Abstract
Cells assemble stress granules (SGs) to protect their RNAs from exposure to harmful chemical reactions induced by environmental stress. These SGs release RNAs, which resume translation once the stress is relieved. During stem cell differentiation, gene expression is altered to allow cells to adopt various functional and morphological features necessary to differentiate. This process induces stress within a cell, and cells that cannot overcome this stress die. Here, we investigated the role of SGs in the progression of stem cell differentiation. SGs aggregated during the neuronal differentiation of human bone marrow-mesenchymal stem cells, and not in cell lines that could not undergo differentiation. SGs were observed between one and three hours post-induction; RNA translation was restrained at the same time. Immediately after disassembly of SGs, the expression of the neuronal marker neurofilament-M (NF-M) gradually increased. Assembled SGs that persisted in cells were exposed to salubrinal, which inhibited the dephosphorylation of eukaryotic translation initiation factor 2 subunit 1 (eIF2α), and in eIF2α/S51D mutant cells. When eIF2α/S51A mutant cells differentiated, SGs were not assembled. In all experiments, the disruption of SGs was accompanied by delayed NF-M expression and the number of neuronally differentiated cells was decreased. Decreased differentiation was accompanied by decreased cell viability, indicating the necessity of SGs for preventing cell death during neuronal differentiation. Collectively, these results demonstrate the essential role of SGs during the neuronal differentiation of stem cells.
Keywords: eukaryotic translation initiation factor 2 alpha, gene expression, mesenchymal stem cells, neuronal differentiation, stem cells, stress granule
INTRODUCTION
Cells are often exposed to a variety of stressful conditions, including oxidative stress, energy depletion, and infection. The cells must respond appropriately to survive this period (Yamasaki and Anderson, 2008). Cells require an intrinsic response system to overcome these stresses. One cellular strategy is the formation of stress granules (SGs) to protect RNAs from various stresses, such as oxidative, endoplasmic reticulum (ER), and hyperosmotic, as well as ultraviolet damage (Aulas et al., 2017; Liu and Qian, 2014; Nover et al., 1989). During these stressful conditions, some messenger RNAs (mRNAs) that encode housekeeping proteins and other essential proteins required for cellular function experience delayed translation. This promotes the expression of mRNAs related to repair until the cell adapts to the environmental stress (Anderson and Kedersha, 2009; Yamasaki and Anderson, 2008). The mRNA whose translation has been inhibited, is not destroyed but rather, it is incorporated into the SG cytoplasmic foci, where it remains until the stress is controlled (Buchan and Parker, 2009; Gutierrez-Beltran et al., 2015; Nover et al., 1989). Therefore, SGs are believed to regulate cellular metabolism via modulating mRNA translation in stress-inducing environmental conditions.
SGs are ribonucleoprotein (RNP) complexes consisting of arrested translation initiation complexes (Anderson and Kedersha, 2008), RNA binding proteins (RBP), microRNAs, and various proteins involved in cell metabolism signaling (Anderson and Kedersha, 2009). The assembly of SGs results in the phosphorylation of the serine 51 (S51) residue of eukaryotic translation initiation factor 2 alpha (eIF2α) (Holcik and Sonenberg, 2005) by kinases that include general control nonderepressible 2 (GCN2), protein kinase R (PKR), heme-regulated inhibitor kinase (HRI), and protein kinase R-like ER-localized eIF2a kinase (PERK), which are activated by various environmental stresses (Aulas et al., 2017; Holcik and Sonenberg, 2005). Phosphorylated eIF2α disrupts the eIF2-GTP-tRNAiMet complex, which results in the accumulation of translationally stalled messenger ribonucleoproteins (mRNPs). mRNPs contain T-cell intracellular antigen 1 (TIA-1), TIA-1 related protein (TIAR), Ras-GAP SH3 domain binding protein (G3BP), and U6 snRNA-associated Sm-like protein (Lsm4). mRNPs contribute to the assembly of SGs (Anderson and Kedersha, 2006; Gilks et al., 2004; Kedersha et al., 2005; Yamasaki and Anderson, 2008). Previous studies have shown that the eIF2α mutants, in which the S51 residue is replaced with an alanine (S51A; which cannot be phosphorylated) interrupts the assembly of SGs and inhibits translation (Gerlitz et al., 2002; Sudhakar et al., 2000). Another mutant (S51D; phosphomimetic eIF2α) retains assembled SGs (Costache et al., 2012). Thus, the phosphorylation of eIF2α is critical to induce the assembly of SGs and repress translation.
In the stem cell differentiation process, overall gene expression patterns are converted into tissue-specific gene profiles (Black, 2003; Zimmer et al., 2011). This gene expression transition involves a variety of factors, such as transcription factors that regulate tissue-specific gene expression, RBPs (Linares et al., 2015), RNA granules, and non-coding RNAs (Sim et al., 2014), which are responsible for post-transcriptional regulation. Thus, changes in gene expression may depend on the level of stress involved in differentiating stem cells, which can also result in cell death.
Stem cells have the potential to differentiate into various cell types (Caplan, 2007; Jeong et al., 2013). In this study, we investigated the factors that regulate gene expression during stem cell differentiation. The results revealed that SGs are the factors that regulate neuronal differentiation.
MATERIALS AND METHODS
Cell culture
Human bone marrow-mesenchymal stem cells (hBM-MSCs) were purchased from Cell Engineering For Origin (CEFO) (Korea) and cultured in T75 flasks (Falcon; Corning, USA) according to the supplier’s recommendations. hBM-MSCs were cultivated in mesenchymal stem cell growth medium (Gibco, Grand Island, NY, USA) as previously described (Jeong and Cho, 2015). Passage seven (P-7) hBM-MSCs were used. Rat fetal neural stem cells (NSCs) were purchased from Gibco (Grand Island, NY, USA) and cultured in T75 flasks (Falcon) according to the supplier's recommendations. NSCs were cultivated in growth medium containing 2% StemPro® NSC supplement (Gibco, Grand Island, NY, USA), 2 mM GlutaMaxTM I supplement (Gibco, Grand Island, NY, USA), 20 ng/ml basic fibroblast growth factor (bFGF) (Gibco, Carlsbad, CA, USA), and 20 ng/ml epidermal growth factor (Gibco, Carlsbad, CA, USA) in KnockOutTM Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Grand Island, NY, USA). P-3 NSCs were used for the experiments. Human osteosarcoma cells (U2OS) were cultured in DMEM/F12 (Gibco, Grand Island, NY, USA) with 10% fetal bovine serum (FBS) (Origin Canada; Gibco, Grand Island, NY, USA), 2 mM L-glutamine, and 1% penicillin/streptomycin antibiotics at 37°C in a 5% CO2 incubator.
Plasmids and transient transfection
hBM-MSCs were transfected with pCMV-Wild/eIF2α, pCMV-eIF2α/S51A, or pCMV-eIF2α/S51D plasmid in Lipofectamine transfection reagent (Thermo Fisher Scientific, Carlsbad, CA, USA) containing Opti-MEM medium at a final concentration of 500 ng in a 24-well plate or 100 ng in a 96-well plate. At 24 h post-transfection, media was changed and the cells were further incubated for 12 h.
Neuronal differentiation
Neuronal differentiation of hBM-MSCs was performed as previously described (Jeong et al., 2013; Woodbury et al., 2000). Briefly, hBM-MSCs were incubated in pre-induction medium (10% FBS, 10 ng/ml bFGF, and 500 µM β-mercaptoethanol) for 24 h, and further incubated with induction medium (100 µM butylated hydroxyanisole, and 2% dimethylsulfoxide [DMSO]) for 1 to 24 h. Cells were imaged with an ECLIPSE TS100 light microscope (Nikon, Japan) and an IMTcam3 digital camera (i-Solution, Japan). Cells were considered neuronally differentiated when each cell body had more than two dendrites longer than 60 µm (Jeong et al., 2013). NSCs were differentiated into neuronal cells with differentiation induction medium (2% StemPro® NSC supplement, 2 mM GlutaMaxTM-I supplement in KnockOutTM DMEM) for 7 days.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
hBM-MSCs (8 × 103) were seeded in 96-well plates. The next day, hBM-MSCs were treated with 10 µM salubrinal for 24 h or transfected with eIF2α/S51A, and, then, differentiated into neurons. Cell viability was determined by MTT assay (Sigma-Aldrich, USA), according to the manufacturer’s instructions.
Nascent protein synthesis
Newly synthesized proteins were measured using the Click-iT AHA protein kit (Invitrogen, USA) according to the manufacturer’s instructions. hBM-MSCs were incubated with methionine-free medium for 1 h at 37°C to deplete methionine reserves. For metabolic labeling, cells were further incubated with 25 µM Click-iT AHA reagent (l-azidohomoalanine) for 4 h at 37°C. Cells were harvested and total protein was extracted using the lysis buffer. The prepared total proteins were subjected to electrophoresis and blotting. Finally, Azide-modified proteins were visualized using a biotin-azide and streptavidin conjugate.
Immunocytochemical staining
hBM-MSCs were grown on poly L-lysine-coated coverslips (Thermo Fisher Scientific, St. Louis, MO, USA). The cells were treated with 10 µM salubrinal for 24 h or transfected with eIF2α cDNA (wild type, mutants S51A or S51D), and induced to differentiate into neuron-like cells. Cells were then subjected to immunocytochemical staining with primary antibodies against G3BP, TIA-1, or NF-M (Santa Cruz Biotechnology, USA) diluted 1:200 in blocking buffer overnight at 4°C, and secondary antibodies Alexa 488-conjugated donkey anti-mouse IgG or donkey anti-goat IgG antibody (Molecular Probes, USA) diluted 1:500 in Hoechst 33342 (Molecular Probes) for 1 h at room temperature. Cells were washed with phosphate-buffered saline and mounted with a drop of mounting solution (ProLong Gold antifade reagent; Molecular Probes). The cells were visualized with fluorescent microscopy using an Eclipse 80Ti microscope (Nikon). Cell images were taken with a model DS-RI1 digital camera (Nikon).
Immunoblot analysis
hBM-MSCs were treated with 10 µM salubrinal for 24 h or transfected with mutant eIF2α cDNA (S51A) and induced to differentiate into neuron-like cells. Total protein was extracted with 400 µl RIPA buffer containing protease and dephosphatase inhibitors (Santa Cruz Biotechnology). Proteins were quantified using Pierce BCA Protein Assay kit (Thermo Fisher Scientific, Rockford, IL, USA). SDS loading buffer was added and boiled for 5 min. Later the proteins were analyzed by SDS-PAGE. After electrophoresis proteins were transferred onto a PVDF membrane (GE Healthcare, Germany). Membranes were then blocked with 5% Normal horse serum in Tris-buffer saline and Tween-20. The membrane was then incubated in 4°C for overnight with primary antibodies against eIF2α (1:1,000 dilution; Santa Cruz Biotechnology), phosphorylated-eIF2α (p-eIF2α; 1:500 dilution; Santa Cruz Biotechnology), ATF-4 (1:500 dilution; Cell Signaling Technology, USA), NF-M (1:1,000 dilution; Santa Cruz Biotechnology), Musashi (1:1,000 dilution; Santa Cruz Biotechnology), or β-actin (1:1,000 dilution; Sigma-Aldrich), followed by the appropriate horseradish peroxidase-conjugated secondary antibody (1:20,000; Jackson Immuno Research Laboratories, USA). Blots were then developed using Amersham ECL Western Blotting Detection Reagents (GE Healthcare, UK) according to manufacturer’s protocol.
RESULTS
SGs assemble during neuronal differentiation
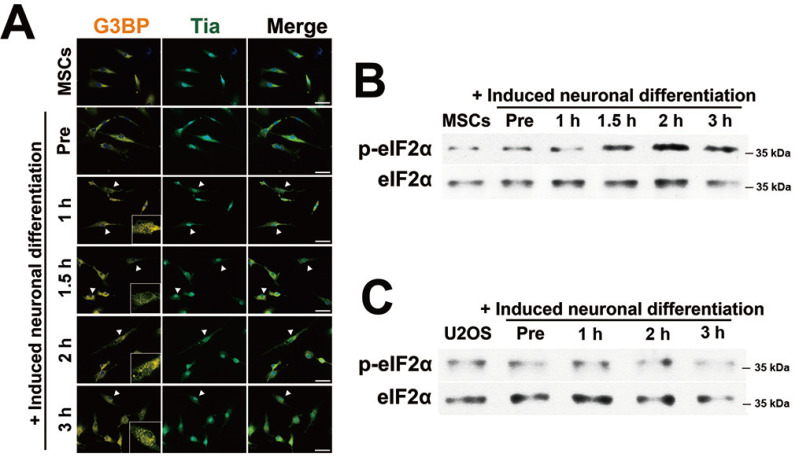
To examine whether SGs assembled during the neuronal differentiation of stem cells, SG markers G3BP and TIA-1 were examined by immunocytochemical staining. SGs assembled between 1 h and 3 h after incubation with neuronal induction medium (Fig. 1A). The levels of phosphorylated eIF2α (p-eIF2α; translation initiation factor 2α) were also markedly increased in the immunoblot analysis, which was consistent with Fig. 1A (Fig. 1B). Similarly, human U2OS osteosarcoma cells were incubated in the same medium and under the same conditions. Unlike in hBM-MSCs, SGs were not observed in U2OS cells (Supplementary Fig. S1) and the expression of p-eIF2α was not changed (Fig. 1C). These results indicated that SG assembly occurs only during the neuronal differentiation of stem cells.
Fig. 1. SGs aggregation during neuronal differentiation.
(A) Human bone marrow-mesenchymal stem cells (hBM-MSCs) were incubated with pre-induction medium for one day and exposed to neuronal induction medium for 0, 1, 1.5, 2, and 3 h prior to fixation and immunocytochemical staining for G3BP (orange) and TIA-1 (green). SG-positive cells are indicated by arrowheads. Scale bars = 50 μm. (B) The protein expression of eIF2α and p-eIF2α was measured by immunoblot analysis after hBM-MSCs were incubated with neuronal induction medium for the indicated time. (C) U2OS cells were incubated with differentiation medium as shown in panel A. Total protein from induced U2OS cells was measured by immunoblot analysis using antibodies for eIF2α and p-eIF2α. “Pre,” indicates hBM-MSCs incubated with pre-induction medium for one day.
SGs are associated with neuronal differentiation
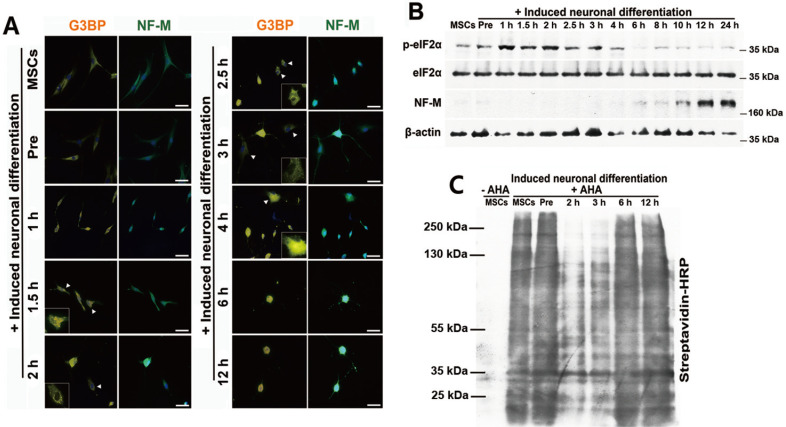
To examine the role of SG assembly during the differentiation process, neuronally differentiating cells were stained using antibodies against G3BP and NF-M, which is a neuronal specific marker. SGs were observed after 1 h incubation with neuronal induction medium, while the expression of NF-M gradually increased in the absence of SGs and reached a maximal level after 12 h of incubation (Fig. 2A). Immunoblot analysis with elF2α and NF-M demonstrated that p-eIF2α levels were significantly increased within the first 4 h of neuronal differentiation, then gradually decreased, whereas NF-M expression began to increase after 6 h (Fig. 2B).
Fig. 2. SGs are associated with neuronal differentiation of stem cells.
(A) hBM-MSCs were treated with neuronal induction medium for 0, 1, 1.5, 2, 2.5, 3, 4, 6, and 12 h. Cells were fixed and immunostained for G3BP (orange) and NF-M (green). Arrowheads indicate SG-positive cells. Scale bars = 50 μm. (B) hBM-MSCs were induced to differentiate into neurons. Total proteins were extracted from the differentiating cells at the indicated times and p-eIF2α and NF-M expression levels were measured by immunoblot analysis. (C) The cells were labeled with 25 μM AHA for 4 h and harvested at the indicated times. Newly synthesized proteins were measured by immunoblot analysis for biotin-azide and streptavidin conjugation. “–AHA” indicates not labeled with AHA.
SG assembly and p-eIF2α expression can lead to the repression of translation (Yamasaki and Anderson, 2008). Since SGs and p-eIF2α were observed during the differentiation process, global translation was examined using the Click-iT AHA detection method. Translation did not change during pre-induction, whereas it was markedly reduced after 2 to 3 h of incubation with induction medium (Fig. 2C). The findings were consistent with the timeline of the appearance of SGs, indicating that SGs may modulate the translation of mRNA involved in neuronal differentiation and influence the expression of neuronal specific markers.
Disruption of SG formation impedes neuronal differentiation
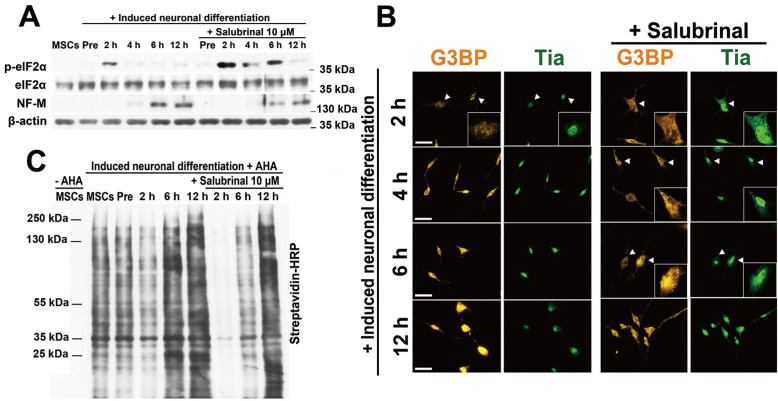
To examine the role of SGs in the differentiation process, 10 μM salubrinal was administered to block SG disaggregation by inhibiting dephosphorylation of eIF2α. The levels of phosphorylated eIF2α were increased 2 h after neuronal induction. The increase was sustained for up to 6 h in salubrinal-treated cells (Fig. 3A). The expression of NF-M was effectively impeded 6 h after neuronal induction by blocking the dephosphorylation of eIF2α (Fig. 3A). When cells were stained with G3BP and TIA-1, SGs appeared at 2 h and remained for up to 6 h of neuronal induction in salubrinal-treated cells (Fig. 3B). Translation in salubrinal treated cells was also markedly reduced after 2 h and up to 6 h of incubation with induction medium, consistent with Figs. 3A and 3B (Fig. 3C).
Fig. 3. Disrupting SG assembly using salubrinal impedes neuronal differentiation.
(A) hBM-MSCs treated with/without salubrinal were induced to differentiate into neurons for the indicated times, and total protein was extracted. p-eIF2α and NF-M expression levels were measured with their specific antibodies. (B) Differentiating cells were fixed and stained with G3BP (orange) and TIA-1 (green) antibodies. SG-positive cells are indicated by an arrowheads. Scale bars = 50 μm. (C) Nascent proteins were measured using the AHA labeling in differentiating hBM-MSCs treated with/without salubrinal.
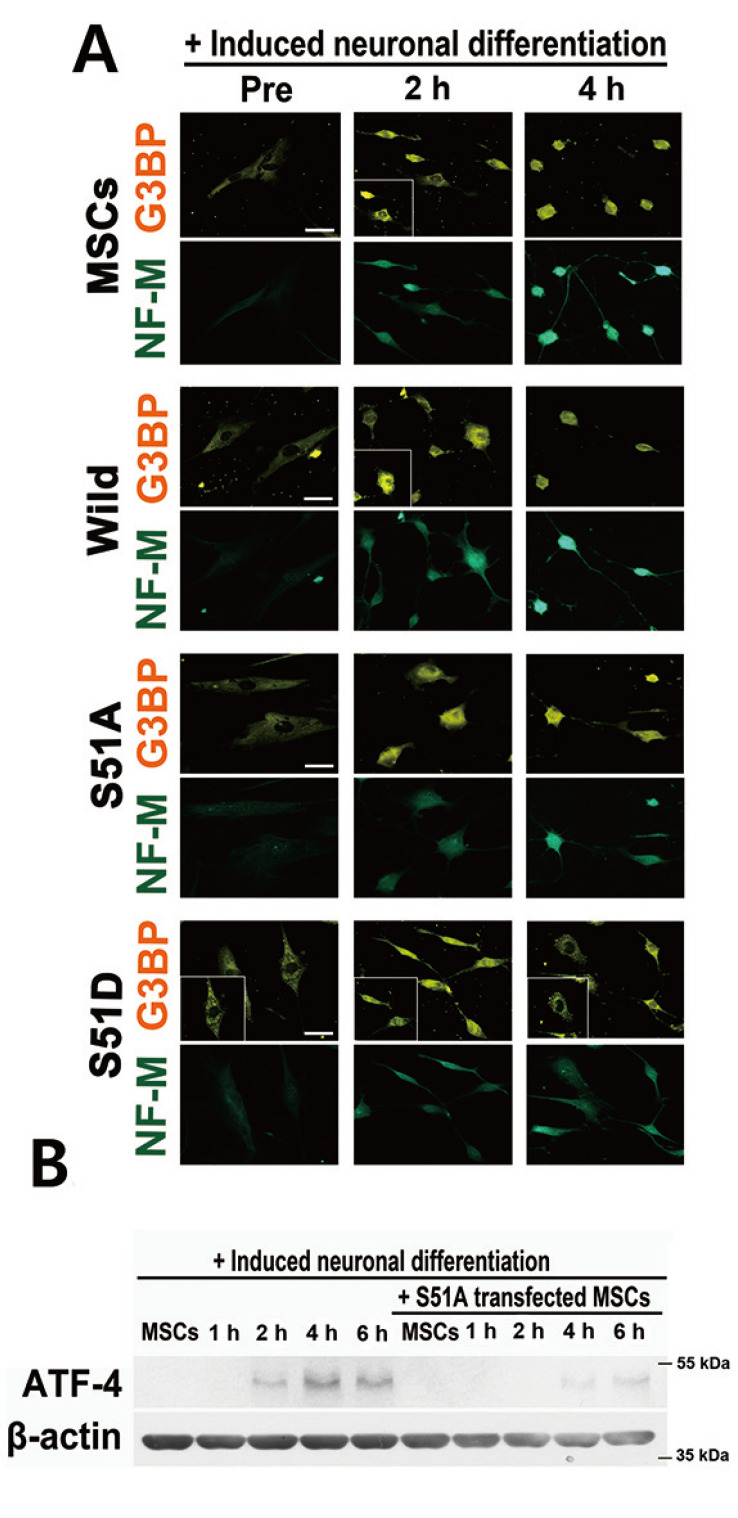
The dominant negative form (eIF2α/S51A) and constitutively active form (eIF2α/S51D) of eIF2α cDNA were used to further confirm the functional role of SGs during neuronal differentiation. hBM-MSCs were transfected with wild type or mutant eIF2α and neuronally differentiated. SGs appeared 2 h after neuronal induction and NF-M was present at 4 h in non-transfected and wild type eIF2α-transfected cells. In S51A-transfected cells, SGs did not form during neuronal induction (Fig. 4A). SGs were present in S51D-transfected cells at all tested time points. Both eIF2α mutants delayed the expression of neuronal markers (Fig. 4A). Several studies have reported that the unfolded protein response is induced during the neuronal differentiation process (Cho et al., 2009; Zhang et al., 2019). Moreover, ATF-4 expression is induced and promotes osteoclastic differentiation with bone marrow MSCs (Yu et al., 2013). Hence, we tested whether ATF-4 is induced during neuronal differentiation under our induction conditions. ATF-4 levels were significantly increased during the neuronal differentiation process but were markedly reduced in S51A-transfected cells (Fig. 4B). These results suggested that the disruption of differentiation signal mediated eIF2α phosphorylation and SG assembly dynamics might inhibit the effective timely expression of neural-specific proteins and interfere with the differentiation process.
Fig. 4. Disrupting SGs with mutant eIF2α cDNA interrupts neuronal differentiation.
(A) hBM-MSCs were transfected with mutant eIF2α DNA (S51A and S51D) and incubated with neuronal induction media for the indicated times. Cells were immunostained for G3BP (orange) and NF-M (green). Scale bars = 50 μm. (B) S51A-transfected hBM-MSCs were incubated with induction media for the indicated times and ATF-4 protein was measured using specific antibody.
Cells with disrupted SG formation fail to differentiate and undergo cell death
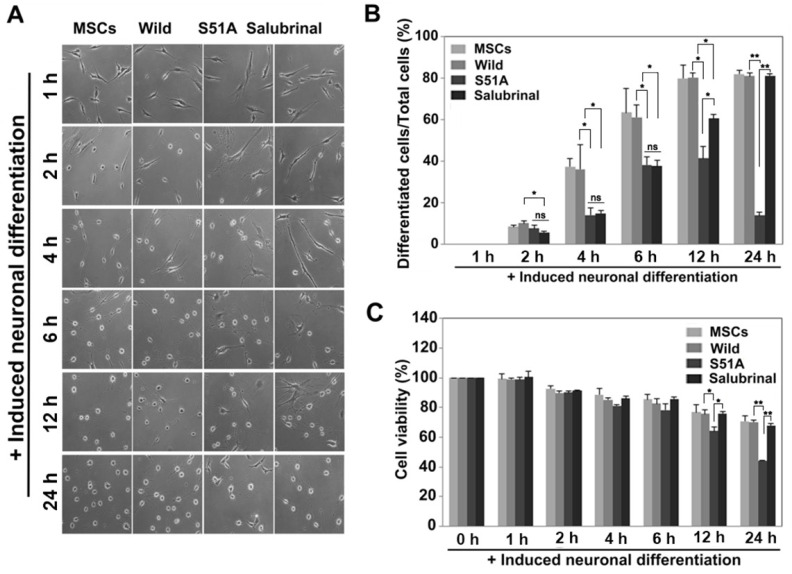
If the formation of SGs is crucial for neuronal differentiation, then the rate of differentiation should decrease upon disrupting SG formation. To assess this idea, hBM-MSCs were transfected with wild type or mutant eIF2α/S51A and treated with salubrinal. They were then induced to differentiate into neuronal induction medium for up to 24 h (Fig. 5A). Neuronally differentiated cells were counted (Fig. 5B). The rate of differentiation gradually increased in cells which are non-treated with salubrinal and wild type eIF2α-transfected cells, and significantly decreased in S51A cells. The inhibition of neuronal differentiation in salubrinal-treated cells was effective until 6 h of treatment. The effectiveness gradually decreased thereafter (Fig. 5B).
Fig. 5. Cells that fail to form SGs die at the late stage of neuronal differentiation.
(A) hBM-MSCs, hBM-MSC which are transfected with either wild type or mutant eIF2α and salubrinal treated hBM-MSCs were differentiated into neurons. Cells were observed at the indicated times. Scale bars = 100 μm. (B) Neuronally differentiated cells were counted (n = 4, *P < 0.05, **P < 0.005, Student’s t-test; ns, not significant). (C) Treated cells were incubated with induction media for the indicated times and the cell viability was measured (n = 3, *P < 0.05, **P < 0.005, Student’s t-test).
Cell viability was examined to determine the fate of the cells with disrupted SG formation. The overall survival rates gradually decreased during the differentiation process, which is a normal phenomenon. The mutant eIF2α/S51A-transfected cells showed significantly reduced viability compared to wild type cells, while salubrinal-treated cells showed delayed differentiation with only marginal effect on cell death (Fig. 5C). These data suggested that cells with disrupted SG formation fail to differentiate and undergo cell death, whereas SG formation prevents cell death and plays an important role in neuronal differentiation.
SGs are associated with NSC differentiation
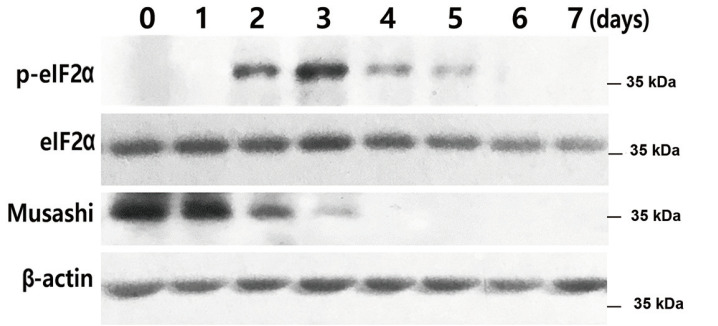
Since all the experiments were performed for the neural differentiation of hBM-MSCs, we examined whether the effects of SGs were associated during the neuronal differentiation of NSCs. Immunoblot analysis with elF2α and Musashi as stem cell markers demonstrated that p-eIF2α levels increased at 2 to 3 days of neuronal differentiation, then gradually decreased. In contrast, Musashi expression was high at the beginning, then gradually decreased until no expression was detected after 3 days (Fig. 6). The findings suggested that SG formation also plays a role in NSC differentiation.
Fig. 6. SGs are associated with NSC differentiation.
NSCs were treated with neuronal induction media for 7 days and total proteins were extracted at the indicated times (0, 1, 2, 3, 4, 5, 6, and 7 days). eIF2a, p-eIF2α, and Musashi expression levels were measured by immunoblot analysis.
DISCUSSION
When cells are exposed to stress, SGs composed of several proteins and RNA form to protect RNAs from harmful chemical reactions (Gutierrez-Beltran et al., 2015; Nover et al., 1989). Once the stress is removed or is under control, SGs release the RNAs, which resume protein synthesis necessary for vital activities (Unsworth et al., 2010; Yague and Raguz, 2010). When stem cells are induced to differentiate, the differentiation process exposes the cells to stress, especially ER stress (Cho et al., 2009). Once cells overcome this stress, they alter their identity to become the differentiated cell type. In this study, we investigated whether SGs play an important role in cell differentiation.
First, the aggregation of SGs was observed every hour during the neuronal differentiation process in hBM-MSCs. SGs were observed 1 h after neural differentiation, were dramatically increased at 2 h, and gradually decreased after 3 h (Figs. 1A and 1B). SGs were not observed in U2OS human osteosarcoma cells under the same conditions (Supplementary Fig. S1, Fig. 1C). The observation of SGs confirmed that this phenomenon occurs only in stem cells that have the potential to differentiate.
Next, to understand the role of SGs in the neuronal differentiation process, cells were probed with the neuronal protein marker NF-M and the SG protein marker G3BP and examined at various time points (Fig. 2A). SGs were observed between 1 and 3 h after induction of differentiation and NF-M expression was observed to gradually increase (Figs. 2A and 2B). In addition, SG formation led to the inhibition of protein translation, which then resumed once the SGs expression started to disappear (Fig. 2B). These observations implied that the formation of SGs takes place during the differentiation process. In fact, it was confirmed that NF-M expression increased as did the rate of neuronally differentiated cells immediately after SG disassembly (Figs. 5A and 5B).
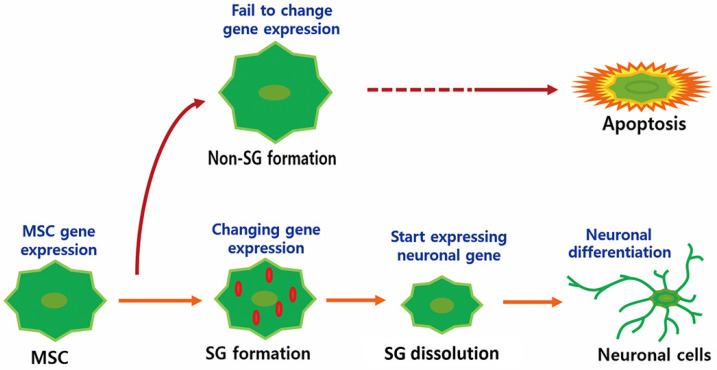
In the experiment with salubrinal-treated cells, SG formation was maintained for up to 6 h following the induction of differentiation, resulting in the delayed expression of NF-M (Figs. 3A and 3B) and decreased neuronal differentiation (Figs. 5A and 5B). In the eIF2α transfection experiment, SG formation was inhibited in eIF2α/S51A cells, whereas SGs were maintained at all observed time points in eIF2α/S51D cells (Fig. 4). In both cases, the expression of NF-M was delayed (Fig. 4) and the rate of neuronal differentiation was decreased (Figs. 5A and 5B). The perturbation of neuronal differentiation by overexpression of eIF2a mutants or by salubrinal treatment possibly reflected the dysregulation of ATF-4 expression, which depends on eIF2a phosphorylation status induced by unfolded protein stress (Cho et al., 2009; Zhang et al., 2019). Interestingly, the inhibition of SG formation in eIF2α/S51A cells resulted in a significant increase in cell death and a decrease in neuronal differentiation (Fig. 5C). Therefore, SG formation is important for the cellular transition during neuronal differentiation. Blocking the formation of SGs limits neural differentiation and leads to apoptosis (Fig. 7).
Fig. 7. Schematic diagram of the neuronal differentiation process.
The formation of SGs is a mechanism adopted by cells as a way to protect themselves against stress (Aulas et al., 2017; Liu and Qian, 2014). Once the stress is removed, SGs disappear and cells switch to routine cellular processes, such as cell division and energy metabolism. The induction of differentiation in stem cells can be a cause of stress in cells. Once stress is overcome, cells can adopt different capabilities. However, cells that fail to adapt to the stress may undergo cell death. We investigated the role of SGs during neuronal differentiation in stem cells. The findings support the hypothesis that SGs are important in the stem cell differentiation process. There are two possibilities for the effect of SG on stem cell differentiation. First, global gene expression profiles need to be changed for stem cell differentiation, which causes unfolded protein stress because of heavy loading with protein synthesis. SGs might act as a center for cells to achieve optimal gene expression to promote stem cell differentiation. Second, SGs might also act as a survival mediator that inhibits cell death during the differentiation process. Furthermore, we confirmed this effect in NSCs (Fig. 6). However, the formation of SGs was only observed in vitro cultured stem cells. Further studies using animal models are needed to confirm the implications of this research and to determine whether these events may be associated with neurological diseases.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This work was supported by a grant from the Basic Science Research Program of the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2017R1D1A1B03034527 and NRF-2019R1C1C1011040).
Footnotes
AUTHOR CONTRIBUTIONS
S.G.J., C.H.J., and G.W.C. designed the study. S.G.J. performed the research. C.H.J., T.O., and G.W.C. analyzed the data. S.G.J., C.H.J., K.V., and G.W.C. wrote the manuscript. C.H.J. and G.W.C. supervised the project.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Anderson P., Kedersha N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- Aulas A., Fay M.M., Lyons S.M., Achorn C.A., Kedersha N., Anderson P., Ivanov P. Stress-specific differences in assembly and composition of stress granules and related foci. J. Cell Sci. 2017;130:927–937. doi: 10.1242/jcs.199240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Buchan J.R., Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan A.I. Adult mesenchymal stem cells for tissue engineering versus regenerative medicine. J. Cell. Physiol. 2007;213:341–347. doi: 10.1002/jcp.21200. [DOI] [PubMed] [Google Scholar]
- Cho Y.M., Jang Y.S., Jang Y.M., Chung S.M., Kim H.S., Lee J.H., Jeong S.W., Kim I.K., Kim J.J., Kim K.S., et al. Induction of unfolded protein response during neuronal induction of rat bone marrow stromal cells and mouse embryonic stem cells. Exp. Mol. Med. 2009;41:440–452. doi: 10.3858/emm.2009.41.6.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costache V., Bilotto S., Laguerre L., Belle R., Cosson B., Cormier P., Morales J. Dephosphorylation of eIF2alpha is essential for protein synthesis increase and cell cycle progression after sea urchin fertilization. Dev. Biol. 2012;365:303–309. doi: 10.1016/j.ydbio.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Gerlitz G., Jagus R., Elroy-Stein O. Phosphorylation of initiation factor-2 alpha is required for activation of internal translation initiation during cell differentiation. Eur. J. Biochem. 2002;269:2810–2819. doi: 10.1046/j.1432-1033.2002.02974.x. [DOI] [PubMed] [Google Scholar]
- Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L.M., Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 2004;15:5383–5398. doi: 10.1091/mbc.e04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Beltran E., Moschou P.N., Smertenko A.P., Bozhkov P.V. Tudor staphylococcal nuclease links formation of stress granules and processing bodies with mRNA catabolism in Arabidopsis. Plant Cell. 2015;27:926–943. doi: 10.1105/tpc.114.134494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M., Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- Jeong S.G., Cho G.W. Endogenous ROS levels are increased in replicative senescence in human bone marrow mesenchymal stromal cells. Biochem. Biophys. Res. Commun. 2015;460:971–976. doi: 10.1016/j.bbrc.2015.03.136. [DOI] [PubMed] [Google Scholar]
- Jeong S.G., Ohn T., Kim S.H., Cho G.W. Valproic acid promotes neuronal differentiation by induction of neuroprogenitors in human bone-marrow mesenchymal stromal cells. Neurosci. Lett. 2013;554:22–27. doi: 10.1016/j.neulet.2013.08.059. [DOI] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares A.J., Lin C.H., Damianov A., Adams K.L., Novitch B.G., Black D.L. The splicing regulator PTBP1 controls the activity of the transcription factor Pbx1 during neuronal differentiation. Elife. 2015;4:e09268. doi: 10.7554/eLife.09268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Qian S.B. Translational reprogramming in cellular stress response. Wiley Interdiscip. Rev. RNA. 2014;5:301–315. doi: 10.1002/wrna.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L., Scharf K.D., Neumann D. Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol. Cell. Biol. 1989;9:1298–1308. doi: 10.1128/mcb.9.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S.E., Bakes J., Kaang B.K. Neuronal activity-dependent regulation of MicroRNAs. Mol. Cells. 2014;37:511–517. doi: 10.14348/molcells.2014.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhakar A., Ramachandran A., Ghosh S., Hasnain S.E., Kaufman R.J., Ramaiah K.V. Phosphorylation of serine 51 in initiation factor 2 alpha (eIF2 alpha) promotes complex formation between eIF2 alpha(P) and eIF2B and causes inhibition in the guanine nucleotide exchange activity of eIF2B. Biochemistry. 2000;39:12929–12938. doi: 10.1021/bi0008682. [DOI] [PubMed] [Google Scholar]
- Unsworth H., Raguz S., Edwards H.J., Higgins C.F., Yague E. mRNA escape from stress granule sequestration is dictated by localization to the endoplasmic reticulum. FASEB J. 2010;24:3370–3380. doi: 10.1096/fj.09-151142. [DOI] [PubMed] [Google Scholar]
- Woodbury D., Schwarz E.J., Prockop D.J., Black I.B. Adult rat and human bone marrow stromal cells differentiate into neurons. J. Neurosci. Res. 2000;61:364–370. doi: 10.1523/JNEUROSCI.5060-03.2004. [DOI] [PubMed] [Google Scholar]
- Yague E., Raguz S. Escape from stress granule sequestration: another way to drug resistance? Biochem. Soc. Trans. 2010;38:1537–1542. doi: 10.1042/BST0381537. [DOI] [PubMed] [Google Scholar]
- Yamasaki S., Anderson P. Reprogramming mRNA translation during stress. Curr. Opin. Cell Biol. 2008;20:222–226. doi: 10.1016/j.ceb.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Zhu K., Lai Y., Zhao Z., Fan J., Im H.J., Chen D., Xiao G. atf4 promotes beta-catenin expression and osteoblastic differentiation of bone marrow mesenchymal stem cells. Int. J. Biol. Sci. 2013;9:256–266. doi: 10.7150/ijbs.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Wang M., Li Y., Li C., Tang S., Qu X., Feng N., Wu Y. The PERK-EIF2alpha-ATF4 signaling branch regulates osteoblast differentiation and proliferation by PTH. Am. J. Physiol. Endocrinol. Metab. 2019;316:E590–E604. doi: 10.1152/ajpendo.00371.2018. [DOI] [PubMed] [Google Scholar]
- Zimmer B., Kuegler P.B., Baudis B., Genewsky A., Tanavde V., Koh W., Tan B., Waldmann T., Kadereit S., Leist M. Coordinated waves of gene expression during neuronal differentiation of embryonic stem cells as basis for novel approaches to developmental neurotoxicity testing. Cell Death Differ. 2011;18:383–395. doi: 10.1038/cdd.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.