Abstract
Background
The distribution of β-lactam resistance genes in P. aeruginosa is often closely related to the distribution of certain high-risk international clones. We used whole-genome sequencing (WGS) to identify the predominant sequence types (ST) and β-lactamase genes in clinical isolates of multidrug-resistant (MDR)-P. aeruginosa from Qatar
Methods
Microbiological identification and susceptibility tests were performed by automated BD Phoenix™ system and manual Liofilchem MIC Test Strips.
Results
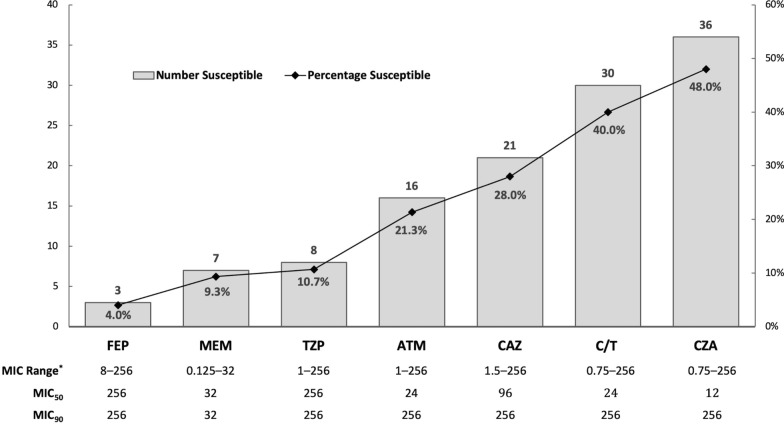
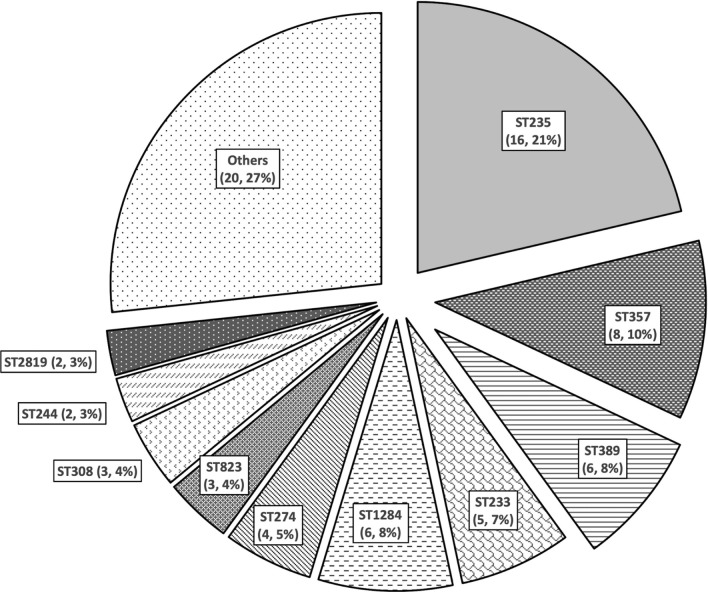
Among 75 MDR-P. aeruginosa isolates; the largest proportions of susceptibility were to ceftazidime-avibactam (n = 36, 48%), followed by ceftolozane-tazobactam (30, 40%), ceftazidime (n = 21, 28%) and aztreonam (n = 16, 21.3%). All isolates possessed Class C and/or Class D β-lactamases (n = 72, 96% each), while metallo-β-lactamases were detected in 20 (26.7%) isolates. Eight (40%) metallo-β-lactamase producers were susceptible to aztreonam and did not produce any concomitant extended-spectrum β-lactamases. High risk ST235 (n = 16, 21.3%), ST357 (n = 8, 10.7%), ST389 and ST1284 (6, 8% each) were most frequent. Nearly all ST235 isolates (15/16; 93.8%) were resistant to all tested β-lactams.
Conclusion
MDR-P. aeruginosa isolates from Qatar are highly resistant to antipseudomonal β-lactams. High-risk STs are predominant in Qatar and their associated MDR phenotypes are a cause for considerable concern.
Keywords: Beta-lactamases, MDR, P. aeruginosa, ST235
Background
Due to their established efficacy and safety, anti-pseudomonal β-lactam antibiotics play a vital role in the clinical management of P. aeruginosa infections [1]. Key antimicrobial resistance mechanisms in P. aeruginosa include over-expression of efflux pumps, impermeability through porin modification or loss, target modification, and enzyme-mediated antimicrobial inactivation (e.g., β-lactamases). Multiple resistance mechanisms are frequently present in concert resulting in simultaneous resistance to multiple agents [1]. The epidemiology of β-lactamases is often closely related to the distribution of certain high-risk international clones [2]. In this study, we used whole-genome sequencing (WGS) to identify the predominant sequence types (STs) and β-lactamase genes in multi-drug resistant (MDR) P. aeruginosa clinical isolates from Qatar.
Methods
The study setting, bacterial identification, antimicrobial susceptibility testing, whole genome sequencing, and statistical analysis details are provided in Additional File 1. MDR status was defined as in-vitro resistance to at least one agent from three or more classes of anti-pseudomonal agents [3]. β-lactamases were classified according to their molecular groups [4]. Clinical data were retrieved from the electronic healthcare system.
Results
Seventy-five MDR-P. aeruginosa isolates were included (Additional file 1: Table S1). The largest proportions of susceptibility were to ceftazidime-avibactam [36, 48%; minimum inhibitory concentration (MIC)50/90 12/256 µg/ml] and ceftolozane-tazobactam (30, 40%; MIC50/90 24/256 µg/ml) (Fig. 1). Four (5.3%) isolates were resistant to all tested β-lactams except ceftazidime-avibactam, while only one (1.3%) isolate was only susceptible to ceftolozane-tazobactam (Additional file 1: Table S1).
Fig. 1.
Antimicrobial susceptibility testing results for 75 MDR-P. aeruginosa isolates from Qatar. Columns represent number (Y axis) of isolates susceptible and line represents percentage (Z axis) of isolates susceptible to the corresponding antipseudomonal β-lactam. Reporting is based on CLSI breakpoint recommendations (M100, 30th edition—January 2020). ATM, aztreonam; CAZ, ceftazidime; CZA, ceftazidime-avibactam; C/T, ceftolozane-tazobactam; FEP, cefepime; MEM, meropenem; MIC, minimum inhibitory concentration in µg/mL, TZP, piperacillin-tazobactam
Almost all isolates possessed Class C and Class D β-lactamases (72, 96% each). All 4 β-lactamase classes were present in 3 (4%) isolates. Metallo-β-lactamases (MBL) were detected in 20 (26.7%) isolates. Eight (40%) MBL producers were susceptible to aztreonam and did not produce any concomitant extended-spectrum β-lactamases (ESBL) (Additional file 1: Table S1).
The most frequent STs identified were ST235 (16, 21.3%) and ST357 (8, 10.7%) (Fig. 2). All but one ST235 isolate were resistant to all tested β-lactam agents. Furthermore, amongst the 16 ST235 MDR-P. aeruginosa isolates included in this study, MBL were detected in nine (56.3%), blaVEB-9 in 8 (50%), blaPDC-2 in 15 (93.8%), and blaOXA-10 and blaOXA-50 in all 16 (100%). There were five ST233 MDR-P. aeruginosa isolates; all possessed blaVIM-2, blaPDC-3, blaOXA-4 and blaOXA-486, and four (80%) of them were resistant to all tested β-lactams except aztreonam. Different patterns of β-lactamase genes and β-lactam susceptibility were observed in other STs (Additional file 1: Table S2).
Fig. 2.
Distribution of sequence types of MDR-P. aeruginosa (n = 75) isolated in Qatar from 2014 to 2017
Discussion
This study included data representative of the whole country, as it analyzed isolates from a national diagnostic laboratory. Notably, MDR-P. aeruginosa in Qatar are highly resistant to β-lactam agents. The most active β-lactam antibiotics in this study were those in combination with β-lactamase inhibitors, ceftazidime-avibactam and ceftolozane-tazobactam, were not available for clinical use at the time of the study. Yet, less than half of the isolates were susceptible. Given their recent availability for patients in Qatar, the results reported demonstrate the importance of their appropriate clinical use to minimize further loss of activity [5].
This report included 20 (26.67%) isolates that possessed 21 MBL-encoding genes (16 blaVIM-2, 2 blaVIM-5, and 3 blaIMP-2) (Additional file 1: Tables S1 and S2). This is consistent with the known predominance of Verona integron-encoded metallo-β-lactamases (VIM), and to a lesser extent imipenemases (IMP), in P. aeruginosa from the Middle East [6–8]. Unlike other geographic settings, New Delhi metallo-β-lactamases (NDM) have not been detected in P. aeruginosa from the Arabian Peninsula [7, 9].
Apart from areas with a high prevalence of MBL in P. aeruginosa, the presence of Class A ESBL β-lactamases can result in resistance to ceftolozane-tazobactam [1]. Avibactam is an efficient inhibitor of Class A β-lactamases and hence ceftazidime-avibactam combination retains its activity in this situation but not ceftolozane-tazobactam [10, 11]. In a report from Spain of 24 extremely-drug resistant ST235 P. aeruginosa isolates, 13% were susceptible to ceftolozane-tazobactam and 58% to ceftazidime-avibactam and the predominant β-lactamases identified were VIM-2 (42%) and the Class A ESBL Guiana-Extended-Spectrum (GES)-5 (46%) [12]. Consistent with this, five out of seven ceftolozane-tazobactam-resistant, ceftazidime-avibactam-susceptible MDR-P. aeruginosa isolates in our study possessed class A blaSHV-11 and ESBL-encoding genes such as blaVEB-9 and blaTEM-116. Interestingly, those 7 isolates belonged to seven different STs (Additional file 1: Table S1).
The β-lactamase blaVEB-9 (19, 25.33%), formerly known as blaVEB-1a, was the most frequent ESBL gene identified in the present study [11]. blaVEB-1 is one of the most frequently reported ESBLs in P. aeruginosa from the Middle East including Kuwait, Saudi Arabia and Iran [13–15]. Though blaVEB-9 was reported from Thailand and Eastern Europe, to the best of our knowledge, it has not been previously reported from the Middle East [11, 16]. In this study, MDR-P. aeruginosa producing Vietnamese extended-spectrum beta-lactamase-9 (VEB-9) belonged to ST235 (8/16), ST357 (7/8), ST308 (1/3) and ST3022 (1/1) (Additional file 1: Tables S1 and S2). This pattern suggests dissemination within specific P. aeruginosa STs in Qatar that may be different from neighboring countries.
An interesting observation in this study was that 16 (21.33%) MDR-P. aeruginosa isolates were susceptible to aztreonam but resistant to several other antipseudomonal β-lactams tested (Additional file 1: Table S1). Aztreonam is a weak inducer of Class C enzymes and is not a substrate for Class B and narrow-spectrum Class D β-lactamases [17]. The retained aztreonam activity in these isolates despite resistance to other antipseudomonal β-lactams may be explained by the absence of Class A ESBL in those isolates. Therefore, aztreonam should be included in routine antimicrobial susceptibility testing of clinical P. aeruginosa isolates.
Most MDR-P. aeruginosa isolates included in this study belonged to five STs and had consistent β-lactamase genetic profiles and β-lactam susceptibility patterns (Additional file 1: Table S2). ST235, ST233, and ST357 are already known as high-risk clones in Qatar, Saudi Arabia, Bahrain, and the United Arab Emirates [7]. These three STs are globally disseminated MDR-P. aeruginosa clones [2]. Often, these strains cause regional or nationwide outbreaks, express MDR phenotypes, and are associated with high mortality [12, 18, 19]. VIM-producing ST1284 P. aeruginosa have been described from Brazil, and ST389 from cystic fibrosis patients in Italy [20, 21]. Both sequence types have otherwise limited geographic distribution.
Conclusion
MDR-P. aeruginosa isolates from Qatar are highly resistant to antipseudomonal β-lactams. Global high-risk STs predominate in Qatar and their associated multi-resistant phenotype is a cause for considerable concern.
Supplementary information
Additional file 1. Supplementary data file.
Abbreviations
- CLSI
Clinical Laboratory Standards Institute
- ESBL
Extended-spectrum β-lactamase
- GES
Guiana-Extended-Spectrum
- IMP
Imipenemase
- MBL
Metallo-β-lactamase
- MIC
Minimum inhibitory concentration
- MDR
Multi-drug resistant
- NDM
New Delhi metallo-β-lactamase
- ST
Sequence type
- VEB
Vietnamese extended-spectrum β-lactamase
- VIM
Verona integron-encoded metallo-β-lactamase
- WGS
Whole-genome sequencing
Authors’ contributions
Conception and design of study: M.A.S., E.B.I.; Acquisition of data: M.A.S., F.A.K.; Data analysis and interpretation: M.A.S., A.S.O.; Drafting of manuscript: M.A.S., A.S.O. All authors critically reviewed the manuscript and agreed to its publication. All authors read and approved the final manuscript.
Funding
The study was funded by an internal research grant (IRGC-01-51-033 to EI) from the Medical Research Centre at Hamad Medical Corporation, Doha, Qatar. Support was also provided by NPRP grant (NPRP12S-0219-190109 to AAS) from the Qatar National Research Fund (a member of Qatar Foundation) and the Swedish Research Council for Environment, Agricultural Sciences, and Spatial Planning (Formas) (Grant 219-2014-837 to JJ and BS). The funders were not involved in the conduct of the study, the preparation of the manuscript, or the decision to submit the manuscript for publication.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was approved by the Research Ethics Committee (Protocol Number IRGC-01-51-033) at Hamad Medical Corporation, Doha, Qatar.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13756-020-00838-y.
References
- 1.Horcajada JP, Montero M, Oliver A, Sorli L, Luque S, Gomez-Zorrilla S, et al. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant pseudomonas aeruginosa infections. Clin Microbiol Rev. 2019;32(4):e00031–e119. doi: 10.1128/CMR.00031-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oliver A, Mulet X, Lopez-Causape C, Juan C. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat. 2015;21–22:41–59. doi: 10.1016/j.drup.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 4.Bush K. The ABCD’s of β-lactamase nomenclature. J Infect Chemother. 2013;19(4):549–559. doi: 10.1007/s10156-013-0640-7. [DOI] [PubMed] [Google Scholar]
- 5.Haidar G, Philips NJ, Shields RK, Snyder D, Cheng S, Potoski BA, et al. Ceftolozane-tazobactam for the treatment of multidrug-resistant pseudomonas aeruginosa infections: clinical effectiveness and evolution of resistance. Clin Infect Dis. 2017;65(1):110–120. doi: 10.1093/cid/cix182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yezli S, Shibl AM, Memish ZA. The molecular basis of beta-lactamase production in Gram-negative bacteria from Saudi Arabia. J Med Microbiol. 2015;64(Pt 2):127–136. doi: 10.1099/jmm.0.077834-0. [DOI] [PubMed] [Google Scholar]
- 7.Zowawi HM, Syrmis MW, Kidd TJ, Balkhy HH, Walsh TR, Al Johani SM, et al. Identification of carbapenem-resistant Pseudomonas aeruginosa in selected hospitals of the Gulf Cooperation Council States: dominance of high-risk clones in the region. J Med Microbiol. 2018;67(6):846–853. doi: 10.1099/jmm.0.000730. [DOI] [PubMed] [Google Scholar]
- 8.Ghasemian A, Salimian Rizi K, Rajabi Vardanjani H, Nojoomi F. Prevalence of clinically isolated metallo-beta-lactamase-producing Pseudomonas aeruginosa, coding genes, and possible risk factors in Iran. Iran J Pathol. 2018;13(1):1–9. [PMC free article] [PubMed] [Google Scholar]
- 9.Khan AU, Maryam L, Zarrilli R. Structure, genetics and worldwide spread of New Delhi Metallo-β-lactamase (NDM): a threat to public health. BMC Microbiol. 2017;17(1):101. doi: 10.1186/s12866-017-1012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ortiz de la Rosa J-M, Nordmann P, Poirel L. ESBLs and resistance to ceftazidime/avibactam and ceftolozane/tazobactam combinations in Escherichia coli and Pseudomonas aeruginosa. J Antimicrob Chemother. 2019:74(7):1934–1939. [DOI] [PubMed]
- 11.Lahiri SD, Alm RA. Identification of novel VEB β-lactamase enzymes and their impact on avibactam inhibition. Antimicrob Agents Chemother. 2016;60(5):3183–3186. doi: 10.1128/AAC.00047-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Recio R, Villa J, Viedma E, Orellana MÁ, Lora-Tamayo J, Chaves F. Bacteraemia due to extensively drug-resistant Pseudomonas aeruginosa sequence type 235 high-risk clone: facing the perfect storm. Int J Antimicrob Agents. 2018;52(2):172–179. doi: 10.1016/j.ijantimicag.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 13.Poirel L, Rotimi VO, Mokaddas EM, Karim A, Nordmann P. VEB-1-like extended-spectrum ß-lactamases in pseudomonas aeruginosa. Kuwait Emerg Infect Dis. 2007;7(3):468–470. doi: 10.3201/eid0703.010322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Agamy MH, Shibl AM, Tawfik AF, Elkhizzi NA, Livermore DM. Extended-spectrum and metallo-beta-lactamases among ceftazidime-resistant Pseudomonas aeruginosa in Riyadh. Saudi Arabia J Chemother. 2012;24(2):97–100. doi: 10.1179/1120009X12Z.00000000015. [DOI] [PubMed] [Google Scholar]
- 15.Amirkamali S, Naserpour-Farivar T, Azarhoosh K, Peymani A. Distribution of the bla OXA, bla VEB-1, and bla GES-1 genes and resistance patterns of ESBL-producing Pseudomonas aeruginosa isolated from hospitals in Tehran and Qazvin. Iran Rev Soc Bras Med Trop. 2017;50:315–320. doi: 10.1590/0037-8682-0478-2016. [DOI] [PubMed] [Google Scholar]
- 16.Laudy AE, Róg P, Smolińska-Król K, Ćmiel M, Słoczyńska A, Patzer J, et al. Prevalence of ESBL-producing Pseudomonas aeruginosa isolates in Warsaw, Poland, detected by various phenotypic and genotypic methods. PLoS ONE. 2017;12(6):e0180121. doi: 10.1371/journal.pone.0180121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karlowsky JA, Kazmierczak KM, de Jonge BLM, Hackel MA, Sahm DF, Bradford PA. In vitro activity of aztreonam-avibactam against enterobacteriaceae and Pseudomonas aeruginosa Isolated by clinical laboratories in 40 Countries from 2012 to 2015. Antimicrob Agents Chemother. 2017;61(9):e00472–e517. doi: 10.1128/AAC.00472-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hrabák J, Červená D, Izdebski R, Duljasz W, Gniadkowski M, Fridrichová M, et al. Regional spread of Pseudomonas aeruginosa ST357 producing IMP-7 metallo-β-lactamase in Central Europe. J Clin Microbiol. 2011;49(1):474–475. doi: 10.1128/JCM.00684-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zafer MM, Al-Agamy MH, El-Mahallawy HA, Amin MA, El Din AS. Dissemination of VIM-2 producing Pseudomonas aeruginosa ST233 at tertiary care hospitals in Egypt. BMC Infect Dis. 2015;15(1):122. doi: 10.1186/s12879-015-0861-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balero de Paula S, Cayô R, Streling AP, Silva Nodari C, Pereira Matos A, Eches Perugini MR, et al. Detection of blaVIM-7 in an extensively drug-resistant Pseudomonas aeruginosa isolate belonging to ST1284 in Brazil. Diagn Microbiol Infect Dis. 2017;89(1):80–2. [DOI] [PubMed]
- 21.Marvig RL, Dolce D, Sommer LM, Petersen B, Ciofu O, Campana S, et al. Within-host microevolution of Pseudomonas aeruginosa in Italian cystic fibrosis patients. BMC Microbiol. 2015;15(1):218. doi: 10.1186/s12866-015-0563-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary data file.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.