Significance
The outer membrane (OM) of Gram-negative bacteria serves as a barrier that protects cells from harmful chemical compounds, including many antibiotics. Understanding how bacteria build this barrier is an important step in engineering strategies to circumvent it. A long-standing mystery in the field is how phospholipids (PLs) are transported from the inner membrane (IM) to the OM. We previously discovered that a mutation in the gene mlaA that causes rapid flow of PLs to the OM, eventually resulting in IM rupture. Here, we found that deletion of the gene yhdP delayed cell death in the mlaA mutant by slowing flow of PLs to the OM. These findings reveal a high-flux diffusive pathway for PL transport in Gram-negative bacteria modulated by YhdP.
Keywords: MlaA, AsmA, lipid transport, Gram-negative cell envelope, cell death
Abstract
The outer membrane (OM) of Gram-negative bacteria is a selective permeability barrier that allows uptake of nutrients while simultaneously protecting the cell from harmful compounds. The basic pathways and molecular machinery responsible for transporting lipopolysaccharides (LPS), lipoproteins, and β-barrel proteins to the OM have been identified, but very little is known about phospholipid (PL) transport. To identify genes capable of affecting PL transport, we screened for genetic interactions with mlaA*, a mutant in which anterograde PL transport causes the inner membrane (IM) to shrink and eventually rupture; characterization of mlaA*-mediated lysis suggested that PL transport can occur via a high-flux diffusive flow mechanism. We found that YhdP, an IM protein involved in maintaining the OM permeability barrier, modulates the rate of PL transport during mlaA*-mediated lysis. Deletion of yhdP from mlaA* reduced the rate of IM transport to the OM by 50%, slowing shrinkage of the IM and delaying lysis. As a result, the weakened OM of ∆yhdP cells was further compromised and ruptured before the IM during mlaA*-mediated death. These findings demonstrate the existence of a high-flux diffusive pathway for PL flow in Escherichia coli that is modulated by YhdP.
The outer membrane (OM) of Gram-negative bacteria is an asymmetric bilayer composed of lipopolysaccharides (LPS) in the outer leaflet and phospholipids (PLs) in the inner leaflet (1). Strong lateral interactions between LPS molecules in the outer leaflet result in a bilayer that is impermeable to both hydrophobic and large hydrophilic compounds (2). In addition to its role as a permeability barrier, β-barrel proteins and lipoproteins in the OM play key roles in a variety of other important processes, including motility, pathogenesis, and cell division (3). Because the periplasm lacks conventional sources of energy, such as adenosine triphosphate (ATP), Gram-negative bacteria face a significant challenge in transporting and assembling OM components. To circumvent this challenge, cells utilize ATP hydrolysis in the inner membrane (IM) to transport LPS molecules across a protein bridge that spans the periplasm (4, 5). β-barrel proteins and lipoproteins also use ATP hydrolysis to cross the IM but are escorted across the periplasm by soluble carriers (6, 7).
While relatively little is known about the transport of PLs to the OM, current understanding points to a mechanism that is highly distinct from the known OM transport pathways. Liposome fusion experiments in Salmonella Typhimurium demonstrated that, unlike proteins and LPS, PL transport is bidirectional and indiscriminate (8). Rapid transfer from the OM to the IM was observed for all major and minor species of Salmonella PLs and even for cholesteryl oleate, which is not a normal component of bacterial membranes (8). One explanation consistent with these findings is that PLs can be transported by diffusive flow. Diffusive PL transport could occur at zones of hemifusion that form spontaneously. Diffusion could also require protein facilitators, for instance to encourage formation of hemifusions or to form protein channels through which PLs flow.
Although the bacterial PL transport pathway is currently unknown, the mechanisms by which cells maintain asymmetry in the OM are much better understood. When the integrity of the outer leaflet is disrupted, PLs from the inner leaflet migrate to fill gaps in the LPS, creating zones that are newly permeable to toxic hydrophobic compounds. The cell remedies this problem using the maintenance of the lipid asymmetry (Mla) pathway, which removes mislocalized PLs from the outer leaflet and shuttles them to the IM (9). MlaA is a donut-shaped lipoprotein that sits in the OM (10), removes PLs from the outer leaflet, and delivers them to the soluble carrier, MlaC. MlaC then transports them across the periplasm to the MlaFEDB complex, an ABC transport system that unloads MlaC and returns PLs to the IM.
In E. coli, a dominant negative mutation in mlaA, called mlaA*, reverses the protein’s normal function (11). Instead of removing surface-exposed PLs, MlaA* allows properly localized PLs to flow through its pore into the outer leaflet (10, 11). Accumulation of PLs in the outer leaflet triggers a cell death pathway that results in lysis during stationary phase (11). First, the presence of PLs in the outer leaflet activates the OM phospholipase PldA, which cleaves surface-exposed PLs, generating breakdown products that signal to increase production of LPS (12). Hyperproduction of LPS destabilizes the OM, resulting in loss of OM material through blebbing. PLs then flow from the IM to the OM to replace the lost material. In stationary phase, cells can no longer synthesize new PLs to replace those lost from the IM. As a result, PL flow causes the IM to shrink and ultimately rupture.
We hypothesized that changing the rate of PL flow from the IM to the OM would affect the rate of lysis in mlaA* cells since PL flow to the OM is what eventually causes the IM to rupture. Hence, we should be able to identify genes that affect PL transport through genetic interactions with mlaA*. Our screen identified yhdP, a gene already known to play a role in maintaining the barrier function of the OM (13). Deletion of yhdP slowed lysis but did not restore wild-type LPS levels, indicating that it affects a step in the pathway after LPS levels have already increased. Single-cell microscopy showed that the IM of mlaA* ΔyhdP cells shrank more slowly, implying slower anterograde flow. In mlaA* cells, PL flow ultimately leads to IM rupture in stationary phase but also compensates for loss of OM material while cells are actively growing. By contrast, without YhdP, the OM ruptured before the IM, suggesting that these cells cannot efficiently compensate for OM loss through anterograde flow.
Results
mlaA* Causes High-Flux Passive Phospholipid Flow.
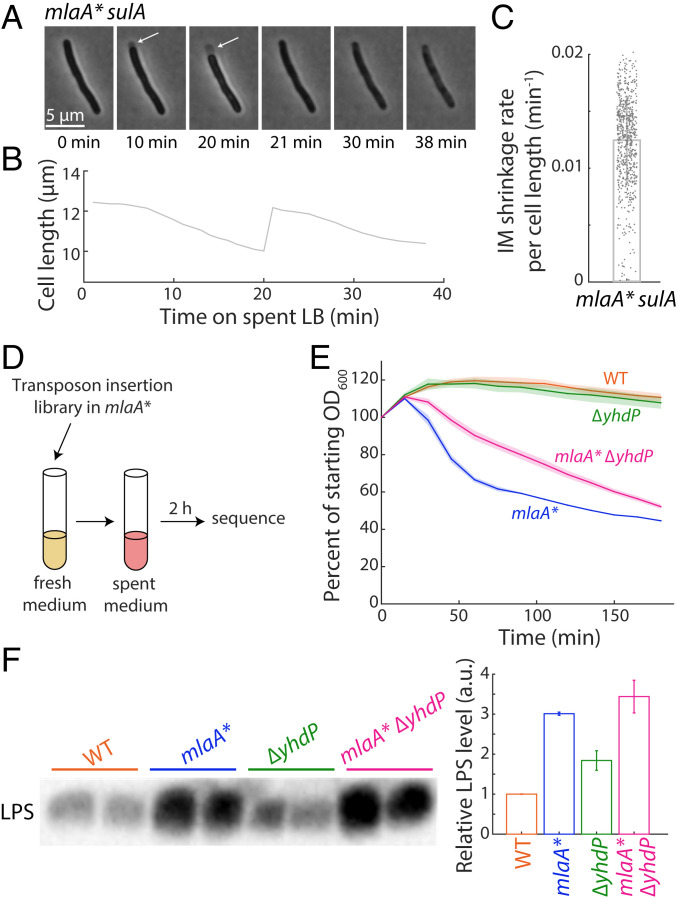
It was previously shown that PL flow in mlaA* cells is not affected by membrane depolarization or ATP synthase mutations, indicating that flow occurs via a passive mechanism (11). To further characterize this pathway, we quantified the rate of anterograde flow. We induced the cell division inhibitor SulA (14) and then transitioned exponentially growing cells onto agarose pads containing spent medium to cause the mlaA* death phenotype. The SulA-induced cells became filamentous, and hence we could quantify the IM shrinkage from one of the poles prior to cell death (Fig. 1A, white arrows) more easily than in nonfilamentous cells. Since IM shrinkage in mlaA* cells is the result of PL transport to the OM (11), we measured the rate of shrinkage as a proxy for the PL transport rate.
Fig. 1.
Deletion of yhdP slows the rate of mlaA* lysis. (A) Exponentially growing mlaA* sulA cells were transitioned onto an agarose pad containing spent medium. In a typical cell, the IM shrank away from the cell wall (white arrows) before the cell eventually popped (t = 21 min) and died. (B) Cell length of the cell in A initially decreased, then rapidly snapped back to approximately the initial size at the time of transition to spent medium, and finally decreased due to leakage. (C) During the initial 20 min in spent medium, the IM length shrank ∼1% per minute. Each dot represents a single cell (total n = 677 cells), and the bar represents mean ± SD. (D) Schematic of TraDIS selection. Libraries were grown into late exponential phase and transitioned to spent medium for 2 h to induce lysis. The resulting library was subsequently sequenced for enrichment of mutants. (E) Cultures were grown to late exponential phase at optical density at 600 nm (OD600) ∼0.8, spun down, and resuspended in spent medium to induce lysis. OD600 was measured to determine the rate of lysis. Deletion of yhdP slowed down mlaA*-mediated lysis. Data points are mean ± SD with n = 3 replicates. (F) Overnight cultures were normalized by OD600 and assayed for LPS abundance by immunoblotting. (F, Left) Immunoblotting gel image. (F, Right) Quantification of LPS abundances. Data points are mean ± SD with n = 2 biological replicates. Deletion of yhdP did not affect LPS levels either alone or in combination with mlaA*.
The IM shrunk by ∼20% in ∼20 min (Fig. 1 B and C), corresponding to a PL flow rate of 1.2 ± 0.4% of the cell length per minute. That a substantial fraction of the IM can be transported quickly even under the energetic limitations that occur upon entry into stationary phase provides further evidence that PL flow can occur via a diffusive mechanism. It also shows that the diffusive pathway is high flux, permitting transport of a large proportion of the IM within a short period of time.
Genetic Interactions with mlaA* Depend on the Length of Time in Spent Medium.
To identify genetic interactions with mlaA*, we constructed transposon insertion libraries in mlaA* and ΔmlaA cells. We grew the libraries to late exponential phase and incubated them in spent medium overnight to induce lysis. We repeated this process three times successively, inferring that the survival of any mutant that suppressed mlaA*-mediated cell death would be amplified by the repeated incubations.
As expected, by far the most abundant hit was mlaA since null mutations in mlaA* prevent production of the mutant protein, completely suppressing cell death (11). The next most abundant hit was pldA, again expected as without PldA there is no signal to increase production of LPS (11). After three rounds of incubation, insertions in mlaA and pldA accounted for 96.3% of all reads. Among the other hits (Table 1), several were known to affect LPS levels, corresponding to the results of a previous low-throughput screen, which also identified several suppressor mutations that lowered LPS levels (11). Since overproduction of LPS is a critical step in the cell-death pathway, mutations that restore wild-type LPS levels are expected to suppress lysis independent of any potential impact on PL transport (11). We therefore sought to find a genetic disruption that suppressed mlaA* without lowering LPS levels.
Table 1.
Percentage of reads in the mlaA* library mapping to suppressor genes following three successive overnight incubations in spent medium
| Gene name | Percentage of total reads, incubation 1 | Percentage of total reads, incubation 2 | Percentage of total reads, incubation 3 |
| mlaA | 48.1 | 84.1 | 86.0 |
| pldA | 14.7 | 9.8 | 10.3 |
| lptC | 8.6 | 0.9 | 0 |
| dsbA | 4.5 | 0 | 0 |
| yaiP | 1.3 | 2.7 | 1.7 |
| acs | 0.7 | 0 | 0 |
| fadE | 0.6 | 0 | 0 |
| secA | 0.5 | 0 | 0 |
| asmA | 0.3 | 0 | 0 |
| yejM | 0.2 | 0 | 0 |
Genomic DNA was extracted from the mlaA* library following overnight incubation, which induces lysis, and transposon junctions were sequenced. Reads were mapped to the E. coli MC4100 genome and open reading frames were quantified to identify gene disruptions that were enriched after each incubation and hence were potential suppressors. The two known strongest suppressors of mlaA*, mlaA and pldA, quickly predominated in the culture.
Since the most potent suppressors of mlaA* block the earliest steps of the pathway, we hypothesized that slowing PL flow, the final step in the pathway, would only slow lysis. Hence, we carried out a similar experiment in which cells were only incubated for 2 h in spent medium rather than overnight to identify partial suppressors of mlaA* (Fig. 1D and Table 2). Now, the most abundant hit in the mlaA* library was yhdP, which encodes a large (1,266 amino acid) IM protein. Interestingly, another member of its protein family, asmA, was identified as a suppressor in the previous screen (Table 1). YhdP has been shown to enhance the OM permeability barrier function during stationary phase, but its mechanism is currently unknown (13). Deletion of yhdP causes sensitivity to sodium dodecyl sulfate/(ethylenedinitrilo)tetraacetic acid (SDS/EDTA) and vancomycin regardless of growth phase, indicating that it plays a role in maintaining OM integrity (15). To confirm that disruption of yhdP inhibits lysis, we grew mlaA* ΔyhdP cells to late exponential phase, resuspended them in spent medium, and measured OD600 over time; deletion of yhdP slowed the rate of lysis of mlaA* cells (Fig. 1E).
Table 2.
Number of reads in the mlaA* and ∆mlaA libraries mapping to various genes after a 2-h incubation in spent medium
| Gene name | No. of reads – mlaA* | No. of reads – ΔmlaA | log2(mlaA*/ΔmlaA) |
| yhdP | 2,512,727 | 49,893 | 5.7 |
| cyaA | 1,153,018 | 83,061 | 3.8 |
| mlaA | 680,035 | N/A | N/A |
| cysG | 348,756 | 13,056 | 4.7 |
| rbsD | 215,556 | 13,903 | 4.0 |
| sdhA | 139,279 | 16,305 | 3.1 |
| rssB | 138,738 | 3885 | 5.2 |
Genomic DNA was extracted from the mlaA* and ΔmlaA libraries following 2 h of incubation in spent medium, and transposon junctions were sequenced. Reads were mapped to the E. coli MC4100 genome, and open reading frames were quantified. The most abundant gene disruption in the mlaA* library was yhdP.
Deletion of yhdP Slows mlaA* Lysis without Lowering LPS Levels.
Since modulating PL flow would affect a step in the cell death pathway after LPS levels have already increased, we expected that inhibiting PL flow would slow mlaA* lysis without restoring wild-type LPS levels. To test the effects of yhdP deletion on mlaA* cells, we measured LPS levels using immunoblotting (Methods). Deletion of yhdP had no effect on LPS levels either alone or in combination with mlaA*, suggesting that it affects a later step in the pathway (Fig. 1F). In addition, this finding suggests that yhdP does not slow lysis by affecting LPS transport as it has been shown that slowing transport of LPS also reduces LPS levels in mlaA* cells (12).
Deletion of yhdP Slows Shrinking of the IM.
In mlaA* cells, shrinking of the IM away from the cell pole is thought to reflect anterograde PL flow to the OM (11). We therefore expected that a mutation that slows PL flow would also slow IM shrinking. To determine whether yhdP deletion affects PL flow, we imaged mlaA* or mlaA* ∆yhdP cells during incubation in a microfluidic flow cell. Cells were first kept in lysogeny broth (LB) until they reached steady-state growth and then rapidly switched into spent medium. With continuous flow of spent medium, all mlaA* cells died within 20–30 min (11). Deletion of yhdP delayed cell death (Fig. 2A), consistent with the dynamics in bulk culture (Fig. 1E).
Fig. 2.
Deletion of yhdP slows shrinking of the IM during transition to spent medium. (A) mlaA* and mlaA* ∆yhdP cells were separately incubated in a microfluidic flow cell and transitioned from fresh LB to spent medium to induce cell death. Consistent with bulk measurements, deletion of yhdP slowed down cell death. Data points are mean ± SD with n = 3 replicates of at least 50 cells in each experiment. (B) Representative single-cell traces after switching to spent medium. (C) Deletion of yhdP reduced total shrinkage in mlaA* cells (P < 10−10, n > 100 cells, and two-tailed Student’s t test). (D) During the popping immediately preceding lysis, mlaA* and mlaA* ∆yhdP cells returned to approximately their initial length prior to the transition to spent medium (compare length expansion to the shrinkage in C; mlaA* cells exhibited more expansion than mlaA* ∆yhdP cells, P < 10−10, n > 100 cells, and two-tailed Student’s t test). (E) Deletion of yhdP slowed down the shrinkage rate of mlaA* cells by ∼50% (P < 10−10, n > 100 cells, and two-tailed Student’s t test). In C–E, each dot represents a single cell, and the bar plots represent mean ± SD.
For both strains, the cytoplasm started to shrink immediately upon the transition to spent medium. The cytoplasm of mlaA* cells shrank by an average of ∼0.7 µm (20%, Fig. 2 B and C) over ∼20 min (Fig. 2A) and appeared to increase in density, followed by a “popping” expansion and then gradual loss of phase contrast (Fig. 2B) that we previously characterized as typical of mlaA*-mediated death (11). mlaA* ∆yhdP cells displayed a qualitatively similar death trajectory (Fig. 2B). The average time to lysis was longer (29 min, Fig. 2A) and yet less shrinkage occurred (0.5 µm, 15%, Fig. 2 B and C) before popping than in mlaA* cells. In both strains, the expansion at cell death roughly restored cell length to the preshrinkage size (Fig. 2 C and D), suggesting that the cell envelope returned to a relaxed state after the expansion. Shrinkage rate prior to popping was also slowed down in mlaA* ∆yhdP cells by 50% (Fig. 2E). Taken together, these data indicate that YhdP plays an important role in PL transport during mlaA*-mediated lysis.
The Effect of YhdP on Lysis Is Cyclic Enterobacterial Common Antigen Independent.
It was previously shown that the OM permeability phenotypes of ΔyhdP cells can be suppressed by preventing synthesis of cyclic enterobacterial common antigen (ECA), indicating that YhdP regulates cyclic ECA (15). To test whether the effect of yhdP deletion on mlaA*-mediated lysis also depends on cyclic ECA, we constructed strains lacking wzzE. WzzE is the ECA chain length regulator, and in its absence cyclic ECA is not synthesized. If the effect of yhdP deletion on the lysis rate also depends on cyclic ECA, we would expect that deleting wzzE in mlaA* ΔyhdP cells would reverse the effect of yhdP deletion, resulting in dynamics upon transition to spent medium similar to those of mlaA* alone.
To quantify the effect of cyclic ECA in mlaA* cells, we imaged mlaA* ΔwzzE or mlaA* ∆yhdP ∆wzzE cells in a microfluidic device during the transition to spent medium. Deletion of wzzE did not restore mlaA*-like death dynamics (Fig. 3A) nor did it change the shrinkage rate of the mlaA* ∆yhdP strain (Fig. 3B). Deletion of wzzE did not affect the death (Fig. 3A) or shrinkage (Fig. 3B) of mlaA* cells, indicating that the effect of YhdP on PL transport during mlaA*-mediated lysis does not require cyclic ECA.
Fig. 3.
Cyclic ECA is not responsible for suppression of death by ΔyhdP. (A) Cells were incubated in a microfluidic flow cell and transitioned from fresh LB to spent medium to induce cell death. Deletion of the cyclic ECA biosynthesis gene wzzE did not restore mlaA*-like lysis dynamics to mlaA* ∆yhdP. mlaA* ∆yhdP ∆wzzE cells exhibited distinct and slower death dynamics compared to mlaA* cells, while deletion of wzzE from mlaA* slightly accelerated cell death. Data points are mean ± SD with n = 3 replicates. (B) Deletion of wzzE did not alter the shrinkage rate of mlaA* ∆yhdP cells (n > 100 cells) and only slightly reduced the rate in mlaA* cells, indicating that the effect of YhdP on lysis is cyclic ECA-independent. Each dot represents a single cell (n > 100 cells for each strain), and the bar plots represent mean ± SD. P values are from two-tailed Student’s t tests.
Deletion of yhdP Weakens the OM Chemically and Mechanically.
Another explanation for how deletion of yhdP could slow lysis is by preventing loss of OM material. To test whether deleting yhdP improves OM integrity in mlaA* cells, we assayed OM permeability by plating on vancomycin or SDS/EDTA. It was previously shown that cells lacking yhdP are vancomycin sensitive (15). However, by plating on a low concentration of vancomycin such that wild-type, mlaA*, and ΔyhdP cells all grew to the same dilution as on LB without drugs, we observed that mlaA* ΔyhdP cells had a synthetic OM permeability defect (Fig. 4A). On SDS/EDTA, mlaA* and ΔyhdP were both sensitive; combining the two mutations did not relieve the defect (Fig. 4A). These results demonstrate that deletion of yhdP does not slow lysis by enhancing OM integrity.
Fig. 4.
Deletion of yhdP chemically and mechanically disrupts the OM. (A) Overnight cultures were normalized by OD600, serially diluted, and plated on LB, LB + 20 µg/mL vancomycin, and LB + 0.5% SDS/0.5 mM EDTA. mlaA* and ∆yhd had a synthetic permeability defect with vancomycin, and neither mlaA* nor mlaA* ∆yhdP cells grew with SDS/EDTA. (B) Exponentially growing cells were loaded into a microfluidic device and allowed to grow in LB before being exposed to a large hyperosmotic shock with 3 M sorbitol, then treated with EDTA in the presence of sorbitol. The length of the fluorescently labeled cell wall was tracked. Sorbitol ("sorb") treatment relieved turgor pressure and reduced cell-wall length. EDTA treatment disrupted the OM and led to a further decrease in cell length. In both conditions, ∆yhdP cells shrank more compared to wild-type cells. (C and D) Length contraction upon sorbitol (C) and EDTA (D) treatment for cells in B. In both conditions, ∆yhdP cells shrank more than wild type, indicating a mechanically weakened OM. Individual dots are data from single cells (n > 50 for each strain), and bar plots represent mean ± SD. P values are from two-tailed Student’s t tests. (E) Spheroplasts were generated overnight in the presence of cefsulodin, washed, and plated on fresh media. Both mlaA* and ∆yhdP exhibited a mechanically weakened OM and reduced spheroplast survival rates. Deletion of wzzE partially rescued the mechanical defect in mlaA* ∆yhdP cells. Dots represent biological replicates (n >= 3 replicates for each strain), and the bar plots are mean ± SD. P values are from one-tailed Student’s t tests.
Since deleting yhdP increased OM permeability in mlaA* cells, we wondered whether inhibition of anterograde flow might be due to destabilization of the OM. To further characterize the effect of YhdP on the OM, we investigated its impact on OM mechanical strength. In a previous study, we showed that the mechanical stiffness of the E. coli OM is greater than or comparable to that of the cell wall and that genetic or chemical perturbations to the OM can reduce the overall stiffness of cells (16). To determine if YhdP plays a role in determining OM stiffness, we utilized an assay in which exponentially growing cells are first exposed to a large hyperosmotic shock with 3 M sorbitol and then treated with EDTA. We used a microfluidic flow cell to precisely control the timing of treatments and track single cells throughout (Methods). Upon the shock, wild-type cells experienced a large decrease in the length of the fluorescently labeled cell wall (Fig. 4B) as expected since turgor pressure was relieved and hence the cell wall-OM envelope complex was no longer under stress. EDTA treatment, which disrupts the OM by rapidly inducing loss of LPS molecules (17, 18), led to a further decrease in cell length (Fig. 4B), signifying that the stiff OM was holding the cell wall out beyond its rest length before its removal. Application of this assay to ∆yhdP cells showed greater contraction of the cell wall after the osmotic shock (Fig. 4 B and C) and after EDTA treatment (Fig. 4 B and D), indicating that the overall stiffness of ∆yhdP cells was lower than that of wild type.
To further test whether deletion of yhdP weakened cells mechanically, we quantified the yield of viable cells after breaking down the cell wall using β-lactam antibiotics to form wall-less spheroplasts with intact IM and OM (Methods). We previously showed that survival of spheroplasts is strongly correlated with the stiffness of the OM across chemical and genetic perturbations (16). In this assay, spheroplasts were generated overnight in the presence of cefsulodin and then were washed and plated on fresh medium without antibiotics after the cell wall was removed. Survival in the absence of a cell wall relies on having a stiff OM to bear the stress of turgor. We observed that mlaA* and yhdP deletion each caused a dramatic (>1,000-fold) decrease in spheroplast viability in comparison with wild type (Fig. 4E). The mlaA* ΔyhdP double mutant exhibited a further decrease in spheroplast viability, highlighting the importance of YhdP in determining OM stiffness. However, deletion of wzzE partially suppressed the decrease in spheroplast viability due to ∆yhdP (Fig. 4E), demonstrating that the effect of YhdP on OM mechanical strength is cyclic ECA-dependent.
Taken together, these results suggest that deleting yhdP does not slow lysis by preventing loss of OM material. Deletion of yhdP severely disrupts OM integrity, which is more likely to promote loss of OM material than to prevent it. Furthermore, yhdP deletion still slows lysis even when its effect on the mechanical strength of the OM is suppressed (Fig. 3 A and B), indicating that yhdP’s effect on lysis is not a result of its effect on OM mechanics.
Impairment of Phospholipid Flow Leads to OM Rupture.
We observed that the IM of mlaA* ∆yhdP cells shrank more slowly and less relative to mlaA* (Fig. 2 B–E). We would expect that a mutation that decreases PL flow would cause the IM to shrink more slowly. To explain why the IM shrank less before lysis, we wondered whether, in these cells, lysis occurs for a reason other than IM rupture. In mlaA*, anterograde flow leads to rupture of the IM, followed shortly by OM rupture (11). We surmised that impairing PL flow in mlaA* cells would increase the stress on the OM, potentially causing the OM to rupture before the IM.
To test this hypothesis, we constructed mlaA* and mlaA* ∆yhdP strains expressing both a cytoplasmic and a periplasmic fluorescent protein. When the mlaA* strain was shifted into spent medium, shrinkage of the IM led to a large periplasmic space with a high mCherry signal (Fig. 5A, white arrow). The mCherry signal remained intact throughout shrinkage, and when the cells popped and lysed, periplasmic mCherry and cytoplasmic GFP signals were lost simultaneously in every cell (Fig. 5 A and B), presumably because rupture of the IM led to rapid OM rupture (11). By contrast, in mlaA* ∆yhdP cells, the extent of IM shrinkage was much smaller (Fig. 2C), and the periplasmic mCherry signal remained largely uniform around cell periphery rather than intensified at a cell pole(s) (Fig. 5C). During the transition to spent medium, the mCherry signal was lost tens of minutes before popping (Fig. 5 C and D), while the cytoplasmic YFP signal remained intact until popping occurred (Fig. 5C). Taken together, these data indicate that disruption of anterograde flow caused by yhdP deletion in mlaA* ∆yhdP cells leads to rupture of the OM before the IM (Fig. 5E).
Fig. 5.
Deletion of yhdP from mlaA* cells causes the OM to rupture before the IM in spent medium. (A) Death trajectory of mlaA* cells on an agarose pad with spent medium. Cells were labeled with periplasmic mCherry and cytoplasmic green fluorecent protein (GFP). During shrinkage, PLs flowed from the IM to the OM, causing the IM to shrink away from the cell wall and OM. As a result, periplasmic mCherry was enriched at one cell pole (white arrow). At the time of popping, both fluorescence signals were lost in the same frame. (Scale bar: 1 µm.) (B) During the transition to spent medium, mCherry and GFP signals were lost simultaneously in all mlaA* cells (n = 27). The dots represent single cells, and the black line is x = y. The dots are slightly jittered to visualize overlapping data. (C) Death trajectory of mlaA* ∆yhdP cells on an agarose pad with spent medium. Cells were labeled with periplasmic mCherry and cytoplasmic yellow fluorescent protein (YFP). During the period of shrinkage (52–87 min), the IM did not shrink away from cell wall and OM, as shown by the uniform mCherry signal around the cell periphery. The cell also lost its periplasmic mCherry signal tens of minutes before losing cytoplasmic YFP signal, suggesting that the OM ruptured before the IM. (Scale bar: 1 µm.) (D) During the transition to spent medium, the mCherry signal was lost at least 2 min before the YFP signal in n = 74 (out of 110) mlaA* ∆yhdP cells, indicating that deletion of yhdP leads to rupture of the OM before the IM. In all other cells, both signals were lost simultaneously. The dots represent single cells, and the black line is x = y. The dots are slightly jittered to visualize overlapping data. (E) Model of ∆yhdP-mediated death. The mlaA* mutation leads to membrane loss via OM vesicles and disrupts PL homeostasis during the transition into stationary phase. In the mlaA* background (Top), PLs flow from the IM to the OM to replenish the membrane loss, causing the IM to shrink away from OM and eventually leading to cell death through IM rupture. By contrast, in mlaA* ∆yhdP cells (Bottom), deletion of yhdP suppresses PL flow, leading to further weakening of an already compromised OM that ruptures before the IM.
Discussion
The existence of fusion junctions facilitating PL flow between the IM and the OM has been a matter of controversy for some time. In the 1960s, electron microscopy showed sites of contact between the two membranes, but improved microscopy methods called into question the existence of these “Bayer’s junctions” (19, 20). While it may be the case that the junctions observed in those early images were indeed artifacts, several lines of evidence now suggest that intermembrane PL transport can occur via diffusion.
Previous studies showed that PL transport is bidirectional and can involve even nonnative lipids (8, 21). In the mlaA* mutant, PL flow does not require either ATP or proton motive force (11). In addition, in this mutant ∼20% of the IM is lost by transport even under the nutrient limitations that trigger entry into stationary phase. These data are strong evidence that PL flow in mlaA* cells is passive and occurs through a high-flux pathway. It remains to be seen whether this pathway functions in normal PL transport or is active only in certain conditions.
In this study, we provide evidence that YhdP is involved in modulating the high-flux PL transport pathway. Time-lapse imaging showed that deleting yhdP slowed shrinkage of the IM in mlaA* cells (Fig. 2), implying that PLs flowed more slowly from the IM to the OM. In mlaA*, loss of lipids from the IM ultimately causes it to rupture (11). As a result, slowing PL flow delays cell death. However, since PL flow also compensates for loss of OM material, slowing flow from the IM comes at the cost of OM integrity. Thus, while lysis takes longer in mlaA* ∆yhdP cells, when it does occur, the OM rather than the IM ruptures first (Fig. 5).
Cells survive without yhdP, suggesting that YhdP functions specifically in high-flux PL transport. If it does play a role in normal PL transport, then there must be multiple, redundant pathways. How YhdP modulates PL transport is still unknown, but an intriguing possibility is suggested by its protein family. YhdP belongs to a family of six “AsmA-like” proteins (AsmA, TamB, YdbH, YicH, YhjG, and YhdP). Two members of this family, AsmA and TamB, are predicted to share homology with the eukaryotic PL transporter, Vps13 (22). Vps13 forms a hydrophobic channel through which PLs are transported between membranes (23, 24). The structure of TamB also includes a channel with a highly hydrophobic interior (25). Interestingly, it has been suggested that due to its ability to accommodate many lipids at once, Vps13 functions specifically in high-flux PL transport (26).
While our study does not determine YhdP’s molecular mechanism, it does rule out certain possibilities. Deleting yhdP does not lower LPS levels (Fig. 1F), hence it must affect a step in the mlaA* death pathway after LPS levels have already increased. Moreover, the effect of yhdP deletion on mlaA* lysis cannot be explained by slowed transport of LPS to the OM as it has previously been shown that slowing LPS transport also decreases LPS levels in mlaA* cells (12). It is also unlikely that deleting yhdP slowed lysis (Fig. 2 A and B) by preventing loss of OM material as yhdP deletion has a severe negative impact on OM integrity (Fig. 4). Of the remaining options, a direct role in transport is certainly the simplest. YhdP is a large (1,266 amino acid) IM protein with one clear N-terminal and possibly a second C-terminal transmembrane domain. Given the size of its periplasmic domain, it is plausible that YhdP can span the periplasm, but further structural and biochemical studies are needed to determine its precise role in anterograde PL transport. Regardless, our data provide new insight into the process of PL flow and cell lysis caused by the dominant negative mlaA* allele and shed light on the multiple roles played by YhdP in the maintanence of OM integrity. The fact that YhdP changes both OM stiffness and permeability suggests an intriguing link between these two properties. Our discovery of a mutant capable of slowing PL transport should provide a useful foothold in the investigation of this poorly understood pathway.
Methods
Bacterial Strains.
The strains used in this study are listed in SI Appendix, Table S1. Strains were constructed by generalized P1 transduction with all deletions originating from the Keio collection (27, 28). Kanamycin resistance cassettes were removed using the Flp recombinase system as previously described (29). Overnight cultures were grown at 37 °C in LB medium supplemented with 10 mM MgSO4 to prevent mlaA* lysis and diluted into unsupplemented LB for subsequent experiments. When necessary, media were supplemented with 25 µg/mL kanamycin or 25 µg/mL tetracycline.
TraDIS Sample Preparation.
Transposon mutant libraries were constructed using the EZ-Tn5 <KAN-2>TnP Transposome Kit (Epicentre) according to the manufacturer’s instructions. When preparing electrocompetent cells, overnight cultures were grown in LB supplemented with 5 mM MgSO4 to prevent lysis of mlaA* and then subcultured in 2xYT medium. Following electroporation, cells were plated on LB+25 µg/mL kanamycin plates supplemented with 5 mM MgSO4. Approximately 300,000 and 150,000 colonies were pooled to construct the mlaA* and ∆mlaA libraries, respectively. Genomic DNA was extracted from samples of 2 × 109 cells after lysis using the DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer’s instructions. Libraries were prepared according to the TraDIS method (30) and sequenced on Illumina HiSeq 2500 rapid flowcells as single-end 75-nucleotide reads.
TraDIS Data Analysis.
Sequencing reads were mapped to the E. coli K12 genome using BWA v. 1.2.3. Mapped reads were quantified using HTSeq-count v. 0.6.0. The integrative genomics viewer was used to visualize the mapped reads.
Lysis Curves.
To generate spent medium, wild-type (MC4100) cultures were grown for 24 h in LB at 37 °C, cells were pelleted, and the supernatant was filter-sterilized using a 0.2-µm filter. All experiments were conducted using wild-type spent medium. To assay the rate of lysis, cultures were grown until OD600∼0.8, pelleted, and resuspended in spent medium. Cultures were then incubated at 37 °C, and OD600 was measured at 15-min intervals.
Immunoblot Analyses.
The equivalent of 1 mL of culture at OD600∼1 was taken from overnight cultures, pelleted, and resuspended in LDS sample buffer (Invitrogen). Samples were boiled for 10 min and allowed to cool. Samples were loaded on 4–12% SDS/polyacrylamide gel electrophoresis (PAGE) gels and run at 100 V. LPS was then transferred to nitrocellulose membranes and blocked in 5% nonfat dried milk for 1 h at room temperature. Membranes were then incubated overnight at 4 °C with anti-LPS antibody (1:400,000; Hycult Biotech) in milk. Membranes were washed and incubated with a secondary antibody for 1 h at room temperature (1:20,000; goat anti-mouse immunoglobulin G (H+L)-horseradish peroxidase conjugate; Bio-Rad).
Efficiency of Plating Assay.
Cultures were grown overnight in LB+10 mM MgSO4, standardized by OD600, and serially diluted. Dilutions were then transferred to plates using a 96-well-plate replica plater and incubated overnight at 37 °C.
Single-Cell Imaging.
Cells were imaged on a Nikon Eclipse Ti-E inverted fluorescence microscope with a 100X (numerical aperture [NA] 1.40) oil-immersion objective (Nikon Instruments). Images were collected on a DU885 electron-multiplying charged couple device camera (Andor Technology) or a Neo scientific complementary metal–oxide–semiconductor camera (Andor Technology) using µManager version 1.4 (https://micro-manager.org) (31). Cells were maintained at 37 °C during imaging with an active-control environmental chamber (HaisonTech).
For experiments conducted on agarose pads, 1 µL of cells was spotted onto a pad of 1% agarose in fresh LB or spent medium. For transition experiments, exponentially growing cells were washed three times in spent medium before spotting. Flow-cell experiments were performed in ONIX B04A microfluidic chips (CellASIC), and medium was exchanged using the ONIX microfluidic platform (CellASIC).
Imaging in Microfluidic Devices.
Overnight cultures were diluted 100-fold into 1 mL of fresh LB and incubated for 2 h with shaking at 37 °C. ONIX B04A microfluidic plates (CellASIC) were loaded with medium and prewarmed to 37 °C. Cells were loaded into the plate, which was incubated at 37 °C without shaking for 30 min before imaging. As necessary, the cell wall was stained with wheat germ agglutinin-AlexaFluor488 (WGA-AF488, Life Technologies), which was added to the loading well to a final concentration of 10 µg/mL prior to loading cells into the imaging chamber. The osmolarity of the growth medium was modulated with sorbitol (Sigma-Aldrich).
During plasmolysis/lysis experiments to quantify the effect of yhdP deletion on cell stiffness, cells were allowed to grow for 5 min in medium in the imaging chamber before being plasmolyzed with LB+3 M sorbitol and exposed to LB+3 M sorbitol + 10 mM EDTA 5 min later.
Image Analysis.
Time-lapse images were first segmented with the software DeepCell (32), and the resulting segmented images were analyzed using Morphometrics (33) to obtain cell contours at subpixel resolution. Static images were directly segmented using Morphometrics (33). Cell width and length were calculated using the MicrobeTracker meshing algorithm (34).
Quantification of Spheroplast Viability.
Overnight cultures of the appropriate strains were diluted 1:100 into LFLB (LB supplemented with 3.6% sucrose and 10 mM MgSO4). Cultures were incubated at 37 °C for 1 h, normalized to OD600∼0.08, at which point cefsulodin was added to a final concentration of 60 µg/mL. Cells were further incubated for 12 h with shaking at 30 °C. Ten microliters of serial 10-fold dilutions were plated on LFLB plates. Plates were incubated at 30 °C for 24 h, and colony forming units were counted manually.
Supplementary Material
Acknowledgments
We thank the K.C.H. and T.J.S. laboratories for helpful suggestions and the Genomics Core Facility of Princeton University for help with next-generation sequencing experiments. This research was supported by the National Institute of General Medical Sciences of the NIH under Grants 5R35GM118024 (to T.J.S.) and T32-GM007388 (to J.G.). We acknowledge partial support from NIH Grant R01 GM082938 (N.S.W.) and support from a James S. McDonnell Postdoctoral Fellowship (to H.S.). K.C.H. is a Chan Zuckerberg Biohub Investigator.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015556117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Nikaido H., Outer membrane of Salmonella Typhimurium. Transmembrane diffusion of some hydrophobic substances. Biochim. Biophys. Acta 433, 118–132 (1976). [DOI] [PubMed] [Google Scholar]
- 2.Nikaido H., Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kovacs-Simon A., Titball R. W., Michell S. L., Lipoproteins of bacterial pathogens. Infect. Immun. 79, 548–561 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okuda S., Freinkman E., Kahne D., Cytoplasmic ATP hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli. Science 338, 1214–1217 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okuda S., Sherman D. J., Silhavy T. J., Ruiz N., Kahne D., Lipopolysaccharide transport and assembly at the outer membrane: The PEZ model. Nat. Rev. Microbiol. 14, 337–345 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konovalova A., Silhavy T. J., Outer membrane lipoprotein biogenesis: Lol is not the end. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20150030 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rigel N. W., Silhavy T. J., Making a beta-barrel: Assembly of outer membrane proteins in Gram-negative bacteria. Curr. Opin. Microbiol. 15, 189–193 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones N. C., Osborn M. J., Translocation of phospholipids between the outer and inner membranes of Salmonella Typhimurium. J. Biol. Chem. 252, 7405–7412 (1977). [PubMed] [Google Scholar]
- 9.Malinverni J. C., Silhavy T. J., An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc. Natl. Acad. Sci. U.S.A. 106, 8009–8014 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abellón-Ruiz J.et al., Structural basis for maintenance of bacterial outer membrane lipid asymmetry. Nat. Microbiol. 2, 1616–1623 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Sutterlin H. A.et al., Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc. Natl. Acad. Sci. U.S.A. 113, E1565–E1574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.May K. L., Silhavy T. J., The Escherichia coli phospholipase PldA regulates outer membrane homeostasis via lipid signaling. MBio 9, e00379-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell A. M., Wang W., Silhavy T. J., Novel RpoS-dependent mechanisms strengthen the envelope permeability barrier during stationary phase. J. Bacteriol. 199, e00708-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bi E., Lutkenhaus J., Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J. Bacteriol. 175, 1118–1125 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchell A. M., Srikumar T., Silhavy T. J., Cyclic enterobacterial common antigen maintains the outer membrane permeability barrier of Escherichia coli in a manner controlled by YhdP. MBio 9, e01321-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rojas E. R.et al., The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature 559, 617–621 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leive L., Shovlin V. K., Mergenhagen S. E., Physical, chemical, and immunological properties of lipopolysaccharide released from Escherichia coli by ethylenediaminetetraacetate. J. Biol. Chem. 243, 6384–6391 (1968). [PubMed] [Google Scholar]
- 18.Amro N. A.et al., High-resolution atomic force microscopy studies of the Escherichia coli outer membrane: Structural basis for permeability. Langmuir 16, 2789–2796 (2000). [Google Scholar]
- 19.Bayer M. E., Areas of adhesion between wall and membrane of Escherichia coli. J. Gen. Microbiol. 53, 395–404 (1968). [DOI] [PubMed] [Google Scholar]
- 20.Kellenberger E., The “Bayer bridges” confronted with results from improved electron microscopy methods. Mol. Microbiol. 4, 697–705 (1990). [DOI] [PubMed] [Google Scholar]
- 21.Jones N. C., Osborn M. J., Interaction of Salmonella Typhimurium with phospholipid vesicles. Incorporation of exogenous lipids into intact cells. J. Biol. Chem. 252, 7398–7404 (1977). [PubMed] [Google Scholar]
- 22.Levine T. P., Remote homology searches identify bacterial homologues of eukaryotic lipid transfer proteins, including Chorein-N domains in TamB and AsmA and Mdm31p. BMC Mol. Cell Biol. 20, 43 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar N.et al., VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 217, 3625–3639 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li P., Lees J. A., Lusk C. P., Reinisch K. M., Cryo-EM reconstruction of a VPS13 fragment reveals a long groove to channel lipids between membranes. J. Cell Biol. 219, e202001161 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Josts I.et al., The structure of a conserved domain of TamB reveals a hydrophobic β taco fold. Structure 25, 1898–1906.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lees J. A., Reinisch K. M., Inter-organelle lipid transfer: A channel model for Vps13 and chorein-N motif proteins. Curr. Opin. Cell Biol. 65, 66–71 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baba T.et al., Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2, 0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silhavy T. J., Berman M. L., Enquist L. W., Experiments with Gene Fusions, (Cold Spring Harbor Laboratory, 1984). [Google Scholar]
- 29.Datsenko K. A., Wanner B. L., One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langridge G. C., et al., Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 19, 2308–2316 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edelstein A., Amodaj N., Hoover K., Vale R., Stuurman N., Computer control of microscopes using µManager. Curr. Protoc. Mol. Biol. 14, Unit14.20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Valen D. A.et al., Deep learning automates the quantitative analysis of individual cells in live-cell imaging experiments. PLoS Comput. Biol. 12, e1005177 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ursell T.et al., Rapid, precise quantification of bacterial cellular dimensions across a genomic-scale knockout library. BMC Biol. 15, 17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sliusarenko O., Heinritz J., Emonet T., Jacobs-Wagner C., High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol. Microbiol. 80, 612–627 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.