Abstract
The Rosetta software for macromolecular modeling, docking, and design is extensively used in laboratories worldwide. During two decades of development by a community of laboratories at more than 60 institutions, Rosetta has been continuously refactored and extended. Here we review tools developed in the last five years, including over 80 methods. We discuss improvements to the score function, user interfaces, and usability. Rosetta is available at www.rosettacommons.org.
Editorial summary
Tools developed over the past five years in the macromolecular modeling, docking and design software Rosetta are reviewed in this Perspective.
Introduction
The understanding that molecular structure determines biological function has motivated decades of experimental determination of protein structure and function. Many computational packages have been developed to guide experimental methods and elucidate macromolecular structure, including Rosetta. Rosetta offers capabilities spanning many bioinformatics and structural-bioinformatics tasks. Computational structural biology frameworks with similarly comprehensive scope are few, but key to progress in biology. Schrodinger1, the Molecular Operating Environment2, and Discovery Studio3 are computational chemistry platforms for advanced modeling and design for structural biology, drug discovery and material science, based on molecular mechanics, molecular dynamics and quantum mechanics calculations. The HHSuite4 includes tools for bioinformatics, sequence alignments, structure prediction and modeling. The BioChemicalLibrary5 (BCL) includes tools for structure prediction, drug discovery, and several sequence-to-structure methods using machine learning approaches. The Integrative Modeling Platform6 (IMP) models large macromolecular complexes by incorporating various types of experimental data. OpenBabel7 is a ChemInformatics toolbox supporting molecular mechanics calculations, being most heavily used for interconversion of file formats.
Molecular dynamics packages like CHARMM8, AMBER9, GROMACS10 and others simulate most atoms explicitly with a physics-based energy function that relies on solving Newton’s equation of motion. These methods can be used for folding small proteins, model refinement, modeling phenomena such as ion flow through membrane channels, and modeling interactions with small molecules and are therefore highly complementary to Rosetta. OpenMM11 is an API (application programming interface) for setting up molecular simulations and can be used as a library or standalone application.
Many other tools are available for more specialized tasks, for instance for de novo modeling (AlphaFold12,13, QUARK14, RaptorX15), homology modeling (Modeller16, SwissModel17), fold recognition (iTasser18), protein-protein docking (HADDOCK19, Zdock20, ClusPro21), ligand docking (AutoDock22, FlexX23, Glide24) and numerous other tasks requiring molecular modeling. As the focus here is on Rosetta developments, a comprehensive list of related methods is listed in the Supplementary Note.
One of Rosetta’s advantages is inter-operability of its large number of applications; however, this makes it challenging to track the scope of functionality available to scientists who wish to use the software. This Perspective is meant to guide new, returning, or seasoned users; to help them find the right protocol hiding in the Rosetta haystack.
Development of Rosetta started in the mid-1990s; it was initially aimed at protein structure prediction and protein folding25. Over time, the number of applications grew to address diverse modeling tasks, from protein–protein or –small molecule docking to incorporating NMR data, loop modeling, protein design, and interaction with peptides and nucleic acids (Figure 1). Over more than 20 years, the community of developers and scientists, the RosettaCommons, grew from a single academic laboratory to laboratories at over 60 institutions wordwide26. The software has undergone several transitions, including in programming language and implementation, with the latest protocols based on Rosetta3, first released in 200827. The score function has been continuously improved and has been described in 28 and 29. As part of our sustained focus on accessibility, usability, and scientific reproducibility, we developed several interfaces (PyRosetta30, RosettaScripts31, Foldit32), and emphasized publishing protocol captures33 to accompany manuscripts. As those interfaces have grown more versatile and modular, development has accelerated and branched in many directions. However, this interoperability, extensibility and modularity enable scientists to combine modules in a wide variety of combinations, making it difficult to keep up with all the developments within the software and the scientific community. Here we have compiled the latest method developments in Rosetta from the past five years, divided into several categories; we provide direction on where to find further information for specific modeling problems. The Supplementary Note contains more details on the protocols with extensive links to documentation, resources on the web, limitations, and competitors.
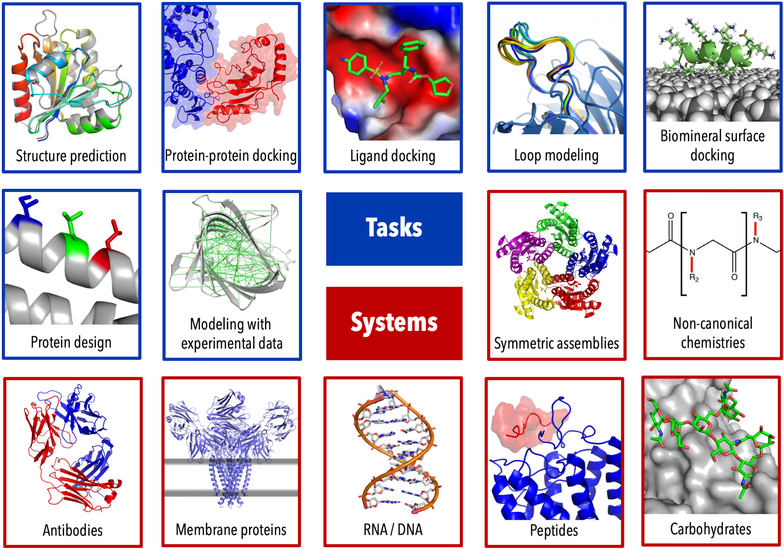
Figure 1: Capabilities of the Rosetta macromolecular modeling suite.
Some popular tasks that can be addressed in Rosetta (blue) and major systems that can be modeled (red). Note this is an incomplete list of Rosetta’s broad modeling capabilities.
1. General overview and challenges
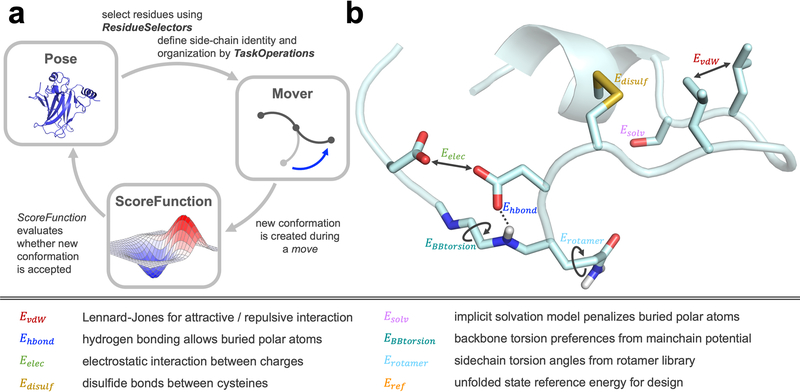
A typical Rosetta protocol is outlined in Figure 2A: the conformation of a biomolecule (the Pose) is altered, either deterministically or stochastically, via a Mover and the resulting conformation is evaluated by a ScoreFunction. The Move is accepted based on the Metropolis criterion and the energy difference between the original and the new conformation:
Figure 2: Main elements of Rosetta are scoring and sampling.
(A) Three main elements are required in a Rosetta protocol. The Pose is the biomolecule, such as a protein, RNA, DNA, small molecule, or glycan, in a specific conformation. Residues in the Pose can be selected via ResidueSelectors and the behavior for side-chain optimization or mutation can be defined by TaskOperations. Specific Movers then control how the conformation of the Pose is changed, and the new conformation is subsequently evaluated by a ScoreFunction. The Metropolis criterion decides whether the new conformation is accepted during sampling. Many independent sampling trajectories are generated, and the final models are evaluated based on the purpose of the protocol. (B) The score function consists of a weighted linear combination of various score terms, highlighted in the figure and described above.
Many independent trajectories are generated, and the final models are evaluated based on the scientific objective. This setup highlights common limitations in Rosetta protocols involving sampling, scoring (discussed in the score function section), or technical challenges. Many protocols suffer from under-sampling34, especially when flexibility is involved. Sampling is a limitation for structure prediction (especially for large structures), protein design and unconstrained global protein-protein docking. For example, even with local docking we are limited by backbone flexibility and performance deteriorates with larger flexibility in the binding interface. Small molecule docking similarly relies on correct identification of the binding interface and is limited by flexibility between unbound and bound states. Enormous conformational search spaces are also prohibitive for RNA modeling due to the size and combinatorics of their torsion space (see RNA section), membrane proteins due to their size, and carbohydrates because of branching and flexibility.
Some Rosetta applications suffer from (1) technical challenges in implementation, (2) a lack of documentation, protocol captures, or support, and (3) a need for more diverse chemistries for biomolecules. Technical challenges are either historical or due to lack of interest in the community to develop and advance methods in these unique areas.
2. Rosetta’s score function
Rosetta’s score function has been continuously improved over many years35 with guiding principles including: improving speed of computation, increasing extensibility, and improving accuracy across multiple tasks. The main score function is a linear combination of weighted score terms that balances physics-based and statistically derived potentials describing van der Waals energies, hydrogen bonds, electrostatics, disulfide bonds, residue solvation, backbone torsion angles, sidechain rotamer energies, and an average unfolded state reference energy (Figure 2B):
Some energy terms are decomposed into several components to parameterize each of them separately. For instance, the van der Waals energy is split into attractive and repulsive terms between different residues, in addition to an intra-residue repulsive term. A detailed account of the all-atom score function was published recently28.
The newest score function REF201529 reproduces thermodynamic observables (such as liquid-phase properties36 and liquid-to-vapor transfer free energies37) in addition to structure38-based tests. It also utilizes a new, derivative-free optimization technique, which is suitable for robust optimization of >100 parameters. Further, a new energy term was added that takes into consideration non-ideality of bond lengths and angles in cartesian space39. The cartesian term39 is also the basis for a cartesian_ddG method that has been used to calculate ΔΔGs of mutations to assess changes in protein stability. Only the backbones and side chains of residues near the mutation site are allowed to move40. Due to the local optimization, this protocol is much faster than the previous gold-standard ddg_monomer41, while retaining the same level of accuracy. REF2015 is now compatible with an expanded palette of chemical building-blocks: canonical and non-canonical L-α-amino acids and their D-amino acid counterparts, exotic achiral amino acids, peptoids, and oligoureas, and can model metalloproteins42. Score functions that enable simultaneous modeling of protein and RNA are being explored43. REF2015 is now thread-safe and fully mirror symmetric, i.e. enantiomers in mirror conformations score identically. Guidance energy terms for design have been added to encourage certain features, such as specific amino acid compositions44,45, hydrogen bonding networks, or global or local net charges, and discourage others, such as repeat sequences that hinder NMR assignments, buried unsatisfied hydrogen bond donors and acceptors, or voids within the protein46.
Hydrogen bond networks are important for biomolecular structure and catalysis but have been challenging to design because of pairwise interactions that have multi-body, cooperative properties. The HBNet protocol47 has been used to design de novo coiled coils with interaction specificity mediated by designed hydrogen bond networks, including homo-oligomers47, membrane proteins48, and large sets of orthogonal heterodimers49. An improvement to HBNet uses a Monte Carlo search to sample hydrogen bond networks with drastically improved performance50. We further developed a statistical potential to place highly-coordinated water molecules on the surface of biomolecules. On a data set of 153 high-resolution protein-protein interfaces, the method predicts 17% of native interface waters with 20% precision within 0.5 Å of the crystallographic water positions51. The potential is accessible through the ExplicitWaterMover (former: WaterBoxMover) in RosettaScripts.
There are still several limitations to the score function: (1) it does not directly estimate entropy52, which has been shown to improve sampling efficiency53. However, rotamer bond angles, solvation, fragments and pair terms all implicitly model this component of the free energy, which at these temperatures and solvation densities account for more than half of the entropy. (2) In most cases, knowledge-based score terms are derived from high-resolution crystal structures, representing a single state on the energy landscape and do not represent flexibility well (compared to solution NMR); (3) knowledge-based terms are less interpretable and transferable than physics-based terms; (4) scoring performance scales with the number of score terms and has become slower, yet more accurate, over time; (5) the solvation model is implicit, hence fast, but hinders explicit modeling of ions, water molecules, or lipid environments; (6) several score functions for specific applications (RNA, membrane proteins, carbohydrates, non-canonical amino acids) are still developing.
3. Major applications
Predicting protein structures
Rosetta was originally developed for de novo protein structure prediction, assembling fragments from known protein structures via a Monte Carlo procedure and evaluating the models with the score function. While the community’s main goals have moved to macromolecular design over the past decade, performance in the CASP13 blind prediction challenge remains respectable54, with ranking for refinement and prediction of multimeric complexes among the top three groups. Meanwhile, other groups have refined their tools exploiting evolutionary couplings and machine learning, for instance Google’s DeepMind developed AlphaFold12,13 (which uses Rosetta for refinement) with outstanding performance in the recent CASP1354. Another highly ranking method is the Zhang server built on iTasser14, and QUARK14.
Homology modeling was improved by using multiple templates in RosettaCM55 (now available on the new Robetta56,57 server), which hybridizes the most homologous portions from multiple templates into a single model, while modeling missing residues de novo55. Without a template, predicting protein structures de novo, remains one of the most challenging tasks in structural biology, even though the incorporation of evolutionary coupling constraints (for instance from GREMLIN58) has led to enormous improvements in model quality. An iterative hybridize approach improves sampling and uses a genetic algorithm that recombines models from an input pool to create models that have features from their parents but are also distinct. Creating several child models in each iteration, updating the input pool, and performing 30–50 iterations led to improved model accuracy because features that are scored favorably are repeatedly used in the recombination, such that the models in the pool converge over time. Iterative hybridization has been used to improve model quality of de novo predicted models59 as well as homology models60. Model refinement or generating ensembles of structures (useful for design) can be accomplished by several algorithms in Rosetta: FastRelax61, Backrub62, or vicinity sampling using KIC/Next-Generation-KIC loop modeling 63,64. Loop modeling65 was implemented early in Rosetta66,67, with initial approaches relying on fragments sampling and iterative Cyclic Coordinate Descent (CCD)68 for chain closure. Later, a kinematic closure (termed “KIC”) approach relied on polynomial resultants to analytically solve for closed conformations, producing more native-like loops69,70. Next-Generation KIC (NGK)64 is a recent innovation that improves sampling by employing diversification (i.e. wider range of conformations) and intensification (i.e. focus around previously generated conformations), substantially increasing the fraction of near-native models64 and modeling longer loops. A related method, GeneralizedKIC44 (GenKIC) samples loop geometries between fixed endpoints including non-standard peptide chemistries or chemistries that conventional loop-modelling algorithms do not typically handle.
Modeling protein–protein complexes
Another early expansion of Rosetta’s functionality was RosettaDock, a method for predicting the structure of protein-protein complexes. The latest version, RosettaDock4.074 incorporates protein flexibility from pre-generated protein ensembles, mimicking conformer selection. This has improved sampling efficiency by automatically adjusting the sampling rate based on the diversity of the input ensembles. Scoring has been improved by a six-dimensional coarse-grained scoring scheme called motif_dock_score, employing score grids generated from known complexes in the Protein Data Bank (PDB). In local docking benchmarks with backbone deviations of up to 2.2 Å, RosettaDock4.0 successfully docked ~50% of complexes74. For symmetric homomers, Rosetta SymDock275 uses the same six-dimensional scoring scheme as RosettaDock. Symmetry information can be extracted from a homologous complex, or from a global docking search for a given point symmetry using our symmetry framework152. An induced-fit based all-atom refinement relieves clashes in tightly-packed complexes to give physically realistic models. On a benchmark set of 43 complexes with different cyclic and dihedral symmetries, global docking on homology models had accuracies of 61% and 42% for cyclic and dihedral symmetries, respectively75. These accuracies can be dramatically improved when adding restraints.
Docking small molecule ligands into proteins
Structure-based drug design has become a key drug optimization tool and leverages the vast array of knowledge contained in the increasing numbers of deposited structures in the PDB. RosettaLigand76 has demonstrated success in predicting small molecule-protein interactions. Later in the drug development process, medicinal chemists optimize ligands based on structure-activity relationships (SAR) by synthesizing different ligands that share a core chemical scaffold and are assumed to bind to their target in a similar fashion153. RosettaLigandEnsemble79 improves sampling during ligand docking by taking advantage of ligand similarities and docking a congeneric series of ligands simultaneously, allowing for a placement that works for all considered ligands while optimizing the binding interface for each ligand independently. Experimental SARs can help identify preferred binding modes. Small molecule ligands can also be used as competitive inhibitors of protein-protein interactions. However, a protein’s inhibitor-bound conformation often differs from the unbound or protein-protein bound conformation, thus Rosetta’s ability to model protein conformational flexibility is key. Rosetta’s pocket optimization approach identifies protein surface pockets and uses their volume as an additional scoring term: this allows the user to start from an unbound protein structure and bias sampling such that low-energy pocket-containing states are preferentially explored80,81. The sampled conformations match “druggable” alternate conformations observed in ligand-bound structures80,81, making these states excellent starting points for virtual screening. Pockets sampled on a protein surface can then be matched to complementary ligands by using the pocket as the starting point for pharmacophore-based screening154.
Modeling and designing antibodies and immune system proteins
Due to the therapeutic significance of antibodies, several antibody-specific and immune-specific protocols have been developed for structure prediction, docking and design (with specific protocols targeting IgG, T-cell receptors, displayed antigens of the Major Histocompatibility Complex (MHC) and other soluble antigens and immunogens). RosettaAntibody85–88 is a protocol for modeling of antibodies88. It identifies homologous templates, assembles them into a single structure and then models CDR H3 loops de novo while refining the VH-VL orientation155. Recent advances use multiple templates155, incorporate key structural constraints156,157 into CDR H3 modeling, model camelid antibodies87 and antibodies on the scale of the human repertoire158,159. AbPredict89 predicts antibody structures without homologous templates. Instead, it samples backbone fragments and rigid-body orientations from known antibody structures, without relying on sequence homology, therefore accurately modeling cases with sequence identity as low as 10%. AbPredict2 is available as a webserver90. SnugDock93 is a related method for antibody-antigen docking, taking as input a plausible starting conformation and optionally an ensemble of antibodies/antigens. SnugDock then runs local docking to refine both the antibody–antigen interface and the heavy–light chain interface (within the antibody) and re-models the CDR H2/H3 loops at the interface. Recent advances include a CDR H3 structural constraint156,157 and docking camelid antibodies160. Limitations in antibody modeling depend on the task: docking is limited by knowledge of the binding site (global vs. local docking); structure prediction, design and refinement are limited by protein flexibility, and modeling of CDRs or other loops is challenging if they are longer than 12 to 15 residues.
RosettaAntibodyDesign94 (RAbD) is based on RosettaAntibody87 (see below) and allows design of specific CDRs of different clusters and lengths, sequence design using cluster-based CDR profiles or conservative mutations, or de novo design of whole antibodies. RAbD uses North-Dunbrack CDR clustering161, reducing deleterious sequence mutations, and was benchmarked on 60 diverse antibody-antigen interfaces from complexes including both λ and κ light chains. Experimental benchmarking of two antibody-antigen complexes showed affinity improvements between 10 and 50-fold. Rosetta has been integrated with experimental immunogenic epitope data, MHC epitope prediction tools, and host genomic data to design proteins with reduced immunogenicity while retaining function and stability95. The approach implements machine learning-based epitope prediction for 28 different alleles, restricts design to select 15mer epitope regions, and uses a greedy stepwise protein design96 to eliminate the most immunogenic epitopes with the least mutations, avoiding disruptive core mutations likely to destabilize the protein. Another method, AbDesign, splits experimentally determined antibody structures along conserved positions to create interchangeable segments and then recombines them to produce a diverse set of novel antibody models97,98. The models are docked to a target of interest, either locally to a specific epitope, or globally, followed by an optimization step comprised of rigorous backbone sampling and sequence design for improving model stability and binding affinity.
Designing new proteins and functions
Protein design162 relies on several of the same core functionalities needed for structure prediction, and synergy and interoperability between design and prediction models has always been a core Rosetta design principle. For example, this synergy is well illustrated by the biased forward folding method: During de novo protein design163, a test for the consistency of the designed sequence is whether ab initio structure prediction will yield the same structure that was used as a starting point for the design. However, computationally testing a large number of designs is prohibited by the vast conformational search space for ab initio structure prediction. To limit that space and test more designs, biased forward folding72 uses three (instead of 200) fragments per residue position with fragments being chosen based on the RMSD to the native structure used to instantiate the design process. Protein design is easier when starting from known structures and when redesigning for well understood objectives like thermostability 164. More difficult design objectives include de novo design (without a template structure) and design for novel folds or functions. Successes in these cases require sampling of enormous conformational spaces, depending on the protein size. Another simplification of de novo design is thermostabilization of the protein, essentially creating rigid structures that are mostly non-functional, by expanding the energy gap between folded and unfolded designs to facilitate structural characterization. To date, novel functional designs mostly exploit known structures and the next frontier is the design of novel functions onto de novo scaffolds. Moreover, nature typically does not design for the global minimum energy conformation (in terms of stability) because proteins require flexibility to carry out their functions.
Design of novel protein structures and functions towards therapeutic intervention is addressed by various methods in Rosetta: SEWING creates de novo designs by recombining parts of protein structures from randomly-selected helical building blocks99. SEWING’s requirement-driven approach allows users to specify features that should be incorporated into their designs during backbone generation without requiring a certain size or three-dimensional fold. New features include incorporation of functional motifs such as protein-binding peptides for protein interface design and ligand binding sites for ligand-binding protein design100. A similar algorithm has been implemented for antibody design (AbDesign, see above), which was generalized for enzyme design165. A more general approach is RosettaRemodel, performing protein design by rebuilding parts or all of the structure101 from fragments of known proteins structures. RosettaRemodel uses a blueprint file in which the user defines secondary and supersecondary structure of the desired fold. Remodel interfaces with various Rosetta protocols and allows de novo modeling, fixed-backbone sequence design, refinement, loop insertion, deletion, and remodeling, disulfide engineering, domain assembly, and motif grafting.
A common task is not only design towards a certain goal (positive design), but additionally, design away from undesired features (negative design). Such a Multi-State Design166 (MSD) approach evaluates strengths and weaknesses of a single sequence on multiple backbones, for instance binding to one but not another protein partner. REstrained CONvergence103 (RECON) allows each state to sample multiple sequences during the design process, which is iteratively applied by increasing the restraint weight to encourage sequence convergence. RECON achieves on average 70% sequence recovery (a 30% increase compared to MSD) for large multi-state design problems, such as antibody affinity maturation or predicting evolutionary sequence profiles of flexible backbones167,168.
Protein function can be designed by motif grafting, i.e. grafting a known motif or predicted active- or binding-site from a template structure onto a new protein. This approach has been used for antibodies and vaccine design104 using the fold_from_loops application, where the functional motif is used as a starting point of an extended structure that is folded following the constraints of a target topology. Iterative refinement is carried out via sequence design and structural relaxation before filtering and human-guided optimization. This protocol has been extended into the Functional Folding and Design (FunFolDes) protocol, including multi-segment motif grafting, different residue length motif insertion, incorporating restraints, and folding in the presence of a binding target105. Performance of the folding stage can be improved by selecting fragments according to the target topology via the StructFragmentMover.
Designing interfaces between proteins and interaction partners
Protein design problems include interface design of proteins with proteins or small molecule ligands and predicting ΔΔGs of mutation (e.g. alanine scanning). Predicting ΔΔGs of mutations for protein stability or protein-protein interactions is difficult with low correlation coefficients (0.5–0.7)169, because the effect of the mutation is small compared to the total energy in the system, and because protein flexibility adds noise to the energies that can mask the effect of mutations. In alanine scanning (mutating into Ala), methods that use a “soft-repulsive” score function without modeling backbone flexibility170,171 typical outperform methods that allow protein flexibility and use hard-repulsive score functions172. FlexDDG106 improves protein-protein interface ΔΔG predictions and generalizes them to residues other than Ala. The protocol creates conformational ensembles using backrub sampling173, then repacks sidechains, minimizes torsions and computes change in protein-protein interaction ΔΔG by averaging across the ensembles. On 1240 interface mutants, FlexDDG outperforms the earlier ddg_monomer application, which was created to predict changes in stability upon mutation, not interfaces.
Symmetric protein assemblies modeled using parametric design. Nature created super-helical coiled-coils that are well-described by geometric equations using Crick parameters174, including variables for the radius of the bundle, major helical twist, minor helix rotation about the primary axis, etc. Several Movers such as MakeBundle, PerturbBundle, and BundleGridSampler allow designing helical bundles48,108 and β-barrels based on pre-defined or sampled parameters. These parametric methods do not rely on fragments libraries and can be applied to non-canonical coiled-coil heteropolymers.
Modeling peptides and peptidomimetics
The inherent flexibility of peptides imparts a large conformational search space to them, leading to challenging modeling problems; when peptide modeling is combined with another simulation, e.g. docking, the increase in conformational space makes the modeling task quite challenging by any method. PIPER-FlexPepDock111 is Rosetta’s global peptide docking protocol. It rigid-body docks fragments using PIPER FFT-based docking175, and refines the complex using FlexPepDock109. PIPER-FlexPepDock can generate peptide-protein complexes from a peptide sequence and a free receptor structure (Figure 3F). Performance decreases in case of receptor flexibility.
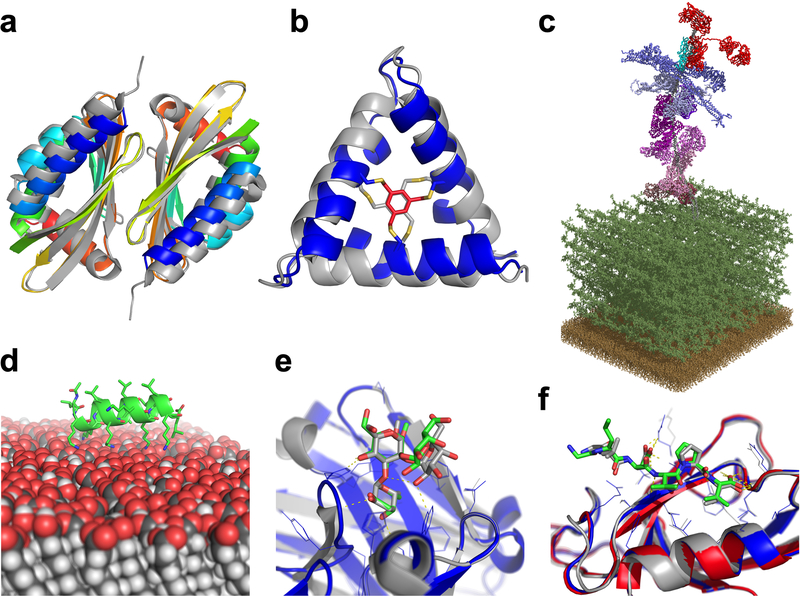
Figure 3: Rosetta can successfully address diverse biological questions.
(A) Curved β-sheet design: overlay of the designed homo-dimeric curved β-sheet (dcs-E_4_dim_cav3) in rainbow and the crystal structure in gray (PDBID 5u35). The protein is designed de novo and features a curved β-sheet, a large pocket, and a homodimer interface72. (B) Parametric design: overlay of the de novo designed macrocycle 3H1 in blue and the NMR structure in gray (PDBID 5v2g). This “CovCore” (covalent core) miniprotein is held together covalently by a hydrophobic cross-linker at its core (in red for the design and gray for the NMR structure)108. (C) PyTXMS: the interactome of M1 protein (virulence factor of Group A streptococcus) and 15 human plasma proteins on the surface of bacteria (peptidoglycan layer (dark green), and the membrane (brown)). This 1.8MDa structure contains over 200 chemical cross-links128 and is measured in a complex mixture of intact bacteria and human plasma. All models are provided by Rosetta: M1 protein (gray), IgG (red), four fibrinogens (dark to light blue), six albumins (dark to light pink), coagulation factor XIII A [F13A] (purple), C4bPa (cyan), haptoglobin [HP] (brown), and alpha-1-antitrypsin [SerpinA1] (plum). (D) RosettaSurface: model of an LK-α peptide (LKKLLKLLKKLLKL with a periodicity of 3.5 assuming a helical conformation) on a hydrophilic self-assembled monolayer surface. The peptide is unstructured in solution and assumes helical structure115 when on the surface, as experiments show. (E) RosettaCarbohydrate: flexible docking of a carbohydrate antigen to an antibody. The crystal structure is in gray (PDBID 1mfa) and the model in blue, with the carbohydrate in green. Antibody coordinates were taken from the PDB and glycan coordinates started from a randomized backbone conformation and rigid-body orientation143. (F) PIPER-FlexPepDock: high-resolution model of a peptide-protein complex (model: blue; solved structure in gray, PDBID 1mfg). The model was generated from a peptide sequence (LDVPV, derived from the C-terminal tail of ErbB2R) and the unbound structure of the receptor (Erbin PDZ domain, PDBID 2h3l, colored in red)111.
Cyclic peptide conformations can be sampled with simple_cycpep_predict, restricting the conformational search space through cyclization44,45,108 via the Generalized Kinematic Closure (GenKIC) algorithm (see “loop modeling” above). Simple_cycpep_predict does not rely on protein fragments and can model non-canonical chemistries (Figure 3B), being a generalization of earlier protocols. Experimental protein structure determination is challenging for proteins on solid surfaces such as biominerals, self-assembled monolayers, inorganic catalysts, and nanomaterials. RosettaSurface114 samples protein conformations ab initio in both the solution and adsorbed states (Figure 3D) to account for adsorption-induced conformational changes. Experimental data can be incorporated115 to improve scoring.
Using experimental data to direct modeling
Using experimental data in modeling can vastly restrict the conformational space, allowing the modeling of larger, more complex biomolecules to greater accuracy. Electron density maps generated by cryo-electron microscopy (cryoEM) or X-ray crystallography have improved in quality and become substantially more available in the past decade and methods to incorporate them can produce high-resolution structures. To deal with variations in the resolution of these methods RosettaES118 samples enumeratively, not requiring initial assignment of densities; it gradually extends the model one residue at a time until all residues are assigned. At each iteration, short fragments are used to sample the nearby conformational space of the growing model, while undergoing a series of clustering and filtering steps based on the energy and fit to the density. If assignment is complete but the data are low-resolution, refinement into density maps is necessary. Several methods have been developed for density maps in the 3.0–4.5Å resolution range. More recently, an automated fragment-guided refinement pipeline121 splits the density map into independent training and validation maps. It finds regions with poor density fit, iteratively rebuilds them with fragments using the training map, filters the models based on their fit to the validation map, model geometry from MolProbity and fit to the full map, and then optimizes against the full map. Further, the frameworks for electron density maps and carbohydrate modeling143 (below) were connected144, allowing refinement of carbohydrates into low-resolution density maps.
NMR data were incorporated into de novo structure prediction early on, embodied in RosettaNMR. Chemical shifts were used for fragment picking using CS-Rosetta122, which could be used with Nuclear Overhauser Enhancements (NOEs), Residual Dipolar Couplings (RDCs)176, Pseudo-Contact Shifts (PCSs)123,124,177 and Paramagnetic Relaxation Enhancement (PRE) data. Improvements, for instance through RASREC resampling178 allowed the use of sparse179 or unassigned data180, easier-to-obtain data (backbone-only181), modeling larger and more complex proteins182, membrane proteins183, symmetric systems184, and combination with data from SAXS185, cryoEM186, distance restraints from homologous proteins187 and evolutionary couplings188. CS-Rosetta also has the AutoNOE189,190 module for automated assignment of NOESY data for use in structure calculations. RosettaNMR was recently overhauled and reconciled with CS-Rosetta and PCS-Rosetta to seamlessly integrate several types of NMR restraints (CS, RDC, PCS, PRE, NOE) in one consistent framework191 for structure prediction, protein-protein docking, protein-ligand docking, and symmetric assemblies.
Covalent labeling mass spectrometry data provides information on relative solvent exposure of residues, yielding information on protein tertiary structure. A low-resolution score term that allows for use of hydroxyl radical foot-printing has been implemented that can improve model quality in structure prediction126,127. Moreover, data from chemical cross-linking mass spectrometry has been incorporated into an automated workflow to identify protein-protein interactions. The PyTXMS128 protocol combines the sensitivity of mass spectrometry to analyze complex samples with the power of Rosetta structural modeling and protein-protein docking to efficiently sample the vast conformational space and identify interactions (Figure 3C). A machine learning algorithm based on high resolution MS1 data guides the potential binding interface selection, being validated and adjusted by a repository of structural models and MS2 (data-dependent acquisition (DDA)) samples.
Modeling nucleic acids and their interactions with proteins
DNA and RNA modeling requires addressing a multitude of challenges due to a lack of structures leading to under-developed score functions, low quality alignments, and a much larger sampling torsion space than for proteins (70 residue RNA comparable to 200 residue protein). In contrast to protein helices where side-chains display sequence information on the helix exterior, helical RNA sidechains point inwards, therefore hiding sequence information from the environment, making prediction of tertiary or non-local contacts more difficult. Non-local contacts are mediated by loops, challenging for prediction algorithms. Several advances have been made in the representation of nucleic acids in Rosetta. The StepWise Monte Carlo protocol (SWM) has achieved RNA structure predictions reaching atomic accuracy131; the approach provides an acceleration over the original enumerative StepWise Assembly (SWA) method129,130. A version of SWA that rebuilds one nucleotide at a time enables fine-grained correction of errors in RNA coordinates fit into crystallographic or cryo-EM maps by Enumerative Real-space Refinement ASsisted by Electron density under Rosetta135,136 (ERRASER).
The most recent advances in RNA tools expand the fragment assembly protocol to support modeling RNA-protein complexes through simultaneous folding and docking134. RNA-protein interactions are handled via additional knowledge-based score terms that supplement the low-resolution RNA score function. Free energy perturbations from RNA or protein mutations can be modeled with the Rosetta-Vienna ΔΔG protocol43. Structure coordinates can further be built into cryo-EM density maps for large RNA-protein complexes with DRRAFTER (De novo Ribonucleoprotein modeling in Real space through Assembly of Fragments Together with Experimental density in Rosetta)138. Redesign and prediction of protein-DNA interfaces192,193 has been accomplished with flexible protein backbones194, genetic algorithms192,194,195 and motif-biased rotamer sampling196,197. A potential limitation is the reliance on fixed DNA backbone conformations, which can be flexible. Key to successful protein-DNA design is a score function optimized197,198 for these highly charged and solvated interfaces. Rosetta supports prediction of specificity and affinity199, the prediction of DNA binding preferences of homologous proteins and multi-template modeling in RosettaCM55,200.
Modeling membrane proteins
Membrane proteins constitute about 30% of all proteins and are targets for over 60% of pharmaceuticals on the market201. However, experimental difficulties have limited our understanding of their structures202. Previously, Yarov-Yarovoy203 and Barth204 implemented tools for low- and high-resolution structure prediction of membrane proteins, termed RosettaMembrane. These tools were re-engineered for compatibility with Rosetta327 into a platform called RosettaMP139. RosettaMP implements core modules for representing, sampling, and scoring proteins in the context of an implicit membrane. RosettaMP is compatible with key modeling protocols including docking, design, ΔΔG prediction169, PyMOL visualization205, and assembly of symmetric proteins. Additionally, a set of basic modeling tools140 allows scoring, transforming a membrane protein into the membrane coordinate frame, de novo modeling for single transmembrane span helices, introducing mutations, and visualization in the membrane. RosettaMP has enabled rapid development of new tools including structure-based detection of lipid exposed residues in the membrane141 and domain assembly of full-length protein models from structures of transmembrane and soluble domains142. The RosettaCM protocol for multi-template homology modeling has also been adapted to membrane proteins33.
Describing membrane protein energetics is challenging as these proteins reside in an anisotropic environment and bury polar solvent molecules (e.g. water, ions) that stabilize the structure and participate in important conformational transitions. Implicit membrane models often fail to reliably model membrane protein interiors. The method SPaDES is based on a hybrid explicit-implicit solvent model that enhances the prediction and design of membrane protein structures206. Limitations to membrane protein modeling are similar but less severe than for RNA modeling: there are fewer structures in databases, fewer method developers in this field and hence fewer available tools. Consequently, the score function is less mature compared to the latest score functions for soluble proteins: the implicit solvent hydrophobic slab model is a coarse-gained representation of the membrane. Ongoing efforts expand this model by including pores, lipid specificity and different thicknesses207, yet many effects remain to be acknowledged such as measurement-specific or observed membrane geometries (micelles, bicelles, nanodiscs, vesicles, different pore types, fusion and fission of multiple membranes) and macroscopic physical phenomena like membrane tension and fluidity. Challenges in including these effects are experimental measurements for parameterization of these models and adaptation of a multitude of score terms.
Adding carbohydrates to the modeling process
Carbohydrates are fundamental to life208,209, but because of challenges in experimental characterization and computational sampling and scoring, their structures have been historically under-studied. The RosettaCarbohydrate framework143 models carbohydrate structures and complexes such as glycosylated proteins or protein–sugar complexes (Figure 3F) with the same algorithms one would use for proteins. RosettaCarbohydrate can handle commonly studied and uncommon carbohydrate structures, including linear, cyclic, and branched structures, sugar modifications, and conjugations. Methods exist for sampling ring conformations, packing substituents, refining glycosidic linkages, sampling from linkage “fragments”, and extending glycan chains. Scoring of saccharide-containing sugars includes a quantum-mechanically derived intrinsic backbone term210. Because saccharide residues are stored as distinct data structures, we can integrate bioinformatic and statistical data into these algorithms, opening the door for glycoengineering and design applications. RosettaCarbohydrate has been integrated with other frameworks, such as loop modeling (GenKIC and Stepwise Assembly), refinement (GlycanTreeModeler), symmetry, and RosettaScripts-accessible classes such as MoveMaps and ResidueSelectors. Linkages are automatically determined during PDB read-in. Carbohydrates work with Cartesian minimization, and can be refined into electron density maps144. Limitations in the carbohydrate framework include the increased sampling space due to carbohydrate flexibility and branching, and need to model many different chemistries with possible branching and cyclization. Developments in this area have only recently started and much work has yet to be done.
4. User interfaces and usability
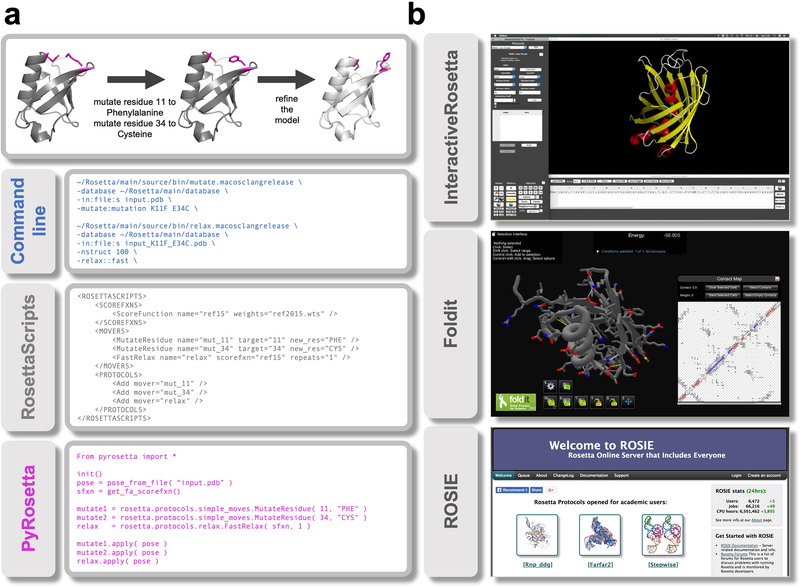
Advances have also focused on improving usability of Rosetta through several user interfaces to suit different use cases and workflow styles (Figure 4). The command line was the first and is still the most-often used interface to Rosetta methods. Additionally, Rosetta features two popular scripting interfaces: RosettaScripts and PyRosetta. RosettaScripts31 uses Extensible Markup Language (XML) to build complex protocols using core machinery27, without requiring knowledge of the codebase. PyRosetta30,145 is a collection of Python bindings to the source code, allowing flexible and fast custom protocol development, but requires familiarity with the underlying codebase. Other interfaces are InteractiveRosetta146 and the gaming interface Foldit Standalone147,149 (see Supplementary Note).
Figure 4: User interfaces to the codebase.
(A) Rosetta can be run from a terminal and offers three interfaces to the codebase. The top panel outlines the task to be accomplished: making two mutations in a protein and then refining the structure. The panels underneath show how this task can be accomplished in the different interfaces. The command line panel shows the executable, input files and options to run two specific applications. RosettaScripts is an XML-based scripting language that offers more flexibility by combining Movers and ScoreFunctions into a custom Protocol. PyRosetta offers direct access to the underlying code objects but requires knowledge of the codebase. (B) Point-and-click interfaces to the codebase. InteractiveRosetta is a graphical user-interface (GUI) to PyRosetta. It offers controls to the most popular protocols, file formats and options. Foldit is a videogame primarily used to crowd-source real-world scientific puzzles but can also be used on custom proteins of interest. It can run some popular applications via a game interface. ROSIE hosts a multitude of servers each executing a particular protocol. It currently includes servers for 21 Rosetta methods. [The InteractiveRosetta and Foldit panels were originally published in 213 and 147 under Creative Commons licenses that allows reproduction as is.]
We devoted an enormous effort to rewrite and add documentation (Figure 5). A public-facing Gollum wiki (https://www.rosettacommons.org/docs/latest/Home) houses various levels of documentation, such as application documentation, tutorials for beginning users, and static protocol captures that accompany manuscripts for scientific reproducibility (see Supplementary Note for links). The Gollum wiki is easily editable by members of the RosettaCommons which has drastically improved the quantity and quality of documentation.
Figure 5: Main external documentation page.
In 2015, our community performed a complete overhaul of our documentation. Documentation is now hosted on a Gollum wiki, which is version controlled and easily editable by members of our community. Accessibility and ability to edit the documentation has drastically improved the user-experience of the software.
A limitation of Rosetta is the need for a local installation and compilation in a Unix-like environment. Webservers provide a user-friendly alternative and a number of independent servers have emerged in our community. However, implementing and maintaining such servers comes at a substantial cost. To make it easier to provide protocol webservers, ROSIE (Rosetta Online Server that Includes Everyone)150,151 (http://rosie.rosettacommons.org/) implements a simple framework for “serverification” of protocols. ROSIE currently contains 24 webservers, with additional protocols continually being added.
Conclusion
The Rosetta software is developed by a large, global community aiming to solve complex problems through real-time collaborative code development. In the last five years, great strides have been made in our software. More protocols enable modeling a broader range of biological and chemical macromolecular systems. Prediction accuracies have improved through advances in the score function, which is a combination of physics-based and knowledge-based potentials that were fit against known structures and thermodynamic observables. Incorporating experimental data into modeling has been facilitated and improved. Further, our community now develops more general, reusable, user-friendly, and scientifically reproducible protocols. This was motivated by the growth of the software and the developer community, the various user interfaces, the diversity of the community26, and the complexities of the protocols used to solve real-world problems. The improvements to documentation allow users to quickly start using or developing custom protocols, while facilitating user support for the various interfaces (command line, RosettaScripts, PyRosetta, etc.). Over the years, these applications have moved beyond tackling basic science questions (i.e. the protein folding and design challenges) to more application-based scientific developments. The myriad advances described above have made integration of Rosetta into existing experimental and computational scientific workflows increasingly useful and standard, as evidenced by the large number of licenses (~30,000 academic and ~70 commercial including most of the largest pharmaceutical companies), 11 spin-off companies that were created from the RosettaCommons26, and the ever-increasing number of citations from labs beyond those affiliated with RosettaCommons.
Rosetta development is ongoing and will continue to focus on expanding the scope of protein design and modeling by integrating high-throughput experimental data with high-throughput computation, impacting score function development and aiding in developing novel therapeutic interventions211; restructuring the software for massively parallel computing architectures (e.g. GPUs, TPUs) and quantum computers212; greater use of machine-learning (e.g. deep-learning) approaches (e.g. for score function development); modeling more realistic cellular environments; and improving user interfaces to make Rosetta accessible to more scientists. The predictive powers that we have reviewed above can be leveraged not only to analyze and verify existing data but to inform experiments that will galvanize engineering industrial enzymes, enable the creation of novel biomaterials, and accelerate the discovery of new potent therapeutics.
Code availability
Rosetta is licensed and distributed through www.rosettacommons.org. Licenses for academic, non-profit and government laboratories are free of charge, there is a license fee for industry users.
Supplementary Material
Table 1:
Overview of recent methods developed in the Rosetta software
| Method | Lab developed |
|---|---|
| Score function | |
| REF2015 score function28,29 | Frank DiMaio, David Baker |
| cartesian_ddG29 | Frank DiMaio, Phil Bradley |
| HBNet47,50 | David Baker, Brian Kuhlman |
| HBNetEnergy47 | Richard Bonneau, David Baker* |
| AACompositionEnergy | Richard Bonneau, David Baker* |
| AARepeatEnergy | Richard Bonneau, David Baker* |
| VoidsPenaltyEnergy | Richard Bonneau, David Baker* |
| NetChargeEnergy | Richard Bonneau, David Baker* |
| BuriedUnsatPenalty | Richard Bonneau, David Baker* |
| Protein structure prediction | |
| fragment picker71 | Dominik Gront*,** |
| RosettaCM55 | David Baker |
| iterative hybridize59,60 | David Baker, Sergey Ovchinnikov* |
| Loop modeling | |
| NGK (next-generation KIC) 64 | Tanja Kortemme |
| GenKIC (generalized KIC) 44 | Richard Bonneau, David Baker* |
| LoopHashKIC | Tanja Kortemme |
| Consensus_Loop_Design72,73 | David Baker |
| Protein-protein docking | |
| RosettaDock4.074 | Jeffrey Gray |
| Rosetta SymDock275 | (Ingemar André), Jeffrey Gray |
| Small molecule ligand docking | |
| RosettaLigand76–78 | Jens Meiler |
| RosettaLigandEnsemble79 | Jens Meiler |
| pocket optimization80,81 | John Karanicolas |
| DARC82–84 | John Karanicolas |
| Modeling of antibodies and immune system proteins | |
| RosettaAntibody85–88 | Jeffrey Gray |
| AbPredict89,90 | Sarel Fleishman |
| RosettaMHC91 | Nik Sgourakis |
| TCRModel92 | Brian Pierce |
| SnugDock93 | Jeffrey Gray |
| Design of antibodies and immune system proteins | |
| RAbD94 (Rosetta AntibodyDesign) | Bill Schief, Roland Dunbrack |
| Epitope removal95,96 | David Baker, Cyrus Biotechnology |
| AbDesign97,98 | Sarel Fleishman |
| Protein design | |
| SEWING99,100 | Brian Kuhlmann |
| RosettaRemodel101 | Possu Huang*,** |
| LooDo102 | Sagar Khare |
| RECON103 | Jens Meiler |
| curved β-sheet design72 | David Baker |
| biased forward folding72 | David Baker |
| fold_from_loops104 | Bruno Correia*,** |
| FunFolDes105 | Bruno Correia |
| Protein interface design | |
| FlexDDG106 | Tanja Kortemme |
| Coupled Moves107 | Tanja Kortemme & DSM Biotechnology Center |
| Parametric design48,108 | Richard Bonneau* |
| Peptides and peptidomimetics | |
| FlexPepDock109,110 | Ora Schueler-Furman |
| PIPER-FlexPepDock111 | Ora Schueler-Furman |
| PeptiDerive112 | Ora Schueler-Furman |
| simple_cycpep_predict44,45,108 | Richard Bonneau, David Baker* |
| MFPred113 | Sagar Khare |
| RosettaSurface114–116 | Jeffrey Gray |
| Modeling with experimental data | |
| cryoEM de novo117 | Frank DiMaio, David Baker |
| cryoEM: RosettaES118 | Frank DiMaio |
| cryoEM: iterative refinement119,120 | (formerly David Baker) Frank DiMaio |
| cryoEM: automated refinement121 | Frank DiMaio |
| NMR: CS-Rosetta122 | Nik Sgourakis |
| NMR: PCS-Rosetta, GPS-Rosetta123,124 | Thomas Huber |
| RosettaNMR framework125: using RDC/PRE/PCS/NOE/CS for ab initio, protein-protein docking, ligand docking, symmetric assembly | Jens Meiler, Richard Bonneau (Jeffrey Gray) |
| mass-spec: HRF hydroxyl radical footprinting126,127 | Steffen Lindert |
| mass-spec: PyTXMS128 | Lars Malmstroem |
| RNA modeling | |
| SWA (stepwise assembly) 129,130 | Rhiju Das |
| SWM (stepwise Monte-Carlo) 131 | Rhiju Das |
| FARFAR (fragment assembly medium resolution structure prediction) 132–134 | Rhiju Das |
| ERRASER (refinement into EM density maps) 135,136 | Rhiju Das |
| CS-Rosetta-RNA (modeling with NMR data) 137 | Rhiju Das |
| RECCES (Reweighting of Energy-function Collection with Conformational Ensemble Sampling) | Rhiju Das |
| DRRAFTER (de novo modeling of protein-RNA complexes into EM densities) 138 | Rhiju Das |
| Membrane proteins | |
| RosettaMP framework139: mp_ddg, mp_dock, mp_relax, mp_symdock | Jeffrey Gray, Richard Bonneau |
| RosettaMP toolkit140: mp_score, mp_transform, mp_mutate_relax, helix_from_sequence | Jeffrey Gray, Richard Bonneau |
| mp_lipid_acc141 | Richard Bonneau |
| mp_domain_assembly142 | Richard Bonneau |
| RosettaCM for membrane proteins33 | Jens Meiler |
| Carbohydrates | |
| RosettaCarbohydrate framework143,144 | Jeffrey Gray, William Schief |
| User interfaces | |
| PyRosetta30,145 | Jeffrey Gray |
| RosettaScripts31,33 | Sarel Fleishman*,** |
| InteractiveRosetta146 | Chris Bystroff |
| Foldit Standalone32,147–149 | Seth Cooper*,**, Firas Khatib*,**, Justin Siegel, Scott Horowitz, David Baker |
| ROSIE server150,151 | Jeffrey Gray |
| Miscellaneous | |
| Metalloproteins42 | David Baker, Richard Bonneau* |
| Waters51 | Frank DiMaio |
| SimpleMetrics | William Schief |
| AmbRose | Sagar Khare |
| RosettaRC | William Schief |
the main developer(s) in this lab was/were formerly in the lab of David Baker when this application was developed
the main developer now has their own lab
Acknowledgements
RosettaCommons is supported by NIH R01 GM073151 to B. Kuhlman, NSF, the Packard Foundation, the Beckman Foundation, the Alfred P. Sloan Foundation, and the Simons Foundation. This work was also supported by 100,000,000 CPU-hour donation from Google Inc to P. Conway; 125,760,000 CPU-hour allocation on the Mira and Theta supercomputers through the Innovative and Novel Computational Impact on Theory and Experiment (INCITE) program to D. Baker, F. DiMaio, A. Leaver-Fay, and V. Mulligan. This research used resources of the Argonne Leadership Computing Facility, which is a DOE Office of Science user facility supported under Contract DE-AC02-06CH11357. AHA 18POST34080422 to G. Kuenze; AMED J-PRIDE JP18fm0208022h to D. Kuroda; Biltema Foundation to B. Correia; Boehringer Ingelheim Fonds to C. Norn; Computing was performed using resources of the Argonne Leadership Computing Facility at Argonne National Laboratory which is supported by the Office of Science of the US to P. Conway; DFG KU 3510/1-1 to G. Kuenze; DP120100561 to T. Huber; DP150100383 to T. Huber, K. Pilla; Defense Threat Reduction Agency to N. King; EMBO long-term fellowship ALTF 698-2011 to A. Stein; EPFL-Fellows - H2020 Marie Sklodowska-Curie to J. Bonet; European Research Council Grant 310873 to O. Schueler-Furman, N. Alam; European Research Council Grant 310873 to Y. Sedan, O. Marcu; European Research Council Starting grant - 716058 to B. Correia, A. Scheck; FT0991709 to T. Huber; Foundation of Knut and Alice Wallenberg 20160023 to L. Malmström; Hertz Foundation Fellowship to R. Alford; Howard Hughes Medical Institute to D. Baker; Hyak supercomputer system supported in part by the University of Washington eScience Institute to D. Baker and F. DiMaio labs; Israel Science Foundation 2017717 to O. Schueler-Furman, N. Alam; Japan Society for the Promotion of Science JP17K18113 to D. Kuroda; MCB1330760 to S. Khare; Marie Curie International Outgoing Fellowship FP7-PEOPLE-2011-IOF 298976 to E. Marcos; National Science Centre, Poland, 2018/29/B/ST6/01989 to D. Gront; NIAID T32AI007244 to J. Adolf-Bryfogle; NIAID U19 AI117905 to A. Sevy; NIEHS P42ES004699 to J. Siegel; NIGMS Ruth L Kirschstein National Research Service Award T32GM008268 to P. Conway; NIGMS T32 GM007628 to B. Bender; NIH 1R35 GM122579 to R. Das; NSF DMREF award # 1728858 and DMR-0820341 to R. Bonneau, NIH 1UH2CA203780 to S. Cooper, F. Khatib; NIH 5F32GM110899-02 to T. Linsky; NIH F31GM123616 to J. Jeliazkov; NIH F32CA189246 to J. Labonte; NIH P01 U19AI117905, R01 AI113867, UM1 Al100663 to W. Schief; NIH R00 GM120388 to S. Horowitz; NIH R01 AI143997 to N. Sgourakis; NIH R01 DK097376 to J. Meiler; NIH R01 GM073960 to B. Kuhlman; NIH R01 GM076324 to J. Siegel; NIH R01 GM078221 to J. Gray; NIH R01 GM080403 to J. Meiler; NIH R01 GM084453 to R, Dunbrack; NIH R01 GM088277 to P. Bradley; NIH R01 GM092802 to D. Baker; NIH R01 GM092802 to D. Baker; NIH R01 GM098101 to T. Kortemme; NIH R01 GM099842 to J. Meiler; NIH R01 GM099959 to J. Karanicolas; NIH R01 GM110089 to T. Kortemme; NIH R01 GM117189 to T. Kortemme; NIH R01 GM117968 to B. Kuhlman; NIH R01 GM121487 to P. Bradley; NIH R01 GM123089 to F. DiMaio; NIH R01 GM126299 to B. Pierce; NIH R01 GM127578 to J. Gray; NIH R01 GM099827 to C. Bystroff; NIH R01 GM092802 to D. Baker; NIH R01 HL122010 to J. Meiler; NIH R01067553 to B. Kuhlman; NIH R01 GM078221 to J. Gray; NIH R01084433 to D. Baker; NIH R01088277 to S. Thyme; NIH R21 AI121799 to J. Meiler; NIH R21 CA219847 to R. Das; NIH R21 GM102716 to R. Das; NIH R35 GM122517 to R. Dunbrack; NIH R35 GM125034 to N. Sgourakis; NIH RL1CA133832 to D. Baker; NIH U19 AI117905 to J. Meiler; NIH/NCI Cancer Center Support Grant P30 CA006927 to J. Karanicolas; NSF 1507736 to J. Gray; NSF 1627539 to J. Siegel; NSF 1629879 to S. Cooper; NSF 1805510 to J. Siegel; NSF 1827246 to J. Siegel; NSF CHE 1305874 to J. Meiler; NSF CHE 1750666 to S. Lindert; NSF CISE 1629811 to J. Meiler; NSF CNS-1629811 to J. Meiler; NSF DBI-1262182 to T. Kortemme; NSF DBI-1564692 to T. Kortemme; NSF DMR 1507736 to J. Gray; NSF GRF DGE-1433187 to A. Rubenstein; NSF Graduate Research Fellowship to R. Alford, K. Kappel, B. Koepnick, S. Thyme; NSF MCB1330760 to S. Khare; NSF MCB1716623 to S. Khare; Open Philanthropy to B. Coventry; PhRMA Informatics Pre-Doctoral Fellowship U22879-001 to S. Smith; PhRMA foundation Predoctoral Fellowship to D. Fu; RosettaCommons to L. Goldschmidt, A. Rubenstein, F. DiMaio, S. Cooper, A. Watkins, M. Szegedy, C. Geniesse, K. Blacklock, R. Das, S. Khare, J. Koehler Leman, K. Kappel; Career Award at the Scientific Interface from Burroughs Wellcome Fund to S. Boyken; Simons Foundation to V. Mulligan, R. Bonneau, D. Renfrew, J. Koehler Leman; Stanford Graduate Fellowship to K. Kappel; Starter Grant from the European Research Council to G. Lapidoth; Swiss National Science Foundation - NCCR Molecular Systems Engineering 51NF40-141825 to B. Correia; Swiss National Science Foundation 310030_163139 to B. Correia; Swiss National Science Foundation SNF 200021 160188 to L. Malmström, H. Khakzad; UCSF/UCB Graduate Program in Bioengineering to X. Pan; USA-Israel Binational Science Foundation 2009418 to B. Raveh, L. Zimmerman, N. London; USA-Israel Binational Science Foundation 2009418, 2015207 to O. Schueler-Furman, N. Alam; USA-Israel Binational Science Foundation 2015207 to A. Khramushin; Washington Research Foundation Innovation Postdoctoral Fellowship to B. Weitzner; XSEDE, which is supported by NSF ACI-1548562; NIH R01 GM097207 to P. Barth; MCB120101 XSEDE allocation to P. Barth.
Competing Interests:
Rosetta software has been licensed to numerous non-profit and for-profit organizations. Rosetta Licensing is managed by UW CoMotion, and royalty proceeds are managed by the RosettaCommons. Under institutional participation agreements between the University of Washington, acting on behalf of the RosettaCommons, their respective institutions may be entitled to a portion of revenue received on licensing Rosetta software including programs described here. Baker, Malmström, Gront, Meiler, Schueler-Furman, Gray, Sgourakis, Lindert, Karanicolas, Bonneau, Kortemme, and Bradley are unpaid board members of the RosettaCommons. As members of the Scientific Advisory Board of Cyrus Biotechnology, Baker and Gray are granted stock options. Yifan Song, Indigo C. King, Steven M. Lewis, Brandon Frenz, Karen Khar and Ryan Pavlovicz are currently employed at Cyrus Biotechnology with granted stock options. Cyrus Biotechnology distributes the Rosetta software. Brian D. Weitzner and Scott E. Boyken hold equity in Lyell Immunopharma. Vikram K. Mulligan is a co-founder of and shareholder in Menten Biotechnology Labs, Inc. The content of this manuscript is relevant to work performed at Lyell and Menten. Justin B. Siegel is a co-founder and shareholder of Digestiva, Inc. and PvP Biologics Inc. David Baker is a co-founder, shareholder, or advisor to the following companies: ARZEDA, PvP Biologics, Cyrus Biotechnology, Cue Biopharma, Icosavax, Neoleukin Therapeutics, Lyell Immunotherapeutics, Sana Biotechnology, and A-Alpha Bio.
References
- 1.Schrodinger - Biologics Design. at <https://www.schrodinger.com/science-articles/biologics-design>
- 2.Molecular Operating Environment (MOE) | MOEsaic | PSILO. at <https://www.chemcomp.com/Products.htm>
- 3.Ref. Dassault Systèmes BIOVIA, Discovery Studio Modeling Environment, Release 2017, San Diego: Dassault Systèmes, 2016. at <https://www.3dsbiovia.com/products/collaborative-science/biovia-discovery-studio/> [Google Scholar]
- 4.Steinegger M, Meier M, Mirdita M, Vöhringer H, Haunsberger SJ & Söding J HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinformatics 20, 473 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vu O, Mendenhall J, Altarawy D & Meiler J BCL::Mol2D—a robust atom environment descriptor for QSAR modeling and lead optimization. J. Comput. Aided. Mol. Des. 33, 477–486 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Webb B, Viswanath S, Bonomi M, Pellarin R, Greenberg CH, Saltzberg D & Sali A Integrative structure modeling with the Integrative Modeling Platform. Protein Sci. 27, 245–258 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T & Hutchison GR Open Babel: An open chemical toolbox. J. Cheminform. 3, 33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM & Karplus M CHARMM: The biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Wolf RM, Caldwell JW, Kollman PA & Case DA Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–74 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE & Berendsen HJC GROMACS: fast, flexible, and free. J. Comput. Chem. 26, 1701–18 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Eastman P, Swails J, Chodera JD, McGibbon RT, Zhao Y, Beauchamp KA, Wang L-P, Simmonett AC, Harrigan MP, Stern CD, Wiewiora RP, Brooks BR & Pande VS OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PLOS Comput. Biol. 13, e1005659 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, Qin C, Zidek A, Nelson A, Bridgland A, Penedones H, Petersen S, Simonyan K, Crossan S, Jones D, Silver D, Kavukcuoglu K, Hassabis D & Senior A De novo structure prediction with deep-learning based scoring. Thirteen. Crit. Assess. Tech. Protein Struct. Predict December, (2018). [Google Scholar]
- 13.Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, Qin C, Žídek A, Nelson AWR, Bridgland A, Penedones H, Petersen S, Simonyan K, Crossan S, Kohli P, Jones DT, Silver D, Kavukcuoglu K & Hassabis D Improved protein structure prediction using potentials from deep learning. Nature (2020). doi: 10.1038/s41586-019-1923-7 [DOI] [PubMed] [Google Scholar]
- 14.Zheng W, Li Y, Zhang C, Pearce R, Mortuza SM & Zhang Y Deep-learning contact-map guided protein structure prediction in CASP13. Proteins Struct. Funct. Bioinforma 87, 1149–1164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu J & Wang S Analysis of distance-based protein structure prediction by deep learning in CASP13. Proteins Struct. Funct. Bioinforma. (2019). doi: 10.1002/prot.25810 [DOI] [PubMed] [Google Scholar]
- 16.Fiser A & Sali A MODELLER: Generation and Refinement of Homology-Based Protein Structure Models. Methods Enzymol. 374, 461–491 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Bienert S, Waterhouse A, de Beer TAP, Tauriello G, Studer G, Bordoli L & Schwede T The SWISS-MODEL Repository—new features and functionality. Nucleic Acids Res. 45, D313–D319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J, Yan R, Roy A, Xu D, Poisson J & Zhang Y The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Zundert GCP, Rodrigues JPGLM, Trellet M, Schmitz C, Kastritis PL, Karaca E, Melquiond ASJ, van Dijk M, de Vries SJ & Bonvin AMJJ The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 428, 720–725 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T & Weng Z ZDOCK server: Interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 30, 1771–1773 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Padhorny D, Kazennov A, Zerbe BS, Porter KA, Xia B, Mottarella SE, Kholodov Y, Ritchie DW, Vajda S & Kozakov D Protein-protein docking by fast generalized Fourier transforms on 5D rotational manifolds. Proc. Natl. Acad. Sci. U. S. A. 113, E4286–93 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trott O & Olson AJ AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, NA-NA (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.FlexX version 4.1; BioSolveIT GmbH, Sankt Augustin, Germany, 2019, www.biosolveit.de/FlexX. [Google Scholar]
- 24.Tubert-Brohman I, Sherman W, Repasky M & Beuming T Improved Docking of Polypeptides with Glide. J. Chem. Inf. Model. 53, 1689–1699 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Sorenson JM & Head-Gordon T Matching simulation and experiment: a new simplified model for simulating protein folding. J. Comput. Biol. 7, 469–81 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Koehler Leman J, Weitzner BD, Renfrew PD, Lewis SM, Moretti R, Watkins AM, Mulligan VK, Lyskov S, Adolf-Bryfogle J, Labonte JW, Consortium R, Bystroff C, Schief W, Schueler-Furman O, Baker D, Bradley P, Dunbrack R, Kortemme T, Leaver-Fay A, Strauss CE, Meiler J, Kuhlman B, Gray JJ & Bonneau R Better together: Elements of successful scientific software development in distributed collaborative community. Accept. PlosCompBio (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson JM, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban Y-EA, Fleishman SJ, Corn JE, Kim DE, Berrondo M, Mentzer S, Popovic Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D & Bradley P ROSETTA3: An Object-Oriented Software Suite for the Simulation and Design of Macromolecules. Methods Enzymol. 487, 545–74 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alford RF, Leaver-Fay A, Jeliazkov JR, O’Meara MJ, Dimaio FP, Park H, Shapovalov MV, Renfrew PD, Mulligan VK, Kappel K, Labonte JW, Pacella MS, Bonneau R, Bradley P, Dunbrack RL, Das R, Baker D, Kuhlman B, Kortemme T & Gray JJ The Rosetta all-atom energy function for macromolecular modeling and design. J. Chem. Theory Comput. 13, 1–35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park H, Bradley P, Greisen P, Liu Y, Mulligan VK, Kim DE, Baker D & DiMaio F Simultaneous Optimization of Biomolecular Energy Functions on Features from Small Molecules and Macromolecules. J. Chem. Theory Comput. 12, 6201–6212 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaudhury S, Lyskov S & Gray JJ PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics 26, 689–691 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fleishman SJ, Leaver-Fay A, Corn JE, Strauch E-MM, Khare SD, Koga N, Ashworth J, Murphy P, Richter F, Lemmon G, Meiler J & Baker D RosettaScripts: A scripting language interface to the Rosetta Macromolecular modeling suite. PLoS One 6, 1–10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooper S, Khatib F, Treuille A, Barbero J, Lee J, Beenen M, Leaver-Fay A, Baker D, Popović Z & Players F Predicting protein structures with a multiplayer online game. Nature 466, 756–760 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bender BJ, Cisneros A, Duran AM, Finn JA, Fu D, Lokits AD, Mueller BK, Sangha AK, Sauer MF, Sevy AM, Sliwoski G, Sheehan JH, Dimaio F, Meiler J & Moretti R Protocols for Molecular Modeling with Rosetta3 and RosettaScripts . Biochemistry acs.biochem.6b00444 (2016). doi: 10.1021/acs.biochem.6b00444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simoncini D, Allouche D, de Givry S, Delmas C, Barbe S & Schiex T Guaranteed Discrete Energy Optimization on Large Protein Design Problems. J. Chem. Theory Comput. 11, 5980–9 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Leaver-Fay A, O’Meara MJ, Tyka M, Jacak R, Song Y, Kellogg EH, Thompson J, Davis IW, Pache RA, Lyskov S, Gray JJ, Kortemme T, Richardson JS, Havranek JJ, Snoeyink J, Baker D & Kuhlman B Scientific benchmarks for guiding macromolecular energy function improvement. Methods Enzymol. 523, 109–43 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jorgensen WL, Jorgensen WL, Maxwell DS & Tirado-rives J Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. AM. CHEM. SOC 11225–11236 (1996). at <http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.334.2959> [Google Scholar]
- 37.Radzicka A & Wolfenden R Comparing the polarities of the amino acids: side-chain distribution coefficients between the vapor phase, cyclohexane, 1-octanol, and neutral aqueous solution. Biochemistry 27, 1664–1670 (1988). [Google Scholar]
- 38.O’Meara MJ, Leaver-Fay A, Tyka MD, Stein A, Houlihan K, DiMaio F, Bradley P, Kortemme T, Baker D, Snoeyink J & Kuhlman B Combined Covalent-Electrostatic Model of Hydrogen Bonding Improves Structure Prediction with Rosetta. J. Chem. Theory Comput. 11, 609–622 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conway P, Tyka MD, DiMaio F, Konerding DE & Baker D Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci. 23, 47–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park H, Lee H & Seok C High-resolution protein-protein docking by global optimization: recent advances and future challenges. Curr. Opin. Struct. Biol. 35, 24–31 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Kellogg EH, Leaver-Fay A & Baker D Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins Struct. Funct. Bioinforma. 79, 830–838 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mills JH, Khare SD, Bolduc JM, Forouhar F, Mulligan VK, Lew S, Seetharaman J, Tong L, Stoddard BL & Baker D Computational Design of an Unnatural Amino Acid Dependent Metalloprotein with Atomic Level Accuracy. J. Am. Chem. Soc. 135, 13393–13399 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kappel K, Jarmoskaite I, Vaidyanathan PP, Greenleaf WJ, Herschlag D & Das R Blind tests of RNA–protein binding affinity prediction. Proc. Natl. Acad. Sci. 116, 8336–8341 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhardwaj G, Mulligan VK, Bahl CD, Gilmore JM, Harvey PJ, Cheneval O, Buchko GW, Pulavarti SVSRK, Kaas Q, Eletsky A, Huang P-S, Johnsen WA, Greisen PJ, Rocklin GJ, Song Y, Linsky TW, Watkins A, Rettie SA, Xu X, Carter LP, Bonneau R, Olson JM, Coutsias E, Correnti CE, Szyperski T, Craik DJ & Baker D Accurate de novo design of hyperstable constrained peptides. Nature 538, 329–335 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hosseinzadeh P, Bhardwaj G, Mulligan VK, Shortridge MD, Craven TW, Pardo-Avila F, Rettie SA, Kim DE, Silva D-A, Ibrahim YM, Webb IK, Cort JR, Adkins JN, Varani G & Baker D Comprehensive computational design of ordered peptide macrocycles. Science (80-. ). 358, 1461–1466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leaver-Fay A, Butterfoss GL, Snoeyink J & Kuhlman B Maintaining solvent accessible surface area under rotamer substitution for protein design. J. Comput. Chem. 28, 1336–41 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Boyken SE, Chen Z, Groves B, Langan RA, Oberdorfer G, Ford A, Gilmore JM, Xu C, DiMaio F, Pereira JH, Sankaran B, Seelig G, Zwart PH & Baker D De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity. Science 352, 680–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu P, Min D, DiMaio F, Wei KY, Vahey MD, Boyken SE, Chen Z, Fallas JA, Ueda G, Sheffler W, Mulligan VK, Xu W, Bowie JU & Baker D Accurate computational design of multipass transmembrane proteins. Science (80-. ). 359, 1042–1046 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Z, Boyken SE, Jia M, Busch F, Flores-Solis D, Bick MJ, Lu P, VanAernum ZL, Sahasrabuddhe A, Langan RA, Bermeo S, Brunette TJ, Mulligan VK, Carter LP, DiMaio F, Sgourakis NG, Wysocki VH & Baker D Programmable design of orthogonal protein heterodimers. Nature 565, 106–111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maguire JB, Boyken SE, Baker D & Kuhlman B Rapid Sampling of Hydrogen Bond Networks for Computational Protein Design. J. Chem. Theory Comput. 14, 2751–2760 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pavlovicz RE, Park H & DiMaio F Efficient consideration of coordinated water molecules improves computational protein-protein and protein-ligand docking. bioRxiv 618603 (2019). doi: 10.1101/618603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhowmick A, Sharma SC, Honma H & Head-Gordon T The role of side chain entropy and mutual information for improving the de novo design of Kemp eliminases KE07 and KE70. Phys. Chem. Chem. Phys. 18, 19386–19396 (2016). [DOI] [PubMed] [Google Scholar]
- 53.König R & Dandekar T Solvent entropy-driven searching for protein modeling examined and tested in simplified models. Protein Eng. 14, 329–35 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Kryshtafovych A, Schwede T, Topf M, Fidelis K & Moult J Critical assessment of methods of protein structure prediction (CASP)—Round XIII. Proteins Struct. Funct. Bioinforma. (2019). doi: 10.1002/prot.25823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song Y, Dimaio F, Wang RY-RR, Kim DE, Miles C, Brunette T, Thompson J & Baker D High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.New Robetta server - http://new.robetta.org/.
- 57.Park H, Kim DE, Ovchinnikov S, Baker D & DiMaio F Automatic structure prediction of oligomeric assemblies using Robetta in CASP12. Proteins Struct. Funct. Bioinforma. 86, 283–291 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kamisetty H, Ovchinnikov S & Baker D Assessing the utility of coevolution-based residue-residue contact predictions in a sequence- and structure-rich era. Proc. Natl. Acad. Sci. U. S. A. 110, 15674–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ovchinnikov S, Park H, Varghese N, Huang P-S, Pavlopoulos GA, Kim DE, Kamisetty H, Kyrpides NC & Baker D Protein structure determination using metagenome sequence data. Science (80-. ). 355, 294–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park H, Ovchinnikov S, Kim DE, Dimaio F & Baker D Protein homology model refinement by large-scale energy optimization. Proc. Natl. Acad. Sci. U. S. A. 115, 3054–3059 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tyka MD, Keedy DA, André I, Dimaio F, Song Y, Richardson DC, Richardson JS & Baker D Alternate states of proteins revealed by detailed energy landscape mapping. J. Mol. Biol. 405, 607–18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Friedland GD, Linares AJ, Smith CA & Kortemme T A simple model of backbone flexibility improves modeling of side-chain conformational variability. J. Mol. Biol. 380, 757–74 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kapp GT, Liu S, Stein A, Wong DT, Remenyi A, Yeh BJ, Fraser JS, Taunton J, Lim WA & Kortemme T Control of protein signaling using a computationally designed GTPase/GEF orthogonal pair. Proc. Natl. Acad. Sci. 109, 5277–5282 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stein A & Kortemme T Improvements to robotics-inspired conformational sampling in rosetta. PLoS One 8, e63090 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin MS & Head-Gordon T Improved Energy Selection of Nativelike Protein Loops from Loop Decoys. J. Chem. Theory Comput. 4, 515–21 (2008). [DOI] [PubMed] [Google Scholar]
- 66.Rohl CA, Strauss CEM, Chivian D & Baker D Modeling structurally variable regions in homologous proteins with rosetta. Proteins 55, 656–77 (2004). [DOI] [PubMed] [Google Scholar]
- 67.Wang C, Bradley P & Baker D Protein-Protein Docking with Backbone Flexibility. J. Mol. Biol. 373, 503–519 (2007). [DOI] [PubMed] [Google Scholar]
- 68.Canutescu AA & Dunbrack RL Cyclic coordinate descent: A robotics algorithm for protein loop closure. Protein Sci. 12, 963–72 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mandell DJ, Coutsias EA & Kortemme T Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat. Methods 6, 551–2 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mandell DJ & Kortemme T Backbone flexibility in computational protein design. Curr. Opin. Biotechnol. 20, 420–8 (2009). [DOI] [PubMed] [Google Scholar]
- 71.Gront D, Kulp DW, Vernon RM, Strauss CEM & Baker D Generalized Fragment Picking in Rosetta: Design, Protocols and Applications. PLoS One 6, e23294 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marcos E, Basanta B, Chidyausiku TM, Tang Y, Oberdorfer G, Liu G, Swapna GVT, Guan R, Silva D-A, Dou J, Pereira JH, Xiao R, Sankaran B, Zwart PH, Montelione GT & Baker D Principles for designing proteins with cavities formed by curved β sheets. Science 355, 201–206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marcos E, Chidyausiku TM, McShan AC, Evangelidis T, Nerli S, Carter L, Nivón LG, Davis A, Oberdorfer G, Tripsianes K, Sgourakis NG & Baker D De novo design of a non-local β-sheet protein with high stability and accuracy. Nat. Struct. Mol. Biol. 25, 1028–1034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marze NA, Roy Burman SS, Sheffler W & Gray JJ Efficient flexible backbone protein–protein docking for challenging targets. Bioinformatics 34, 3461–3469 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roy Burman SS, Yovanno RA & Gray JJ Flexible Backbone Assembly and Refinement of Symmetrical Homomeric Complexes. Structure (2019). doi: 10.1016/j.str.2019.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meiler J & Baker D RosettaLigand: protein-small molecule docking with full side-chain flexibility. Proteins 65, 538–48 (2006). [DOI] [PubMed] [Google Scholar]
- 77.DeLuca S, Khar K & Meiler J Fully Flexible Docking of Medium Sized Ligand Libraries with RosettaLigand. PLoS One 10, e0132508 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Davis IW & Baker D RosettaLigand Docking with Full Ligand and Receptor Flexibility. J. Mol. Biol. 385, 381–392 (2009). [DOI] [PubMed] [Google Scholar]
- 79.Fu DY & Meiler J RosettaLigandEnsemble: A Small-Molecule Ensemble-Driven Docking Approach. ACS Omega 3, 3655–3664 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnson DK & Karanicolas J Druggable Protein Interaction Sites Are More Predisposed to Surface Pocket Formation than the Rest of the Protein Surface. PLoS Comput. Biol. 9, e1002951 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johnson DK & Karanicolas J Selectivity by Small-Molecule Inhibitors of Protein Interactions Can Be Driven by Protein Surface Fluctuations. PLOS Comput. Biol. 11, e1004081 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gowthaman R, Miller SA, Rogers S, Khowsathit J, Lan L, Bai N, Johnson DK, Liu C, Xu L, Anbanandam A, Aubé J, Roy A & Karanicolas J DARC: Mapping Surface Topography by Ray-Casting for Effective Virtual Screening at Protein Interaction Sites. J. Med. Chem. 59, 4152–4170 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Khar KR, Goldschmidt L & Karanicolas J Fast Docking on Graphics Processing Units via Ray-Casting. PLoS One 8, e70661 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gowthaman R, Lyskov S & Karanicolas J DARC 2.0: Improved Docking and Virtual Screening at Protein Interaction Sites. PLoS One 10, e0131612 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sircar A, Kim ET & Gray JJ RosettaAntibody: antibody variable region homology modeling server. Nucleic Acids Res. 37, W474–479 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weitzner BD, Kuroda D, Marze N, Xu J & Gray JJ Blind prediction performance of RosettaAntibody 3.0: Grafting, relaxation, kinematic loop modeling, and full CDR optimization. Proteins Struct. Funct. Bioinforma. 82, 1611–1623 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Weitzner BD, Jeliazkov JR, Lyskov S, Marze N, Kuroda D, Frick R, Adolf-Bryfogle J, Biswas N, Dunbrack RL & Gray JJ Modeling and docking of antibody structures with Rosetta. Nat. Protoc 12, 401–416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sivasubramanian A, Sircar A, Chaudhury S & Gray JJ Toward high-resolution homology modeling of antibody F v regions and application to antibody-antigen docking. Proteins Struct. Funct. Bioinforma. 74, 497–514 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Norn CH, Lapidoth G & Fleishman SJ High-accuracy modeling of antibody structures by a search for minimum-energy recombination of backbone fragments. Proteins 85, 30–38 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lapidoth G, Parker J, Prilusky J & Fleishman SJ AbPredict 2: a server for accurate and unstrained structure prediction of antibody variable domains. Bioinformatics (2018). doi: 10.1093/bioinformatics/bty822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Toor JS, Rao AA, McShan AC, Yarmarkovich M, Nerli S, Yamaguchi K, Madejska AA, Nguyen S, Tripathi S, Maris JM, Salama SR, Haussler D & Sgourakis NG A Recurrent Mutation in Anaplastic Lymphoma Kinase with Distinct Neoepitope Conformations. Front. Immunol. 9, 99 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gowthaman R & Pierce BG TCRmodel: high resolution modeling of T cell receptors from sequence. Nucleic Acids Res. 46, W396–W401 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sircar A & Gray JJ SnugDock: paratope structural optimization during antibody-antigen docking compensates for errors in antibody homology models. PLoS Comput. Biol. 6, e1000644 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Adolf-Bryfogle J, Kalyuzhniy O, Kubitz M, Weitzner BD, Hu X, Adachi Y, Schief WR & Dunbrack RL RosettaAntibodyDesign (RAbD): A general framework for computational antibody design. PLOS Comput. Biol. 14, e1006112 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.King C, Garza EN, Mazor R, Linehan JL, Pastan I, Pepper M & Baker D Removing T-cell epitopes with computational protein design. Proc. Natl. Acad. Sci. U. S. A. 111, 8577–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nivón LG, Bjelic S, King C & Baker D Automating human intuition for protein design. Proteins 82, 858–66 (2014). [DOI] [PubMed] [Google Scholar]
- 97.Lapidoth GD, Baran D, Pszolla GM, Norn C, Alon A, Tyka MD & Fleishman SJ AbDesign: An algorithm for combinatorial backbone design guided by natural conformations and sequences. Proteins 83, 1385–406 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baran D, Pszolla MG, Lapidoth GD, Norn C, Dym O, Unger T, Albeck S, Tyka MD & Fleishman SJ Principles for computational design of binding antibodies. Proc. Natl. Acad. Sci. U. S. A. 114, 10900–10905 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jacobs TM, Williams B, Williams T, Xu X, Eletsky A, Federizon JF, Szyperski T & Kuhlman B Design of structurally distinct proteins using strategies inspired by evolution. 352, 687–90 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Guffy SL, Teets FD, Langlois MI & Kuhlman B Protocols for Requirement-Driven Protein Design in the Rosetta Modeling Program. J. Chem. Inf. Model. 58, 895–901 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huang P-S, Ban Y-EA, Richter F, Andre I, Vernon R, Schief WR & Baker D RosettaRemodel: A Generalized Framework for Flexible Backbone Protein Design. PLoS One 6, e24109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Blacklock KM, Yang L, Mulligan VK & Khare SD A computational method for the design of nested proteins by loop-directed domain insertion. Proteins Struct. Funct. Bioinforma. 86, 354–369 (2018). [DOI] [PubMed] [Google Scholar]
- 103.Sevy AM, Jacobs TM, Crowe JE & Meiler J Design of Protein Multi-specificity Using an Independent Sequence Search Reduces the Barrier to Low Energy Sequences. PLoS Comput. Biol. 11, e1004300 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Correia BE, Bates JT, Loomis RJ, Baneyx G, Carrico C, Jardine JG, Rupert P, Correnti C, Kalyuzhniy O, Vittal V, Connell MJ, Stevens E, Schroeter A, Chen M, MacPherson S, Serra AM, Adachi Y, Holmes MA, Li Y, Klevit RE, Graham BS, Wyatt RT, Baker D, Strong RK, Crowe JE, Johnson PR & Schief WR Proof of principle for epitope-focused vaccine design. Nature 507, 201–206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bonet J, Wehrle S, Schriever K, Yang C, Billet A, Sesterhenn F, Scheck A, Sverrisson F, Veselkova B, Vollers S, Lourman R, Villard M, Rosset S, Krey T & Correia BE Rosetta FunFolDes - A general framework for the computational design of functional proteins. PLoS Comput. Biol. 14, e1006623 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Barlow KA, Ó Conchúir S, Thompson S, Suresh P, Lucas JE, Heinonen M & Kortemme T Flex ddG: Rosetta Ensemble-Based Estimation of Changes in Protein-Protein Binding Affinity upon Mutation. J. Phys. Chem. B 122, 5389–5399 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ollikainen N, de Jong RM & Kortemme T Coupling Protein Side-Chain and Backbone Flexibility Improves the Re-design of Protein-Ligand Specificity. PLOS Comput. Biol. 11, e1004335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dang B, Wu H, Mulligan VK, Mravic M, Wu Y, Lemmin T, Ford A, Silva D-A, Baker D & DeGrado WF De novo design of covalently constrained mesosize protein scaffolds with unique tertiary structures. Proc. Natl. Acad. Sci. U. S. A. 114, 10852–10857 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Raveh B, London N & Schueler-Furman O Sub-angstrom modeling of complexes between flexible peptides and globular proteins. Proteins 78, 2029–40 (2010). [DOI] [PubMed] [Google Scholar]
- 110.Raveh B, London N, Zimmerman L & Schueler-Furman O Rosetta FlexPepDock ab-initio: Simultaneous Folding, Docking and Refinement of Peptides onto Their Receptors. PLoS One 6, e18934 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Alam N, Goldstein O, Xia B, Porter KA, Kozakov D & Schueler-Furman O High-resolution global peptide-protein docking using fragments-based PIPER-FlexPepDock. PLoS Comput. Biol. 13, e1005905 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sedan Y, Marcu O, Lyskov S & Schueler-Furman O Peptiderive server: derive peptide inhibitors from protein-protein interactions. Nucleic Acids Res. 44, W536–41 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rubenstein AB, Pethe MA & Khare SD MFPred: Rapid and accurate prediction of protein-peptide recognition multispecificity using self-consistent mean field theory. PLOS Comput. Biol. 13, e1005614 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pacella MS, Koo DCE, Thottungal RA & Gray JJ Using the RosettaSurface algorithm to predict protein structure at mineral surfaces. Methods Enzymol. 532, 343–366 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lubin JH, Pacella MS & Gray JJ A Parametric Rosetta Energy Function Analysis with LK Peptides on SAM Surfaces. Langmuir 34, 5279–5289 (2018). [DOI] [PubMed] [Google Scholar]
- 116.Pacella MS & Gray JJ A Benchmarking Study of Peptide–Biomineral Interactions. Cryst. Growth Des. 18, 607–616 (2018). [Google Scholar]
- 117.Wang RY-R, Kudryashev M, Li X, Egelman EH, Basler M, Cheng Y, Baker D & DiMaio F De novo protein structure determination from near-atomic-resolution cryo-EM maps. Nat. Methods 12, 335–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Frenz B, Walls AC, Egelman EH, Veesler D & DiMaio F RosettaES: a sampling strategy enabling automated interpretation of difficult cryo-EM maps. Nat. Methods 14, 797–800 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.DiMaio F, Echols N, Headd JJ, Terwilliger TC, Adams PD & Baker D Improved low-resolution crystallographic refinement with Phenix and Rosetta. Nat. Methods 10, 1102–4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.DiMaio F, Song Y, Li X, Brunner MJ, Xu C, Conticello V, Egelman E, Marlovits TC, Cheng Y & Baker D Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nat. Methods 12, 361–5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang RY-R, Song Y, Barad BA, Cheng Y, Fraser JS & DiMaio F Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. Elife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nerli S & Sgourakis NG CS-ROSETTA. Methods Enzymol. (2018). doi: 10.1016/BS.MIE.2018.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yagi H, Pilla KB, Maleckis A, Graham B, Huber T & Otting G Three-dimensional protein fold determination from backbone amide pseudocontact shifts generated by lanthanide tags at multiple sites. Structure 21, 883–890 (2013). [DOI] [PubMed] [Google Scholar]
- 124.Schmitz C, Vernon R, Otting G, Baker D & Huber T Protein structure determination from pseudocontact shifts using ROSETTA. J. Mol. Biol. 416, 668–77 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kuenze G, Bonneau R, Leman JK & Meiler J Integrative Protein Modeling in RosettaNMR from Sparse Paramagnetic Restraints. Structure 27, 1721–1734.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Aprahamian ML, Chea EE, Jones LM & Lindert S Rosetta Protein Structure Prediction from Hydroxyl Radical Protein Footprinting Mass Spectrometry Data. Anal. Chem. 90, 7721–7729 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Aprahamian ML & Lindert S Utility of Covalent Labeling Mass Spectrometry Data in Protein Structure Prediction with Rosetta. J. Chem. Theory Comput. acs.jctc.9b00101 (2019). doi: 10.1021/acs.jctc.9b00101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hauri S, Khakzad H, Happonen L, Teleman J, Malmström J & Malmström L Rapid determination of quaternary protein structures in complex biological samples. Nat. Commun. 10, 192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sripakdeevong P, Kladwang W & Das R An enumerative stepwise ansatz enables atomic-accuracy RNA loop modeling. Proc. Natl. Acad. Sci. 108, 20573–20578 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Das R Atomic-Accuracy Prediction of Protein Loop Structures through an RNA-Inspired Ansatz. PLoS One 8, e74830 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Watkins AM, Geniesse C, Kladwang W, Zakrevsky P, Jaeger L & Das R Blind prediction of noncanonical RNA structure at atomic accuracy. Sci. Adv. 4, eaar5316 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Das R, Karanicolas J & Baker D Atomic accuracy in predicting and designing noncanonical RNA structure. Nat. Methods 7, 291–294 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cheng CY, Chou F-C & Das R Modeling Complex RNA Tertiary Folds with Rosetta. Methods Enzymol. 553, 35–64 (2015). [DOI] [PubMed] [Google Scholar]
- 134.Kappel K & Das R Sampling Native-like Structures of RNA-Protein Complexes through Rosetta Folding and Docking. Structure 27, 140–151.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chou F-C, Sripakdeevong P, Dibrov SM, Hermann T & Das R Correcting pervasive errors in RNA crystallography through enumerative structure prediction. Nat. Methods 10, 74–76 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chou F-C, Echols N, Terwilliger TC & Das R in 269–282 (Humana Press, New York, NY, 2016). doi: 10.1007/978-1-4939-2763-0_17 [DOI] [Google Scholar]
- 137.Sripakdeevong P, Cevec M, Chang AT, Erat MC, Ziegeler M, Zhao Q, Fox GE, Gao X, Kennedy SD, Kierzek R, Nikonowicz EP, Schwalbe H, Sigel RKO, Turner DH & Das R Structure determination of noncanonical RNA motifs guided by 1H NMR chemical shifts. Nat. Methods 11, 413–416 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kappel K, Liu S, Larsen KP, Skiniotis G, Puglisi EV, Puglisi JD, Zhou ZH, Zhao R & Das R De novo computational RNA modeling into cryo-EM maps of large ribonucleoprotein complexes. Nat. Methods 15, 947–954 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Alford RF, Koehler Leman J, Weitzner BD, Duran AM, Tilley DC, Elazar A & Gray JJ An Integrated Framework Advancing Membrane Protein Modeling and Design. PLoS Comput. Biol. 11, e1004398 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Koehler Leman J, Mueller BK & Gray JJ Expanding the toolkit for membrane protein modeling in Rosetta. Bioinformatics 11, 1–3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Koehler Leman J, Lyskov S & Bonneau R Computing structure-based lipid accessibility of membrane proteins with mp_lipid_acc in RosettaMP. BMC Bioinformatics 18, 115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Koehler Leman J & Bonneau R A novel domain assembly routine for creating full-length models of membrane proteins from known domain structures. Biochemistry acs.biochem.7b00995 (2017). doi: 10.1021/acs.biochem.7b00995 [DOI] [PubMed] [Google Scholar]
- 143.Labonte JW, Adolf-Bryfogle J, Schief WR & Gray JJ Residue-centric modeling and design of saccharide and glycoconjugate structures. J. Comput. Chem. 38, 276–287 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Frenz B, Rämisch S, Borst AJ, Walls AC, Adolf-Bryfogle J, Schief WR, Veesler D & DiMaio F Automatically Fixing Errors in Glycoprotein Structures with Rosetta. Structure 0, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gray JJ, Chaudhury S, Lyskov S, and Labonte JW The PyRosetta Interactive Platform for Protein Structure Prediction and Design: A Set of Educational Modules. (2014). at <http://www.amazon.com/PyRosetta-Interactive-Platform-Structure-Prediction/dp/1500968277>
- 146.Schenkelberg CD & Bystroff C InteractiveROSETTA: A graphical user interface for the PyRosetta protein modeling suite. Bioinformatics (2015). doi: 10.1093/bioinformatics/btv492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kleffner R, Flatten J, Leaver-Fay A, Baker D, Siegel JB, Khatib F & Cooper S Foldit Standalone: a video game-derived protein structure manipulation interface using Rosetta. Bioinformatics 33, 2765–2767 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Khatib F, Cooper S, Tyka MD, Xu K, Makedon I, Popovic Z, Baker D & Players F Algorithm discovery by protein folding game players. Proc. Natl. Acad. Sci. U. S. A. 108, 18949–53 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cooper S, Sterling ALR, Kleffner R, Silversmith WM & Siegel JB Repurposing citizen science games as software tools for professional scientists. in Proc. 13th Int. Conf. Found. Digit. Games - FDG ‘18 1–6 (ACM Press, 2018). doi: 10.1145/3235765.3235770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lyskov S, Chou F-C, Conchúir SÓ, Der BS, Drew K, Kuroda D, Xu J, Weitzner BD, Renfrew PD, Sripakdeevong P, Borgo B, Havranek JJ, Kuhlman B, Kortemme T, Bonneau R, Gray JJ & Das R Serverification of molecular modeling applications: the Rosetta Online Server that Includes Everyone (ROSIE). PLoS One 8, e63906 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Moretti R, Lyskov S, Das R, Meiler J & Gray JJ Web-accessible molecular modeling with Rosetta: The Rosetta Online Server that Includes Everyone (ROSIE). Protein Sci. 27, 259–268 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.DiMaio F, Leaver-Fay A, Bradley P, Baker D & André I Modeling Symmetric Macromolecular Structures in Rosetta3. PLoS One 6, e20450 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fu DY & Meiler J Predictive Power of Different Types of Experimental Restraints in Small Molecule Docking: A Review. J. Chem. Inf. Model. 58, 225–233 (2018). [DOI] [PubMed] [Google Scholar]
- 154.Johnson DK & Karanicolas J Ultra-High-Throughput Structure-Based Virtual Screening for Small-Molecule Inhibitors of Protein–Protein Interactions. J. Chem. Inf. Model. 56, 399–411 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Marze NA, Lyskov S & Gray JJ Improved prediction of antibody V L –V H orientation. Protein Eng. Des. Sel. 29, 409–418 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Finn JA, Koehler Leman J, Willis JR, Cisneros A, Crowe JE & Meiler J Improving Loop Modeling of the Antibody Complementarity-Determining Region 3 Using Knowledge-Based Restraints. PLoS One 11, e0154811 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Weitzner BD & Gray JJ Accurate Structure Prediction of CDR H3 Loops Enabled by a Novel Structure-Based C-Terminal Constraint. J. Immunol. 198, 505–515 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.DeKosky BJ, Lungu OI, Park D, Johnson EL, Charab W, Chrysostomou C, Kuroda D, Ellington AD, Ippolito GC, Gray JJ & Georgiou G Large-scale sequence and structural comparisons of human naive and antigen-experienced antibody repertoires. Proc. Natl. Acad. Sci. 113, E2636–E2645 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jeliazkov JR, Sljoka A, Kuroda D, Tsuchimura N, Katoh N, Tsumoto K & Gray JJ Repertoire Analysis of Antibody CDR-H3 Loops Suggests Affinity Maturation Does Not Typically Result in Rigidification. Front. Immunol. 9, 413 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sircar A, Sanni KA, Shi J & Gray JJ Analysis and modeling of the variable region of camelid single-domain antibodies. J. Immunol. 186, 6357–67 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.North B, Lehmann A & Dunbrack RL A New Clustering of Antibody CDR Loop Conformations. J. Mol. Biol. 406, 228–256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Vaissier Welborn V & Head-Gordon T Computational Design of Synthetic Enzymes. Chem. Rev. 119, 6613–6630 (2019). [DOI] [PubMed] [Google Scholar]
- 163.Marcos E & Silva D-A Essentials of de novo protein design: Methods and applications. Wiley Interdiscip. Rev. Comput. Mol. Sci. 8, e1374 (2018). [Google Scholar]
- 164.Zhou J, Panaitiu AE & Grigoryan G A general-purpose protein design framework based on mining sequence-structure relationships in known protein structures. bioRxiv 431635 (2018). doi: 10.1101/431635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lapidoth G, Khersonsky O, Lipsh R, Dym O, Albeck S, Rogotner S & Fleishman SJ Highly active enzymes by automated combinatorial backbone assembly and sequence design. Nat. Commun. 9, 2780 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Leaver-Fay A, Jacak R, Stranges PB & Kuhlman B A Generic Program for Multistate Protein Design. PLoS One 6, e20937 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sevy AM, Wu NC, Gilchuk IM, Parrish EH, Burger S, Yousif D, Nagel MBM, Schey KL, Wilson IA, Crowe JE & Meiler J Multistate design of influenza antibodies improves affinity and breadth against seasonal viruses. Proc. Natl. Acad. Sci. U. S. A. 116, 1597–1602 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sevy AM, Crowe JE & Meiler J Multi-State Design of Flexible Proteins Predicts Sequences Optimal for 2 Conformational Change 3 4 Marion Sauer. (2019). doi: 10.1101/741454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kroncke BM, Duran AM, Mendenhall JL, Meiler J, Blume JD & Sanders CR Documentation of an Imperative To Improve Methods for Predicting Membrane Protein Stability. Biochemistry 55, 5002–5009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kortemme T & Baker D A simple physical model for binding energy hot spots in protein-protein complexes. Proc. Natl. Acad. Sci. U. S. A. 99, 14116–21 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kortemme T, Kim DE & Baker D Computational alanine scanning of protein-protein interfaces. Sci. STKE 2004, pl2 (2004). [DOI] [PubMed] [Google Scholar]
- 172.Ó Conchúir S, Barlow KA, Pache RA, Ollikainen N, Kundert K, O’Meara MJ, Smith CA & Kortemme T A Web Resource for Standardized Benchmark Datasets, Metrics, and Rosetta Protocols for Macromolecular Modeling and Design. PLoS One 10, e0130433 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Smith CA & Kortemme T Backrub-like backbone simulation recapitulates natural protein conformational variability and improves mutant side-chain prediction. J. Mol. Biol. 380, 742–56 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Crick FHC The Fourier transform of a coiled-coil. Acta Crystallogr. 6, 685–689 (1953). [Google Scholar]
- 175.Kozakov D, Brenke R, Comeau SR & Vajda S PIPER: an FFT-based protein docking program with pairwise potentials. Proteins 65, 392–406 (2006). [DOI] [PubMed] [Google Scholar]
- 176.Rohl CA & Baker D De novo determination of protein backbone structure from residual dipolar couplings using Rosetta. J. Am. Chem. Soc. 124, 2723–9 (2002). [DOI] [PubMed] [Google Scholar]
- 177.Pilla KB, Otting G & Huber T Pseudocontact Shift-Driven Iterative Resampling for 3D Structure Determinations of Large Proteins. J. Mol. Biol. 428, 522–532 (2016). [DOI] [PubMed] [Google Scholar]
- 178.Lange OF & Baker D Resolution-adapted recombination of structural features significantly improves sampling in restraint-guided structure calculation. Proteins Struct. Funct. Bioinforma. 80, 884–895 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bowers PM, Strauss CEM & Baker D De novo protein structure determination using sparse NMR data. 311–318 (2000). [DOI] [PubMed] [Google Scholar]
- 180.Meiler J & Baker D Rapid protein fold determination using unassigned NMR data. Proc. Natl. Acad. Sci. U. S. A. 100, 15404–9 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Raman S, Raman S, Lange OF, Rossi P, Tyka M, Wang X, Aramini J, Liu G, Ramelot TA, Eletsky A, Szyperski T, Kennedy MA, Prestegard J, Montelione GT & Baker D NMR Structure Determination for Larger Proteins Using Backbone-Only Data. 1014, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lange OF, Rossi P, Sgourakis NG, Song Y, Lee H-W, Aramini JM, Ertekin a., Xiao R, Acton TB, Montelione GT & Baker D Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples. Proc. Natl. Acad. Sci. 109, 10873–10878 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Reichel K, Fisette O, Braun T, Lange OF, Hummer G & Schäfer LV Systematic evaluation of CS-Rosetta for membrane protein structure prediction with sparse NOE restraints. Proteins 85, 812–826 (2017). [DOI] [PubMed] [Google Scholar]
- 184.Sgourakis NG, Lange OF, DiMaio F, André I, Fitzkee NC, Rossi P, Montelione GT, Bax A & Baker D Determination of the structures of symmetric protein oligomers from NMR chemical shifts and residual dipolar couplings. J. Am. Chem. Soc. 133, 6288–98 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Rossi P, Shi L, Liu G, Barbieri CM, Lee HW, Grant TD, Luft JR, Xiao R, Acton TB, Snell EH, Montelione GT, Baker D, Lange OF & Sgourakis NG A hybrid NMR/SAXS-based approach for discriminating oligomeric protein interfaces using Rosetta. Proteins Struct. Funct. Bioinforma. 83, 309–317 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Demers J-P, Habenstein B, Loquet A, Kumar Vasa S, Giller K, Becker S, Baker D, Lange A & Sgourakis NG High-resolution structure of the Shigella type-III secretion needle by solid-state NMR and cryo-electron microscopy. Nat. Commun. 5, 4976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Thompson JM, Sgourakis NG, Liu G, Rossi P, Tang Y, Mills JL, Szyperski T, Montelione GT & Baker D Accurate protein structure modeling using sparse NMR data and homologous structure information. Proc. Natl. Acad. Sci. U. S. A. 109, 9875–9880 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Braun T, Koehler Leman J & Lange OF Combining Evolutionary Information and an Iterative Sampling Strategy for Accurate Protein Structure Prediction. PLoS Comput. Biol. 11, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Evangelidis T, Nerli S, Nováček J, Brereton AE, Karplus PA, Dotas RR, Venditti V, Sgourakis NG & Tripsianes K Automated NMR resonance assignments and structure determination using a minimal set of 4D spectra. Nat. Commun. 9, 384 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lange OF Automatic NOESY assignment in CS-RASREC-Rosetta. J. Biomol. NMR 59, 147–159 (2014). [DOI] [PubMed] [Google Scholar]
- 191.Kuenze G, Bonneau R, Koehler Leman J & Meiler J Integrative protein modeling in RosettaNMR from sparse paramagnetic restraints. bioRxiv 597872 (2019). doi: 10.1101/597872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Thyme SB, Jarjour J, Takeuchi R, Havranek JJ, Ashworth J, Scharenberg AM, Stoddard BL & Baker D Exploitation of binding energy for catalysis and design. Nature 461, 1300–1304 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ashworth J, Havranek JJ, Duarte CM, Sussman D, Monnat RJ, Stoddard BL & Baker D Computational redesign of endonuclease DNA binding and cleavage specificity. Nature 441, 656–659 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ashworth J, Taylor GK, Havranek JJ, Quadri SA, Stoddard BL & Baker D Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs. Nucleic Acids Res. 38, 5601–5608 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Havranek JJ & Harbury PB Automated design of specificity in molecular recognition. Nat. Struct. Biol. 10, 45–52 (2003). [DOI] [PubMed] [Google Scholar]
- 196.Thyme SB, Boissel SJS, Arshiya Quadri S, Nolan T, Baker DA, Park RU, Kusak L, Ashworth J & Baker D Reprogramming homing endonuclease specificity through computational design and directed evolution. Nucleic Acids Res. 42, 2564–2576 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Thyme SB, Baker D & Bradley P Improved Modeling of Side-Chain–Base Interactions and Plasticity in Protein–DNA Interface Design. J. Mol. Biol. 419, 255–274 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yanover C & Bradley P Extensive protein and DNA backbone sampling improves structure-based specificity prediction for C2H2 zinc fingers. Nucleic Acids Res. 39, 4564–76 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ashworth J & Baker D Assessment of the optimization of affinity and specificity at protein-DNA interfaces. Nucleic Acids Res. 37, e73 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Thyme SB, Song Y, Brunette TJ, Szeto MD, Kusak L, Bradley P & Baker D Massively parallel determination and modeling of endonuclease substrate specificity. Nucleic Acids Res. 42, 13839–13852 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Overington JP, Al-Lazikani B & Hopkins AL How many drug targets are there? Nat. Rev. Drug Discov. 5, 993–6 (2006). [DOI] [PubMed] [Google Scholar]
- 202.Koehler Leman J, Ulmschneider MB & Gray JJ Computational modeling of membrane proteins. Proteins Struct. Funct. Bioinforma. 83, 1–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yarov-Yarovoy V, Schonbrun J & Baker D Multipass membrane protein structure prediction using Rosetta. Proteins Struct. Funct. Bioinforma. 62, 1010–1025 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Barth P, Schonbrun J & Baker D Toward high-resolution prediction and design of transmembrane helical protein structures. 2007, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Baugh EH, Lyskov S, Weitzner BD & Gray JJ Real-time PyMOL visualization for Rosetta and PyRosetta. PLoS One 6, e21931 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lai JK, Ambia J, Wang Y & Barth P Enhancing Structure Prediction and Design of Soluble and Membrane Proteins with Explicit Solvent-Protein Interactions. Structure 25, 1758–1770.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Alford RF, Fleming PJ, Fleming KG & Gray JJ Protein structure prediction and design in a biologically-realistic implicit membrane. bioRxiv 630715 (2019). doi: 10.1101/630715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Varki A Biological roles of oligosaccharides: all of the theories are correct. Glycobiology 3, 97–130 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW & Etzler ME Essentials of Glycobiology. Essentials Glycobiol. (Cold Spring Harbor Laboratory Press, 2009). [PubMed] [Google Scholar]
- 210.Nivedha AK, Thieker DF, Makeneni S, Hu H & Woods RJ Vina-Carb: Improving Glycosidic Angles during Carbohydrate Docking. J. Chem. Theory Comput. 12, 892–901 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Project Audacious - Institute for Protein Design. (2019). at <https://www.ipd.uw.edu/audacious/>
- 212.Mulligan VK, Melo H, Merritt HI, Slocum S, Weitzner BD, Watkins AM, Renfrew PD, Pelissier C, Arora PS & Bonneau R Designing Peptides on a Quantum Computer . bioRxiv 752485 (2019). doi: 10.1101/752485 [DOI] [Google Scholar]
- 213.Hooper WF, Walcott BD, Wang X & Bystroff C Fast design of arbitrary length loops in proteins using InteractiveRosetta. BMC Bioinformatics 19, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.