Significance
Novel antibiotics are urgently needed to treat the ever-increasing number of drug-resistant infections. Venoms constitute a treasure trove of novel potential medicines. Here, we converted a peptide derived from venom into potent antimicrobials capable of resolving otherwise lethal infections in mice. We demonstrate that the peptide acts directly on bacteria by targeting their membrane, while also modulating the host immune response and dampening unwanted inflammation. Venom-derived molecules such as the ones described here represent an exciting new source of antibiotics.
Keywords: venoms, antimicrobial peptides, immunomodulatory peptides, antiinfectives, structure–activity relationship
Abstract
Novel antibiotics are urgently needed to combat multidrug-resistant pathogens. Venoms represent previously untapped sources of novel drugs. Here we repurposed mastoparan-L, the toxic active principle derived from the venom of the wasp Vespula lewisii, into synthetic antimicrobials. We engineered within its N terminus a motif conserved among natural peptides with potent immunomodulatory and antimicrobial activities. The resulting peptide, mast-MO, adopted an α-helical structure as determined by NMR, exhibited increased antibacterial properties comparable to standard-of-care antibiotics both in vitro and in vivo, and potentiated the activity of different classes of antibiotics. Mechanism-of-action studies revealed that mast-MO targets bacteria by rapidly permeabilizing their outer membrane. In animal models, the peptide displayed direct antimicrobial activity, led to enhanced ability to attract leukocytes to the infection site, and was able to control inflammation. Permutation studies depleted the remaining toxicity of mast-MO toward human cells, yielding derivatives with antiinfective activity in animals. We demonstrate a rational design strategy for repurposing venoms into promising antimicrobials.
The excessive use and misuse of antibiotics has led to increasing rates of resistance in pathogenic bacteria (1). According to the Centers for Disease Control and Prevention in 2019, 2.8 million antibiotic-resistant infections occur in the United States each year, leading to ∼35,000 deaths annually (2). In addition, 19.7% (∼11 million) of all global deaths in 2017 were caused by sepsis, a syndrome most commonly linked to bacterial infections and for which antibiotics are the primary treatment (3). Drug-resistant infections are not only serious on their own; secondary infections are also highly relevant and represent a major cause of death in global pandemics, being responsible for 95% of deaths during the 1918 flu pandemic (4), and also playing a role in the current COVID-19 pandemic (5).
The emergence of multidrug-resistant bacterial pathogens has coincided with a severe decline in the development and approval of new antibacterial drugs (1). Thus, there is an urgent need for novel antibiotics effective against drug-resistant bacteria. Toxins represent a previously untapped source of potential antimicrobials. They are contained in venoms and have evolved through the tree of life (e.g., in plants, animals, and microbes) as part of defensive strategies. Typically, crude venoms contain a wide range of different peptide toxins, many of which are small in size, easy to synthesize, structurally stable, and highly biodiverse (6). Antimicrobial peptides (AMPs), a promising class of antibiotic candidates (7), have been found in venoms (8). AMPs have an almost unlimited sequence space and multiple mechanisms of action against microorganisms (1, 8–14). These agents can present broad- or narrow-spectrum activity and rapid antimicrobial (8, 10, 11, 15) and immunomodulatory (12, 16–18) properties, which allow them to act against a range of microorganisms, including gram-negative and gram-positive bacteria, fungi, viruses, and parasites. Relevant to their potential clinical translatability, bacteria exposed to AMPs have been shown to develop resistance at a much lower rate than when exposed to conventional antibiotics (19).
Cationic peptides displaying an α-helical structure constitute a class of AMPs found in venoms. This class consists of small amphipathic peptides with net positive charge that tend to adopt an α-helical structure upon contact with hydrophilic/hydrophobic interfaces, such as the membrane of microorganisms (9). One such example is mast-L, an AMP from the mastoparan family (20–22), derived from the venom of the social wasp Vespula lewisii (23). However, the use of this peptide as a therapeutic has been hindered by its toxicity profile, as it displays high hemolytic activity (14) and promotes massive mast cell degranulation (24, 25). Here, using rational design, we increased the therapeutic properties of mast-L, while decreasing its inherent cytotoxicity. Through this approach, novel antiinfective synthetic peptides were generated that lacked toxicity and displayed potent antimicrobial and immunomodulatory activities in clinically relevant sepsis and skin infection animal models.
Results and Discussion
Design and Structural Analyses of Wasp Venom-Derived Peptides.
Despite the promise of mast-L as an antimicrobial, its activity must be improved to surpass that of standard-of-care antibiotics in order to enter clinical trials. In order to increase its antimicrobial and immunomodulatory properties, we performed a comprehensive search and analysis of the Antimicrobial Peptides Database (26) looking for conserved motifs associated with such biological functions. The pentapeptide motif (FLPII) was identified to be conserved in the N-terminal extremity of peptides with high antimicrobial and immunomodulatory (e.g., chemotactic) activities, including brevenins (27, 28), temporins (28), vespid chemotactic peptides (29) (Fig. 1A and SI Appendix, Table S1), and mediating interactions with membranes in plant lipocalins (30). In order to design a synthetic peptide with increased biological activity, we engineered the motif FLPII into the N-terminal extremity of mast-L (Fig. 1B), as changes in this part of the sequence are known to lead to loss of toxicity toward neutral vesicles mimicking human cell membranes (SI Appendix, Table S1).
Fig. 1.
Repurposing a toxic mastoparan peptide from wasp venom into a viable antimicrobial. (A) Schematic representation of the design approach based on (B) the insertion into the mast-L sequence of a pentapeptide motif (FLPII) conserved in naturally occurring peptides (e.g., brevenin, temporin, and vespid chemotactic peptide families) with potent antimicrobial, immunomodulatory, and chemotactic properties. Such insertion yielded the synthetic peptide mast-MO.
Mastoparans, like other helical AMPs, are unstructured in aqueous solution and tend to structure into an α-helix upon contact with hydrophobic/hydrophilic interfaces, such as the one between bacterial membranes and the surrounding environment. The antimicrobial activity of such AMPs correlates with their ability to undergo the structural conformation change from unstructured to helical (8), and even small modifications to their sequence impact their structural tendency and biological activity. In order to assess whether insertion of the FLPII motif affected peptide structure, we elucidated the 3D structure of both mast-L and mast-MO through circular dichroism (CD) (SI Appendix, Fig. S1A) and of mast-MO using NMR (Table 1 and SI Appendix, Fig. S1 B and C). The CD experiments indicated that both peptides (SI Appendix, Fig. S1) presented well-defined helical structures in the presence of trifluoroethanol (TFE)/water (1:1; v:v), SDS (100 mmol L−1) micelles, and phospholipid vesicles. The positive band around 192 to 193 nm and two negative bands at 208 and 222 nm (SI Appendix, Fig. S1A) observed for both mast-L and mast-MO are characteristic profiles of α-helical structures (31). The highest α-helical content values, recorded for both peptides, was in SDS micelles (75.0% for mast-L and 79.5% for mast-MO), indicating that they undergo coil-helical transition upon contact with hydrophobic/hydrophilic interfaces, such as bacterial membranes. The same behavior was observed when peptides were added to other hydrophobic/hydrophilic interfaces, such as dodecylphosphocholine (DPC) vesicles, TFE/water mixture (3:2, v:v), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC): 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG) (3:1, 10 mmol L−1) vesicle.
Table 1.
Cytotoxic activity profile of mast-MO and mast-L
| Cell line | LC100 (µmol L−1) | ||||
| Mast- L | Mast-MO | LL-37 | Gentamicin | Imipenem | |
| hRBCs | 7 | >400 | 87 | >400 | >400 |
| HEK293 | 50 | 25 | NT | NT | NT |
| L929 | 10 | >400 | 95 | >400 | >400 |
| RAW 264.7 | 10 | >400 | 100 | >400 | >400 |
The cytotoxicity of the peptides was evaluated against human red blood cells (hRBCs), murine fibroblasts (L929), murine macrophages monocytes (RAW 264.7), and human embryonic kidney (HEK293) cells. Results indicate the LC100, which is the concentration at which 100% of cells are killed. NT, not tested.
The α-helical structure of mast-MO was confirmed by 2D-NMR spectroscopy in deuterated SDS-d25 micelles (100 mmol L−1). The 1H resonance assignments were performed by simultaneous analyses of the TOCSY and NOESY contour maps (SI Appendix, Fig. S1 B and C), as previously described (32). The structural statistics for low-energy structures are detailed in SI Appendix, Table S2. The secondary structure of the peptides was predicted by chemical-shift index (CSI) using NMRView, and the experimental chemical shifts of Hα, Cα, HN, and N. CSI showed a well-defined α-helical structure throughout the whole sequence (between Leu3 and Leu14 residues) of mast-L and the maintenance of this helical behavior in mast-MO even with the addition of the pentapeptide motif (between Ile5 and Ile17 residues). We confirmed the α-helical structure using the TALOS+ software, in which we estimated the dihedral angles, phi (ϕ) and psy (ψ) torsion angles (SI Appendix, Table S3) of the peptide backbone and classified the values obtained as consistent with the predicted structures by CSI.
Mastoparan peptides depend on their helical structure to exert antimicrobial activity (8). After initial electrostatic interactions with the membrane, this class of peptides tends to adopt an amphipathic helical structure with well-defined hydrophobic and hydrophilic faces upon contact with the phospholipids present on the membrane lipid bilayer, leading to membrane destabilization. Thus, our structural data show that mast-MO preserved the helical tendency of mast-L, a critical requirement for these peptides to display antimicrobial activity.
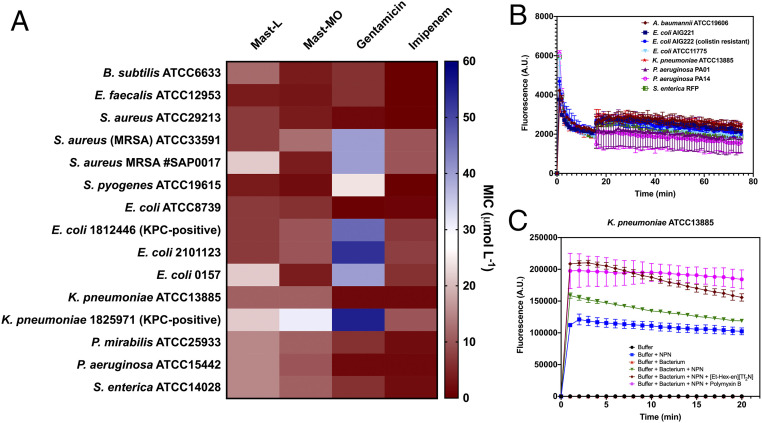
Synthetic peptide mast-MO exhibits decreased toxicity and increased antimicrobial and immunomodulatory activities compared to parent wasp venom toxin mast-L. Helical conformations are known to be crucial for the antimicrobial activity of mastoparans (8). Once we confirmed that insertion of the conserved pentapeptide motif FLPII did not alter the helical tendency of the peptide, we performed a series of bacterial growth inhibition assays in vitro to evaluate whether the motif influenced biological function. First, we assessed the minimal inhibitory concentration (MIC) of the peptide against 15 pathogens (Fig. 2A and SI Appendix, Fig. S2). Pathogenic bacteria, including members of the ESKAPE list of the World Health Organization (2) and antibiotic-resistant strains, were exposed to increasing concentrations of the peptides for 24 h. Overall, mast-MO presented a similar activity profile, although slightly superior, compared to its predecessor mast-L, and increased activity with respect to two standard-of-care antibiotics (gentamicin and imipenem) (Fig. 2A). Briefly, mast-MO presented increased activity against gram-positive bacteria compared to gram-negatives, and it was slightly more effective than mast-L against the majority of strains tested, with the exceptions of Staphylococcus aureus methicillin-resistant S. aureus (MRSA), Escherichia coli (2101123), E. coli 0157, and Klebsiella pneumoniae 1825971 (KPC971). Mast-MO (4 μg mL−1) also exhibited increased antibiofilm properties against E. coli ATCC0157 and S. aureus ATCC33591 compared to mast-L both in vitro (SI Appendix, Fig. S3) and in vivo (see, for example, Fig. 5F).
Fig. 2.
Antimicrobial activity and mechanism of action of wasp venom-derived peptide. (A) Antimicrobial activity of mast-L, mast-MO, and standard-of-care antibiotics gentamicin and imipenem against 15 clinically relevant pathogens. (B) Cytoplasmic membrane depolarization effects of mast-MO against eight different gram-negative pathogens determined using the DiSC3(5) assay and (C) NPN assay demonstrating the ability of mast-MO to potently damage and permeabilize the bacterial outer membrane compared to the controls in the absence of mast-MO.
Fig. 5.
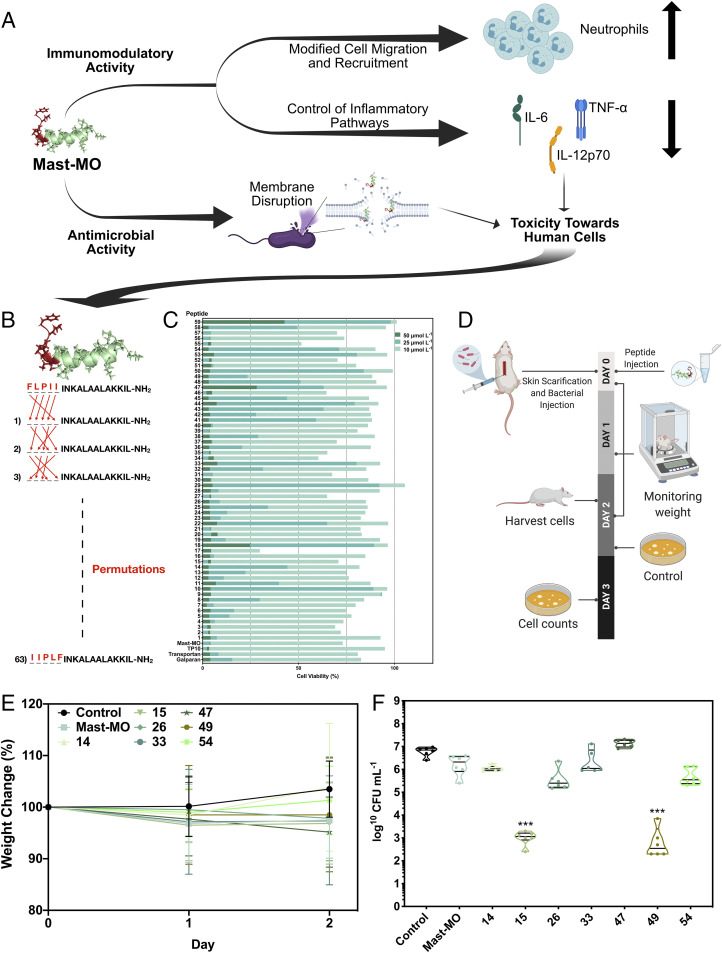
Permutations of the conserved motif lead to mast-MO variants that lack toxicity against human cells and display antiinfective activity in a mouse model. (A) Schematic showing the dual immunomodulatory and antimicrobial activities of mast-MO demonstrated in this work. The peptide killed bacterial cells by destabilizing their outer membrane and displayed immunomodulatory properties by attracting leukocytes to the site of infection and antiinflammatory activity by repressing proinflammatory mediators (e.g., TNF-α and IL-12). Despite these therapeutic properties, mast-MO retained some cytotoxicity toward human cells at concentrations immediately above its antimicrobial and immunomodulatory activities, hindering its use as a potential therapy in humans. (B) Design of mast-MO derivatives through permutation of its FLPII motif. (C) Cytotoxic activity of mast-MO and its permutation variants against human embryonic kidney cells (i.e., HEK293). (D) Schematic of the experimental design. Briefly, the back of mice was shaved, and an abrasion was generated to damage the stratum corneum and the upper layer of the epidermis. Subsequently, an aliquot of 50 μL containing 5 × 107 CFU/20 μL of P. aeruginosa in PBS was inoculated over each defined area. One day postinfection, peptides (16 μmol L−1) were administered to the infected area. (E) Mouse body weight measurements throughout the experiment were taken and normalized by the body weight of noninfected mice as a proxy for potential toxicity. (F) Antiinfective activity of the permutation-based synthetic analogs compared to mast-MO–treated and negative control groups. Briefly, mice were anesthetized with isoflurane, had their backs shaved, and a superficial linear skin abrasion was made with a needle in order to damage the stratum corneum and upper layer of the epidermis. A bacterial load 1 × 107 CFU/20 μL of bacteria (in 20 μL) in PBS was inoculated over each defined area containing the scratch with a pipette tip. One day after the infection, a single dose of each peptide (16 μmol L−1) was administered to the infected area. Six animals per group were killed and the area of scarified skin was excised 2 d postinfection, homogenized using a bead beater for 20 min (25 Hz), and serially diluted for CFU quantification (statistical significance was determined using two-way ANOVA followed by Dunnett’s test, ***P < 0.001).
Mastoparans are known to act through several mechanisms of action (14, 21, 23–25, 33, 34), and even minor modifications to their sequence have been shown to lead to different activity profiles (14, 21, 34). In order to gain insights into the mechanisms of action by which these peptides kill bacterial cells, we used the probes 3,3′-dipropylthiadicarbocyanine iodide [DiSC3(5)] and 1-(N-phenylamino)-naphthalene (NPN) to assess whether mast-MO was capable of depolarizing the cytoplasmic membrane and permeabilizing the outer membrane of bacteria, respectively. DiSC3(5) is a potentiometric fluorophore that accumulates in the cytoplasmic membrane of bacterial cells. When the accumulation of DiSC3(5) molecules reaches a certain threshold, those molecules aggregate, quenching their fluorescence. Upon disbalance on the transmembrane potential of the membrane, the fluorophore migrates to the cytoplasm or the outer environment, fluorescing again. In these assays, mast-MO only mildly depolarized the cytoplasmic membrane of eight different clinically relevant gram-negative bacterial strains (Fig. 2B), against which we had observed antimicrobial activity (SI Appendix, Fig. S2). On the other hand, mast-MO potently permeabilized the outer membrane of the same set of eight bacterial strains (Fig. 2C and SI Appendix, Fig. S4), as determined by increased fluorescence of the lipophilic dye NPN in the NPN assay, indicating that this is one of the mechanisms of action of the peptide. NPN generates weak fluorescence in aqueous environments and is unable to permeate bacterial membranes unless they are damaged, but its fluorescence increases upon contact with lipidic environments, such as the interior of the lipid bilayers of bacterial membranes, therefore indicating that the membrane has been permeabilized and is thus compromised.
Next, we assessed whether mast-MO was capable of synergistically interacting with different classes of commonly used antibiotics. First, we screened 12 antibiotics with different mechanisms of action against three bacterial strains (colistin-resistant E. coli AIG222, Acinetobacter baumannii ATCC19606 and Pseudomonas aeruginosa PAO1) (SI Appendix, Fig. S5). Then, we selected four antibiotics with moderate to high activity against each of the selected strains and screened, in checkerboard assays, serial dilutions of the antibiotics in the presence of increasing concentrations of mast-MO (SI Appendix, Fig. S6). Mast-MO presented its highest synergistic interactions (fractional inhibitory concentration index = 0.1) when combined with antibiotics that block protein synthesis, such as kanamycin, chloramphenicol, and gentamicin, against colistin-resistant E. coli and A. baumannii cells (SI Appendix, Fig. S6). We speculate that mast-MO permeabilizes the outer membrane, thus allowing these antibiotics to penetrate into the cell reaching their target (i.e., blocking the protein synthesis machinery).
Once we elucidated the synergistic interactions of mast-MO with a range of antibiotics, in addition to the mechanisms by which the peptide targeted bacterial cells, we evaluated the effect of the motif insertion on peptide stability and cytotoxicity against mammalian cells. Mast-MO was as stable as mast-L (SI Appendix, Fig. S7) when exposed to proteolytic enzymes contained in fetal bovine serum. Mast-MO did not present hemolytic activity in human red blood cell assays at the concentration range tested (1 to 400 µmol L−1), but it was toxic at 25 µmol L−1 against the human embryonic cell line HEK293, whereas mast-L was both hemolytic and toxic toward the different human and murine cell lines tested at 7 to 50 µmol L−1 (Table 1). Interestingly, mast-MO was not cytotoxic when incubated with mouse cell lines, such as murine fibroblasts (L929 cells) and murine macrophages (RAW 264.7 cells) (Table 1). Therefore, we proceeded to test the antiinfective activity of mast-MO in preclinical mouse models. First, we analyzed the toxicity of a single dose of either mast-L or mast-MO administered peritoneally to mice in order to determine the range of concentrations to be used in subsequent animal experiments. We also tested the hemolytic activity of mast-MO (up to 200 µmol L−1) against murine red blood cells (SI Appendix, Table S5) prior to conducting animal experiments. In preliminary mouse experiments, mast-L was highly toxic at concentrations higher than 10 mg kg−1, whereas mast-MO lacked toxicity (SI Appendix, Table S4), in line with our results using mouse cell lines (Table 1). Based on these results, we used concentrations ranging from 1 to 10 mg kg−1 of mast-MO and 1 to 5 mg kg−1 of mast-L, which are not cytotoxic, in subsequent experiments.
To test whether the FLPII motif within the mast-MO sequence led to increased immunomodulatory and chemotactic properties with respect to mast-L, we performed ELISA experiments using the peritoneal fluid collected from mice (see for example, Fig. 4A). We infected C57BL/6 mice with both gram-negative and gram-positive pathogens and administered either mast-L or mast-MO intraperitoneally. For mice infected with E. coli ATCC8739, we monitored the peritoneal cavity to detect and quantify the release and presence of several immune mediators (Fig. 3E). Mast-MO–treated groups exhibited a pronounced ability to induce leukocyte migration to the site of infection, a key step into resolving infections, compared to the mast-L–treated group (Fig. 3B). This effect was similar to the positive control group of mice treated with transglutaminase (TGA) (Fig. 3B). Whereas mast-L did not significantly induce leukocyte recruitment over the time period tested, mast-MO stimulated recruitment at all time points with a peak of ∼108 observed 3 h postinfection (Fig. 3B). After 24 h, the leukocyte recruitment levels in mast-MO–treated mice dropped to ∼106, likely attributed to the resolution of the infection by the peptide (Fig. 4 B and C). These results are consistent with the intrinsic ability of the pentapeptide motif FLPII to act as a chemoattractant (Fig. 1A), in line with our initial design, and highlight the immune boosting capability of our designed peptide.
Fig. 4.
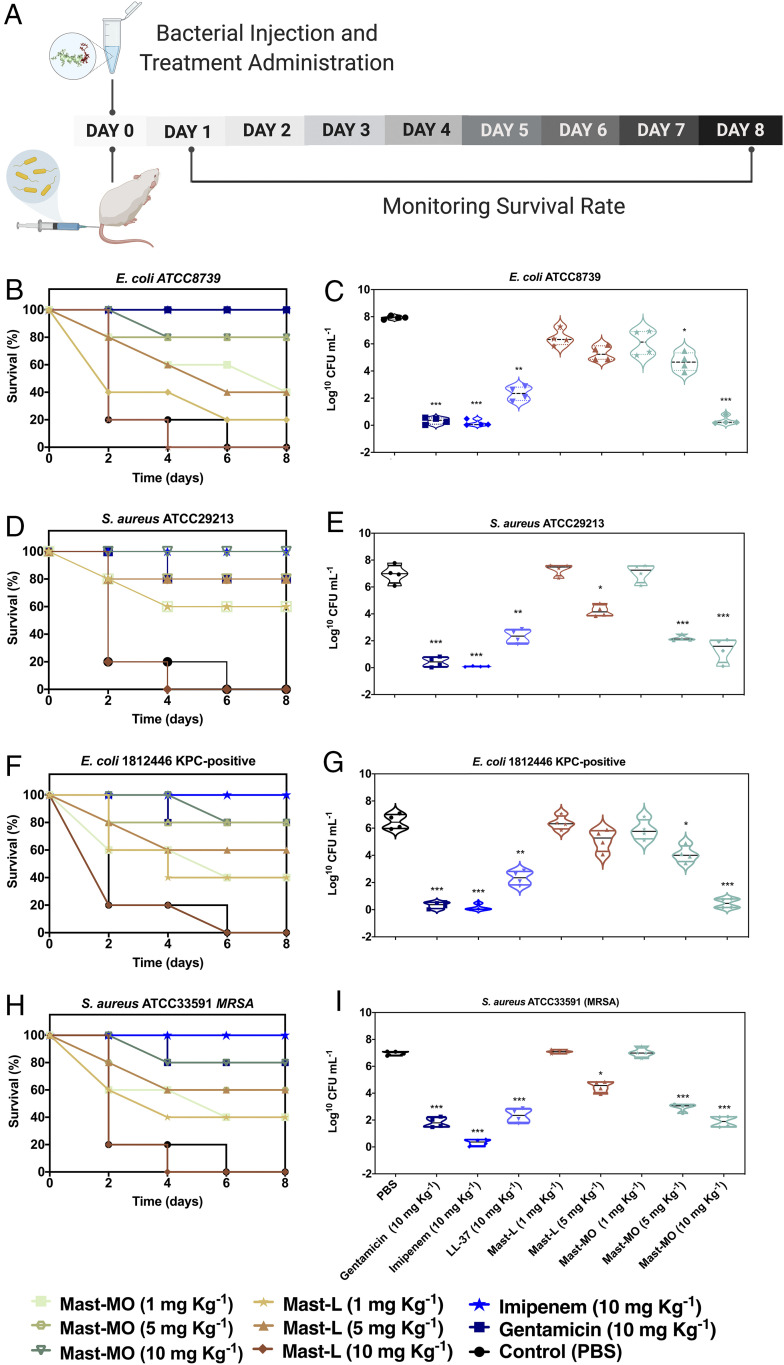
Antiinfective activity of mast-MO against gram-negative and gram-positive pathogens in a lethal sepsis infection model. (A) Mice were infected with bacteria, treated with each peptide or antibiotic, and monitored for 8 d. Survival and cell counts after 24 h, respectively, are shown for E. coli ATCC8739 (2 × 104 CFU/mL inoculum) (B and C), S. aureus ATCC29213 (2 × 109 CFU/mL inoculum) (D and E), E. coli 1812446 (KPC+) (2 × 104 CFU inoculum) (F and G), and S. aureus (MRSA) ATCC33591 (2 ×109 CFU inoculum) (H and I). Mice were treated intraperitoneally with 1, 5, and 10 mg kg−1 of mast-L, mast-MO, or 10 mg kg−1 of the antibiotics gentamicin and imipenem. PBS was used as a control for the untreated group of mice. Treatments were administered 3 h postinfection. Data are expressed as the mean ± SD. Statistical analysis was performed using Bonferroni test. *P < 0.1, **P < 0.01, ***P < 0.001 compared to the untreated control.
Fig. 3.
Engineered peptide mast-MO triggers immune cell migration and represses inflammation pathways in vivo. (A) Schematic of different interactions displayed by mastoparan peptides on the surface of biological membranes leading to internalization into the cell and subsequent immunomodulatory effects. (B) Mast-MO (10 mg kg−1) triggered leukocyte recruitment to the E. coli ATCC8739 infection site to levels comparable to the positive control TGA, (C) release over time (48 h) of IL-12, and (D) TNF-α into the peritoneal cavity of mice. Cytokines monitored in mice for 24 h after infection with (E) E. coli ATCC8739 and (F) S. aureus ATCC29213. Cytokine release (i.e., IFN-γ, IL-6, IL-10, IL-12p70, and TNF-α) into the peritoneal cavity of C57BL/6 mice was detected and quantified 24 h after infection with E. coli ATCC8739 and S. aureus ATCC29213. In all experiments, mice were treated with 10 mg kg−1 of mastoparan-MO and 5 mg kg−1 of mastoparan-L. PBS and imipenem (10 mg kg−1) were used as negative and positive controls, respectively. Data were expressed as mean ± SD. Statistical analysis was performed using Bonferroni test. *P < 0.1, ***P < 0.001 as significant compared to the control.
We also longitudinally tracked for 48 h the concentration of IL-12, a protein produced mainly by macrophages and neutrophils in response to antigenic stimulation, and the proinflammatory cytokine TNF-α. Mast-MO and mast-L repressed both proinflammatory cytokines, in addition to IL-6, thus showing their ability to repress inflammation in vivo (Fig. 3 C–F). We monitored, via ELISAs, the cytokine profiles present within the peritoneal cavity of mice infected with E. coli ATCC8739 and S. aureus ATCC29213 (Figs. 3E and 4F, respectively) and treated with a single dose of peptide. Treatment with peptides in both cases led to a significant decrease in the inflammatory response within the peritoneal fluid resulting from the bacterial infection, thus highlighting the potential of the peptides as antiinflammatory agents. These results confirmed that mast-MO appeared to control inflammation in addition to acting as a chemoattractant for leukocytes. Overall, our data suggest that mast-MO helps balance host responses by reducing the prolonged production of proinflammatory mediators while enhancing the activity and recruitment of leukocytes, which are important for baseline immune function.
Synthetic Peptide Mast-MO Confers Protection to Mice Against Lethal Septicemia Caused by Bacterial Infections.
To determine the antiinfective activity of mast-MO in a preclinical mouse model, we tested its ability to kill pathogenic and drug-resistant E. coli and S. aureus strains in a systemic sepsis infection mouse model (Fig. 4). We first determined the minimal bacterial load resulting in a lethal dose (LD100) 24 h postinjection. The following LD100 were determined: ∼2 × 104 for E. coli ATCC8739 and E. coli 1812446 and ∼2 × 109 for S. aureus ATCC29213 and S. aureus (MRSA) ATCC33591. Mice were infected with the LD100 of bacteria and treated 3 h postinfection with either mast-L (1 and 5 mg kg−1) or mast-MO (1, 5, and 10 mg kg−1), which were also administered intraperitoneally. Mice infected with E. coli ATCC8739 and clinical isolate E. coli 1812446 (KPC+) and subsequently treated with mast-L (5 mg kg−1) displayed decreased survival compared to groups treated with mast-MO (5 and 10 mg kg−1) (Figs. 4 B and C and 5 E and F, respectively). Overall, mice treated with mast-MO exhibited 80% survival, whereas those treated with mast-L displayed 40 to 60% survival. Mouse groups treated with mast-L at 10 mg kg−1 had to be killed due to observed toxic effects when the peptide was injected systemically. Conversely, mice treated with mast-MO (10 mg kg−1) were completely protected from infection by S. aureus ATCC29213 (Fig. 4 D and E), while those treated with mast-L (5 mg kg−1) presented 80% survival. Finally, mice infected with S. aureus MRSA ATCC33591 and treated with mast-MO (10 mg kg−1) exhibited 80% survival (Fig. 4 H and I). Only 60% of mice infected with S. aureus MRSA ATCC33591 and treated with mastoparan-L (5 mg kg−1) survived. Once again, mice treated with 10 mg kg−1 of mast-L had to be killed.
In summary, in all systemic infection experiments performed, 10 mg kg−1 of mast-MO led to significant reduction of bacterial counts and high survival rates, comparable to standard-of-care antibiotics, such as the aminoglycoside gentamicin and the β-lactam imipenem, which are usually administered at 1 to 30 mg kg−1 in mouse models (35). Our results further confirmed in vivo that mast-MO did not present toxicity in the mouse model tested, whereas the wasp venom peptide mast-L led to toxicity when injected systemically at 10 mg kg−1. Mast-MO, even at relatively low doses (10 mg kg−1), proved to be an excellent antiinfective agent, leading to complete inhibition of otherwise lethal bacterial infections caused by drug-resistant and clinically relevant gram-negative and gram-positive pathogens. Our in vivo data also demonstrated the dual activity of our engineered synthetic peptide mast-MO to both modulate the immune response and directly kill bacteria (Fig. 5A).
Rational Design Yields Mast-MO Derivatives Lacking Toxicity against Human Cells and with Increased Antimicrobial and Antiinfective Activity in a Murine Model.
In order to deplete the remaining toxicity observed in mast-MO toward certain human cells (e.g., HEK293) (Table 1), we designed and synthesized a permutation library of 59 novel mast-MO derivatives (SI Appendix, Table S6) composed of single amino acid permutations of the FLPII motif (Fig. 5B). The FLPII motif was chosen for the permutations to preserve the natural antimicrobial activity of mast-L and also conserve the hydrophobic content inserted in the N-terminal extremity for the initial design of mast-MO. Next, we assessed the antimicrobial (SI Appendix, Fig. S8) and cytotoxic activities of the mast-MO analog library against HEK293 cells (Fig. 5C). This specific cell line was chosen as it derives from human embryonic kidney cells and potential antimicrobial (i.e., AMPs, antibiotics) toxicity affects the kidneys, which are responsible for clearing such molecules (36). We also measured whether the peptides aggregated at the highest concentration tested (128 µmol L−1) (SI Appendix, Table S6), since the motif inserted is composed of residues with aliphatic or aromatic side chain groups that may be prone to aggregation when in proximity within the primary peptide sequence, thus altering biological activity. Minor changes to the first five residues inserted in the mast-MO primary sequence led to a pronounced decrease in cytotoxic activity (2.5- to 5-fold) compared to mast-MO (Fig. 5C).
The most active peptides obtained, which were not cytotoxic and did not aggregate in aqueous solution, were selected and tested in a skin scarification mouse model that mimics an epithelial abscess infection (8, 13, 37). Mice were infected topically with the gram-negative pathogen P. aeruginosa and subsequently treated with a single low dose (16 μmol L−1) of the seven lead synthetic peptides (analogs 14, 15, 26, 33, 47, 49, and 54) (Fig. 5 D–F and SI Appendix, Table S5). The peptides did not cause any side effects or adverse toxicity (i.e., hemolysis or cytotoxicity) toward mouse or human cell lines (Fig. 5E). Synthetic analogs 15 (IFLPIINLKALAALAKKIL-NH2) and 49 (FPIILINLKALAALAKKIL-NH2) displayed increased antiinfective activity compared to their parent peptide mast-MO (Fig. 5E). Analog 15 is very similar to mast-MO as the permutation used to generate this molecule involved insertion an Ile residue at the beginning of the sequence while maintaining the last four residues of mast-MO. On the other hand, analog 49 presented all its aliphatic residues next to each other in positions three to five of the added motif, contrary to mast-MO that has a leucine residue in position two. Overall, we designed several bactericidal mast-MO analogs by permuting the 5 amino acid residues that compose the FLPII and two of these analogs presented high antiinfective activity with a single dosage in a skin abscess formation mouse model at low micromolar concentration (16 μmol L−1), comparable to the most active antiinfective peptides (8, 13).
Conclusions
Multidrug-resistant pathogens are becoming increasingly prevalent and no new classes of antibiotics have been discovered for decades. Therefore, strategies enabling the discovery of new strategies for treating infections are urgently needed. Toxins from the venom of insects and arthropods are rich in antimicrobials that represent an untapped source of template molecules for the design of novel drug candidates. Here, we leverage a sequential rational design strategy to convert a highly toxic peptide derived from wasp venom into nontoxic synthetic derivatives with drug-like properties and antiinfective activity in preclinical animal models. Initially, we engineer into mast-L a pentapeptide motif highly conserved in existing chemotactic and antimicrobial peptides. In relation to mast-L, the resulting synthetic peptide, mast-MO, presented enhanced antimicrobial activity by destabilizing the bacterial outer membrane, and exhibited immunomodulatory properties by increasing leukocyte migration to the infection site and repressing proinflammatory factors, crucial to allow infection clearance. Mast-MO was also shown to act as a potentiator of conventional antibiotics, thus suggesting future applications of this molecule as an adjuvant. In animal models, mast-MO treatment caused a decrease in proinflammatory cytokines IL-12, TNF-α, and IL-6 at the site of infection and increased leukocyte migration required for infection resolution. Since mast-MO retained some toxicity against human cells, we built, via permutation design, a second generation of analogs (Fig. 5). Seven identified synthetic peptides were no longer toxic against human cells and presented increased antiinfective activity against clinically relevant bacteria both in vitro and in vivo (Fig. 5).
The dual antimicrobial and immunomodulatory actions of mast-MO enable it to directly kill bacterial pathogens and selectively enhance host immune responses to clear infections while maintaining control of the inflammatory response. Multifunctional antimicrobials, such as the ones presented here, represent a new paradigm for treating infections by targeting both bacteria and the host. Moreover, the peptide did not induce any apparent immunotoxicities in any of the mouse models tested. Our study is an example of how leveraging template-based design strategies coupled with sequential rational design approaches can be used to modulate multiple biological functions leading to promising antimicrobial leads. We envision that the principles and approaches exploited here can be applied to better understand the role of motifs and sequence determinants underlying the immunomodulatory and antimicrobial activities of these molecules, and to successfully repurpose venom peptides into potential therapeutics.
Materials and Methods
Peptide Synthesis.
Mast-L, mast-MO, and LL-37 were purchased from Shanghai Hanhong Chemical and mast-MO analogs were purchased from Biopolymers (Massachusetts Institute of Technology). The peptides were synthesized using the solid-phase peptide synthesis and N-9-fluoromethyloxycarbonyl (Fmoc) strategy and purified by HPLCγ (HPLC). Peptide purity used in biologic assays was higher than 95%.
CD Spectroscopy.
The CD experiments were performed to determine the ideal conditions for carrying out the NMR experiments, including conformational preferences and the stability ratio at different pH and media concentrations. Mast-L at 70 µmol L−1 and mast-MO at 50 µmol L−1 were analyzed in SDS (100 mmol L−1), DPC (15 mmol L−1), TFE/water (1:1, v:v), and PCPG (1 mmol L−1) phospholipid vesicles, at 20 °C. The spectra were recorded on a Jasco J-815 spectropolarimeter coupled to a Peltier Jasco PTC-423L, using a 1.0-mm path length rectangular quartz cuvette (NSG), six accumulations from 260 to 190 nm, 1.0-nm spectral bandwidth, 0.2-nm step resolution, 100-nm min −1 scan speed, and 1-s response time. Similar experiments with the respective blank solutions were performed for background subtraction. The spectra were analyzed using the CDPro software package (38, 39). The relative helix content (fH) was calculated from the ellipticity values at 222 nm, as described by Chen et al. (40)
NMR Spectroscopy.
The samples were prepared by dissolving the peptide stock solution (1 mmol L−1) in 500 μL H2O/D2O (9/1, vol/vol) in a micellar solution containing 100 mmol L−1 of SDS (SDS-d25), pH 4.0 and 25 °C. All spectra were recorded on a Bruker Avance III 500 spectrometer equipped with a 5-mm broadband inverse probehead. Proton chemical shifts were referenced to trimethylsilyl-2,2,3,3-d4-propionate sodium (TMSP-d4). The conditions for the 1H-1H TOCSY and 1H-1H NOESY experiments are detailed in SI Appendix.
Bacterial Strains and Media.
Strains used included clinical isolates E. coli KPC+ ID 1812446, E. coli multidrug-resistant ID 2101123, and carbapenemase-producing K. pneumoniae KPC+ ID 1825971, as well as reference strains Bacillus subtilis ATCC6633, Enterococcus faecalis ATCC12953, S. aureus ATCC29213, MRSA ATCC33591, Streptococcus pyogenes ATCC19615, E. coli ATCC8739, K. pneumoniae ATCC13885, Proteus mirabilis ATCC25933, P. aeruginosa ATCC 15442, Salmonella enterica ATCC14028, A. baumannii ATCC19606, E. coli ATCC11775, E. coli AIG221, E. coli AIG222, E. coli BL21, K. pneumoniae ATCC133883, P. aeruginosa PAO1, P. aeruginosa PA14, and S. enterica RFP. Bacteria were plated on brain heart infusion broth (BHI; HiMedia) from a frozen stock. Following 24 h of incubation of the agar plate, three isolated colonies were transferred to 1 mL of BHI. The broth culture was incubated overnight (12 to 16 h) at 37 °C with shaking.
Antibacterial Assays.
MIC of peptides and antibiotics were evaluated using the broth microdilution technique in Mueller-Hinton Broth medium (MHB) with an initial inoculum of 5 × 105 cells in nontreated Polystyrene microtiter plates (Corning) in accordance with Wiegand et al. (41). The MIC was interpreted as the lowest concentration of peptide or antibiotic that completely inhibited the visible growth of bacteria after 12 h of incubation of the plates at 37 °C. Each agent was tested in triplicate in at least three independent experiments. The MIC assays were also performed using the broth microdilution method in sterile 96-well polypropylene microtiter plates. Peptides were added to the plate as solutions in BM2 minimal medium in concentrations ranging from 0 to 128 μg⋅mL−1, and the bacteria (E. coli BL21, S. aureus ATCC12600, P. aeruginosa PA01 and PA14) were grown overnight at 37 °C and inoculated at a final concentration of 5 × 105 colony forming units (CFU) mL−1 per well. The plates were incubated at 37 °C for 24 h and read in a plate reader at 610 nm. All assays were done in triplicate.
Bacterial Killing Assays.
The bacterial killing experiments involved performing 1:100 dilutions of overnight cultures of E. coli BL21, S. aureus ATCC12600, P. aeruginosa PAO1 and PA14 in the absence or presence of increasing concentrations of the samples (0 to 200 μg mL−1). After 24 h of treatment, 10-fold serial dilutions were performed, bacteria were plated on LB agar plates (E. coli BL21 and S. aureus ATCC12600), and Pseudomonas Isolation Agar (P. aeruginosa PAO1 and PA14) and allowed to grow overnight at 37 °C after which CFU counts were recorded. All experiments were done in triplicates.
Antibiofilm Assays.
Experiments were performed as described previously. Biofilms were grown in BM2 glucose medium for 72 h, at 37 °C in flow cell chambers with channel dimensions of 1 × 4 × 40 mm, as previously described by de la Fuente-Nunez et al. (42). For the treatment of preformed biofilms, bacteria were allowed to develop structured 2-d-old biofilms prior to treatment with peptides for the following 24 h. Biofilm cells were then stained using the Live/Dead BacLight bacterial viability kit (Molecular Probes) and subsequently examined using a confocal laser scanning microscope (Olympus, Fluoview FV1000); 3D reconstructions were generated using the Imaris software package (Bitplane).
Hemolysis Assays.
Hemolytic activity of peptides was determined by using fresh mouse red blood cells; by measuring the peptide-induced changes of the optical density (OD) at 540 nm (Victor X, Perkin-Elmer), 100% lysis was determined by analyzing the supernatant of erythrocytes that had been incubated with Triton X-100 (1%).
Mammalian Cell Toxicity Assays.
L929 mice fibroblasts (Rio de Janeiro Cell Bank) and SPCs were seeded in 96-well microtiter plates in a concentration of 1.0 × 105 cells per well, in DMEM, supplemented with different concentrations of tested peptides (1 to 600 μM). After 48-h incubation, a thiazolyl blue tetrazolium bromide (MTT) protocol was performed. Briefly, 60% of the medium was removed, and 10 μL of MTT (5 mg mL−1; Sigma) solution was added to each well and the plate was incubated for 4 h, in 5% CO2, at 37 °C. The blue formazan product generated was dissolved by the addition of 100 μL of 100% DMSO (Mallinckrodt) per well. Plates were then gently swirled for 5 min, at room temperature, to dissolve the precipitate. The absorbance was monitored at 575 nm using a microplate spectrophotometer (Bio-Tek). Cytotoxicity was determined as a percentage of the maximum value after subtracting the background. The results were expressed as the percentage of each sample compared to the negative control (PBS buffer, pH 7.4) and cell culture was incubated in a lysis buffer (10 mM Tris, pH 7.4, 1 mM EDTA, and 0.1% Triton X-100).
Membrane Permeabilization Assays.
The membrane permeability of the mast-MO (20 μmol L−1) was determined by using the NPN uptake assay. All of the bacterial strains were grown to OD600 of 0.4, centrifuged (10,000 rpm at 4 °C for 10 min), washed, and resuspended in buffer (5 mmol L−1 Hepes, 5 mmol L−1 glucose, pH 7.4). Next, 4 μL of NPN solution (0.5 mmol L−1; working concentration of 10 μmol L−1 after dilutions) was added to 100 μL of the bacterial solution in a white 96-wells plate. The background fluorescence was recorded at λex = 350 nm and λem = 420 nm. Mast-MO samples in water (100 μL solution at 1.2 μmol L−1) were added to the 96-wells plate, and fluorescence was recorded as a function of time until no further increase in fluorescence was observed (20 min).
Membrane Depolarization Assays.
The cytoplasmic membrane depolarization activity of mast-MO (20 μmol L−1) was determined by the membrane potential-sensitive dye, DiSC3(5). Briefly, the bacterial strains were grown at 37 °C with agitation to the midlog phase (OD600 = 0.5). The cells were centrifuged and washed twice with washing buffer (20 mmol L−1 glucose, 5 mmol L−1 Hepes, pH 7.2) and resuspended to an OD600 of 0.05 in the same buffer (20 mmol L−1 glucose, 5 mmol L−1 Hepes, pH 7.2) but containing 0.1 mol L−1 KCl. Thereafter, the cells (100 μL) were incubated for 15 min with 20 nmol L−1 of DiSC3(5) until a stable reduction of fluorescence was achieved, indicating the incorporation of the dye into the bacterial membrane. Membrane depolarization was then monitored by observing the change in the fluorescence emission intensity of the membrane potential-sensitive dye, DiSC3(5) (λex = 622 nm, λem = 670 nm).
Synergy Assays.
Colistin-resistant E. coli, P. aeruginosa PAO1, and A. baumannii ATCC19606 were chosen for the synergy assays because of their relevance as clinical pathogens. After determination of the MIC for each antibiotic, those ranging from most active to moderately active were orthogonally diluted using the microdilution technique at concentrations ranging from 2-fold MIC to 0.03-fold MIC. Plates were incubated at 37 °C for 24 h. All assays were done in three independent replicates.
Stability Assays.
The resistance to degradation assay is described in detail by Powell et al. (43). Here, the peptides were exposed to 25% fetal bovine serum in water. The reaction kinetics were followed by reverse-phase liquid chromatography and the percentage of peptide remaining was calculated by integrating the peptide peak area.
Bacterial Toxicity Mouse Model.
Six-week-old female C57BL/6 mice were used in the murine systemic infection model. Animals were provided by the Central Bioterium of the Universidade de São Paulo, Ribeirão Preto. All animals were housed in individual cages under a constant temperature (22 °C) and humidity with a 12-h light/dark cycle and had access to food and water ad libitum throughout the study. The mice were killed by CO2 at the end of the experiments. All procedures, care, and handling of the animals were approved by the Ethics Committee of the Catholic University of Brasilia number 005/13. CD-1 IGS female mice (6-wk-old) were used for the skin scarification model and maintained in accordance with the Guide for the Care and Use of Laboratory Animals (44) in an American Association for Accreditation of Laboratory Animal Care-accredited facility.
Systemic Bacterial Infection Mouse Model.
Acute toxicity assay was performed based on the work of Navon-Venezia et al. (45). The experiment was performed by intraperitoneal injection of the tested peptides to groups of 10 C57BL/6 mice. Each mouse was injected with a 0.5-mL solution of freshly prepared of peptides in PBS. The doses of peptide administered per mouse were 0, 10, 30, 50, 70, and 90 mg kg−1 of body weight. Animals were directly inspected for adverse effects for 6 h, and mortality was monitored for 7 d thereafter. Differences between groups were analyzed using the Fisher exact test (differences were considered to be statistically significant when the P value was <0.05). Experimental conditions are detailed in SI Appendix.
Isolation of Leukocytes from the Peritoneal Cavity of Mice.
Leukocyte migration into the peritoneal cavity of uninfected C57BL/6 mice treated with 10 mg kg−1 of peptides was evaluated to assess the chemotactic activity of the peptides, as previously described (12). Specifically, 10 mg kg−1 of peptides suspended in sterile saline were injected intraperitoneally in C57BL/6 mice. Mice were subsequently killed, and their peritoneal lavage was collected over time. Leukocyte counts present in the in peritoneal cavity of mice infected with E. coli ATCC 8739 were quantified. Animals infected and treated with peptides were killed over time (0.5 to 24 h) posttreatment, and a peritoneal lavage was carried out to quantify leukocyte numbers present in the peritoneal cavity (46). TGA 3% was used as a positive control for leukocyte migration. Animals were killed, and cells present in the peritoneal cavity were harvested in 3 mL of PBS containing 1 mmol L−1 EDTA. Total cell counts were quantified using a cell counter (Coulter AC T series analyzer), and differential cell counts were conducted on cytocentrifuge slides (Cytospin 3; Thermo Shandon) stained by the May–Grünwald–Giemsa (Rosenfeld) method. The results are expressed as the number of cells per cavity (47).
Skin Abscess Infection Mouse Model.
P. aeruginosa strain PAO1 was used for inducing a skin infection in the mouse model as described by Pane et al. (37). Briefly, bacteria were grown in tryptic soy broth medium. Subsequently, cells were washed twice with sterile PBS (pH 7.4, 13,000 rpm for 1 min), and resuspended to a final concentration of 1 × 107 CFU/20 μL. Female CD-1 mice (6-wk-old) were anesthetized with isoflurane and had their backs shaved and a superficial linear skin abrasion was made with a needle in order to damage the stratum corneum and upper-layer of the epidermis. An aliquot of 20 μL containing 1 × 107 CFU of bacteria in PBS was inoculated over each defined area containing the scratch with a pipette tip. One day after the infection, peptides (16 μmol L−1) were administered to the infected area. Animals were killed and the area of scarified skin was excised 2 and 4 d postinfection, homogenized using a bead beater for 20 min (25 Hz), and serially diluted for CFU quantification. Two independent experiments were performed with three mice per group in each condition (n = 6 per group).
Supplementary Material
Acknowledgments
We thank Robin M. Kramer for help establishing the mouse scarification model protocol. C.d.l.F.-N. holds a Presidential Professorship at the University of Pennsylvania, is a recipient of the Langer Prize by the AIChE Foundation, and acknowledges funding from the Institute for Diabetes, Obesity, and Metabolism and the Penn Mental Health AIDS Research Center of the University of Pennsylvania. This work was also supported by Fundação de Amparo à Pesquisa do Estado de Goiás (#201710267000062); Comissao de Aperfeiçoamento de Pessoal de Nival Superior, Conselho Nacional de Pesquisas (Process: 407181/2013-0); and Financiadora de Estudos e Projetos. All figures were prepared using the Biorender drawing toolkit.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2012379117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.de la Fuente-Nunez C., Torres M. D., Mojica F. J., Lu T. K., Next-generation precision antimicrobials: Towards personalized treatment of infectious diseases. Curr. Opin. Microbiol. 37, 95–102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention , “2019 Antimicrobial Resistant Threats Report” (2019), https://www.cdc.gov/drugresistance/biggest-threats.html. Accessed 1 June 2020.
- 3.Rudd K. E.et al., Global, regional, and national sepsis incidence and mortality, 1990-2017: Analysis for the Global Burden of Disease Study. Lancet 395, 200–211 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morens D. M., Taubenberger J. K., Fauci A. S., Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: Implications for pandemic influenza preparedness. J. Infect. Dis. 198, 962–970 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou F.et al., Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 395, 1054–1062 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis R. J., Garcia M. L., Therapeutic potential of venom peptides. Nat. Rev. Drug Discov. 2, 790–802 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Mookherjee N., Anderson M. A., Haagsman H. P., Davidson D. J., Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 19, 311–332 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Torres M. D. T.et al., Structure-function-guided exploration of the antimicrobial peptide polybia-CP identifies activity determinants and generates synthetic therapeutic candidates. Commun. Biol. 1, 221 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torres M. D. T., Sothiselvam S., Lu T. K., de la Fuente-Nunez C., Peptide design principles for antimicrobial applications. J. Mol. Biol. 431, 3547–3567 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Cardoso M. H.et al., A computationally designed peptide derived from Escherichia coli as a potential drug template for antibacterial and antibiofilm therapies. ACS Infect. Dis. 4, 1727–1736 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Cândido E. S.et al., Short cationic peptide derived from Archaea with dual antibacterial properties and anti-infective potential. ACS Infect. Dis. 5, 1081–1086 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Silva O. N.et al., An anti-infective synthetic peptide with dual antimicrobial and immunomodulatory activities. Sci. Rep. 6, 35465 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porto W. F.et al., In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design. Nat. Commun. 9, 1490 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oshiro K. G. N.et al., Computer-aided design of mastoparan-like peptides enables the generation of nontoxic variants with extended antibacterial properties. J. Med. Chem. 62, 8140–8151 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Torres M. D. T.et al., Decoralin analogs with increased resistance to degradation and lower hemolytic activity. ChemistrySelect 2, 18–23 (2017). [Google Scholar]
- 16.Nijnik A., Hancock R., Host defence peptides: Antimicrobial and immunomodulatory activity and potential applications for tackling antibiotic-resistant infections. Emerg. Health Threats J. 2, e1 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lima S. M. F.et al., Antimicrobial and immunomodulatory activity of host defense peptides, clavanins and LL-37, in vitro: An endodontic perspective. Peptides 95, 16–24 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Kim W. H., Lillehoj H. S., Min W., Evaluation of the immunomodulatory activity of the chicken NK-Lysin-Derived peptide cNK-2. Sci. Rep. 7, 45099 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lázár V.et al., Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nat. Microbiol. 3, 718–731 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi M. B., Lee Y., The structure and antimicrobial potential of wasp and hornet (Vespidae) mastoparans: A review. Entomol. Res. 50, 369–376 (2020). [Google Scholar]
- 21.Chen X.et al., Evaluation of the bioactivity of a mastoparan peptide from wasp venom and of its analogues designed through targeted engineering. Int. J. Biol. Sci. 14, 599–607 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hilchie A. L.et al., Mastoparan is a membranolytic anti-cancer peptide that works synergistically with gemcitabine in a mouse model of mammary carcinoma. Biochim. Biophys. Acta 1858, 3195–3204 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higashijima T., Uzu S., Nakajima T., Ross E. M., Mastoparan, a peptide toxin from wasp venom, mimics receptors by activating GTP-binding regulatory proteins (G proteins). J. Biol. Chem. 263, 6491–6494 (1988). [PubMed] [Google Scholar]
- 24.Mousli M.et al., Activation of rat peritoneal mast cells by substance P and mastoparan. J. Pharmacol. Exp. Ther. 250, 329–335 (1989). [PubMed] [Google Scholar]
- 25.Higashijima T., Burnier J., Ross E. M., Regulation of Gi and Go by mastoparan, related amphiphilic peptides, and hydrophobic amines. Mechanism and structural determinants of activity. J. Biol. Chem. 265, 14176–14186 (1990). [PubMed] [Google Scholar]
- 26.Wang G., Li X., Wang Z., APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 44, D1087–D1093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conlon J. M.et al., Peptides with potent cytolytic activity from the skin secretions of the North American leopard frogs, Lithobates blairi and Lithobates yavapaiensis. Toxicon 53, 699–705 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Goraya J.et al., Peptides with antimicrobial activity from four different families isolated from the skins of the North American frogs Rana luteiventris, Rana berlandieri and Rana pipiens. Eur. J. Biochem. 267, 894–900 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Yang X., Wang Y., Lee W.-H., Zhang Y., Antimicrobial peptides from the venom gland of the social wasp Vespa tropica. Toxicon 74, 151–157 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Hernández-Gras F., Boronat A., A hydrophobic proline-rich motif is involved in the intracellular targeting of temperature-induced lipocalin. Plant Mol. Biol. 88, 301–311 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenfield N. J., Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 1, 2876–2890 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wüthrich K., NMR of Proteins and Nucleic Acids, (John Wiley and Sons, New York, 1986). [Google Scholar]
- 33.Irazazabal L. N.et al., Selective amino acid substitution reduces cytotoxicity of the antimicrobial peptide mastoparan. Biochim. Biophys. Acta 1858, 2699–2708 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Souza B. M.et al., Structure-activity relationship of mastoparan analogs: Effects of the number and positioning of Lys residues on secondary structure, interaction with membrane-mimetic systems and biological activity. Peptides 72, 164–174 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Pletzer D., Mansour S. C., Wuerth K., Rahanjam N., Hancock R. E. W., New mouse model for chronic infections by gram-negative bacteria enabling the study of anti-infective efficacy and host-microbe interactions. mBio 8, e00140-e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brouwer C. P. J. M., Wulferink M., Welling M. M., The pharmacology of radiolabeled cationic antimicrobial peptides. J. Pharm. Sci. 97, 1633–1651 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Pane K.et al., Identification of novel cryptic multifunctional antimicrobial peptides from the human stomach enabled by a computational-experimental platform. ACS Synth. Biol. 7, 2105–2115 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Sreerama N., Woody R. W., Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Sreerama N., Woody R. W., On the analysis of membrane protein circular dichroism spectra. Protein Sci. 13, 100–112 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y. H., Yang J. T., Chau K. H., Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry 13, 3350–3359 (1974). [DOI] [PubMed] [Google Scholar]
- 41.Wiegand I., Hilpert K., Hancock R. E. W., Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175, 10.1038/nprot.2007.521 (2008). [DOI] [PubMed] [Google Scholar]
- 42.de la Fuente-Núñez C., et al., D-Enantiomeric Peptides that Eradicate Wild-Type and Multidrug-Resistant Biofilms and Protect against Lethal Pseudomonas aeruginosa Infections. Cell Chem. Biol. 22, 196–205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Powell M. F.et al., Peptide stability in drug development. II. Effect of single amino acid substitution and glycosylation on peptide reactivity in human serum. Pharm. Res. 10, 1268–1273 (1993). [DOI] [PubMed] [Google Scholar]
- 44.National Research Council , Guide for the Care and Use of Laboratory Animals, (National Academies Press, Washington, DC, 8th Ed., 2011). [Google Scholar]
- 45.Navon-Venezia S., Feder R., Gaidukov L., Carmeli Y., Mor A., Antibacterial properties of dermaseptin S4 derivatives with in vivo activity. Antimicrob. Agents Chemother. 46, 689–694 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ray A., Dittel B. N., Isolation of mouse peritoneal cavity cells. J. Vis. Exp., e1488 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moreno S. E.et al., IL-12, but not IL-18, is critical to neutrophil activation and resistance to polymicrobial sepsis induced by cecal ligation and puncture. J. Immunol. 177, 3218–3224 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.