Abstract
Histone acetylation and deacetylation play central roles in the regulation of chromatin structure and transcription by RNA polymerase II (RNA Pol II). Although Hda1 histone deacetylase complex (Hda1C) is known to selectively deacetylate histone H3 and H2B to repress transcription, previous studies have suggested its potential roles in histone H4 deacetylation. Recently, we have shown that Hda1C has two distinct functions in histone deacetylation and transcription. Histone H4-specific deacetylation at highly transcribed genes negatively regulates RNA Pol II elongation and H3 deacetylation at inactive genes fine-tunes the kinetics of gene induction upon environmental changes. Here, we review the recent understandings of transcriptional regulation via histone deacetylation by Hda1C. In addition, we discuss the potential mechanisms for histone substrate switching by Hda1C, depending on transcriptional frequency and activity.
Keywords: gene induction, Hda1C, histone deacetylation, substrate switching, transcription elongation
INTRODUCTION
Chromatin is composed of eukaryotic DNA and histone proteins and its primary function is to package long DNA molecules into the cell. In addition, chromatin plays important roles in the regulation of many DNA-based processes including transcription by RNA polymerase II (RNA Pol II), DNA replication, and DNA damage responses (Kim et al., 2019a; Kouzarides, 2007; Li et al., 2007a). The fundamental repeating unit of chromatin is the nucleosome, comprised of a histone octamer with two copies of each of four histone proteins, H3, H4, H2A, and H2B, as well as 147 base pairs of DNA (Luger et al., 1997; Richmond et al., 1984). At least 20% of the residues in histone proteins are positively charged amino acids, including lysine and arginine, which stabilize the interactions with the negatively charged DNA. Nucleosomes function as a barrier of transcription and DNA synthesis, and should be evicted and reincorporated during and after RNA Pol II transcription and DNA replication (Petesch and Lis, 2012).
Covalent modifications on histone tails, including acetylation, methylation, ubiquitination, and phosphorylation, play critical roles in RNA Pol II transcription (Kouzarides, 2007). Acetylation on histone tails can directly activate transcription by neutralizing positively charged lysines, thereby reducing the interaction between DNA and histones. This histone mark also creates a binding site for bromodomain proteins that are often associated with large protein complexes that positively regulate transcription (Kouzarides, 2007; Li et al., 2007a). The histone acetylation pattern is dynamically regulated by the opposing functions of histone acetyltransferases (HATs) and histone deacetylase complexes (HDACs). Multiple mechanisms for targeting of HATs and HDACs to specific regions of a genome have been proposed (Woo et al., 2017). Site-specific histone methylations recruit and/or stimulate HATs or HDACs to control local histone acetylation. H3K4me3 enriched at active promoters acts as a binding site for multiple HATs and HDACs including NuA3 and NuA4 HATs, Rdp3L HDACs in yeast as well as, HBO HAT and the mSin3a-HDAC1 complex in mammals (Lee et al., 2018; Saksouk et al., 2009; Shi et al., 2006; 2007; Taverna et al., 2006; Woo et al., 2017). Set3 HDAC binds to H3K4me2 and modulates the kinetics of gene induction upon environmental changes (Kim and Buratowski, 2009; Kim et al., 2012). H3K36me3 peaking at 3’ regions of genes enhances histone deacetylation by Rpd3S HDAC to repress transcription initiation from internal cryptic promoters (Carrozza et al., 2005; Keogh et al., 2005; Li et al., 2007b). Transcription machinery also contributes to recruitment of multiple HATs and HDACs to optimize RNA Pol II elongation (Govind et al., 2010).
The Hda1 histone deacetylase complex (Hda1C) includes three subunits, Hda1, Hda2, and Hda3, and is known to selectively deacetylate histone H3 and H2B to repress transcription (Carmen et al., 1996; Rundlett et al., 1996; Wu et al., 2001a; 2001b). Although this complex has HDAC activity toward histone H4 in vitro, and mutants for Hda1C show a global increase in H4 acetylation (Carmen et al., 1996; Rundlett et al., 1996), previous studies have focused on the functions of Hda1C-mediated H3 deacetylation for the regulation of transcription and cellular responses. We recently found that Hda1C mediated histone H4-sepcific deacetylation at hyperactive genes, indicating two distinct functions of Hda1C in histone deacetylation (Ha et al., 2019). Here we review recent findings on the roles of Hda1C in transcription regulation, deacetylation of distinct histone substrates, and potential mechanisms for controlling substrate switching by Hda1C.
COMPOSITION AND STRUCTURE OF Hda1C
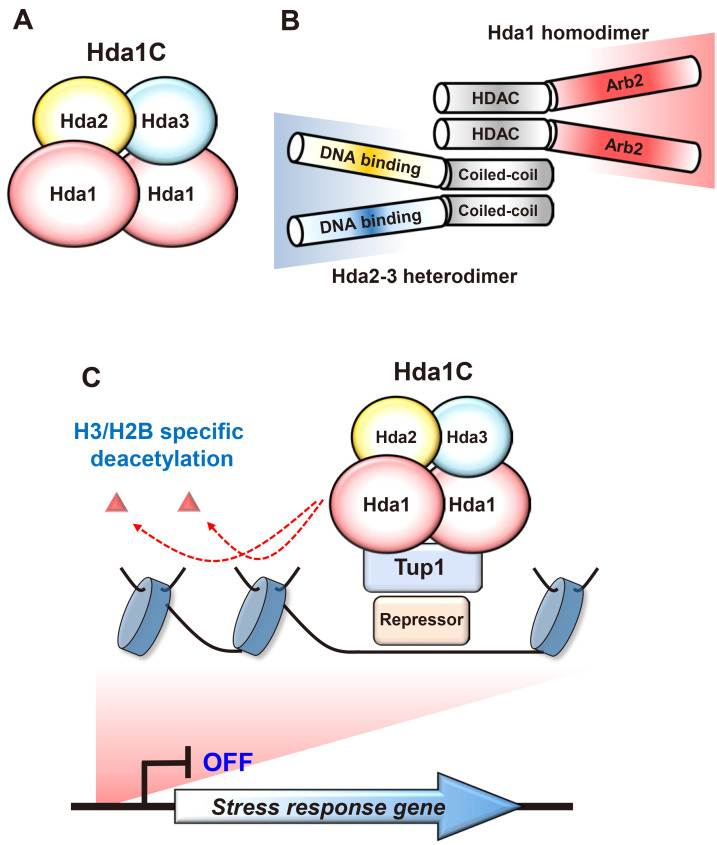
Hda1C is a conserved fungal HDAC initially identified in budding yeast. This complex includes three components, Hda1, Hda2, and Hda3, and the catalytic subunit, Hda1 belongs to a class II HDAC sensitive to trichostatin-A (Carmen et al., 1996; Rundlett et al., 1996; Wu et al., 2001a). The active form of Hda1C is a tetrameric complex composed of two copies of Hda1 and a single copy of Hda2 and Hda3 (Lee et al., 2009) (Fig. 1A). Hda1 has two important domains, a HDAC domain and an Arb2 domain at its N-terminus and C-terminus, respectively, and it forms a homodimer via its N-terminal domain (Fig. 1B). The Arb2 domain was initially identified in the Arb2 protein, a subunit of a fission Argonaute complex (Argonaute siRNA chaperone, ARC) that regulates H3K9 methylation, heterochromatin assembly and siRNA generation (Buker et al., 2007). Hda2 and Hda3 are structurally and functionally similar and interact with each other to form a heterodimeric complex through their C-termini (Fig. 1B). Both proteins have a potential nucleic acid-binding domain and a coiled-coil domain at their N-terminus and C-terminus, respectively (Lee et al., 2009). The Hda1 N-terminal region directly interacts with the C-terminal coiled-coil domains of Hda2 and Hda3 to form an active Hda1C (Fig. 1B). Truncated Hda1C, including the N-terminal part of Hda1 and the coiled-coil domains of Hda2 and Hda3, showed comparable HDAC activities with the full-length complex in vitro, suggesting that other domains are likely involved in targeting of this complex in vivo (Lee et al., 2009).
Fig. 1. Composition of Hda1C and its role in transcription.
(A) Hda1C is a class II HDAC identified in yeast and is composed of a Hda1 homodimer and Hda2-3 heterodimer. Hda1 is the catalytic subunit and both Hda2 and Hda3 are essential for the HDAC activity of Hda1C. (B) Hda1 has a HDAC domain and an Arb2 domain at its N-terminus and C-terminus, respectively. Although the Arb2 domain directly interacts with histone octamers in vitro, its function in vivo remains unknown. Both Hda2 and Hda3 show sequence similarity and have a potential DNA binding domain and a coiled-coil domain at their N- and C-terminal regions, respectively. The HDAC domain of Hda1 and coiled-coil domains of Hda2 and Hda3 directly interact to form a stable complex. (C) Hda1C deacetylates histone H3 and H2B to repress transcription. Hda1C is recruited to stress response genes via gene-specific repressors and a general corepressor, Tup1, and selectively deacetylates histone H3 and H2B to repress RNA Pol II transcription.
Recent studies have revealed that co-transcriptional methylations on histone tails recruit and/or activate multiple HATs and HDACs to regulate transcription by RNA Pol II (Carrozza et al., 2005; Ha et al., 2019; Keogh et al., 2005; Kim et al., 2012; 2016; Martin et al., 2006; Woo et al., 2017). However, Hda1C lacks domains recognizing co-transcriptional histone methylations, suggesting that this complex is likely recruited to target genes via distinct mechanisms (Ha et al., 2019; Lee et al., 2009). Although the function of the Arb2 domain of Hda1 in vivo remains elusive, this domain is known to interact with histone octamers in vitro (Shen et al., 2016). Hda2 and Hda3 may also contribute to targeting of this complex as N-terminal regions of both proteins act as binding modules to single-stranded or double-stranded DNA, with no sequence specificity (Fig. 1B) (Lee et al., 2009).
CONTROL OF GENE INDUCTION VIA HISTONE H3-SPECIFIC DEACETYLATION BY Hda1C
Tup1 is a general corepressor in yeast regulating many physiological responses, and has sequence similarity to other mammalian transcriptional repressors (Malave and Dent, 2006). This protein is recruited to target genes by sequence-specific DNA binding repressors, but how Tup1 represses target gene was not well understood. A previous study has shown that Hda1C functionally and physically interacts with Tup1 to repress transcription of stress response genes (Fig. 1C) (Wu et al., 2001b). Loss of Hda1C or Tup1 resulted in hyperacetylation of histone H3 and H2B tails at an inactive promoter. However, no increase in acetylation of histone H2A and H4 tails was seen in mutants for Hda1C, indicating that Hda1C preferentially deacetylates histone H3 and H2B to repress transcription (Wu et al., 2001b). Consistent with these findings, we also observed increased H3 acetylation upon loss of Hda1C at inactive genes. At inducible GAL genes, hyperacetylation of histone H3 was seen in mutants for Hda1C when these genes were repressed in media containing glucose (Ha et al., 2019). However, this pattern was no longer observed upon activation of GAL genes by transferring the cells to media containing galactose (Ha et al., 2019). Genome-wide ChIP-seq analyses for histone acetylation further revealed that Hda1C preferentially deacetylates histone H3 at inactive genes but not at highly transcribed regions (Ha et al., 2019). Interestingly, a mutant for Tup1 showed slightly different patterns of histone acetylation. Whereas histone H3 acetylation at inactive genes was increased in mutants for both Hda1 and Tup1, only TUP1 deleting cells showed hyperacetylation of histone H4 at inactive promoters, suggesting that Tup1 might functionally interact with an additional HDAC for histone H4 deacetylation (Ha et al., 2019).
Although Hda1C plays important roles in deacetylation of histone H3 at inactive genes, mutants for this complex had no obvious effects on genome-wide transcript levels in a steady-state condition or under optimal laboratory culture conditions (Lenstra et al., 2011). Many chromatin regulators also have little effect on global gene expression in a steady-state condition. Instead, they play essential roles in regulating transcriptional responses to support cellular adaptation and survival upon environmental changes (Lenstra et al., 2011; Woo et al., 2017). For example, the Set2-Rpd3S HDAC pathway and Set3 HDAC reduced the kinetics of gene induction upon carbon source shifts (Kim et al., 2012; 2016). Furthermore, Rpd3L HDAC mainly functions at actively transcribed genes to enhance gene repression and transcriptional repression memory upon environmental changes (Kim et al., 2019b; Lee et al., 2018). Hda1C is also likely involved in regulation of the kinetics of transcriptional responses upon environmental changes, because mutants for Hda1C had increased expression of genes during diamide stress (Weiner et al., 2012).
We previously identified approximately 1,000 genes that differentially respond during carbon source shifts and most of them were induced or repressed by galactose exposure (Kim et al., 2012). RNA-seq analyses of wild type and mutant cells for Hda1C under the same conditions revealed that although global gene expression was not affected in media containing raffinose, the induction of 331 genes was significantly delayed by Hda1C during galactose incubation (Fig. 2A) (Ha et al., 2019). Analysis of histone acetylation patterns revealed that mutants for Hda1C had increased histone H3 acetylation at the promoters and within coding regions of Hda1C-regulated genes. In contrast, no change in histone H4 acetylation was observed (Ha et al., 2019). Therefore, Hda1C preferentially deacetylates histone H3 to delay the rate and kinetics of gene induction upon environmental shifts.
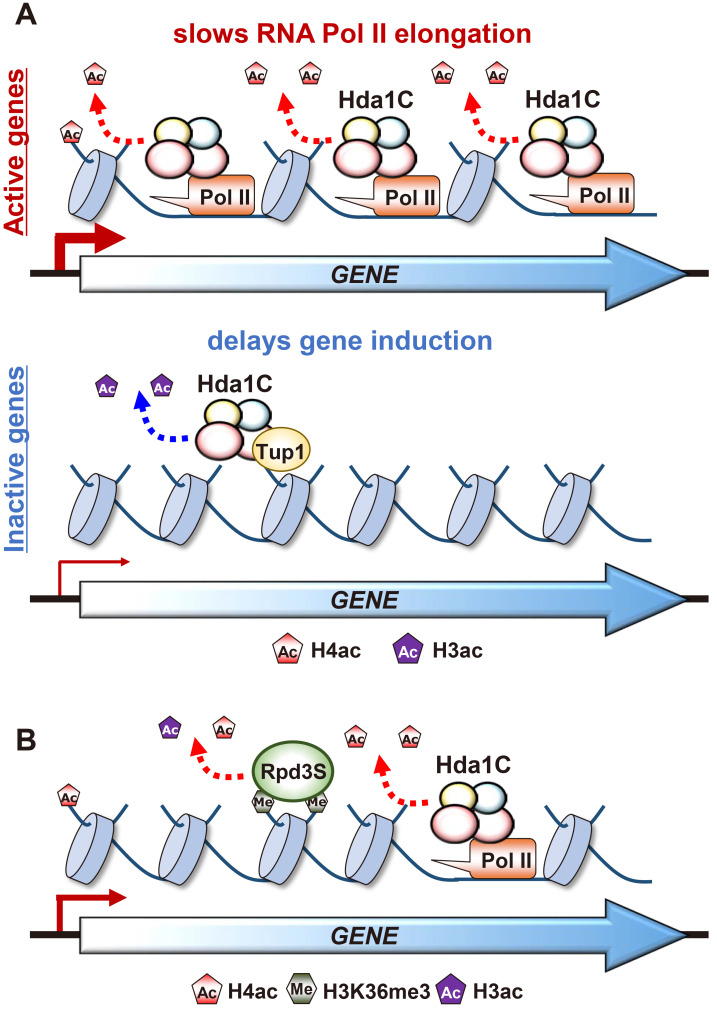
Fig. 2. Hda1C differentially deacetylates histone H4 and H3, depending on transcription frequency.
(A) Hda1C is recruited to hyperactive genes via its interaction with elongating RNA Pol II, RNA transcripts, and histones. This complex selectively deacetylates histone H4 at coding regions of hyperactive genes to negatively regulate RNA Pol II elongation. In contrast, Hda1C functionally and physically interacts with Tup1 corepressor to preferentially deacetylate histone H3 and repress transcription of stress response genes. (B) Hda1C and the Set2-Rpd3S pathway collaborate to maintain optimal histone acetylation within coding regions and may function at different times on the same genes. Hda1C deacetylates histone H4 at actively transcribing genes through the interaction with elongating RNA Pol II. In contrast, Rpd3S HDAC may deacetylate histones via recognition of H3K36 methylation when the genes are not currently transcribing by RNA Pol II.
LONG NONCODING RNA-DEPENDENT REGULATION OF TRANSCRIPTION BY Hda1C
Long noncoding RNA (lncRNA) transcription and lncRNA itself play important roles in the regulation of chromatin structure and transcription (Pelechano and Steinmetz, 2013). In yeast, approximately 12% of genes are overlapped with lncRNA transcription and a majority of them are inactive or inducible genes, suggesting the importance of overlapping lncRNA transcription in the regulation of RNA Pol II transcription (Xu et al., 2011). We and other groups have shown that lncRNA transcription places specific histone methylation at core promoter regions to target histone deacetylation by Set3 HDAC or Rpd3S HDAC resulting in gene repression (Kim et al., 2012; 2016; Venkatesh et al., 2016).
Previous works on Hda1C also suggested the functional interplay between a lncRNA and Hda1C (Camblong et al., 2007; 2009). Hda1C bound to the PHO84 locus to deacetylate histone H3. Interestingly, this binding was stimulated by PHO84 antisense RNA. In wild type cells, Hda1C binding was not seen but this binding was significantly increased when PHO84 antisense RNA was stabilized by loss of Rrp6, a component of nuclear exosome. In addition, Hda1C specifically deacetylates histone H3 to enhance repression of the PHO84 gene (Camblong et al., 2007; 2009). Our recent work has shown that Hda1C delays the induction of 331 genes during carbon source shifts by deacetylating histone H3 (Ha et al., 2019). Most of these genes are overlapped by lncRNA transcription, suggesting that Hda1C may interact with lncRNAs to modulate the kinetics of gene induction.
RECRUITMENT OF Hda1C TO ACTIVE GENES BY ELONGATING RNA Pol II
Whereas Hda1C is known to be targeted to inactive promoters via a gene-specific repressor and Tup1 corepressor, we and other groups have shown that this complex preferentially binds to active coding regions (Govind et al., 2010; Ha et al., 2019). Hda1C strongly bound to highly transcribed genes, YEF3 and PMA1, when compared to an inactive gene, TKL2. At inducible GAL genes, Hda1C binding was only slightly higher than the background signal when these genes were inactive in media containing glucose. However, this binding was significantly increased when RNA Pol II actively transcribed these genes during galactose incubation. Furthermore, genome-wide ChIP-seq analyses further confirmed that Hda1C strongly associated with highly transcribed coding regions (Fig. 2A) (Ha et al., 2019). In higher eukaryotes, HDAC6 but not other HDACs is enriched at coding regions of actively transcribed genes (Wang et al., 2009).
An important question is how Hda1C is recruited to active coding regions. Multiple factors likely contribute to Hda1C binding to actively transcribed regions. Both Hda2 and Hda3 enhance Hda1 chromatin binding, as loss of these proteins partially reduced the interaction between Hda1 and active coding regions (Ha et al., 2019). These proteins have a potential nucleic acid binding domain at their N-terminus (Fig. 1B) (Lee et al., 2009). Interestingly, when the RNA was removed by RNaseA and T1 treatment, the Hda1 binding pattern was similar to that of cells lacking both Hda2 and Hda3. In addition, the effect of RNA removal was not seen in the absence of Hda2 and Hda3 indicating that these two proteins may enhance Hda1 chromatin binding via the interaction with RNAs being transcribed by RNA Pol II (Ha et al., 2019). In addition, the Arb2 domain of Hda1 was essential for its chromatin binding. This domain only partially affected Hda1 stability, but Hda1 binding to actively transcribed regions was completely absent in mutants lacking the Arb2 domain. Interestingly, we found that Hda1C directly interacted with RNA Pol II and this interaction required the Arb2 domain of Hda1 (Figs. 1B and 2A). Consistent with this finding, a previous study suggested that Hda1C associated with phosphorylated C-terminal domain of Rpb1, the largest subunit of RNA Pol II (Govind et al., 2010). Taken together, our findings suggested that elongating RNA Pol II initially recruits Hda1C to active coding regions, and then multiple interactions including the RNA Pol II-Arb2 domain of Hda1, nucleic acids-Hda2 and -Hda3, and Arb2 domain-histones, stabilize Hda1C binding to actively transcribed regions.
DEACETYLATION OF HISTONE H4 AT HYPERACTIVE GENES BY Hda1C
Since Hda1C deacetylates only histone H3 and H2B at inactive genes, it has been suggested that this complex is a H3- and H2B-specific HDAC (Wu et al., 2001b). However, loss of this complex showed a global increase in acetylation of both histone H4 and H3 (Rundlett et al., 1996). Furthermore, purified Hda1C exhibited a HDAC activity toward histone H4 further indicating that this complex likely contributes to H4 deacetylation (Carmen et al., 1996). Mutants for Hda1C had increased acetylation of histone H3 but not H4 at inactive GAL genes, but this pattern was reversed when GAL genes are active, indicating that Hda1C also strongly associates with active GAL genes to preferentially deacetylate histone H4 (Ha et al., 2019). Increased acetylation of H4 but not H3 was also observed at two highly active genes, YEF3 and PMA1. Factors that recruit Hda1C to active genes are required for deacetylation of histone H4. Loss of Hda2 or Hda3, and of the Arb2 domain of Hda1 resulted in hyperacetylation of histone H4 at actively transcribed genes. Genome-wide ChIP-seq analyses for H4 acetylation showed that approximately 47% of yeast genes were deacetylated by Hda1C (Fig. 2A) (Ha et al., 2019). Analyses of the relationships between changes in H4 acetylation and transcription frequency or gene length indicated that this complex is mainly targeted to actively transcribed long genes. Interestingly, Tup1 had no effect on histone H4 acetylation patterns, indicating that Hda1C and Tup1 are functionally separated at actively transcribed regions (Ha et al., 2019).
Histone deacetylation within coding regions is also mediated by the Set2-Rpd3S HDAC pathway (Carrozza et al., 2005; Joshi and Struhl, 2005; Keogh et al., 2005). Set2 associated with elongating RNA Pol II methylates K36 of histone H3 (Krogan et al., 2003; Schaft et al., 2003; Strahl et al., 2002). The Eaf3 chromodomain recognizing H3K36 methylation and the Rco1 PHD finger enhance histone deacetylation and chromatin binding of Rpd3S HDAC (Carrozza et al., 2005; Joshi and Struhl, 2005; Keogh et al., 2005; Li et al., 2007b). Although both Hda1C and Rpd3S HDAC deacetylate histones within coding regions, they appear to be functionally distinct. An obvious difference is that whereas Hda1C targets hyperactive genes, Rpd3S HDAC preferentially deacetylates histones at infrequently transcribed genes (Ha et al., 2019; Li et al., 2007c). Furthermore, Rpd3S HDAC deacetylates both histone H3 and H4 but Hda1C is active at only histone H4 within coding regions (Fig. 2B).
Although Hda1C and the Set2-Rpd3S pathway act independently, approximately 1,800 genes were deacetylated by both HDACs. This raises an interesting possibility that they function at different times on the same genes. Whereas Hda1C may deacetylate histone H4 at actively transcribing genes through the interaction with elongating RNA Pol II, Rpd3S HDAC may deacetylate histones when the genes are not currently transcribing by RNA Pol II by recognizing H3K36 methylation deposited by Set2 (Fig. 2B). It is also possible that Hda1C acts as a major HDAC when a gene is hyperactive, but is replaced by Rpd3S if it is less active upon environmental changes. It will be of interest to determine how these two HDACs collaborate each other to maintain optimal histone acetylation within coding regions.
THE ROLES OF H4-SPECIFIC DEACETYLATION BY Hda1C IN TRANSCRIPTION
What is the role of Hda1C-mediated H4 deacetylation in transcription? The Set2-Rpd3S HDAC pathway is known to repress transcription initiation from cryptic promoters within open reading frames, and this inhibits RNA Pol II elongation by deacetylating histones (Carrozza et al., 2005; Keogh et al., 2005). In addition, targeting of H3K36 methylation and histone deacetylation by Rpd3S HDAC to mRNA promoters via overlapping lncRNA transcription fine-tunes the kinetics of transcriptional responses upon environmental changes (Kim et al., 2016; Venkatesh et al., 2016). We recently showed that loss of Hda1C partially bypassed the requirement for Bur1, a positive elongation factor for RNA Pol II (Ha et al., 2019). Consistent with this finding, mutants for Hda1C showed hyperacetylation of histone H4 within active coding regions. These findings suggest that Hda1C may negatively regulate RNA Pol II elongation by deacetylating histone H4 (Fig. 2A).
POTENTIAL MECHANISMS FOR SUBSTRATE SWITCHING BY Hda1C
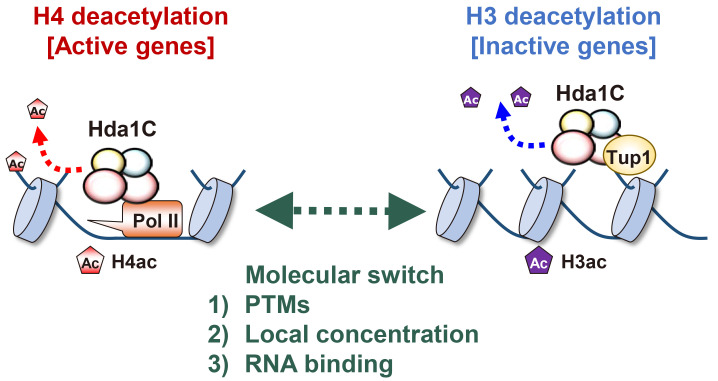
Hda1C apparently deacetylates distinct histones on the same gene or at different loci depending on the transcription frequency. This complex preferentially deacetylates histone H3 at inactive genes, but histone H4 at actively transcribed genes (Ha et al., 2019). An obvious question is how does Hda1C identify distinct histone substrates depending on transcription levels. One possibility is that covalent modifications of Hda1C subunits contribute to its specificity. Both Hda1 and Hda3 are sumoylated upon stresses and are phosphorylated by unknown kinases (Fig. 3) (Lewicki et al., 2015; Swaney et al., 2013). Whether these modifications affect the specificity of Hda1C needs to be addressed. Alternatively, it is also possible that the local concentration of Hda1C at target genes might contribute to substrate switching. Hda1C is more highly enriched at actively transcribed genes than at inactive genes. Accumulation of Hda1C might switch its main target from histone H3 to H4. Finally, Hda1C specificity can be modulated via its interaction with RNAs (Fig. 3). Our recent work and previous studies suggested that Hda1C functionally interacted with RNA transcripts (Camblong et al., 2007; 2009; Ha et al., 2019). In yeast, most inactive or inducible genes are overlapped with lncRNA transcription (Xu et al., 2011). As Hda1C preferentially deacetylates histone H3 at these genes, these noncoding RNAs might function to stimulate the histone H3-specific deacetylase activity of Hda1C. In contrast, the interaction between Hda1C and mRNAs from actively transcribed genes may enhance the histone H4-speicific deacetylase activity of this complex (Fig. 3). Determining whether the interaction of Hda1C with coding or noncoding RNAs affects the substrate switching will be of future interest.
Fig. 3. Potential mechanisms for the substrate switching by Hda1C.
Hda1C specifically deacetylates histone H4 and H3 at hyperactive and inactive genes, respectively. The specificity of this complex may be regulated by posttranslational modifications of Hda1 and Hda3 including sumoylation and phosphorylation. In addition, Hda1C is highly enriched at hyperactive genes but not at inactive genes. This difference in local concentrations of this complex may also affect the substrate switching of Hda1C. Finally, both Hda2 and Hda3 have the ability to bind to RNAs. At hyperactive genes, Hda1C may interact with mRNAs, resulting in H4-specific deacetylation. As most inactive genes are associated with lncRNA transcription from either an upstream or an antisense promoter, Hda1C may directly bind to lncRNAs transcribed from inactive genes to enhance deacetylation of histone H3.
PERSPECTIVES
Most chromatin regulators are known to have specific substrates, and none of the known HDACs or HATs has been shown to change their substrate specificity, depending on transcription frequency. Hda1C is the first example of such a regulator that differentially deacetylates histone H3 and H4 at inactive and at highly transcribed genes, respectively. This finding suggests that other chromatin regulators may also switch their substrate specificity depending on local environments. This should be considered in future studies on the function of chromatin regulators.
A previous study has shown that Hda1C strongly associates with RNA Pol III genes including tRNA genes (Venters et al., 2011). It will be important to understand whether Hda1C differentially deacetylates histones at RNA Pol II versus RNA Pol III genes. Recruitment of Hda1C to actively transcribed RNA Pol II genes requires multiple interactions between Hda1C and histones, RNA transcripts, and RNA Pol II (Fig. 2A). Determining whether Hda1C also binds to RNA Pol III genes via similar or distinct mechanisms will be important in understanding a general mechanism for Hda1C targeting to highly transcribed genes, including both RNA Pol II and RNA Pol III genes.
ACKNOWLEDGMENTS
We thank all members of the Kim lab for helpful discussions. This work was supported by grants from the National Research Foundation (NRF-2017M3A9B5060887, NRF-2017M3A9G7073033, NRF-2017M3C9A5029980, and NRF-2019R1A5A6099645 to T.K.).
Footnotes
AUTHOR CONTRIBUTIONS
M.K.L. and T.K. wrote the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Buker S.M., Iida T., Buhler M., Villen J., Gygi S.P., Nakayama J., Moazed D. Two different Argonaute complexes are required for siRNA generation and heterochromatin assembly in fission yeast. Nat. Struct. Mol. Biol. 2007;14:200–207. doi: 10.1038/nsmb1211. [DOI] [PubMed] [Google Scholar]
- Camblong J., Beyrouthy N., Guffanti E., Schlaepfer G., Steinmetz L.M., Stutz F. Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev. 2009;23:1534–1545. doi: 10.1101/gad.522509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camblong J., Iglesias N., Fickentscher C., Dieppois G., Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Carmen A.A., Rundlett S.E., Grunstein M. HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J. Biol. Chem. 1996;271:15837–15844. doi: 10.1074/jbc.271.26.15837. [DOI] [PubMed] [Google Scholar]
- Carrozza M.J., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Govind C.K., Qiu H., Ginsburg D.S., Ruan C., Hofmeyer K., Hu C., Swaminathan V., Workman J.L., Li B., Hinnebusch A.G. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol. Cell. 2010;39:234–246. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S.D., Ham S., Kim M.Y., Kim J.H., Jang I., Lee B.B., Lee M.K., Hwang J.T., Roh T.Y., Kim T. Transcription-dependent targeting of Hda1C to hyperactive genes mediates H4-specific deacetylation in yeast. Nat. Commun. 2019;10:4270. doi: 10.1038/s41467-019-12077-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A.A., Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Keogh M.C., Kurdistani S.K., Morris S.A., Ahn S.H., Podolny V., Collins S.R., Schuldiner M., Chin K., Punna T., Thompson N.J., et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Lee B.B., Oh Y.M., Zhu C., Steinmetz L.M., Lee Y., Kim W.K., Lee S.B., Buratowski S., Kim T. Modulation of mRNA and lncRNA expression dynamics by the Set2-Rpd3S pathway. Nat. Commun. 2016;7:13534. doi: 10.1038/ncomms16122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Eom J., Jung I. Characterization of structural variations in the context of 3D chromatin structure. Mol. Cells. 2019a;42:512–522. doi: 10.14348/molcells.2019.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.Y., Lee J.E., Kim L.K., Kim T. Epigenetic memory in gene regulation and immune response. BMB Rep. 2019b;52:127–132. doi: 10.5483/BMBRep.2019.52.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T., Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5' transcribed regions. Cell. 2009;137:259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T., Xu Z., Clauder-Munster S., Steinmetz L.M., Buratowski S. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell. 2012;150:1158–1169. doi: 10.1016/j.cell.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Krogan N.J., Kim M., Tong A., Golshani A., Cagney G., Canadien V., Richards D.P., Beattie B.K., Emili A., Boone C., et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2003;23:4207–4218. doi: 10.1128/mcb.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.B., Choi A., Kim J.H., Jun Y., Woo H., Ha S.D., Yoon C.Y., Hwang J.T., Steinmetz L., Buratowski S., et al. Rpd3L HDAC links H3K4me3 to transcriptional repression memory. Nucleic Acids Res. 2018;46:8261–8274. doi: 10.1093/nar/gky573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Maskos K., Huber R. Structural and functional studies of the yeast class II Hda1 histone deacetylase complex. J. Mol. Biol. 2009;391:744–757. doi: 10.1016/j.jmb.2009.06.059. [DOI] [PubMed] [Google Scholar]
- Lenstra T.L., Benschop J.J., Kim T., Schulze J.M., Brabers N.A., Margaritis T., van de Pasch L.A., van Heesch S.A., Brok M.O., Groot Koerkamp M.J., et al. The specificity and topology of chromatin interaction pathways in yeast. Mol. Cell. 2011;42:536–549. doi: 10.1016/j.molcel.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewicki M.C., Srikumar T., Johnson E., Raught B. The S. cerevisiae SUMO stress response is a conjugation-deconjugation cycle that targets the transcription machinery. J. Proteomics. 2015;118:39–48. doi: 10.1016/j.jprot.2014.11.012. [DOI] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L. The role of chromatin during transcription. Cell. 2007a;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Li B., Gogol M., Carey M., Lee D., Seidel C., Workman J.L. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007b;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- Li B., Gogol M., Carey M., Pattenden S.G., Seidel C., Workman J.L. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 2007c;21:1422–1430. doi: 10.1101/gad.1539307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Malave T.M., Dent S.Y. Transcriptional repression by Tup1-Ssn6. Biochem. Cell Biol. 2006;84:437–443. doi: 10.1139/o06-073. [DOI] [PubMed] [Google Scholar]
- Martin D.G., Baetz K., Shi X., Walter K.L., MacDonald V.E., Wlodarski M.J., Gozani O., Hieter P., Howe L. The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol. Cell. Biol. 2006;26:7871–7879. doi: 10.1128/MCB.00573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V., Steinmetz L.M. Gene regulation by antisense transcription. Nat. Rev. Genet. 2013;14:880–893. doi: 10.1038/nrg3594. [DOI] [PubMed] [Google Scholar]
- Petesch S.J., Lis J.T. Overcoming the nucleosome barrier during transcript elongation. Trends Genet. 2012;28:285–294. doi: 10.1016/j.tig.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond T.J., Finch J.T., Rushton B., Rhodes D., Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984;311:532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- Rundlett S.E., Carmen A.A., Kobayashi R., Bavykin S., Turner B.M., Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. U. S. A. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksouk N., Avvakumov N., Champagne K.S., Hung T., Doyon Y., Cayrou C., Paquet E., Ullah M., Landry A.J., Cote V., et al. HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol. Cell. 2009;33:257–265. doi: 10.1016/j.molcel.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaft D., Roguev A., Kotovic K.M., Shevchenko A., Sarov M., Shevchenko A., Neugebauer K.M., Stewart A.F. The histone 3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation. Nucleic Acids Res. 2003;31:2475–2482. doi: 10.1093/nar/gkg372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Zhu Y., Wang C., Yan H., Teng M., Li X. Structural and histone binding ability characterization of the ARB2 domain of a histone deacetylase Hda1 from Saccharomyces cerevisiae. Sci. Rep. 2016;6:33905. doi: 10.1038/srep33905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Hong T., Walter K.L., Ewalt M., Michishita E., Hung T., Carney D., Pena P., Lan F., Kaadige M.R., et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Kachirskaia I., Walter K.L., Kuo J.H., Lake A., Davrazou F., Chan S.M., Martin D.G., Fingerman I.M., Briggs S.D., et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 2007;282:2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B.D., Grant P.A., Briggs S.D., Sun Z.W., Bone J.R., Caldwell J.A., Mollah S., Cook R.G., Shabanowitz J., Hunt D.F., et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney D.L., Beltrao P., Starita L., Guo A., Rush J., Fields S., Krogan N.J., Villen J. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods. 2013;10:676–682. doi: 10.1038/nmeth.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna S.D., Ilin S., Rogers R.S., Tanny J.C., Lavender H., Li H., Baker L., Boyle J., Blair L.P., Chait B.T., et al. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S., Li H., Gogol M.M., Workman J.L. Selective suppression of antisense transcription by Set2-mediated H3K36 methylation. Nat. Commun. 2016;7:13610. doi: 10.1038/ncomms13610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters B.J., Wachi S., Mavrich T.N., Andersen B.E., Jena P., Sinnamon A.J., Jain P., Rolleri N.S., Jiang C., Hemeryck-Walsh C., et al. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol. Cell. 2011;41:480–492. doi: 10.1016/j.molcel.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zang C., Cui K., Schones D.E., Barski A., Peng W., Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A., Chen H.V., Liu C.L., Rahat A., Klien A., Soares L., Gudipati M., Pfeffner J., Regev A., Buratowski S., et al. Systematic dissection of roles for chromatin regulators in a yeast stress response. PLoS Biol. 2012;10:e1001369. doi: 10.1371/journal.pbio.1001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H., Dam Ha S., Lee S.B., Buratowski S., Kim T. Modulation of gene expression dynamics by co-transcriptional histone methylations. Exp. Mol. Med. 2017;49:e326. doi: 10.1038/emm.2017.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Carmen A.A., Kobayashi R., Suka N., Grunstein M. HDA2 and HDA3 are related proteins that interact with and are essential for the activity of the yeast histone deacetylase HDA1. Proc. Natl. Acad. Sci. U. S. A. 2001a;98:4391–4396. doi: 10.1073/pnas.081560698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Suka N., Carlson M., Grunstein M. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell. 2001b;7:117–126. doi: 10.1016/s1097-2765(01)00160-5. [DOI] [PubMed] [Google Scholar]
- Xu Z., Wei W., Gagneur J., Clauder-Munster S., Smolik M., Huber W., Steinmetz L.M. Antisense expression increases gene expression variability and locus interdependency. Mol. Syst. Biol. 2011;7:468. doi: 10.1038/msb.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]