Significance
One of the central regulators of plant growth and development is the phytohormone cytokinin. Likewise, it has become increasingly apparent that autophagy is a critical regulatory process in both plants and animals. Our work provides a mechanistic link between these regulatory processes, demonstrating that autophagy can fine-tune cytokinin perception by modulating the level of the type-A ARRs, which negatively regulate the cellular response to cytokinin. Further, our work sheds light on the mechanism of selective autophagy and on how a particular cargo is targeted for autophagic degradation by a novel autophagy receptor belonging to the EXO70D gene family.
Keywords: cytokinin signaling, selective autophagy, concanamycin A, carbon starvation
Abstract
The phytohormone cytokinin influences many aspects of plant growth and development, several of which also involve the cellular process of autophagy, including leaf senescence, nutrient remobilization, and developmental transitions. The Arabidopsis type-A response regulators (type-A ARR) are negative regulators of cytokinin signaling that are transcriptionally induced in response to cytokinin. Here, we describe a mechanistic link between cytokinin signaling and autophagy, demonstrating that plants modulate cytokinin sensitivity through autophagic regulation of type-A ARR proteins. Type-A ARR proteins were degraded by autophagy in an AUTOPHAGY-RELATED (ATG)5-dependent manner, and this degradation is promoted by phosphorylation on a conserved aspartate in the receiver domain of the type-A ARRs. EXO70D family members interacted with type-A ARR proteins, likely in a phosphorylation-dependent manner, and recruited them to autophagosomes via interaction of the EXO70D AIM with the core autophagy protein, ATG8. Consistently, loss-of-function exo70D1,2,3 mutants exhibited compromised targeting of type-A ARRs to autophagic vesicles, have elevated levels of type-A ARR proteins, and are hyposensitive to cytokinin. Disruption of both type-A ARRs and EXO70D1,2,3 compromised survival in carbon-deficient conditions, suggesting interaction between autophagy and cytokinin responsiveness in response to stress. These results indicate that the EXO70D proteins act as selective autophagy receptors to target type-A ARR cargos for autophagic degradation, demonstrating modulation of cytokinin signaling by selective autophagy.
Autophagy is a major catabolic pathway that maintains cellular homeostasis in response to intrinsic developmental changes and environmental cues. It mediates the degradation of protein complexes, misfolded and aggregated proteins, and damaged organelles by targeting proteins to proteases localized in the vacuole or lysosome, with a subsequent retrotransport of cellular building blocks back into the cytosol (1). There are three main types of autophagy: macro, micro, and chaperone-mediated autophagy (2). Macroautophagy (hereafter referred to simply as autophagy) is mediated by conserved AUTOPHAGY-RELATED GENEs (ATGs) that coordinate the de novo biogenesis of the autophagy organelle, the autophagosome (3, 4). Autophagosomes are double membrane vesicles that sequester various cargoes and ultimately deliver them to the lytic vacuole, resulting in their degradation and subsequent recycling. Although basal autophagy occurs in cells under steady-state conditions (5), it is enhanced in response to biotic and abiotic stresses (6, 7) and can result in selective or bulk (nonselective) degradation of proteins (2). Typically, bulk autophagy randomly degrades cytosolic content, while selective autophagy requires unique receptors to target specific cargos for degradation (8, 9). These receptors contain the unique ATG8-INTERACTING MOTIFs (AIM) or LIR-INTERACTING MOTIFs that allow them to interact with the ATG proteins on the autophagosomal membrane (10). They also contain cargo-binding domains that mediate selective recruitment of specific cargo to the growing autophagosome. The presence of an AIM in a protein can suggest a link to autophagy. For example, EXO70B1 contains an AIM domain and has been linked to autophagy as it colocalizes with ATG8 proteins in autophagosomes, and disruption of EXO70B1 resulted in fewer vacuolar autophagic vesicles (11). Multiple other EXO70 isoforms also contain AIM domains (12, 13), suggesting that they may also act as receptors for the autophagic regulation of various cellular components.
The identification of receptors with their corresponding cargos has improved our understanding of the role of selective autophagy in regulating plant growth and development (9). For example, in response to sulfur stress, Joka2, a tobacco member of the family of selective autophagy cargo receptors, triggers the autophagic degradation of the sulfur responsive protein UPC9 by interacting with both UPC9 and ATG8f (14). Selective autophagy also regulates rubisco degradation during leaf senescence (15), brassinosteroid responses via targeting of BRI1-EMS SUPPRESSOR 1 (BES) by DOMINANT SUPPRESSOR OF KAR2 (DSK2) (16), and ATG8-INTERACTING PROTEIN 1 (ATI1)–mediated turnover of plastid proteins (17).
Autophagy has been linked to various phytohormones (18), including cytokinins, which are N6-substituted adenine derivatives. Cytokinin and autophagy affect an overlapping set of plant developmental and physiological processes, including leaf senescence, nutrient remobilization, root meristem function, lateral root development, vascular development, and the response to biotic and abiotic stresses (18). For example, cytokinin negatively regulates leaf senescence and plays a positive role in nitrogen uptake (19). Disruption of autophagy in rice (via the osatg7 mutant) results in early leaf senescence and compromised nitrogen reutilization (20). Moreover, nitrogen remobilization between organs is reduced in autophagy-deficient mutants of Arabidopsis and maize (21). ATG genes are generally up-regulated during leaf senescence (22) and in response to biotic and abiotic stresses (23). Consistent with a link between cytokinin and autophagy, overexpression of a GFP-ATG8f fusion protein in Arabidopsis results in altered sensitivity to exogenous cytokinin, and cytokinin reduces the incorporation of this fusion protein into vacuolar structures that are likely autophagic vesicles (24). Further, transcriptomic analyses identified overlapping genes differentially expressed in atg5-1 and cytokinin signaling mutants (25). Despite these links, it is unclear how autophagy is integrated into cytokinin signaling.
The cytokinin signaling cascade in Arabidopsis is similar to bacterial two-component signaling systems (TCS) and begins with the binding of cytokinin to histidine kinase receptors, proceeds through a series of phosphotransfers, and culminates in the phosphorylation of conserved Asp residues in type-A and type-B response regulators (ARRs) (19). Type-B ARRs are DNA-binding transcription factors that mediate the transcriptional response to cytokinin, including the induction of type-A ARRs (26, 27). The Arabidopsis genome encodes 10 type-A ARRs which, unlike type-B ARRs, lack a DNA binding domain and act as negative regulators of cytokinin signaling (28). Type-A ARR proteins generally have a short half-life and their levels are tightly regulated. Phosphorylation at the conserved Asp in response to cytokinin stabilizes a subset of the type-A ARRs (29). Some type-A ARRs are degraded by the ubiquitin proteasome system (30, 31), though there may be alternative ubiquitin-independent pathways regulating type-A ARRs turnover (29, 32). For example, the nuclear-localized periplasmic degradation protein 9 (DEG9) selectively binds to and targets ARR4 (but not other type-A ARRs) for ubiquitin-independent protein degradation (33).
Here, we show that type-A ARRs are targeted for degradation by autophagy, at least in part through their interaction with members of the EXO70D subfamily. EXO70D3 interacts with several ATG8 isoforms via an AIM located in the N-terminal domain. Consistently, the exo70D1,2,3 triple mutant has increased type-A ARR protein levels and reduced sensitivity to cytokinin. Our results suggest that these EXO70Ds target type-A ARRs for autophagic degradation and thus mediate the interplay between cytokinin signaling and autophagy.
Results
Type-A ARR4 Protein Levels Are Regulated by Autophagy.
The level of type-A ARRs proteins plays a critical role in modulating the responsiveness to cytokinin in multiple developmental and physiological processes. Type-A ARRs levels are controlled both by regulation of their transcription by multiple inputs (34) and by regulation of their protein stability, at least in part by phosphorylation of the conserved Asp residue (29–32). Given the potential links between autophagy and cytokinin function, we tested if autophagy plays a role in the turnover of type-A ARR signaling elements. Inhibiting vacuolar degradation in transgenic Arabidopsis seedlings with the vacuolar proton pump inhibitor concanamycin A (ConcA) resulted in the rapid accumulation of various type-A ARR:CFP/eGFP (cyan fluorescent protein/enhanced green fluorescent protein) fusion proteins (ARR3, ARR4, ARR5, ARR7, and ARR16) that were constitutively expressed from a UBIQUITIN10 (UBQ10) promoter (Fig. 1A and SI Appendix, Fig. S1A) but did not affect type-A ARR transcript levels (SI Appendix, Fig. S1B). The effect of ConcA on type-A ARR proteins was independent of the epitope tag or its location as N-terminally tagged ARR7 (myc:ARR7) also accumulated in response to ConcA (SI Appendix, Fig. S1C). ConcA treatment induced the accumulation of multiple type-A ARR:CFP fusion proteins into vacuolar vesicles (SI Appendix, Fig. S1D), which suggests type-A ARR proteins could be degraded via autophagy.
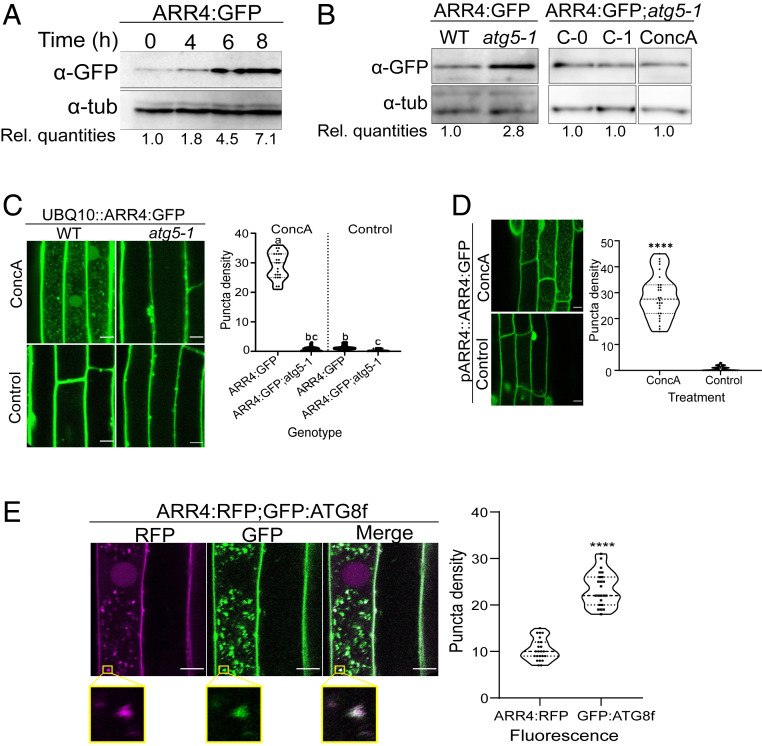
Fig. 1.
ARR4 is degraded by the autophagy pathway in Arabidopsis roots. (A) Response of root-expressed ARR4 to ConcA. Seedlings carrying pUBQ10::ARR4:GFP constructs were treated with ConcA and transferred to dark to induce carbon starvation. ARR4 protein quantities in the roots analyzed by immunoblot assays using anti-GFP (α-GFP) antibodies. Anti-tubulin (α-tub) served as loading control. Rel. quantities represent ratio of intensity of α-GFP to α-tub band relative to ratio at 0 h. (B) Disruption of autophagosome-forming ATG5 gene elevated the levels of ectopically expressed ARR4 proteins. The progeny of a cross between pUBQ10::ARR4:GFP-expressing plants described in A and atg5-1 and wild-type plants were grown under normal conditions for 9 d and for additional 1 d under carbon starvation conditions. Seedlings were either untreated (C-0) or treated with DMSO mock control (C-1) and ConcA (ConcA) as described above, prior to immunoblot assay. Rel. quantities represent ratio of intensity of α-GFP to α-tub band relative to ratio of WT band. For immunoblot assay of ARR4:GFP;atg5-1, Rel. quantities represent intensity ratio relative to bands of C-0. (C) Representative confocal micrograph of root of 5-d-old seedlings described in B, treated with ConcA or DMSO as mock control and exposed to carbon starvation for 18 h prior to imaging. (Right) Quantification of the number of GFP-containing vacuolar puncta. Data represent the average number of puncta per 100 µm2, n = 23, analyzed with one-way ANOVA followed by Tukey–Kramer multiple comparison analyses; P < 0.05. Different letters represent statistically different means. (D) Representative confocal micrograph of root of T1 seedlings of wild-type Col-0 transformed with pARR4::ARR4:GFP construct. Seedlings were treated as described in C. (Right) Graph represents average number of puncta per 5,000-μm2 area (n = 60, from 20 independent T1 seedlings). (E) Intracellular colocalization of RFP signal resulting from ARR4:RFP with the GFP signal from GFP:ATG8f. Seedling coexpressing both constructs were treated with ConcA as described in C. (Scale bar, 10 µm.) The magnified area enclosed by the yellow box indicates section of vacuole showing colocalization of ARR4:RFP- and GFP:ATG8f-containing vesicles. Graph represents mean (n = 23) number of RFP- and GFP-containing puncta per 50 µm2 of vacuole area. (For C, D, E: Scale bar, 10 μm.) For D, E, ****statistical differences at P < 0.0001 using Student’s t test.
To further explore the role of autophagy in type-A ARR turnover, we focused on a transgenic line expressing an ARR4:eGFP fusion protein driven by the UBQ10 promoter. Similar to the effect of ConcA, genetic disruption of autophagy (via the atg5 mutation) resulted in elevated ARR4:GFP protein levels, without affecting ARR4 transcript levels (Fig. 1B and SI Appendix, Fig. S1E). Further, the accumulation of ARR4:GFP into vesicles was nearly eliminated in atg5-1 mutants (Fig. 1C), indicating that these vesicles are indeed autophagic vesicles. The level of ARR4:GFP protein in the atg5-1 background was unaffected by ConcA treatment, consistent with the increased level of ARR4 in response to ConcA being the result of disrupted autophagy (Fig. 1B). An ARR4:GFP fusion protein expressed from its native promoter (pARR4::ARR4:GFP) also accumulated in autophagic vesicles in response to ConcA treatment (Fig. 1D), indicating that ARR4 trafficking into autophagic vesicles was not the result of overexpression.
To further confirm that the ConcA-dependent ARR4-containing vesicles are autophagic vesicles, we analyzed the intracellular colocalization of ARR4:red fluorescent protein (RFP)- with GFP:ATG8f-containing vesicles in roots of stable transgenic Arabidopsis plants. Overall, there were more than twice as many GFP:ATG8f vesicles as ARR4:RFP vesicles (Fig. 1E). Postthresholded Manders’ colocalization coefficient (35) values indicate that 81% of the ARR4:RFP vesicles overlapped with GFP:ATG8f vesicles, while 52% of GFP:ATG8f associate with compartments containing ARR4:RFP. This analysis indicates that the ARR4:RFP signal significantly colocalized with GFP:ATG8f, consistent with ARR4’s being targeted to a subset of autophagic vesicles.
Type-A ARRs Interact with the EXO70D Proteins.
To explore the mechanism of autophagic regulation of type-A ARRs and to identify potential receptors involved in targeting type-A ARR proteins to autophagic vesicles, we screened for proteins that interact with an activated form of type-A ARR protein. Previous studies indicate that mutating the aspartic acid residue (D) that is the target of phosphorylation in the receiver domain of ARRs to a glutamic acid (E) partially mimics the activated, phosphorylated form of the protein (29). Using an ARR5D87E bait in a yeast two-hybrid screen, we identified three independent preys that corresponded to the EXO70D3 gene (AT3G14090). EXO70D3 belongs to the 23-member QR-motif-containing Arabidopsis EXO70 gene family (36) and is most closely related to two other paralogs, EXO70D1 (AT1G72470) and EXO70D2 (AT1G54090) (SI Appendix, Fig. S2A). EXO70s are subunits of the octomeric exocyst complex, which is involved in exocytosis and other cellular trafficking processes. Recent studies have shown that EXO70 paralogs could also play roles in autophagy (11, 12, 37). EXO70D3 interacted with ARR5D87E but not with a wild-type ARR5 or ARR5D87A bait in a yeast two-hybrid assay (Fig. 2A), suggesting it may interact with ARR5 in a phospho-Asp–dependent manner. We examined the interactions among the EXO70D isoforms and multiple type-A ARRs using a yeast two-hybrid assay (Fig. 2A). The three EXO70D paralogs interacted differentially with a subset of type-A ARRs, generally preferentially with their phosphomimic forms. The D→E version of ARR4 (ARR4D95E) interacted with all three EXO70D paralogs, but neither the wild type nor the D→A version of ARR4 interacted appreciably with any EXO70D isoform in this assay. All three versions of ARR5 (ARR5, ARR5D87A, and ARR5D87E) interacted strongly with EXO70D1, but only ARR5D87E interacted with EXO70D2 and EXO70D3. The D→E version of ARR7 (ARR7D85E) interacted with EXO70D1, but EXO70D2 and EXO70D3 did not appreciably interact with any version of ARR7. The D→E version of ARR16 (ARR16D93E) interacted very weakly with EXO70D1 but not with EXO70D2 or EXO70D3. These results suggest that all three paralogs of EXO70D may play a role in regulating type-A ARR function, with some degree of specificity.
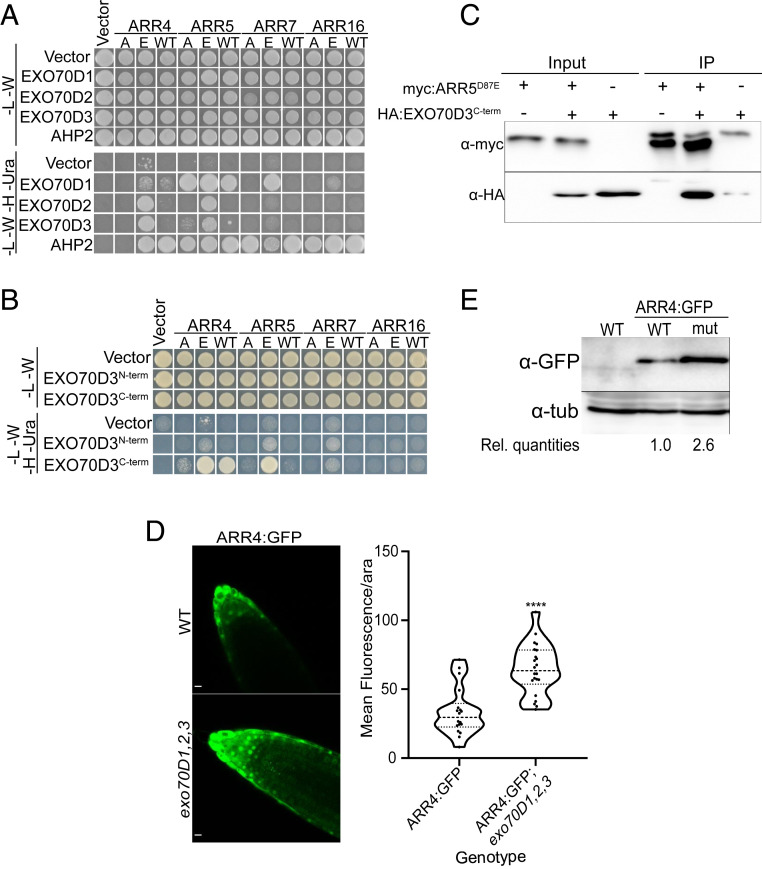
Fig. 2.
Interaction between EXO70Ds and type-A ARRs results in the destabilization of the type-A ARRs. (A) Yeast two-hybrid (Y2H) assay showing pairwise interactions between members of the EXO70D subclade and representative type-A ARRs. The EXO70D paralogs were cloned into the Gal4 DNA activation domain prey vector while the type-A ARRs (WT) with their respective phosphor-dead (A) and phosphor-mimic (E) mutations were cloned as baits in Gal4 DNA binding domain vectors. Interactions were accessed on -Leu-Trp-His-Ura (-L -W -H -Ura) quadruple dropout media. The -Leu-Trp (-L -W) served as control media. Interactions between the type-A ARRs and Arabidopsis Histidine Phosphotransfer Protein 2 (AHP2) served as positive control. (B) Y2H assay of interactions of N-terminal (EXO70D3N-term) or C-terminal (EXO70D3C-term) domains of EXO70D3 with the representative type-A ARRs described in A. See SI Appendix, Fig. S3A for details on N-term and C-term. Interaction assays are similar to A. (C) EXO70D3C-term interacts with phosphor-mimic mutants of ARR5 (ARR5D87E) in coimmunoprecipitation assay. Leaves of N. benthamiana were infiltrated with plasmids expressing myc:EXO70D3C-term or HA:ARR5D87E. The input extracts and the myc-immunoprecipitated proteins (IP) were analyzed by immunoblotting with anti-HA and anti-myc. (D) Ectopically expressed ARR4:GFP accumulates in roots tips of exo70D1,2,3 triple loss-of-function mutant. Confocal microscopy images indicating the expression of pUBQ10::ARR4:GFP in wild-type (Top) and exo70D1,2,3 mutant (Bottom) plants. (Scale bars, 10 µm.) (Right) Images are quantified by measuring signal intensity of individual nuclei, after background normalization. The graph represents the average signal measurement from 22 images of each genotype. Data were analyzed by unpaired Student’s t test. ****Statistical difference at P < 0.001 (4.025e-06). (E) Anti-GFP immunoblot analyses of roots of 10-d-old Arabidopsis seedlings, transgenic seedlings expressing pUBQ10::ARR4:GFP in wild type (WT) or exo70D1,2,3 (mut) described in D. Rel. quantities represent ratio of intensity of α-GFP to α-tub band relative to ratio of bands from WT (ARR4:GFP).
We further confirmed the phospho-Asp–dependent interaction of ARR5 and EXO70D3 in planta using bimolecular fluorescence complementation (BiFC) (SI Appendix, Fig. S2B). Transient coexpression of cYFP:ARR5D87E and nYFP:EXO70D3 in leaves of Nicotiana benthamiana resulted in stronger fluorescence as compared to coexpressed cYFP:ARR5D87A and nYFP:EXO70D3. These BiFC interactions occurred in the cytosol and are consistent with the localization of a subset of type-A ARRs partially in the cytosol (38) and the exclusive localization of UBQ10::EXO70D3:GFP in the cytosol of Arabidopsis roots (SI Appendix, Fig. S2C) and in transfected Arabidopsis protoplasts (39).
Analysis of EXO70D3 using the iLIR database (40) suggested xLIR (L232-V237) as the principal AIM, located in the N-terminal domain (SI Appendix, Fig. S3A). This AIM has a high position-specific scoring matrix score and with only two mismatches has significantly high sequence similarity to experimentally determined mammalian AIMs (41). The presence of an AIM motif suggests EXO70D3 may interact with ATG8 proteins and act as a receptor to mediate the targeting of type-A ARRs to autophagosomes. Both EXO70D1 and EXO70D2 also contain EXO70 domains (EXO70D1: R247-D617 and EXO70D2: R239-D607) and putative AIMs (EXO70D1: WxxL motif: L237-L242, and “anchor”: D90-I95; EXO70D2: xLIR: L229-V234) (SI Appendix, Fig. S3B), which are suggestive of roles as autophagy receptors.
To identify the domain of EXO70D3 that interacts with type-A ARRs, we performed pairwise yeast two-hybrid assays using the N terminus (EXO70D3N-term; amino acids 1 to 298; includes the AIM domain) or C terminus (EXO70D3C-term; amino acids 299 to 623) (SI Appendix, Fig. S3A) of EXO70D3 with wild-type or phosphomutant forms of type-A ARR paralogs (Fig. 2B). While an EXO70D3N-term bait did not interact with type-A ARRs, an EXO70D3C-term bait interacted most strongly with the phosphomimic versions of ARR4 and ARR5 but weakly or not at all with ARR7 and ARR16, consistent with the results observed using the full-length protein. We confirmed the interaction of ARR5 and EXO70D3 in vivo using coimmunoprecipitation with ARR5D87E and EXO70D3C-term (Fig. 2C). We conclude that the C-terminal domain of EXO70Ds interacts preferentially with the phosphorylated form of multiple type-A ARRs.
EXO70D3 Regulates Type-A ARR Protein Levels.
We examined the consequence of the interaction of EXO70Ds and type-A ARRs by analyzing the effect of modulating EXO70D function on type-A ARR protein levels. When transiently coexpressed in N. benthamiana leaves, EXO70D3 reduced ARR5D87E protein levels (SI Appendix, Fig. S3C). Coexpression with a negative control plasmid had no effect on ARR levels. Although they did not interact in our yeast two-hybrid assay, EXO70D3 reduced ARR5WT proteins in a concentration-dependent manner when coinfiltrated in N. benthamiana leaves (SI Appendix, Fig. S3D), suggesting that these proteins may interact in planta, possibly reflecting a weak interaction insufficient for the yeast two-hybrid assay or in planta phosphorylation of ARR5WT in this system. We examined the effect of separating the N- and C-terminal domains of EXO70D3 on degradation of ARR5 in planta. Coexpressing either EXO70D3N-term or EXO70D3C-term with ARR5D87E did not significantly affect its stability (SI Appendix, Fig. S3E), indicating that both the type-A ARR-interacting (EXO70D3C-term) and AIM-containing domains (EXO70D3N-term) are required for effective destabilization of ARR5. This is consistent with EXO70D3 acting as a receptor that recruits type-A ARR cargos for autophagic degradation, requiring both type-A ARR and ATG8 (i.e., AIM motif) binding domains.
All three paralogs EXO70Ds are expressed in Arabidopsis roots (SI Appendix, Fig. S3F), and thus we examined the effect of disruption of the EXO70Ds on type-A ARR stability in this tissue. Disruption of EXO70D1, EXO70D2, and EXO70D3 resulted in a significant increase in fluorescence in roots of an Arabidopsis line expressing GFP-tagged ARR4 from a UBQ10 promoter (Fig. 2D and SI Appendix, Fig. S3G). We confirmed this result using an independent line carrying a UBQ10::ARR4:RFP transgene in a wild-type and exo70D1,2,3 triple-mutant background (SI Appendix, Fig. S3H). Further, immunoblotting revealed a ∼2.5-fold increase in ARR4:GFP levels in exo70D1,2,3 as compared to a wild-type background (Fig. 2E).
EXO70D3 Promotes Targeting of ARR4 to Autophagic Vesicles.
The results described above suggest that the EXO70D paralogs play a role in the autophagic turnover of at least a subset of type-A ARRs. Indeed, while EXO70Ds are likely not recruited to the protein secretory exocyst-positive organelle (EXPO) (39, 42), other members of EXO70 gene family have been linked to autophagy and the formation of autophagosomes (11, 13). Thus, we tested the hypothesis that the EXO70Ds target type-A ARRs to autophagic vesicles. In the roots of stable transgenic Arabidopsis plants, disrupting all three members of the EXO70D gene family resulted in a 50% reduction in the number of ConcA-dependent vacuolar vesicles containing the ARR4:GFP proteins driven by UBQ10 promoter (Fig. 3A). Similar results were obtained with GFP-tagged ARR4 fusion proteins expressed from its own promoter (pARR4::ARR4:GFP) (SI Appendix, Fig. S4A). Accordingly, the increase in ARR4:GFP protein levels in response to ConcA was largely dependent on functional EXO70D genes (Fig. 3B). These results suggest that EXO70Ds regulate ARR4 protein levels via autophagy.
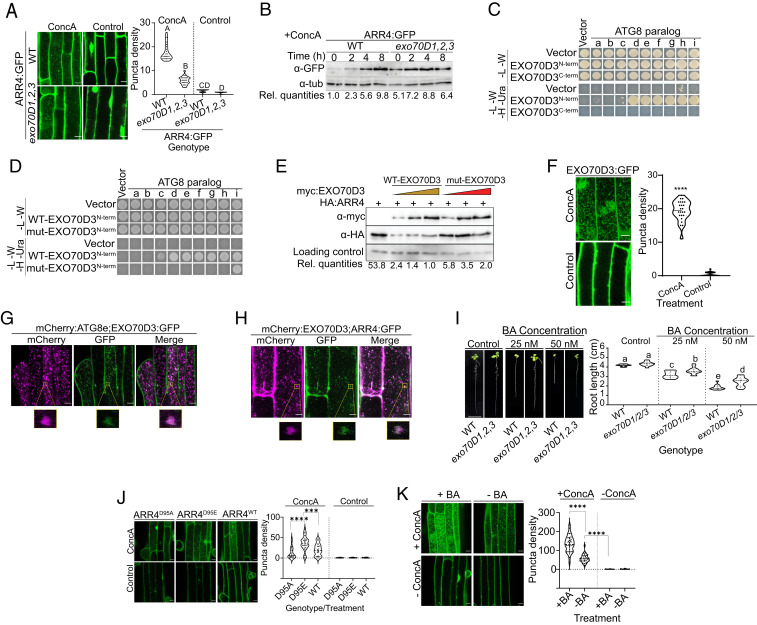
Fig. 3.
EXO70Ds mediate cytokinin responses by recruiting type-A ARRs to the ATG8-tagged autophagy machinery for degradation. (A) Representative confocal micrographs of the root elongation zones of ectopically expressed ARR4:GFP in wild type (WT) and exo70D1,2,3 mutants incubated with ConcA or DMSO (as control). Quantification of the number of GFP-containing puncta is as described in Fig. 1C. Different letters represent values that are statistically different (n = 23) at the indicated P value. (B) Disruption of EXO70D genes inhibits autophagy-mediated destabilization of type-A ARR proteins. Seedlings were treated with ConcA followed by immunoblot assays using anti-GFP. α-tub was a loading control. Rel. quantities represent the ratio of intensity of α-GFP to α-tub band relative to ratio of bands in WT (ARR4:GFP) at 0 h. (C) Yeast two-hybrid (Y2H) interaction of ATG8 paralogs with N-terminal (EXO70D3N-term) or C-terminal (EXO70D3C-term) domains of EXO70D3. (D) Y2H interaction of prey ATG8 paralogs with N-terminal domains of WT (WT-EXO70D3N-term) or W234A/V237A AIM-mutated (mut-EXO70D3N-term) EXO70D3 as bait. (E) Functional AIM is required for the EXO70D3-dependent destabilization of ARR4 proteins. N. benthamiana leaves were transiently cotransformed with fixed amount of HA:ARR4 and increasing titer of myc-tagged full-length WT (WT-EXO70D3) or W234A/V237A mutant (mut-EXO70D3) EXO70D3. α-GFP served as transfection control. Rel. quantities represent the ratio of band intensities of α-HA:α-myc:α-GFP (α-HA/α-myc/α-GFP) relative to the ratio of lane 4 which was set at 1.0. (F) Representative confocal microscopy image of root of Arabidopsis pUBQ10::EXO70D3:GFP seedlings treated with ConcA or DMSO (as control). Quantification of the number of EXO70D3:GFP-containing puncta. Data represent mean of 24 independent images. ****Values that are statistically different at P < 5.066 × 10−0321. (G) Colocalization of EXO70D3:GFP and mCherry:ATG8e in roots of Arabidopsis seedlings carrying plasmids expressing both genes. (H) Colocalization of mCherry:EXO70D3 and ARR4:GFP in elongation zone of 5-d-old Arabidopsis seedlings ectopically expressing both genes. For G and H, seedling treatment and imaging is as described in Fig. 1E. The magnified areas enclosed by the yellow boxes show colocalization of vesicles containing mCherry:ATG8e and EXO70D3:GFP, and Cherry:EXO70D3 and ARR4:GFP, respectively. (I) Root elongation assay showing the cytokinin response of exo70D1,2,3 and WT seedling roots. Seedlings were grown on MS media supplemented with BA (cytokinin) or NaOH (control). (Scale bar, 1 cm.) Different letters represent statistically different means (n ≥ 13) at P < 0.05. (J) The autophagic flux of ARR4 is dependent on the phosphorylation status of the conserved aspartate. Representative confocal micrograph of root of seedlings expressing the indicated GFP-tagged proteins, treated with ConcA (Upper) or DMSO (Lower). Graph represents average number of puncta per 5,000-μm2 area (n = 48, from 8 independent transformants). *** and **** represent statistically different values at P = 0.0001 and P < 0.0001, respectively. (K) Cytokinin enhances the autophagic flux of ARR4. Representative confocal micrograph of roots of seedlings treated with ConcA (+ConcA) (Upper) or DMSO (-ConcA) (Lower) under carbon starvation conditions for 10 h. Seedlings were then treated with 5 µM BA (+BA) (Left) or NaOH as vehicle control (-BA) (Right) for 4 h prior to imaging. Graph represents average number of puncta per 66,000-pixel2 area (for n ≥ 30). ****Values that are statistically different between treatments. Note that the BA × ConcA interaction was also significant. (Scale bar in A, F, G, H, J, K, 10 μm.)
If the EXO70D proteins act as autophagy receptors, they should interact with various ATG8 isoforms and should themselves be targeted to autophagic vesicles. Indeed, EXO70D3N-term interacted with multiple members of the ATG8 gene family in a yeast two-hybrid assay (Fig. 3C), consistent with the presence of AIMs in this domain. Substituting the conserved W and L/V residues with A in the LIR domain disrupts the interaction of receptors with ATG homologs and inhibits autophagy in mammals and maize (43, 44). In Arabidopsis, similar mutations in DSK2 abolished its interaction with ATG8 proteins (16). To test whether the L232-V237 domain of EXO70D3 functions as an AIM, we examined the impact of W234A and V237A substitutions on the ATG8–EXO70D3 interaction and on the EXO70D3-mediated destabilization of type-A ARRs. These mutations in EXO70D3N-term (mut-EXO70D3N-term) significantly reduced its interactions with nearly all isoforms of ATG8 in a pairwise yeast two-hybrid assay (Fig. 3D). In leaves of N. benthamiana, coexpression with full length mut-EXO70D3 had a reduced effect on ARR4 protein levels compared to wild-type EXO70D3 (Fig. 3E). The residual type-A ARR-destabilization activity of the mutant EXO70D3 may reflect other less conserved, partially functional AIMs in EXO70D3.
We observed ConcA-dependent EXO70D3:GFP-containing vesicles in Arabidopsis roots (Fig. 3F). In Arabidopsis seedlings coexpressing EXO70D3:GFP and mCherry:ATG8e, these ConcA-dependent EXO70D3-containing puncta colocalized with mCherry:ATG8e-tagged puncta (Fig. 3G). To test whether these EXO70D3-containing puncta are related to the ARR4-containing vesicles, we analyzed the localization of these proteins in stable transgenic Arabidopsis plants. The overlap between the mCherry and GFP-containing puncta indicates that both EXO70D3 and ARR4 are recruited into the autophagic vesicles (Fig. 3H). These observations are consistent with the EXO70D acting as receptors for the selective autophagy of ARR4 by recruiting the ARR4 to the autophagosome via the interaction between EXO70D3 and ATG8.
Perturbation of EXO70Ds Alters Cytokinin Response.
In agreement with previous reports (36), we did not observe any substantial morphological changes in seedlings or adult plants of single (exo70D1, exo70D2, or exo70D3), double (exo70D1,2, exo70D1,3, or exo70D2,3), or triple (exo70D1,2,3) mutants as compared to the wild type (Fig. 3I and SI Appendix, Fig. S4B).
We investigated the link between EXO70D function and cytokinin. The EXO70D genes are not transcriptionally regulated by cytokinin (SI Appendix, Fig. S5A). Prior studies revealed that overexpression of type-A ARRs results in hyposensitivity of primary roots to cytokinin (29, 32), and as type-A ARR protein accumulates in the exo70D1,2,3 triple mutant (Fig. 2 D and E), we examined if the exo70D1,2,3 mutant has altered response to exogenous cytokinin. The exo70D1,2,3 triple mutant showed reduced sensitivity to low doses of exogenous cytokinin in a root elongation assay (Fig. 3I). The single (exo70D1, exo70D2, and exo70D3) and double (exo70D1,2, exo70D1,3, and exo70D2,3) mutants did not exhibit altered cytokinin responsiveness (SI Appendix, Fig. S5 B and C). These results demonstrate that members of the EXO70D gene family act redundantly to positively regulate cytokinin responsiveness.
To investigate the effect of disruption of EXO70Ds on the spatial pattern of the cytokinin response, we examined the expression of a TCSn::GFP cytokinin reporter in roots of wild type and exo70D1,2,3 mutants grown in the presence of cytokinin. In the absence of exogenous cytokinin, TCSn::GFP expression was reduced in the exo70D1,2,3 mutant as compared to wild-type roots (SI Appendix, Fig. S5D). When grown in the presence of exogenous cytokinin, TCSn::GFP expression in the exo70D1,2,3 mutant was significantly lower compared to wild type. Taken together, these results suggest that disruption of the EXO70Ds results in reduced responsiveness to endogenous and exogenous cytokinin, likely as a consequence of elevated type-A ARR protein levels.
Our yeast two-hybrid results suggest that some EXO70D isoforms interact preferentially with the phosphorylated forms of some type-A ARRs (Fig. 2A). To further explore the role of phosphorylation in the autophagic turnover of type-A ARR proteins, we examined the autophagic flux of GFP-tagged ARR4WT, ARR4D95E, and ARR4D95A expressed from the CAMV promoter in stably transformed Arabidopsis seedlings (Fig. 3J). In the presence of ConcA, there were significantly more ARR4D95E-containing vesicles as compared to ARR4WT and ARR4D95A. The number of ARR4WT-containing vesicles was ∼70% more than those containing ARR4D95A, though the difference was not statistically significant (Fig. 3J) and may reflect a fairly low level of phosphorylation of ARR4 at endogenous cytokinin levels in this tissue. We tested this by analyzing the autophagic flux of ARR4:GFP in the presence of cytokinin. Indeed, exogenous cytokinin markedly enhanced the autophagic flux of ARR4 (Fig. 3K), consistent with phosphorylation of the conserved Asp promoting the autophagic turnover of type-A ARR proteins. To gain insight into the role of EXO70Ds in the cytokinin-mediated stabilization of type-A ARRs, we analyzed the effect of cytokinin on type-A ARR protein levels in an exo70D1,2,3 triple mutant. Interestingly, although exogenous cytokinin resulted in accumulation of ARR4:GFP protein in the roots of both wild-type and exo70D1,2,3 seedlings, the rate of accumulation in the triple mutant was slower than in the wild type (SI Appendix, Fig. S5E). These results suggest that phosphorylation of the conserved Asp can increase targeting of type-A ARR to EXO70D-mediated autophagic degradation.
As our results indicate that autophagy can modulate type-A ARR levels, we examined if mutants affecting autophagy altered cytokinin sensitivity using a root elongation assay (SI Appendix, Fig. S6). Consistent with prior results (24), overexpression of ATG8f (35S::GFP:ATG8f) resulted in hypersensitivity to cytokinin, which is consistent with increased autophagic targeting of type-A ARR proteins in this line. We also examined lines with loss-of-function alleles for various ATG8 genes. While most atg8 mutants displayed wild-type sensitivity to cytokinin, the atg8c mutant displayed hyposensitivity to cytokinin, consist with an increased level of type-A ARRs in this line. Surprisingly, the atg5-1 mutant was hypersensitive to cytokinin. This may reflect the fact that ATG5 encodes a nonredundant regulator of autophagy, and thus its disruption may result in pleiotropic effects that may affect the response to cytokinin in unanticipated ways.
EXO70Ds and Type-A ARRs Enhance Response to Fixed-Carbon Starvation.
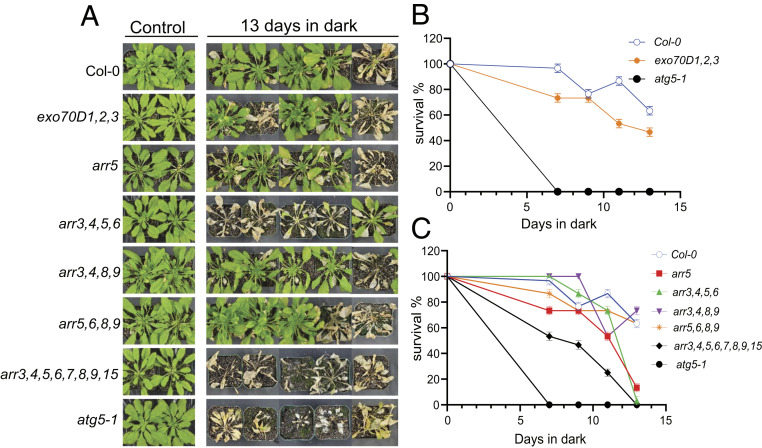
Mutants that disrupt some ATG genes or receptors of autophagy are hypersensitive to or have reduced survival in nitrogen or carbon starvation assays (11, 16, 45). Thus, we assayed the response of the single, double, and triple mutants of EXO70Ds to carbon starvation. Consistent with a role in autophagy, exo70D1,2,3 is hypersensitive to carbon starvation; After 7 d of dark treatment, the survival rate of exo70D1,2,3 was 75% compared to 100% for wild-type plants. After 11 d of dark treatment, the survival decreased to 50% for exo70D1,2,3, while it was greater than 80% for wild-type plants (Fig. 4 A and B). Responses of single mutants were generally comparable to that of wild-type plants after 7 and 11 d of dark treatment (SI Appendix, Fig. S7A). Among the double mutants, 11 d of dark treatment resulted in 50% and 80% survival for exo70D1,3 and exo70D2,3, respectively, compared to about 90% for exo70D1,2 and wild-type plants (SI Appendix, Fig. S7B). Overall, these results suggest that members of the EXO70D gene family redundantly function in regulating autophagic responses during carbon starvation.
Fig. 4.
EXO70Ds and type-A ARRs modulate plant response to fixed-carbon starvation. Seedlings were grown for 6 wk on potted soil under short-day conditions of 16/8 h day/night at 22 °C. Pots were transferred to dark for 7, 9, 11, and 13 d and recovered in the light for 7 d. (A) Representative images of exo70D1,2,3 triple loss-of-function mutants and arr5-1 and some high-order type-A ARR loss-of-function mutants (arr3,4,5,6; arr3,4,8,9; arr5,6,8,9; arr3,4,5,6,7,8,9,15) following 13-d-dark treatment. Col-0 and atg5-1 served as wild-type and autophagy deficient controls, respectively. (B and C) Quantification of survival of exo70D1,2,3 (B) and type-A ARR mutants (C) in response to carbon starvation. Survival was estimated as the percentage of plants with new leaves after the dark treatment. Values represent mean ± SEM percentage survival of three biological replicates. Each biological replicate consisted of eight plants per genotype per treatment.
We tested if the type-A ARRs also play a role in the response to carbon starvation by examining the phenotypes of high-order type-A ARR loss-of-function mutants in fixed-carbon starvation survival assays. Compared to wild-type plants, the type-A ARR higher-order mutants were significantly more sensitive to carbon starvation (Fig. 4C), with the arr3,4,5,6,7,8,9,15 octuple mutant displaying the strongest phenotype, suggesting at least partial functional redundancy among the type-A ARRs in this response.
Discussion
Autophagy and cytokinin regulate overlapping plant growth and developmental processes, including senescence, root and vascular development, and nutrient remobilization (2, 7, 19). Further links between autophagy and cytokinin include the observations that atg7 mutants in rice have decreased levels of cytokinins (46) and overexpression of ATG8 enhances cytokinin perception in Arabidopsis (24). Here, we demonstrate that plants modulate responses to cytokinin, at least in part, via the autophagic regulation of type-A ARRs. This input into cytokinin function is mediated by members of the EXO70D, a subclade of the EXO70 gene family, which act as receptors to recruit type-A ARRs to the autophagosome for subsequent degradation. This conclusion is based on: 1) the interaction of EXO70Ds with a subset of type-A ARRs and with various ATG8 isoforms; 2) the colocalization of EXO70D3, type-A ARRs, and ATG8 in vacuolar vesicles; 3) the elevated level of type-A ARR proteins in atg5 and exo70D3 mutants; and 4) the reduction in the number of ConcA-dependent ARR-containing vesicles in exo70D1,2,3 mutants. The elevated levels of type-A ARR proteins in atg5 and exo70D1,2,3 mutants result in hyposensitivity to cytokinin, consistent with previous studies on transgenic lines overexpressing type-A ARRs (29, 32), which, like the exo70D1,2,3 triple mutant, are largely aphenotypic when grown under normal laboratory conditions. Thus, autophagy regulates the sensitivity to cytokinin, at least in part via EXO70Ds.
The preferential interaction of EXO70D isoforms to ARR4 and ARR5, but not ARR7 or ARR16, in the yeast two-hybrid interaction suggests some specificity, though this may not fully reflect in planta interactions. Indeed, both ARR7 and ARR16 accumulated in autophagic vesicles despite their lack of interaction in the yeast two-hybrid assay, which could reflect heterodimerization of type-A ARRs in planta or the presence of additional receptors for trafficking type-A ARR into the autophagic vesicles. The residual trafficking of ARR4 into autophagic vesicles in exo70D1,2,3 mutants is consistent with this notion of additional receptors involved in the selective autophagy of type-A ARRs. Alternatively, bioinformatic analysis revealed the presence of four putative WxxL-containing AIMs in ARR4, suggesting it may interact weakly with ATG8 to promote limited trafficking to autophagic vesicles. The ARR–EXO70D–ATG8 interactions and the responses to cytokinin and carbon-deficient conditions demonstrated in this study provide a molecular mechanism linking autophagy and cytokinin responses.
The EXO70 genes are highly expanded in Arabidopsis and other land plants (36, 47), are expressed in specific tissue types (48), and regulate diverse physiological processes (49). For example, Exo70A1 is implicated in the recycling of PIN-FORMED proteins (PINs) at the plasma membrane and polar growth (36, 50), development of tracheary elements (51), cell cycling (52), Casparian strip development (53), and cell plate formation (54). EXO70C2 regulates pollen tube growth (55) and Exo70B1 and Exo70H1 regulate pathogen defense responses (56, 57). Here we demonstrate a role for the EXO70D clade of this expanded gene family.
While the results presented here indicate that the phosphorylation of a subset of type-A ARRs, which occurs in response to cytokinin, increases their targeting to autophagic degradation, prior studies indicated that cytokinin stabilizes a subset of type-A ARR proteins, also likely through phosphorylation of the conserved Asp in their receiver domain (29). Further, while our findings show that type-A ARR proteins are regulated by autophagy, other reports indicate that the 26S proteasome also plays a role in the turnover of type-A ARRs (31, 32, 58). Similar proteasome-dependent and -independent regulator pathways have been reported for BRI1-EMS SUPPRESSOR 1 (16) and for a number of ABA signaling components (59).
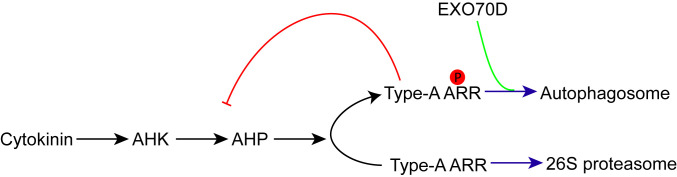
We propose a model (Fig. 5) in which the unphosphorylated type-A ARRs are targeted for degradation by the 26S proteasome. In response to cytokinin, type-A transcript levels rise and the type-A ARR proteins become phosphorylated, reducing their targeting to the 26S proteasome, but ultimately increasing their degradation by autophagy. The relative contributions of these two mechanisms to type-A ARR protein turnover is likely influenced by tissue/cell type, the particular type-A ARR isoform involved, and other regulatory inputs. Together, these two mechanisms likely complement each other to optimally tune cytokinin responsiveness in response to various developmental and environmental cues.
Fig. 5.
Model of autophagic and proteasome regulation of type-A ARRs. In the presence of cytokinin, type-A ARRs are phosphorylated on the conserved aspartate in the receiver domain. Phosphorylated type-A ARRs are stabilized and negatively regulate cytokinin responses. To regulate this constitutive type-A ARR action, EXO70Ds recruit the phosphorylated type-A ARR to the autophagosome by interacting with ATG8 isoforms. This EXO70D-mediated autophagic mechanism presents a more rapid degradation pathway for type-A ARRs. In the absence of cytokinin, unphosphorylated type-A ARRs are ubiquitinated and shuttled to the 26S proteasome for degradation. This 26S proteasome pathway is probably activated for a prolong degradation of type-A ARRs. Black arrow: phosphate transfer; green line: protein–protein interaction; purple arrow: protein degradation; red line: negative feedback regulation.
Although exo70D1,2,3 is morphologically indistinguishable from wild-type plants under normal growing conditions, it is significantly compromised in survival under carbon-deficient conditions. Consistent with this, mutations in autophagy receptors, such as exo70B1 and dsk2, result in reduced survival in nutrient-deficient conditions (16, 60). The observation that disruption of all three EXO70Ds resulted in a less severe effect as compared to mutations disrupting bulk autophagy regulators (i.e., ATG5) is consistent with a role for EXO70Ds acting as receptors for selective autophagy.
The observation that disruption of type-A ARRs significantly compromises plant survival under carbon-limiting conditions suggests there is feedback between autophagy and cytokinin signaling. As cytokinin regulation has a pleiotropic effect on plant development, including senescence and nutrient partitioning (19), it is possible that disrupting a cytokinin signaling component such as type-A ARRs may significantly affect responses to carbon limitation indirectly, rather than by directly modulating autophagy. For example, cytokinin promotes localized sink activity (19), and thus disruption of type-A ARRs may affect nutrient partitioning, which in turn would likely impact survival in response to carbon starvation. Cytokinin also plays a key role in regulating leaf senescence (19), which may impact the response to carbon starvation. Alternatively, it is possible that cytokinin signaling has a direct effect on autophagy via an as-yet-unidentified mechanism.
In conclusion, our findings have uncovered additional pathway regulating type-A ARR turnover. As type-A ARRs are primary cytokinin response genes and negatively regulate the cytokinin signaling pathway, it is perhaps not surprising that they are regulated through multiple pathways. The relative contributions of these regulatory mechanisms, the cellular and environmental conditions under which they are triggered, the cell types in which they predominantly occur, and the specific type-A ARR isoforms that are regulated by each mechanism remain to be determined. As type-A and type-B ARRs both possess receiver domains that are phosphorylated by AHPs (19) and given that only a subset of the ATG8 paralogs interact with EXO70D3, it will be interesting to determine if phosphoaspartate-dependent EXO70D-mediated autophagic degradation also plays a role in the regulation of type-B ARRs.
Materials and Methods
All Arabidopsis thaliana lines used in this study are of the Col-0 ecotype. The GFP:ATG8f:HA autophagic marker lines were previously described (61). Transfer DNA (T-DNA) insertion mutants of the Exo70D gene family (exo70D1 (SALK_074650), exo70D2 (WiscDsLox450H08), and the previously-described exo70D3 (SAIL_175_D08) (36), atg8c-1 (SALK_003706C), atg8f-1 (SALK_057021C), atg8g-1 (SALK_205553C), and atg5-1 (CS39993) (45, 62) were obtained from the Arabidopsis Biological Resources Centre, Ohio State University. Double (exo70D1,2, exo70D1,3, and exo70D1,2) and triple (exo70D1,2,3) mutants were generated by crossing. TCSn::GFP;exo70D1,2,3 was generated by crossing exo70D1,2,3 to plants carrying a synthetic reporter for type-B ARR activity, TCSn::GFP (63). Stable transgenic Arabidopsis lines expressing ARR4:GFP, EXO70D3:GFP, mCherry:ATG8E, and type-A ARR:CFP were generated by Agrobacterium-mediated transformation. The ARR4:GFP;exo70D1,2,3, ARR4:RFP;GFP:ATG8f, EXO70D3:GFP;mCherry:ATG8E, and ARR4:GFP;atg5-1lines were generated by crossing. All crosses were confirmed by PCR-based genotyping using T-DNA and gene-specific primers (SI Appendix, Table S1).
For growth of Arabidopsis on plates, seeds were surface-sterilized, plated on Murashige and Skoog (MS) media (MS basal salts, plus 1% sucrose and 0.8% Phytagel or phytoagar, pH. 5.8), and stratified at 4 °C for 3 d. Unless otherwise stated, seedlings were grown for 10 d at 22 °C under long-day (LD, 16/8 h day/night) conditions. Adult plants were grown in soil at 22 °C in either LD or short-day (8 h light and 16 h dark) photoperiod conditions as noted.
Cloning and Vector Construction.
High-fidelity DNA polymerase was used for all PCR amplifications. Primers used for gene cloning are detailed in SI Appendix, Table S1. Phospho-mimic (D-E) and phospho-dead (D-A) versions of the type-A ARRs were generated through site-directed mutagenesis of the conserved phosphorylation sites (28). We generated the W234A and V237A mutations in EXO70D3 by site-directed mutagenesis using a QuikChange Site-Directed Mutagenesis kit (Stratagene). To create entry clones, blunt-end PCR products with directional overhangs were cloned into pENTR/D-TOPO vector using the pENTR Directional TOPO Cloning Kit (Thermo Fisher Scientific). A 3-kbp genomic ARR4 fragment, including a 1.85-kbp sequence upstream of the translational start site, was amplified and cloned into pENTR/D-TOPO entry vector. The entry clones were inserted into respective Gateway-based destination vectors using Gateway LR Clonase II Enzyme Mix (Thermo Fisher Scientific). mCherry-ATG8E was generated by N-terminal tagging of the full-length ATG8E CDS sequence, using Greengate cloning.
The CFP, GFP, and RFP-tagged overexpression vectors were created by recombining the relevant entry clones into pUBC-CFP-Dest, pUBC-GFP-Dest, and pUBC-RFP-Dest (64), respectively. Other overexpression vectors used were the GFP-tagged pGWB4 and pGWB6 (65). The genomic ARR4 entry vector was recombined with the GFP-tagged pGWB4 to generate the pARR4::ARR4:GFP clones. ARR4D95A, ARR4D95E, and ARR4WT fragments were also cloned into the PGWB6 vectors. For mCherry-tagged vectors, the yellow fluorescent protein (YFP) cassette of pUBN-YFP-Dest was excised by Spe1 and Mun1 restriction enzymes and replaced with mCherry CDS flanked by Spe1 and Mun1 restriction sites. mCherry_Spe1 and mCherry_Mun1 primers were used to introduce 5′ Spe1 and 3′ Mun1 sites by PCR.
Yeast two-hybrid vectors were generated by the Gateway-based recombination of respective entry clones into the bait, pBMTN116c-D9, and prey, pACT2, vectors (66). For BiFC, the type-A ARR and EXO70D3 entry vectors were cloned into pUBN-cYFP-Dest and pUBN-nYFP-Dest (64) destination vectors, respectively. For coexpression and coimmunoprecipitation assays in N. benthamiana, type-A ARRs, AHP2, and full- and partial-length EXO70D3 (wild-type or the W234A/V237A mutation) amplicons were cloned into myc-tagged pEarleyGate203 and HA-tagged pEarleyGate201 binary vectors (67). Destination clones were transformed into Agrobacterium tumefaciens strain, GV3101 and transformed in N. benthamiana using the leaf infiltration method (68).
Chemical Treatment.
For autophagic induction, seedlings were transferred in liquid MS (without sucrose) supplemented with 1 μM ConcA (11050; Cayman Chemical) (69). Dimethyl sulfoxide (DMSO) served as vehicle control. T1 and T2 generation of seedlings, with segregating transgenes, were first grown on appropriate selection prior to treatment and confocal microscopy. For cytokinin treatment, seedlings were grown in MS media supplemented various concentrations of 6-benzylamino purine (BA) (B3408; Sigma) or sodium hydroxide as vehicle control.
Seedling Cytokinin Response Assay.
Responses to exogenous cytokinin was determined by a root elongation assay (28). To determine the effect of cytokinin treatment on stability of ARR4:GFP, 10-d-old seedlings were incubated in liquid MS medium supplemented with 5 μM BA for 0 to 12 h. Protein extracts from whole seedlings or roots were analyzed by an immunoblot assay. The effect of cytokinin on autophagic flux of ARR4:GFP was analyzed by confocal microcopy. Briefly, surface-sterilized seeds were plated on MS agar, stratified at 4 °C for 3 d, and moved to growth chambers at 22 °C, under 24 h light for 4 d. Seedlings were transferred to liquid MS medium with or without ConcA (1 μM) for 10 h. Seedlings were treated with 5 μM BA for 4 h prior to imaging. DMSO served as vehicle control.
Yeast Two-Hybrid Analysis.
Both bait and prey constructs were transformed into the L40ccαU strain (66) using the lithium acetate transformation protocol (70). Interactions were tested as previously decribed (66). Transformants were selected on -Leu-Trp Synthetic Complete double dropout media. To test interactions, cell suspensions of positive transformants were grown to an optical density at 600 nm of 0.5 and 10 µL were plated on -Leu-Trp-Ura-His media. Plates were incubated at 30 °C for 4 d and analyzed for interacting genes. Negative and positive controls for each interaction included cotransforming with either the bait or prey vectors and previously described interactors, respectively.
BiFC Assay.
Agrobacterium carrying pUBN-cYFP-Dest–expressing EXO70D3 and phospho-mimic or phospho-dead ARR cloned into pUBN-nYFP-Dest were infiltrated into leaves of N. benthamiana (68). Plasmids expressing the tomato bushy stunt virus suppressor p19 (71) were added to each infiltration mix to suppress host silencing of transgenes. The infiltrated plants were grown for 3 d in constant light. Leaf epidermal cells were imaged for YFP and CFP signals using a Zeiss LSM710 confocal scanning microscope. To determine strength of interacting protein pairs, the fluorescent intensities of interacting proteins were quantified by Fiji software (72).
Coimmunoprecipitation Assays.
ARR5 and EXO70D3 was transiently expressed in leaf epidermal cells of N. benthamiana as described above. Infiltrated leaves were ground in liquid nitrogen and total protein extracted in lysis buffer (1% glycerol, 25 mM Tris⋅HCl, pH 7.5, 1 mM ethylenediaminetetraacetic acid [EDTA], 150 mM NaCl, 10 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride, and 2% polyvinyl polyvinyl-polypyrolidone) supplemented with cOmplete ULTRA Tablets, Mini, EDTA‐free, EASYpack Protease Inhibitor Mixture (Sigma). Coimmunoprecipitation was conducted using the μMACS Epitope Tag Protein Isolation Kits according to the manufacturer’s instructions (Miltenyi Biotec) and purified on a magnetic μ Column (Miltenyi Biotec). Columns were washed with wash buffer (1% glycerol, 25 mM Tris⋅HCl, pH 7.5, 1 mM EDTA, 250 mM NaCl, 10 mM DTT, and cOmplete ULTRA Protease inhibitor Mixture tablet), and proteins eluted with Elution Buffer (Miltenyi Biotec). Protein extracts were the subjected to immunoblot analyses using α-myc or α-hemagglutinin (α-HA) antibodies.
Immunoblot Analyses.
Proteins were resolved on sodium dodecyl sulfate polyacrylamide gel electrophoresis followed by immunoblotting using α-HA high-affinity antibody 3F10 (Sigma) and α-c‐Myc antibody (9E10) (sc‐40: Santa Cruz Biotechnology) monoclonal primary antibodies, followed by goat anti‐rat immunoglobulin G–horseradish peroxidase (IgG–HRP):sc‐2006 (Santa Cruz Biotechnology) or chicken anti‐mouse IgG–HRP:sc‐2954 (Santa Cruz Biotechnology) secondary antibody. For loading controls, membranes were probed with mouse α-tubulin sc-5286 (Santa Cruz Biotechnology) or α-GFP (11814460001; Roche) monoclonal primary antibody and anti-mouse secondary antibody. Signals were detected after incubating with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) and band intensities were quantified by Fiji software (72).
Fluorescence Microscopy Analysis.
Whole Arabidopsis roots and N. benthamiana leaf epidermal cells were imaged using a Zeiss LSM710 confocal scanning microscope equipped with 40× 1.2 W C-Apochromat objective, an X-Cite 120 light-emitting diode fluorescent lamp, and narrow-band fluorescent filter cubes. CFP, GFP, RFP, and YFP were excited by 439-nm, 488-nm, 584-nm, and 514-nm argon lasers, respectively, and collected by 485/20-nm, 516/20-nm, 610/10-nm and 540/25-nm emission filters. Fluorescent signals were detected using two HyD detectors in photon-counting mode (single sections) and normal mode (z-series). Bright-field images were taken using differential interference contrast optics and overlaid with fluorescence. To detect colocalization of autophagic vesicles, both fluorescence signals were sequentially line-captured using similar settings but a with narrower detection window. Images were collected and processed using ZEN 2009 software and edited using Fiji software (72). To calculate the colocalization of fluorescent signals, the images were thresholded using the Coste’s method (73), and Manders’ colocalization coefficient (35) for overlays was calculated using Fiji software (72).
qRT-PCR Analyses.
Whole roots were isolated and total RNA extracted using the RNeasy Plus kit (Qiagen). Complemetary DNA was synthesized from the DNase-treated RNA. qRT-PCR was performed with 2x PowerUP SYBRGreen Master Mix in the QuantStudio 6 Flex Real‐Time PCR System (Applied Biosystems). GAPDH (AT1G13440) was used as housekeeping gene for normalization in all reactions. Primer sequences are provided in SI Appendix, Table S1. Each sample was analyzed six times, including three biological replicates and two technical replicates each. Gene expression was determined using the ∆∆Ct method of Pfaffl and presented as relative quantitation of target genes compared to the housekeeping genes.
Supplementary Material
Acknowledgments
This work was supported by grants from the NSF (IOS-1856431 to J.J.K. and MCB-1856248 to J.J.K. and G.E.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. H.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013161117/-/DCSupplemental.
Data Availability.
All study data are included in the paper and SI Appendix.
References
- 1.Dikic I., Proteasomal and autophagic degradation systems. Annu. Rev. Biochem. 86, 193–224 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Marshall R. S., Vierstra R. D., Autophagy: The master of bulk and selective recycling. Annu. Rev. Plant Biol. 69, 173–208 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N., et al., A protein conjugation system essential for autophagy. Nature 395, 395–398 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Thompson A. R., Vierstra R. D., Autophagic recycling: Lessons from yeast help define the process in plants. Curr. Opin. Plant Biol. 8, 165–173 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Wang P., Mugume Y., Bassham D. C., New advances in autophagy in plants: Regulation, selectivity and function. Semin. Cell Dev. Biol. 80, 113–122 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshimoto K., et al., Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21, 2914–2927 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Signorelli S., Tarkowski Ł. P., Van den Ende W., Bassham D. C., Linking autophagy to abiotic and biotic stress responses. Trends Plant Sci. 24, 413–430 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stolz A., Ernst A., Dikic I., Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 16, 495–501 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Stephani M., Dagdas Y., Plant selective autophagy—Still an uncharted territory with a lot of hidden gems. J. Mol. Biol. 432, 63–79 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Noda N. N., Ohsumi Y., Inagaki F., Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 584, 1379–1385 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Kulich I., et al., Arabidopsis exocyst subcomplex containing subunit EXO70B1 is involved in autophagy-related transport to the vacuole. Traffic 14, 1155–1165 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Pecenková T., Marković V., Sabol P., Kulich I., Žárský V., Exocyst and autophagy-related membrane trafficking in plants. J. Exp. Bot. 69, 47–57 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Cvrčková F., Zárský V., Old AIMs of the exocyst: Evidence for an ancestral association of exocyst subunits with autophagy-associated Atg8 proteins. Plant Signal. Behav. 8, e27099 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zientara-Rytter K., et al., Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy 7, 1145–1158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osugi A., et al., Systemic transport of trans-zeatin and its precursor have differing roles in Arabidopsis shoots. Nat. Plants 3, 17112 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Nolan T. M., et al., Developmental cell selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev. Cell 41, 33–46.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michaeli S., Honig A., Levanony H., Peled-Zehavi H., Galili G., Arabidopsis ATG8-INTERACTING PROTEIN1 is involved in autophagy-dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell 26, 4084–4101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gou W., et al., Autophagy in plant: A new orchestrator in the regulation of the phytohormones homeostasis. Int. J. Mol. Sci. 20, 2900 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kieber J. J., Schaller G. E., Cytokinins. Arabidopsis Book 12, e0168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wada S., et al., Autophagy supports biomass production and nitrogen use efficiency at the vegetative stage in rice. Plant Physiol. 168, 60–73 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guiboileau A., et al., Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol. 194, 732–740 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Breeze E., et al., High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23, 873–894 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y., et al., Autophagy regulates programmed cell death during the plant innate immune response. Cell 121, 567–577 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Slavikova S., Ufaz S., Avin-Wittenberg T., Levanony H., Galili G., An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. J. Exp. Bot. 59, 4029–4043 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masclaux-Daubresse C., et al., Stitching together the multiple dimensions of autophagy using metabolomics and transcriptomics reveals impacts on metabolism, development, and plant responses to the environment in Arabidopsis. Plant Cell 26, 1857–1877 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakai H., et al., ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294, 1519–1521 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Hwang I., Sheen J., Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413, 383–389 (2001). [DOI] [PubMed] [Google Scholar]
- 28.To J. P. C., et al., Type-A ARRs are partially redundant negative regulators of cytokinin signaling in Arabidopsis. Plant Cell 16, 658–671 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.To J. P., et al., Cytokinin regulates type-A Arabidopsis response regulator activity and protein stability via two-component phosphorelay. Plant Cell 19, 3901–3914 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryu M. Y., Cho S. K., Kim W. T., RNAi suppression of RPN12a decreases the expression of type-A ARRs, negative regulators of cytokinin signaling pathway, in Arabidopsis. Mol. Cells 28, 375–382 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Li Y., Kurepa J., Smalle J., AXR1 promotes the Arabidopsis cytokinin response by facilitating ARR5 proteolysis. Plant J. 74, 13–24 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Ren B., et al., Genome-wide comparative analysis of type-A Arabidopsis response regulator genes by overexpression studies reveals their diverse roles and regulatory mechanisms in cytokinin signaling. Cell Res. 19, 1178–1190 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Chi W., et al., DEG9, a serine protease, modulates cytokinin and light signaling by regulating the level of ARABIDOPSIS RESPONSE REGULATOR 4. Proc. Natl. Acad. Sci. U.S.A. 113, E3568–E3576 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brenner W. G., Ramireddy E., Heyl A., Schmülling T., Gene regulation by cytokinin in Arabidopsis. Front. Plant Sci. 3, 8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manders E. M. M., Verbeek F. J., Aten J. A., Measurement of co‐localization of objects in dual‐colour confocal images. J. Microsc. 169, 375–382 (1993). [DOI] [PubMed] [Google Scholar]
- 36.Synek L., et al., AtEXO70A1, a member of a family of putative exocyst subunits specifically expanded in land plants, is important for polar growth and plant development. Plant J. 48, 54–72 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teh O.-K., et al., Phosphorylation of the exocyst subunit Exo70B2 contributes to the regulation of its function. bioRxiv:10.1101/266171 (19 February 2018).
- 38.Dortay H., et al., Toward an interaction map of the two-component signaling pathway of Arabidopsis thaliana. J. Proteome Res. 7, 3649–3660 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Ding Y., et al., Exo70E2 is essential for exocyst subunit recruitment and EXPO formation in both plants and animals. Mol. Biol. Cell 25, 412–426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalvari I., et al., iLIR: A web resource for prediction of Atg8-family interacting proteins. Autophagy 10, 913–925 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alemu E. A., et al., ATG8 family proteins act as scaffolds for assembly of the ULK complex: Sequence requirements for LC3-interacting region (LIR) motifs. J. Biol. Chem. 287, 39275–39290 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J., et al., EXPO, an exocyst-positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in Arabidopsis and tobacco cells. Plant Cell 22, 4009–4030 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ichimura Y., et al., Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 283, 22847–22857 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Zhang X., et al., Reticulon proteins modulate autophagy of the endoplasmic reticulum in maize endosperm. eLife 9, e51918 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson A. R., Doelling J. H., Suttangkakul A., Vierstra R. D., Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138, 2097–2110 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kurusu T., et al., Autophagy-mediated regulation of phytohormone metabolism during rice anther development. Plant Signal. Behav. 12, e1365211 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Žárský V., Sekereš J., Kubátová Z., Pečenková T., Cvrčková F., Three subfamilies of exocyst EXO70 family subunits in land plants: Early divergence and ongoing functional specialization. J. Exp. Bot. 71, 49–62 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Li S., et al., Expression and functional analyses of EXO70 genes in Arabidopsis implicate their roles in regulating cell type-specific exocytosis. Plant Physiol. 154, 1819–1830 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zárský V., Kulich I., Fendrych M., Pečenková T., Exocyst complexes multiple functions in plant cells secretory pathways. Curr. Opin. Plant Biol. 16, 726–733 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Drdová E. J., et al., The exocyst complex contributes to PIN auxin efflux carrier recycling and polar auxin transport in Arabidopsis. Plant J. 73, 709–719 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Li S., et al., EXO70A1-mediated vesicle trafficking is critical for tracheary element development in Arabidopsis. Plant Cell 25, 1774–1786 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hála M., et al., An exocyst complex functions in plant cell growth in Arabidopsis and tobacco. Plant Cell 20, 1330–1345 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kalmbach L., et al., Transient cell-specific EXO70A1 activity in the CASP domain and Casparian strip localization. Nat. Plants 3, 17058 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Fendrych M., et al., The Arabidopsis exocyst complex is involved in cytokinesis and cell plate maturation. Plant Cell 22, 3053–3065 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Synek L., et al., EXO70C2 is a key regulatory factor for optimal tip growth of pollen. Plant Physiol. 174, 223–240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pecenková T., et al., The role for the exocyst complex subunits Exo70B2 and Exo70H1 in the plant-pathogen interaction. J. Exp. Bot. 62, 2107–2116 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang W., Liu N., Gao C., Rui L., Tang D., The Pseudomonas syringae effector AvrPtoB associates with and ubiquitinates Arabidopsis exocyst subunit EXO70B1. Front. Plant Sci. 10, 1027 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kurepa J., Li Y., Smalle J. A., Proteasome-dependent proteolysis has a critical role in fine-tuning the feedback inhibition of cytokinin signaling. Plant Signal. Behav. 8, e23474 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu F., Xie Q., Non-26S proteasome endomembrane trafficking pathways in ABA signaling. Trends Plant Sci. 22, 976–985 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Kulich I., et al., Arabidopsis exocyst subunits SEC8 and EXO70A1 and exocyst interactor ROH1 are involved in the localized deposition of seed coat pectin. New Phytol. 188, 615–625 (2010). [DOI] [PubMed] [Google Scholar]
- 61.Sláviková S., et al., The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J. Exp. Bot. 56, 2839–2849 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Le Bars R., Marion J., Le Borgne R., Satiat-Jeunemaitre B., Bianchi M. W., ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat. Commun. 5, 4121 (2014). [DOI] [PubMed] [Google Scholar]
- 63.Zürcher E., et al., A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol. 161, 1066–1075 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grefen C., et al., A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64, 355–365 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Nakagawa T., et al., Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Dortay H., Mehnert N., Bürkle L., Schmülling T., Heyl A., Analysis of protein interactions within the cytokinin-signaling pathway of Arabidopsis thaliana. FEBS J. 273, 4631–4644 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Earley K. W., et al., Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629 (2006). [DOI] [PubMed] [Google Scholar]
- 68.Yang Y., Li R., Qi M., In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 22, 543–551 (2000). [DOI] [PubMed] [Google Scholar]
- 69.Dröse S., et al., Inhibitory effect of modified bafilomycins and concanamycins on P- and V-type adenosinetriphosphatases. Biochemistry 32, 3902–3906 (1993). [DOI] [PubMed] [Google Scholar]
- 70.Gietz D., Woods R., “Yeast transformation by the LiAc/SS carrier DNA/PEG method” in Methods in Molecular Biology, Xiao W., Ed. (Humana Press, Totowa, NJ, 2006), pp. 107–120. [DOI] [PubMed] [Google Scholar]
- 71.Chiba M., et al., Diverse suppressors of RNA silencing enhance agroinfection by a viral replicon. Virology 346, 7–14 (2006). [DOI] [PubMed] [Google Scholar]
- 72.Schindelin J., et al., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Costes S. V., et al., Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 86, 3993–4003 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the paper and SI Appendix.