Abstract
Background
Platinum-based chemotherapy plays an antitumor role by damaging DNA. X-ray repair crosscomplementing protein 1 (XRCC1) participates in DNA repair and thus affects the sensitivity to platinum drugs. Two polymorphisms of XRCC1, rs25487 (Arg399Gln) and rs1799782 (Arg194Trp), have been widely studied for the association with clinical outcomes of platinum-based chemotherapy in Asian patients with non-small-cell lung cancer (NSCLC), but the results remain inconclusive. Thus, we performed the present meta-analysis.
Methods
Literature search was performed in PubMed, Web of Science, and EMBASE up to June 2019. Odds ratios (ORs) for objective response ratio (ORR), Cox proportional hazard ratios (HRs) of overall survival (OS) and progression-free survival (PFS), and the corresponding 95% confidence intervals (95% CIs) were calculated to assess the association strengths between XRCC1 polymorphisms and clinical outcomes. Comparisons were performed in homozygous, heterozygous, dominant, and recessive models.
Results
Finally, a total of 23 studies involving 5567 patients were included in the meta-analysis. Compared to ArgArg of rs25487, GlnGln (OR = 1.71, 95% CI: 1.16-2.52, p = .007, I2 = 56.8%) and GlnArg (OR = 1.23, 95% CI: 1.07-1.40, p = .003, I2 = 29.0%) were associated with higher ORR. Meanwhile, GlnGln indicated a favorable OS (HR = 0.60, 95% CI: 0.40-0.88) and PFS (HR = 0.64, 95% CI: 0.46-0.90). We also found positive associations between rs1799782 and ORR in all comparison models with low between-study heterogeneity. The association strength increased with the number of variant alleles (TrpTrp vs. ArgArg: OR = 1.73, 95% CI:1.31-2.27; TrpArg vs. ArgArg: OR = 1.28, 95% CI: 1.06-1.55), suggesting a gene dosage effect. In addition, TrpTrp predicted a longer OS.
Conclusion
Our results showed that rs25487 and rs1799782 of XRCC1 are potential markers to predict clinical outcomes of platinum-based chemotherapy in Asian patients with NSCLC.
1. Introduction
Lung cancer is the most prevalence malignant tumor and the leading cause of cancer deaths worldwide. It accounts for about 13% of newly diagnosed cancers and is responsible for 17.6% of cancer-related deaths each year1. Lung cancer can be histologically classified as non-small-cell lung cancer (NSCLC) and small cell lung cancer (SCLC). NSCLC, accounting for ~85% of all cases, includes several subtypes: squamous cell carcinoma, adenocarcinoma, large cell carcinoma, and the other subtypes2. The prognosis of lung cancer is usually poor since only a few patients can be early diagnosed and surgically treated with a good prognosis3. Approximately 70% of lung cancer patients develop to advanced stage upon diagnosis4.
Platinum-based chemotherapy has been the standard treatment of advanced NSCLC for decades5. This chemotherapy is usually in combination with gemcitabine, vinorelbine, paclitaxel, pemetrexed, and so on, yielding a response rate of 30%, a median overall survival of 12 months, and a median disease-free survival of 6 months6. Although molecule-targeted drugs have been recommended as the first choice for advanced patients with driver gene mutations in recent years, platinum-based chemotherapy is still the first-line therapy option for those with wildtype genes7. However, there is great variability in terms of response to platinum-based chemotherapy among NSCLC patients, even among those at the same stage. Knowing the prognostic factors can help schedule individualized treatment and improve the clinical outcomes of platinum-based chemotherapy.
Platinum drugs, including cisplatin and carboplatin, exert the antitumor effect by binding to DNA, forming DNA adducts that lead to intrastrand or interstrand crosslink and finally inducing severe dislocation of DNA double helix8,9. Treatment failure is mainly caused by resistance to platinum agents. Therefore, the DNA repair pathway, which detects and repairs these damaged DNA, plays a pivotal role in modifying the treatment efficacy of platinum-based chemotherapy in NSCLC.
There are four mainly different DNA repair mechanisms: nucleotide excision repair (NER), base excision repair (BER), mismatch repair (MMR), and double-strain break repair (DSB)10. X-ray repair crosscomplementing protein 1 (XRCC1), belonging to the BER pathway, interacts with DNA polymerase-beta, DNA ligase III, and PARP (poly ADP-ribose polymerase) to repair damaged DNA, including platinum-induced damage11,12. Thus, increasing the DNA repair capacity of XRCC1 decreases the clinical response13.
There are two polymorphisms of XRCC1, rs25487 (Arg399Gln) and rs1799782 (Arg194Trp), that are believed to change the DNA repair activity and affect the sensitivity of tumor cells to platinum drugs14. Thus, they have been widely investigated for the association with response to platinum-based chemotherapy in NSCLC patients15–17, mostly in Asian populations18–22. However, the results remain inconclusive. In the present study, we aim to evaluate the association between these two polymorphisms in XRCC1 and clinical outcomes of NSCLC treated with platinum-based chemotherapy in Asian populations.
2. Materials and Methods
2.1. Literature Search
This meta-analysis was in accordance with the PRISMA checklist. We performed literature search on electronic databases including PubMed, Web of Science, and EMBASE using the following keywords and their combinations: (“X-ray repair cross complementing group 1” or “XRCC1” or “rs25487” or “rs1799782”) AND (“platinum” or “cisplatin” or “carboplatin”) AND (“lung cancer” or “NSCLC”) prior to June 2019. Studies investigating the association between XRCC1 polymorphisms and chemotherapy efficacy of NSCLC were retrieved. The language was restricted to English. Additional articles from the reference lists of reviews and retrieved articles were manually searched to avoid missing eligible studies.
2.2. Inclusion and Exclusion Criteria
Eligible studies should fulfil the following criteria: all patients were confirmed as NSCLC (1); all patients received platinum-based chemotherapy (2); all patients were Asians (3); genotype data of rs25487 or rs1799782 were provided (4); and objective response rate (ORR), overall survival (OS), or progression-free survival (PFS) was reported as the outcomes of efficacy assessment, and adequate data were given for the present meat-analysis (5). Studies involving small-cell lung cancer (SCLC) patients or duplicated with other studies or performed in animals or cell lines were excluded. Reviews and case reports were discarded.
2.3. Data Extraction
The following information for each eligible study was extracted: first author, year of publication, country, tumor stage, chemotherapy regimens, genotyping method, sample size (male and female), age of participants, genotype distribution, and clinical outcomes. Literature search, filtering, and data extraction were performed by two independent researchers (NZ and YO), and any discrepancy was solved by further discussion with a third researcher.
2.4. Quality Assessment
We assessed the quality of all eligible studies by using the Newcastle-Ottawa Scale (NOS, http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp). NOS contains 8 items from 3 domains (selection, comparability and outcome) and has a total of 9 stars. Studies with 6 or more stars were considered of high quality.
2.5. Definition of Outcomes
ORR was classified into four categories according to RECIST (Response Evaluation Criteria in Solid Tumors)23: complete response (CR), partial response (PR), stable disease (SD), and progressive disease (PD). The responding group included CR and PR while the nonresponding group included SD and PD. OS was defined as the time from starting treatment to the death from any cause or the last follow-up. PFS was defined as the period from the date of treatment to disease progression or death from any cause.
2.6. Statistics
Odds ratio (OR) and its 95% confidence interval (95% CI) were calculated to assess the strength of association between XRCC1 polymorphisms and ORR of NSCLC receiving platinum-based chemotherapy. Hazard ratio (HR) with corresponding 95% CI was calculated to assess the association strength of polymorphisms with OS or PFS. I2 test and Q test were performed for the assessment of between-study heterogeneity. If no obvious heterogeneity existed (I2 < 50% and Q test, p > 0.1), a fixed-effect model was applied; otherwise, a random-effect model was used. Meta-regression regarding year of publication, sample size, median age of participants, percent of male, smoking rate, percent of stage IV patients and histological types, and sensitivity analysis were performed to identify the potential source of heterogeneity. Publication bias was assessed by funnel plots and Egger's test. All of the above analyses were performed in the following genetic models of rs25487 or rs1799782: homozygous model (homozygous variant vs. wildtype), heterozygous model (heterozygous variant vs. wildtype), dominant model (homozygous+heterozygous variant vs. wildtype), and recessive model (homozygous variant vs. heterozygous variant+wildtype). STATA v11.0 (STATA Corporation, College Station, TX, USA) was used for meta-analysis, and a p value<.05 was considered statistically significant.
3. Results
3.1. Basic Characteristics
A total of 121 potentially relevant articles were retrieved from literature search, and finally, 23 studies18–22,24–41 comprised of 5567 NSCLC patients were eligible and included in our analysis (Figure 1). Among them, all studies investigated rs25487 and 11 investigated rs1799782 on their association with chemotherapy efficacy. Twenty-one studies only recruited advanced NSCLC patients while 2 included patients with all stages29,33. ORR, OS and PFS were reported in 21, 14, and 5 studies, respectively. Thirteen and ten studies were awarded 6 and 7 stars, respectively, according to NOS, and they were considered of high quality. The basic characteristics of all included studies were listed in Table 1.
Figure 1.
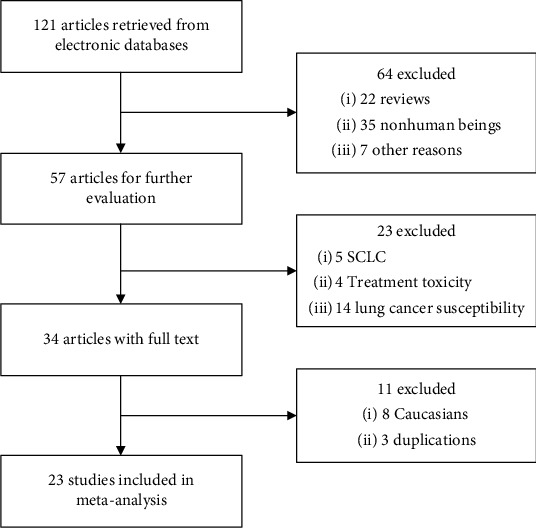
Flowchart of literature search of the present meta-analysis.
Table 1.
Basic characteristics of all included studies.
| Author | Year | Country | Tumor stage | Sample size (male/female) | Median age (year) | Smoking rate (%) | Genotyping method | SNPs | Clinical outcomes | NOS |
|---|---|---|---|---|---|---|---|---|---|---|
| Sun | 2009 | China | IV | 82 (53/29) | 59 | NR | DNA microarray | rs25487, rs1799782 | ORR | 6 |
| Yao | 2009 | China | IIIB-IV | 108 (71/38) | 61 | NR | PCR-RFLP | rs25487 | ORR, OS | 6 |
| Yuan | 2010 | China | III-IV | 199 (129/70) | 56 | 59.3 | PCR-RFLP | rs25487, rs1799782 | OS, PFS | 7 |
| Shiraishi | 2010 | Japan | III-IV | 201 (136/65) | 57.2 | 63.2 | TaqMan | rs25487 | ORR | 6 |
| Li | 2011 | China | III-IV | 89 (64/25) | 59.1 | NR | PCR-RFLP | rs25487 | ORR | 7 |
| Zhou | 2011 | China | IV | 111 (67/44) | 57 | NR | Direct sequencing | rs25487 | ORR | 7 |
| Liao | 2012 | China | IIIB-IV | 62 (35/27) | 57 | NR | SNPstream | rs25487 | ORR, OS | 6 |
| Ke | 2012 | China | I-IV | 460 (334/126) | 59.5 | 67.4 | PCR-CTTP | rs25487, rs1799782 | OS | 6 |
| Zhao | 2013 | China | IIIB-IV | 147 (92/55) | 60 | 45.9 | TaqMan | rs25487, rs1799782 | ORR, OS, PFS | 7 |
| Lee | 2013 | South Korea | III-IV | 382 (311/71) | NR | 83.2 | MALDI-TOF | rs25487 | ORR, OS | 6 |
| Zhang | 2014 | China | III-IV | 375 (249/126) | 60.9 | NR | MALDI-TOF | rs25487, rs1799782 | ORR, OS, PFS | 6 |
| Peng | 2014 | China | III-IV | 235 (180/55) | 58 | 61.3 | PCR-CTTP | rs25487 | ORR | 6 |
| Liu | 2014 | China | I-IV | 378 (297/81) | 62.4 | 56.1 | MALDI-TOF | rs25487, rs1799782 | ORR, OS, PFS | 7 |
| Du | 2014 | China | III-IV | 161 (108/53) | 60 | 54.7 | qPCR | rs25487 | ORR | 7 |
| Deng | 2015 | China | IIIB-IV | 97 (66/31) | 57 | 40.2 | Direct sequencing | rs25487 | ORR, PFS | 6 |
| Han | 2015 | China | IIIB-IV | 325 (116/209) | 57.6 | 68.3 | PCR-RFLP | rs25487, rs1799782 | ORR, OS | 7 |
| Liu | 2015 | China | IIIB-IV | 322 (226/96) | 62.5 | 43.5 | PCR-RFLP | rs25487, rs1799782 | ORR, OS | 6 |
| Zhao | 2015 | China | III-IV | 206 (124/82) | 56.1 | 65.5 | PCR-RFLP | rs25487, rs1799782 | ORR, OS | 7 |
| Chen | 2016 | China | III-IV | 1024 (724/300) | NR | 55.6 | MALDI-TOF | rs25487 | ORR | 6 |
| Bu | 2016 | China | III-IV | 141 (100/41) | 55.9 | 66.0 | PCR-RFLP | rs25487, rs1799782 | ORR, OS | 6 |
| Liu | 2016 | China | IIIB-IV | 252 (104/148) | 56.3 | 75.4 | PCR-RFLP | rs25487, rs1799782 | ORR, OS | 7 |
| Liao | 2018 | China | IV | 58 (39/19) | 58 | 44.8 | TaqMan | rs25487 | ORR, OS | 7 |
| Dong | 2018 | China | IIIB-IV | 152 (101/51) | 52 | 52.0 | MALDI-TOF | rs25487 | ORR | 6 |
PCR-RFLP: polymerase chain reaction restriction fragment length polymorphism; PCR-CTTP: PCR with the confronting-two-pair primer; MALDI-TOF: matrix-assisted laser desorption/ionization time-of-flight mass spectrometry; ORR: objective response rate; OS: overall survival; PFS: progression-free survival; NR: not reported. NOS: Newcastle-Ottawa Scale.
3.2. Rs25487
3.2.1. Objective Response Rate
Twenty-one studies involving 4708 NSCLC patients were included in the association analysis between rs25487 (Arg399Gln) and ORR, of which 17, 17, 20, and 18 were for homozygous, heterozygous, dominant, and recessive models, respectively. There was high between-study heterogeneity in most of the comparisons (Table 2). Compared to the ArgArg genotype, GlnGln (OR = 1.71, 95% CI:1.16-2.52, p = .007, I2 = 56.8%, Figure 2(a)) or GlnArg (OR = 1.23, 95% CI:1.07-1.40, p = .003, I2 = 29.0%, Figure 2(b)) genotypes were associated with higher ORR. In the recessive model, GlnGln carriers had higher probability to respond to platinum-based chemotherapy (OR = 1.45, 95% CI:1.02-2.06, p = .037, I2 = 54.2%).
Table 2.
Meta-analysis of association between rs25487 or rs1799782 and efficacy of NSCLC treated with platinum-based chemotherapy in Asians.
| XRCC1 | ORR | OS | PFS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | I 2 (%) | OR (95% CI) | p | No. | I 2 (%) | HR (95% CI) | p | No. | I 2 (%) | HR (95% CI) | p | |
| rs25487 (Arg399Gln)$ | ||||||||||||
| Homozygous model | 17 | 56.8 | 1.71 (1.16-2.52) | .007 | 12 | 63.2 | 0.60 (0.40-0.88) | .009 | 4 | 24.4 | 0.64 (0.46-0.90) | .010 |
| Heterozygous model | 17 | 29.0 | 1.23 (1.07-1.40) | .003 | 12 | 63.2 | 0.81 (0.64-1.03) | .083 | 4 | 0 | 0.87 (0.71-1.07) | .193 |
| Dominant model | 20 | 69.3 | 1.11 (0.87-1.41) | .386 | 5 | 39.4 | 1.00 (0.79-1.27) | .980 | 4 | 0 | 0.87 (0.71-1.08) | .215 |
| Recessive model | 18 | 54.2 | 1.45 (1.02-2.06) | .037 | 3 | 13.8 | 1.14 (0.82-1.59) | .490 | — | — | — | — |
| rs1799782 (Arg194Trp)# | ||||||||||||
| Homozygous model | 9 | 34.0 | 1.73 (1.31-2.27) | <.001 | 10 | 53.4 | 0.63 (0.43-0.91) | .013 | 4 | 0 | 0.93 (0.68-1.26) | .627 |
| Heterozygous model | 9 | 0 | 1.28 (1.06-1.55) | .007 | 10 | 0 | 0.89 (0.70-1.05) | .173 | 4 | 0 | 1.02 (0.83-1.26) | .838 |
| Dominant model | 9 | 25.5 | 1.38 (1.16-1.65) | <.001 | 4 | 0 | 0.91 (0.71-1.16) | .446 | 3 | 0 | 1.05 (0.83-1.34) | .662 |
| Recessive model | 9 | 12.5 | 1.56 (1.20-2.03) | <.001 | — | — | — | — | — | — | — | — |
$Homozygous, heterozygous, dominant, and recessive models indicated GlnGln vs. ArgArg, GlnArg vs. ArgArg, GlnGln+GlnArg vs. ArgArg ,and GlnGln vs. GlnArg+ArgArg, respectively. #Homozygous, heterozygous, dominant, and recessive models indicated TrpTrp vs. ArgArg, TrpArg vs. ArgArg, TrpTrp+TrpArg vs. ArgArg, and TrpTrp vs. TrpArg+ArgArg, respectively. Abbreviations: NSCLC: non-small-cell lung cancer; ORR: objective response rate; OS: overall survival; PFS: progression-free survival; OR: odds ratio; HR: hazard ratio.
Figure 2.
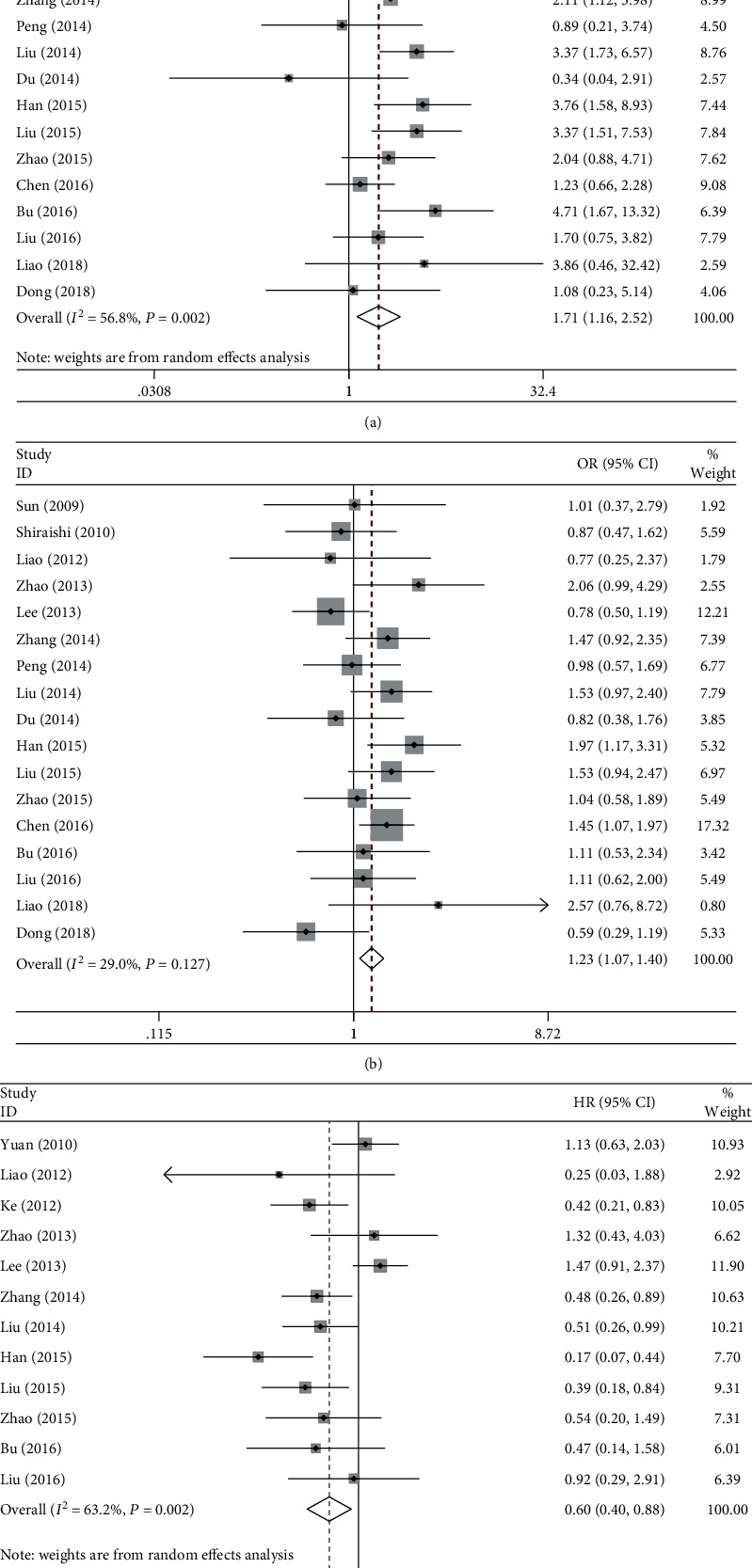
Forest plots of rs25487 (Arg399Gln) and clinical outcomes in NSCLC patients under platinum-based chemotherapy. (a) ORR in the homozygous model. (b) ORR in the heterozygous model. (c) OS in the homozygous model. NSCLC: non-small-cell lung cancer; ORR: objective response rate; OS: overall survival.
3.2.2. Overall Survival and Progression-Free Survival
We included 12 and 4 studies in the meta-analysis regarding the association of rs25487 with OS or PFS, respectively. In the homozygous model, carriers of GlnGln had a favorable OS (HR = 0.60, 95% CI:0.40-0.88, p = .009, I2 = 63.2%, Figure 2(c)) and PFS (HR = 0.64, 95% CI:0.46-0.90, p = .010, I2 = 24.4%). However, no significant associations were found in the other comparisons (Table 2).
3.3. Rs1799782
3.3.1. Objective Response Rate
Nine studies, comprising 2228 NSCLC patients, with respect to the association between rs1799782 (Arg194Trp) and ORR of platinum-based chemotherapy, were included. Low or no heterogeneity was found, and fixed-effect model was used. After meta-analysis of all comparisons, Trp194 was found to be significantly associated with higher ORR (Table 2). Comparing to the homozygous wildtype, carrying at least one Trp allele was more likely to respond completely or partially to the chemotherapy (OR = 1.38, 95% CI:1.16-1.65, p < .001, I2 = 25.5%, Figure 3(a)). Significant associations were also found in homozygous, heterozygous, and recessive models Figure 3(b).
Figure 3.
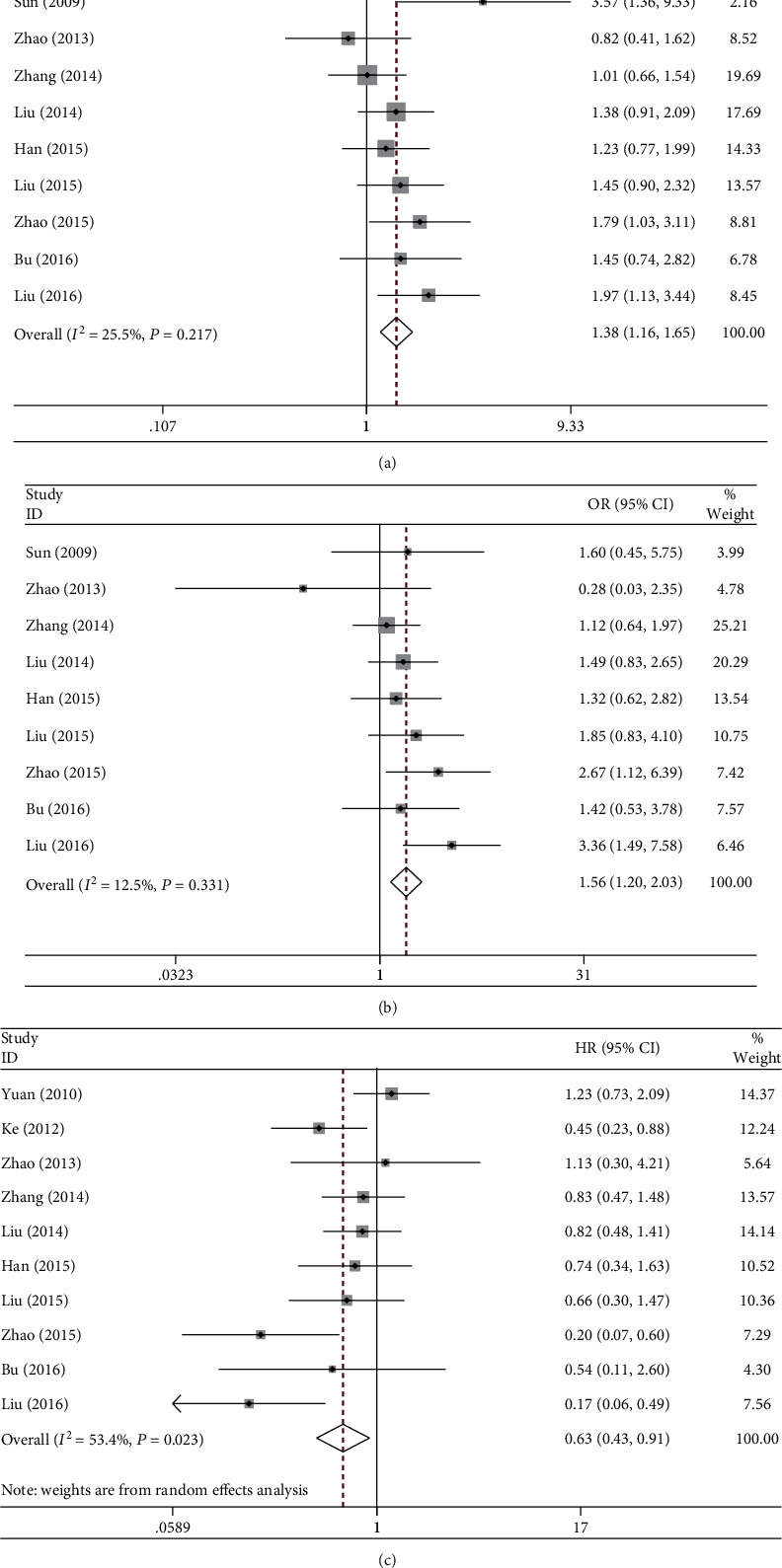
Forest plots of rs1799782 (Arg194Trp) and clinical outcomes in NSCLC patients under platinum-based chemotherapy. (a) ORR in the dominant model. (b) ORR in the recessive model. (c) OS in the homozygous model. NSCLC: non-small-cell lung cancer; ORR: objective response rate; OS: overall survival.
3.3.2. Overall Survival and Progression-Free Survival
Compared to ArpArp, TrpTrp carriers had a favorable OS (HR = 0.63, 95% CI:0.43-0.91, p = .013, I2 = 53.4%, Figure 3(c)) after pooling 10 studies together, while TrpArg was not associated with OS (p = .173, I2 = 0). Only a few studies reported the association of rs1799782 with PFS, and none of the genotypes indicated a favorable PFS in pooling analysis (p > .05).
3.4. Sensitivity Analysis and Metaregression
Sensitivity analysis, by excluding one study each time and pooling the rest studies together, showed that none of a single study had significant impact on the pooled effect sizes of meta-analysis. Even after excluding two studies that comprised all stages of NSCLC patients 29,33, the effect sizes remained unchanged.
For 25487, meta-regression analysis revealed an influence of age of participants in the heterozygous model (coefficient = 0.071, p = .029) and year of publication in the recessive model (coefficient = 0.18, p = .025), with ORR increasing with age and time (Figure 4). No similar associations were identified in other variables, particularly in the smoking rate (p > .05).
Figure 4.
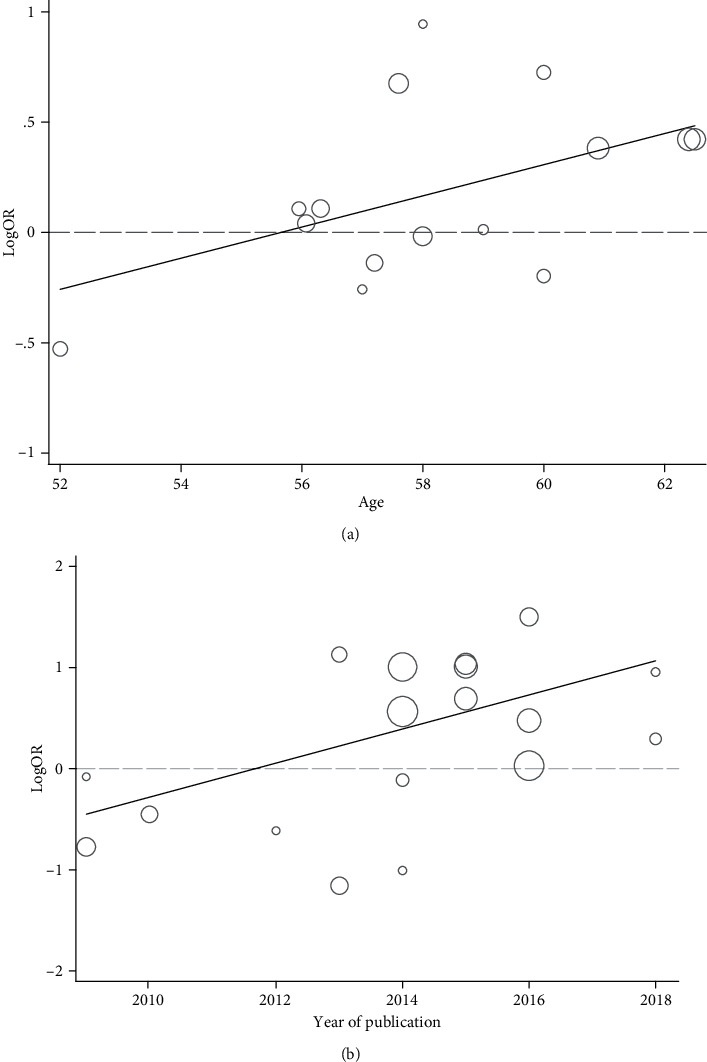
Meta-regression analysis with significant findings. (a) The effect of age on association of the rs25487 genotype with ORR in the heterozygous model. (b) The effect of year of publication on association of the rs25487 genotype with ORR in the recessive model. ORR: objective response rate.
3.5. Publication Bias
The funnel plots in most of the meta-analyses were symmetric, and Egger's test indicated no evidence of publication bias (p > .10), except in the heterozygous model of 1799782 with ORR.
4. Discussion
XRCC1 is the major component of the BER pathway, and its enhanced capacity to repair platinum-induced DNA damage may be correlated with the sensitivity to platinum drugs. Two mostly studied functional polymorphisms of XRCC1, rs25487 (Arg399Gln) and rs1799782 (Arg194Trp), are both located in the exons and modify the DNA repair activity. The Arg399Gln polymorphism occurs in the PARP-binding domain and may affect complex assembly or repair efficiency. The 399Gln allele was associated with elevated levels of aflatoxin B1-DNA adducts and glycophorin A somatic mutations42 and with a higher frequency of sister chromatid exchange43. Another polymorphism, Arg194Trp, resides in the linking region between the DNA polymerase β domain and PARP domain and was reported to disrupt the functionality of XRCC1 in hamsters44. Although the mechanisms have not been fully elucidated, the carriers of 399Gln and 194Trp are inferred to have deficient DNA repair activity and are more sensitive to platinum-based regimen. In accordance with this, several studies reported positive associations of rs25487 and rs1799782 with increased objective response rate and longer overall survival27,29,45. However, some other studies identified insignificant even negative associations15,17,25.
The present meta-analysis pooled all studies performed in Asian populations to further assessing the associations of both polymorphisms with platinum-based chemotherapy efficacy. For rs25487, carrying one or two 399Gln alleles had better ORR compared with the homozygous wildtype (OR = 1.23 and 1.71, respectively, p < .01). Meanwhile, the homozygous variant had longer overall survival and progression-free survival (HR = 0.60 and 0.64, respectively, p ≤ .010). However, obvious heterogeneity was found in most of the comparisons, and thus the results should be interpreted in caution. For rs1799782, significant correlations with ORR were identified in homozygous, heterozygous, dominant, and recessive models (all p values <.010), suggesting a gene dosage effect of positive association. Additionally, the homozygous 194Trp was associated with a favorable OS (HR = 0.63, p = .013).
The present study mainly focused on the Asian populations. Several studies have reported controversial associations with ORR, OS, or PFS in the Caucasians15–17,45–49. We found, in subanalysis of Caucasians, significant associations of rs25487 with a shorter OS in the homozygous model (HR = 2.72, p = .001, I2 = 49.4%) by pooling 3 studies and with a decreased ORR in the dominant model (OR = 0.68, p = .031, I2 = 0) by pooling 5 studies. However, there seems a contradictory role of rs25487 in clinical outcomes between different ethnicities since it predicted longer OS and increased ORR in Asians. This may be caused by various factors, such as genetic background, sample size, and dietary or living habits. This discrepancy needs further confirmation by including more investigations with larger sample size in Caucasians. Since only a few studies in Caucasians were available, we gave up the subanalysis of Caucasians in the present study. Compared with previous meta-analyses50–52, we included more eligible studies and a larger sample size of more than 5500 patients under similar genetic background, indicating a stronger robustness of our results.
Our results revealed that two functional polymorphisms of XRCC1 may have prognostic value of NSCLC patients under platinum-based chemotherapy, but they are still far away from clinical practice, since many genes in other pathways, apart from the BER pathway, also contribute to the sensitivity to platinum drugs 53. The NER pathway, through ERCC147,54, ERCC255, and ERCC533, also played a pivotal role in repairing DNA lesions and was reported to be associated with clinical outcomes. The other genes, including XRCC346 in the DSB pathway, TP5356 and MDM257 in the p53 pathway, AKT158 in the PI3K/PTEN/AKT pathway, SMAD359 in the TGF-β pathway, and MTHFR60 involved in folate metabolism, were also found in association with ORR, OS, or PFS in NSCLC patients using platinum-based regimen. The combinations of various markers should be furtherly investigated for the prognostic value in NSCLC patients under platinum-based chemotherapy.
There are several limitations in our mate-analysis. First, an ideal meta-analysis should be based on individual-level data, especially in the analysis of OS and PFS, although it is usually quite difficult to obtain these data. The interpretation of our results in terms of OS and PFS should be cautious. Secondly, we did not analyze the interaction of both polymorphisms in predicting the clinical outcomes because of a lack of sufficient data. Thirdly, there were substantial between-study heterogeneities in the pooling analysis of rs25487, and thus more studies are needed in the future.
5. Conclusion
In conclusion, our meta-analysis suggested that two functional polymorphisms of XRCC1, rs25487 and rs1799782, can be used as prognostic factors for platinum-based chemotherapy in Asian patients with NSCLC.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA: A Cancer Journal for Clinicians. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Santarpia M., Rolfo C., Peters G. J., Leon L. G., Giovannetti E. On the pharmacogenetics of non-small cell lung cancer treatment. Expert Opinion on Drug Metabolism & Toxicology. 2016;12(3):307–317. doi: 10.1517/17425255.2016.1141894. [DOI] [PubMed] [Google Scholar]
- 3.Kanou T., Okami J., Tokunaga T., Ishida D., Kuno H., Higashiyama M. Prognostic factors in patients with postoperative brain recurrence from completely resected non-small cell lung cancer. Thoracic Cancer. 2015;6(1):38–42. doi: 10.1111/1759-7714.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Socinski M. A., Crowell R., Hensing T. E., et al. Treatment of non-small cell lung cancer, stage IV. Chest. 2007;132(3):277S–289S. doi: 10.1378/chest.07-1381. [DOI] [PubMed] [Google Scholar]
- 5.Pfister D. G., Johnson D. H., Azzoli C. G., et al. American Society of Clinical Oncology treatment of unresectable non-small-cell lung cancer guideline: update 2003. Journal of Clinical Oncology. 2004;22(2):330–353. doi: 10.1200/JCO.2004.09.053. [DOI] [PubMed] [Google Scholar]
- 6.Ardizzoni A., Boni L., Tiseo M., et al. Cisplatin- versus carboplatin-based chemotherapy in first-line treatment of advanced non-small-cell lung cancer: an individual patient data meta-analysis. Journal of the National Cancer Institute. 2007;99(11):847–857. doi: 10.1093/jnci/djk196. [DOI] [PubMed] [Google Scholar]
- 7.Ettinger D. S., Wood D. E., Akerley W., et al. NCCN guidelines insights: non-small cell lung cancer, version 4.2016. Journal of the National Comprehensive Cancer Network. 2016;14(3):255–264. doi: 10.6004/jnccn.2016.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jordan P., Carmo-Fonseca M. Molecular mechanisms involved in cisplatin cytotoxicity. Cellular and Molecular Life Sciences. 2000;57(8):1229–1235. doi: 10.1007/PL00000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin L. P., Hamilton T. C., Schilder R. J. Platinum resistance: the role of DNA repair pathways. Clinical Cancer Research. 2008;14(5):1291–1295. doi: 10.1158/1078-0432.CCR-07-2238. [DOI] [PubMed] [Google Scholar]
- 10.Sakthivel K. M., Hariharan S. Regulatory players of DNA damage repair mechanisms: role in cancer chemoresistance. Biomedicine & Pharmacotherapy. 2017;93:1238–1245. doi: 10.1016/j.biopha.2017.07.035. [DOI] [PubMed] [Google Scholar]
- 11.Nazarkina Z. K., Khodyreva S. N., Marsin S., Lavrik O. I., Radicella J. P. XRCC1 interactions with base excision repair DNA intermediates. DNA Repair. 2007;6(2):254–264. doi: 10.1016/j.dnarep.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Whitehouse C. J., Taylor R. M., Thistlethwaite A., et al. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104(1):107–117. doi: 10.1016/S0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 13.Simon G. R., Sharma S., Cantor A., Smith P., Bepler G. ERCC1 expression is a predictor of survival in resected patients with non-small cell lung cancer. Chest. 2005;127(3):978–983. doi: 10.1378/chest.127.3.978. [DOI] [PubMed] [Google Scholar]
- 14.Matullo G., Palli D., Peluso M., et al. XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32)P-DNA adducts in a sample of healthy subjects. Carcinogenesis. 2001;22(9):1437–1445. doi: 10.1093/carcin/22.9.1437. [DOI] [PubMed] [Google Scholar]
- 15.Gurubhagavatula S., Liu G., Park S., et al. XPDandXRCC1Genetic polymorphisms are prognostic factors in advanced non-small-cell lung cancer patients treated with platinum chemotherapy. Journal of Clinical Oncology. 2004;22(13):2594–2601. doi: 10.1200/JCO.2004.08.067. [DOI] [PubMed] [Google Scholar]
- 16.Sullivan I., Salazar J., Majem M., et al. Pharmacogenetics of the DNA repair pathways in advanced non-small cell lung cancer patients treated with platinum-based chemotherapy. Cancer Letters. 2014;353(2):160–166. doi: 10.1016/j.canlet.2014.07.023. [DOI] [PubMed] [Google Scholar]
- 17.Pérez-Ramírez C., Cañadas-Garre M., Alnatsha A., et al. Pharmacogenetics of platinum-based chemotherapy: impact of DNA repair and folate metabolism gene polymorphisms on prognosis of non-small cell lung cancer patients. The Pharmacogenomics Journal. 2019;19(2):164–177. doi: 10.1038/s41397-018-0014-8. [DOI] [PubMed] [Google Scholar]
- 18.Sun X., Li F., Sun N., et al. Polymorphisms in XRCC1 and XPG and response to platinum-based chemotherapy in advanced non-small cell lung cancer patients. Lung Cancer. 2009;65(2):230–236. doi: 10.1016/j.lungcan.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 19.Shiraishi K., Kohno T., Tanai C., et al. Association of DNA repair gene polymorphisms with response to platinum-based doublet chemotherapy in patients with non-small-cell lung cancer. Journal of Clinical Oncology. 2010;28(33):4945–4952. doi: 10.1200/JCO.2010.30.5334. [DOI] [PubMed] [Google Scholar]
- 20.Lee S. Y., Kang H. G., Yoo S. S., et al. Polymorphisms in DNA repair and apoptosis-related genes and clinical outcomes of patients with non-small cell lung cancer treated with first-line paclitaxel-cisplatin chemotherapy. Lung Cancer. 2013;82(2):330–339. doi: 10.1016/j.lungcan.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 21.Chen J., Wang Z., Zou T., et al. Pharmacogenomics of platinum-based chemotherapy response in NSCLC: a genotyping study and a pooled analysis. Oncotarget. 2016;7(34):55741–55756. doi: 10.18632/oncotarget.9688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong J., Wang X., Yu Y., Yan X., Cui J. W. Association of base excision repair gene polymorphisms with the response to chemotherapy in advanced non-small cell lung cancer. Chinese Medical Journal. 2018;131(16):1904–1908. doi: 10.4103/0366-6999.238141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eisenhauer E. A., Therasse P., Bogaerts J., et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) European Journal of Cancer. 2009;45(2):228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 24.Yao C. Y., Huang X. E., Li C., et al. Lack of influence of XRCC1 and XPD gene polymorphisms on outcome of platinum-based chemotherapy for advanced non small cell lung cancers. Asian Pacific Journal of Cancer Prevention. 2009;10(5):859–864. [PubMed] [Google Scholar]
- 25.Yuan P., Liu L., Wu C., et al. No association between XRCC1 polymorphisms and survival in non-small-cell lung cancer patients treated with platinum-based chemotherapy. Cancer Biology & Therapy. 2010;10:854–859. doi: 10.4161/cbt.10.9.13238. [DOI] [PubMed] [Google Scholar]
- 26.Li D., Zhou Q., Liu Y., Yang Y., Li Q. DNA repair gene polymorphism associated with sensitivity of lung cancer to therapy. Medical Oncology. 2012;29(3):1622–1628. doi: 10.1007/s12032-011-0033-7. [DOI] [PubMed] [Google Scholar]
- 27.Zhou F., Yu Z., Jiang T., Lv H., Yao R., Liang J. Genetic polymorphisms of GSTP1 and XRCC1: prediction of clinical outcome of platinum-based chemotherapy in advanced non-small cell lung cancer (NSCLC) patients. Swiss Medical Weekly. 2011;141, article w13275 doi: 10.4414/smw.2011.13275. [DOI] [PubMed] [Google Scholar]
- 28.Liao W. Y., Shih J. Y., Chang G. C., et al. Genetic polymorphism of XRCC1 Arg399Gln is associated with survival in non-small-cell lung cancer patients treated with gemcitabine/platinum. Journal of Thoracic Oncology. 2012;7(6):973–981. doi: 10.1097/JTO.0b013e31824fe98c. [DOI] [PubMed] [Google Scholar]
- 29.Ke H. G., Li J., Shen Y., et al. Prognostic significance of GSTP1, XRCC1 and XRCC3 polymorphisms in non-small cell lung cancer patients. Asian Pacific Journal of Cancer Prevention. 2012;13(9):4413–4416. doi: 10.7314/APJCP.2012.13.9.4413. [DOI] [PubMed] [Google Scholar]
- 30.Zhao W., Hu L., Xu J., et al. Polymorphisms in the base excision repair pathway modulate prognosis of platinum-based chemotherapy in advanced non-small cell lung cancer. Cancer Chemotherapy and Pharmacology. 2013;71(5):1287–1295. doi: 10.1007/s00280-013-2127-8. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L., Ma W., Li Y., Wu J., Shi G. Y. Pharmacogenetics of DNA repair gene polymorphisms in non-small-cell lung carcinoma patients on platinum-based chemotherapy. Genetics and Molecular Research. 2014;13(1):228–236. doi: 10.4238/2014.January.14.2. [DOI] [PubMed] [Google Scholar]
- 32.Peng Y., Li Z., Zhang S., et al. Association of DNA base excision repair genes (OGG1, APE1 and XRCC1) polymorphisms with outcome to platinum-based chemotherapy in advanced nonsmall-cell lung cancer patients. International Journal of Cancer. 2014;135(11):2687–2696. doi: 10.1002/ijc.28892. [DOI] [PubMed] [Google Scholar]
- 33.Liu D., Wu J., Shi G. Y., Zhou H. F., Yu Y. Role of XRCC1 and ERCC5 polymorphisms on clinical outcomes in advanced non-small cell lung cancer. Genetics and Molecular Research. 2014;13(2):3100–3107. doi: 10.4238/2014.April.17.6. [DOI] [PubMed] [Google Scholar]
- 34.Du Y., Su T., Zhao L., et al. Associations of polymorphisms in DNA repair genes and MDR1 gene with chemotherapy response and survival of non-small cell lung cancer. PLoS One. 2014;9(6, article e99843) doi: 10.1371/journal.pone.0099843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng J. H., Deng J., Shi D. H., Ouyang X. N., Niu P. G. Clinical outcome of cisplatin-based chemotherapy is associated with the polymorphisms of GSTP1 and XRCC1 in advanced non-small cell lung cancer patients. Clinical & Translational Oncology. 2015;17(9):720–726. doi: 10.1007/s12094-015-1299-6. [DOI] [PubMed] [Google Scholar]
- 36.Han B., Guo Z., Ma Y., et al. Association of GSTP1 and XRCC1 gene polymorphisms with clinical outcome of advanced non-small cell lung cancer patients with cisplatin-based chemotherapy. International Journal of Clinical and Experimental Pathology. 2015;8(4):4113–4119. [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J. Y., Li L. R., Liu Q. M. Association of GSTP1 and XRCC1 gene polymorphisms with clinical outcomes of patients with advanced non-small cell lung cancer. Genetics and Molecular Research. 2015;14(3):10331–10337. doi: 10.4238/2015.August.28.19. [DOI] [PubMed] [Google Scholar]
- 38.Zhao R., Chen G. Role of GSTP1 Ile105Val and XRCC1 Arg194Trp, Arg280His and Arg399Gln gene polymorphisms in the clinical outcome of advanced non-small cell lung cancer. International Journal of Clinical and Experimental Pathology. 2015;8(11):14909–14916. [PMC free article] [PubMed] [Google Scholar]
- 39.Bu L., Zhang L. B., Mao X., Wang P. GSTP1 Ile105Val and XRCC1 Arg399Gln gene polymorphisms contribute to the clinical outcome of patients with advanced non-small cell lung cancer. Genetics and Molecular Research. 2016;15(2) doi: 10.4238/gmr.15027611. [DOI] [PubMed] [Google Scholar]
- 40.Liu H. F., Liu J. S., Deng J. H., Wu R. R. Role of XRCC1 gene polymorphisms in non-small cell lung cancer cisplatin-based chemotherapy, and their effect on clinical and pathological characteristics. Genetics and Molecular Research. 2016;15(4) doi: 10.4238/gmr15049084. [DOI] [PubMed] [Google Scholar]
- 41.Liao W.-Y., Ho C.-C., Tsai T.-H., Chen K.-Y., Shih J.-Y., Yu C.-J. Combined effect of ERCC1 and ERCC2 polymorphisms on overall survival in non-squamous non-small-cell lung cancer patients treated with first-line pemetrexed/platinum. Lung Cancer. 2018;118:90–96. doi: 10.1016/j.lungcan.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 42.Lunn R. M., Langlois R. G., Hsieh L. L., Thompson C. L., Bell D. A. XRCC1 polymorphisms: effects on aflatoxin B1-DNA adducts and glycophorin a variant frequency. Cancer Research. 1999;59(11):2557–2561. [PubMed] [Google Scholar]
- 43.Duell E. J., Wiencke J. K., Cheng T. J., et al. Polymorphisms in the DNA repair genes XRCC1 and ERCC2 and biomarkers of DNA damage in human blood mononuclear cells. Carcinogenesis. 2000;21(5):965–971. doi: 10.1093/carcin/21.5.965. [DOI] [PubMed] [Google Scholar]
- 44.Thompson L. H., West M. G. XRCC1 keeps DNA from getting stranded. Mutation Research. 2000;459(1):1–18. doi: 10.1016/S0921-8777(99)00058-0. [DOI] [PubMed] [Google Scholar]
- 45.Kalikaki A., Kanaki M., Vassalou H., et al. DNA repair gene polymorphisms predict favorable clinical outcome in advanced non-small-cell lung cancer. Clinical Lung Cancer. 2009;10(2):118–123. doi: 10.3816/CLC.2009.n.015. [DOI] [PubMed] [Google Scholar]
- 46.de las Peñas R., Sanchez-Ronco M., Alberola V., et al. Polymorphisms in DNA repair genes modulate survival in cisplatin/gemcitabine-treated non-small-cell lung cancer patients. Annals of Oncology. 2006;17(4):668–675. doi: 10.1093/annonc/mdj135. [DOI] [PubMed] [Google Scholar]
- 47.Joerger M., Burgers S. A., Baas P., et al. Germline polymorphisms in patients with advanced nonsmall cell lung cancer receiving first-line platinum-gemcitabine chemotherapy: a prospective clinical study. Cancer. 2012;118(9):2466–2475. doi: 10.1002/cncr.26562. [DOI] [PubMed] [Google Scholar]
- 48.Butkiewicz D., Drosik A., Suwiński R., et al. Influence of DNA repair gene polymorphisms on prognosis in inoperable non-small cell lung cancer patients treated with radiotherapy and platinum-based chemotherapy. International Journal of Cancer. 2012;131(7):E1100–E1108. doi: 10.1002/ijc.27596. [DOI] [PubMed] [Google Scholar]
- 49.Kalikaki A., Voutsina A., Koutsopoulos A., et al. ERCC1SNPs as potential predictive biomarkers in non-small cell lung cancer patients treated with platinum-based chemotherapy. Cancer Investigation. 2015;33(4):107–113. doi: 10.3109/07357907.2014.1001897. [DOI] [PubMed] [Google Scholar]
- 50.Li D. J., Xiao D. Association between the XRCC1 polymorphisms and clinical outcomes of advanced NSCLC treated with platinum-based chemotherapy: a meta-analysis based on the PRISMA statement. BMC Cancer. 2017;17(1):p. 501. doi: 10.1186/s12885-017-3487-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu S.-N., Liu G.-F., Li X.-F., Fu B.-H., Dong L.-X., Zhang S.-H. Evaluation of prediction of polymorphisms of DNA repair genes on the efficacy of platinum-based chemotherapy in patients with non-small cell lung cancer: a network meta-analysis. Journal of Cellular Biochemistry. 2017;118(12):4782–4791. doi: 10.1002/jcb.26147. [DOI] [PubMed] [Google Scholar]
- 52.Tan L. M., Qiu C. F., Zhu T., et al. Genetic polymorphisms and platinum-based chemotherapy treatment outcomes in patients with non-small cell lung cancer: a genetic epidemiology study based meta-analysis. Scientific Reports. 2017;7(1):p. 5593. doi: 10.1038/s41598-017-05642-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pérez-Ramírez C., Cañadas-Garre M., Molina M. Á., Robles A. I., Faus-Dáder M. J., Calleja-Hernández M. Á. Contribution of genetic factors to platinum-based chemotherapy sensitivity and prognosis of non-small cell lung cancer. Mutation Research/Reviews in Mutation Research. 2017;771:32–58. doi: 10.1016/j.mrrev.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 54.Isla D., Sarries C., Rosell R., et al. Single nucleotide polymorphisms and outcome in docetaxel-cisplatin-treated advanced non-small-cell lung cancer. Annals of Oncology. 2004;15(8):1194–1203. doi: 10.1093/annonc/mdh319. [DOI] [PubMed] [Google Scholar]
- 55.Li X. D., Han J. C., Zhang Y. J., Li H. B., Wu X. Y. Common variations of DNA repair genes are associated with response to platinum-based chemotherapy in NSCLCs. Asian Pacific Journal of Cancer Prevention. 2013;14(1):145–148. doi: 10.7314/APJCP.2013.14.1.145. [DOI] [PubMed] [Google Scholar]
- 56.Zheng D., Chen Y., Gao C., et al. Polymorphisms of p53 and MDM2 genes are associated with severe toxicities in patients with non-small cell lung cancer. Cancer Biology & Therapy. 2014;15(11):1542–1551. doi: 10.4161/15384047.2014.956599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dong J., Ren B., Hu Z., et al. MDM2SNP309 contributes to non-small cell lung cancer survival in Chinese. Molecular Carcinogenesis. 2011;50(6):433–438. doi: 10.1002/mc.20727. [DOI] [PubMed] [Google Scholar]
- 58.Kim M. J., Kang H. G., Lee S. Y., et al. AKT1 polymorphisms and survival of early stage non-small cell lung cancer. Journal of Surgical Oncology. 2012;105(2):167–174. doi: 10.1002/jso.22071. [DOI] [PubMed] [Google Scholar]
- 59.Lin M., Stewart D. J., Spitz M. R., et al. Genetic variations in the transforming growth factor-beta pathway as predictors of survival in advanced non-small cell lung cancer. Carcinogenesis. 2011;32(7):1050–1056. doi: 10.1093/carcin/bgr067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tiseo M., Giovannetti E., Tibaldi C., et al. Pharmacogenetic study of patients with advanced non-small cell lung cancer (NSCLC) treated with second-line pemetrexed or pemetrexed-carboplatin. Lung Cancer. 2012;78(1):92–99. doi: 10.1016/j.lungcan.2012.07.009. [DOI] [PubMed] [Google Scholar]


