Abstract
Three-dimensional (3D) cell cultures are rapidly emerging as a promising tool to model various human physiologies and pathologies by closely recapitulating key characteristics and functions of in vivo microenvironment. While high-throughput 3D culture is readily available using multi-well plates, assessing the internal microstructure of 3D cell culture models yet remains extremely slow because of manual, laborious and time-consuming histological procedures. Here we present a four-dimensional (4D) printed transformable tube array (TTA) using a shape memory polymer (SMP) to enable massively parallel histological analysis of 3D cultures. The interconnected TTA can be programmed to be expanded by 3.6 times of its printed dimension to match the size of a multi-well plate, with the ability to restore its original dimension for transferring all cultures to a histology cassette in order. Being compatible with microtome sectioning, the TTA allows for parallel histology processing for the entire samples cultured in a multi-well plate. The test result with human neural progenitor cell (hNPC) spheroids suggests a remarkable reduction in histology processing time by an order of magnitude. High-throughput analysis of 3D cultures enabled by our TTA has great potential to further accelerate innovations in various 3D culture applications such as high-throughput/content screening, drug discovery, disease modeling, and personalized medicine.
Keywords: 3D cell culture, 4D printing, high-throughput/content screening, parallel histology, shape memory polymers
Graphical Abstract
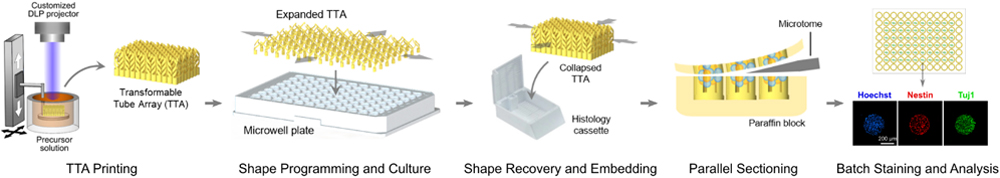
4D printed transformable culture tube array (TTA) can directly transfer a large number of 3D culture models from a microwell plate to a histology cassette, enabling rapid and parallel histological analysis. A seamless workflow from cell culture to histological analysis is demonstrated with human neural progenitor cell (hNPC) spheroids cultured in the TTA.
Cell-based assays and 2D/3D culturing cells in a controlled microenvironment have received considerable attention in the areas of high-throughput/content drug screening and stem cell-based personalized medicine, as these research tools are critical to investigate the formation, functionality, and pathophysiology of biological tissues’ development.[1,2] To better recapitulate cellular behavior in vivo under specific physiological conditions, research efforts have been shifting from traditional two-dimensional (2D) culture where cells spread and adhere to a flat substrate to three-dimensional (3D) cell culture where tissue morphology and cell-to-cell interactions are closer to those found in biology.[3–5] Emerging 3D cell culture models include spheroids and organoids, which are cell aggregates derived from one particular cell type or mixtures of multiple cell types, respectively. Because of their ability to emulate morphological and functional characteristics of in vivo biology, 3D culture models show great potential to provide better insight into cell differentiation, disease processes, and drug discovery, delivery, and efficacy.[6–8] In addition, the use of a standard multi-well plate with compatible tools and instruments such as multichannel pipettes and robotic dispensing systems[9] has enabled parallel culture of a massive number of spheroids and organoids.
While high-throughput culture is viable using widely used multi-well plates, analysis yet remains extremely slow because of the need for assessing the internal microstructure of 3D cell assemblies using histological processes. Histological methods used for studying the cellular microstructures of biological tissues entail obtaining thin slices of tissues, which reveals cell morphology, spatial arrangement, biological heterogeneity, and their relationships to tissue functionality.[10] The histological analysis of 3D cell culture models typically requires a series of laborious, time-consuming, and mostly manual procedures of sample fixation, paraffin embedding, repetitive microtome sectioning, and staining, which usually takes many hours of tedious work for a specialist to complete a single specimen.[11] In addition, harvesting 3D culture models from a microwell plate and transferring them to a histology cassette is a challenging process due to the small physical dimension, sticky formaldehyde-fixed surface protein layer,[12,13] and delicate nature of specimens.[14,15] Furthermore, this demanding process has to be repeated for each specimen when a large number of 3D culture assays are to be investigated. There have been some efforts to speed up histology analysis using specially designed arrays,[16–19] but they are not compatible with standard culture devices such as multi-well plates, thus still requiring harvesting and transferring steps. These painfully slow and labor-intensive histological processes have been a significant bottleneck to rapid and efficient analysis of 3D cell culture models.
To advance beyond this long-standing barrier and enable high-throughput analysis of 3D cell cultures, we report a transformable tube array (TTA) that can directly transfer a large number of 3D culture models from a microwell plate to a much smaller histology cassette in the same configuration as a single specimen. In order to address the size difference between the two standard devices,[20,21] we employed four-dimensional (4D) printing to create a TTA with a temperature-responsive shape memory polymer (SMP). 4D printing refers to 3D printing of smart materials whose shapes, properties, and functions evolve with time in a programmed way.[22,23] In particular, a temperature-responsive SMP can be programmed into a temporary shape (shape programming) which can return to the original shape (shape recovery) when heated,[24] which has been utilized in various biomedical applications.[25–28] As illustrated in Figure 1, the TTA consists of an array of cell culture tubes, each of which is comprised of a lower cell chamber for cell culture and upper helical connecting arms for transformation. After 3D printing with an SMP in the size of a histology cassette, it is biaxially stretched and fixed in the size of a standard microwell plate. The SMP helical arms are unwound to facilitate large deformation and lock the TTA in the expanded shape. The TTA is then mounted on a multiwell plate, followed by typical high-throughput 3D cell culture. Once the culture is completed, the TTA can transform back to the original size through shape memory recovery, bringing the entire array of samples into a small area that fits into a standard histology cassette. Since the printed SMP can be sectioned by a microtome, all culture samples within the TTA are cut through together, generating an array of histology sections on a single microscopy slide while maintaining the registry of specimens in the microwell plate. By eliminating the need for a series of harvesting, transferring, and repetitive sectioning of individual specimens, the TTA enables rapid and parallel histology of the entire cell culture array.
Figure 1.
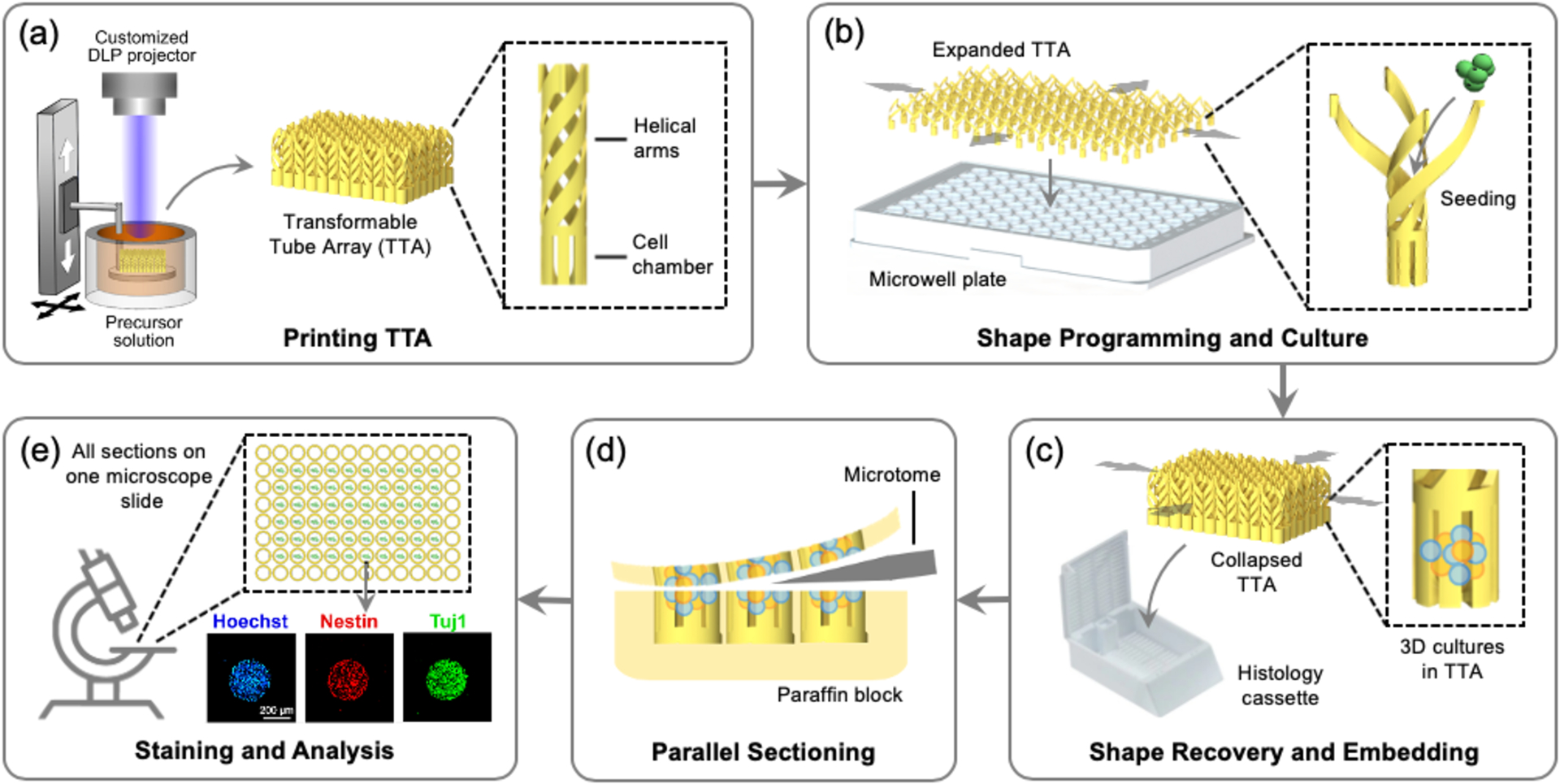
High-throughput cell culture and histology with a 4D printed transformable tube array (TTA). (a) The TTA is printed using projection micro-stereolithography. (b) The TTA is expanded and mounted on a microwell plate. Cell are seeded and cultured in the TTA. (c) The TTA, together with the entire 3D culture, collapses into its original size and inserted into a histology cassette for embedding. (d) All 3D culture models are sectioned together. (e) The sectioned samples are deposited on one microscope slide for analysis.
For the 3D printable SMP, we prepared a photo-curable precursor solution with poly(ethylene glycol) diacrylate (average Mn 250) (PEGDA250) and bisphenol A ethoxylate dimethacrylate (average Mn 1700) (BPA) (see details in Supporting Information). The glass transition temperature Tg of the SMP can be tailored by a weight ratio of each polymer.[29,30] While a higher Tg of the SMP is advantageous in terms of retaining the expanded shape of the TTA after stretching, biological samples may be degraded if the temperature is raised too high to trigger shape memory recovery. Based on the paraffin wax melting temperature of 54 oC[31] and processing temperature of ~60 oC in the histology process, we tuned Tg of the SMP to 60 oC by using a 9:1 weight ratio of PEGDA:BMA (Figure S1). Note that cultured biological samples are already fixed (not alive) when the TTA undergoes shape memory recovery at Tg. Figure 2a shows the dynamic mechanical analysis (DMA) results of 3D printed SMP samples. The high stiffness of 1.2 GPa at room temperature keeps the TTA rigid for stable handling and operation. The shape memory behavior of an SMP is characterized by shape-fixity and shape-recovery ratios, respectively (see details in Supporting Information).[32] The shape programming and recovery of the SMP are shown in Figure 2b (see details in Supporting Information). First, an SMP film was uniaxially stretched to a strain of 8% at 60 oC. While maintaining the strain, the temperature was dropped down to and maintained at 25 oC for 10 min. The programmed strain was 7% after stress was released, which gives a shape-fixity ratio of 87%. Shape recovery triggered at 60 oC brought the strain down to 0.5%, which gives a shape-recovery ratio of 94%. The strain at failure of the SMP was also measured from uniaxial tensile testing at five different temperatures between 25 oC and 60 oC, as shown in Figure 2c. While stiffness decreases with temperature, as also shown in the DMA result, strain at failure is relatively constant at around 10% across all temperatures tested (Figure 2d), which determines the maximum deformation limit of the TTA without mechanical failure.
Figure 2.
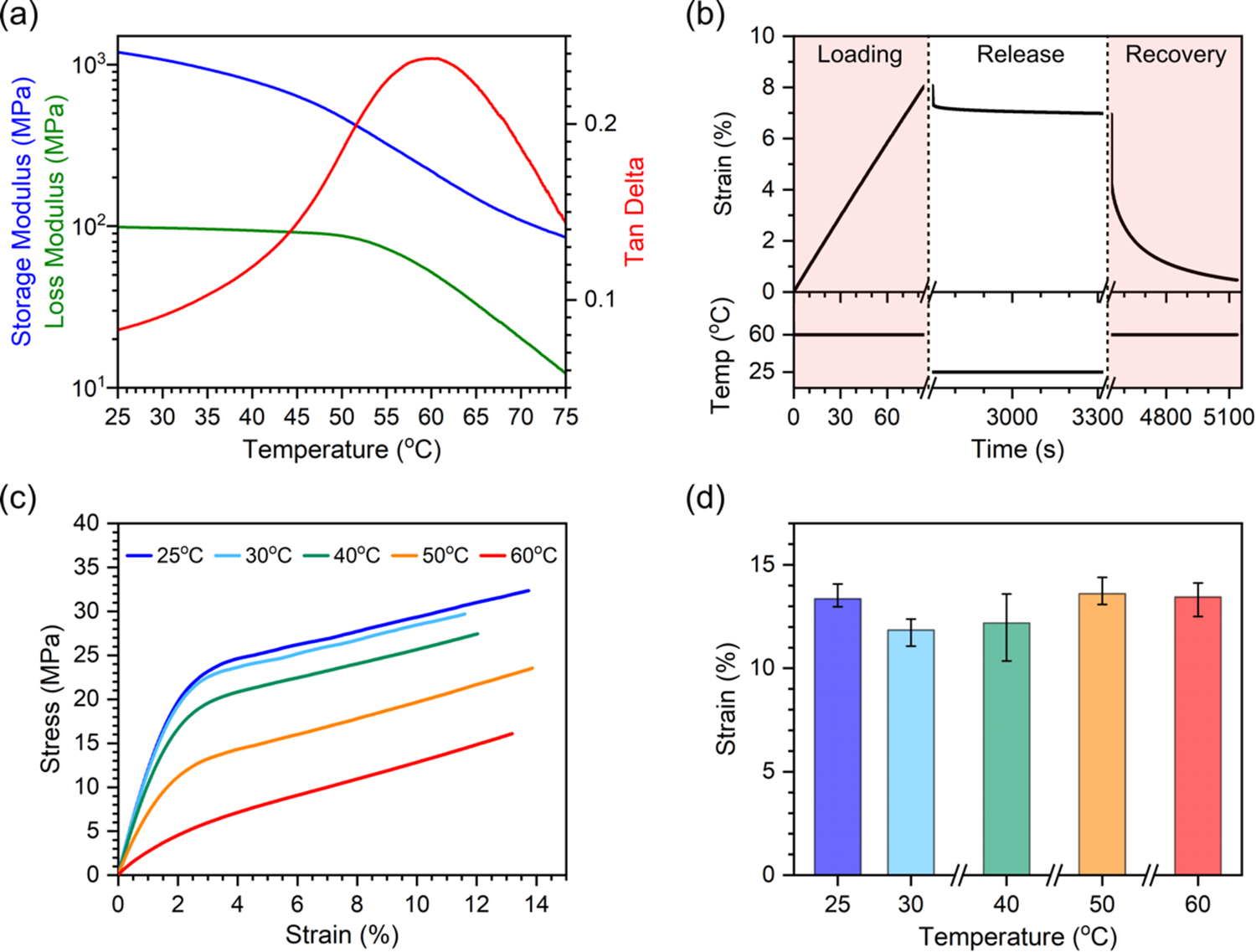
Thermomechanical properties of the SMP. (a) Storage modulus, loss modulus and tanδ of the SMP. (b) Shape programming and recovery of the SMP. (c) Stress-strain curves of the SMP at 5 different temperatures. (d) Failure strain of SMP at 5 different temperatures. (n = 3 for each temperature. Error bars represent maximum and minimum values.)
While the TTA can be designed for various standard microwell plates having 6, 24, 96, and 384 wells, we used the most widely used 96-well plate in this work. A 96-well plate has 12 × 8 wells with an overall lateral dimension of 101 × 65 mm and a center-to-center distance between neighboring wells of 9 mm.[20] A histology cassette, however, can fit an object with a lateral size only up to 30 × 25 mm inside.[21] To fit the interconnected 12 × 8 array of culture tubes in a histology cassette in its original dimension, the culture tube was designed to have a diameter of 2.5 mm as shown in Figure 3a (detailed dimensions in Figure S3a). When biaxially stretched, the helical arms unwind to increase the extent of the culture tube to 9 mm, as shown in Figure 3b, thereby matching the size of the expanded TTA to that of a 96-well plate. Finite element simulation showed that the maximum local strain in the stretched state is 8.4% occurring at the connecting points of the arms, as shown in Figure 3c, which falls within the material’s ability to stretch without failure shown in Figure 2d (see details in Supporting Information). While mounted on a 96-well plate for cell culture, each culture tube in the TTA is inserted in the corresponding microwell of the 96-well plate filled with culture media. To facilitate culture media exchange, the cell chamber was designed to have vertical slits with a width of 150 μm (Figure S3a). The slit width was determined by the size of the 3D cell culture models used in this work, human neural progenitor cells (hNPCs). They have a diameter of 400 ~ 500 μm, so they can be retained in the cell chamber throughout culture and histology processes.
Figure 3.
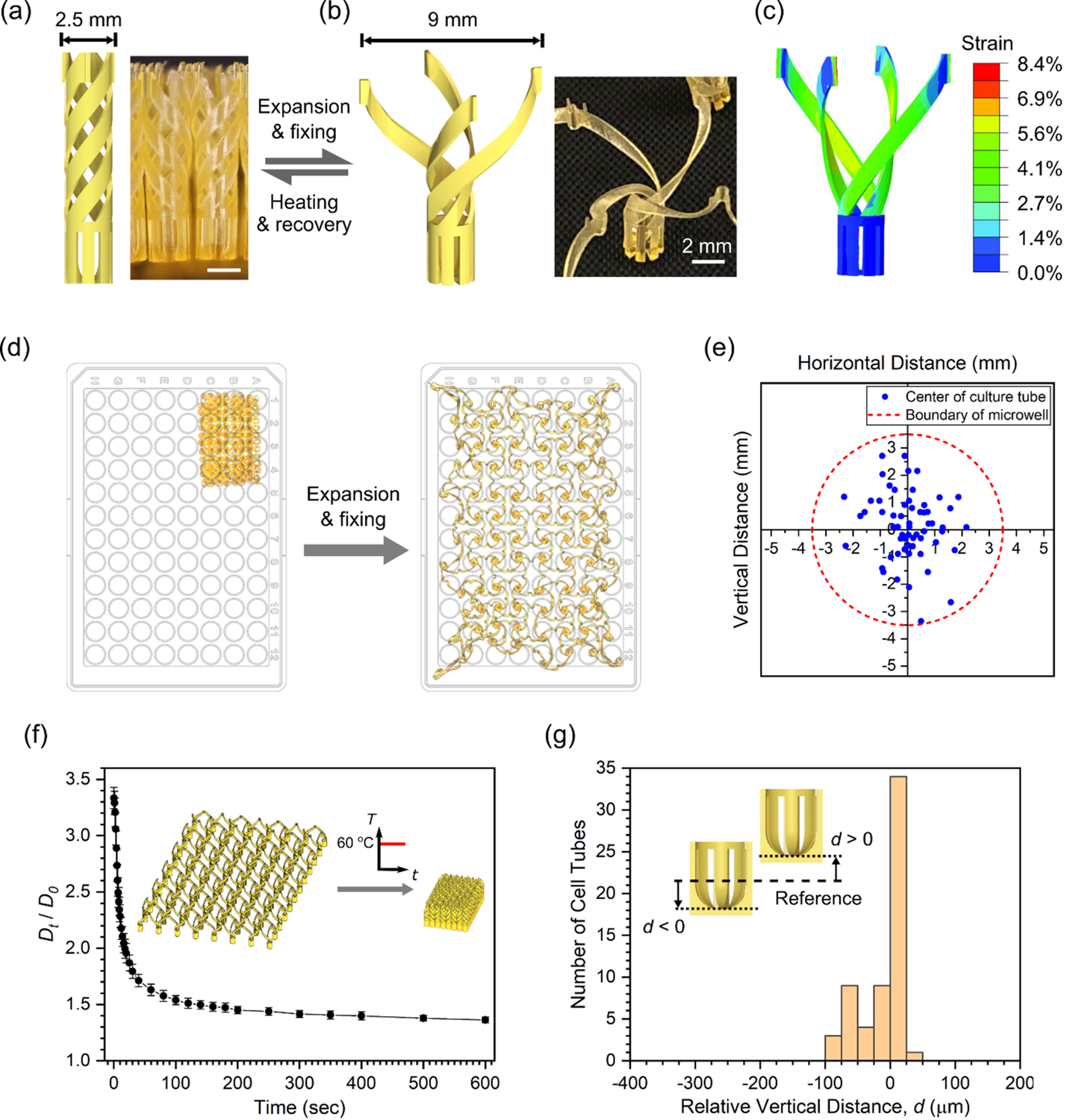
Design of the TTA and characterization of TTA performance. (a) Culture tube in the collapsed state. (b) Culture tube in the expanded state. (c) Numerical simulation of the culture tube expansion. Scale bars in (a) and (b) are 2 mm. (d) Stretching and shape fixing of the TTA to match a 96-well plate. Actual photos of the TTA are overlaid on a 96-well plate for size comparison. (e) Positions of the center of culture tubes relative to corresponding microwells in the stretched state. (f) Shape recovery of the TTA upon heating at 60 °C. (n = 6 for each time point. Error bars represent a standard deviation.) (g) Distribution of relative vertical positions of cell chambers from a reference sectioning plane.
To realize this intricate 3D design of the interconnected TTA using the SMP, we used projection micro-stereolithography (PμSL). PμSL is a high precision additive manufacturing technique where the digital projection of UV light is used to lithographically solidify layers in series to build a 3D object.[33–35] After printing, post-processing procedures were taken to crosslink the SMP fully and to remove residual cytotoxic materials (see details in Supporting Information). For shape programming of the TTA to the expanded state, the printed TTA was stretched using a custom-made biaxial stretcher (Figure S6). Figure 3d shows images of the TTA overlaid on a 96-well plate before and after stretching, showing the significant lateral expansion of the TTA. To assess how accurately culture tubes are registered to the corresponding microwells in a 96-well plate after shape programming, we measured the location of each culture tube of a TTA relative to the center of the microwell using image analysis. Relative positions of the culture tubes in the expanded state (excluding those around edges to be used for the fixture, as shown in Figure S7) are plotted in Figure 3e, where a dotted line indicates the boundary of each microwell with a radius of 3.5 mm. The result shows that all of the 60 cell culture tubes of the TTA fall within the boundary, validating that biaxial stretching and shape programming transforms the TTA in such a way that each culture tube is positioned at its intended location, and thus easily inserted into the microwells when the TTA is mounted on a 96-well plate. This result was consistently reproduced with different TTA samples (Figure S10a, see details in Supporting Information).
The transformable TTA utilizes temperature-responsive shape recovery to transfer the culture samples from a 96-well plate to a smaller histology cassette (see Movie S1). Figure 3f shows the time evolution of the TTA shape recovery performed at 60 oC (see details in Supporting Information). Dt/D0 is the normalized expansion with respect to the original size, where D0 and Dt are averaged sizes of culture tubes in the printed state and at each time point, respectively. In the experiment, 95% of the shape recovery took place in the initial 120 sec of heating, followed by notably slow but continuous recovery. Although recovery did not reach 100% due to friction from the surface, the TTA collapsed further when displaced from the surface and fitted into a histology cassette. Similar shape recovery deformation was consistently reproduced by different TTA samples (Figure S10b, see details in Supporting Information). To minimize excessive evaporation of water, a 2 min of recovery time was given at 60 oC for the functional testing with cell culture models in the TTA.
Once transferred to a histology cassette, 3D cell culture models in the TTA go through dehydration and paraffin processing using an automated tissue processor, followed by typical paraffin embedding. Molten paraffin wax is poured into an embedding mold and cooled to generate a paraffin block, which is sliced into 5–15 μm thick sections using a microtome. Since 3D models are in suspensions near the bottom of the cell chambers, it is important that the cell chambers are vertically positioned at the same height so that all 3D models in the TTA are cut through together into one section. An embedding routine for the TTA was developed (see details in Supporting Information) and evaluated by tracking vertical locations of the bottom of each cell chamber from consecutive 5 μm-thick sections. When sectioning starts from the bottom of the cell chamber, a snowflake-shape cross-section with a solid center appears first (Figure S9b). As the sectioning proceeded upward, a hollow center starts to show when the inner side of the cell chamber is reached. With a reference plane (d = 0) defined by a section with the half of the cell chambers showing a hollow center (Figure S9c), we measured relative vertical positions of the sectioned cell chambers in the TTA. The distribution in Figure 3g shows that all cell chambers in the TTA are sectioned together with a vertical position variation not exceeding 150 μm. This result was consistently reproduced from different TTA samples (Figure S10c, see details in Supporting Information), suggesting that the TTA can enable parallel sectioning and deposition of the entire 3D culture array from one paraffin block onto one microscope slide for any 3D cultures larger than 150 μm, including typical spheroids and organoids. This condition is also consistent with the design criterion for 150 μm-wide slits in the cell chamber (Figure S3a).
The parallel histology processing capability of the TTA was first demonstrated using a non-biological material. After a printed TTA was stretched and programmed to the expanded configuration, the TTA was mounted onto a 96-well plate. The cell chambers were then filled with polymer precursor solutions with two different fluorescent dyes, rhodamine B (λex=540 nm, λem=625 nm) and DiOC2 (λex=482 nm, λem=497 nm), in an alternating arrangement, followed by UV curing, as shown in Figure 4a. The TTA was removed from the 96-well plate, shape recovered at 60 oC, and inserted into a histology cassette. After standard tissue processing, paraffin embedding and microtome sectioning, a thin section was obtained, as shown in Figure 4b (see details in Figure S11 and Supporting Information). This section clearly shows that the TTA can enable a parallel histology examination for the entire array of samples while maintaining the registry of the samples in the 96-well plate configuration, which can potentially lead to rapid combinatorial studies for disease modeling and drug screening.
Figure 4.
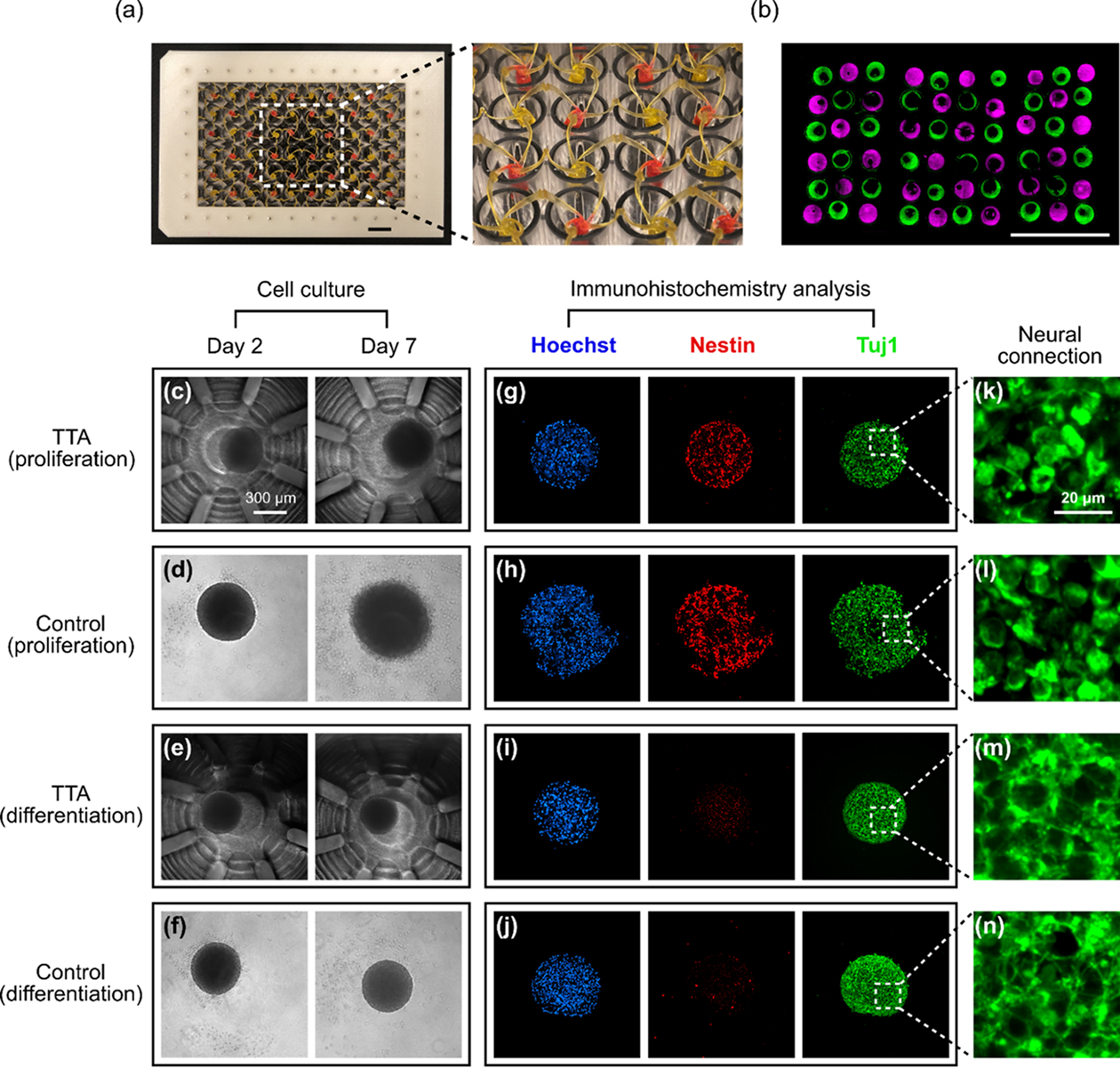
Parallel histological processing with the TTA and 3D cell culture and histological analysis of hNPC spheroids using the TTA and a ULA plate (control). (a) The TTA filled with alternating dyes (green: DiOC2, magenta: Rhodamine B). Red signal from Rhodamine B was replaced with magenta for readers with red-green color-blindness. (b) A sectioned slide showing alternating color under a fluorescent microscope. Scale bars in (a) and (b) are 10 mm. (c-f) hNPC spheroids culture. Scale bar in (c) is 300 μm. (g-j) Immunohistochemistry analysis of hNPC spheroids (Hoechst: nucleus marker; Nestin: neural stem cell marker; Tuj1: early neural differentiation marker). (k-n) Magnified images of Tuj1 staining. Stronger dendrites-synapse connections are observed in the differentiation group, (m) and (n). Scale bar in (k) is 20 μm.
To demonstrate the use of the TTA for 3D cell culture, human neural progenitor cells (hNPCs) were assembled into neurospheres and differentiated through growth factor withdrawal.[36,37] Cell viability test results confirmed the biocompatibility of the TTA with a variety of cells including hNPCs (Figure S12). After routine culture passage, hNPCs were seeded into a 96-well, non-treated, PCR reaction plate (~500 μm diameter) to self-assemble into neurospheres overnight. The newly formed spheroids were then transferred to the TTA and cultured in either proliferation or differentiation (i.e. growth factor-withdrawn) media. As a control set, some hNPC neurospheres were transferred to a commercially available ultra-low attachment (ULA) 96-well plate to examine whether TTA affected proliferation, differentiation, or spheroid morphology. Neurospheres were examined daily via phase-contrast microscopy to monitor gross morphological changes. Images from Day 2 (transfer from self-assembly plate to TTA or ULA plate) and Day 7 are shown in Figure 4c to 4f. Neurosphere morphology remained largely unchanged from Day 2 to Day 7, though a small increase in size was seen in neurospheres cultured in proliferation media as reflected in Figure 4c and 4d. At the end of the 7-day spheroid culture, neurospheres were formaldehyde-fixed, embedded in paraffin wax, sectioned, and immunostained for characterization, as shown in Figure 4g to 4j. Spheroid shape distortions from control groups seen in the immunostained images in Figure 4h and 4j were caused by spheroid transfer after fixing in the ULA plate. Other than that, we observed negligible morphological differences between neurospheres cultured in the TTA or the control for each media condition under Hoechst (nucleus) staining. Most strikingly, culturing neurospheres with differentiation media, regardless of TTA or ULA plate culture, resulted in a significant decrease in Nestin (neural stem cell marker), as shown in Figure 4g to 4j. Concurrently, Tuj1 (early neural differentiation marker) staining images showed stronger dendrites-synapse connections when neurospheres were cultured in differentiation media, indicated by an increased number of web-like connected microstructures as shown in the magnified images in Figure 4m and 4n, compared to those that remained more discrete when cultured in proliferation media as shown in Figure 4k and 4l (see Supporting Information and Figure S13 for the difference in connectedness highlighted by image processing). Since neurospheres cultured in the TTA followed expected protein expression patterns (decreased stem cell markers Nestin and increased neural Tuj1 density in response to differentiation media) and did not differ from neurospheres cultured in commercially available multiwell-plates, we can conclude that the TTA is a suitable high-throughput culture and histological analysis tool for 3D cultures. Based on the estimated processing time for spheroids, it would take only 8 hours for preparation of 60 immunohistology samples, while conventional approach using the ULA plate can take up to 100 hours in total, suggesting more than a 10-fold reduction in processing time as well as associated labor costs (Table S3). Based on the biocompatibility with other cell lines (Figure S12), the TTA can also be used for other spheroids and 3D cell culture models (e.g. organoids).
This work presents a 4D printed transformable tube array that can enable massively parallel histological analysis of 3D cell culture models. Through shape programming and recovery of the interconnected helical arms, the TTA can morph between two different configurations for a 96-well plate and a histology cassette, which are 3.6 times different in lateral size. The shape recovery temperature of the 3D printable SMP was tailored to match the existing histology processing temperature. We successfully demonstrated the rapid transfer of all culture models from a microwell plate to a histology cassette without the repetition of manual operations. Using the TTA, an array of 3D biological models can be cultured in a microwell plate, and later sectioned together and deposited onto a microscope slide as a whole array for parallel examination. Further, hNPC spheroids cultured in differentiation media using the TTA showed significantly stronger dendrites-synapse connections, similar to those cultured using a commercial ULA plate. Our estimation suggests that the TTA can reduce the histology processing time by an order of magnitude when a large number of 3D culture assays are analyzed. Being biocompatible with multiple cell lines, the TTA has a great potential to accelerate the pace of innovations in a broad range of 3D culture applications including disease modeling, drug development and testing in both high-throughput and high-content manner. In addition to a remarkable reduction in time and labor costs, the unprecedented high-throughput parallel histology using our TTA may also enable rapid targeted and personalized therapy for aggressive tumors and cancers.
Supplementary Material
Acknowledgements
This work was supported in part with Federal funds from the National Cancer Institute (NCI), National Institutes of Health (NIH), under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. H.L. acknowledges partial financial support from the National Science Foundation (NSF) (CBET-1804591). H.E.S. acknowledges partial financial support from Rutgers Cancer Institute of New Jersey Shared Resources with funding from NCI-CCSG (P30CA072720), NCI (5R01CA226746). K.-B. Lee acknowledges partial financial support from the NSF (CHE-1429062) and NIH R01 (1R01DC016612, and 3R01DC016612-01S1). S.A. Chester acknowledges partial financial support from the NSF (CMMI-1751520).
Footnotes
Competing interests
Rutgers University has filed a patent application related to this work (US62/664,740). H.E.S. is the scientific founder of Celvive, Inc. The authors declare no other competing interest.
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Chen Yang, Department of Mechanical and Aerospace Engineering, Rutgers University-New Brunswick, 98 Brett Road, Piscataway, NJ 08854, USA.
Jeffrey Luo, Department of Chemistry and Chemical Biology, Rutgers University-New Brunswick, 123 Bevier Rd, Piscataway, NJ 08854, USA.
Marianne Polunas, Research Pathology Services, Rutgers University-New Brunswick, 41 Gordon Road, Suite B, Piscataway, NJ 08854, USA.
Nikola Bosnjak, Department of Mechanical Engineering, New Jersey Institute of Technology, 200 Central Ave, Newark, NJ 07102, USA.
Sy-Tsong Dean Chueng, Department of Chemistry and Chemical Biology, Rutgers University-New Brunswick, 123 Bevier Rd, Piscataway, NJ 08854, USA.
Shawn A. Chester, Department of Mechanical Engineering, New Jersey Institute of Technology, 200 Central Ave, Newark, NJ 07102, USA
Ki-Bum Lee, Department of Chemistry and Chemical Biology, Rutgers University-New Brunswick, 123 Bevier Rd, Piscataway, NJ 08854, USA.
Howon Lee, Department of Mechanical and Aerospace Engineering, Rutgers University-New Brunswick, 98 Brett Road, Piscataway, NJ 08854, USA.
References
- [1].Harrison RG, Greenman MJ, Mall FP, Jackson CM, Anat. Rec 1907, 1, 116. [Google Scholar]
- [2].Villa‐Diaz LG, Ross AM, Lahann J, Krebsbach PH, STEM CELLS 2013, 31, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Huh D, Hamilton GA, Ingber DE, Trends Cell Biol. 2011, 21, 745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Breslin S, O’Driscoll L, Drug Discov. Today 2013, 18, 240. [DOI] [PubMed] [Google Scholar]
- [5].Haycock JW, in 3D Cell Cult. Methods Protoc (Ed.: Haycock JW), Humana Press, Totowa, NJ, 2011, pp. 1–15. [Google Scholar]
- [6].Hirschhaeuser F, Menne H, Dittfeld C, West J, Mueller-Klieser W, Kunz-Schughart LA, J. Biotechnol 2010, 148, 3. [DOI] [PubMed] [Google Scholar]
- [7].Bates RC, Edwards NS, Yates JD, Crit. Rev. Oncol. Hematol 2000, 36, 61. [DOI] [PubMed] [Google Scholar]
- [8].Stansley B, Post J, Hensley K, J. Neuroinflammation 2012, 9, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kempner ME, Felder RA, JALA J Assoc. Lab. Autom 2002, 7, 56. [Google Scholar]
- [10].Ross MH, Pawlina W, Histology, Lippincott Williams & Wilkins, 2006. [Google Scholar]
- [11].Gartner LP, Hiatt JL, Color Textbook of Histology E-Book, Elsevier Health Sciences, 2006. [Google Scholar]
- [12].Solt P, Konnerth J, Gindl-Altmutter W, Kantner W, Moser J, Mitter R, van Herwijnen HWG, Int. J. Adhes. Adhes 2019, 94, 99. [Google Scholar]
- [13].Brüningk SC, Rivens I, Box C, Oelfke U, ter Haar G, Sci. Rep 2020, 10, 1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Clemson University, Alexis F, Cytol. Tissue Biol 2014, 1, 1. [Google Scholar]
- [15].Massai D, Isu G, Madeddu D, Cerino G, Falco A, Frati C, Gallo D, Deriu MA, Falvo D’Urso Labate G, Quaini F, Audenino A, Morbiducci U, PloS One 2016, 11, e0154610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kononen J, Bubendorf L, Kallionimeni A, Bärlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallionimeni O-P, Nat. Med 1998, 4, 844. [DOI] [PubMed] [Google Scholar]
- [17].Sabaliauskas NA, Foutz CA, Mest JR, Budgeon LR, Sidor AT, Gershenson JA, Joshi SB, Cheng KC, Methods 2006, 39, 246. [DOI] [PubMed] [Google Scholar]
- [18].Gabriel J, Brennan D, Elisseeff JH, Beachley V, Sci. Rep 2019, 9, 16287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Parker RN, Cairns DM, Wu WA, Jordan K, Guo C, Huang W, Martin‐Moldes Z, Kaplan DL, Adv. Healthc. Mater 2020, 2000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Microplate Dimensions, Working Volumes and Packaging, https://www.perkinelmer.com/lab-products-and-services/application-support-knowledgebase/microplates/plate-dimensions.html, accessed: 06, 2020. [Google Scholar]
- [21].Tissue Embedding, Tissue Processing, and Biopsy Cassettes, https://www.emsdiasum.com/microscopy/products/histology/cassettes.aspx, accessed: 06, 2020. [Google Scholar]
- [22].Tibbits S, Archit. Des 2012, 82, 68. [Google Scholar]
- [23].Ge Q, Qi HJ, Dunn ML, Appl. Phys. Lett 2013, 103, 131901. [Google Scholar]
- [24].Lendlein A, Kelch S, Angew. Chem. Int. Ed 2002, 41, 2034. [PubMed] [Google Scholar]
- [25].Lendlein A, Langer R, Science 2002, 296, 1673. [DOI] [PubMed] [Google Scholar]
- [26].Small W IV, Singhal P, Wilson TS, Maitland DJ, J. Mater. Chem 2010, 20, 3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Miao S, Zhu W, Castro NJ, Nowicki M, Zhou X, Cui H, Fisher JP, Zhang LG, Sci. Rep 2016, 6, 27226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Miao S, Cui H, Nowicki M, Xia L, Zhou X, Lee S-J, Zhu W, Sarkar K, Zhang Z, Zhang LG, Adv. Biosyst 2018, 2, 1800101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gordon M, Taylor JS, J. Appl. Chem 1952, 2, 493. [Google Scholar]
- [30].Yakacki CM, Shandas R, Safranski D, Ortega AM, Sassaman K, Gall K, Adv. Funct. Mater 2008, 18, 2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].ParaProTM LMP, https://www.statlab.com/histology/tissue-processing-embedding/paraffin/pplmp.html, accessed: 06, 2020.
- [32].Tobushi H, Shimada D, Hayashi S, Endo M, Proc. Inst. Mech. Eng. Part J. Mater. Des. Appl 2003, 217, 135. [Google Scholar]
- [33].Sun C, Fang N, Wu DM, Zhang X, Sens. Actuators Phys 2005, 121, 113. [Google Scholar]
- [34].Ge Q, Sakhaei AH, Lee H, Dunn CK, Fang NX, Dunn ML, Sci. Rep 2016, 6, srep31110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yang C, Boorugu M, Dopp A, Ren J, Martin R, Han D, Choi W, Lee H, Mater. Horiz 2019, 6, 1244. [Google Scholar]
- [36].Palmer TD, Ray J, Gage FH, Mol. Cell. Neurosci 1995, 6, 474. [DOI] [PubMed] [Google Scholar]
- [37].Kuhn HG, Winkler J, Kempermann G, Thal LJ, Gage FH, J. Neurosci 1997, 17, 5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


