Significance
To ensure immunity, and yet limit pathology, inflammatory responses must be confined within the proverbial “Goldilocks zone.” TLR4 is the prototypical sensor that orchestrates inflammatory responses through a series of well-characterized downstream cascades. How TLR4 signals are confined remains incompletely understood. Using trans-scale approaches ranging from disease modeling in live animals, through cell-based interventional studies, to structure-guided biochemical studies that offer an atomic-level resolution, this study unravels the existence of a “brake” within the TLR4-signaling cascade, i.e., GIV; the latter is a prototypical member of an emerging class of scaffold proteins. By showing that GIV uses conserved mechanisms to impact multi-TLR signaling, this work unravels a multiscale point of convergence of immune signaling of broader impact beyond TLR4.
Keywords: Girdin, TIR domain, inflammation, macrophages, ccdc88a
Abstract
Sensing of pathogens by Toll-like receptor 4 (TLR4) induces an inflammatory response; controlled responses confer immunity but uncontrolled responses cause harm. Here we define how a multimodular scaffold, GIV (a.k.a. Girdin), titrates such inflammatory response in macrophages. Upon challenge with either live microbes or microbe-derived lipopolysaccharides (a ligand for TLR4), macrophages with GIV mount a more tolerant (hypo-reactive) transcriptional response and suppress proinflammatory cytokines and signaling pathways (i.e., NFkB and CREB) downstream of TLR4 compared to their GIV-depleted counterparts. Myeloid-specific gene-depletion studies confirmed that the presence of GIV ameliorates dextran sodium sulfate-induced colitis and sepsis-induced death. The antiinflammatory actions of GIV are mediated via its C-terminally located TIR-like BB-loop (TILL) motif which binds the cytoplasmic TIR modules of TLR4 in a manner that precludes receptor dimerization; such dimerization is a prerequisite for proinflammatory signaling. Binding of GIV’s TILL motif to TIR modules inhibits proinflammatory signaling via other TLRs, suggesting a convergent paradigm for fine-tuning macrophage inflammatory responses.
Macrophages are sentinel cells of the innate immune system; their location varies from peripheral blood to various organs including lungs, liver, brain, kidneys, skin, testes, and vascular endothelium. Consequently, dysregulated activation of macrophages impacts the outcome of diverse organ systems in a multitude of diseases (1).
Of the signaling pathways that modulate macrophage function, Toll-like receptors (TLRs) constitute a key signaling system; they recognize a wide variety of pathogen associated molecular patterns (PAMPS) and initiate acute inflammation through the production of inflammatory cytokines (2). Specialized TLRs for each class of PAMP allow fine-tuning of the inflammatory response for efficient removal of the pathogen. The prototypic member, TLR4, efficiently senses Gram-negative bacterial infections through recognition of the bacterial membrane component, lipopolysaccharides (LPS). Binding of LPS to TLR4 triggers signaling cascades (e.g., NFkB and MAPK) that culminate in the production of proinflammatory cytokines (TNFα, IL-1β, IL-6, IL-12) and type-I interferons required for propagation of the inflammatory response and ultimately pathogen destruction (3). Although many components of the TLR-signaling pathway have been well characterized, regulatory mechanisms that intricately balance proinflammatory and antiinflammatory responses remain incompletely understood.
In this work, we reveal an unexpected role of GIV, a multimodular scaffold protein and the prototypical member of the nonreceptor Guanine nucleotide Exchange Modulator (GEM) family of proteins (4), as a key determinant of macrophage polarization and inflammatory cytokine expression. Because GIV binds and modulates G-protein activity downstream from a diverse variety of ligand-activated receptors, e.g., growth factor and integrins (reviewed in refs. 5, 6), here we studied if and how GIV may impact the LPS/TLR4 signaling in the most relevant cell line, i.e., macrophages. We dissect the relevance of those findings in murine disease models and its broader relevance among other TLRs.
Results and Discussion
GIV Is Preferentially Expressed in the Myeloid Cells of Our Immune System.
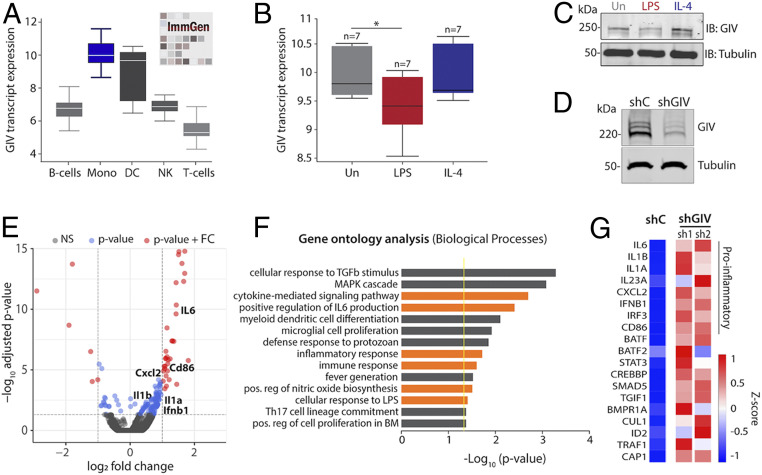
Using a publicly available protein expression database (The Human Protein Atlas), we noted that GIV is highly expressed in several immune tissues including lymph nodes, appendix, spleen, bone marrow, and tonsil (SI Appendix, Fig. S1A). An analysis of RNA-seq datasets curated by the NIH/National Institute of Allergy and Infectious Diseases-supported Immunological Genome Project (http://immgen.org) further confirmed that GIV (gene CCDC88A) is most highly expressed in macrophages and dendritic cells, moderately expressed in B-cells and natural killer (NK) cells, and least expressed in T cells (Fig. 1A).
Fig. 1.
GIV/Gidrin expression is associated with proinflammatory gene programs in macrophages. (A) Box plots representing the GIV (gene: CCDC88A) transcript level in various immune cell populations. (Mono: monocyte; DC: dendritic cell; NK: natural killer cell) (B) Box plots representing GIV transcript levels in a dataset (GSE35449) comprised of polarized human CD14+ monocytes isolated from peripheral blood mononuclear cells stimulated with either LPS (10 ng/mL) and IFNγ (200 U/mL) or IL-4 (1,000 U/mL). (C) Immunoblot of GIV protein expression in polarized bone marrow-derived macrophages stimulated with either LPS (10 ng/mL) or IL-4 (20 ng/mL) for 24 h. (D) Immunoblot of RAW 264.7 macrophages depleted of GIV using shRNA. (E) Volcano plot of significantly (red) up-regulated and down-regulated gene transcripts in GIV-depleted (shRNA) RAW 264.7 macrophages stimulated with LPS (100 ng/mL, 6 h) compared to controls (scrambled shRNA). Significance was determined using a P value of <0.05 and log2 fold change (FC) cutoffs. (F) Bar graph of significantly enriched biological processes determined by GO analysis. Yellow line designates the P = 0.05 cutoff. Orange bars highlight biological processes relevant to macrophage inflammatory responses. (G) Heatmap of selected inflammatory gene transcript expression in GIV-depleted RAW 264.7 macrophages stimulated with LPS (100 ng/mL for 6 h). shC: scrambled shRNA control; sh1 and sh2: two different GIV-targeting shRNA.
To explore possible functions of GIV in the immune cell type where it is most highly expressed, i.e., macrophages, we asked how its expression changes during macrophage polarization, which has classically been studied using a simplified nomenclature of reactive (a.k.a. M1, proinflammatory) vs. tolerant M2 (antiinflammatory/healing). While the reactive state is important for engulfing and clearing invading pathogens and damaged cells and for mounting tailored inflammatory responses, the tolerant state is critical for restoring tissue homeostasis (7). An analysis of numerous human and mouse RNA-seq datasets (8) revealed that GIV expression significantly decreased in LPS-stimulated [a widely used approach to induce M1 polarization (9)] but not in IL-4–stimulated [a widely used approach to induce M2 polarization (10)] macrophages compared to controls (Fig. 1B and SI Appendix, Fig. S1 B and C). These findings were validated in both murine bone marrow-derived macrophages and RAW 264.7 macrophages stimulated either with LPS or IL-4 and subsequently assessed for GIV expression by immunoblotting (Fig. 1C and SI Appendix, Fig. S1D). We conclude that GIV is expressed in tissues with immune function and that high expression is seen in macrophages. In addition, GIV’s expression changes during macrophage polarization; i.e., it is suppressed in reactive macrophages but not in the tolerant ones.
GIV Dampens Macrophage Reactivity to Live Microbes and LPS.
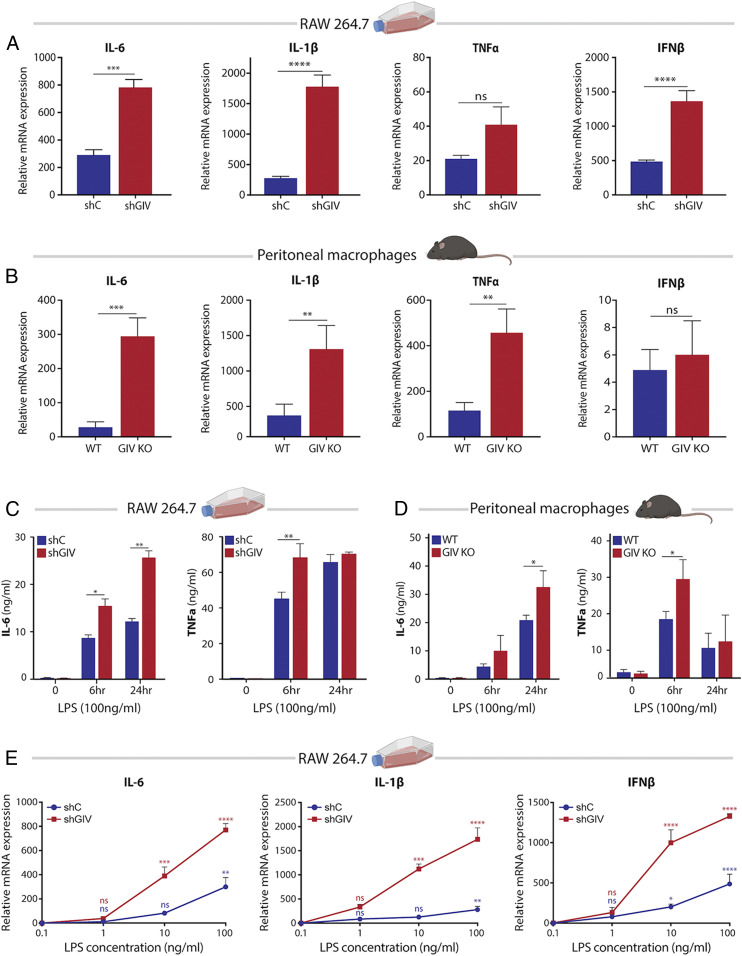
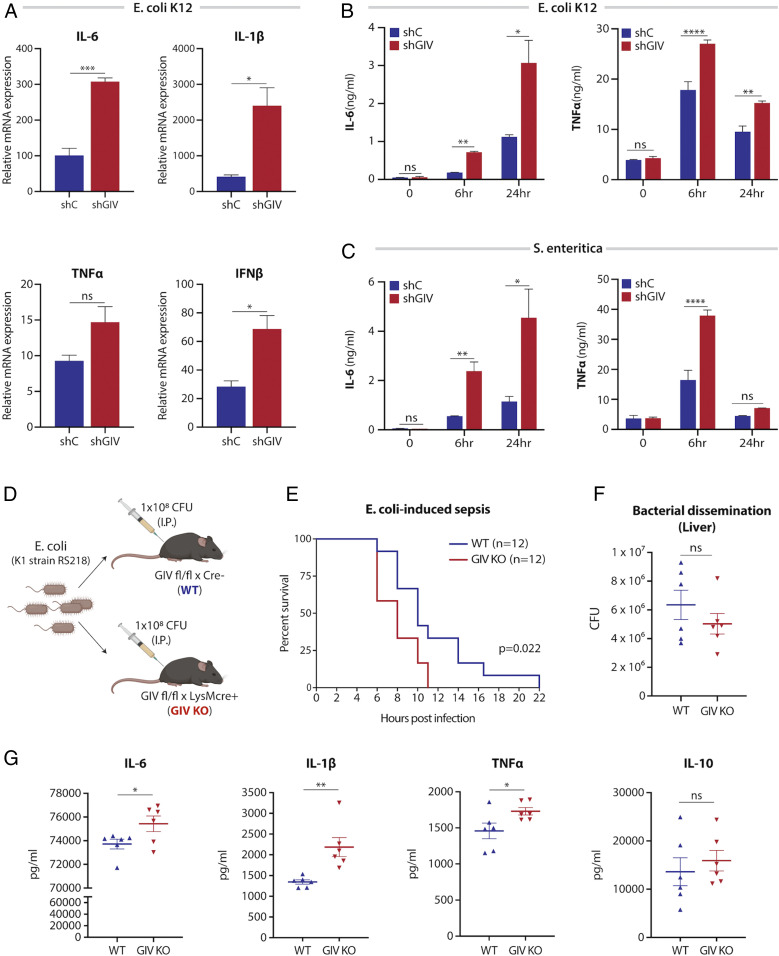
To study the role of GIV in macrophage inflammatory responses, we generated two model systems: 1) a GIV-depleted RAW 264.7 macrophage cell line using short-hairpin RNA (shRNA) (Fig. 1D) and 2) a myeloid-specific conditional GIV knockout mouse, generated by crossing previously generated Girdin floxed mice (11) to LysMcre mice (SI Appendix, Fig. S2). We asked if the presence or absence of GIV impacts macrophage responses and began by seeking insights from the transcriptome. GIV-depleted and control RAW 264.7 macrophages were stimulated with LPS, and the relative levels of transcript expression were analyzed by RNA sequencing. We found that 150 genes were significantly up-regulated, and 26 genes were significantly down-regulated in GIV-depleted macrophages compared to controls (Fig. 1E). Gene ontology (GO) analysis performed using DAVID GO (https://david.ncifcrf.gov/) on the set of up-regulated genes revealed 29 significantly enriched biological processes, whereas down-regulated genes did not show such enrichment (Fig. 1F). Most enriched pathways were involved in proinflammatory signaling and cytokine responses, including up-regulation of IL-6, IL-1b, IL-1a, IL-23a, IL-17A, IL-12A, CXCL2, and IFNb1 (Fig. 1G). These findings were confirmed by quantitative PCR and enzyme-linked immunosorbent assay (ELISA) studies using GIV-depleted RAW 264.7 macrophages and peritoneal macrophages harvested from GIV knockout (KO) mice (LysMcre) (Fig. 2 A–D). Increased expression of proinflammatory cytokines was consistently observed in both RAW 264.7 cells and peritoneal macrophages depleted of GIV (Fig. 2 A and B). Identical findings were observed in assays where we replaced LPS with the live microbes, Escherichia coli K12 strain, and Salmonella enteritica serovar Typhimurium; both microbes induced a higher proinflammatory response in GIV-depleted macrophages compared to controls (Fig. 3 A–C and SI Appendix, Fig. S3C).
Fig. 2.
GIV depletion increases the magnitude and sensitivity of cytokine responses to LPS. (A and B) Bar graphs displaying cytokine transcript levels (qPCR) in GIV-depleted RAW 264.7 macrophages (A) or GIV KO peritoneal macrophages (B) stimulated with LPS (100 ng/mL, 6 h) compared to controls. (C and D) Bar graphs showing levels of secreted proinflammatory cytokines (ELISA) in GIV-depleted RAW 264.7 (C) or GIV KO peritoneal (D) macrophages stimulated with LPS. (E) Line graphs comparing sensitivity of cytokine transcript response to increasing doses of LPS (6 h stimulation) in GIV-depleted RAW 264.7 macrophages compared to controls. All qPCR and ELISA results are from three independent experiments and displayed as mean ± SEM. Student’s t test was used for two-parameter statistical analysis (A and B), and two-way ANOVA using Sidak’s multiple comparisons test was used for multiparameter statistical analysis (C–E). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001; ns, not significant.
Fig. 3.
GIV depletion enhances proinflammatory cytokine response during live-microbe infection. (A) Bar graphs displaying cytokine transcript levels (qPCR) in GIV-depleted RAW 264.7 macrophages infected with live E. coli K12 (MOI: 1) for 6 h compared to controls. (B and C) Bar graphs showing levels of secreted proinflammatory cytokines (ELISA) in GIV-depleted or control RAW 264.7 macrophages infected with either E. coli K12 (MOI: 1) or S. enteritica (MOI: 10) for 6 h. (D) Schematic of sepsis-induced-death mouse model. (E) Survival curve of GIV KO or WT mice infected with E. coli K1 strain RS218 (i.p.). Values are expressed as percentage of survival. (F) Scatter plot of bacterial dissemination to liver following E. coli infection. (G) Scatter plots showing serum cytokine levels following E. coli infection. All qPCR and ELISA results are from at least three independent experiments and displayed as mean ± SEM. Student’s t test was used for two-parameter statistical analysis (A, B, F, and G), and two-way ANOVA using Sidak’s multiple comparisons test was used for multiparameter statistical analysis (B and C). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001; ns, not significant.
Intriguingly, compared to controls, the GIV-depleted RAW 264.7 and GIV KO peritoneal macrophages showed a reduction in the messenger RNA for IL-10 (SI Appendix, Fig. S3A), a potent antiinflammatory cytokine that promotes healing (12, 13). Thus, what emerged as a consistent finding is that, compared to macrophages without GIV, those with GIV selectively suppress the proinflammatory responses to both forms of infectious stimuli, LPS and live microbes.
GIV may inhibit macrophage inflammatory responses either by reducing sensitivity or by inducing anergy (i.e., becoming refractive to repeated stimulation); to distinguish between the two, we carried out two commonly used assays: sensitivity at lower doses of LPS and anergy during repeated LPS challenges. Macrophages without GIV displayed increased reactivity to lower doses of LPS compared to controls (Fig. 2E), indicating that GIV reduces sensitivity (or increases tolerance) of macrophages to LPS. When exposed to repeat doses of LPS, both control and GIV-depleted macrophages displayed LPS-induced anergy; even though GIV-depleted macrophages mounted a higher response than control cells during both exposures, repeat exposures elicited a weaker response than the first exposure in both cells (SI Appendix, Fig. S3B). These findings indicate that the presence or absence of GIV may not impact anergy.
Taken together, these findings demonstrate that the presence of GIV in macrophages suppresses the proinflammatory gene signature, the production of proinflammatory cytokines (but not the antiinflammatory cytokine IL-10), and reduces sensitivity to LPS without inducing anergy. We conclude that the physiologic role of GIV is to dampen proinflammatory macrophage responses.
GIV Ameliorates Inflammation in Murine Models of Colitis and Sepsis.
To explore the consequences of GIV’s role in macrophage polarization in vivo, we first utilized a sepsis-induced death model. Wild-type (WT) or myeloid-specific GIV KO mice were infected with a lethal dose of E. coli (1 × 108 colony-forming units [CFUs]/mouse) and monitored for survival, levels of serum cytokines, and bacterial dissemination to spleen and liver (Fig. 3D). GIV-depleted mice succumbed to sepsis-induced death significantly faster than controls (Fig. 3E) and produced more proinflammatory cytokines (TNFα, IL-6, and IL1-β, but not the healing-associated IL-10 cytokine) despite comparable amounts of microbial dissemination to peripheral organs (Fig. 3 F and G). Findings demonstrate that two major parameters in sepsis, i.e., “cytokine storm” and death, are suppressed in the presence of GIV. That the microbial counts were comparable in both groups suggests that the protective effect of GIV on sepsis-induced mortality is likely to be due to its ability to suppress the cytokine storm and unlikely to be confounded by overt changes in bacterial replication and/or defective clearance.
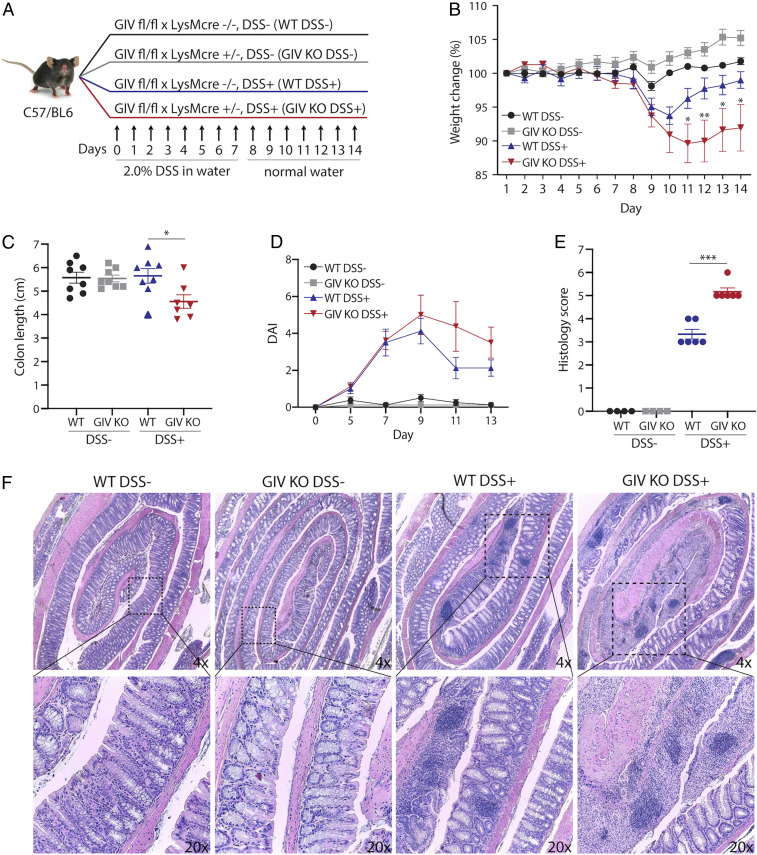
Next we assessed the role of GIV in colitis. We chose this as a disease model because of two reasons: 1) the gut, and more specifically, the colon is the largest reservoir for LPS-producing Gram-negative bacteria (14); and (2) hyperreactive immune responses by macrophages to gut microbes have been implicated in the initiation and perpetuation of colitis-associated syndromes such as inflammatory bowel disease (IBD), Crohn’s disease, and ulcerative colitis (15–17). We used the dextran sodium sulfate (DSS) mouse model of colitis because prior studies using this model have documented the importance of macrophage polarization states in limiting disease severity (18, 19). Because our prior observations demonstrate that GIV dampens macrophage inflammatory responses, we hypothesized that depletion of GIV in macrophages might exacerbate DSS-induced colitis. Mice were treated with DSS and monitored for changes in weight, stool consistency, rectal bleeding, colon length, colon tissue destruction, and immune infiltrates (Fig. 4A). We found that GIV KO mice had increased weight loss, fibrotic shortening of the colon, and disease activity index (DAI) compared to WT controls (Fig. 4 B–D). Histomorphological analysis of colon tissue sections revealed increased destruction of crypt architecture and immune infiltrates in GIV KO mice compared to WT controls (Fig. 4 E and F). Findings demonstrate that all of the major parameters of severity of colitis were suppressed in the presence of GIV.
Fig. 4.
Myeloid cell-specific GIV depletion exacerbates disease in DSS colitis. (A) Schematic outlining experimental design of DSS colitis model. (B) Line graph showing body weight change monitored daily during the course of acute DSS colitis. (C) Scatter plot of colon length assessed at day 14 of DSS experiment. (D) Line graph of DAI using stool consistency (0 to 4), rectal bleeding (0 to 4), and weight loss (0 to 4) as scoring criteria. (E) Scatter plot of histomorphological evaluation of inflammation in hematoxylin and eosin (H&E)-stained colon tissues using inflammatory cell infiltrate (1 to 3) and epithelial architecture (1 to 3) as scoring criteria. (F) Representative images of colon tissue stained with H&E. Data displayed as mean ± SEM and either one-way or two-way ANOVA using Tukey’s or Sidak’s multiple comparisons test was used to determine significance. *P ≤ 0.05, ***P ≤ 0.001.
These observations provide in vivo evidence for GIV’s role in restricting macrophage proinflammatory responses during microbial infection. Although the use of LysMcre for targeted depletion in macrophages is widely accepted, target protein depletion in other cell types including granulocytes, neutrophils, and dendritic cells (20), all of which are known to express GIV and play a role in the setting of sepsis (21) and in the pathogenesis of IBD (22, 23), cannot be ruled out. However, taken together, the results from DSS colitis, acute sepsis, and in vitro cell stimulation assays (Figs. 1–4) suggest that the phenotypes that we observe are at least in part due to GIV-depleted macrophages that are hyperreactive.
GIV Suppresses LPS/TLR4-Induced Proinflammatory Signaling Pathways.
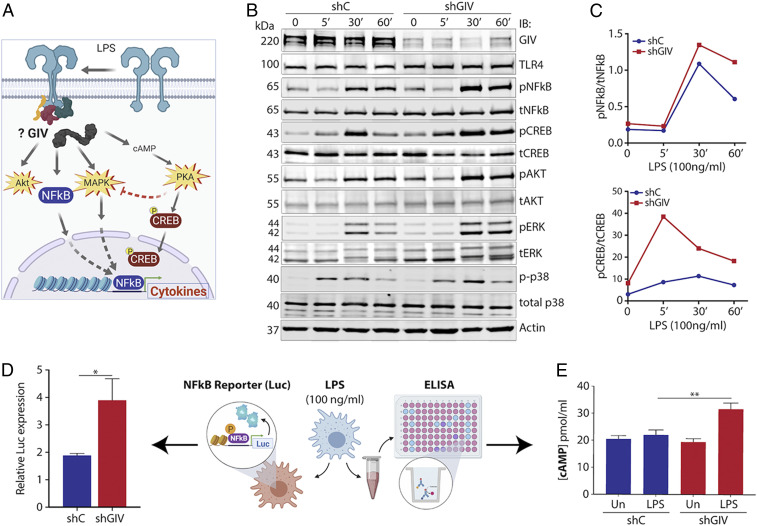
Extensive work has gone into elucidating the signaling pathways downstream of LPS/TLR4 in macrophages (3) (summarized in Fig. 5A). GIV is known to modulate several of those pathways by linking G-protein signaling, via its GEM motif, to a multitude of cell-surface receptors (reviewed in ref. 24). To investigate if GIV may also regulate TLR4 and/or the proinflammatory signaling pathways, control or GIV-depleted macrophages were stimulated with LPS, and the signaling dynamics of key pathways (i.e., phosphorylation of NFkB, CREB, AKT, and MAPK) were assessed by immunoblot (Fig. 5 B and C). NFkB and CREB pathways, but not Akt, showed increased activation in GIV-depleted macrophages compared to controls, as examined by the ratio of phospho/total proteins. As for the MAPK pathway, GIV depletion caused increased phosphorylation of p38 MAPK and increases in both phospho- and total ERK1/2 proteins. No appreciable difference was observed in JNK activation (SI Appendix, Fig. S4). The pathways that were enhanced remained so even at 60 min, suggesting that GIV may inhibit negative feedback mechanisms required to dampen TLR4 signaling. We also observed enhanced CREB signaling in GIV-depleted macrophages as early as 5 min post stimulation, which is in agreement with the increase in sensitivity that we observed in cytokine production (Fig. 2E). Because the expression of GIV did not decrease in control cells within 60 min of LPS stimulation (as it did after 24 h stimulation [Fig. 1 B and C]), results indicate that GIV is required to suppress the pathways that were up-regulated in GIV-depleted cells.
Fig. 5.
GIV depletion in macrophages enhances proinflammatory signaling pathways during LPS response. (A) Schematic highlighting GIV’s potential role in modulating signaling pathways downstream of TLR4. (B) Immunoblot of whole-cell lysates from GIV-depleted or control RAW 264.7 macrophages stimulated with LPS (100 ng/mL) and probed for activation of indicated signaling pathways. (C) Line graphs of representative densitometry values taken from signaling immunoblots. (D) Bar graph of relative NFkB activity in GIV-depleted or control RAW 264.7 macrophages stimulated with LPS (100 ng/mL, 1 h) using NFkB luciferase reporter assay. (E) Bar graph of intracellular cAMP levels in LPS-stimulated (100 ng/mL, 30 min), GIV-depleted, or control RAW 264.7 macrophages. Results are from three independent experiments and displayed as mean ± SEM. Student’s t test was used for two-parameter statistical analysis (D), and two-way ANOVA using Sidak’s multiple comparisons test was used for multiparameter statistical analysis (E). *P ≤ 0.05, **P ≤ 0.01.
We also found that GIV suppresses NFkB activity; upon LPS stimulation, NFkB activity increased approximately two-fold higher in GIV-depleted macrophages compared to controls, as determined by a well-established luciferase reporter assay (NanoLuc Promega) (Fig. 5D). As for the observed increases in CREB phosphorylation, prior studies have implicated three parallel pathways downstream of TLR4 that are known to converge on CREB phosphorylation (25–28): 1) the cAMP→PKA pathway, 2) the cAMP→Epac pathway, and 3) the p38-MAPK cascade. Because GIV inhibits cAMP production by activating Gαi (29, 30), we hypothesized that cellular cAMP levels may increase during LPS stimulation in the absence of GIV. We found that cAMP levels were indeed elevated in GIV-depleted macrophages responding to LPS compared to controls (Fig. 5E), indicating that GIV suppresses cellular cAMP in macrophages responding to LPS. We conclude that GIV-dependent suppression of cAMP downstream of TLR4 stimulation may represent a G-protein–coupled receptor (GPCR)-independent pathway for modulation of cellular cAMP in macrophages responding to infections.
Taken together, these findings indicate that, in LPS-challenged macrophages, GIV specifically suppresses signaling within major proinflammatory signaling cascades, e.g., the NFkB→cytokine, cAMP→CREB, and the MAPK/ERK pathways, but does not seem to significantly impact others, e.g., JNK and PI3K→Akt. Because the PI3K→Akt pathway promotes antiinflammatory responses (31) and because JNK regulates macrophage development and survival (32), we conclude that GIV’s impact on TLR4 signaling is limited to those pathways that are directly related to cytokine production, but not cell fate.
GIV Binds the Cytoplasmic TIR Module of TLR4 and Couples TLR4 to G-Protein Pathways.
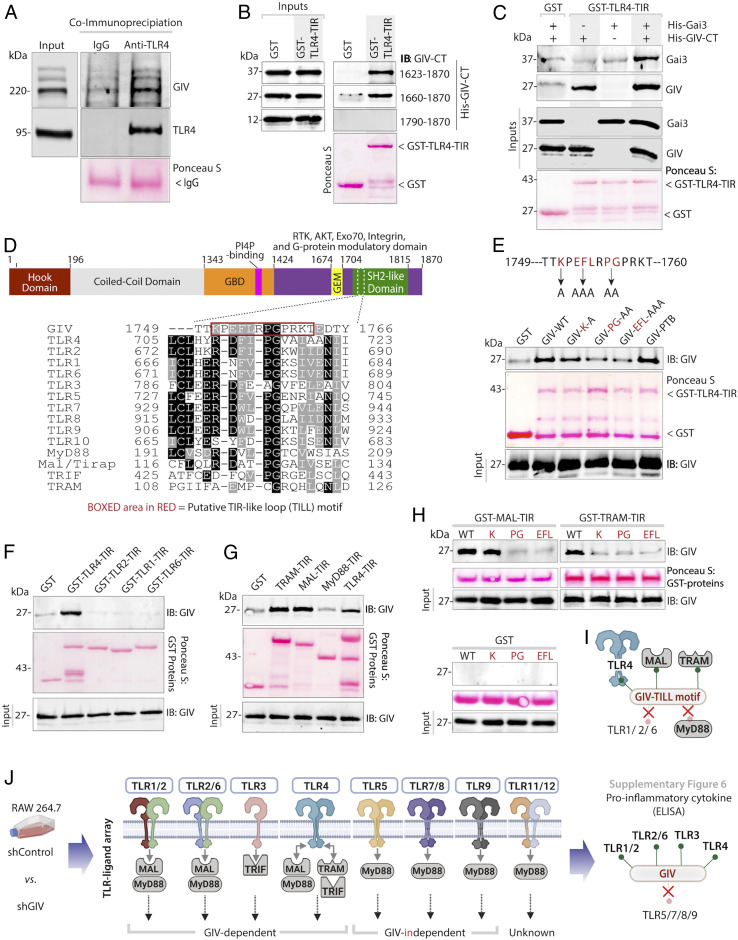
To explore the mechanisms by which GIV modulates LPS/TLR4 signaling, and to pinpoint where within the TLR4-signaling cascade GIV acts, we used a combination of biochemical assays. Published work from us and others has identified three critical features within the C terminus of GIV: 1) a GEM motif that is required for interaction with and modulation of G proteins; 2) distinct short linear interaction motifs (SLIMs) that couple GIV to receptor tyrosine kinases (RTKs) and integrins (reviewed in ref. 24); and 3) all these SLIMs are packed within an ∼210-amino-acid (aa)-long GIV C terminus, which is an intrinsically disordered protein (IDP) (33, 34) (SI Appendix, Fig. S5). Because IDPs that fold/unfold thereby exposing/hiding SLIMs are known to impart plasticity to protein-protein interaction networks during signal transduction (35), we hypothesized that GIV may do something similar in TLR4 signaling. It may directly bind TLR4 through one SLIM, facilitate through another SLIM the assembly-disassembly of ternary receptor•GIV•Gαi complexes, and thereby dynamically shape postreceptor signaling. Immunoprecipitation of endogenous full-length proteins from RAW 264.7 macrophages revealed that GIV forms a complex with TLR4 at steady state (Fig. 6A) and that such complexes were detected even ∼30 min after LPS stimulation (SI Appendix, Fig. S7A). These findings show that the GIV•TLR4 interaction is constitutive; i.e., it is not significantly altered by ligand stimulation. In vitro pulldown assays using various fragments of recombinant His-GIV-CT and GST-tagged cytoplasmic Toll/interleukin-1 receptor (TIR) module of TLR4 (TLR4-TIR; aa 676 to 835) confirmed that the constitutive interaction between GIV and TLR4 observed in cells is direct. An ∼110-aa stretch in GIV’s C terminus (Fig. 6B) and the cytoplasmic TIR module of TLR4 are sufficient for the interaction.
Fig. 6.
GIV directly interacts with TLR4 using a TILL motif within its C terminus and physically links TLR4 and Gαi. (A) Endogenous TLR4 was immunoprecipitated from RAW 264.7 lysates. Bound complex of TLR4 and GIV was visualized by immunoblot. Equal loading of IgG and anti-TLR4 were confirmed by Ponceau S staining. (B) Various constructs of recombinant His-GIV-CT (∼3 μg) was used in GST pulldown assays with GST or GST-TLR4-TIR, and bound GIV was visualized by immunoblot. (C) Recombinant His-GIV-CT (∼3 μg) and His-Gαi3 (∼3 μg) were used in a GST pulldown assay with GST or GST-TLR4-TIR, and bound GIV and Gαi were visualized by immunoblot. (D) Sequence alignment showing a short linear TILL motif that is conserved between GIV- and TIR-containing proteins. (E) Recombinant His-GIV-CT or TILL mutants were used in a GST pulldown assay with GST or GST-TLR4-TIR, and bound GIV was visualized by immunoblot. (F and G) Recombinant His-GIV-CT (∼3 μg) was used in a GST pulldown assay with various GST-TLR proteins (F) and GST-TIR adaptors (G), and bound GIV was detected by immunoblot. (H) Recombinant His-GIV-CT TILL mutants were used in GST pulldown assays with TIR adaptor proteins, and bound GIV was visualized by immunoblot. For all recombinant GST pulldown assays, equal loading of GST proteins was confirmed by Ponceau S staining. (I) Schematic summarizing GIV•TIR interactions. (J) Schematic summarizing data from SI Appendix, Fig. S6, investigating the impact of GIV on various TLR inflammatory responses.
Furthermore, using an in vitro pulldown assay between recombinant TLR4-TIR (bait) and Gαi3 (prey) proteins in the presence or absence of GIV-CT, we confirmed that GIV facilitates the formation of a TLR4•GIV•Gαi ternary complexes (Fig. 6C). Gαi3 bound TLR4 exclusively in the presence on GIV, suggesting that GIV acts as a physical link between TLR4 and Gαi, as it has been demonstrated to do for multiple RTKs (24) and for β1-integrin (34).
A Short Linear Motif (TILL) within GIV’s C Terminus Binds Multiple TIR Modules.
Because TLR4 signaling relies on the ability of its cytoplasmic TIR module to assemble multimeric postreceptor homo- and heterotypic dimers via key contact sites, we hypothesized that the ∼100-aa-long stretch within GIV’s C terminus may contain a SLIM that bears homology to one or more of such contact sites. Sequence alignment revealed an ∼12-aa stretch with sequence homology to the BB-loop region of TIR domains (Fig. 6D), which is also evolutionarily conserved (SI Appendix, Fig. S5). The BB-loop is essential both for homo- and heterodimerization of TLRs and for the recruitment of TIR-domain–containing adaptors, and mutations in this loop inhibit TLR signaling (36). Additionally, a TIR-like BB loop (henceforth, TILL) defined first in the cytoplasmic tail of IL17RA has been shown to be essential for NFkB and MAPK activation (37), adding support to the idea that TILL motifs can modulate inflammatory responses. To test if the newly identified putative TILL motif in GIV is required for binding TLR4, we replaced critical residues within GIV-TILL with alanines (K1749A, EFL1751-53AAA, PG1754-55AA) and found that all three mutants showed decreased binding to TLR4. This motif appears to be specific because disruption of a neighboring SLIM that is ∼13 aa upstream (PTB-binding motif, which enables binding to integrins) had no effect (34) (Fig. 6E). These findings demonstrate that the direct interaction between GIV and TLR4-TIR is mediated via GIV’s C-terminally located TILL motif.
To assess what other TIR modules GIV may bind, we conducted GST pulldown assays with His-GIV-CT (prey) and GST-tagged TIR modules found in the cytoplasmic tails of other TLRs (baits: TLR2, TLR1, TLR6) and in TLR-associated adaptor proteins (Mal, TRAM, TRIF, MyD88). Among the TLRs tested, TLR4 was the only one that could bind GIV (Fig. 6F). GIV also bound the TIR adaptors MAL and TRAM, but not MyD88 (Fig. 6G), indicating that GIV’s C terminus can directly bind multiple TIR modules. Mutations within the TILL motif of GIV disrupted them all (Fig. 6H), indicating that the TILL motif is necessary for these GIV•TIR interactions.
GIV•TIR Interactions Aid in Postreceptor Signal Integration and Ligand Specificity.
Because GIV’s TILL motif bound multiple TIR modules, but demonstrated selectivity by interacting with some (TLR4, MAL, TRAM) but not others (TLR1/2/3, MyD88) (Fig. 6I), we conclude that this pattern of binding may support two fundamental properties in immune response and signal transduction: 1) multi-TIR binding could mediate signal convergence/integration and receptor cross-talk to mount a consistent response regardless of the ligand, whereas 2) selectivity between TLRs could impact ligand specificity. To study these two properties, we conducted a TLR-ligand screen where GIV-depleted or control RAW 264.7 macrophages were stimulated with ligands for various TLRs (Fig. 6J). We observed a consistent increase in the production of proinflammatory cytokines in GIV-depleted macrophages stimulated with ligands for TLR1/2, TLRR2/6, and TLR3; however, the presence or absence of GIV did not impact responses to ligands for TLR5, TLR7/8, or TLR9 (Fig. 6J and SI Appendix, Fig. S6), indicating that the latter are GIV-independent. We noted that the GIV-independent TLRs are capable of directly engaging MyD88, whereas those that are GIV-dependent engage with MAL, TRAM, and TRIF. Because GIV binds MAL and TRAM, but not MyD88, these findings raise the possibility that GIV inhibits TLR signaling either by directly interacting with the cytoplasmic domain and preventing TIR-mediated receptor dimerization or blocks the recruitment of other TIR adaptors, or both. Alternatively, GIV could interact with the TIR-adaptor proteins MAL and TRAM and sequester them from the cytoplasmic domain of other TLRs. Regardless of the exact mechanism(s) involved, it appears that GIV•TIR interactions aid in signal integration while maintaining specificity.
Binding of GIV’s TILL Motif to TLR4-TIR Precludes TIR•TIR Homotypic Homodimerization and Inhibits Proinflammatory Macrophage Activation.
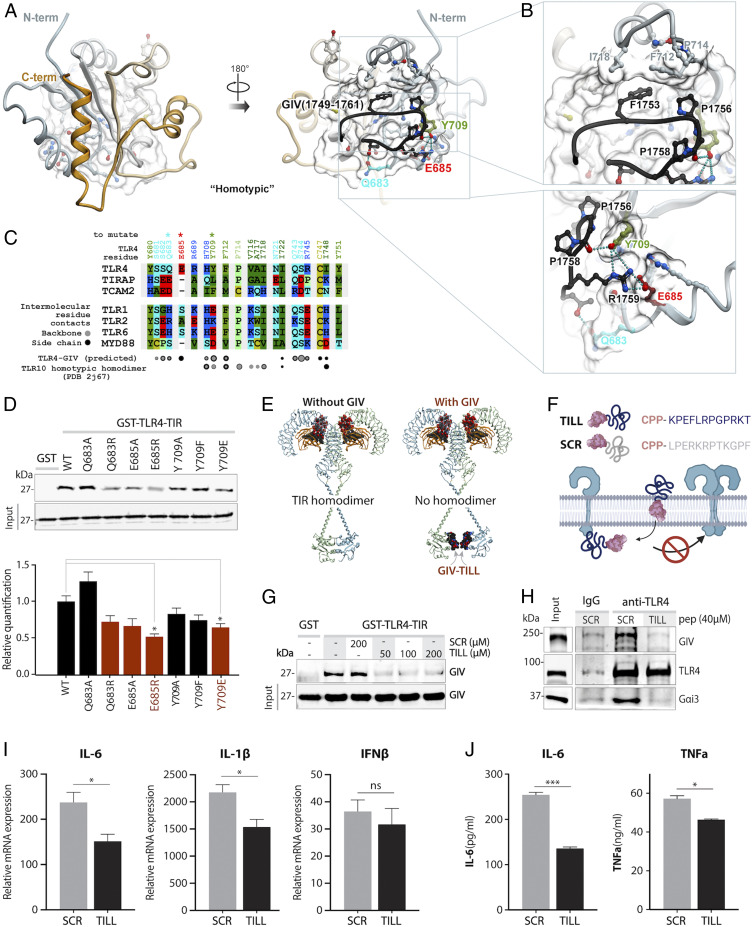
We next sought a structural explanation of GIV’s binding to the TLR4 TIR domain and its effects on macrophage inflammatory signaling. Structural, biochemical, and computational studies have delineated two types of TIR•TIR assemblies: 1) a “homotypic” assembly involves interlocking of the BB-loop regions of two TIR-modules during receptor homodimerization and 2) a “heterotypic” assembly involves binding of the BB-loop of one TIR module to the C-terminal helix of the other during the recruitment of TIR adaptors (38–40). We hypothesized that the binding of the GIV-TILL motif to TLR4-TIR may occur either at the BB-loop, mimicking and competitively disrupting homotypic TLR4-TIR dimers, or at the C-terminal helix, preventing the recruitment of TIR-domain–containing adaptors. Approximate models of GIV TILL complexes with TLR4-TIR were built in both geometries (Fig. 7A and SI Appendix, Fig. S7B); interface residues were identified and compared across a panel of TIR domains that were shown experimentally to bind (or not) GIV-CT. The “homotypic” binding hypothesis provided a better explanation for the specificity of GIV’s interactions with TIR proteins (Fig. 6 F–H): the alignment of interface residues from TLR4, MyD88, Mal, TRAM, TLR1, TLR2, and TLR6 revealed Q683, E685, and Y709 as contact sites specific to the GIV•TLR4 interface and closely conserved in other TIR domains that bind GIV but not in TIR domains that do not (Fig. 7 B and C). By contrast, the heterotypic binding mode showed compatibility (SI Appendix, Fig. S7 B and C) but failed to explain the binding specificity. Thus, the homotypic, but not heterotypic, binding mode is consistent with the observed selectivity of GIV for some TIRs (and not others). Model-guided mutagenesis confirmed that to be true because replacement of contact-site residues on the homotypic TLR4 dimer with corresponding residues found in TLR1/2/6 (i.e., TLRs that do not bind GIV) reduced GIV’s ability to bind TLR4 (Fig. 7D).
Fig. 7.
Identification of key contact sites between GIV’s TILL and BB-loop of TLR4. (A) A homology model of GIV’s TILL motif (black) bound to the BB-loop of human TLR4 (gray) using a homotypic interface. (B) Magnified view of the predicted homotypic interface between GIV’s TILL and TLR4’s BB-Loop. (C) Sequence alignment of TLR4 with other TLRs and TIR adaptors highlighting residues in the GIV-TLR4 interaction interface that may confer binding specificity. (D, Top) Recombinant His-GIV-CT was used in a GST pulldown assay with either WT GST-TLR4-TIR or GIV•TLR4 interface mutants, and bound GIV was visualized by immunoblot. (D, Bottom) Bar graph of densitometry values from three independent experiments. (E) A model of TLR-bound GIV-TILL peptide showing the structural basis for obstruction of TIR•TIR homodimerization by GIV. (F) Schematic of cell-penetrating TILL peptide design. (G) Recombinant His-GIV-CT was used in GST pulldown assay with GST or GST-TLR4-TIR and increasing amounts of TILL peptide or scrambled (SCR) control peptide. Bound GIV was visualized by immunoblot. Equal loading of GST proteins was confirmed by Ponceau S staining (SI Appendix, Fig. S7). (H) Endogenous TLR4 was immunoprecipitated from RAW 264.7 lysates in the presence of either TILL peptide or scrambled control. Bound complex of TLR4, GIV, and Gαi was visualized by immunoblot. Equal loading of IgG and anti-TLR4 were confirmed by Ponceau S staining (SI Appendix, Fig. S7). (I and J) Bar graphs displaying cytokine transcript levels (qPCR) (I) or secreted protein (ELISA) (J) in RAW 264.7 macrophages stimulated with LPS (100 ng/mL, 6 h) in the presence of either TILL peptide or scrambled control. Results are from three independent experiments and displayed as mean ± SEM. Student’s t test was used to determine significance. *P ≤ 0.05, ***P ≤ 0.001; ns, not significant.
The homotypic model revealed key residues on GIV and TLR4 that facilitate binding: GIV’s K1750, P1756, and R1759 made contacts with TLR4’s Q683, E685, and Y709, respectively (Fig. 7 B and C). Identification of K1750 and P1756 as binding residues is supported by our biochemical assays (Fig. 6E). It is also in keeping with prior literature supporting the essential role of the proline P1756 (which corresponds to proline 714 in TLR4 in humans) in the BB-loop of TLR4-TIR which is essential for TIR•TIR interactions (41). Tyrosine 709 is also known to form essential contacts in the TIR•TIR-binding interface (42, 43), raising the possibility that the GIV•TLR4 interaction may be phospho-regulated by tyrosine-based signals. The binding of GIV to TLR4 in a homotypic mode would preclude the assembly of homotypic homodimers of TLR4-TIR modules (Fig. 7E). Because the latter is a prerequisite for adaptor recruitment and for the initiation of proinflammatory signaling cascades (44), such binding is consistent with the experimental findings in this work.
To determine if the binding of GIV-TILL to TLR4 is sufficient to recapitulate the observed antiinflammatory role of GIV in macrophages, we tested the ability of synthetic peptides mimicking this region to exogenously modulate macrophage inflammatory responses. Cell-penetrable peptides containing the TILL motif of GIV (KPEFLRPGPRKT) fused to the C terminus of the Antennapedia peptide [RQIKIWFQNRRMKWKK (45); Fig. 7F] were synthesized commercially. These peptides, representing the minimal required segment of GIV’s C terminus, were predicted to have two measurable consequences if they bound TLR4: 1) such binding should “displace” GIV•Gαi complexes from TLR4, and 2) binding should inhibit TLR activation and the production of proinflammatory cytokines. As expected, the GIV-TILL peptide, but not a control peptide with a scrambled sequence, but of the same charge and residue composition, disrupted the interaction between His-GIV-CT and GST-TLR4-TIR in in vitro protein interaction assays (Fig. 7G). The GIV-TILL peptide also displaced both full-length GIV and Gαi from immunoprecipitated TLR4 in RAW 264.7 macrophages (Fig. 7H). When we investigated LPS-triggered inflammatory responses, macrophages incubated with GIV-TILL peptide had reduced expression of proinflammatory cytokines compared to control peptide with scrambled sequence (Fig. 7 I and J).
These results demonstrate that the GIV-TILL motif is both necessary and sufficient for binding to TLR4; such binding is sufficient to inhibit proinflammatory responses upon LPS stimulation in macrophages. These findings add support to the working model that GIV’s TILL motif binds and disrupts TLR4 signaling, thereby inhibiting proinflammatory cascades (see legend, Fig. 8).
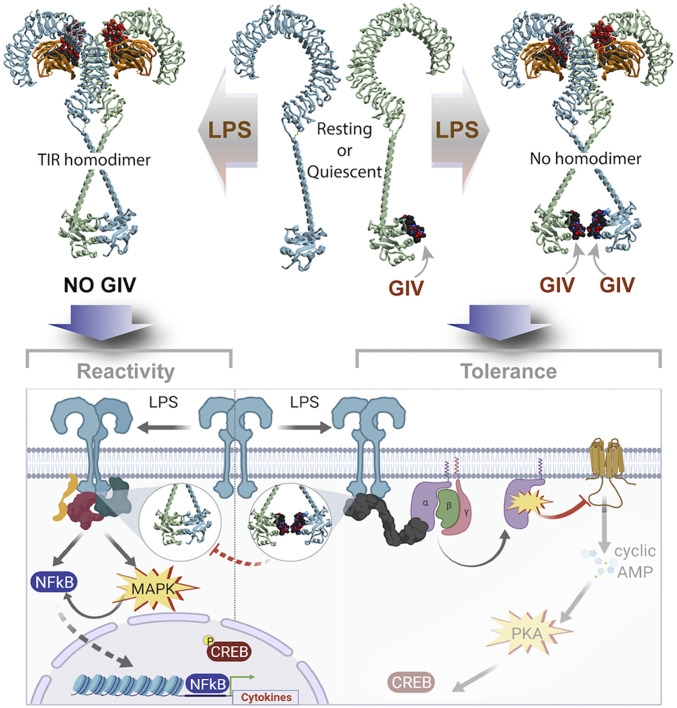
Fig. 8.
Summary of findings and working model. In macrophages expressing GIV (Right), GIV constitutively and directly binds the cytoplasmic TIR domains of TLR4. Binding occurs under resting conditions (Center, Top), which reduces the sensitivity of macrophages to low concentrations of LPS stimuli. Binding of GIV to TLR4 also continues after ligand stimulation (Right, Top), and their mode of binding precludes receptor dimerization (via TIR•TIR homodimerization). GIV-mediated inhibition of receptor dimerization may serve as a potential mechanism for the inhibition of proinflammatory signaling cascades during an LPS/TLR4 inflammatory response at the immediate postreceptor level (Right, Bottom). In cells without GIV (Left), or when GIV levels are reduced after prolonged LPS stimulation, TLR4-TIR modules readily homodimerize, even at low concentrations of LPS stimulation. Consequently, macrophages mount proinflammatory signals (NFkB, CREB, and MAPK) (Left, Bottom) and are sensitive to stimuli.
Conclusion
To ensure immunity, but avoid diseases and limit pathology, inflammatory responses must be fine-tuned through continuous feedback that confines responses to the proverbial “Goldilocks zone” (46). The major discovery we report here is how macrophages confine TLR4 signaling and prevent an overzealous inflammatory response using a multimodular scaffold, GIV. Prior work has suggested that balanced immune responses are brought about when the immune system dynamically responds to cues from both host and pathogen across multiple scales (46). Our findings showing that binding of GIV to TIR modules, thereby impacting multi-TLR signaling, could represent one such example of a multiscale point of convergence for cues from host (levels of expression and immunomodulatory actions of GIV) and from diverse pathogens (ligands of multiple TLRs). GIV’s ability to bind some TIR modules (TLR4, TRAM, MAL) but not others (other TLRs, MyD88) via a single conserved SLIM suggests that GIV may function as a signal convergence/integration point for TLR cross talk during pathogen-sensing. Whether other immune sensors and/or receptors (i.e., cytokine receptors, nucleotide-binding oligomerization domain [NOD]-containing receptors, etc.) also converge similarly, and if so, whether such convergence requires the same SLIM, remain to be studied.
By elucidating the role of GIV, a noncanonical GEF for Gαi proteins, our findings also provide insights into a hitherto unknown mechanism of signal integration and convergence between TLRs and heterotrimeric G proteins. The role of the GPCR→G protein→cAMP signaling cascade in macrophage differentiation and in shaping macrophage responses to pathogens and injury-related danger molecules is well recognized (47). It has also been widely accepted that cross talk between the GPCR→cAMP pathway and the TLRs impacts LPS-triggered inflammatory responses (27, 48–50). By demonstrating that GIV modulates the Gαi→cAMP→CREB cascade during LPS/TLR4 signaling, we define a GPCR-independent regulation of cAMP in macrophages that may serve as a noncanonical mechanism for cross talk between the LPS/TLR and G protein/cAMP pathways. Our finding that cAMP increased in the hyperreactive GIV-depleted macrophages is consistent with GIV’s previously described biochemical function (i.e., a GEF for Gαi) and with studies that have shown that elevated levels of cAMP in response to LPS are responsible for the rapid induction of IL-6 production (27, 28). However, the impact of enhanced cAMP and CREB phosphorylation in GIV-depleted macrophages on downstream signaling and transcriptional responses remains unresolved. We propose that GIV-dependent suppression of cAMP in response to LPS may represent a noncanonical, antiinflammatory cascade in macrophages responding to infectious stimuli (see legend, Fig. 8).
In addition to revealing the mechanisms underlying GIV’s immunomodulatory role, we have also demonstrated that GIV-derived peptides may serve as effective tools for mimicking such an immunomodulatory role. In doing so, we have provided proof-of-concept that mechanistic insights gained herein can be exploited for the development of TLR4-inhibitory peptides, which could inspire immunomodulatory peptide-mimetic therapies.
Overall, this work ushers in important insights into how macrophage inflammatory responses are fine-tuned by the nature of protein complexes at the immediate postreceptor level and opens the door for the development of an interface to target for immunomodulatory therapies.
Materials and Methods
All methods are detailed in SI Appendix and briefly mentioned here.
LPS Stimulation and Bacterial Infection.
Cells were seeded (12-well plate: 2.5 × 105 cells; 6-well plate: 5 × 105 cells) and incubated overnight at 37 °C before stimulation with LPS at indicated doses. Bacteria were maintained and cultured in accordance with ATCC protocols (51) prior to infecting cells at a multiplicity of infection (MOI) of 1 for E. coli and 10 for Salmonella. For RNA readouts, cells were washed once with 1x PBS and stored a −80 °C in TRIzol. For supernatant cytokine analysis (ELISA), absolute levels of IL-6, IL-10, and TNFα were quantified using ELISA MAX or OptEIA ELISA kits (SI Appendix, Materials and Methods, Key Resource Table).
Bacterial Infection of Mice.
Bacterial sepsis in mice was induced by injection of E. coli K1 strain RS218. For survival experiments, 9-wk-old female Girdin floxed x LysMcre and littermate control WT mice were injected intraperitoneally (i.p.) with 1 × 108 colony forming units (cfu) of E. coli, and mouse survival was recorded for 24 h following injection. For measurement of serum IL-6, IL-10, and IL-1β levels, serum was collected 3 h after injection and cytokines were quantified by ELISA (R&D Systems) following the manufacturer’s protocol.
DSS Colitis.
Seven- to 8-wk-old GIV fl/fl x LysMcre or GIV fl/fl littermate controls were given either normal drinking water or 2% DSS for 7 d, followed by 7 d of recovery with normal drinking water. DAI was calculated as done previously (52). Histology scoring was carried out as previously described (51).
Computational Modeling of GIV-TLR Interactions.
Models were built by homology using the TIR-domain structures of TLR1 (53), TLR6 (54), and TLR10 (55) as templates. Homology modeling was performed in Internal Coordinate Mechanics (ICM-Pro) software (56, 57). The position of the conserved Pro-Gly motif of the GIV (aa 1,749 to 1,761) peptide was inherited from the corresponding BB-loop motif in the homotypic TIR-domain homodimer of TLR10 (55) and the heterotypic TIR-domain homodimer of MAL/TIRAP (39). The peptide was built ab initio and tethered to the respective Pro-Gly positions, and its conformations were extensively sampled (>108 steps) by biased probability Monte Carlo sampling in internal coordinates, with the TLR4 TIR domain represented as a set of energy potentials precalculated on a 0.5-Å three-dimensional grid and including Van der Waals potential, electrostatic potential, hydrogen-bonding potential, and surface energy. Following such grid-based docking, the peptide poses were merged with full-atom models of the TLR4 TIR domain, and further sampling was conducted for the peptide and surrounding side chains of the TLR4 residues.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants AI141630, CA100768, and CA160911 (to P.G.), grants DK107585, a pilot award program from UL1TR001442 and a NIDDK DiaComp Pilot and Feasibility Award (to S.D.), and by AI118985 (to I.K.). P.G. and S.D. were also supported by the NIH Grant AI155696 and by the Helmsley Charitable Trust and Foundation (Crohn’s disease project). L.S. was supported by NIH Training Grant DK 0070202.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2011667117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Wynn T. A., Chawla A., Pollard J. W., Macrophage biology in development, homeostasis and disease. Nature 496, 445–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moynagh P. N., TLR signalling and activation of IRFs: Revisiting old friends from the NF-kappaB pathway. Trends Immunol. 26, 469–476 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Lu Y. C., Yeh W. C., Ohashi P. S., LPS/TLR4 signal transduction pathway. Cytokine 42, 145–151 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Ghosh P., Rangamani P., Kufareva I., The GAPs, GEFs, GDIs and…now, GEMs: New kids on the heterotrimeric G protein signaling block. Cell Cycle 16, 607–612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Marcos M., Ghosh P., Farquhar M. G., GIV/Girdin transmits signals from multiple receptors by triggering trimeric G protein activation. J. Biol. Chem. 290, 6697–6704 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghosh P., G protein coupled growth factor receptor tyrosine kinase: No longer an oxymoron. Cell Cycle 14, 2561–2565 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murray P. J., Macrophage polarization. Annu. Rev. Physiol. 79, 541–566 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Beyer M.et al., High-resolution transcriptome of human macrophages. PLoS One 7, e45466 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ying W., Cheruku P. S., Bazer F. W., Safe S. H., Zhou B., Investigation of macrophage polarization using bone marrow derived macrophages. J. Vis. Exp., 50323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orecchioni M., Ghosheh Y., Pramod A. B., Ley K., Macrophage polarization: Different gene signatures in M1(LPS+) vs. classically and M2(LPS-) vs. alternatively activated macrophages. Front. Immunol. 10, 1084 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asai M.et al., Similar phenotypes of Girdin germ-line and conditional knockout mice indicate a crucial role for Girdin in the nestin lineage. Biochem. Biophys. Res. Commun. 426, 533–538 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Ip W. K. E., Hoshi N., Shouval D. S., Snapper S., Medzhitov R., Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science 356, 513–519 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Couper K. N., Blount D. G., Riley E. M., IL-10: The master regulator of immunity to infection. J. Immunol. 180, 5771–5777 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Canny G. O., McCormick B. A., Bacteria in the intestine, helpful residents or enemies from within? Infect. Immun. 76, 3360–3373 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kühl A. A., Erben U., Kredel L. I., Siegmund B., Diversity of intestinal macrophages in inflammatory bowel diseases. Front. Immunol. 6, 613 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Na Y. R., Stakenborg M., Seok S. H., Matteoli G., Macrophages in intestinal inflammation and resolution: A potential therapeutic target in IBD. Nat. Rev. Gastroenterol. Hepatol. 16, 531–543 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Zhu Y.et al., Control of intestinal inflammation, colitis-associated tumorigenesis, and macrophage polarization by fibrinogen-like protein 2. Front. Immunol. 9, 87 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones G. R.et al., Dynamics of colon monocyte and macrophage activation during colitis. Front. Immunol. 9, 2764 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang F. C., Chiu P. Y., Chen Y., Mak T. W., Chen N. J., TREM-1-dependent M1 macrophage polarization restores intestinal epithelium damaged by DSS-induced colitis by activating IL-22-producing innate lymphoid cells. J. Biomed. Sci. 26, 46 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clausen B. E., Burkhardt C., Reith W., Renkawitz R., Förster I., Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8, 265–277 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Efron P., Moldawer L. L., Sepsis and the dendritic cell. Shock 20, 386–401 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Bernardo D., Chaparro M., Gisbert J. P., Human intestinal dendritic cells in inflammatory bowel diseases. Mol. Nutr. Food Res. 62, e1700931 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Wéra O., Lancellotti P., Oury C., The dual role of neutrophils in inflammatory bowel diseases. J. Clin. Med. 5, 118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aznar N., Kalogriopoulos N., Midde K. K., Ghosh P., Heterotrimeric G protein signaling via GIV/Girdin: Breaking the rules of engagement, space, and time. BioEssays 38, 379–393 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters-Golden M., Putting on the brakes: Cyclic AMP as a multipronged controller of macrophage function. Sci. Signal. 2, pe37 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Avni D., Ernst O., Philosoph A., Zor T., Role of CREB in modulation of TNFalpha and IL-10 expression in LPS-stimulated RAW264.7 macrophages. Mol. Immunol. 47, 1396–1403 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Song J.et al., A novel TLR4-mediated signaling pathway leading to IL-6 responses in human bladder epithelial cells. PLoS Pathog. 3, e60 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song J.et al., TLR4-mediated expulsion of bacteria from infected bladder epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 106, 14966–14971 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Getz M., Swanson L., Sahoo D., Ghosh P., Rangamani P., A predictive computational model reveals that GIV/girdin serves as a tunable valve for EGFR-stimulated cyclic AMP signals. Mol. Biol. Cell 30, 1621–1633 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta V.et al., GIV/Girdin activates Gαi and inhibits Gαs via the same motif. Proc. Natl. Acad. Sci. U.S.A. 113, E5721–E5730 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vergadi E., Ieronymaki E., Lyroni K., Vaporidi K., Tsatsanis C., Akt signaling pathway in macrophage activation and M1/M2 polarization. J. Immunol. 198, 1006–1014 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Himes S. R.et al., The JNK are important for development and survival of macrophages. J. Immunol. 176, 2219–2228 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Ghosh P.et al., A Galphai-GIV molecular complex binds epidermal growth factor receptor and determines whether cells migrate or proliferate. Mol. Biol. Cell 21, 2338–2354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohena C.et al., GIV•Kindlin interaction is required for kindlin-mediated integrin recognition and activation. iScience 23, 101209 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wright P. E., Dyson H. J., Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 16, 18–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohnishi H.et al., Structural basis for the multiple interactions of the MyD88 TIR domain in TLR4 signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 10260–10265 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaffen S. L., Structure and signalling in the IL-17 receptor family. Nat. Rev. Immunol. 9, 556–567 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bovijn C.et al., Identification of interaction sites for dimerization and adapter recruitment in Toll/interleukin-1 receptor (TIR) domain of Toll-like receptor 4. J. Biol. Chem. 287, 4088–4098 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ve T.et al., Structural basis of TIR-domain-assembly formation in MAL- and MyD88-dependent TLR4 signaling. Nat. Struct. Mol. Biol. 24, 743–751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bovijn C.et al., Identification of binding sites for myeloid differentiation primary response gene 88 (MyD88) and Toll-like receptor 4 in MyD88 adapter-like (Mal). J. Biol. Chem. 288, 12054–12066 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poltorak A.et al., Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science 282, 2085–2088 (1998). [DOI] [PubMed] [Google Scholar]
- 42.Basith S., Manavalan B., Govindaraj R. G., Choi S., In silico approach to inhibition of signaling pathways of Toll-like receptors 2 and 4 by ST2L. PLoS One 6, e23989 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ronni T.et al., Common interaction surfaces of the toll-like receptor 4 cytoplasmic domain stimulate multiple nuclear targets. Mol. Cell. Biol. 23, 2543–2555 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Núñez Miguel R.et al., A dimer of the Toll-like receptor 4 cytoplasmic domain provides a specific scaffold for the recruitment of signalling adaptor proteins. PLoS One 2, e788 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones S. W.et al., Characterisation of cell-penetrating peptide-mediated peptide delivery. Br. J. Pharmacol. 145, 1093–1102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cicchese J. M.et al., Dynamic balance of pro- and anti-inflammatory signals controls disease and limits pathology. Immunol. Rev. 285, 147–167 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X., Iyer A., Lyons A. B., Körner H., Wei W., Emerging roles for G-protein coupled receptors in development and activation of macrophages. Front. Immunol. 10, 2031 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan H., et al., Lipopolysaccharide- and gram-positive bacteria-induced cellular inflammatory responses: Role of heterotrimeric Galpha(i) proteins. Am. J. Physiol. Cell Physiol. 289, C293–C301 (2005). Erratum in: Am. J. Physiol. Cell Physiol.289, C1360 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Ferlito M.et al., Implication of Galpha i proteins and Src tyrosine kinases in endotoxin-induced signal transduction events and mediator production. J. Endotoxin Res. 8, 427–435 (2002). [PubMed] [Google Scholar]
- 50.Dauphinee S. M., Voelcker V., Tebaykina Z., Wong F., Karsan A., Heterotrimeric Gi/Go proteins modulate endothelial TLR signaling independent of the MyD88-dependent pathway. Am. J. Physiol. Heart Circ. Physiol. 301, H2246–H2253 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Das S.et al., ELMO1 has an essential role in the internalization of Salmonella Typhimurium into enteric macrophages that impacts disease outcome. Cell. Mol. Gastroenterol. Hepatol. 1, 311–324 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim J. J., Shajib M. S., Manocha M. M., Khan W. I., Investigating intestinal inflammation in DSS-induced model of IBD. J. Vis. Exp., 3678 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu Y.et al., Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature 408, 111–115 (2000). [DOI] [PubMed] [Google Scholar]
- 54.Jang T. H., Park H. H., Crystal structure of TIR domain of TLR6 reveals novel dimeric interface of TIR-TIR interaction for toll-like receptor signaling pathway. J. Mol. Biol. 426, 3305–3313 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Nyman T.et al., The crystal structure of the human toll-like receptor 10 cytoplasmic domain reveals a putative signaling dimer. J. Biol. Chem. 283, 11861–11865 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Abagyan R.et al., Homology modeling with internal coordinate mechanics: Deformation zone mapping and improvements of models via conformational search. Proteins 29 (suppl. 1), 29–37 (1997). [DOI] [PubMed] [Google Scholar]
- 57.Abagyan R., Totrov M., Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J. Mol. Biol. 235, 983–1002 (1994). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.