while biofilms from the other four plates were washed with 0.9% NaCl to remove planktonic bacteria. Bacteria were then exposed to either M9 or M9 supplemented with 25 µg/mL of colistin, i.e., much higher than the minimal inhibitory concentration (MIC) for the planktonic growth of P. aeruginosa (4 µg/mL), allowing us to focus specifically on the biofilm bacteria. After 24 h treatment, the medium was removed, biofilms were washed with 0.9% NaCl to remove all traces of antibiotics, and bacteria were allowed to recover in fresh colistin-free M9 medium. After 24 h incubation, the recovery of biofilm bacteria was measured by turbidity (OD600) to reveal if the mutation influences the resistance attributed by the biofilm. To confirm the reliability of our screening, promising mutants were analyzed independently in triplicate. Cell suspensions of each mutant were prepared in M9 medium (5 × 106 CFU/mL) and biofilm biomass was quantified by crystal violet after 24 h incubation at 37 °C. Biofilm cell resistance was quantified by measuring the turbidity of biofilm suspension after 24 h treatment with different concentrations of colistin and after 24 h recovery in fresh M9 medium. Selected mutants of interest were further characterized to assess biofilm formation (as described above), MIC, and MBIC (see Supplementary Fig. 3 for detail).
Protein extraction from MPAO1 planktonic and biofilm cultures
For planktonic protein extractions, 10 mL MPAO1 was grown overnight (~18 h) in M9 medium under gentle rotation (150 rpm), centrifuged at 4000×g/5 min/RT, and the pellet resuspended in 1 mL Hanks’ Balanced Salt Solution (HBSS). Biofilms were grown for 72 h using the microfluidic device as previously described, the PDMS device removed from the glass coverslip, and the combined biofilm biomass from 3 lanes harvested into 1 mL HBSS. Cells from both populations were washed twice in HBSS at 10,000×g/5 min/RT and the pellets resuspended in 1 mL lysis buffer (7 M urea, 2 M thiourea, 35 mM CHAPS, 20 mM DTT, 1 M NaCl). Samples were frozen at −80 °C for 30 min and then thawed at 34 °C for 20 min. Trichloroacetic acid (TCA) precipitation was performed by adding the bacterial samples to 100% ice-cold acetone and 100% trichloroacetic acid in a 1:8:1 ratio and precipitating at −20 °C for 1 h. Samples were then centrifuged (18,000×g/10 min/4 °C), the supernatant discarded, and the pellet washed twice with 1 mL ice-cold acetone (18,000 × g/10 min/4 °C). Acetone was removed, the pellet air-dried at room temperature, and resuspended in 0.1 M Triethylammonium bicarbonate (TEAB) plus 0.1% Rapigest. Protein sample validation was performed by 1DE gel electrophoresis. 19.5 μL sample was added to 7.5 μL NuPAGE LDS buffer and 3 μL NuPAGE reducing reagent, heated at 70 °C for 10 min, then run on a NuPAGE 4–12% Bis-Tris gel with MOPS buffer at 200 V for 50 min alongside a Novex Sharp standard. The gel was stained with SimplyBlue Safe Stain for 1 h, then destained with dH2O.
Protein processing, mass spectrometry, and database search
Protein samples were heated at 80 °C for 10 min, then DTT added at a final concentration of 2 mM and incubated at 60 °C for 45 min. Samples were then briefly vortexed, pulse spun, and cooled to room temperature before adding iodoacetamide to a final concentration of 6 mM. Samples were incubated at room temperature for 45 min (protected from light), vortexed and pulse spun briefly, then trypsin added at a final concentration of 1.3 µg/mL. Following incubation overnight at 37 °C (protected from light), trifluoroacetic acid (TFA) was added to a final concentration of 0.5% then incubated at 37 °C for 30 min. Samples were centrifuged at 13,000×g for 10 min at RT, the supernatants removed and vacuum concentrated. The resultant pellets were resuspended in 3% acetonitrile + 0.1% trifluoroacetic acid and peptide quantification performed using the Direct Detect system (Merck Millipore). Protein samples were normalized then vacuum concentrated in preparation for mass spectrometry.
Peptide extracts (1 μg on column) were separated on an Ultimate 3000 RSLC nano system (ThermoScientific) using a PepMap C18 EASY-Spray LC column, 2 μm particle size, 75 μm × 75 cm column (ThermoScientific) over a 140 min (single run) linear gradient of 3–25% buffer B (0.1% formic acid in acetonitrile (v/v)) in buffer A (0.1% formic acid in water (v/v)) at a flow rate of 300 nL/min. Peptides were introduced using an EASY‐Spray source at 2000 V to a Fusion Tribrid Orbitrap mass spectrometer (ThermoScientific). The ion transfer tube temperature was set to 275 °C. Full MS spectra were recorded from 300 to 1500 m/z in the Orbitrap at 120,000 resolution using TopSpeed mode at a cycle time of 3 s. Peptide ions were isolated using an isolation width of 1.6 amu and trapped at a maximal injection time of 120 ms with an AGC target of 300,000. Higher‐energy collisional dissociation (HCD) fragmentation was induced at an energy setting of 28 for peptides with a charge state of 2–4. Fragments were analyzed in the Orbitrap at 30,000 resolution. Analysis of raw data was performed using Proteome Discoverer software (ThermoScientific) and the data processed to generate reduced charge state and deisotoped precursor and associated product ion peak lists. These peak lists were searched against the P. aeruginosa MPAO1 protein database (a max. of one missed cleavage was allowed for tryptic digestion, the variable modification was set to contain oxidation of methionine and N-terminal protein acetylation, and carboxyamidomethylation of cysteine was set as a fixed modification). The FDR was estimated with randomized decoy database searches and was filtered to below 1% FDR at the protein level. Differentially abundant proteins were identified using DESeq289; significantly differentially abundant proteins had an adjusted (multiple testing corrected) p value ≤ 0.05 and a log2 fold change of ≥ 1 or ≤−1.
Proteogenomics
An iPtgxDB was created for P. aeruginosa MPAO1 as described previously31, using the NCBI annotation as anchor annotation. Ab initio gene predictions from Prodigal90 and ChemGenome91 and a modified in silico prediction that considers alternative start codons (TTG, GTG, CTG) and ORFs above 6 amino acids (aa) in length were integrated into a step-wise fashion. Proteomics data from MPAO1 cells grown planktonically or as biofilm were searched against this iPtgxDB with MS-GF+ (v2019.04.18)92 using Cysteine carbamidomethylation as fixed, and oxidation of methionine as variable modifications. Using the target-decoy approach of MS-GF+, the FDR at the PSM level was estimated and filtered below 0.2%. Only unambiguous peptides as identified by a PeptideClassifier analysis55, using the extension that supports proteogenomics for prokaryotes31, were considered.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank Jürg Frey and Daniel Frei (Agroscope) for generating Illumina MiSeq data. The authors acknowledge funding from the Joint Programming Initiative on AntiMicrobial Resistance (JPIAMR) and national grants to J.S.W. for R.N.A. (MRC MR/R005621/1), HvdM for OCO (ZonMW grant #547001003), Q.R. and M.T.B. for J.V. (SNSF grant 40AR40_173611), and to F.S. for F.P. (BMBF #01KI1710). C.H.A. acknowledges funding for A.R.V. through grants 156320 and 197391 from the SNSF. J.S.W. and R.N.A. would also like to thank Mark Willett and the Southampton Imaging and Microscopy Centre for support in performing biofilm imaging.
Author contributions
V.S. and A.R.V. carried out genome assembly. A.R.V. performed comparative genomics analyses, remapped existing Tn-seq data, created the iPtgxDB, performed proteogenomics analyses, and created figures with C.H.A. M.T.B. grew cells and extracted gDNA. J.D.P.V. devised and carried out the screening approach, overseen by Q.R. O.C.O. and H.C.v.d.M. designed the mold for the microfluidic flow chamber and J.W. provided device replication expertise. Microfluidic-confined biofilm growth and confirmed reproducibility were undertaken by R.N.A., J.S.W., O.C.O., H.C.v.d.M., F.P., and F.S. P.S. generated shotgun proteomics data from planktonic and biofilm cells provided by R.N.A. R.N.A. and J.S.W. carried out biofilm growth in the mold for selected mutants from the transposon mutant collection and analyzed proteomics data. C.H.A. oversaw genome sequencing and assembly, comparative genomics and proteogenomics, and wrote the manuscript together with input from R.N.A., F.S., J.V., A.R.V., and all other authors. A.R.V., R.N.A., and J.D.P.V. share co-first authorship.
Data availability
The MPAO1 genome sequence is available at NCBI Genbank (acc# CP027857; Bioproject: PRJNA438597, Biosample: SAMN08722738). Read data are available under SRR10153205 (Illumina) and SRR10153206 (PacBio). Proteomics data are available from PRIDE (acc# PXD017122) upon acceptance of the manuscript. The iPtgxDBs for P. aeruginosa MPAO1 and PAO1 are available from https://iptgxdb.expasy.org, both as a searchable protein database (FASTA format) and a GFF file, which can be loaded in a genome viewer and overlaid with experimental evidence. Biofilm growth data from the microfluidic chamber will be made available at 10.21253/DMU.c.4851483. To support technology dissemination, the polyurethane master molds of the microfluidic chambers are available upon request from the UoS/NBIC; a CAD file can be found as Supplementary Data 5. Code availability: all analyses presented rely on open source software or published code that is referenced.
Code availability
Open-source R code used for genomic, Tn-seq, and proteomics data processing is available from the authors upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Adithi R. Varadarajan, Raymond N. Allan, Jules D. P. Valentin.
Contributor Information
Adithi R. Varadarajan, Email: vadithi.ethz@gmail.com
Christian H. Ahrens, Email: christian.ahrens@agroscope.admin.ch
Supplementary information
Supplementary information is available for this paper at 10.1038/s41522-020-00154-8.
References
- 1.Moradali MF, Ghods S, Rehm BH. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front. Cell Infect. Microbiol. 2017;7:39. doi: 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stover CK, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 3.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 4.Mah TF, O’Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 5.Livermore DM. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 2002;34:634–640. doi: 10.1086/338782. [DOI] [PubMed] [Google Scholar]
- 6.Partridge SR, Kwong SM, Firth N, Jensen SO. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018;31:e00088-00017. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 2009;22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez L, Hancock RE. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 2012;25:661–681. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li XZ, Plesiat P, Nikaido H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 2015;28:337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boucher HW, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 11.Rice LB. Progress and challenges in implementing the research on ESKAPE pathogens. Infect. Control Hosp. Epidemiol. 2010;31(Suppl 1):S7–S10. doi: 10.1086/655995. [DOI] [PubMed] [Google Scholar]
- 12.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall CW, Mah TF. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017;41:276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- 14.Mulcahy LR, Burns JL, Lory S, Lewis K. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 2010;192:6191–6199. doi: 10.1128/JB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crabbe A, Jensen PO, Bjarnsholt T, Coenye T. Antimicrobial tolerance and metabolic adaptations in microbial biofilms. Trends Microbiol. 2019;27:850–863. doi: 10.1016/j.tim.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Kolpen M, et al. Hyperbaric oxygen sensitizes anoxic Pseudomonas aeruginosa biofilm to ciprofloxacin. Antimicrob. Agents Chemother. 2017;61:e01024–01017. doi: 10.1128/AAC.01024-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conibear TC, Collins SL, Webb JS. Role of mutation in Pseudomonas aeruginosa biofilm development. PLoS ONE. 2009;4:e6289. doi: 10.1371/journal.pone.0006289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McElroy KE, et al. Strain-specific parallel evolution drives short-term diversification during Pseudomonas aeruginosa biofilm formation. Proc. Natl Acad. Sci. USA. 2014;111:E1419–E1427. doi: 10.1073/pnas.1314340111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandler CE, et al. Genomic and phenotypic diversity among ten laboratory isolates of Pseudomonas aeruginosa PAO1. J. Bacteriol. 2019;201:e00595–00518. doi: 10.1128/JB.00595-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klockgether J, et al. Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J. Bacteriol. 2010;192:1113–1121. doi: 10.1128/JB.01515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobs MA, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallagher LA, Shendure J, Manoil C. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. MBio. 2011;2:e00315–00310. doi: 10.1128/mBio.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray JL, Kwon T, Marcotte EM, Whiteley M. Intrinsic antimicrobial resistance determinants in the superbug Pseudomonas aeruginosa. mBio. 2015;6:e01603–e01615. doi: 10.1128/mBio.01603-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SA, et al. General and condition-specific essential functions of Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA. 2015;112:5189–5194. doi: 10.1073/pnas.1422186112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc. Natl Acad. Sci. USA. 2015;112:4110–4115. doi: 10.1073/pnas.1419677112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poulsen BE, et al. Defining the core essential genome of Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA. 2019;116:10072–10080. doi: 10.1073/pnas.1900570116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gray AN, et al. High-throughput bacterial functional genomics in the sequencing era. Curr. Opin. Microbiol. 2015;27:86–95. doi: 10.1016/j.mib.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmid M, et al. Pushing the limits of de novo genome assembly for complex prokaryotic genomes harboring very long, near identical repeats. Nucleic Acids Res. 2018;46:8953–8965. doi: 10.1093/nar/gky726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmid M, et al. Comparative genomics of completely sequenced Lactobacillus helveticus Genomes provides insights into strain-specific genes and resolves metagenomics data down to the strain level. Front. Microbiol. 2018;9:63. doi: 10.3389/fmicb.2018.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Omasits U, et al. An integrative strategy to identify the entire protein coding potential of prokaryotic genomes by proteogenomics. Genome Res. 2017;27:2083–2095. doi: 10.1101/gr.218255.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olivas AD, et al. Intestinal tissues induce an SNP mutation in Pseudomonas aeruginosa that enhances its virulence: possible role in anastomotic leak. PLoS ONE. 2012;7:e44326. doi: 10.1371/journal.pone.0044326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winsor GL, et al. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016;44:D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ross MG, et al. Characterizing and measuring bias in sequence data. Genome Biol. 2013;14:R51. doi: 10.1186/gb-2013-14-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manz B, Volke F, Goll D, Horn H. Measuring local flow velocities and biofilm structure in biofilm systems with magnetic resonance imaging (MRI) Biotechnol. Bioeng. 2003;84:424–432. doi: 10.1002/bit.10782. [DOI] [PubMed] [Google Scholar]
- 36.MacCallum N, et al. Liquid-infused silicone as a biofouling-free medical material. ACS Biomater. Sci. Eng. 2014;1:43–51. doi: 10.1021/ab5000578. [DOI] [PubMed] [Google Scholar]
- 37.Nishijyo T, Haas D, Itoh Y. The CbrA-CbrB two-component regulatory system controls the utilization of multiple carbon and nitrogen sources in Pseudomonas aeruginosa. Mol. Microbiol. 2001;40:917–931. doi: 10.1046/j.1365-2958.2001.02435.x. [DOI] [PubMed] [Google Scholar]
- 38.Clarke DM, Loo TW, Gillam S, Bragg PD. Nucleotide sequence of the pntA and pntB genes encoding the pyridine nucleotide transhydrogenase of Escherichia coli. Eur. J. Biochem. 1986;158:647–653. doi: 10.1111/j.1432-1033.1986.tb09802.x. [DOI] [PubMed] [Google Scholar]
- 39.Colvin KM, et al. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 2012;14:1913–1928. doi: 10.1111/j.1462-2920.2011.02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodman AL, et al. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 41.Petrova OE, Sauer K. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 2009;5:e1000668. doi: 10.1371/journal.ppat.1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fernandez L, et al. Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob. Agents Chemother. 2010;54:3372–3382. doi: 10.1128/AAC.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McPhee JB, Lewenza S, Hancock RE. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 2003;50:205–217. doi: 10.1046/j.1365-2958.2003.03673.x. [DOI] [PubMed] [Google Scholar]
- 44.Filloux A, Hachani A, Bleves S. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology. 2008;154:1570–1583. doi: 10.1099/mic.0.2008/016840-0. [DOI] [PubMed] [Google Scholar]
- 45.Duong F, et al. The AprX protein of Pseudomonas aeruginosa: a new substrate for the Apr type I secretion system. Gene. 2001;262:147–153. doi: 10.1016/S0378-1119(00)00541-2. [DOI] [PubMed] [Google Scholar]
- 46.McGuffie BA, Vallet-Gely I, Dove SL. sigma factor and anti-sigma factor that control swarming motility and biofilm formation in Pseudomonas aeruginosa. J. Bacteriol. 2015;198:755–765. doi: 10.1128/JB.00784-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang W, et al. Extracellular matrix-associated proteins form an integral and dynamic system during Pseudomonas aeruginosa biofilm development. Front. Cell Infect. Microbiol. 2015;5:40. doi: 10.3389/fcimb.2015.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petrova OE, Schurr JR, Schurr MJ, Sauer K. Microcolony formation by the opportunistic pathogen Pseudomonas aeruginosa requires pyruvate and pyruvate fermentation. Mol. Microbiol. 2012;86:819–835. doi: 10.1111/mmi.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuchma SL, et al. Cyclic-di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa: the pilY1 gene and its impact on surface-associated behaviors. J. Bacteriol. 2010;192:2950–2964. doi: 10.1128/JB.01642-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borlee BR, et al. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 2010;75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Allsopp LP, et al. RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA. 2017;114:7707–7712. doi: 10.1073/pnas.1700286114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hachani A, et al. Type VI secretion system in Pseudomonas aeruginosa: secretion and multimerization of VgrG proteins. J. Biol. Chem. 2011;286:12317–12327. doi: 10.1074/jbc.M110.193045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaille C, Reimmann C, Haas D. Isochorismate synthase (PchA), the first and rate-limiting enzyme in salicylate biosynthesis of Pseudomonas aeruginosa. J. Biol. Chem. 2003;278:16893–16898. doi: 10.1074/jbc.M212324200. [DOI] [PubMed] [Google Scholar]
- 54.Kang D, Kirienko NV. Interdependence between iron acquisition and biofilm formation in Pseudomonas aeruginosa. J. Microbiol. 2018;56:449–457. doi: 10.1007/s12275-018-8114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qeli E, Ahrens CH. PeptideClassifier for protein inference and targeted quantitative proteomics. Nat. Biotechnol. 2010;28:647–650. doi: 10.1038/nbt0710-647. [DOI] [PubMed] [Google Scholar]
- 56.Flemming HC, et al. Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 57.Fraser CM, Eisen JA, Nelson KE, Paulsen IT, Salzberg SL. The value of complete microbial genome sequencing (you get what you pay for) J. Bacteriol. 2002;184:6403–6405. doi: 10.1128/JB.184.23.6403-6405.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Vrieze M, et al. Linking comparative genomics of nine potato-associated pseudomonas isolates with their differing biocontrol potential against late blight. Front. Microbiol. 2020;11:857. doi: 10.3389/fmicb.2020.00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Felnagle EA, et al. Nonribosomal peptide synthetases involved in the production of medically relevant natural products. Mol. Pharm. 2008;5:191–211. doi: 10.1021/mp700137g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gulick AM. Nonribosomal peptide synthetase biosynthetic clusters of ESKAPE pathogens. Nat. Prod. Rep. 2017;34:981–1009. doi: 10.1039/C7NP00029D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cotter PD, Hill C, Ross RP. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 2005;3:777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 62.Ghequire, M. G. K. & Ozturk, B. A colicin M-type bacteriocin from Pseudomonas aeruginosa targeting the HxuC heme receptor requires a novel immunity partner. Appl. Environ. Microbiol.84, 10.1128/AEM.00716-18 (2018). [DOI] [PMC free article] [PubMed]
- 63.Page R, Peti W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016;12:208–214. doi: 10.1038/nchembio.2044. [DOI] [PubMed] [Google Scholar]
- 64.Klausen M, et al. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 2003;48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- 65.Weiss Nielsen M, Sternberg C, Molin S, Regenberg B. Pseudomonas aeruginosa and Saccharomyces cerevisiae biofilm in flow cells. J. Vis. Exp. 2011 doi: 10.3791/2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yeung AT, Bains M, Hancock RE. The sensor kinase CbrA is a global regulator that modulates metabolism, virulence, and antibiotic resistance in Pseudomonas aeruginosa. J. Bacteriol. 2011;193:918–931. doi: 10.1128/JB.00911-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dietrich LE, et al. Bacterial community morphogenesis is intimately linked to the intracellular redox state. J. Bacteriol. 2013;195:1371–1380. doi: 10.1128/JB.02273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang F, et al. The Pseudomonas aeruginosa Type VI secretion PGAP1-like effector induces host autophagy by activating endoplasmic reticulum stress. Cell Rep. 2016;16:1502–1509. doi: 10.1016/j.celrep.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 69.Cheng Y, et al. Population dynamics and transcriptomic responses of Pseudomonas aeruginosa in a complex laboratory microbial community. NPJ Biofilms Microbiomes. 2019;5:1. doi: 10.1038/s41522-018-0076-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Varadarajan AR, et al. A proteogenomic resource enabling integrated analysis of listeria genotype-proteotype-phenotype relationships. J. Proteome Res. 2020;19:1647–1662. doi: 10.1021/acs.jproteome.9b00842. [DOI] [PubMed] [Google Scholar]
- 71.Hood RD, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Storz G, Wolf YI, Ramamurthi KS. Small proteins can no longer be ignored. Annu Rev. Biochem. 2014;83:753–777. doi: 10.1146/annurev-biochem-070611-102400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Impens F, et al. N-terminomics identifies Prli42 as a membrane miniprotein conserved in Firmicutes and critical for stressosome activation in Listeria monocytogenes. Nat. Microbiol. 2017;2:17005. doi: 10.1038/nmicrobiol.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Omasits U, et al. Directed shotgun proteomics guided by saturated RNA-seq identifies a complete expressed prokaryotic proteome. Genome Res. 2013;23:1916–1927. doi: 10.1101/gr.151035.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chin CS, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 76.Somerville V, et al. Long-read based de novo assembly of low-complexity metagenome samples results in finished genomes and reveals insights into strain diversity and an active phage system. BMC Microbiol. 2019;19:143. doi: 10.1186/s12866-019-1500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Okonechnikov K, Conesa A, Garcia-Alcalde F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2016;32:292–294. doi: 10.1093/bioinformatics/btv566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sedlazeck FJ, et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods. 2018;15:461–468. doi: 10.1038/s41592-018-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tatusova T, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arndt D, et al. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fernandez N, et al. An integrated systems approach unveils new aspects of microoxia-mediated regulation in Bradyrhizobium diazoefficiens. Front. Microbiol. 2019;10:924. doi: 10.3389/fmicb.2019.00924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Page AJ, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Darling AE, Mau B, Perna N. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997 (2013).
- 86.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heydorn A, et al. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 88.Mah, T. F. Establishing the minimal bactericidal concentration of an antimicrobial agent for planktonic cells (MBC-P) and biofilm cells (MBC-B). J. Vis. Exp. e50854, 10.3791/50854 (2014). [DOI] [PMC free article] [PubMed]
- 89.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hyatt D, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinforma. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Singhal P, Jayaram B, Dixit SB, Beveridge DL. Prokaryotic gene finding based on physicochemical characteristics of codons calculated from molecular dynamics simulations. Biophys. J. 2008;94:4173–4183. doi: 10.1529/biophysj.107.116392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim S, Pevzner PA. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MPAO1 genome sequence is available at NCBI Genbank (acc# CP027857; Bioproject: PRJNA438597, Biosample: SAMN08722738). Read data are available under SRR10153205 (Illumina) and SRR10153206 (PacBio). Proteomics data are available from PRIDE (acc# PXD017122) upon acceptance of the manuscript. The iPtgxDBs for P. aeruginosa MPAO1 and PAO1 are available from https://iptgxdb.expasy.org, both as a searchable protein database (FASTA format) and a GFF file, which can be loaded in a genome viewer and overlaid with experimental evidence. Biofilm growth data from the microfluidic chamber will be made available at 10.21253/DMU.c.4851483. To support technology dissemination, the polyurethane master molds of the microfluidic chambers are available upon request from the UoS/NBIC; a CAD file can be found as Supplementary Data 5. Code availability: all analyses presented rely on open source software or published code that is referenced.
Open-source R code used for genomic, Tn-seq, and proteomics data processing is available from the authors upon request.