Abstract
Inadequate representation of evidence and knowledge about potential drug-drug interactions is a major factor underlying disagreements among sources of drug information that are used by clinicians. In this paper we describe the initial steps toward developing a foundational domain representation that allows tracing the evidence underlying potential drug-drug interaction knowledge. The new representation includes biological and biomedical entities represented in existing ontologies and terminologies to foster integration of data from relevant fields such as physiology, anatomy, and laboratory sciences.
Keywords: Potential drug-drug interactions, ontologies, knowledge management
1. Introduction
Every year, many thousands of people are harmed by exposure to two or more drugs for which there exists a known interaction potential. Exposure to such “potential drug-drug interactions” (PDDIs), are a significant source of preventable drug-related harm, leading to clinically important events in 5.3% – 14.3% of inpatients, and accounting for 0.02% to 0.17% of the 129 million emergency department visits that occur in the U.S. each year[1][2]. Multiple defenses exist in the healthcare system to prevent patient harm from PDDIs including clinician knowledge, computer screening, and monitoring. Each defense depends on complete, accurate, and current knowledge of what drugs have the potential to interact, and the most appropriate methods for managing patients when exposure to a PDDI is unavoidable [3]. However, most sources of clinically-oriented PDDI knowledge disagree substantially in their content, including about which drug combinations should never be never co-administered. For example, only one quarter of 59 contraindicated drug pairs were listed in three PDDI information sources[4], only 18 (28%) of 64 pharmacy information and clinical decisions support systems correctly identified 13 PDDIs considered clinically significant by a team of drug interaction experts[5], and four clinically oriented drug information compendia agreed on only 2.2% of 406 PDDIs considered to be “major” by at least one source[6].
A key factor underlying the existing disagreements among sources of drug information that are used by clinicians is the inadequate representation of PDDI evidence and knowledge. In practice, organizations that provide PDDI information as part of their information services employ an expert or panels of experts (editorial boards) to search, evaluate, synthesize, and stay current with evidence. The process involves applying some criteria to judge whether a drug combination could lead to an interaction, what impact it might have on exposed patients, and how to best manage patient exposure. In the current paradigm, these individuals or groups must search across multiple information sources, including the scientific literature, drug product labeling, and documents submitted to regulatory groups during the drug development/approval process. There is significant variation across drug knowledge bases with respect to ratings of specific drug pairs and currency. Moreover, the available sources rarely include first-hand clinical experience, information that can help contextualize management recommendations[7]. In addition, those multiple sources are currently not created in a way that fosters semantic integration of their data at a later stage. This leads to inefficient and discordant approaches to the acquisition of PDDI evidence and synthesis of that evidence into knowledge. The result is that there is general disagreement among drug information systems about what PDDI exist and their clinical importance.
A goal of the “Addressing gaps in clinically useful evidence on drug-drug interactions” project is to identify the core components of a new PDDI knowledge representation paradigm that addresses these issues. As we describe below, the project makes a fundamental distinction between assertions of PDDI knowledge and the evidence that supports or refutes such assertions. The central thesis of the project is that a framework for representing PDDI assertions and evidence as interoperable Linked Data[8] will enable a more integrated approach to the acquisition and synthesis of PDDI evidence into knowledge. Linked Data methodologies should be used to semantically integrate the various relevant sources of PDDI evidence so that experts can more easily retrieve all relevant evidence items. This will lead to more complete, accurate, and current PDDI information provision to any single evidence board than is possible with current resources.
The proposed framework requires a new foundational representation of PDDIs that covers the material entities and processes in the domain of discourse for PDDI evidence and knowledge claims. The representation will enable the integration of drug interaction mechanisms, effects, risk factors, severity, and management options with the chemical and pharmacological properties (e.g., chemical structure, function, pharmacokinetic and pharmacodynamic properties) of the interacting drugs. This paper specifies the design requirements for such a foundational representation that we are calling the Drug-drug Interaction and Drug-drug Interaction Evidence Ontology (DIDEO). Section 2 provides clinical background. Section 3 discusses the basic design principles and decisions for the new ontology. Finally in section 4, we show that the classes in DIDEO are sufficient to represent a concrete example of PDDI evidence selected from the Drug Interaction Knowledge Base.
2. Background
2.1. A conceptual framework for clinically useful PDDI Knowledge and Evidence
There is a rather complex relationship between the evidence that establishes a PDDI, and information that can help clinicians accurately assess the risk of exposure within a given patient[7]. The foundational model we envision would benefit from an explicit conceptual model of that relationship. Eric van Roon et al. proposed a conceptual model of PDDI information using the definition that clinically-useful PDDI information is that which helps discern whether some action should be taken with respect to a PDDI (Figure 1)[9]. Evidence for, or against, the existence of a PDDI is an important component in that model, along with consideration of patient risk factors, the potential severity of an adverse event that could be caused by exposure, and prior experience with exposure in relevant patient populations. While the van Roon model is not considered a standard for representing PDDI knowledge, it captures the essence of recommendations by other PDDI experts[10][11], including developers of PDDI databases in the United States and Europe[12][13].
Fig. 1.
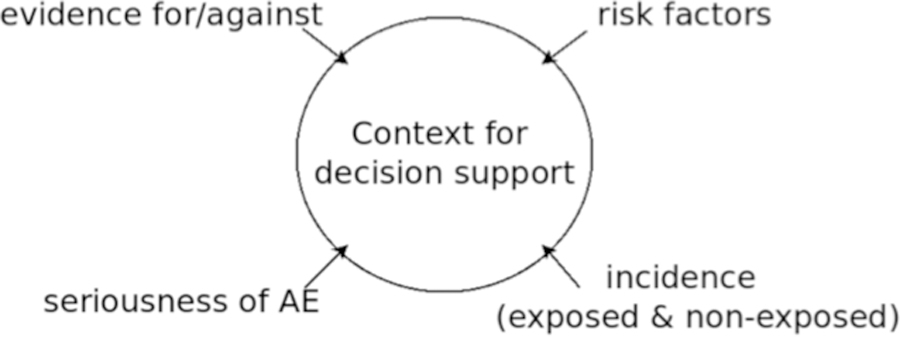
The four types of information used by van Roon et al. to determine if a PDDI warrants clinical action. AE – Adverse Event
The van Roon model helps to conceptually outline the principal information domains for clinically-useful PDDI knowledge. We think that it is also important to consider the relationship between PDDI evidence and claims of PDDI knowledge established by evidence. The evidence for, or against, PDDI assertions is dynamic and of varying robustness to various forms of bias. For example, in prior work on the Drug Interaction Knowledge Base (DIKB) [14][[15][16][17], the editorial board considered certain pharmacologic assertions written in a FDA guidance to industry[18][19][20] useful as PDDI evidence. The assertions reflected the state of science at the time the documents were published. The guidance has been updated, from 1999 to 2006 to 2012, each update leading to changes in the DIKB evidence base.
Based on these observations, it’s possible to conceive of PDDI evidence board as sociotechnical reasoning system that manages both an evidence base and a knowledge base (Figure 2)[21]. This implies that the foundational PDDI knowledge representation we envision would find application within three specific contexts:
within the evidence base, by defining the four types of information from van Roon’s model (Figure 1): evidence that can be used to establish the existence of a PDDI, patient risk factors, the potential severity of an adverse event that could be caused by exposure, and prior experience with exposure in relevant patient populations;
within the knowledge base, by representing the entities explicit within PDDI assertions (e.g., “drug X interacts with drug Y”) and pharmacologic assertions that can be used to infer PDDIs (e.g., “drug X inhibits enzyme Q which is important for the clearance of drug Y from the body”); and
within the reasoning system, by constraining the inference activities of the evidence board so that inferred knowledge is logically consistent with all of the other assertions in the knowledge base.
Fig. 2.
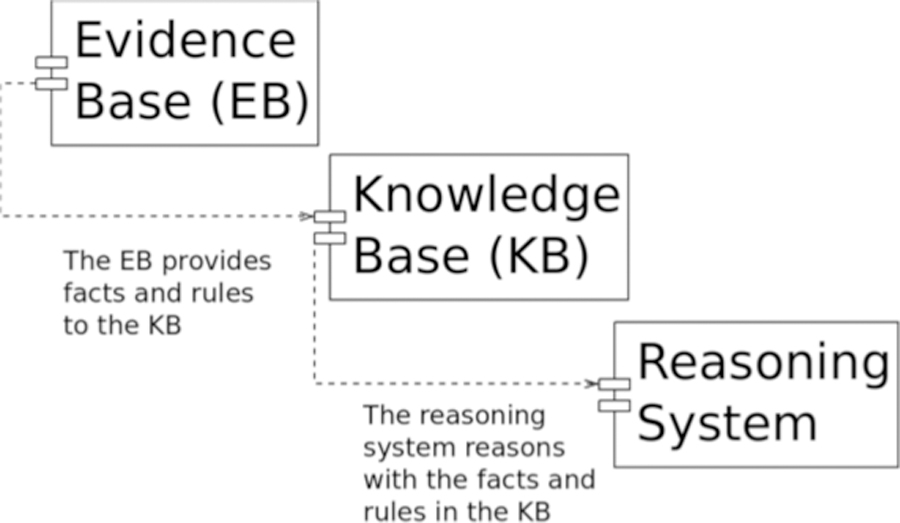
Distinctions between evidence, knowledge, and reasoning with respect to PDDIs
These distinctions can be illustrated using the artifacts used to support the DIKB. Underlying the DIKB’s evidence base are specific examples of evidence types; these evidence types are outlined in a draft online document[22]. Meanwhile the DIKB’s knowledge base contains PDDI and pharmacologic assertions; these assertions address competency questions that were identified during prior work on the DIKB, which can be found in a different draft online document[23].
2.2. Related work
Currently, there are two ontologies built specifically for the domain of drug-drug interactions: the Drug Interaction Ontology (DIO)[24] and the Drug-drug Interaction Ontology (DINTO)[25]. Both provide insights that are valuable for representing the domain. However, neither was designed with the perspective outlined above. Nor do they allow for a consistent and scalable representation of the ontological distinctions relevant to representing clinically useful drug-drug interaction assertions, the drugs involved, and the supporting or refuting evidence. We introduce these ontological distinctions in the course of discussing the existing ontologies.
The DIO is an ontology of drug interactions developed with the goal of predicting drug interactions[24]. While DIO[24] is inspired by both Basic Formal Ontology (BFO) and the NCI Thesaurus (via UMLS), it is not aligned with either one. Although the DIO [24] specifically refers to BFO’s distinction between continuants and processes (occurrents)[26], the BFO’s representation of process (its definition and entity URI), is not reused in the OWL implementation of DIO accompanying the aforementioned paper[27]. Rather, within the DIO OWL file, process is defined as: “A sequence of events which produces some outcome” [27], reusing the CUI and definition from the National Cancer Institute Thesaurus (NCIT)[28]. But the DIO OWL file includes several axioms that are inconsistent with both the NCIT definition of process and the BFO’s representation of process. In particular, DIO specifies necessary conditions for processes, such as
hasEnableTriggerParticipant min 1 Thing
hasResultantPopulationChange min 1 Thing
hasResultantPopulationChange only Increased
A possible explanation for this inconsistency is that the developers of DIO intended to map their domain representation to those found in the UMLS (NCIT) and other terminologies and ontologies rather than actually reuse them. However, the reuse of terms should always be accompanied by ensuring that the intended meaning and the ontological commitments of source and target resource match. (We will elaborate on the role of ontological commitments in Section 3.)
Another shortcoming of DIO is that it does not represent roles. Each instance of a chemical is a drug, regardless of whether its dosage or formulation allows it to act as a drug. We find, for instance, that:
Capecitabine rdfs:subClassOf Drugs rdfs:subClassOf DrugOrMetabolite rdfs:subClassOf Chemicals
But active ingredients can only bear a role as a drug in a specific dose and in conjunction with excipients[10]. However, it is by now standard accepted practice in numerous drug terminologies and ontologies to carefully distinguish among drug products, their ingredients, and the molecules that constitute those ingredients[29][30][31][32]. Most recently, Hogan et al. in this regard showed that assigning therapeutic properties to active ingredients disregards the effect of dose form and therefore leads to mistakes that contradict scientific knowledge (e.g., oral vancomycin treating bacterial endocarditis)[30].
The second ontology that we took into consideration is the Drug Interaction Ontology (DINTO). DINTO is intended “to represent all possible mechanisms that can lead to a drug-drug interaction. The ontology provides the general pharmacological principles of the domain”[25]. The developers have provided a version of DINTO that is an extension of BFO[33]. The key limitation of DINTO with respect to our goals is that DINTO does not represent potential drug-drug interactions at all, but only drug-drug interactions (DDIs). Representing PDDIs, as we aim to do, is quite different from representing a DDI. For each individual instance of a drug-drug interaction it is possible to specify the individual patient who suffered from its effects. However, this is not possible for all instances of PDDIs, because some of them are not actualized. DIDEO will be based on a novel definition of PDDI (Section 3).
While DINTO does not represent PDDIs at all, the way it represents the actual occurrences of DDIs and information about those occurrences is problematic. DINTO specifies a subclass of DDIs named DDI described in a database. The members of this class are intended to be DDIs1 that are linked to a database by the is described in-relation. According to the DINTO OWL file[33] the class ‘DDI in database’ is intended to “represent those DDIs imported in DINTO from the DrugBank database[34][35] with the purpose of distinguishing them from those inferred from the ontology”. Notably, the information loaded from DrugBank will not be about drug-drug interactions, but about PDDIs, as [35] clearly indicates. This demonstrates that DINTO does not provide ways to distinguish information about actual DDIs from evidence pointing at drug co-medications that are suspected to lead to unwanted effects.
3. Methods
Building from the pioneering work of the DIO and DINTO we propose to develop a new ontology, DIDEO. The DIDEO will address the aforementioned limitations of the two ontologies while being in alignment with the van Roon conceptual model. Moreover, the ontology will comply with principles of good practice in ontology development, such as formulated by the OBO Foundry[36], for instance:
Reuse of pre-existing resources –
Because integration of data is among the key rationales for using ontologies, it is crucial to use Unique Resource Identifiers to refer to the same entities even across domains. We strive to reuse entities from pre-existing ontologies wherever reasonable. Reuse of entities may be limited by the fact that the basic ontological commitments of the source and the target ontology need to be the same. For instance, an ontology that defines ‘drug’ as ‘a chemical entity that bears a drug role that is realized by its use in a pharmacotherapy’ cannot import an individual drug, for instance ‘acetaminophen’ from an ontology that defines drugs as chemicals that are used in pharmacotherapy2. In our example both ontologies represent ‘drug’, but each representation comes with a different ontological commitment. One way to assure consistency of ontological commitment is to select entities from ontologies using the same upper ontology.
Use of an upper ontology relevant in biomedical informatics –
The DIDEO should support the integration of drug-drug interaction data with data on other biomedically relevant phenomena, for example proteins, protein interactions, laboratory methods and clinical studies. We reuse entities from the Drug Ontology (DRON) [29][30][37] the Ontology of Biomedical Investigation (OBI)[38][39], the Gene Ontology (GO)[40][41] and the Information Artifact Ontology (IAO)[42] These ontologies are all listed on the OBO Foundry[43] webpage. The reuse of DRON, OBI and IAO commits the DIDEO to use the Basic Formal Ontology (BFO) [26][44] as the upper ontology. The Gene Ontology is not using any upper ontology, but multiple classes from GO have been subsumed under BFO classes in the aforementioned ontologies. Representation of biological and biomedical entities was one of the use cases in the development of BFO[26]. In addition, BFO provides well-documented representations for roles, functions and dispositions[45], which are also relevant for biological and biomedical phenomena. Hence we plan to use BFO 2.0 as our upper level and we will import existing OWL entities for reuse using the MIREOT methodology[46] implemented in the MIREOT Protégé plugin[47]. One open question when using MIREOT to import terms from pre-existing ontologies is how to track changes in the source ontologies. MIREOT [46] relies on OBO Foundry’s internal (non-automatic) monitoring process, which might not be an optimal solution, especially when the methodology is applied outside the Foundry.
Compliancy with relevant standards of drug representation –
The initial development of DIDEO re-uses the drug representation of the Drug Ontology[29] (DRON) which is based on RxNorm[32]. For a PDDI ontology, active ingredients must not be assigned the status of drugs, because the excipients, route of administration, and dose impact the potential for, likelihood of, and severity of interactions. DRON provides ontologically sound representations of Clinical Drugs and Branded Drugs. These representations contain information about dosage and intended route of administration. In addition, DRON provides information about drug ingredients that is linked to the Chemical Entities of Biological Interest ontology [29][30][48].
Community-driven development –
Once the initial OWL version of DIDEO is created it will be made publicly available and the project will be continued as an open source project. In addition, we aim to build a community of consumers/contributors to help us create, expand and maintain the ontology. We have already reached out to the developers of DINTO. Since DINTO is using BFO as the upper ontology just as DIDEO we want to investigate possibilities to align our efforts.
4. Results
To appropriately represent PDDI knowledge and its evidence, a definition of PDDIs is crucial. We start our definition by making a basic ontological categorization based on BFO 2.0[49]. The most basic ontological categories of BFO are independent continuant (such as material entities), generically dependent continuants (such as information content entities), specifically dependent continuants (such as qualities and dispositions) and occurrents (such as processes)[26]. Consider an individual PDDI. We might be tempted to categorize it as an occurrent, since the term “interaction” points to a process. However, a PDDI is not an actual process. It is also not a potential or disposition inhering in a substance that may or may not be realized. Representing PDDI in that way, would neglect the fact, that in PDDI research we are collecting information about occurrences that could be drug-drug interactions. Rather, a PDDI is a piece of information about the possible effects of a certain event, for instance the co-administration of azithromycin and ergot alkaloids. We propose the following definition for PDDI:
“A potential drug-drug interaction (PDDI) is an information content entity that specifies the possibility of a drug-drug interaction based on either reasonable extrapolation about drug-drug interaction mechanisms or a data item created by clinical studies, clinical observation or physiological experiments.”
From this starting point it is crucial to represent the informational bases of PDDIs, namely a) reasonable extrapolation, b) physiological observations from clinical studies and c) drug-drug interaction observational data and d) mechanistic assertions that are useful for inferring drug-drug interactions (derived either from clinical studies or from various experiments, such as inhibition and transport protein experiments)
The aim of DIDEO is to adequately represent all types of PDDI evidence, as well as their differing bases. Not all instances of PDDI evidence data are based on actually observing a drug-drug interaction. In many instances, it is unclear from the evidence whether any actual interaction between the object and precipitant drug has occurred. Hence, it would not be appropriate for us to code the ontology in a way that implies, from the existence of the PDDI evidence, the existence of at least one instance of the specific drug-drug interaction. These bases imply the existence of specific physiological processes, drugs, drug components, and in some cases even DDIs. The following example describes a case of PDDI without evidence for an instance of the actual DDI: Assume the class ‘potential azithromycin-ergot alkaloid interaction evidence data’ exists in an OWL ontology. To link the data item to actual physiological processes we could axiomatize that each element of this class is about some element in the class ‘azithromycin-ergot alkaloid interaction’. But our axiom will then imply that at least one element of the latter class exists.
This existential import can be avoided by using a feature novel in OWL2 called ‘punning’. ‘Punning’ enables users to assign the same name to an OWL class and an OWL individual, allowing the use of the individual when referring to the type and the use of the class when referring to individuals or aggregate of individuals. Despite the two entities bearing the same name, no cross-inferences are made when reasoning[50]. Thus, punning would allow stating that a PDDI is about at least two types of drugs, without affirming that each individual entity is about one individual portion of that drug.
Figure 3 shows a DIDEO representation (without punning) of a clinical study potentially useful as evidence for a mechanistic assertion that could be used for inferring drug-drug interactions. The figure shows how information about the drug and enzyme involved in the study can be traced from the study data item. It would also be possible to track the type of clinical study. Some ontological commitments and design decision depicted in Figure 3 warrant more detail:
A particular simvastatin metabolism process is the proper occurrent part of a drug metabolism assay. In natural language we might say that the metabolism participates in the assay. But one of the ontological commitments of BFO is that the ‘participates in’-relation only holds between a continuant and an occurrent[51]. To simply say that the metabolism and the assay temporarily overlap would not be sufficient here. Many processes overlap in time, without being interrelated in any other way.
When we represent substances that are referred to by mass nouns, we talk about a portion of that substance (e.g. the simvastatin metabolism has a portion of simvastatin as a participant). This is inline with the practice used in numerous OBO Foundry ontologies, to distinguish between a specific instance of a portion of the entity and the term denoting its type[52].
Both the portion of simvastatin and the portion of CYP3A4 are affirmed to participate in the process at some times. Claiming their participation at all times we would exclude the possibility that there is no CYP3A4 available to participate in the metabolism, while the final part of the process are still occurring. Moreover, ‘participate at some time’ is not a negation of ‘participate at all time’, but, entails the latter [53].
Assays of a simvastatin metabolism establish that CYP3A4 is the bearer of a disposition called ‘drug metabolism enabler disposition’ that enables the metabolism, and is realized by that metabolism.
The outcome of the assay is a simvastatin metabolism data item. Data items are defined as “an information content entity that is intended to be a truthful statement about something (modulo, e.g., measurement precision or other systematic errors) and is constructed/acquired by a method which reliably tends to produce (approximately) truthful statements”[42]. The simvastatin metabolism data item is a member of the class of data items that are specified output of some drug metabolism assays.
Fig. 3:
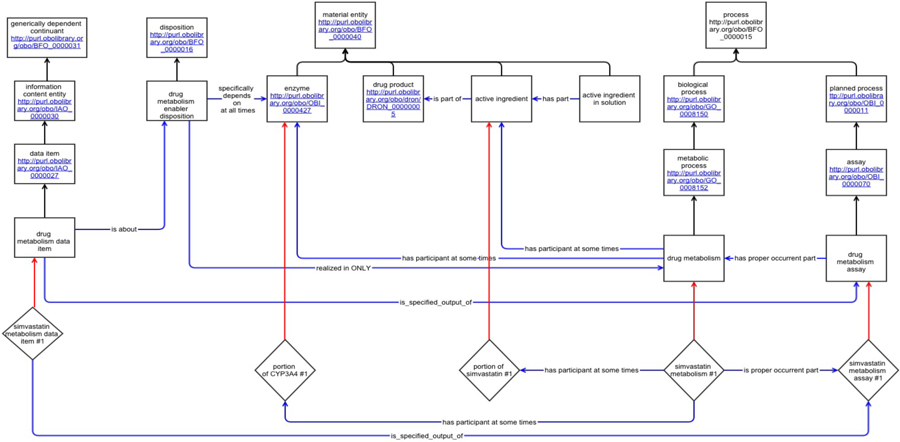
Representation of instances derived from DIKB in the new semantic model, DIDEO. (Boxes represent OWL named classes; Text in boxes gives the label for the class and for imported terms the class URI, diamonds represent individuals, arrows represent object properties or rdfs:subClassOf (i.e., the “is a” relation)).
5. Conclusion
This paper offers justification for a new foundational domain representation for PDDI knowledge and describes the initial steps toward its development. This new semantic model, the Drug-drug Interaction and Drug-drug Interaction Evidence Ontology (DIDEO), is motivated by the needs of experts who must search, evaluate, and synthesize PDDI evidence into knowledge claims. The results reported in this paper form a foundation for the further development of DIDEO. We will now start to implement DIDEO in OWL in order to test its applicability with respect to competency questions [23] specified in as part of the “Addressing gaps in clinically useful evidence on drug-drug interactions” project. During implementation we will seek to coordinate our efforts with the developers of DINTO and DRON.
Acknowledgments:
For all authors: This project is supported by a grant from the National Library of Medicine: “Addressing gaps in clinically useful evidence on drug-drug interactions” (R01LM011838-01) and the National Institute of Aging “Improving medication safety for nursing home residents prescribed psychotropic drugs” (K01 AG044433-01). The authors thank Michel Dumontier and Alan Ruttenberg for their valuable comments, which have significantly improved the paper.
For PE: This work is supported by the National Center For Advancing Translational Sciences of the National Institutes of Health under Award Number KL2TR000146.
For DM: This work is partially supported by the Agency for Healthcare Research and Quality (AHRQ) Grant No. 1R13HS021826-01 (Malone DC-PI)
For JS: This work was carried out during the tenure of an ERCIM “Alain Bensoussan” Fellowship Programme. The research leading to these results has received funding from the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement no 246016.
Footnotes
The axiomatization of the class actually falls short of specifying that, since being a DDI is not part of its necessary and sufficient condition.
The aspirin in my medicine cabinet is not a drug according to the second definition given above since it is not participating in any pharmacotherapy, yet it is most certainly a drug according to the first definition.
References
- 1.Magro L, Moretti U, Leone R: Epidemiology and characteristics of adverse drug reactions caused by drug-drug interactions. Expert Opin. Drug Saf 11, 83–94 (2012). DOI: 10.1517/14740338.2012.631910 [DOI] [PubMed] [Google Scholar]
- 2.FastStats – Emergency Department Visits, http://www.cdc.gov/nchs/fastats/emergency-department.htm. Last accessed: 07/29/2014.
- 3.Horn JR, Hansten PD: Sources of error in drug interactions: the Swiss cheese model. Pharm. Times 70, 53–55 (2004). [Google Scholar]
- 4.Wang LM, Wong M, Lightwood JM, Cheng CM: Black box warning contraindicated comedications: concordance among three major drug interaction screening programs. Ann. Pharmacother 44, 28–34 (2010). DOI: 10.1345/aph.1M475 [DOI] [PubMed] [Google Scholar]
- 5.Saverno KR, Hines LE, Warholak TL, Grizzle AJ, Babits L, Clark C, Taylor AM, Malone DC: Ability of pharmacy clinical decision-support software to alert users about clinically important drug-drug interactions. J. Am. Med. Inform. Assoc. JAMIA 18, 32–37 (2011). DOI: 10.1136/jamia.2010.007609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abarca J, Malone DC, Armstrong EP, Grizzle AJ, Hansten PD, Van Bergen RC, Lipton RB: Concordance of severity ratings provided in four drug interaction compendia. J. Am. Pharm. Assoc. JAPhA 44, 136–141 (2004). DOI: 10.1331/154434504773062582 [DOI] [PubMed] [Google Scholar]
- 7.Hines LE, Murphy JE, Grizzle AJ, Malone DC: Critical issues associated with drug-drug interactions: highlights of a multistakeholder conference. Am. J. Health-Syst. Pharm. AJHP Off. J. Am. Soc. Health-Syst. Pharm 68, 941–946 (2011). DOI: 10.2146/ajhp100440 [DOI] [PubMed] [Google Scholar]
- 8.Marshall MS, Boyce R, Deus HF, Zhao J, Willighagen EL, Samwald M, Pichler E, Hajagos J, Prud’Hommeaux E, Stephens S: Ontology Paper: Emerging Practices for Mapping and Linking Life Sciences Data Using RDF - A Case Series. Web Semant 14, 2–13 (2012). [Google Scholar]
- 9.Van Roon EN, Flikweert S, le Comte M, Langendijk PNJ, Kwee-Zuiderwijk WJM, Smits P, Brouwers JRBJ: Clinical relevance of drug-drug interactions!: a structured assessment procedure. Drug Saf. Int. J. Med. Toxicol. Drug Exp 28, 1131–1139 (2005). DOI: 10.2165/00002018-200528120-00007 [DOI] [PubMed] [Google Scholar]
- 10.Preskorn SH: Drug-drug interactions: proof of relevance (part I). J. Psychiatr. Pract 11, 116–122 (2005). DOI: 10.1097/00131746-200503000-00006 [DOI] [PubMed] [Google Scholar]
- 11.Preskorn SH: Drug-drug interactions: Proof of relevance (Part II): cause of tolerability problems or noncompliance. J. Psychiatr. Pract 11, 397–401 (2005). DOI: 10.1097/00131746-200511000-00006 [DOI] [PubMed] [Google Scholar]
- 12.Hansten PD, Horn JR, Hazlet TK: ORCA: OpeRational ClassificAtion of drug interactions. J. Am. Pharm. Assoc Washington DC: 1996. 41, 161–165 (2001). DOI: 10.1016/s1086-5802(16)31244-x [DOI] [PubMed] [Google Scholar]
- 13.Ylva Böttiger KL: SFINX-a drug-drug interaction database designed for clinical decision support systems. Eur. J. Clin. Pharmacol 65, 627–33 (2009). DOI: 10.1007/s00228-008-0612-5 [DOI] [PubMed] [Google Scholar]
- 14.Boyce RD: Drug Interaction Knowledge Base, http://purl.org/net/drug-interaction-knowledge-base/. Last accessed: 08/09/2014
- 15.Boyce RD, Collins C, Horn J, Kalet I: Modeling Drug Mechanism Knowledge Using Evidence and Truth Maintenance. IEEE Trans. Inf. Technol. Biomed 11, 386–397 (2007). DOI: 10.1109/titb.2007.890842 [DOI] [PubMed] [Google Scholar]
- 16.Boyce R, Collins C, Horn J, Kalet I: Computing with evidence Part I: A drug-mechanism evidence taxonomy oriented toward confidence assignment. J. Biomed. Inform 42, 979–989 (2009). DOI: 10.1016/j.jbi.2009.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyce R, Collins C, Horn J, Kalet I: Computing with evidence Part II: An evidential approach to predicting metabolic drug-drug interactions. J. Biomed. Inform 42, 990–1003 (2009). DOI: 10.1016/j.jbi.2009.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.US Food and Drug Administration: Guidance for Industry Drug Interaction Studies — Study Design, Data Analysis, Implications for Dosing, and Labeling Recommendations, (1999).
- 19.US Food and Drug Administration: Guidance for Industry Drug Interaction Studies — Study Design, Data Analysis, Implications for Dosing, and Labeling Recommendations, (2006).
- 20.US Food and Drug Administration: Guidance for Industry Drug Interaction Studies — Study Design, Data Analysis, Implications for Dosing, and Labeling Recommendations, (2012).
- 21.Boyce RD, Collins C, Horn J, Kalet I: Modeling drug mechanism knowledge using evidence and truth maintenance. IEEE Trans. Inf. Technol. Biomed. Publ. IEEE Eng. Med. Biol. Soc 11, 386–397 (2007). DOI: 10.1109/titb.2007.890842 [DOI] [PubMed] [Google Scholar]
- 22.Boyce RD: A Draft Evidence Taxonomy and Inclusion Criteria for the Drug Interaction Knowledge Base (DIKB), http://purl.net/net/drug-interaction-knowledge-base/evidence-types-and-inclusion-criteria. Last accessed: 10/01/2014.
- 23.Boyce RD: Draft Competency Questions for a Drug Interaction Knowledge Base Derived from Discussions with Clinical Experts, http://purl.net/net/drug-interaction-knowledge-base/competency-questions. Last accessed: 10/01/2014.
- 24.Arikuma T, Yoshikawa S, Azuma R, Watanabe K, Matsumura K, Konagaya A: Drug interaction prediction using ontology-driven hypothetical assertion framework for pathway generation followed by numerical simulation. BMC Bioinformatics. 9, S11 (2008). DOI: 10.1186/1471-2105-9-S6-S11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrero-Zazo M, Hastings J, Segura-Bedmar I, Croset S, Martinez P, Steinbeck C: An ontology for drug-drug interactions. http://ceur-ws.org/Vol-1114/Session3_Herrero-Zazo.pdf. Last accessed: 10/05/2014
- 26.Grenon P, Smith B, Goldberg L: Biodynamic Ontology: Applying BFO in the Biomedical Domain. Stud. Health Technol. Inform pp. 20–38. IOS Press; (2004). [PubMed] [Google Scholar]
- 27.The Drug Interaction Ontology, http://www.biomedcentral.com/content/supplementary/1471-2105-9-S6-S11-S1.owl. Last accessed: 08/09/2014.
- 28.NCI Thesaurus, http://ncit.nci.nih.gov. Last accessed: 08/09/2014.
- 29.Hanna J, Joseph E, Brochhausen M, Hogan WR: Building a drug ontology based on RxNorm and other sources. J. Biomed. Semant 4, 44 (2013). DOI: 10.1186/2041-1480-4-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hogan WR, Hanna J, Joseph E, Brochhausen M: Towards a Consistent and Scientifically Accurate Drug Ontology. http://www2.unb.ca/csas/data/ws/icbo2013/papers/research/icbo2013_submission_40.pdf. Last accessed: 10/05/2014. [PMC free article] [PubMed]
- 31.Luciano JS, Andersson B, Batchelor C, Bodenreider O, Clark T, Denney CK, Domarew C, Gambet T, Harland L, Jentzsch A, Kashyap V, Kos P, Kozlovsky J, Lebo T, Marshall SM, McCusker JP, McGuinness DL, Ogbuji C, Pichler E, Powers RL, Prud’hommeaux E, Samwald M, Schriml L, Tonellato PJ, Whetzel PL, Zhao J, Stephens S, Dumontier M: The Translational Medicine Ontology and Knowledge Base: driving personalized medicine by bridging the gap between bench and bedside. J. Biomed. Semant 2 Suppl 2, S1 (2011). DOI: 10.1186/2041-1480-2-S2-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson SJ, Zeng K, Kilbourne J, Powell T, Moore R: Normalized names for clinical drugs: RxNorm at 6 years. J. Am. Med. Inform. Assoc 18, 441–448 (2011). DOI: 10.1136/amiajnl-2011-000116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The Drug-Drug Interaction Ontology, https://4625527d7e0f0589865f115fb0c87fc18bef216f.googledrive.com/host/0B-7Po9tR1KLUNkpNdmFEcG44RjA/DINTO_1BFO.owl. Last accessed: 10/01/2014
- 34.Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, Pon A, Banco K, Mak C, Neveu V, Djoumbou Y, Eisner R, Guo AC, Wishart DS: DrugBank 3.0: a comprehensive resource for “omics” research on drugs. Nucleic Acids Res 39, D1035–1041 (2011). DOI: 10.1093/nar/gkq1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y, Maciejewski A, Arndt D, Wilson M, Neveu V, Tang A, Gabriel G, Ly C, Adamjee S, Dame ZT, Han B, Zhou Y, Wishart DS: DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res 42, D1091–1097 (2014). DOI: 10.1093/nar/gkt1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The OBO Foundry: OBO Foundry Principles, http://www.obofoundry.org/crit.shtml. Last accessed: 09/15/2014.
- 37.The Drug Ontology, http://purl.obolibrary.org/obo/dron.owl. Last accessed: 10/05/2014.
- 38.The Ontology of Biomedical Investigation, http://purl.obolibrary.org/obo/obi.owl. Last accessed: 10/05/2014
- 39.Brinkman RR, Courtot M, Derom D, Fostel JM, He Y, Lord P, Malone J, Parkinson H, Peters B, Rocca-Serra P, Ruttenberg A, Sansone S-A, Soldatova LN, Stoeckert CJ, Turner JA, Zheng J, OBI consortium: Modeling biomedical experimental processes with OBI. J. Biomed. Semant 1 Suppl 1, S7 (2010). DOI: 10.1186/2041-1480-1-S1-S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.The Gene Ontology, http://purl.obolibrary.org/obo/go.owl. Last accessed: 10/05/2014
- 41.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sher-lock G: Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet 25, 25–29 (2000). DOI: 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.The Information Artifact Ontology, http://purl.obolibrary.org/obo/iao.owl. Last accessed: 10/05/2014
- 43.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, OBI Consortium, Leontis N, Rocca-Serra P, Ruttenberg A, Sansone S-A, Scheuermann RH, Shah N, Whetzel PL, Lewis S: The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol 25, 1251–1255 (2007). DOI: 10.1038/nbt1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grenon P, Smith B: SNAP and SPAN: Towards Dynamic Spatial Ontology. Spat. Cogn. Comput 4, 69–103 (2004). [Google Scholar]
- 45.Arp R, Smith B: Function, Role, and Disposition in Basic Formal Ontology. Nat. Preced (2008).
- 46.Courtot M, Gibson F, Lister AL, Malone J, Schober D, Brinkman RR, Ruttenberg A: MIREOT: The Minimum Information to Reference an External Ontology Term. Appl Ontol 6, 23–33 (2011). [Google Scholar]
- 47.Hanna J, Chen C, Crow A, Hall R, Liu J, Pendurthi T, Schmidt T, Jennings SF, Brochhausen M, Hogan WR: Simplifying MIREOT: a MIREOT Protégé Plugin. International Semantic Web Conference Posters & Demos, volume 914 of CEUR Workshop Proceedings. CEUR-WS.org, Boston (2012). [Google Scholar]
- 48.Hastings J, de Matos P, Dekker A, Ennis M, Harsha B, Kale N, Muthukrishnan V, Owen G, Turner S, Williams M, Steinbeck C: The ChEBI reference database and ontology for biologically relevant chemistry: enhancements for 2013. Nucleic Acids Res 41, D456–D463 (2013). DOI: 10.1093/nar/gks1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.The Basic Formal Ontology, Release 2014-05-03, http://purl.obolibrary.org/obo/bfo/2014-05-03/bfo.owl. Last accessed: 10/05/2014
- 50.Golbreich C, Wallace EK: OWL 2 Web Ontology Language New Features and Rationale (Second Edition), http://www.w3.org/TR/owl2-new-features/. Last accessed: 09/25/2014.
- 51.Smith B, Almeida M, Bona J, Brochhausen M, Ceusters W, Courtot M, Dipert R, Goldfain A, Grenon P, Hastings J, Hogan WR, Jacuzzo L, Johansson I, Mungall CJ, Natale D, Neuhaus F, Petosa A, Rovetto R, Ruttenberg A, Ressler M, Schulz S: Basic Formal Ontology 2.0 Draft Specification and User’s Guide, https://bfo.googlecode.com/svn/trunk/docs/bfo2-reference/BFO2-Reference.docx, (2014). Last accessed: 10/05/2014
- 52.Kumar A, Smith B, Novotny DD: Biomedical informatics and granularity. Comp. Funct. Genomics 5, 501–508 (2004). DOI: 10.1002/cfg.429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schulz S, Ruttenberg A, Courtot M, Hastings J: Release notes for the BFO 2 2012-07-20 “Graz” OWL Release, http://purl.obolibrary.org/obo/bfo/2012-07-20/ReleaseNotes, (2012). Last accessed: 10/05/2014


