Abstract
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) pandemic was first reported in Wuhan, China in December 2019, moved across the globe at an unprecedented speed, and is having a profound and yet still unfolding health and socioeconomic impacts. SARS-CoV-2, a β-coronavirus, is a highly contagious respiratory pathogen that causes a disease that has been termed the 2019 coronavirus disease (COVID-19). Clinical experience thus far indicates that COVID-19 is highly heterogeneous, ranging from being asymptomatic and mild to severe and causing death. Host factors including age, sex, and comorbid conditions are key determinants of disease severity and progression. Aging itself is a prominent risk factor for severe disease and death from COVID-19. We hypothesize that age-related decline and dysregulation of immune function, i.e., immunosenescence and inflammaging play a major role in contributing to heightened vulnerability to severe COVID-19 outcomes in older adults. Much remains to be learned about the immune responses to SARS-CoV-2 infection. We need to begin partitioning all immunological outcome data by age to better understand disease heterogeneity and aging. Such knowledge is critical not only for understanding of COVID-19 pathogenesis but also for COVID-19 vaccine development.
Keywords: Aging, COVID-19, SARS-CoV-2, Cytokine storm, Immunopathology, Immunosenescence, Inflammaging, Anti-IL-6 therapy, Vaccination
1. Introduction
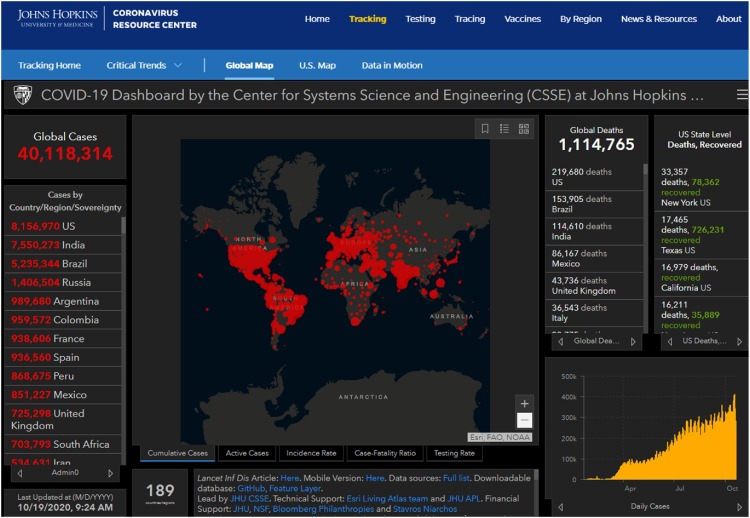
In December 2019, a cluster of novel infectious respiratory syndrome of unknown cause was observed in Wuhan, China. Thanks to the still fairly recent experience gained from the outbreak of severe acute respiratory syndrome (SARS) in 2003, Chinese scientists and clinicians worked together and quickly identified a novel coronavirus, SARS coronavirus 2 (SARS-CoV-2) as the pathogen (Zhou et al., 2020b). A complete lock-down and other quarantine measures (e.g., staying at home, social distancing, wearing mask and gloves, hand washing, etc.) were implemented initially in Wuhan City and then across much of China in the midst of its busy Chinese New Year holiday season. While the outbreak was brought under control in China quite promptly, the virus has quickly spread across the globe, causing significant morbidity and mortality initially in Italy, then other European countries, United States (US), Brazil and the rest of the world. On March 11, 2020, the World Health Organization (WHO) declared COVID-19 a global pandemic. On March 13th, the US government declared a National Emergency. In the US, the New York City-New Jersey area was the epicenter from March to May. By the end of June, COVID-19 resurgence occurred in its Sun Belt States including Arizona, Texas, and Florida, which became the new epicenter. On July 17, the US set a new record of over 77,255 new cases and the world saw over 260,000 new cases in a single day. As of October 19, 2020, there were over 40 million confirmed COVID-19 cases and over 1.1 million deaths worldwide, affecting 189 countries and territories [Fig. 1 , (Johns Hopkins University Coronavirus Resource Center, 2020, https://coronavirus.jhu.edu/map.html].
Fig. 1.
The Johns Hopkins Coronavirus Resource Center webpage. This webpage “Global Map” was taken from its website (https://coronavirus.jhu.edu/map.html) at the time shown at the left lower corner. The website tracks confirmed COVID-19 cases and death across the globe real time. It can be further drilled down to a country or territory for more detailed information. The website also provides other useful information about the ongoing pandemic.
An effective public health and socioeconomic response to control the pandemic must be based on a scientific understanding of COVID-19. In this review, we provide a brief overview of SARS-CoV-2 followed by a detailed assessment of how aging impacts the immune response to the virus and possibly the vaccine.
2. SARS-CoV-2: the virus
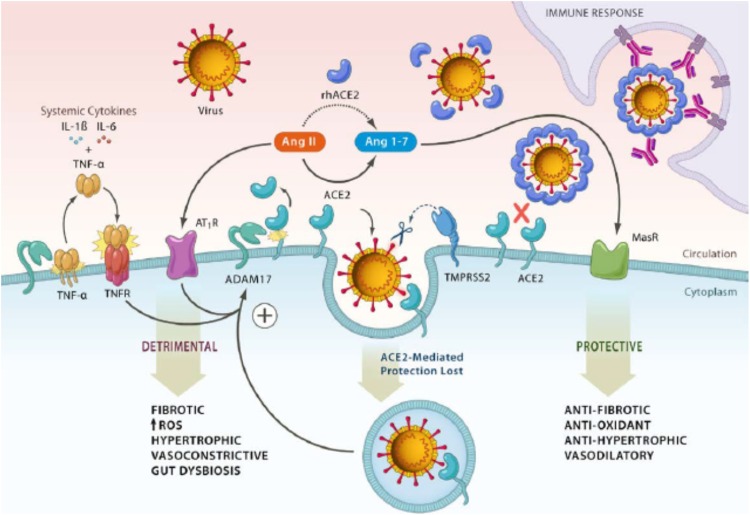
Coronaviruses are a family of enveloped, positive-sense single-stranded RNA viruses described as early as in the 1960s (Holmes, 2003). Common strains including human coronavirus (HCoV)-229E, OC43, HKU1 and NL63 are considered to be mild respiratory pathogens for immunocompetent hosts and can be isolated from many patients with upper respiratory infections (URIs) or the “common cold” in a typical winter season (Cui et al., 2019; Su et al., 2016). However, since the turn of the century, three highly virulent coronavirus strains have emerged. In 2002–2003, SARS-CoV, was initially identified in Guangdong, China and subsequently spread to 17 countries, causing over 8000 cases (de Wit et al., 2016; Zhong et al., 2003). No SARS-CoV infection has been reported since 2004. About 10 years later, however, Middle East Respiratory Syndrome (MERS) caused by MERS coronavirus (MERS-CoV) was reported in 2012–2013, primarily in Middle East countries (Zaki et al., 2012). A MERS outbreak was also reported in South Korea in 2017 (de Wit et al., 2016). Together, MERS caused approximately 2000 cases in total. Bats and camels are known to serve as animal reservoirs for SARS-CoV and MERS-CoV, respectively. While details of the zoonotic aspect of SARS-CoV-2 are still being worked out, bats appear to serve as its animal reservoir as well (Ahmad et al., 2020; Cui et al., 2019). A key feature of these three virulent β coronaviruses is their ability to replicate in epithelial cells and pneumocytes in the lower respiratory track in humans and, thus, cause pneumonia (Cui et al., 2019; de Wit et al., 2016) and, in severe cases, acute respiratory distress syndrome (ARDS). Angiotensin-converting enzyme 2 (ACE2) serves as the functional cellular receptor for both SARS-CoV and SARS-CoV-2 (Bourgonje et al., 2020; Hoffmann et al., 2020). Recent mechanistic and structural analyses indicate that the viral spike (S) protein of SARS-CoV-2 binds to ACE2 in concert with S-protein priming by the host cell transmembrane serine protease TMPRSS2, mediating host cell entry of the virus (Hoffmann et al., 2020; Mittal et al., 2020). This binding of SARS-CoV-2 is shown to be at 10- to 20-fold higher affinity than that of SAR-CoV (Hoffmann et al., 2020), accounting at least partially for the greater infectivity and pathogenicity of SARS-CoV-2. In addition to mediating viral entry, binding of SARS-CoV-2 to ACE2 and subsequent endocytosis dysregulate the angiotensin system, leading to the loss of ACE2-mediated health protection and adverse systemic effects (Gheblawi et al., 2020). Moreover, these molecular events upregulate proteolytic cleavage mediated by a disintegrin and metalloproteinase 17 (ADAM17) of not only ACE2 itself, which further dysregulates the angiotensin system, but also its primary substrate releasing Tumor necrosis factor (TNF)-α along with interleukin (IL)-6 and other cytokine mediators, leading to the cytokine storm in COVID-19 described below [Fig. 2 , (Gheblawi et al., 2020)]. Taken together, ACE2 is the key human cellular receptor and plays a critical role in the pathogenesis of COVID-19 through complex molecular mechanisms against which therapeutic agents can potentially be developed, such as recombinant human ACE2 (rhACE2) and inhibitors against TMPRSS2 and possibly ADAM17 (Bourgonje et al., 2020; Gheblawi et al., 2020; Hoffmann et al., 2020).
Fig. 2.
ACE2 as the key human cellular receptor for SARS-CoV-2, its role and underlying molecular mechanisms in the pathogenesis of COVID-19. In addition to viral entry, binding to ACE2 of SARS-CoV-2 in concert with S-protein priming by TMPRSS2 and subsequent endocytosis result in dysregulation of the angiotensin system, leading to the loss of ACE2-mediated systemic health protection. These molecular events also upregulate ADAM17-mediated proteolytic cleavage of not only ACE2 itself, which further dysregulates the angiotensin system, but also its primary substrate releasing TNF-α along with IL-6 and other cytokine mediators, leading to cytokine storm. TMPRSS2: transmembrane serine protease 2; ADAM17: a disintegrin and metalloproteinase 17. From Gheblawi M, et al. Circulation Research 2020;126:1457–1475 with permission.
Influenza, another zoonotic virus which has a segmented negative-sense RNA genome and is also a significant respiratory pathogen, is well known for its genetic instability, likely due to the lack of proofreading function of its RNA polymerase complex (Te Velthuis and Fodor, 2016). Point mutations or small deletions in individual RNA segments cause relatively minor antigenic changes, termed antigenic drift, leading to new strains each winter season responsible for seasonal influenza. For this reason, annual updates of influenza vaccines are required. Exchange or re-assortment of the viral genome RNA segments between strains from humans and animals (e.g., birds or swine) can lead to major antigenic alterations, termed antigenic shift, so that the novel viruses can escape from existing herd immunity and cause influenza pandemics (Kim et al., 2018; Lyons and Lauring, 2018).
Coronaviruses, on the other hand, are not known for their major genomic re-assortment capacity. Why the aforementioned three virulent novel coronaviruses have emerged in less than 20 years of this young century remains a mystery. Among them, SARS-CoV-2 is the most contagious and SARS-CoV has not recurred since 2004, reasons for which also remain to be determined. Specific mutations are being identified in the SARS-CoV-2 genome during this ongoing pandemic. For example, a mutation that changed the amino acid at position 614 of the spike protein from an aspartic acid to a glycine, known as G614, may increase infectivity of SARS-CoV-2 (Korber et al., 2020; Kupferschmidt, 2020). Unlike influenza and other common coronaviruses, it is now apparent that SARS-CoV-2 spread is not impeded by warm weather despite an early study suggesting distribution of COVID-19 outbreaks along restricted latitudes, temperatures, and humidity (Sajadi et al., 2020); increased international travel by air or cruise-ship may also contribute to the spread of SARS-CoV-2 (Gupta et al., 2020; Tabari et al., 2020). The mechanism for this unique and worrisome feature remains to be elucidated as well. Addressing the challenges of these unknown aspects of SARS-CoV-2 biology is critically important, particularly in considering the current resurgence of new cases and the prospect of ongoing COVID-19 for years to come.
3. Aging
The majority of COVID-19 cases are mild. Some people may not have any clinical manifestation at all after SARS-CoV-2 infection (Gao et al., 2020; Wiersinga et al., 2020). These asymptomatic individuals can serve as a source of virus spread (Gao et al., 2020; Rothe et al., 2020). A report of the data in New York State up to March 31, 2020 showed that 47,326 persons out of 141,495 were tested positive (33 %) for COVID-19, and many of those positives were asymptomatic (Rosenberg et al., 2020). However, more surveillance data are needed to evaluate the extent of asymptomatic infection. Artificial intelligence and machine learning may help address this as emerging evidence suggests the utility of such technologies in the screening and diagnosis of SARS-CoV-2 infection as well as clinical care and other public health measures e.g., contact tracing and developing models for prediction and forecasting, etc.) (Halamka et al., 2020; Lalmuanawma et al., 2020; Minaee et al., 2020). In an infectious disease as heterogeneous as COVID-19, host factors are the key to determine disease severity and progression (Wiersinga et al., 2020). For severe COVID-19 disease, major risk factors include age, male sex, obesity, smoking, and comorbid chronic conditions such as hypertension, type 2 diabetes mellitus, and others (Wu et al., 2020; Zhou et al., 2020a; Garibaldi et al., 2020). Overwhelming evidence from around the world suggests that age itself is the most significant risk factor for severe COVID-19 disease and its adverse health outcomes. The following are supportive data from several individual countries.
China: Early data from China demonstrate that case fatality ratio (CFR) of COVID-19 increases with age, from 0.4 % or lower in patients aged in the 40s or younger, 1.3 % among those in their 50s, 3.6 % in their 60s, 8% in their 70s, to 14.8 % in their 80s or older; the overall CFR is 2.3 % (Wu et al., 2020; Zhu et al., 2020b). In comparison, the overall CFR was approximately 2.8 % worldwide and 2.7 % in the US as of October 19, 2020 (Fig. 1). The rapid growth of the number of older adults in the world’s largest population, termed an "aging tsunami" by some, coupled with the unique socioeconomic context, ongoing healthcare reform, and nascent development of geriatrics, creates significant challenges for China to fight against COVID-19, particularly during the early days of the pandemic (Fang et al., 2015; Li et al., 2018; Yip et al., 2019).
Italy: A more profound effect of aging is shown by COVID-19 CFR data from Italy, the first country affected by the pandemic after China. Again, CFRs are from less than 0.4 % or lower in patients aged in the 40s or younger, 1% among those in their 50s, 3.5 % in their 60s, 12.8 % in their 70s, to 20.2 % in their 80s and above; the overall CFR is 7.2 % (Onder et al., 2020). Of note, the overall CFR is higher in Italy than that in China (7.2 % vs 2.3 %, respectively). This is likely because Italy not only has a higher CFR than China among adults over 70 years of age, but also has a higher proportion of older adults than China (22.8 % vs 11.9 %, respectively).
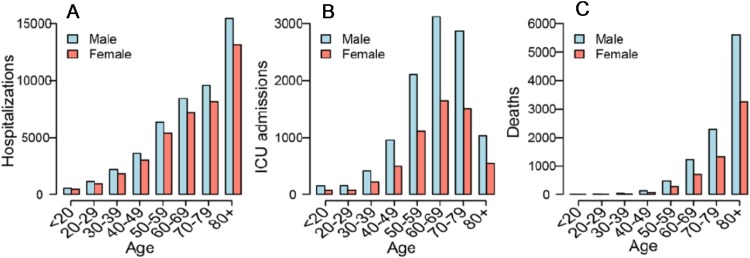
France: Similarly, nationwide data from France up to May 7, 2020 demonstrate age-related increase in hospitalizations Fig. 3 A) and intensive care unit (ICU) admissions (Fig. 3B) in addition to CFR or deaths (Fig. 3C) (Salje et al., 2020).
Fig. 3.
Nation-wide data on number of hospitalizations (A), ICU admissions (B), and deaths (C) of COVID-19 patients by age group and sex reported up to May 7, 2020 in France. From Salje H, et al. Science 10.1126/science.abc3517 (2020) with permission.
The US: The first COVID-19 outbreak reported in the US was at a long-term care facility in Washington State (Arons et al., 2020; McMichael et al., 2020). The first COVID-19 death reported in New York City was an 82-year-old person in Brooklyn. Data from a large case series of 5700 COVID-19 patients who were admitted to hospitals in New York City have shown a strikingly similar trend of age-related increases in COVID-19 deaths. That is, deaths among patients in hospital at the study end point were 3.3 % or lower in patients aged in the 40s or younger, 4.8 % among those in their 50s, 6.4 % in their 60s, 12.6 % in their 70s, to 25.9 % in their 80s and above (Richardson et al., 2020).
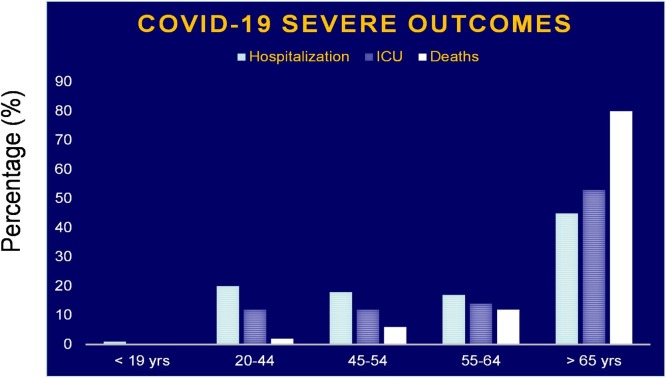
As shown in Fig. 4 , data reported by the US Center for Disease Control and Prevention (CDC) also demonstrate significantly higher rates of hospitalizations, ICU admissions, and deaths secondary to COVID-19 among older adults (> 65 years) than any younger age groups. [(CDC, 2020), https://www.cdc.gov/mmwr/volumes/69/wr/mm6912e2.html].
Fig. 4.
Percentages of hospitalizations (light blue bar), ICU admissions (blue striped bar), and deaths (white bar) of COVID-19 patients by age group in the US. Adapted from the CDC, https://www.cdc.gov/mmwr/volumes/69/wr/mm6912e2.htm.
Perhaps, the most striking evidence is the data on COVID-19 cases and death in nursing homes across the US from a comprehensive analysis updated by The New York Times on September 15th [(New York Times, 2020), https://www.nytimes.com/interactive/2020/us/coronavirus-nursing-homes.html]. Currently, there are up to 1.5 million nursing home residents in the US, less than 0.5 % of its population. However, about 7% of confirmed COVID-19 cases were among these vulnerable elderly individuals. Moreover, they suffered 40 % of COVID deaths in the US.
Taken together, it is unmistakable that aging is an important risk factor for severe COVID-19 disease and its adverse health outcomes including hospitalization, ICU admission, and death.
4. Immunity and aging
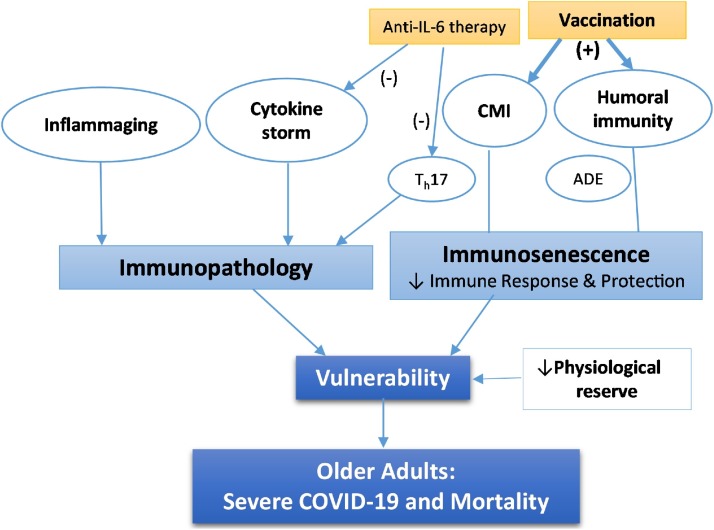
Immunity is a cornerstone of host-pathogen interaction in any infectious disease. It involves three distinct but interrelated key aspects: vulnerability, immune response and protection, and potential immune pathology (Fig. 5 ). In most cases, immune response from prior exposure to the same pathogen or through vaccination with the same dominant antigen can provide at least partial immune protection (i.e., reduction of incidence of infection and/or its severity) via immune memory. The level of vulnerability also involves innate immunity independent of antigen-specific immune responses and other physiological protective mechanisms. If the immune response to the current infection is dysregulated, however, it may cause immune pathology and contribute to the pathogenesis of the disease. Since SARS-CoV-2 is a novel coronavirus with no prior immune response, the entire population is susceptible with essentially no herd immunity. As discussed earlier, this was also the case in influenza pandemics, such as the pandemics in 1918, 1957, 1968, and more recently the swine flu pandemic in 2009 (Petersen et al., 2020). Nonetheless, under certain circumstances, older adults may enjoy better protection than the young against strains that were circulating when they were young, by virtue of immunological memory and/or cross-reactivity. A considerable fraction of healthy individuals not infected with SARS-CoV-2 possess T cells reactive to SARS-CoV-2 antigens, perhaps because of cross-reactivity with other coronaviruses. It remains to be seen whether this provides any protection against COVID-19 (Braun et al., 2020; Grifoni et al., 2020; Mateus et al., 2020; Sette and Crotty, 2020).
Fig. 5.
The immune hypothesis. This hypothesis encompasses age-related impairment of immune response and protection against SARS-CoV-2 and immunopathology. Immune response includes humoral immunity (i.e., antibody response) and cell-mediated immunity (CMI) (right). While age-related immunosenescence is believed to weaken immune protection, vaccination enhances it. Inflammaging and cytokine storm may lead to Immunopathology (left). Not all immune responses are protective as antibody-dependent enhancement (ADE) in humoral immunity may promote SARS-CoV-2 infection while Th17 response in CMI may contribute to cytokine storm. Anti-IL-6 therapy with monoclonal antibodies against either IL-6 or IL-6 receptor currently in clinical trials can block cytokine storm and its downstream event and/or suppress Th17 response. Age-related decrease of physiological reserve in respiratory and other organ systems may also contribute to vulnerability. Together, they lead to disproportionately severe COVID-19 and high mortality in older adults.
In COVID-19, a number of immunological studies were initially reported from clinical observations of COVID-19 patients in Wuhan, China. While more studies are being published almost every day, they often lack breadth and depth in interrogation of the immune system. We will first review some of the published data currently available, and then propose an immune hypothesis for age-related vulnerability to severe COVID-19 and adverse health outcomes. Instead of proposing age-related chronic inflammation (Akbar and Gilroy, 2020) or immunosenescence secondary to cytomegalovirus infection (Kadambari et al., 2020; Moss, 2020) individually as mechanisms underlying this vulnerability, this immune hypothesis integrates immunopathology secondary to cytokine storm and inflammaging and immunosenescence as complex immune mechanisms that could provide a basis for interventional strategies, such as anti-IL-6 therapy and immunization with COVID-19 vaccines (Fig. 5). We will also point out challenges in conducting comprehensive and in-depth immune studies and in interpretation of existing data and their implications for vaccine development.
4.1. “Cytokine storm” and immunopathology
Severe COVID-19 patients typically develop acute respiratory distress syndrome (ARDS) requiring intubation and ventilator support as well as significant involvement of other organ systems. For example, neurological manifestations, termed “neuro−COVID” by some, occur in over one-third of COVID-19 patients (Chiappelli, 2020; Ferrarese et al., 2020; Frontera et al., 2020; Leonardi et al., 2020; Wang et al., 2020b). While SARS-CoV-2 has been characterized regarding its neurotrophic and neuroinvasive properties (Baig et al., 2020; Puelles et al., 2020), inflammatory cytokine release and immunopathology caused by this virus may also play an important role in contributing to neuro−COVID and other systemic manifestations (Jose and Manuel, 2020; Wu and McGoogan, 2020). In fact, earlier clinical observations from Wuhan, China indicated patients with COVID-19 manifest an acute increase of serum levels of inflammatory mediators, such as IL-6 and C-reactive protein (CRP) (Chen et al., 2020a). Levels of other inflammatory mediators including interferon (IFN)-γ induced protein 10 (IP-10, or CXCL-10) and monocyte chemotactic protein-3 (MCP-3) are also acutely elevated in COVID-19 patients, and such elevation is associated with disease severity and progression (Lagunas-Rangel and Chavez-Valencia, 2020; Lin et al., 2020; Yang et al., 2020). Cytokine storm, or cytokine release syndrome (Moore and June, 2020), has also been observed in SARS and MERS and is believed to play an important role in their development and progression (de Wit et al., 2016). Although the sources and regulation of this cytokine storm remain to be elucidated, it is likely derived from dysregulated immune responses to these virulent coronaviruses, leading to immunopathology and severe disease. These observations have led to the ongoing therapeutic development targeting IL-6, a cytokine mediator that is considered as a hallmark of inflammaging (Franceschi et al., 2000, 2017; Maggio et al., 2006). Monoclonal antibodies (MAb) targeting the IL-6 receptor (IL-6R, tocilizumab and sarilumab) or IL-6 itself (siltuximab) are now available. While in-depth discussion of differences in mechanisms regulating IL-6 signaling between these MAbs and implication in their efficacy and/or side effects is beyond the scope of this article, it is worthwhile noting that anti-IL-6 and anti-IL-6R MAbs can all antagonize IL-6 cis signaling (via canonical membrane-bound IL-6Rα and gp130) and trans signaling (via soluble IL-6R), but only anti-IL-6R MAbs can antagonize IL-6 trans presentation, a newly described mode of IL-6 signaling that involves membrane-bound IL-6Rα of dendritic cells (DCs) to generate pathogenic Th17 cells (Heink et al., 2017; Kang et al., 2019). The immediate goal of such therapeutic intervention is to block the dysregulated cascade of immune activation and inflammation downstream of the cytokine storm to ameliorate severe COVID-19 and its further progression. Preliminary results available thus far did not show significant mortality benefit of sarilumab treatment (Della-Torre et al., 2020), further analyses of subgroup patients and studies of other MAbs are needed. Ultimately, effective vaccine and antivirals will be needed to prevent or ameliorate SARS-CoV-2 infection and its induced cytokine storm or immunopathology (Tay et al., 2020).
4.2. Antibody response
Antibody response to a pathogen and the detection of those antibodies are important for both immune protection and evaluation of the infection/exposure. In COVID-19, a number of studies have demonstrated serum antibody responses to SARS-CoV-2 both in severe patients admitted to the ICU and among mild cases who have recovered and are in convalescence (Liu et al., 2020a; Ni et al., 2020; Zhao et al., 2020). One study also reported correlative IgG, IgM, and IgA titers between serum and saliva (Randad et al., 2020). It is not currently known if asymptomatic infection induces detectable antibody responses or whether viral load determines antibody response. In addition, the titer threshold of anti- SARS-CoV-2 antibodies that correlates with clinical protection against COVID-19 has yet to be determined. Studies have shown higher anti-SARS-CoV-2 antibody titers in ICU patients compared to mild patients and prolonged presence of neutralizing antibodies with viral shedding in COVID-19 patients, raising questions about the protective efficiency of such antibody responses (Zhang et al., 2020b; Zhao et al., 2020). Moreover, a phenomenon called antibody-dependent enhancement (ADE) was unexpectedly reported in COVID-19 (Arvin et al., 2020; Wan et al., 2020), further suggesting the complexity of the effects of such antibody responses. Therapeutic use of convalescent plasma collected from recovered COVID-19 patients is a promising passive immune therapy currently in clinical trials. Observational findings suggest improved clinical outcomes in those who are transfused with COVID-19 convalescent plasma (CCP), including radiological resolution, reduction in viral loads, and improved survival (Duan et al., 2020; Harvala et al., 2020; Hegerova et al., 2020; Joyner et al., 2020; Li et al., 2020; Shen et al., 2020; Zhang et al., 2020a). While two randomized trials assessing CCP China and Europe were terminated early and underpowered, they did not find clinically significant differences between the study arms (Li et al., 2020); Gharbharan, et al). Studies aimed at defining factors that impact the quality and titer of antibody, including SARS-CoV-2 neutralization reveal that older age, male sex, and hospitalization with severe COVID-19 are all factors that contribute to greater antiviral antibody responses against SARS-CoV-2 (Klein et al., 2020).
4.3. Cell-mediated immune response
Clinical observations have revealed significant lymphopenia and increased neutrophil counts in severe COVID-19 disease and, therefore, lymphopenia and high neutrophil-lymphocyte ratio (NLR) are considered as useful predictors for COVID-19 death whereas high lymphocyte counts predict better clinical outcomes (Chen et al., 2020; Lagunas-Rangel and Chavez-Valencia, 2020; Qin et al., 2020; Wang et al., 2020a; Zhou et al., 2020a). While neutrophil increases may reflect an acute inflammatory response related to the cytokine storm described above, lymphopenia indicates major impacts on cell-mediate immunity in the early stage of COVID-19. Lymphopenia consists of depletion of both CD4+ and CD8 + T cells (Wang et al., 2020a). The reason for such depletion is not well understood at the present time. One may speculate that T cells are redistributed from the circulation to the site of the infection in the lungs. Sánchez-Cerrillo et al. observed redistribution of activated monocytes and dendritic cells to the lungs in severe COVID-19 patients (Sanchez-Cerrillo et al., 2020). Alternatively, as described in more detail below, viral proteins from SARS-CoV also shared by SARS-CoV-2 can suppress type 1 IFNs leading to a poor CD8 + T cell response (Fung et al., 2020; Welsh et al., 2012). Whether the observed lymphopenia is a general phenomenon of acute viral infections versus unique or more profound in COVID-19 remains to be determined. Regardless, a retrospective study of 548 COVID-19 patients showed that restored lymphocyte counts predicted recovery during hospitalization while lymphopenia persisted in non-survivors (Chen et al., 2020c). This further emphasizes the importance of cell-mediated immunity and its impact on clinical outcomes.
T lymphocytes are a key cellular basis of adaptive immune protection and vaccination and play a critical role in assisting production of neutralizing antibodies and direct virus clearance. A number of studies have shown SARS-CoV-2-specific CD4+ and CD8 + T cell responses in COVID-19 patients (Grifoni et al., 2020; Meckiff et al., 2020; Neidleman et al., 2020; Ni et al., 2020; Weiskopf et al., 2020). In the setting of SARS, antigen-specific memory T cells to SARS-CoV have been shown to persist in convalescent SARS patients at low frequency for up to six years and their responses to ex vivo stimulation with SARS-CoV persist up to 11 years (Ng et al., 2016; Oh et al., 2011). The duration that SARS-CoV-2-specific memory T cells persist is yet to be established.
Not all T cell responses are beneficial. For example, Kang et al. observed hyperactivation of cytotoxic T cell responses as a determinant of COVID-19 severity (Kang et al., 2020). Zhang and colleagues suggest aberrant CD8 T cell activation as a potential mechanism for cardiac injury in severe COVID-19 (Zhang et al., 2020c). Th17 activation is also believed to play a role in contributing to cytokine storm in severe COVID-19, suppression of which is proposed to be a mechanism underlying anti-IL-6R therapy (Lagunas-Rangel and Chavez-Valencia, 2020) and other immunotherapies (Wu and Yang, 2020).
Comprehensive studies of cell-mediated immune responses are few and far between at the present time. Peng et al. observed broad and strong memory CD4+ and CD8 + T cells induced by SARS-CoV-2 in UK convalescent COVID-19 patients, significantly more so in severe compared to mild COVID-19 cases (Peng et al., 2020). A deep immune profiling of COVID-19 patients conducted by Matthew and colleagues, however, revealed great heterogeneity (Mathew et al., 2020).
4.4. Immune hypothesis for age-related vulnerability in older adults
Given the disproportionate burden of severe COVID-19 disease and death in older adults, it is important to understand mechanisms that underlie this age-related vulnerability. Age-related immune system remodeling, or immunosenescence, is considered to be the major reason for increased susceptibility to infection, particularly respiratory infections such as influenza, as well as impaired immune responses to vaccination (Li et al., 2011; Pawelec, 2018). Here we propose an immune hypothesis for COVID-19 vulnerability of older adults. It involves age-related impairment of immune defense against SARS-CoV-2 infection, or immunosenescence, and increased risk for immunopathology (Fig. 5).
Although age-related change in innate and adaptive immunity against SARS-CoV-2 infection are yet to be investigated in detail, its impairment and dysregulation in older adults can be inferred. For example, senescence-related impairment of the type 1 IFN response is responsible for enhanced influenza viral replication in cell culture (Kim et al., 2016) and older adults manifest impaired type 1 IFN response to influenza vaccination (Thakar et al., 2015). In addition, several SARS-CoV non-structural proteins that are shared by SARS-CoV-2 suppress the type 1 IFN response and such suppression is shown to lead to poor CD8 + T cell response to viral infection (Fung et al., 2020; Manners et al., 2020; Welsh et al., 2012). Therefore, age-associated weaker type 1 IFN responses coupled with direct viral suppression could serve as a critical innate immune mechanism that leads to poor cell mediated immunity and increased vulnerability of older adults to SARS-CoV-2 infection with therapeutic implication (Sallard et al., 2020). Little data is currently available about the impact of aging on CD4+ and CD8 + T cell responses in COVID-19. It is postulated that age-related decline of de novo T cell responsiveness and/or impact from comorbid conditions, particularly persistent viral infections such as chronic cytomegalovirus (CMV) infection could serve as potential causes of COVID-19 vulnerability in older adults (Kadambari et al., 2020; Moss, 2020; Nicoli et al., 2020). Available data on humoral immunity are fascinating and counter-intuitive. Among COVID-19 convalescent plasma donors, Klein et al. observed higher SARS-CoV-2-specific neutralizing and IgG antibody titers in older donors compared with their young counterparts (Klein et al., 2020). In a study cited above, Zhang et al. also described the correlation between greater anti-SARS-CoV-2 IgG titers and older age (Zhang et al., 2020b). The reason for these observations is unknown at the present time and deserves further investigation.
Substantial evidence supports the role of immunopathology in the pathogenesis of COVID-19 overall as described above. Based on their review of data on immune changes published during the early stage of this pandemic, Lin and colleagues proposed a hypothesis for the role of immune and inflammatory factors in contributing to dysregulation of the coagulation system in the pathogenesis of COVID-19 and suggested intravenous immune globulin (IVIg) and low molecular weight heparin (LMWH) anticoagulant therapy (Lin et al., 2020). Studies also suggested a link between complement activation and endothelial dysfunction, likely the key to microvascular thrombosis and multi-organ failure in severe COVID-19 (Mackman et al., 2020; Magro et al., 2020; Noris et al., 2020). However, few studies are available with a focus on aging. Inflammaging is well documented and can be derived from senescence-associated secretory phenotype (SASP), persistent viral infection such as CMV, and other potential sources (Chen et al., 2019; Franceschi et al., 2000; Franceschi and Campisi, 2014). Such an unbalanced pro-inflammatory environment could potentiate further inflammatory responses upon SARS-CoV-2 infection, leading to the development of an exacerbated cytokine storm in older adults. It may also influence ACE2 expression and facilitate viral entry (Radzikowska et al., 2020). Further studies are urgently needed to address this important mechanism with a focus on aging.
Children are overwhelmingly spared from severe COVID-19 disease except for the extremely rare occurrence of multisystem inflammatory syndrome in children (MIS-C), also called Kawasaki disease-like syndrome (Lingappan et al., 2020; Viner and Whittaker, 2020). Insights into children’s defense mechanisms against SARS-CoV-2 infection may shed light on age-related vulnerability in older adults from a different perspective. For example, Chen et al. observed significantly higher counts of total as well as both CD4+ and CD8 + T cells in pediatric COVID-19 cases relative to adult cases (Chen et al., 2020b). On the other hand, children with COVID-19 manifested lower levels of T cell activation than adult COVID-19 patients (Moratto et al., 2020), suggesting better immune system control and regulation in response to SARS-CoV-2 infection in children. Thymic function likely plays an important role in preserving T cells in COVID-19 (Rehman et al., 2020). In fact, Liu et al. showed that thymosin α1 reversed lymphopenia, reversed exhausted T cells and reduced mortality of severe COVID-19 in adults (Liu et al., 2020b). In addition, children demonstrate strong innate immunity despite the fact that the immune system as a whole is yet to be fully developed. One possibility is “trained immunity” through scheduled immunization with many doses of pediatric vaccines (Netea et al., 2020). Whether this can be applicable in older adults (i.e., potential protective effect from immunization with other unrelated vaccines such as influenza vaccine) remain to be investigated.
4.5. Challenges
Despite the large number of research papers including preprints that are published almost every day, it remains a difficult task to obtain reliable knowledge of COVID-19 that is accurate and precise. Conflicting data and results that are not reproducible are not uncommon. Here we discuss a few inherent difficulties that present themselves as great challenges for immune studies of COVID-19, reminding readers to critically evaluate the literature and putting published results in appropriate context. This is especially true for assessing COVID-19 information in social media, particularly in the current environment of rapid dissemination of misinformation and disinformation. First, SARS-CoV-2 infection is highly heterogeneous, from asymptomatic infection to mild, moderate, or severe COVID-19. In addition, the infection can evolve through different stages and progress in either direction (improving and recovery versus worsening and death). Most published immunological studies are cross-sectional and at one time point with a relatively small sample size. Although immunological analysis itself can be cutting edge and in-depth, the results from these studies are only valid in the context of the study with limited generalizability. Asymptomatic SARS-CoV-2 infection can be a challenge in selecting “healthy controls” as well. Due to lack of universal COVID-19 testing, it is difficult to know whether people are truly “healthy” (uninfected) or asymptomatic. Secondly, complex impact of aging on the immune system and influences from comorbid conditions and concomitant use of medications may all contribute to the heterogeneity of the data on T-cell immune responses to SARS-CoV-2 (Mathew et al., 2020). Finally, there is a high-sequence similarity in viral proteins (e.g., nuclear protein) between SARS-CoV-2 and other β coronaviruses (e.g. 229E strain) (Chan et al., 2020). As such, prior coronavirus infections may induce heterotypic cellular immunity against COVID-19. This heterotypic cellular immune response may further complicate COVID-19 immunological studies and interpretation of their results. Therefore, it is important to take these challenges into consideration in interpretation of published data as well as in design and implementation of immunological and interventional studies including COVID-19 vaccine development.
5. Vaccination
Effective and safe vaccination against SARS-CoV-2 is the best strategy to stop viral spread and control the pandemic. Intense worldwide efforts for COVID-19 vaccine development began after the genetic sequence of SARS-CoV-2 was published on January 11th 2020, and has since advanced at previously unimaginable speed with the first vaccine candidate entering human clinical trials on March 16th. The Coalition for Epidemic Preparedness Innovations (CEPI), founded in 2017, works with health authorities and vaccine developers and plays an important role in this global effort. On May 15th, the US federal government announced “Operation Warp Speed”, a public–private partnership involving the National Institutes of Health (NIH), Centers for Disease Control and Prevention (CDC), Food and Drug Administration (FDA), Department of Defense, and other government agencies, whose objective is to facilitate and accelerate the development, manufacturing, and distribution of COVID-19 vaccines, therapeutics, and diagnostics. There are more than 200 vaccine candidates under development, ten of which are in human clinical trials (Lurie et al., 2020).
A striking feature of this global effort is the diverse vaccine development platforms, from the traditional inactivated or live attenuated virus, viral protein subunit, to replicating or non-replicating viral vectors and novel platforms based on DNA or mRNA (Thanh Le et al., 2020). An advantage of the traditional vaccine platform is that this is a mature technology that has already been employed in the production of licensed vaccines with existing large-scale production capacity (recombinant viral protein subunits). Moreover, adjuvant addition is possible, which can enhance immunogenicity and make lower doses viable, thereby enabling vaccination of more people without compromising protection. On the other hand, vaccines based on viral vectors offer a high level of antigenic epitopes expressed by the vector and long-term stability with the likelihood of inducing strong immune responses. Novel vaccine platforms based on DNA or mRNA enable great flexibility in terms of antigen manipulation and potential for speed. In fact, an mRNA-based vaccine (mRNA-1273) made by Moderna entered human clinical trials just two months after the SARS-CoV-2 RNA genome sequence became available and demonstrated promising preliminary results (Jackson et al., 2020). Two phase 1/2 clinical trials of two different adenovirus-vectored COVID-19 vaccines have reported robust antibody and T-cell responses to the vaccines with acceptable side effects in healthy adults (Folegatti et al., 2020; Zhu et al., 2020a). However, the ChAdOx1 nCoV-19 vaccine trial was conducted among adults of 18–55 years of age with no enrollment of older adults (Folegatti et al., 2020).
Despite progress that has been made thus far, significant challenges lie ahead. For example, while focusing on the vaccine itself is important, attention must also be directed to the host. Because of the lack of in-depth knowledge about the immune responses to SARS-CoV-2 infection, no specific immune parameter(s) of vaccine potency, or correlates of protection, are currently available. The phenomenon of ADE discussed above (Arvin et al., 2020), also deserves careful consideration (Lambert et al., 2020).
Aging is a critical host factor to consider in the context of vaccination responses (Pawelec and Weng, 2020). It may be necessary to develop different vaccines on specific vaccine platforms for older adults versus those for children and young adults. In the case of influenza vaccination, the standard dose of trivalent inactivated influenza vaccine (IIV3), the only influenza vaccine available for older adults for many years before the approval of high-dose and adjuvanted influenza vaccines, showed questionable effectiveness, if at all, for the elderly, particularly those who are frail and need vaccine protection the most (Yao et al., 2011). Over a century after the 1918 Spanish flu pandemic, the worst pandemic in human history, we are still in search of a universal influenza vaccine (Paules et al., 2017).
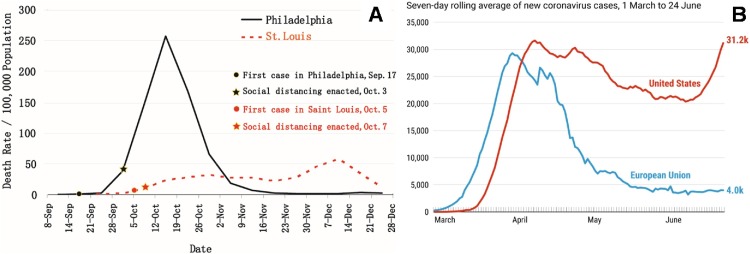
The ongoing pandemic poses special challenges (Diamond and Pierson, 2020). For vaccine trials, levels of pandemic activity as well as various quarantine measures and their implementation significantly impact study enrollment, protection of both research staff and participants, and other trial logistics. Because of the low level of pandemic activity in China, COVID-19 vaccine trials are not feasible at the present time, even for vaccines that were developed in China. When a safe and efficacious COVID-19 vaccine becomes available, the demand of the ongoing pandemic can post huge challenges on large-scale production and distribution of the vaccine. Addressing these and other challenges requires global corporation and coordination among governments, academia, and industry. With rapid progress in COVID-19 vaccine development, it is promising and hopeful that there will be one or several safe and effective COVID-19 vaccines in the near future. But the public should be fully educated about the alternative reality, that is, we may not have such a vaccine for a long time, if ever. To that end, authorities have begun to develop protective programs against COVID-19 outbreaks at nursing homes, the setting with arguably highest risk for older adults as described above. For example, in May 2020, the Pennsylvania State legislature has approved $175 million to establish a statewide Regional Response Health Collaborative Program (RRHCP, https://www.media.pa.gov/pages/DHS_details.aspx?newsid=569). Pennsylvania State University was awarded nearly $23 million to support 244 nursing homes, assisted living and personal care homes in the Southcentral region of Pennsylvania to combat COVID-19 outbreaks and support mitigation measures should COVID-19 be present at these facilities. RRHCP provides a wide range of COVID-19 related clinical and public health supportive services, such as testing performance and laboratory capacity, personal protective equipment (PPE) supplies, infection prevention training and advising, rapid response teams for sites with an active COVID-19 outbreak, staffing support, alternate care settings, mental and behavioral health support for residents and staff, clinical support via telehealth and geriatrician site visits, contact tracing, and education through a statewide learning network [https://news.psu.edu/story/630235/2020/09/01/penn-state-health-receives-grant-mitigate-covid-19-care-facilities]. As to the public, every one of us should adhere to social distancing and other quarantine measures that are known to be effective to prevent virus spread. A historical lesson from the 1918 Spanish flu pandemic is that compared to Philadelphia, St. Louis was able to minimize flu pandemic deaths and flatten the curve through its prompt and strictly enforced quarantine measures (Fig. 6 A) (Hatchett et al., 2007). The contemporary one is that Europe flattened the curve during the summer, 2020, while the US suffers ongoing COVID-19 resurgence (Fig. 6B) (Johns Hopkins University Coronavirus Resource Center, 2020). We should all learn from them.
Fig. 6.
Death curves in Philadelphia and St. Louis during 1918 Spanish flu pandemic (panel A, adapted from Hatchett RJ, et al. Proc. Natl. Acad. Sci. U. S. A. 2007; 104, 7582-7587) and curves of seven-day rolling average of newly confirmed COVID-19 cases in the US (red) and EU (blue) (panel B, Johns Hopkins Coronavirus Resources Center, https://coronavirus.jhu.edu).
Author agreement
All the authors have seen and approved the final version of the manuscript being submitted. All the authors warrant that the article is the authors' original work, hasn't received prior publication and isn't under consideration for publication elsewhere.
Funding
This study was supported in part by funding from National Institutes of Health (NIH) (R01 AI108907 and R42AG054322) and funding from Irma and Paul Milstein Program for Senior Health, Milstein Medical Asian American Partnerhsip (MMAAP) Foundation of USA (www.mmaapf.org) to SXL, NIH R21 AG059742 to SXL and JBM, NIH U54 AG062333 and Department of Defense for COVID-19: W911QY-20−9-0012 to SLK, and Pennsylvania State Regional Response Health Collaborative Program (RRHCP) funding to NO. CJW is an Irma and Paul Milstein Program for Senior Health fellow supported by MMAAP Foundation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Ahmad T., Khan M., Haroon Musa T.H., Nasir S., Hui J., Bonilla-Aldana D.K., Rodriguez-Morales A.J. COVID-19: zoonotic aspects. Travel Med. Infect. Dis. 2020 doi: 10.1016/j.tmaid.2020.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbar A.N., Gilroy D.W. Aging immunity may exacerbate COVID-19. Science. 2020;369:256–257. doi: 10.1126/science.abb0762. [DOI] [PubMed] [Google Scholar]
- Arons M.M., Hatfield K.M., Reddy S.C., Kimball A., James A., Jacobs J.R., Taylor J., Spicer K., Bardossy A.C., Oakley L.P., Tanwar S., Dyal J.W., Harney J., Chisty Z., Bell J.M., Methner M., Paul P., Carlson C.M., McLaughlin H.P., Thornburg N., Tong S., Tamin A., Tao Y., Uehara A., Harcourt J., Clark S., Brostrom-Smith C., Page L.C., Kay M., Lewis J., Montgomery P., Stone N.D., Clark T.A., Honein M.A., Duchin J.S., Jernigan J.A., Public H.-S., King C., Team C.C.-I. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N. Engl. J. Med. 2020;382:2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvin A.M., Fink K., Schmid M.A., Cathcart A., Spreafico R., Havenar-Daughton C., Lanzavecchia A., Corti D., Virgin H.W. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020;584:353–363. doi: 10.1038/s41586-020-2538-8. [DOI] [PubMed] [Google Scholar]
- Baig A.M., Khaleeq A., Ali U., Syeda H. Evidence of the COVID-19 virus targeting the CNS: tissue distribution, host-virus interaction, and proposed neurotropic mechanisms. ACS Chem. Neurosci. 2020;11:995–998. doi: 10.1021/acschemneuro.0c00122. [DOI] [PubMed] [Google Scholar]
- Bourgonje A.R., Abdulle A.E., Timens W., Hillebrands J.L., Navis G.J., Gordijn S.J., Bolling M.C., Dijkstra G., Voors A.A., Osterhaus A.D., van der Voort P.H., Mulder D.J., van Goor H. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19) J. Pathol. 2020;251:228–248. doi: 10.1002/path.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., Baysal E., Mangold M., Henze L., Lauster R., Mall M.A., Beyer K., Rohmel J., Voigt S., Schmitz J., Miltenyi S., Demuth I., Muller M.A., Hocke A., Witzenrath M., Suttorp N., Kern F., Reimer U., Wenschuh H., Drosten C., Corman V.M., Giesecke-Thiel C., Sander L.E., Thiel A. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;2020 doi: 10.1038/s41586-020-2598-9. Jul 29. [DOI] [PubMed] [Google Scholar]
- CDC Severe outcomes among patients with coronavirus disease 2019 (COVID-19) - United States, February 12-March 16, 2020. Morb. Mortal. Wkly. Rep. 2020;69:343–346. doi: 10.15585/mmwr.mm6912e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu S., Leng S.X. Chronic low-grade inflammatory phenotype (CLIP) and senescent immune dysregulation. Clin. Ther. 2019;41:400–409. doi: 10.1016/j.clinthera.2019.02.001. [DOI] [PubMed] [Google Scholar]
- Chen R., Sang L., Jiang M., Yang Z., Jia N., Fu W., Xie J., Guan W., Liang W., Ni Z., Hu Y., Liu L., Shan H., Lei C., Peng Y., Wei L., Liu Y., Hu Y., Peng P., Wang J., Liu J., Chen Z., Li G., Zheng Z., Qiu S., Luo J., Ye C., Zhu S., Zheng J., Zhang N., Li Y., He J., Li J., Li S., Zhong N. Medical Treatment Expert Group for, C., 2020c. Longitudinal hematologic and immunologic variations associated with the progression of COVID-19 patients in China. J. Allergy Clin. Immunol. 2020;146:89–100. doi: 10.1016/j.jaci.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., Zhang X., Zhang M., Wu S., Song J., Chen T., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Zhang Z.Z., Chen Y.K., Long Q.X., Tian W.G., Deng H.J., Hu J.L., Zhang X.X., Pu L., Xiang J.L., Wang D.X., Hu P., Zhou F.C., Li Z.J., Xu H.M., Cai X.F., Wang D.Q., Hu Y., Tang N., Liu B.Z., Wu G.C., Huang A.L. The clinical and immunological features of pediatric COVID-19 patients in China. Genes Dis. 2020;2020 doi: 10.1016/j.gendis.2020.03.008. Apr 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappelli F. Towards Neuro-CoViD-19. Bioinformation. 2020;16:288–292. doi: 10.6026/97320630016288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della-Torre E., Campochiaro C., Cavalli G., De Luca G., Napolitano A., La Marca S., Boffini N., Da Prat V., Di Terlizzi G., Lanzillotta M., Rovere Querini P., Ruggeri A., Landoni G., Tresoldi M., Ciceri F., Zangrillo A., De Cobelli F., Dagna L., Group S.-R.S., members S.-R.S.G. Interleukin-6 blockade with sarilumab in severe COVID-19 pneumonia with systemic hyperinflammation: an open-label cohort study. Ann. Rheum. Dis. 2020;79:1277–1285. doi: 10.1136/annrheumdis-2020-218122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond M.S., Pierson T.C. The challenges of vaccine development against a new virus during a pandemic. Cell Host Microbe. 2020;27:699–703. doi: 10.1016/j.chom.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., Zhou M., Chen L., Meng S., Hu Y., Peng C., Yuan M., Huang J., Wang Z., Yu J., Gao X., Wang D., Yu X., Li L., Zhang J., Wu X., Li B., Xu Y., Chen W., Peng Y., Hu Y., Lin L., Liu X., Huang S., Zhou Z., Zhang L., Wang Y., Zhang Z., Deng K., Xia Z., Gong Q., Zhang W., Zheng X., Liu Y., Yang H., Zhou D., Yu D., Hou J., Shi Z., Chen S., Chen Z., Zhang X., Yang X. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. U. S. A. 2020;117:9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang E.F., Scheibye-Knudsen M., Jahn H.J., Li J., Ling L., Guo H., Zhu X., Preedy V., Lu H., Bohr V.A., Chan W.Y., Liu Y., Ng T.B. A research agenda for aging in China in the 21st century. Ageing Res. Rev. 2015;24:197–205. doi: 10.1016/j.arr.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrarese C., Silani V., Priori A., Galimberti S., Agostoni E., Monaco S., Padovani A., Tedeschi G., Italian Society of, N An Italian multicenter retrospective-prospective observational study on neurological manifestations of COVID-19 (NEUROCOVID) Neurol. Sci. 2020;41:1355–1359. doi: 10.1007/s10072-020-04450-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E.A., Dold C., Faust S.N., Finn A., Flaxman A.L., Hallis B., Heath P., Jenkin D., Lazarus R., Makinson R., Minassian A.M., Pollock K.M., Ramasamy M., Robinson H., Snape M., Tarrant R., Voysey M., Green C., Douglas A.D., Hill A.V.S., Lambe T., Gilbert S.C., Pollard A.J., Oxford C.V.T.G. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C., Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J. Gerontol. A Biol. Sci. Med. Sci. 2014;69(Suppl 1):S4–9. doi: 10.1093/gerona/glu057. [DOI] [PubMed] [Google Scholar]
- Franceschi C., Bonafe M., Valensin S., Olivieri F., De Luca M., Ottaviani E., De Benedictis G. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann. N. Y. Acad. Sci. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- Franceschi C., Garagnani P., Vitale G., Capri M., Salvioli S. Inflammaging and’ Garb-aging’. Trends Endocrinol. Metab. 2017;28:199–212. doi: 10.1016/j.tem.2016.09.005. [DOI] [PubMed] [Google Scholar]
- Frontera J., Mainali S., Fink E.L., Robertson C.L., Schober M., Ziai W., Menon D., Kochanek P.M., Suarez J.I., Helbok R., McNett M., Chou S.H., Study G.C.-N. Global consortium study of neurological dysfunction in COVID-19 (GCS-NeuroCOVID): study design and rationale. Neurocrit. Care. 2020;33:25–34. doi: 10.1007/s12028-020-00995-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung S.Y., Yuen K.S., Ye Z.W., Chan C.P., Jin D.Y. A tug-of-war between severe acute respiratory syndrome coronavirus 2 and host antiviral defence: lessons from other pathogenic viruses. Emerg. Microbes Infect. 2020;9:558–570. doi: 10.1080/22221751.2020.1736644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Xu Y., Sun C., Wang X., Guo Y., Qiu S., Ma K. A systematic review of asymptomatic infections with COVID-19. J. Microbiol. Immunol. Infect. 2020;2020 doi: 10.1016/j.jmii.2020.05.001. May 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garibaldi B.T., Fiksel J., Muschelli J., Robinson M.L., Rouhizadeh M., Perin J., Schumock G., Nagy P., Gray J.H., Malapati H., Ghobadi-Krueger M., Niessen T.M., Kim B.S., Hill P.M., Ahmed M.S., Dobkin E.D., Blanding R., Abele J., Woods B., Harkness K., Thiemann D.R., Bowring M.G., Shah A.B., Wang M.C., Bandeen-Roche K., Rosen A., Zeger S.L., Gupta A. Patient Trajectories Among Persons Hospitalized for COVID-19 : A Cohort Study. Ann Intern Med. 2020:M20–3905. doi: 10.7326/M20-3905. Sep 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.C., Turner A.J., Raizada M.K., Grant M.B., Oudit G.Y. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ. Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Kunte R., Goyal N., Ray S., Singh K. A comparative analysis of control measures on-board ship against COVID-19 and similar novel viral respiratory disease outbreak: quarantine ship or disembark suspects? Med. J. Armed Forces India. 2020;2020 doi: 10.1016/j.mjafi.2020.06.003. Jun 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halamka J., Cerrato P., Perlman A. Redesigning COVID 19 care with network medicine and machine learning: a review. Mayo Clin. Proc. Innov. Qual. Outcomes. 2020;2020 doi: 10.1016/j.mayocpiqo.2020.09.008. Oct 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvala H., Mehew J., Robb M.L., Ijaz S., Dicks S., Patel M., Watkins N., Simmonds P., Brooks T., Johnson R., Gopal R., Roberts D.J., Zambon M., Blood N.H.S., Transplant Convalescent Plasma Testing, G Convalescent plasma treatment for SARS-CoV-2 infection: analysis of the first 436 donors in England, 22 April to 12 May 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.28.2001260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatchett R.J., Mecher C.E., Lipsitch M. Public health interventions and epidemic intensity during the 1918 influenza pandemic. Proc. Natl. Acad. Sci. U. S. A. 2007;104:7582–7587. doi: 10.1073/pnas.0610941104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegerova L., Gooley T.A., Sweerus K.A., Maree C., Bailey N., Bailey M., Dunleavy V., Patel K., Alcorn K., Haley R., Johnsen J.M., Konkle B.A., Lahti A.C., Alexander M.L., Goldman J.D., Lipke A., Lim S.J., Sullivan M.D., Pauk J.S., Pagel J.M. Use of convalescent plasma in hospitalized patients with COVID-19: case series. Blood. 2020;136:759–762. doi: 10.1182/blood.2020006964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heink S., Yogev N., Garbers C., Herwerth M., Aly L., Gasperi C., Husterer V., Croxford A.L., Moller-Hackbarth K., Bartsch H.S., Sotlar K., Krebs S., Regen T., Blum H., Hemmer B., Misgeld T., Wunderlich T.F., Hidalgo J., Oukka M., Rose-John S., Schmidt-Supprian M., Waisman A., Korn T. Trans-presentation of IL-6 by dendritic cells is required for the priming of pathogenic TH17 cells. Nat. Immunol. 2017;18:74–85. doi: 10.1038/ni.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes K.V. SARS-associated coronavirus. N. Engl. J. Med. 2003;348:1948–1951. doi: 10.1056/NEJMp030078. [DOI] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A., Flach B., Doria-Rose N.A., Corbett K.S., Morabito K.M., O’Dell S., Schmidt S.D., Swanson P.A., 2nd, Padilla M., Mascola J.R., Neuzil K.M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J.E., Graham B.S., Beigel J.H., m, R.N.A.S.G An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med. 2020;2020 doi: 10.1056/NEJMoa2022483. Jul 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose R.J., Manuel A. COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir. Med. 2020;8:e46–e47. doi: 10.1016/S2213-2600(20)30216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner M.J., Wright R.S., Fairweather D., Senefeld J.W., Bruno K.A., Klassen S.A., Carter R.E., Klompas A.M., Wiggins C.C., Shepherd J.R., Rea R.F., Whelan E.R., Clayburn A.J., Spiegel M.R., Johnson P.W., Lesser E.R., Baker S.E., Larson K.F., Ripoll J.G., Andersen K.J., Hodge D.O., Kunze K.L., Buras M.R., Vogt M.N., Herasevich V., Dennis J.J., Regimbal R.J., Bauer P.R., Blair J.E., van Buskirk C.M., Winters J.L., Stubbs J.R., Paneth N.S., Verdun N.C., Marks P., Casadevall A. Early safety indicators of COVID-19 convalescent plasma in 5,000 patients. J. Clin. Invest. 2020;130:4791–4797. doi: 10.1172/JCI140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadambari S., Klenerman P., Pollard A.J. Why the elderly appear to be more severely affected by COVID-19: the potential role of immunosenescence and CMV. Rev. Med. Virol. 2020:e2144. doi: 10.1002/rmv.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Tanaka T., Narazaki M., Kishimoto T. Targeting Interleukin-6 signaling in clinic. Immunity. 2019;50:1007–1023. doi: 10.1016/j.immuni.2019.03.026. [DOI] [PubMed] [Google Scholar]
- Kang C.K., Han G.C., Kim M., Kim G., Shin H.M., Song K.H., Choe P.G., Park W.B., Kim E.S., Kim H.B., Kim N.J., Kim H.R., Oh M.D. Aberrant hyperactivation of cytotoxic T-cell as a potential determinant of COVID-19 severity. Int. J. Infect. Dis. 2020;97:313–321. doi: 10.1016/j.ijid.2020.05.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.A., Seong R.K., Shin O.S. Enhanced viral replication by cellular replicative senescence. Immune Netw. 2016;16:286–295. doi: 10.4110/in.2016.16.5.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Webster R.G., Webby R.J. Influenza virus: dealing with a drifting and shifting pathogen. Viral Immunol. 2018;31:174–183. doi: 10.1089/vim.2017.0141. [DOI] [PubMed] [Google Scholar]
- Klein S., Pekosz A., Park H.S., Ursin R., Shapiro J., Benner S., Littlefield K., Kumar S., Naik H.M., Betenbaugh M., Shrestha R., Wu A., Hughes R., Burgess I., Caturegli P., Laeyendecker O., Quinn T., Sullivan D., Shoham S., Redd A., Bloch E., Casadevall A., Tobian A. Sex, age, and hospitalization drive antibody responses in a COVID-19 convalescent plasma donor population. medRxiv. 2020;2020 doi: 10.1172/JCI142004. Aug 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Sheffield C.-G.G., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupferschmidt K. The pandemic virus is slowly mutating. But does it matter? Science. 2020;369:238–239. doi: 10.1126/science.369.6501.238. [DOI] [PubMed] [Google Scholar]
- Lagunas-Rangel F.A., Chavez-Valencia V. High IL-6/IFN-gamma ratio could be associated with severe disease in COVID-19 patients. J. Med. Virol. 2020;2020 doi: 10.1002/jmv.25900. Apr 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalmuanawma S., Hussain J., Chhakchhuak L. Applications of machine learning and artificial intelligence for Covid-19 (SARS-CoV-2) pandemic: a review. Chaos Solitons Fractals. 2020;139 doi: 10.1016/j.chaos.2020.110059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert P.H., Ambrosino D.M., Andersen S.R., Baric R.S., Black S.B., Chen R.T., Dekker C.L., Didierlaurent A.M., Graham B.S., Martin S.D., Molrine D.C., Perlman S., Picard-Fraser P.A., Pollard A.J., Qin C., Subbarao K., Cramer J.P. Consensus summary report for CEPI/BC March 12-13, 2020 meeting: assessment of risk of disease enhancement with COVID-19 vaccines. Vaccine. 2020;38:4783–4791. doi: 10.1016/j.vaccine.2020.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi M., Padovani A., McArthur J.C. Neurological manifestations associated with COVID-19: a review and a call for action. J. Neurol. 2020;267:1573–1576. doi: 10.1007/s00415-020-09896-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Manwani B., Leng S.X. Frailty, inflammation, and immunity. Aging Dis. 2011;2:466–473. [PMC free article] [PubMed] [Google Scholar]
- Li X., Fan L., Leng S.X. The aging tsunami and senior healthcare development in China. J. Am. Geriatr. Soc. 2018;66:1462–1468. doi: 10.1111/jgs.15424. [DOI] [PubMed] [Google Scholar]
- Li L., Zhang W., Hu Y., Tong X., Zheng S., Yang J., Kong Y., Ren L., Wei Q., Mei H., Hu C., Tao C., Yang R., Wang J., Yu Y., Guo Y., Wu X., Xu Z., Zeng L., Xiong N., Chen L., Wang J., Man N., Liu Y., Xu H., Deng E., Zhang X., Li C., Wang C., Su S., Zhang L., Wang J., Wu Y., Liu Z. Effect of convalescent plasma therapy on time to clinical improvement in patients with severe and life-threatening COVID-19: a randomized clinical trial. JAMA. 2020;324:460–470. doi: 10.1001/jama.2020.10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Lu L., Cao W., Li T. Hypothesis for potential pathogenesis of SARS-CoV-2 infection-a review of immune changes in patients with viral pneumonia. Emerg. Microbes Infect. 2020;9:727–732. doi: 10.1080/22221751.2020.1746199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingappan K., Karmouty-Quintana H., Davies J., Akkanti B., Harting M.T. Understanding the age divide in COVID-19: why are children overwhelmingly spared? Am. J. Physiol. Lung Cell Mol. Physiol. 2020;319:L39–L44. doi: 10.1152/ajplung.00183.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Liu W., Zheng Y., Jiang X., Kou G., Ding J., Wang Q., Huang Q., Ding Y., Ni W., Wu W., Tang S., Tan L., Hu Z., Xu W., Zhang Y., Zhang B., Tang Z., Zhang X., Li H., Rao Z., Jiang H., Ren X., Wang S., Zheng S. A preliminary study on serological assay for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 238 admitted hospital patients. Microbes Infect. 2020;22:206–211. doi: 10.1016/j.micinf.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Pang Y., Hu Z., Wu M., Wang C., Feng Z., Mao C., Tan Y., Liu Y., Chen L., Li M., Wang G., Yuan Z., Diao B., Wu Y., Chen Y. Thymosin alpha 1 (Talpha1) reduces the mortality of severe COVID-19 by restoration of lymphocytopenia and reversion of exhausted T cells. Clin. Infect. Dis. 2020;2020 doi: 10.1093/cid/ciaa630. May 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurie N., Sharfstein J.M., Goodman J.L. The development of COVID-19 vaccines: safeguards needed. JAMA. 2020;324:439–440. doi: 10.1001/jama.2020.12461. [DOI] [PubMed] [Google Scholar]
- Lyons D.M., Lauring A.S. 2018. Mutation and Epistasis in Influenza Virus Evolution; p. 10. Viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackman N., Antoniak S., Wolberg A.S., Kasthuri R., Key N.S. Coagulation abnormalities and thrombosis in patients infected with SARS-CoV-2 and other pandemic viruses. Arterioscler. Thromb. Vasc. Biol. 2020;40:2033–2044. doi: 10.1161/ATVBAHA.120.314514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio M., Guralnik J.M., Longo D.L., Ferrucci L. Interleukin-6 in aging and chronic disease: a magnificent pathway. J. Gerontol. A Biol. Sci. Med. Sci. 2006;61:575–584. doi: 10.1093/gerona/61.6.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magro C., Mulvey J.J., Berlin D., Nuovo G., Salvatore S., Harp J., Baxter-Stoltzfus A., Laurence J. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl. Res. 2020;220:1–13. doi: 10.1016/j.trsl.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manners C., Larios Bautista E., Sidoti H., Lopez O.J. Protective adaptive immunity against severe acute respiratory syndrome coronaviruses 2 (SARS-CoV-2) and implications for vaccines. Cureus. 2020;12:e8399. doi: 10.7759/cureus.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., Mallal S., Lammers M., Rubiro P., Quiambao L., Sutherland A., Yu E.D., da Silva Antunes R., Greenbaum J., Frazier A., Markmann A.J., Premkumar L., de Silva A., Peters B., Crotty S., Sette A., Weiskopf D. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K., Manne S., Chen Z., Huang Y.J., Reilly J.P., Weisman A.R., Ittner C.A.G., Kuthuru O., Dougherty J., Nzingha K., Han N., Kim J., Pattekar A., Goodwin E.C., Anderson E.M., Weirick M.E., Gouma S., Arevalo C.P., Bolton M.J., Chen F., Lacey S.F., Ramage H., Cherry S., Hensley S.E., Apostolidis S.A., Huang A.C., Vella L.A., Unit U.P.C.P., Betts M.R., Meyer N.J., Wherry E.J. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael T.M., Currie D.W., Clark S., Pogosjans S., Kay M., Schwartz N.G., Lewis J., Baer A., Kawakami V., Lukoff M.D., Ferro J., Brostrom-Smith C., Rea T.D., Sayre M.R., Riedo F.X., Russell D., Hiatt B., Montgomery P., Rao A.K., Chow E.J., Tobolowsky F., Hughes M.J., Bardossy A.C., Oakley L.P., Jacobs J.R., Stone N.D., Reddy S.C., Jernigan J.A., Honein M.A., Clark T.A., Duchin J.S., Public H.-S., King County, E., Team, C.C.-I Epidemiology of Covid-19 in a Long-term care facility in King County, Washington. N. Engl. J. Med. 2020;382:2005–2011. doi: 10.1056/NEJMoa2005412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckiff B.J., Ramirez-Suastegui C., Fajardo V., Chee S.J., Kusnadi A., Simon H., Grifoni A., Pelosi E., Weiskopf D., Sette A., Ay F., Seumois G., Ottensmeier C.H., Vijayanand P. Single-cell transcriptomic analysis of SARS-CoV-2 reactive CD4 (+) T cells. bioRxiv. 2020;2020 doi: 10.1101/2020.06.12.148916. Jun 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minaee S., Kafieh R., Sonka M., Yazdani S., Jamalipour Soufi G. Deep-COVID: predicting COVID-19 from chest X-ray images using deep transfer learning. Med. Image Anal. 2020;65 doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal A., Manjunath K., Ranjan R.K., Kaushik S., Kumar S., Verma V. COVID-19 pandemic: insights into structure, function, and hACE2 receptor recognition by SARS-CoV-2. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.B., June C.H. Cytokine release syndrome in severe COVID-19. Science. 2020;368:473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- Moratto D., Giacomelli M., Chiarini M., Savare L., Saccani B., Motta M., Timpano S., Poli P., Paghera S., Imberti L., Cannizzo S., Quiros-Roldan E., Marchetti G., Badolato R. Immune response in children with COVID-19 is characterized by lower levels of T-cell activation than infected adults. Eur. J. Immunol. 2020;2020 doi: 10.1002/eji.202048724. Jun 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss P. The ancient and the new": is there an interaction between cytomegalovirus and SARS-CoV-2 infection? Immun. Ageing. 2020;17:14. doi: 10.1186/s12979-020-00185-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidleman J., Luo X., Frouard J., Xie G., Gurjot G., Stein E.S., McGregor M., Ma T., George A.F., Kosters A., Greene W.C., Vasquez J., Ghosn E., Lee S., Roan N.R. SARS-CoV-2-specific T cells exhibit unique features characterized by robust helper function, lack of terminal differentiation, and high proliferative potential. bioRxiv. 2020;2020 doi: 10.1101/2020.06.08.138826. Jun 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea M.G., Dominguez-Andres J., Barreiro L.B., Chavakis T., Divangahi M., Fuchs E., Joosten L.A.B., van der Meer J.W.M., Mhlanga M.M., Mulder W.J.M., Riksen N.P., Schlitzer A., Schultze J.L., Stabell Benn C., Sun J.C., Xavier R.J., Latz E. Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 2020;20:375–388. doi: 10.1038/s41577-020-0285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng O.W., Chia A., Tan A.T., Jadi R.S., Leong H.N., Bertoletti A., Tan Y.J. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34:2008–2014. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L., Ye F., Cheng M.L., Feng Y., Deng Y.Q., Zhao H., Wei P., Ge J., Gou M., Li X., Sun L., Cao T., Wang P., Zhou C., Zhang R., Liang P., Guo H., Wang X., Qin C.F., Chen F., Dong C. Detection of SARS-CoV-2-Specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52:971–977. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoli F., Solis-Soto M.T., Paudel D., Marconi P., Gavioli R., Appay V., Caputo A. Age-related decline of de novo T cell responsiveness as a cause of COVID-19 severity. Geroscience. 2020;42:1015–1019. doi: 10.1007/s11357-020-00217-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noris M., Benigni A., Remuzzi G. The case of complement activation in COVID-19 multiorgan impact. Kidney Int. 2020;98:314–322. doi: 10.1016/j.kint.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H.L., Chia A., Chang C.X., Leong H.N., Ling K.L., Grotenbreg G.M., Gehring A.J., Tan Y.J., Bertoletti A. Engineering T cells specific for a dominant severe acute respiratory syndrome coronavirus CD8 T cell epitope. J. Virol. 2011;85:10464–10471. doi: 10.1128/JVI.05039-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder G., Rezza G., Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA. 2020;323:1775–1776. doi: 10.1001/jama.2020.4683. [DOI] [PubMed] [Google Scholar]
- Paules C.I., Marston H.D., Eisinger R.W., Baltimore D., Fauci A.S. The pathway to a universal influenza vaccine. Immunity. 2017;47:599–603. doi: 10.1016/j.immuni.2017.09.007. [DOI] [PubMed] [Google Scholar]
- Pawelec G. Age and immunity: what is “immunosenescence”? Exp. Gerontol. 2018;105:4–9. doi: 10.1016/j.exger.2017.10.024. [DOI] [PubMed] [Google Scholar]
- Pawelec G., Weng N.P. Can an effective SARS-CoV-2 vaccine be developed for the older population? Immun. Ageing. 2020;17:8. doi: 10.1186/s12979-020-00180-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., Lopez-Camacho C., Slon-Campos J., Zhao Y., Stuart D., Paeson G., Grimes J., Antson F., Bayfield O.W., Hawkins D.E., Ker D.S., Turtle L., Subramaniam K., Thomson P., Zhang P., Dold C., Ratcliff J., Simmonds P., de Silva T., Sopp P., Wellington D., Rajapaksa U., Chen Y.L., Salio M., Napolitani G., Paes W., Borrow P., Kessler B., Fry J.W., Schwabe N.F., Semple M.G., Baillie K.J., Moore S., Openshaw P.J., Ansari A., Dunachie S., Barnes E., Frater J., Kerr G., Goulder P., Lockett T., Levin R., Cornall R.J., Conlon C., Klenerman P., McMichael A., Screaton G., Mongkolsapaya J., Knight J.C., Ogg G., Dong T. Broad and strong memory CD4 (+) and CD8 (+) T cells induced by SARS-CoV-2 in UK convalescent COVID-19 patients. bioRxiv. 2020;2020 doi: 10.1101/2020.06.05.134551. Jun 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen E., Koopmans M., Go U., Hamer D.H., Petrosillo N., Castelli F., Storgaard M., Al Khalili S., Simonsen L. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis. 2020;20:e238–e244. doi: 10.1016/S1473-3099(20)30484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles V.G., Lutgehetmann M., Lindenmeyer M.T., Sperhake J.P., Wong M.N., Allweiss L., Chilla S., Heinemann A., Wanner N., Liu S., Braun F., Lu S., Pfefferle S., Schroder A.S., Edler C., Gross O., Glatzel M., Wichmann D., Wiech T., Kluge S., Pueschel K., Aepfelbacher M., Huber T.B. Multiorgan and renal tropism of SARS-CoV-2. N. Engl. J. Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzikowska U., Ding M., Tan G., Zhakparov D., Peng Y., Wawrzyniak P., Wang M., Li S., Morita H., Altunbulakli C., Reiger M., Neumann A.U., Lunjani N., Traidl-Hoffmann C., Nadeau K., O’Mahony L., Akdis C.A., Sokolowska M. Distribution of ACE2, CD147, CD26 and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors. Allergy. 2020;2020 doi: 10.1111/all.14429. Jun 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randad P.R., Pisanic N., Kruczynski K., Manabe Y.C., Thomas D., Pekosz A., Klein S., Betenbaugh M.J., Clarke W.A., Laeyendecker O., Caturegli P.P., Larman H.B., Detrick B., Fairley J.K., Sherman A.C., Rouphael N., Edupuganti S., Granger D.A., Granger S.W., Collins M., Heaney C.D. COVID-19 serology at population scale: SARS-CoV-2-specific antibody responses in saliva. medRxiv. 2020;2020 doi: 10.1101/2020.05.24.20112300. May 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman S., Majeed T., Azam Ansari M., Ali U., Sabit H., Al-Suhaimi E.A. Current scenario of COVID-19 in pediatric age group and physiology of immune and thymus response. Saudi J. Biol. Sci. 2020;27:2567–2573. doi: 10.1016/j.sjbs.2020.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., Barnaby D.P., Becker L.B., Chelico J.D., Cohen S.L., Cookingham J., Coppa K., Diefenbach M.A., Dominello A.J., Duer-Hefele J., Falzon L., Gitlin J., Hajizadeh N., Harvin T.G., Hirschwerk D.A., Kim E.J., Kozel Z.M., Marrast L.M., Mogavero J.N., Osorio G.A., Qiu M., Zanos T.P. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City Area. JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. and the Northwell, C.R.C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E.S., Dufort E.M., Blog D.S., Hall E.W., Hoefer D., Backenson B.P., Muse A.T., Kirkwood J.N., George K.S., Holtgrave D.R., Hutton B.J., Zucker H.A., New York State Coronavirus Response, T COVID-19 testing, epidemic features, hospital outcomes, and household prevalence, New York state-march 2020. Clin. Infect. Dis. 2020;2020 doi: 10.1093/cid/ciaa549. May 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., Zimmer T., Thiel V., Janke C., Guggemos W., Seilmaier M., Drosten C., Vollmar P., Zwirglmaier K., Zange S., Wolfel R., Hoelscher M. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajadi M.M., Habibzadeh P., Vintzileos A., Shokouhi S., Miralles-Wilhelm F., Amoroso A. Temperature, humidity, and latitude analysis to estimate potential spread and seasonality of coronavirus disease 2019 (COVID-19) JAMA Netw. Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.11834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salje H., Tran Kiem C., Lefrancq N., Courtejoie N., Bosetti P., Paireau J., Andronico A., Hoze N., Richet J., Dubost C.L., Le Strat Y., Lessler J., Levy-Bruhl D., Fontanet A., Opatowski L., Boelle P.Y., Cauchemez S. Estimating the burden of SARS-CoV-2 in France. Science. 2020;369:208–211. doi: 10.1126/science.abc3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallard E., Lescure F.X., Yazdanpanah Y., Mentre F., Peiffer-Smadja N. Type 1 interferons as a potential treatment against COVID-19. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Cerrillo I., Landete P., Aldave B., Sanchez-Alonso S., Sanchez-Azofra A., Marcos-Jimenez A., Avalos E., Alcaraz-Serna A., de Los Santos I., Mateu-Albero T., Esparcia L., Lopez-Sanz C., Martinez-Fleta P., Gabrie L., Del Campo Guerola L., Calzada M.J., Gonzalez-Alvaro I., Alfranca A., Sanchez-Madrid F., Munoz-Calleja C., Soriano J.B., Ancochea J., Martin-Gayo E. Differential redistribution of activated monocyte and dendritic cell subsets to the lung associates with severity of COVID-19. medRxiv. 2020;2020 doi: 10.1101/2020.05.13. May 16. [DOI] [Google Scholar]
- Sette A., Crotty S. Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat. Rev. Immunol. 2020;20:457–458. doi: 10.1038/s41577-020-0389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L., Wei J., Xiao H., Yang Y., Qu J., Qing L., Chen L., Xu Z., Peng L., Li Y., Zheng H., Chen F., Huang K., Jiang Y., Liu D., Zhang Z., Liu Y., Liu L. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabari P., Amini M., Moghadami M., Moosavi M. International public health responses to COVID-19 outbreak: a rapid review. Iran. J. Med. Sci. 2020;45:157–169. doi: 10.30476/ijms.2020.85810.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Renia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te Velthuis A.J., Fodor E. Influenza virus RNA polymerase: insights into the mechanisms of viral RNA synthesis. Nat. Rev. Microbiol. 2016;14:479–493. doi: 10.1038/nrmicro.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakar J., Mohanty S., West A.P., Joshi S.R., Ueda I., Wilson J., Meng H., Blevins T.P., Tsang S., Trentalange M., Siconolfi B., Park K., Gill T.M., Belshe R.B., Kaech S.M., Shadel G.S., Kleinstein S.H., Shaw A.C. Aging-dependent alterations in gene expression and a mitochondrial signature of responsiveness to human influenza vaccination. Aging (Albany N. Y.). 2015;7:38–52. doi: 10.18632/aging.100720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanh Le T., Andreadakis Z., Kumar A., Gomez Roman R., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- Viner R.M., Whittaker E. Kawasaki-like disease: emerging complication during the COVID-19 pandemic. Lancet. 2020;395:1741–1743. doi: 10.1016/S0140-6736(20)31129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Sun S., Tai W., Chen J., Geng Q., He L., Chen Y., Wu J., Shi Z., Zhou Y., Du L., Li F. Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J. Virol. 2020:94. doi: 10.1128/JVI.02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Nie J., Wang H., Zhao Q., Xiong Y., Deng L., Song S., Ma Z., Mo P., Zhang Y. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J. Infect. Dis. 2020;221:1762–1769. doi: 10.1093/infdis/jiaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Shen Y., Li M., Chuang H., Ye Y., Zhao H., Wang H. Clinical manifestations and evidence of neurological involvement in 2019 novel coronavirus SARS-CoV-2: a systematic review and meta-analysis. J. Neurol. 2020;267:2777–2789. doi: 10.1007/s00415-020-09974-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M., Haagmans B.L., de Swart R.L., Sette A., de Vries R.D. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5:2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh R.M., Bahl K., Marshall H.D., Urban S.L. Type 1 interferons and antiviral CD8 T-cell responses. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Wu D., Yang X.O. TH17 responses in cytokine storm of COVID-19: an emerging target of JAK2 inhibitor Fedratinib. J. Microbiol. Immunol. Infect. 2020;53:368–370. doi: 10.1016/j.jmii.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., Huang H., Zhang L., Zhou X., Du C., Zhang Y., Song J., Wang S., Chao Y., Yang Z., Xu J., Zhou X., Chen D., Xiong W., Xu L., Zhou F., Jiang J., Bai C., Zheng J., Song Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020 doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Shen C., Li J., Yuan J., Wei J., Huang F., Wang F., Li G., Li Y., Xing L., Peng L., Yang M., Cao M., Zheng H., Wu W., Zou R., Li D., Xu Z., Wang H., Zhang M., Zhang Z., Gao G.F., Jiang C., Liu L., Liu Y. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J. Allergy Clin. Immunol. 2020;146:119–127. doi: 10.1016/j.jaci.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Hamilton R.G., Weng N.P., Xue Q.L., Bream J.H., Li H., Tian J., Yeh S.H., Resnick B., Xu X., Walston J., Fried L.P., Leng S.X. Frailty is associated with impairment of vaccine-induced antibody response and increase in post-vaccination influenza infection in community-dwelling older adults. Vaccine. 2011;29:5015–5021. doi: 10.1016/j.vaccine.2011.04.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip W., Fu H., Chen A.T., Zhai T., Jian W., Xu R., Pan J., Hu M., Zhou Z., Chen Q., Mao W., Sun Q., Chen W. 10 years of health-care reform in China: progress and gaps in universal health coverage. Lancet. 2019;394:1192–1204. doi: 10.1016/S0140-6736(19)32136-1. [DOI] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang B., Liu S., Tan T., Huang W., Dong Y., Chen L., Chen Q., Zhang L., Zhong Q., Zhang X., Zou Y., Zhang S. Treatment with convalescent plasma for critically ill patients with severe acute respiratory syndrome coronavirus 2 infection. Chest. 2020;158:e9–e13. doi: 10.1016/j.chest.2020.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Zhou X., Zhu C., Song Y., Feng F., Qiu Y., Feng J., Jia Q., Song Q., Zhu B., Wang J. Immune phenotyping based on the neutrophil-to-lymphocyte ratio and igg level predicts disease severity and outcome for patients with COVID-19. Front. Mol. Biosci. 2020;7:157. doi: 10.3389/fmolb.2020.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Wang F.S., Silvestre J.S., Arenzana-Seisdedos F., Tang H. Is aberrant CD8+ T cell activation by hypertension associated with cardiac injury in severe cases of COVID-19? Cell. Mol. Immunol. 2020;17:675–676. doi: 10.1038/s41423-020-0454-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J., Qian S., Hong C., Wang F., Liu Y., Wang Z., He Q., Li Z., He B., Zhang T., Fu Y., Ge S., Liu L., Zhang J., Xia N., Zhang Z. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020;2020 doi: 10.1093/cid/ciaa344. Mar 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong N.S., Zheng B.J., Li Y.M., Poon Xie Z.H., Chan K.H., Li P.H., Tan S.Y., Chang Q., Xie J.P., Liu X.Q., Xu J., Li D.X., Yuen K.Y., Peiris Guan Y. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.C., Guan X.H., Li Y.H., Huang J.Y., Jiang T., Hou L.H., Li J.X., Yang B.F., Wang L., Wang W.J., Wu S.P., Wang Z., Wu X.H., Xu J.J., Zhang Z., Jia S.Y., Wang B.S., Hu Y., Liu J.J., Zhang J., Qian X.A., Li Q., Pan H.X., Jiang H.D., Deng P., Gou J.B., Wang X.W., Wang X.H., Chen W. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396:479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus, I., Research, T A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]