Significance
Upon absorption of light, photoreceptor proteins with bound chromophores undergo conformational changes that enable them to transmit signals over long distances. Such proteins are essential for a wide range of light-regulated biological processes and play an important role in protein engineering efforts aimed at controlling cellular processes with light. Our computer simulations elucidate the complete photocycle of a prototypical photoreceptor protein upon photoexcitation with blue light. These simulations indicate the formation of an unusual glutamine tautomer and the rearrangement of hydrogen-bonding interactions that produce the crucial state capable of long-range signaling. The spectroscopic signatures of this state are consistent with previous experimental measurements. The insights provided by these simulations are important for learning how to engineer photoreceptors with specified characteristics.
Keywords: photoreceptor, proton-coupled electron transfer, molecular dynamics, time-dependent density functional theory
Abstract
Blue light using flavin (BLUF) photoreceptor proteins are critical for many light-activated biological processes and are promising candidates for optogenetics because of their modular nature and long-range signaling capabilities. Although the photocycle of the Slr1694 BLUF domain has been characterized experimentally, the identity of the light-adapted state following photoexcitation of the bound flavin remains elusive. Herein hybrid quantum mechanical/molecular mechanical (QM/MM) molecular dynamics simulations of this photocycle provide a nonequilibrium dynamical picture of a possible mechanism for the formation of the light-adapted state. Photoexcitation of the flavin induces a forward proton-coupled electron transfer (PCET) process that leads to the formation of an imidic acid tautomer of Gln50. The calculations herein show that the subsequent rotation of Gln50 allows a reverse PCET process that retains this tautomeric form. In the resulting purported light-adapted state, the glutamine tautomer forms a hydrogen bond with the flavin carbonyl group. Additional ensemble-averaged QM/MM calculations of the dark-adapted and purported light-adapted states demonstrate that the light-adapted state with the imidic acid glutamine tautomer reproduces the experimentally observed spectroscopic signatures. Specifically, the calculations reproduce the red shifts in the flavin electronic absorption and carbonyl stretch infrared spectra in the light-adapted state. Further hydrogen-bonding analyses suggest the formation of hydrogen-bonding interactions between the flavin and Arg65 in the light-adapted state, providing a plausible explanation for the experimental observation of faster photoinduced PCET in this state. These characteristics of the light-adapted state may also be essential for the long-range signaling capabilities of this photoreceptor protein.
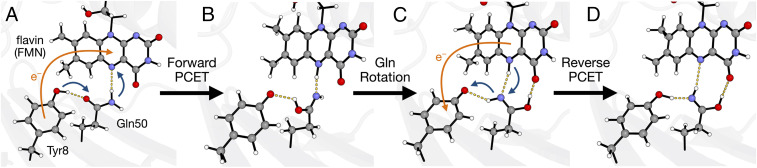
Blue light using flavin (BLUF) photoreceptor proteins play a vital role in a wide range of biological light-regulated processes (1–7). Due to their modular nature, they are also promising candidates for optogenetics, where light is used to manipulate cells in living tissue, such as neurons (8). Moreover, the photocycles of BLUF photoreceptors are thought to involve photoinduced proton-coupled electron transfer (PCET), rendering them prototypical model systems for studying photoinduced PCET in proteins. In particular, experimental and theoretical work has implicated forward and reverse photoinduced PCET processes for the Slr1694 BLUF photoreceptor domain (2, 3, 9–12). When Slr1694 BLUF is in its dark-adapted state (Fig. 1A), photoexcitation of the flavin induces the forward PCET process, which entails electron transfer from Tyr8 to the flavin and then double proton transfer from Tyr8 to the flavin through the intervening Gln50. This forward PCET process, which results in a diradical state (Fig. 1 B and C) (13–15), is followed by a less well-defined reverse PCET process returning the flavin and tyrosine to their original oxidation and protonation states (Fig. 1D). These photoinduced PCET reactions trigger conformational changes, which are thought to involve reorganization of the hydrogen bond network in the active site, thereby yielding the light-adapted state required for biological signaling.
Fig. 1.
(A–D) Schematic of the proposed mechanism for the photocycle in the BLUF Slr1694 photoreceptor, with (A) the dark-adapted state and (D) the purported light-adapted state. Following photoexcitation of the flavin, Tyr8 reduces the flavin to produce a charge-separated state (orange arrow in A), resulting in proton transfer from Tyr8 to the flavin via Gln50 (blue arrows in A) to produce the structure in B. After proton transfer, the Gln50 imidic acid tautomer rotates to form a hydrogen bond with the flavin C4 = O carbonyl, as illustrated by the rotation of Gln50 from B to C. After charge recombination corresponding to electron transfer from the flavin to Tyr8 (orange arrow in C), the proton transfers back from the flavin to Tyr8, again via Gln50 (blue arrows in C) to produce the structure in D, which is the purported light-adapted state with a glutamine imidic acid tautomer. The forward PCET and Gln rotation were studied in ref. 12, and the current work elucidates the reverse PCET process and characterizes the resulting purported light-adapted state.
Despite this level of understanding, the exact nature of the light-adapted state remains unknown in the Slr1694 BLUF domain. The light-adapted state has been fairly well established to arise from hydrogen bond rearrangement around the flavin (1–3, 7, 16, 17) on the basis of the red-shifted flavin UV-visible (UV-vis) absorption spectrum and the red-shifted flavin C4 = O carbonyl infrared (IR) stretch in the light-adapted state (2, 18). However, consensus on the details of these conformational changes has not been reached. In particular, previous studies of other BLUF domains have suggested that the light-adapted state may involve a rotation of the glutamine amide group to form a different hydrogen bond network (19–21). In contrast, other studies have proposed that the light-adapted state involves the formation of a glutamine imidic acid tautomer, either with (16, 22–26) (Fig. 1D) or without (27) rotation of the central glutamine residue. These previous hypotheses have been suggested to be consistent with the experimentally observed spectral shifts.
In this study, we use hybrid quantum mechanical/molecular mechanical (QM/MM) methods to investigate the plausibility of the Gln50 imidic acid tautomer shown in Fig. 1D as an essential component of the light-adapted state in Slr1694. To this end, we show how this purported light-adapted state can be formed. The first part of the proposed mechanism is composed of forward PCET to produce a Gln50 imidic acid tautomer, followed by rotation of the imidic acid group, as shown in our previous work (12). To complete the cycle, the simulations presented herein illustrate the subsequent reverse PCET, yielding an active site with the Gln imidic acid tautomer in conjunction with the tyrosine and flavin in their original oxidation and protonation states. Following the formation and equilibration of this light-adapted state, we perform time-dependent density functional theory (TDDFT) (28) QM/MM calculations to show that the rotated Gln50 imidic acid tautomer results in a red-shifted flavin UV-vis absorption spectrum compared to the original Gln50 amide state, reproducing the experimentally observed ∼10-nm red shift in the light-adapted state. We also show that this same light-adapted state is consistent with the experimentally observed red-shifted flavin C4 = O carbonyl IR stretch. Analysis of the changes in the hydrogen bond network implicates the rotated Gln50 imidic acid tautomer as primarily responsible for these spectral changes and identifies this tautomer as a key characteristic of the Slr1694 light-adapted state.
Results and Discussion
Formation of the Gln50 Imidic Acid Tautomer.
Upon absorption of blue light, the photoexcited flavin chromophore in the Slr1694 BLUF photoreceptor is reduced via electron transfer from Tyr8 (4, 11, 13), yielding a charge-separated state consisting of the positively charged tyrosine and the negatively charged flavin (14). Double proton transfer through the intervening glutamine residue, Gln50, neutralizes these species. Previously, we used a hybrid QM/MM approach to simulate this forward PCET process (12), which is illustrated in Fig. 1 A and B. The QM region used in this previous study, as well as the current study, included Tyr8, Gln50, and the isoalloxazine ring of the flavin shown in Fig. 1, with more specific details provided in SI Appendix, Fig. S1. For the previous simulations, the system was prepared in the ground state, and photoexcitation was modeled by instantaneously placing the system in the locally excited state of the flavin. The nonequilibrium excited-state dynamics of forward PCET, including electron transfer from Tyr8 to the flavin followed by double proton transfer from Tyr8 to the flavin via Gln50, was simulated using Tamm–Dancoff TDA-TDDFT (29) for the QM region until formation of the singlet diradical ground state. At that point, the trajectory was propagated on the singlet diradical ground state using a spin-flip TDA-TDDFT approach (30). In this previous study, we propagated nine independent trajectories that exhibited forward PCET. Given the computational expense of these simulations, we were unable to propagate a larger number of trajectories, and therefore, the results are not statistically meaningful but rather provide qualitative insights into possible reaction pathways.
After forward PCET, which produces a diradical state, we observed rotation of the Gln50 imidic acid tautomer (Fig. 1 B and C) and formation of a hydrogen bond between Gln50 and the C4 = O carbonyl of the flavin (Fig. 1C) for four of the nine trajectories, which will be the focus of this work (12). The other five trajectories either reformed the dark-adapted state or were otherwise unproductive, as described in SI Appendix. Experimentally, this diradical state persists on the order of tens to hundreds of picoseconds until charge recombination and proton transfer return the flavin and the tyrosine to their original oxidation and protonation states in the reverse PCET process (Fig. 1 C and D) (3). Although direct simulation of charge recombination on this timescale is computationally unfeasible with the QM/MM method used previously (12), we simulated the qualitative mechanism of proton transfer after charge recombination by continuing the QM/MM trajectories with closed-shell spin-restricted ground state DFT. This approach has the effect of constraining the QM active site to a closed-shell singlet ground state, thus enforcing charge recombination (31–33).
This procedure for simulating the reverse PCET process (Fig. 1 C and D) was motivated by the experimental characterization of the light-adapted state (9, 20, 34), indicating that the flavin and tyrosine are in their original oxidation and protonation states. To simulate the formation of the light-adapted state, we started with a conformation of the diradical state, constrained the flavin and tyrosine to be in their original oxidation states, and observed the subsequent proton transfer reactions that occurred to return the flavin and tyrosine to their original protonation states. This procedure assumes that the charge recombination occurs prior to the proton transfer reactions and that no additional major conformational changes occur in the diradical state on a longer timescale. As in our previous work, the QM region was described at the LRC-ωPBEh/6-31G* level of theory, and the MM region was treated with the CHARMM36 force field (35) in conjunction with the associated modified TIP3P water model (36). The flavin force field parameters were obtained from a previous study on LOV domains (37). All QM/MM calculations in this work utilized the CHARMM/Q-Chem interface (38–40). Additional computational details are provided in SI Appendix.
The initial conditions for simulating the reverse PCET process were obtained from the endpoints of the trajectories in our previous simulations of the forward PCET process (12). The structure of the active site in the diradical singlet ground state prior to charge recombination is depicted in Fig. 1C. After enforcing the closed-shell singlet spin state, charge recombination results in a negatively charged Tyr8 and a positively charged flavin. To restore charge neutrality, a proton is transferred from the flavin to the intervening Gln50, which subsequently transfers its proton to Tyr8, as depicted by the arrows in Fig. 1C. These proton transfer reactions occurred within 250 fs of the enforced charge recombination for the four trajectories studied, suggesting that the mechanism could be effectively concerted. The structure of the active site following this reverse PCET process is depicted in Fig. 1D. In this structure, the Gln50 imidic acid tautomer has been formed, and the flavin and Tyr8 are in the same oxidation and protonation states as in the dark-adapted state (Fig. 1A). No other significant structural changes were observed up to ∼700 fs following reverse PCET in our QM/MM simulations.
We emphasize that the initial proton transfer from Tyr8 to Gln50 in the forward PCET process is only possible after photoexcitation of the flavin and the subsequent electron transfer from Tyr8 to the flavin. Oxidation of Tyr8 lowers its pKa by ∼12 units, thereby enabling proton transfer from Tyr8 to Gln50. Moreover, the Gln imidic acid tautomer is much higher in energy than the original Gln amide form and is only generated in the forward PCET process and retained in the reverse PCET process because of the double proton transfer mechanisms shown in Fig. 1. Typically, glutamine does not serve as a proton acceptor, and the glutamine imidic acid tautomer is not often observed in biological systems (41). Although the imidic acid tautomer is typically much less stable than the amide form, the imidic acid tautomer may be stabilized by the environment, particularly through the hydrogen-bonding interaction with the C4 = O carbonyl of the flavin, and may persist due to the lack of energetically accessible pathways to regenerate the original amide form. Alternatively, the imidic acid tautomer may be responsible for the initial spectroscopic changes but may relax to the amide form that would have presumably moved or rotated so that it could form a hydrogen bond to the C4 = O carbonyl group of the flavin. The current simulations are not long enough to distinguish between these two possibilities.
Red-Shifted Flavin UV-Vis Absorption Spectrum.
To elucidate the structural changes that occur after equilibration of the purported light-adapted state, we used classical molecular dynamics (MD) to obtain 500 conformations over 100-ns isothermal-isobaric ensemble (NPT, 300 K, 1 atm) trajectories following equilibration of the dark- and light-adapted states. To enable classical MD involving the imidic acid tautomer of Gln50, a force field for the glutamine imidic acid tautomer was parameterized according to the previously reported protocol for the CHARMM force field (42, 43). Details of the force field parameterization, as well as the system preparation and equilibration, are provided in SI Appendix. For comparison, an analogous MD trajectory was propagated for the dark-adapted state to obtain 500 conformations for this state as well.
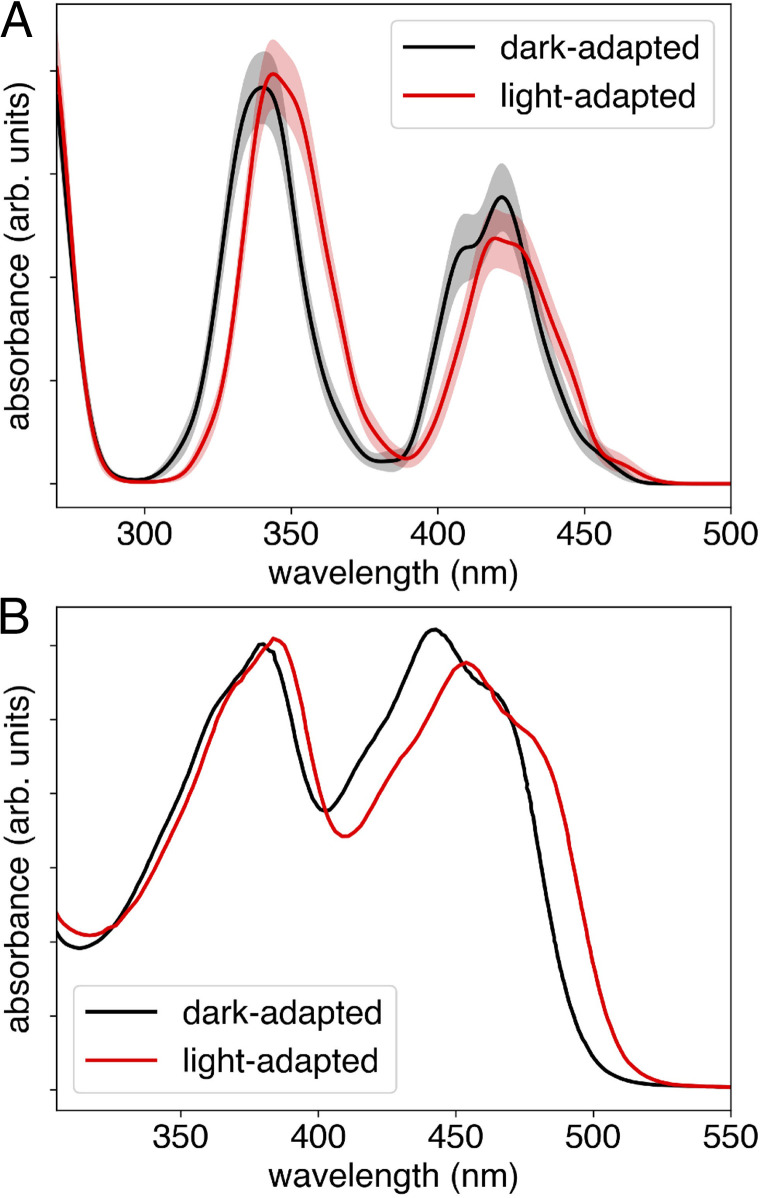
Experimentally, the flavin UV-vis absorption spectrum exhibits a red shift of ∼10 nm in the light-adapted state compared to the dark-adapted state (2, 18). We used electrostatically embedded TDDFT/MM calculations at the LRC-ωPBEh/6-31++G** level of theory (31–33, 44) to compute the ensemble-averaged flavin absorption spectrum. The electrostatic environment included the entire protein with explicit solvent. After computing the 10 lowest excitation energies for each of the 500 conformations with the hybrid TDDFT/MM approach, an ensemble-averaged spectrum was generated by fitting a Gaussian function to each computed peak and summing over all of the conformations. The Gaussian functions were normalized so that each individual peak integrates to its computed oscillator strength. In order to obtain a smooth final spectrum, each individual Gaussian was chosen to have an empirically determined full-width half-maximum of 10 nm. A 95% CI was constructed for each spectrum by computing the SD from 500 bootstrap resamples of the ensemble spectra (45).
The changes in the flavin absorption spectrum between the dark-adapted and purported light-adapted states obtained from our simulations and from experimental measurements are illustrated in Fig. 2. The peak positions in the simulated and experimental spectra are in qualitatively reasonable agreement. According to the simulations, the light-adapted state corresponding to the Gln50 imidic acid tautomer results in a red-shifted absorption spectrum compared to the dark-adapted state corresponding to the Gln50 amide state. This red shifting is primarily due to differences in the hydrogen-bonding interactions between the flavin and Gln50 in its amide versus imidic acid tautomeric form (Fig. 1A versus Fig. 1D). To confirm that the imidic acid tautomer is primarily responsible for the red-shifted absorption spectrum, we also computed the ensemble-averaged gas phase absorption spectra for the two tautomeric states of Gln50 (SI Appendix, Fig. S3), using the same QM conformations of the active site but removing the field of external MM charges from the surrounding protein and solution. Even in the absence of the protein electrostatic environment, the imidic acid tautomer displays the red-shifted spectrum compared to the amide tautomer, highlighting the role it plays in the formation of the purported light-adapted state. Additional tests using TDA-TDDFT (SI Appendix, Fig. S4) and another independently prepared ensemble (SI Appendix, Fig. S5) also reproduce the red-shifted absorption spectra in the light-adapted state.
Fig. 2.
Differences in the flavin absorption spectrum between the dark-adapted and light-adapted states of the Slr1694 BLUF photoreceptor. These calculations were performed at the TDDFT LRC-ωPBEh/6–31++G**/MM level of theory. (A) The computed ensemble spectra with the 95% CI shaded and (B) the experimental absorption spectra (adapted with permission from ref. 18 [Copyright 2004, American Chemical Society]). In both the computed and experimental spectra, the flavin S1 peak at ∼430 nm red shifts by ∼10 nm in the light-adapted state compared to the dark-adapted state.
Red-Shifted Flavin C4 = O IR Stretch.
Experimentally, the flavin C4 = O carbonyl IR stretch red shifts ∼20 cm−1 in the light-adapted state compared to the dark-adapted state. If the light-adapted state involves the rotated imidic acid tautomer of Gln50, this red shift can be rationalized because an additional hydrogen bond is formed to the C4 = O (Fig. 1D) (46). To test if the Gln50 imidic acid tautomer reproduces this vibrational red shift, we used the Fourier grid Hamiltonian (FGH) method (47) to compute the ensemble-averaged C4 = O carbonyl stretch frequencies for the dark- and light-adapted states. These calculations were performed on 50 conformations obtained from 5-ns NPT (300 K, 1 atm) trajectories following equilibration of both the light- and dark-adapted states. Starting from a gas-phase optimized flavin isoalloxazine ring, we generated a potential energy curve corresponding to the C4 = O stretch in normal mode coordinates, sampling the CO distance along a grid in the approximate region between 0.9 and 1.8 Å. These flavin geometries were inserted into the electrostatic environment of each of the 50 solvated protein conformations for each state by minimizing the rmsd between the two flavin rings, and the energy of the full system was calculated for each geometry. Given the relative rigidity of the flavin isoalloxazene ring, this insertion procedure corresponds to moving the flavin carbon, oxygen, and neighboring atoms within the sampled protein conformation along the relevant mode to produce a potential energy curve that allows the calculation of the C = O vibrational frequency without invoking the harmonic approximation. The FGH procedure inherently includes anharmonicity through the numerical solution of the one-dimensional Schrödinger equation associated with this potential energy curve.
The electrostatically embedded single-point energies computed at the DFT/LRC-ωPBEh/6-31++G**/MM level of theory were used to generate a potential energy curve along the C4 = O stretch for each protein conformation. The FGH method was used to solve the one-dimensional Schrödinger equation for the potential energy curve associated with each conformation using the reduced mass of the normal mode, and the C4 = O vibrational frequency was obtained as the energy difference between the lowest two eigenvalues associated with the vibrational energy levels. This procedure resulted in an ensemble of 50 values for the C4 = O vibrational frequency in different protein environments for both the dark- and light-adapted states.
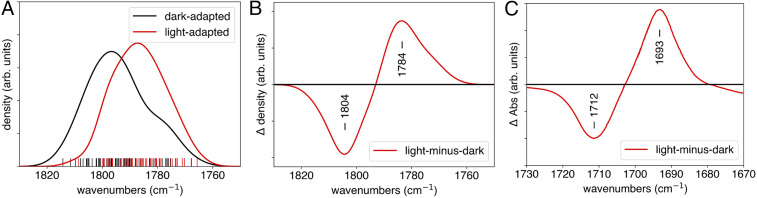
The simulated and experimental vibrational spectra associated with the C4 = O stretch are depicted in Fig. 3. The kernel density estimates (KDEs) of the computed frequencies (black and red curves for the dark- and light-adapted states, respectively) as well as the rug plot of the raw frequencies (black and red sticks) are depicted in Fig. 3A. The KDEs, which provide a continuous representation of the histogram, were generated by fitting a Gaussian function to each computed vibrational frequency and taking the normalized sum of the resulting functions. The difference between the KDEs for the light- and dark-adapted states yields a difference density (Fig. 3B), which is analogous to the FTIR difference spectra (Fig. 3C). As illustrated by the data, the light-adapted state corresponding to the Gln50 imidic acid tautomer reproduces the experimentally observed red shift in the light-adapted state. Quantitatively, the peaks in the difference density in Fig. 3B produce a red shift of 20 cm−1, whereas the experimental counterpart in Fig. 3C produces a red shift of 19 cm−1. Thus, the additional hydrogen bond between the Gln50 imidic acid tautomer and the C4 = O of the flavin is sufficient to produce the red shift of the flavin C4 = O carbonyl stretch.
Fig. 3.
Differences in the flavin C4 = O carbonyl stretch IR spectra between the dark- and light-adapted states of the Slr1694 BLUF photoreceptor. (A) The computed vibrational frequencies for the C4 = O stretch obtained using the FGH method for ensembles of the dark- and light-adapted states (black and red sticks, respectively). The curves denote a KDE of the underlying histogram. (B) The difference between the KDEs for the dark- and light-adapted states, showing a 20 cm−1 red shift between the dark- and light-adapted states. (C) The experimental difference FTIR spectra, showing a 19 cm−1 red shift between the dark- and light-adapted states (adapted with permission from ref. 18 [Copyright 2004, American Chemical Society]).
Active-Site Hydrogen Bond Analysis.
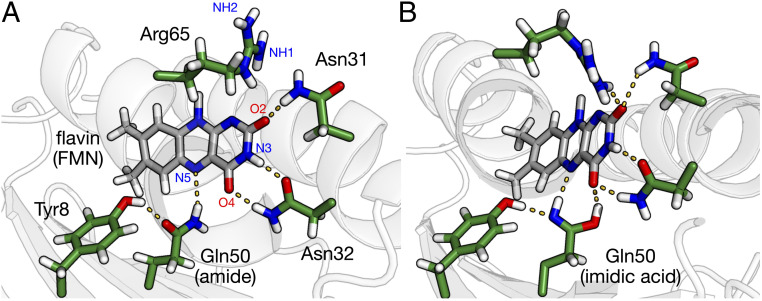
In order to understand the changes in the active site upon formation of the light-adapted state, we carried out a hydrogen bond analysis of the flavin. For this purpose, we analyzed the hydrogen-bonding partners with the isoalloxazine ring along the 100-ns NPT (300 K, 1 atm) trajectories that were propagated for both the dark- and light-adapted states. These were the same trajectories used to compute the simulated flavin UV-vis absorption spectra in Fig. 2 and were sampled every 25 ps to produce 4,000 conformations for each state. A summary of the main hydrogen-bonding partners (e.g., hydrogen-bonding pairs with >5% occupancy) and their relative occupancies during the 100-ns trajectories is given in Table 1. The hydrogen bond occupancy refers to the percentage of the trajectory that a hydrogen bond is formed between a given donor and acceptor. Representative hydrogen-bonding networks for these ensembles are depicted in Fig. 4.
Table 1.
Hydrogen bond occupancy involving the flavin (FMN) in the dark- and light-adapted states of Slr1694 BLUF
| Dark-adapted (Gln50 amide) | Light-adapted (Gln50 imidic acid) | ||||
| Donor | Acceptor | Occupancy* | Donor | Acceptor | Occupancy* |
| FMN:N3 | Asn32:OD1 | 94% | FMN:N3 | Asn32:OD1 | 93% |
| Asn32:ND2 | FMN:O4 | 89% | Asn32:ND2 | FMN:O4 | 85% |
| Asn31:ND2 | FMN:O2 | 86% | Asn31:ND2 | FMN:O2 | 27% |
| Gln50:NE2 | FMN:N5 | 43% | Gln50:NE2 | FMN:N5 | 32% |
| Gln50:NE2 | FMN:O4 | 14% | Gln50:OE1 | FMN:O4 | 79% |
| Arg65:NH2 | FMN:O2 | 63% | |||
| Arg65:NH1 | FMN:O2 | 50% | |||
Percent of time the hydrogen bond was formed for conformations sampled every 25 ps during a 100-ns trajectory. Occupancies less than 5% are not given. A hydrogen bond is defined as having a donor–acceptor distance within 3.2 Å and a donor–hydrogen–acceptor angle between 135° and 180°.
Fig. 4.
Hydrogen-bonding interactions around the flavin binding pocket for (A) the dark-adapted state, corresponding to the Gln50 amide tautomer, and (B) the purported light-adapted state, corresponding to the Gln50 imidic acid tautomer. Only the isoalloxazine ring of the flavin (gray) is shown for clarity. In our simulations, Arg65 hydrogen bonds to the flavin in the purported light-adapted state but not in the dark-adapted state.
Both the dark- and light-adapted states exhibit two hydrogen bonds between Asn32 and FMN with high occupancies (>85%). As expected, the differences between the tautomeric states of Gln50 change the hydrogen-bonding interactions between Gln50 and the flavin. The dark-adapted amide form exhibits hydrogen bonding between the Gln50:NE2 and the FMN:N5 (43% occupancy), with an occasional hydrogen bond formed between the Gln50:NE2 and the FMN:O4 (14% occupancy). For the light-adapted imidic acid form, the hydrogen bond between the Gln50:OE1 and the FMN:O4 has a high occupancy (79%), as expected given the changes observed in both the electronic and vibrational red-shifted absorption spectra. Additionally, another less prevalent hydrogen-bonding interaction is observed between the Gln50:NE2 and the FMN:N5 (32%) in the light-adapted state.
One surprising difference between the dark- and light-adapted states pertains to hydrogen-bonding interactions on the other side of the isoalloxazine flavin ring. The occupancy of the hydrogen-bonding interaction between the Asn31:ND2 and the FMN:O2 is lower for the light-adapted state (27%) compared to the dark-adapted state (86%). In the light-adapted state, we observed that the primary hydrogen bond donor to the FMN:O2 is Arg65 (63 and 50% for Arg65:NH1 and Arg65:NH2, respectively). These percentages suggest that Arg65 can potentially donate two hydrogen bonds to the FMN:O2. This hydrogen-bonding interaction with Arg65 (Fig. 4B) may arise from the additional hydrogen bond to the flavin from the Gln50:OE1 of the imidic acid tautomer, which may pull more strongly on the flavin, disrupting the Asn31:ND2 hydrogen bond, which is replaced by the Arg65 hydrogen bond(s). Although the X-ray crystal structure (PDB 2HFN) (48) used as the starting point for our simulations indicates a hydrogen bond between Arg65 and the C2 = O carbonyl of the flavin in the dark-adapted state, our simulations of the dark-adapted state do not indicate the prevalence of such a hydrogen-bonding interaction (Table 1) and show that Arg65 is a relatively flexible residue. This discrepancy could be due to either limitations of the force field or crystallization effects.
Arg65 does not appear to play a major role in the formation of the red-shifted flavin absorption spectrum, as the red shift is reproduced in gas-phase calculations (SI Appendix, Fig. S3), and is not expected to contribute to the red-shifted C4 = O carbonyl stretch, as it does not donate any hydrogen bonds to the FMN:O4. However, Arg65 could potentially play a role in later changes required for biological signaling through rearrangement of the flavin binding pocket, although further investigation is required. Moreover, the hydrogen bond between Arg65 and the flavin may be partially responsible for the ultrafast (∼1 ps) concerted PCET observed in the light-adapted state through transient pump/probe spectroscopy (21), as Arg65 has previously been shown computationally to stabilize the charge transfer state resulting from electron transfer from Tyr8 to the flavin (10), thereby facilitating the forward PCET process.
Conclusion
Herein we simulated the mechanism for the formation of a potential Slr1694 BLUF light-adapted state involving a rotated Gln50 imidic acid tautomer and showed that the resulting state displays the experimentally observed spectroscopic signatures. The combination of our QM/MM MD simulations presented in ref. 12 and herein illustrates that photoexcitation of the flavin induces a forward PCET process that leads to the formation of the Gln50 imidic acid tautomer, as well as the tyrosyl and FMNH radicals, followed by rotation of Gln50 and a reverse PCET process that retains the Gln50 imidic acid tautomer while returning the tyrosine and flavin to their original oxidation and protonation states. The resulting light-adapted state, which contains the rotated Gln50 imidic acid tautomer, was further characterized and compared to the dark-adapted state, which contains the original Gln50 amide state. Classical MD trajectories initiated after the reverse PCET process allowed sampling of conformations in the purported light-adapted state. We performed ensemble-averaged TDDFT/MM calculations to compute the electronic absorption spectra of the flavin chromophore in the solvated BLUF protein environment, reproducing the experimentally observed ∼10-nm red shift in the flavin absorption spectrum of the light-adapted state. Similarly, we used the FGH method in conjunction with electrostatic embedding to compute the flavin C4 = O carbonyl stretch frequency, reproducing the experimentally observed ∼20 cm−1 red shift in the carbonyl stretch frequency of the light-adapted state.
The red shifts in the flavin UV-vis and C4 = O IR spectra were explained in terms of the new hydrogen bond formed between the flavin and the Gln50 imidic acid tautomer. Hydrogen-bonding analyses of MD trajectories with Gln50 in the amide and imidic acid forms confirmed that the imidic acid tautomer enables a strong, persistent hydrogen bond between the Gln50 hydroxyl group and the C4 = O carbonyl of the flavin. Moreover, these analyses suggested that this new hydrogen-bonding interaction may induce further changes in the hydrogen-bonding network, namely, the formation of hydrogen-bonding interactions between the flavin and Arg65, facilitating electron transfer from Tyr8 to the flavin. This characteristic of the purported light-adapted state provides a plausible explanation for the faster photoinduced PCET reaction observed experimentally in the light-adapted state than in the dark-adapted state. In addition, these changes in the hydrogen-bonding interactions may be essential for the long-range signaling that occurs in the light-adapted state.
Based on the observation of the formation of the rotated Gln50 imidic acid tautomer during QM/MM dynamics, as well as the changes in the flavin UV-vis and IR absorption spectra, we have demonstrated strong computational support for assigning the rotated Gln50 imidic acid tautomer as the dominant structural change in the formation of the light-adapted state. However, we cannot rule out the possibility that the imidic acid tautomer eventually relaxes to the amide form that has reoriented to enable hydrogen bonding to the C4 = O carbonyl of the flavin. Although these results are consistent with the experimentally observed spectral changes occurring upon formation of the light-adapted state, additional studies are required to understand how these changes in the hydrogen-bonding network lead to the conformational changes necessary for long-range biological signaling.
Computational Methods
Details of the computational methodology used in this article are provided in SI Appendix. The computational details are provided for the simulations of the reverse PCET process, the classical MD simulations, and the QM/MM calculations of the flavin absorption spectrum and C4 = O carbonyl stretch frequency. Additional tests for the computed flavin absorption spectra and details of the force field parameterization for the glutamine imidic acid tautomer are also provided in SI Appendix.
Supplementary Material
Acknowledgments
This material is based on work supported by the Air Force Office of Scientific Research (AFOSR) under AFOSR Award FA9550-18-1-0134.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016719117/-/DCSupplemental.
Data Availability.
The computational data collected and analyzed in this paper are available on the Open Science Framework (DOI: 10.17605/OSF.IO/KP4C2) (49).
References
- 1.Kennis J. T. M., Groot M.-L., Ultrafast spectroscopy of biological photoreceptors. Curr. Opin. Struct. Biol. 17, 623–630 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Kennis J. T. M., Mathes T., Molecular eyes: Proteins that transform light into biological information. Interface Focus 3, 20130005 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mathes T., van Stokkum I. H. M., Kennis J. T. M., Photoactivation mechanisms of flavin-binding photoreceptors revealed through ultrafast spectroscopy and global analysis methods. Methods Mol. Biol. 1146, 401–442 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Mathes T., Götze J. P., A proposal for a dipole-generated BLUF domain mechanism. Front. Mol. Biosci. 2, 62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujisawa T., Masuda S., Light-induced chromophore and protein responses and mechanical signal transduction of BLUF proteins. Biophys. Rev. 10, 327–337 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park S.-Y., Tame J. R. H., Seeing the light with BLUF proteins. Biophys. Rev. 9, 169–176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kottke T., Xie A., Larsen D. S., Hoff W. D., Photoreceptors take charge: Emerging principles for light sensing. Annu. Rev. Biophys. 47, 291–313 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Losi A., Gardner K. H., Möglich A., Blue-light receptors for optogenetics. Chem. Rev. 118, 10659–10709 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gauden M.et al., Hydrogen-bond switching through a radical pair mechanism in a flavin-binding photoreceptor. Proc. Natl. Acad. Sci. U.S.A. 103, 10895–10900 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goings J. J., Reinhardt C. R., Hammes-Schiffer S., Propensity for proton relay and electrostatic impact of protein reorganization in Slr1694 BLUF photoreceptor. J. Am. Chem. Soc. 140, 15241–15251 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Sayfutyarova E. R., Goings J. J., Hammes-Schiffer S., Electron-coupled double proton transfer in the Slr1694 BLUF photoreceptor: A multireference electronic structure study. J. Phys. Chem. B 123, 439–447 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Goings J. J., Hammes-Schiffer S., Early photocycle of Slr1694 blue-light using flavin photoreceptor unraveled through adiabatic excited-state quantum mechanical/molecular mechanical dynamics. J. Am. Chem. Soc. 141, 20470–20479 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Mathes T., van Stokkum I. H. M., Stierl M., Kennis J. T. M., Redox modulation of flavin and tyrosine determines photoinduced proton-coupled electron transfer and photoactivation of BLUF photoreceptors. J. Biol. Chem. 287, 31725–31738 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukacs A.et al., BLUF domain function does not require a metastable radical intermediate state. J. Am. Chem. Soc. 136, 4605–4615 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gil A. A.et al., Photoactivation of the BLUF protein PixD probed by the site-specific incorporation of fluorotyrosine residues. J. Am. Chem. Soc. 139, 14638–14648 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stelling A. L., Ronayne K. L., Nappa J., Tonge P. J., Meech S. R., Ultrafast structural dynamics in BLUF domains: Transient infrared spectroscopy of AppA and its mutants. J. Am. Chem. Soc. 129, 15556–15564 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Hall C. R.et al., Site-specific protein dynamics probed by ultrafast infrared spectroscopy of a noncanonical amino acid. J. Phys. Chem. B 123, 9592–9597 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Masuda S., Hasegawa K., Ishii A., Ono T. A., Light-induced structural changes in a putative blue-light receptor with a novel FAD binding fold sensor of blue-light using FAD (BLUF); Slr1694 of synechocystis sp. PCC6803. Biochemistry 43, 5304–5313 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Anderson S.et al., Structure of a novel photoreceptor, the BLUF domain of AppA from Rhodobacter sphaeroides. Biochemistry 44, 7998–8005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonetti C.et al., Hydrogen bond switching among flavin and amino acid side chains in the BLUF photoreceptor observed by ultrafast infrared spectroscopy. Biophys. J. 95, 4790–4802 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathes T.et al., Hydrogen bond switching among flavin and amino acids determines the nature of proton-coupled electron transfer in BLUF photoreceptors. J. Phys. Chem. Lett. 3, 203–208 (2012). [Google Scholar]
- 22.Domratcheva T., Grigorenko B. L., Schlichting I., Nemukhin A. V., Molecular models predict light-induced glutamine tautomerization in BLUF photoreceptors. Biophys. J. 94, 3872–3879 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Udvarhelyi A., Domratcheva T., Glutamine rotamers in BLUF photoreceptors: A mechanistic reappraisal. J. Phys. Chem. B 117, 2888–2897 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Khrenova M. G., Nemukhin A. V., Domratcheva T., Photoinduced electron transfer facilitates tautomerization of the conserved signaling glutamine side chain in BLUF protein light sensors. J. Phys. Chem. B 117, 2369–2377 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Domratcheva T., Hartmann E., Schlichting I., Kottke T., Evidence for tautomerisation of glutamine in BLUF blue light receptors by vibrational spectroscopy and computational chemistry. Sci. Rep. 6, 22669 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grigorenko B. L., Khrenova M. G., Nemukhin A. V., Amide-imide tautomerization in the glutamine side chain in enzymatic and photochemical reactions in proteins. Phys. Chem. Chem. Phys. 20, 23827–23836 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Sadeghian K., Bocola M., Schütz M., A conclusive mechanism of the photoinduced reaction cascade in blue light using flavin photoreceptors. J. Am. Chem. Soc. 130, 12501–12513 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Casida M. E., “Time-dependent density functional response theory for molecules” in Recent Advances in Density Functional Methods, Chong D. P., Ed. (World Scientific, 1995), Vol. 1, pp. 155–192. [Google Scholar]
- 29.Hirata S., Head-Gordon M., Time-dependent density functional theory within the Tamm–Dancoff approximation. Chem. Phys. Lett. 314, 291–299 (1999). [Google Scholar]
- 30.Shao Y., Head-Gordon M., Krylov A. I., The spin–flip approach within time-dependent density functional theory: Theory and applications to diradicals. J. Chem. Phys. 118, 4807–4818 (2003). [Google Scholar]
- 31.Hehre W. J., Ditchfield R., Pople J. A., Self-consistent molecular orbital methods. XII. Further extensions of Gaussian-type basis sets for use in molecular orbital studies of organic molecules. J. Chem. Phys. 56, 2257–2261 (1972). [Google Scholar]
- 32.Hariharan P. C., Pople J. A., The influence of polarization functions on molecular orbital hydrogenation energies. Theor. Chim. Acta 28, 213–222 (1973). [Google Scholar]
- 33.Rohrdanz M. A., Martins K. M., Herbert J. M., A long-range-corrected density functional that performs well for both ground-state properties and time-dependent density functional theory excitation energies, including charge-transfer excited states. J. Chem. Phys. 130, 054112 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Hasegawa K., Masuda S., Ono T.-A., Structural intermediate in the photocycle of a BLUF (sensor of blue light using FAD) protein Slr1694 in a Cyanobacterium Synechocystis sp. PCC6803. Biochemistry 43, 14979–14986 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Best R. B.et al., Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- 37.Freddolino P. L., Gardner K. H., Schulten K., Signaling mechanisms of LOV domains: New insights from molecular dynamics studies. Photochem. Photobiol. Sci. 12, 1158–1170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woodcock H. L. 3rdet al., Interfacing Q-Chem and CHARMM to perform QM/MM reaction path calculations. J. Comput. Chem. 28, 1485–1502 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Brooks B. R.et al., CHARMM: The biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shao Y.et al., Advances in molecular quantum chemistry contained in the Q-Chem 4 program package. Mol. Phys. 113, 184–215 (2015). [Google Scholar]
- 41.Nakamura A.et al., “Newton’s cradle” proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography. Sci. Adv. 1, e1500263 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.MacKerell A. D.et al., All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Vanommeslaeghe K.et al., CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31, 671–690 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clark T., Chandrasekhar J., Spitznagel G. W., Schleyer P. V. R., Efficient diffuse function-augmented basis sets for anion calculations. III. The 3-21+G basis set for first-row elements, Li–F. J. Comput. Chem. 4, 294–301 (1983). [Google Scholar]
- 45.Mai S., Gattuso H., Monari A., González L., Novel molecular-dynamics-based protocols for phase space sampling in complex systems. Front Chem. 6, 495 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kiefer P. M., Pines E., Pines D., Hynes J. T., Solvent-induced red-shifts for the proton stretch vibrational frequency in a hydrogen-bonded complex. 1. A valence bond-based theoretical approach. J. Phys. Chem. B 118, 8330–8351 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Tannor D. J., Introduction to Quantum Mechanics: A Time-Dependent Perspective, (University Science Books, 2007). [Google Scholar]
- 48.Yuan H.et al., Crystal structures of the Synechocystis photoreceptor Slr1694 reveal distinct structural states related to signaling. Biochemistry 45, 12687–12694 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goings J., Li P., Zhu Q., Hammes-Schiffer S., Data for “Formation of an Unusual Glutamine Tautomer in a Blue-Light Using Flavin Photocycle Characterizes the Light-Adapted State.” Open Science Framework. https://osf.io/kp4c2. Deposited 1 September 2020. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The computational data collected and analyzed in this paper are available on the Open Science Framework (DOI: 10.17605/OSF.IO/KP4C2) (49).