Abstract
STUDY QUESTION
Do sperm mitochondrial DNA measures predict probability of pregnancy among couples in the general population?
SUMMARY ANSWER
Those with high sperm mitochondrial DNA copy number (mtDNAcn) had as much as 50% lower odds of cycle-specific pregnancy, and 18% lower probability of pregnancy within 12 months.
WHAT IS KNOWN ALREADY
Semen parameters have been found to poorly predict reproductive success yet are the most prevalent diagnostic tool for male infertility. Increased sperm mtDNAcn and mitochondrial DNA deletions (mtDNAdel) have been associated with decreased semen quality and lower odds of fertilization in men seeking fertility treatment.
STUDY DESIGN, SIZE, DURATION
A population-based prospective cohort study of couples discontinuing contraception to become pregnant recruited from 16 US counties from 2005 to 2009 followed for up to 16 months.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Sperm mtDNAcn and mtDNAdel from 384 semen samples were assessed via triplex probe-based quantitative PCR. Probability of pregnancy within 1 year was compared by mitochondrial DNA, and discrete-time proportional hazards models were used to evaluate the relations with time-to-pregnancy (TTP) with adjustment for covariates.
MAIN RESULTS AND THE ROLE OF CHANCE
Higher sperm mtDNAcn was associated with lower pregnancy probability within 12 months and longer TTP. In unadjusted comparisons by quartile (Q), those in Q4 had a pregnancy probability of 63.5% (95% CI: 53.1% to 73.1%) compared to 82.3% (95% CI: 73.2% to 89.9%) for Q1 (P = 0.002). Similar results were observed in survival analyses adjusting for covariates to estimate fecundability odds ratios (FORs) comparing mtDNAcn in quartiles. Relative to those in Q1 of mtDNAcn, FORs (95% CI) were for Q2 of 0.78 (0.52 to 1.16), Q3 of 0.65 (0.44 to 0.96) and Q4 of 0.55 (0.37 to 0.81), and this trend of decreasing fecundability with increasing mtDNAcn quartile was statistically significant (FOR per log mtDNAcn = 0.37; P < 0.001). Sperm mtDNAdel was not associated with TTP.
LIMITATIONS, REASONS FOR CAUTION
This prospective cohort study consisted primarily of Caucasian men and women and thus large diverse cohorts are necessary to confirm the associations between sperm mtDNAcn and couple pregnancy success in other races/ethnicities.
WIDER IMPLICATIONS OF THE FINDINGS
Our results demonstrate that sperm mtDNAcn has utility as a biomarker of male reproductive health and probability of pregnancy success in the general population.
STUDY FUNDING/COMPETING INTEREST(S)
This work was funded in part by the National Institute of Environmental Health Sciences, National Institutes of Health (R01-ES028298; PI: J.R.P.) and the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, Maryland (Contracts N01-HD-3-3355, N01-HD-3-3356 and N01-HD-3-3358). The authors declare no competing interests.
TRIAL REGISTRATION NUMBER
N/A
Keywords: fecundity, sperm, time-to-pregnancy, mitochondria, mitochondrial DNA copy number, mitochondrial biomarkers
Introduction
Infertility, clinically defined as the inability to achieve pregnancy within 12 months of regular unprotected sexual intercourse, is estimated to affect about 15% of couples globally (Jungwirth et al., 2012; Thoma et al., 2013). The prevalence of male infertility in the USA is reported at 12% (Louis et al., 2013) and accounts for approximately 50% of couple-based infertility (Thonneau et al., 1991). Clinically, male infertility is typically determined using measures of semen quality dictated by World Health Organization (WHO) cut points (Cooper et al., 2010). A recent meta-analysis suggested that sperm concentration and total sperm count among men in developed countries have declined over 50% in the past four decades, raising concerns about temporal trends in male fertility (Levine et al., 2017). However, importantly, semen parameter measures have been shown to poorly predict reproductive success and have minimal association with fecundability (Jungwirth et al., 2012; Buck Louis et al., 2014), raising questions about the significance and interpretation of conventional approaches for measuring male infertility. Thus, the identification of biomarkers that accurately predict male reproductive health and success is of major significance due to the potential to impact clinical care and to improve our understanding of the roots of male factor infertility.
Characteristics of mitochondrial DNA (mtDNA) and results of research to date provide motivation to evaluate the relations between mtDNA copy number (mtDNAcn) and deletions (mtDNAdel) and couple fecundity. Mitochondria are involved in a variety of biological functions, most predominately ATP production via oxidative phosphorylation of the electron transport chain (ETC) but also hormone production, ion homeostasis and apoptosis. Importantly, mtDNA is highly susceptible to damage and dysfunction due to its proximity to the ETC in conjunction with its less robust repair mechanisms compared to those of nuclear DNA (Phillips et al., 2014). Propagation of the mtDNA genome can occur to compensate for defective mitochondria or damaged mtDNA. MtDNAcn and mtDNAdel have been used as biomarkers of mitochondrial function in cancer, in neurodegenerative conditions, and in aging (Taylor and Turnbull, 2005; Greaves and Turnbull, 2009; Coskun et al., 2010).
Mitochondria form tight helices at the mid-piece of sperm during spermatogenesis and contribute to the motility of sperm (Amaral et al., 2013). Sperm mtDNAcn is reduced during spermatogenesis by approximately 8- to 10 folds (Hecht et al., 1984; St John et al., 2010), ensuring sperm mtDNAcn is low upon fertilization because mtDNA is maternally inherited. We (Wu et al., 2019a) and others (Song and Lewis, 2008; Zhang et al., 2016) have shown that retention of mtDNA during spermatogenesis is strongly related to semen quality, such that higher sperm mtDNAcn and mtDNAdel were associated with decreases in sperm concentration, total sperm count and progressive motility. Moreover, we have recently shown that higher sperm mtDNAcn and mtDNAdel were associated with lower odds of fertilization among men seeking fertility treatment and that these mtDNA biomarkers provided incremental predictive ability of fertilization independent of male age and semen parameters (Wu et al., 2019b). Elevated sperm mtDNAcn and mtDNAdel may indicate lower quality spermatozoa resulting from damage due to oxidative stress (Chen et al., 2018), aborted apoptosis or abnormal spermatogenesis (Song and Lewis, 2008). In addition, given that various mechanisms are proposed to prevent the transmission of paternal mtDNA (Sutovsky et al., 1999; St John et al., 2005; Al Rawi et al., 2011), elevated sperm mtDNAcn is not desirable and could negatively impact fertilization and embryo development and these defenses may result in a direct impact of elevated mtDNAcn on fertilization (Darr et al., 2017). Thus, mtDNA biomarkers in sperm may have utility as an indicator of reproductive health. Research to date, however, is largely limited to clinical populations of couples seeking fertility treatment.
To our knowledge, no studies have investigated the relation between sperm mtDNA biomarkers and couple fecundity as measured by time-to-pregnancy (TTP) among couples recruited from the general population. TTP is a functional measure of couples’ individual and combined fecundity in keeping with the couple dependent nature of human reproduction. Using a prospective cohort, we examined the relations between sperm mtDNAcn and mtDNAdel and TTP, and their role as biomarkers of couple fecundity.
Materials and methods
Study population
Whole semen samples were provided by male participants of the Longitudinal Investigation of Fertility and the Environment (LIFE) study, a prospective cohort for which details have been previously published (Buck Louis et al., 2011; Buck Louis et al., 2014). In brief, 501 couples were recruited from 16 counties in Michigan and Texas using a population-based sampling frame (Buck Louis et al., 2011). By design, eligibility criteria were minimal: (i) in a committed relationship and planning to discontinue contraception to become pregnant; (ii) no injectable contraceptive use in past year or off contraception for >2 months; (iii) females aged 18–40 and males aged 18 years or older; (iv) females’ self-reported menstrual cycles between 21 and 42 days; and (v) an ability to communicate in English or Spanish (Buck Louis et al., 2014). Couples reporting prior infertility treatment were not eligible. Previous data has shown that environmental exposure levels among LIFE participants are comparable to those in the general population (Buck Louis et al., 2013). The current study includes 391 couples who had an aliquot of semen available for sperm purification and DNA isolation. Seven samples had limited sperm DNA and were excluded from analyses, which resulted in a final sample size of 384 couples. Study participants gave written informed consent before any data collection, and full institutional review board approval for human subjects was received from all collaborating institutions.
Data collection
Samples and data were collected in the homes of each participant as previously described (Buck Louis et al., 2014). Briefly, whole semen samples were collected in the home of each participant, including a baseline sample at entry into the study and a second sample 1 month later. Both samples were collected after a period of abstinence via masturbation with no lubricant use. Details regarding the sample collection procedures, shipping materials provided to participants and semen parameter quantification methods using computer-assisted semen analysis have been described in detail in prior analyses of the LIFE study (Buck Louis et al., 2014; Bloom et al., 2015). For the current analysis, only semen samples collected at baseline were evaluated. TTP was considered as the number of completed menstrual cycles before an hCG confirmed pregnancy (Buck Louis et al., 2011).
Sample preparation and DNA isolation
Semen samples were subjected to a one-step 40% gradient centrifugation to separate sperm from seminal plasma and somatic cells. Sperm DNA was isolated with our previously published method (Wu et al., 2015), which first homogenizes sperm with 0.2 mm steel beads in RLT buffer (Qiagen, Hilden, Germany) containing 50 mM of tris(2-carboxyethyl)phosphine (Pierce, Rockford, IL, USA) before total sperm DNA is extracted via silica-column purification.
mtDNA quantification
A probe-based triplex quantitative PCR assay was used to simultaneously quantify mtDNAc and mtDNAdel and a nuclear target based on a previously published method (Phillips et al., 2014; Huffman et al., 2018). The minor arc located in the D-Loop of mtDNA was targeted for mtDNAcn assessments because of its high stability and the rarity of deletions in this region. The major arc was targeted for mtDNAdel assessments because of the large prevalence of deletions common to this region. For genomic DNA reference, we used RNAse P (ThermoFisher, cat# 4403326), the standard reference assay for copy number analysis. Sperm mtDNAcn was calculated via the cycle threshold (Ct) ratio of minor arc and nuclear DNA copy number determined by RNAse P: 2(Ct:RNase P − Ct:MinorArc) and the % mtDNAdel was calculated using the Ct ratio of minor arc and major arc: mtDNAdel = 2(CtMinorArc − CtMajorArc) * 100. Reactions were performed in triplicate.
Statistical analysis
Participant characteristics were compared by quartile of mtDNAcn to describe participant characteristics and evaluate relations with mtDNAcn using ANOVA or χ2, as appropriate. Tests for trend were conducted using Spearman correlation analysis. MtDNAcn was right skewed and a log (base 10) transformation was used to meet assumptions for models of continuous mtDNAcn. MtDNAdel followed an approximately normal distribution and was not transformed.
We compared the probability of achieving pregnancy within 12 menstrual cycles between couples grouped by quartile of mtDNAcn, given that couples are classified as infertile after 12 months of unprotected sex. These probabilities were evaluated in unadjusted models using exact tests and the Cochran–Armitage test for trend for P-values.
Discrete-time proportional hazards models were used to evaluate the relation between mtDNA biomarkers and fecundability, as measured by TTP, allowing for covariate adjustment. Factors considered a priori as potential confounders included male and female age, male and female BMI, male race and ethnicity, male smoking status based on serum cotinine, study site of sample collection (i.e. Michigan or Texas), and qPCR batch. No variable was observed to represent a confounder based on bivariate analyses, but these factors were included as covariates in fully adjusted models. Active smoking was determined by a serum cotinine concentration of 10 ng/ml or greater (Bernert et al., 1997). Estimates from discrete time survival models are interpreted as fecundability odds ratios (FORs); estimates <1.0 indicate a lower probability of pregnancy and longer TTP, while FOR >1.0 indicate a higher probability of pregnancy or a shorter TTP. Models were run with mtDNA biomarkers (i.e. log copy number and untransformed deletions) in their continuous form as well as in quartiles to allow for non-linear relations with outcomes. Additionally, survival curves from Kaplan–Meier analysis were generated to display and compare pregnancy probabilities by cycle throughout follow-up between mtDNAcn quartiles.
Results
Men were, on average ± SD, 31.8 ± 4.8 years of age with a BMI of 29.9 ± 5.8. The majority of men were white (n = 311; 81.0%). Mean sperm mtDNAcn and mtDNAdel were 5.55 ± 5.3 and 0.31 ± 0.08, respectively. On average, female partners were 29.9 ± 4.1 years of age with a BMI of 27.5 ± 7.6. A total of 167 (43.5%) women reported never having been pregnant; 182 (47.4%) reported having 1+ live births. Comparisons of participant characteristics by quartiles of sperm mtDNAcn are shown in Table I. Sperm mtDNAcn and mtDNAdel displayed a strong positive association (Table I; P < 0.001). Sperm mtDNAcn was not associated with participant characteristics, otherwise.
Table I.
Characteristics of the study participants (n = 384) by quartile of mtDNAcn.
| Q1 | Q2 | Q3 | Q4 | |||
|---|---|---|---|---|---|---|
| Sperm mtDNAcn | (0.80–2.60) | (2.61–3.96) | (3.98–6.38) | (6.40–61.94) | ||
| Mean (SD) | P-value1 | P-trend2 | ||||
| Male age | 31.8 (4.1) | 31.6 (5.0) | 32.1 (4.7) | 31.4 (5.2) | 0.73 | 0.53 |
| Male BMIa | 30.2 (5.6) | 29.5 (4.9) | 29.7 (6.0) | 30.3 (6.5) | 0.80 | 0.98 |
| Sperm mtDNAdel | 0.27 (0.09) | 0.29 (0.07) | 0.33 (0.06) | 0.37 (0.06) | <0.001 | <0.001 |
| Female age | 29.6 (3.6) | 30.3 (4.5) | 30.0 (3.9) | 29.8 (4.3) | 0.92 | 0.80 |
| Female BMIb | 28.2 (8.5) | 26.4 (6.5) | 26.9 (7.2) | 28.4 (7.9) | 0.77 | 0.56 |
| n (%) | ||||||
| Male current smokingc | 19 (19.8) | 16 (16.7) | 26 (27.1) | 21 (21.9) | 0.35 | 0.14 |
| Male raced | 0.69 | 0.70 | ||||
| White | 80 (83.3) | 76 (79.2) | 75 (78.1) | 80 (83.3) | ||
| Non-white | 15 (15.6) | 19 (19.8) | 21 (21.9) | 16 (16.6) | ||
| Female paritye | 0.87 | 0.71 | ||||
| Never pregnant | 45 (46.9) | 37 (38.5) | 41 (42.7) | 44 (45.8) | ||
| Pregnant, no birth | 8 (8.3) | 7 (7.3) | 10 (10.4) | 9 (9.4) | ||
| Pregnant, live birth | 43 (44.8) | 51 (53.1) | 45 (46.9) | 43 (44.8) | ||
mtDNAcn, mitochondrial DNA copy number; mtDNAdel, mitochondrial DNA deletions.
P-value determined by ANOVA for continuous variables and χ2 for categorical variables.
P-trend determined by Spearman correlation analysis.
Missing n = 3.
Missing n = 1.
Smoking status determined by a serum cotinine >10 ng/ml; Missing n = 4.
Missing n = 2.
Missing n = 1.
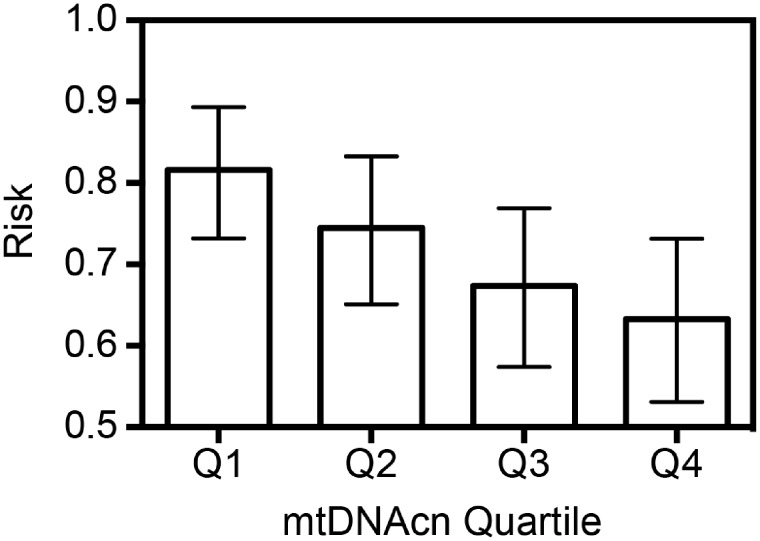
In unadjusted models, a strong inverse association between mtDNAcn and cumulative probability of achieving pregnancy within 12 months of trying was observed, whereby pregnancy probabilities decreased linearly with higher mtDNAcn (Fig. 1). Probabilities (95% CI) were: Q1 = 82.3% (95% CI: 73.2%, 89.3%); Q2 = 75.0% (95% CI: 65.1%, 83.3%); Q3 = 67.7% (95% CI: 57.4%, 76.9%); and Q4 = 63.5% (95% CI: 53.1%, 73.1%). This represents an 18.8% lower probability of pregnancy for couples with male partners in the highest quartile of mtDNAcn than for those in the lowest quartile (P = 0.003). Tests for trend strongly suggest a dose–response, with decreasing pregnancy probability with increasing mtDNAcn (P = 0.002).
Figure 1.
Observed proportions of participants having achieved pregnancy within 12 cycles of trying by quartile (Q) of mitochondrial DNA copy number (mtDNAcn) with 95% CIs. Pregnancy proportions shown above are: Q1 = 0.82 (95% CI: 0.73, 0.89); Q2 = 0.75 (95% CI: 0.65, 0.83); Q3 = 0.68 (95% CI: 0.57, 0.77); and Q4 = 0.63 (95% CI: 0.53, 0.73). Cochran–Armitage P-trend = 0.002.
Sperm mtDNAcn was negatively associated with TTP (FOR = 0.37, 95% CI: 0.22, 0.60; per log unit increase), indicating significantly lower fecundability (and, correspondingly, longer TTP) with higher sperm mtDNAcn (Table II). Similar results were observed when assessing mtDNAcn in quartiles. FOR estimates were: Q2 vs Q1, FOR = 0.78 (95% CI 0.52, 1.16); Q3 vs Q1, FOR = 0.65 (95% CI 0.44, 0.96); and Q4 vs Q1, FOR = 0.55 (95% CI 0.37, 0.81). Sperm mtDNAdel was not associated with fecundability. Adjustment for covariates was observed to have minimal to no impact on model estimates, providing indication of no substantial confounding.
Table II.
Fecundability odds ratios (FOR) estimated using Cox proportional hazards models relating sperm mtDNAcn and mtDNAdel to time-to-pregnancy.
| Biomarker | Unadjusted FOR (95% CI)b | Adjustedc FOR (95% CI) |
|---|---|---|
| mtDNAcna | ||
| Continuous | 0.36 (0.22, 0.58)*** | 0.37 (0.22, 0.60)*** |
| Quartiles | ||
| Q1 | Reference | Reference |
| Q2 | 0.77 (0.53, 1.10) | 0.78 (0.52, 1.16) |
| Q3 | 0.61 (0.42, 0.88)** | 0.65 (0.44, 0.96)* |
| Q4 | 0.53 (0.37, 0.77)*** | 0.55 (0.37, 0.81)** |
| mtDNAdel | ||
| Continuous | 0.25 (0.05, 1.21) | 0.46 (0.06, 3.48) |
| Quartiles | ||
| Q1 | Reference | Reference |
| Q2 | 0.85 (0.59, 1.22) | 0.87 (0.58, 1.31) |
| Q3 | 0.82 (0.56, 1.19) | 0.87 (0.56, 1.35) |
| Q4 | 0.74 (0.51, 1.07) | 0.89 (0.56, 1.40) |
Log10 transformed.
n = 384.
Adjusted for male and female age, male and female BMI, male serum cotinine, male race/ethnicity, study site of sample collection and qualitative PCR batch; n = 374.
P < 0.05;
P < 0.01;
P < 0.001.
Kaplan–Meier survival curves illustrating the adjusted cycle-specific probabilities of pregnancy by quartiles of mtDNAcn are shown in Fig. 2. Couples whose male partners were in Q1 of sperm mtDNAcn were observed to have a higher pregnancy probability as early as cycle two and throughout the study. A dose-dependent relation was observed for Q2, Q3 and Q4 of sperm mtDNAcn and pregnancy probabilities.
Figure 2.
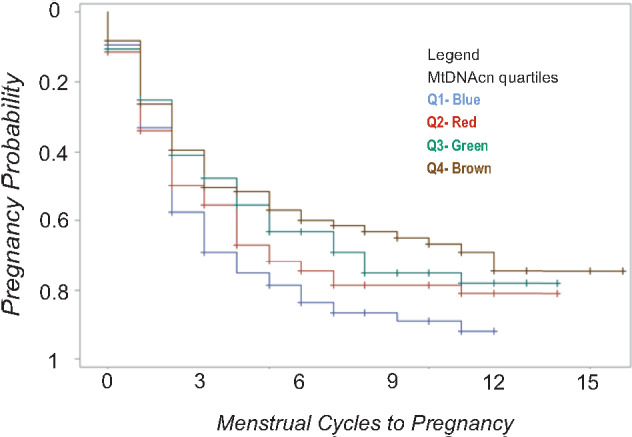
Survival curve indicating time (in menstrual cycles) to pregnancy by quartiles of sperm mtDNAcn. Respective fecundability odds ratios using Q1 as the referent are: Q2 = 0.78 (0.52, 1.16); Q3 = 0.65 (0.44, 0.96)*; Q4 = 0.55 (0.37, 0.81)**. Models adjusted for male and female age, male and female BMI, male serum cotinine, male race/ethnicity, site of sample collection and quantitative PCR batch. *P < 0.05; **P < 0.01.
Discussion
Higher sperm mtDNAcn was strongly associated with diminished fecundability among couples trying to become pregnant. Couples with male partners in the fourth quartile of sperm mtDNAcn had an FOR of 0.55 and 18.8% lower probability of pregnancy within 12 cycles in comparison to couples with male partners in the first quartile of sperm mtDNAcn. This relationship persisted in multivariable models adjusting for potential confounding factors. These findings among pregnancy planners in the general population who are not seeking clinical care are novel and highlight a potential promising biomarker for couple fecundity.
Clinical assessment of male infertility is largely based on WHO 2010 semen reference guidelines and is used to inform decision-making in the setting of ART. Nevertheless, research suggests semen analyses may be limited with regard to prognostic value of successful reproductive outcomes in couples (Esteves et al., 2012; Jungwirth et al., 2012). In a comparison of semen analysis results between fertile and infertile couples, no single semen parameter was observed to strongly predict fertility status (Guzick et al., 2001). Additionally, a 2019 meta-analysis reported a link between leukocytospermia and semen parameters, exhibiting significant decreases in progressive motility and sperm concentration; however, no significant differences in fertilization and pregnancy rates were observed (Castellini et al., 2020). In previous research from the LIFE study that evaluated 36 measures of semen quality parameters and TTP, model estimates were of low magnitude and only one parameter (percent coiled tail) was associated with TTP in multivariable models (Buck Louis et al., 2014). In the current analysis, we observed that sperm mtDNAcn was strongly associated with TTP and with probability of pregnancy within 12 cycles of trying and may serve as a single-measure parameter of sperm quality.
As the mitochondrial genome is maternally inherited in most species including humans, sperm mtDNAcn is depleted, an estimated 8- to 10 folds, during spermiogenesis (Hecht et al., 1984; St John et al., 2010). Thus, sperm mtDNAcn may represent a sensitive molecular marker of abnormal spermatogenesis and serve as an integrated measure of conventional individual semen parameters. Work to date is consistent with this notion; we have previously described that sperm mtDNAcn is correlated with multiple sperm parameters and sperm quality (Zhang et al., 2016; Wu et al., 2019a). Additionally, in an ART cohort of 119 couples, we have observed higher sperm mtDNAcn and mtDNAdel associated with lower rates of fertilization, even after adjustment for male age and semen parameter measures (Wu et al., 2019b). The nature of causal relations among semen parameters is uncertain, raising questions about the causal interpretation of estimates from statistical models adjusting for multiple semen parameters. Nevertheless, the above-described results (Guzick et al., 2001) demonstrate the incremental predictive ability of mtDNA biomarkers beyond semen characteristics alone. The use of a non-IVF cohort in this study suggests that our observations regarding sperm mtDNAcn in clinical infertility populations may have potential applicability to the general population. The biological basis for mtDNAcn as an integrated, global measure of semen quality is unclear. Previous research has speculated that the susceptibility of mtDNA to oxidative damage may provide a mechanistic link between sperm mtDNAcn and sperm quality, yet results have yet to be conclusive (Zhang et al., 2016). Environmental chemicals, among other exposures, can lead to elevated oxidative stress; oxidative stress-related effects on mtDNA may represent a mechanism through which chemical exposures can affect reproductive health. However, this hypothesis remains relatively unexplored. Although the LIFE cohort recruited couples with presumed environmental chemical exposure, we note that persistent chemical levels among participants in the LIFE study have been observed to be comparable to those in the general population (Buck Louis et al., 2013). Additional research is necessary to clarify the mechanism explaining the relation of sperm mtDNAcn with male fecundity.
Conclusion
There is a critical need for accurate measures of male fecundity for assessing overall reproductive health given the inherent limitations associated with conventional semen analysis. Research to date has largely been limited to clinical populations of patients seeking infertility treatment. Though somewhat modest in sample size, our study provides novel data regarding mtDNA biomarkers as predictors of reproductive potential beyond those seeking clinical treatment. Results of our study provide suggestion that sperm mtDNAcn may represent a more accurate, single-measure predictor of male fecundity in the general population. The identification of sperm mtDNAcn as a novel biomarker of male reproductive health and pregnancy success is of potential importance for both clinical care and general population reproductive health.
Authors’ roles
A.J.R. isolated sperm DNA, performed statistical analyses, interpreted results and drafted the manuscript. B.W.W. oversaw study design, statistical analyses, and interpreted results. N.B. generated the sperm mtDNA biomarker data. G.M.B.L., S.L.M. and E.F.S. oversaw study design and interpreted the results. J.R.P. oversaw study design, sperm mtDNA biomarker data and interpreted results.
Funding
This work was funded in part by the National Institute of Environmental Health Sciences, National Institutes of Health (R01-ES028298; PI: J.R.P.) and the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, Maryland (Contracts N01-HD-3-3355, N01-HD-3-3356 and N01-HD-3-3358).
Conflict of interest
The authors declare no competing interests.
Contributor Information
Allyson J Rosati, Department of Environmental Health Sciences, School of Public Health and Health Sciences, University of Massachusetts Amherst, Amherst, MA, USA.
Brian W Whitcomb, Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts Amherst, Amherst, MA, USA.
Nicole Brandon, Department of Environmental Health Sciences, School of Public Health and Health Sciences, University of Massachusetts Amherst, Amherst, MA, USA.
Germaine M Buck Louis, Dean's Office of the College of Health and Human Services, George Mason University, Fairfax, VA, USA.
Sunni L Mumford, Epidemiology Branch, Division of Intramural Population Health Research, Eunice Kennedy Shriver National Institute of Child Health & Human Development, Bethesda, MD, USA.
Enrique F Schisterman, Epidemiology Branch, Division of Intramural Population Health Research, Eunice Kennedy Shriver National Institute of Child Health & Human Development, Bethesda, MD, USA.
J Richard Pilsner, Department of Environmental Health Sciences, School of Public Health and Health Sciences, University of Massachusetts Amherst, Amherst, MA, USA.
References
- Al Rawi S, Louvet-Vallee S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science 2011;334:1144–1147. [DOI] [PubMed] [Google Scholar]
- Amaral A, Lourenco B, Marques M, Ramalho-Santos J. Mitochondria functionality and sperm quality. Reproduction (Cambridge, England) 2013;146:R163–R174. [DOI] [PubMed] [Google Scholar]
- Bernert JT Jr, Turner WE, Pirkle JL, Sosnoff CS, Akins JR, Waldrep MK, Ann Q, Covey TR, Whitfield WE, Gunter EW et al. Development and validation of sensitive method for determination of serum cotinine in smokers and nonsmokers by liquid chromatography/atmospheric pressure ionization tandem mass spectrometry. Clin Chem 1997;43:2281–2291. [PubMed] [Google Scholar]
- Bloom MS, Whitcomb BW, Chen Z, Ye A, Kannan K, Buck Louis GM. Associations between urinary phthalate concentrations and semen quality parameters in a general population. Hum Reprod 2015;30:2645–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck Louis GM, Schisterman EF, Sweeney AM, Wilcosky TC, Gore-Langton RE, Lynch CD, Boyd Barr D, Schrader SM, Kim S, Chen Z et al. Designing prospective cohort studies for assessing reproductive and developmental toxicity during sensitive windows of human reproduction and development—the LIFE study. Paediatr Perinat Epidemiol 2011;25:413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck Louis GM, Sundaram R, Schisterman EF, Sweeney AM, Lynch CD, Gore-Langton RE, Maisog J, Kim S, Chen Z, Barr DB. Persistent environmental pollutants and couple fecundity: the LIFE study. Environ Health Perspect 2013;121:231–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck Louis GM, Sundaram R, Schisterman EF, Sweeney AM, Lynch CD, Kim S, Maisog JM, Gore-Langton R, Eisenberg ML, Chen Z. Semen quality and time to pregnancy: the Longitudinal Investigation of Fertility and the Environment Study. Fertil Steril 2014;101:453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellini C, D'Andrea S, Martorella A, Minaldi E, Necozione S, Francavilla F, Francavilla S, Barbonetti A. Relationship between leukocytospermia, reproductive potential after assisted reproductive technology, and sperm parameters: a systematic review and meta-analysis of case–control studies. Andrology 2020;8:125–135. [DOI] [PubMed] [Google Scholar]
- Chen Y, Liao T, Zhu L, Lin X, Wu R, Jin L. Seminal plasma cell-free mitochondrial DNA copy number is associated with human semen quality. Eur J Obstet Gynecol Reprod Biol 2018;231:164–168. [DOI] [PubMed] [Google Scholar]
- Cooper TG, Noonan E, von Eckardstein S, Auger J, Baker HW, Behre HM, Haugen TB, Kruger T, Wang C, Mbizvo MT et al. World Health Organization reference values for human semen characteristics. Hum Reprod Update 2010;16:231–245. [DOI] [PubMed] [Google Scholar]
- Coskun PE, Wyrembak J, Derbereva O, Melkonian G, Doran E, Lott IT, Head E, Cotman CW, Wallace DC. Systemic mitochondrial dysfunction and the etiology of Alzheimer's disease and down syndrome dementia. J Alzheimers Dis 2010;20(Suppl 2):S293–S310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darr CR, Moraes LE, Connon RE, Love CC, Teague S, Varner DD, Meyers SA. The relationship between mitochondrial DNA copy number and stallion sperm function. Theriogenology 2017;94:94–99. [DOI] [PubMed] [Google Scholar]
- Esteves SC, Zini A, Aziz N, Alvarez JG, Sabanegh ES Jr, Agarwal A. Critical appraisal of World Health Organization's new reference values for human semen characteristics and effect on diagnosis and treatment of subfertile men. Urology 2012;79:16–22. [DOI] [PubMed] [Google Scholar]
- Greaves LC, Turnbull DM. Mitochondrial DNA mutations and ageing. Biochim Biophys Acta 2009;1790:1015–1020. [DOI] [PubMed] [Google Scholar]
- Guzick DS, Overstreet JW, Factor-Litvak P, Brazil CK, Nakajima ST, Coutifaris C, Carson SA, Cisneros P, Steinkampf MP, Hill JA et al. Sperm morphology, motility, and concentration in fertile and infertile men. N Engl J Med 2001;345:1388–1393. [DOI] [PubMed] [Google Scholar]
- Hecht NB, Liem H, Kleene KC, Distel RJ, Ho SM. Maternal inheritance of the mouse mitochondrial genome is not mediated by a loss or gross alteration of the paternal mitochondrial DNA or by methylation of the oocyte mitochondrial DNA. Dev Biol 1984;102:452–461. [DOI] [PubMed] [Google Scholar]
- Huffman AM, Wu H, Rosati A, Rahil T, Sites CK, Whitcomb BW, Richard Pilsner J. Associations of urinary phthalate metabolites and lipid peroxidation with sperm mitochondrial DNA copy number and deletions. Environ Res 2018;163:10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungwirth A, Giwercman A, Tournaye H, Diemer T, Kopa Z, Dohle G, Krausz C. European Association of Urology Guidelines on male fertility: the 2012 update. Eur Urol 2012;62:324–332. [DOI] [PubMed] [Google Scholar]
- Levine H, Jørgensen N, Martino-Andrade A, Mediola J, Weksler-Derri D, Mindlis I, Pinotta R, Swan S. Temporal trends in sperm count: a systematic review and meta-regression analysis. Hum Reprod Update 2017;23:646–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis JF, Thoma ME, Sorensen DN, McLain AC, King RB, Sundaram R, Keiding N, Buck Louis GM. The prevalence of couple infertility in the United States from a male perspective: evidence from a nationally representative sample. Andrology 2013;1:741–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips NR, Sprouse ML, Roby RK. Simultaneous quantification of mitochondrial DNA copy number and deletion ratio: a multiplex real-time PCR assay. Sci Rep 2014;4:3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song GJ, Lewis V. Mitochondrial DNA integrity and copy number in sperm from infertile men. Fertil Steril 2008;90:2238–2244. [DOI] [PubMed] [Google Scholar]
- St John JC, Facucho-Oliveira J, Jiang Y, Kelly R, Salah R. Mitochondrial DNA transmission, replication and inheritance: a journey from the gamete through the embryo and into offspring and embryonic stem cells. Hum Reprod Update 2010;16:488–509. [DOI] [PubMed] [Google Scholar]
- St John JC, Jokhi RP, Barratt CL. The impact of mitochondrial genetics on male infertility. Int J Androl 2005;28:65–73. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C, Schatten G. Ubiquitin tag for sperm mitochondria. Nature 1999;402:371–372. [DOI] [PubMed] [Google Scholar]
- Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet 2005;6:389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma ME, McLain AC, Louis JF, King RB, Trumble AC, Sundaram R, Buck Louis GM. Prevalence of infertility in the United States as estimated by the current duration approach and a traditional constructed approach. Fertil Steril 2013;99:1324–1331.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thonneau P, Marchand S, Tallec A, Ferial ML, Ducot B, Lansac J, Lopes P, Tabaste JM, Spira A. Incidence and main causes of infertility in a resident population (1,850,000) of three French regions (1988-1989). Hum Reprod 1991;6:811–816. [DOI] [PubMed] [Google Scholar]
- Wu H, de Gannes MK, Luchetti G, Pilsner JR. Rapid method for the isolation of mammalian sperm DNA. Biotechniques 2015;58:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Huffman AM, Whitcomb BW, Josyula S, Labrie S, Tougias E, Rahil T, Sites CK, Pilsner JR. Sperm mitochondrial DNA measures and semen parameters among men undergoing fertility treatment. Reprod Biomed Online 2019. a;38:66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Whitcomb BW, Huffman A, Brandon N, Labrie S, Tougias E, Lynch K, Rahil T, Sites CK, Pilsner JR. Associations of sperm mitochondrial DNA copy number and deletion rate with fertilization and embryo development in a clinical setting. Hum Reprod 2019. b;34:163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Wang Z, Ling X, Zou P, Yang H, Chen Q, Zhou N, Sun L, Gao J, Zhou Z et al. Mitochondrial biomarkers reflect semen quality: results from the MARCHS study in Chongqing, China. PLoS One 2016;11:e0168823. [DOI] [PMC free article] [PubMed] [Google Scholar]