Significance
HORMA domain proteins play integral roles in several cellular processes such as mitosis, meiosis, and DNA repair. Intriguingly, despite their disparate cellular functions, the HORMA proteins MAD2, REV7, and HORMAD1/2 are controlled by a common regulatory circuit involving the ATPase TRIP13. Here, we extend this paradigm, showing that the adaptor protein p31comet, known to mediate TRIP13 activity toward MAD2, is also involved in REV7 regulation. Notably, TRIP13 is overexpressed in many cancers, and TRIP13 hyperactivity inactivates the REV7-Shieldin complex, a critical determinant of sensitivity to Poly-ADP-ribose polymerase (PARP) inhibitors, causing resistance to this clinically important class of drugs. In this work, we present evidence that p31comet overexpression is also common in cancer and largely mirrors the effects of TRIP13 overexpression.
Keywords: homologous recombination, translesion synthesis, Fanconi anemia, PARP inhibitor, REV7
Abstract
The repair of DNA double strand breaks (DSBs) that arise from external mutagenic agents and routine cellular processes is essential for life. DSBs are repaired by two major pathways, homologous recombination (HR) and classical nonhomologous end joining (C-NHEJ). DSB repair pathway choice is largely dictated at the step of 5′-3′ DNA end resection, which is promoted during S phase, in part by BRCA1. Opposing end resection is the 53BP1 protein, which recruits the ssDNA-binding REV7-Shieldin complex to favor C-NHEJ repair. We recently identified TRIP13 as a proresection factor that remodels REV7, causing its dissociation from the Shieldin subunit SHLD3. Here, we identify p31comet, a negative regulator of MAD2 and the spindle assembly checkpoint, as an important mediator of the TRIP13-REV7 interaction. p31comet binds to the REV7-Shieldin complex in cells, promotes REV7 inactivation, and causes PARP inhibitor resistance. p31comet also participates in the extraction of REV7 from the chromatin. Furthermore, p31comet can counteract REV7 function in translesion synthesis (TLS) by releasing it from REV3 in the Pol ζ complex. Finally, p31comet, like TRIP13, is overexpressed in many cancers and this correlates with poor prognosis. Thus, we reveal a key player in the regulation of HR and TLS with significant clinical implications.
Cells need to maintain the integrity of their genomes in the face of attack by endogenous and exogenous DNA damaging agents (1). One of the most challenging lesions is the double strand break (DSB), the repair of which depends on cell cycle phase (2–4). In G1 phase, classical nonhomologous end joining (C-NHEJ) is quick and efficient, entailing the religation of broken ends with minimal processing (3). However, in S, G2, or M phases, cells invoke the more complicated but high fidelity homologous recombination (HR) repair pathway that uses the sister chromatid as repair template for synthesizing DNA across the break (5, 6). The first step of HR is resection of the DSB ends and is highly regulated because it commits repair to HR, thus dictating repair pathway choice and outcome (4).
End resection is normally repressed by a signaling cascade comprising the 53BP1, RIF1, REV7-Shieldin (SHLD1-3), and CST-Pol α proteins (7–17). The ssDNA binding of SHLD2 and the fill-in activity of CST-Pol α impedes and counteracts end resection, respectively (11, 14, 18). This brake on end resection is relieved specifically in S and G2 phases by the BRCA1-BARD1 complex, at least partly by repositioning and dephosphorylating 53BP1, and recruiting CtIP, a component of the end resection machinery (5). Thus, cells deficient for BRCA1 cannot mount an HR response, resulting in sensitivity to the PARP inhibitor, Olaparib (19). However, when these cells concurrently lose any member of the 53BP1-RIF1-REV7-Shieldin anti-end resection cascade, they reacquire the ability to perform HR and become resistant to olaparib (7–17). We recently revealed another route to olaparib resistance through up-regulation of the ATPase TRIP13, an enzyme that inactivates REV7 through a conformational change (20).
REV7 is a founding member of the HORMA protein family, also comprising the meiotic factors Hop1 and HORMAD1/2, the spindle assembly checkpoint (SAC) protein MAD2, and the autophagy proteins Atg101 and Atg13 (21, 22). This family is defined by the high structural similarity of its members, a key feature of which is their C-terminal seatbelt region (21). This “seatbelt” clamps down on an interacting partner in a so-called active “closed” form (21, 23). In the Shieldin complex, the REV7 seatbelt engages SHLD3 upon closure, which is required for the recruitment of SHLD1 and SHLD2 (11–13, 20, 24).
Another commonality among HORMA family members, albeit demonstrated to different degrees, is their inactivation by the highly conserved ATPase TRIP13 (23). Shown most convincingly for MAD2, TRIP13 binds to the N terminus of closed MAD2, destabilizing critical contacts and causing its C-terminal seatbelt to unlatch and transition to an “open” conformation, thereby releasing its seatbelt-binding partner (25–27). The critical seatbelt-binding partner of MAD2 is CDC20, such that MAD2 closed over CDC20 is proficient for SAC activation, while remodeled open-MAD2 without CDC20 associated with it cannot activate the SAC.
TRIP13 is aided in this structural sleight of hand by yet another HORMA-like protein, p31comet (referred to as p31 henceforth), whose C-terminal seatbelt folds back onto itself (Fig. 1A) (25, 26, 28, 29). p31 acts as a bridge between TRIP13 and MAD2, stimulating TRIP13 to remodel MAD2 and release CDC20 (25, 27). p31 also plays an important TRIP13-independent role in MAD2 inhibition, namely, it binds to the MAD2 homodimerization interface by virtue of its structural similarity with MAD2, effectively preventing MAD2 homodimerization which is required for its activation (30). Whether p31 works with TRIP13, or independently, to similarly inactivate REV7 and favor HR is unknown.
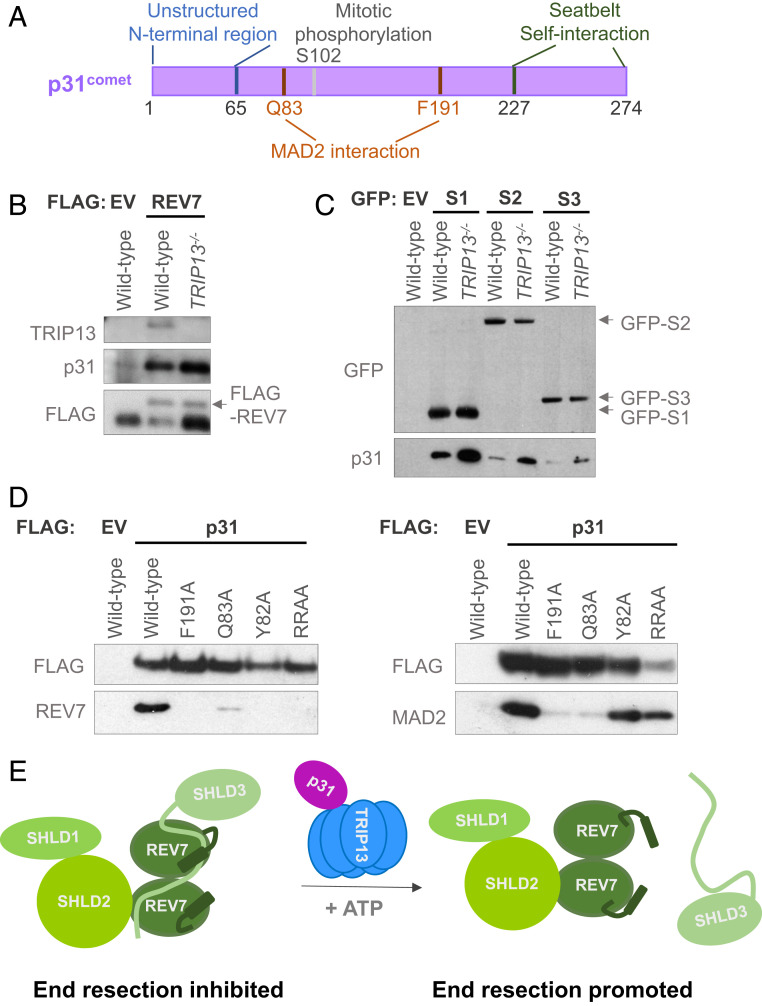
Fig. 1.
p31 physically interacts with REV7-Shieldin. (A) Schematic of p31 protein showing its various functional regions and modification sites. (B) Western blot showing FLAG immunoprecipitation of FLAG-empty vector (EV) or FLAG-REV7 in wild-type and TRIP13−/− U2OS cells and the coimmunoprecipitation of endogenous p31. (C) Western blot showing GFP immunoprecipitation of GFP-tagged: empty vector (EV), SHLD1 (S1), SHLD2 (S2), and SHLD3 (S3) in wild-type and TRIP13−/− U2OS cells and the coimmunoprecipitation of endogenous p31. (D) Western blot showing the FLAG immunoprecipitation of EV, wild-type p31, and various mutant forms of p31, and the coimmunoprecipitation of endogenous REV7 (Left) or MAD2 (Right) in p31−/− HEK293T cells. RRAA denotes the RR188,189AA mutant. (E) Schematic of our proposed model of p31 function as mediating the interaction between REV7 and TRIP13, and hence the dissolution of REV7-Shieldin, through the remodeling of the REV7 seatbelt that is bound to SHLD3. Here, we depict the seatbelts of both REV7 monomers as being unlatched by the action of TRIP13-p31, but it is possible that only the seatbelt closed over SHLD3 is opened.
Here, we reveal that p31 is a regulator of end resection and repair pathway choice. It physically interacts with REV7 and engenders the release of the seatbelt interactor SHLD3 from the REV7-Shieldin complex, thereby promoting HR. p31 also attenuates REV7 association with its other seatbelt partner REV3 in the Pol ζ complex to inhibit their function in interstrand cross-link (ICL) repair. The clinical importance of these findings is underscored by the observation that p31 is frequently overexpressed in cancers and is a marker of poor prognosis in BRCA1-deficient settings. Thus, p31 emerges as an important player in multiple arenas of genome maintenance.
Results
p31 Physically Interacts with the REV7-Shieldin Complex.
The TRIP13 ATPase inactivates the HORMA protein REV7 by unlatching its seatbelt to release SHLD3 and promote HR (20). As TRIP13 has been shown to work with the adaptor protein p31, we asked whether p31 plays any role in this function. p31 has several features of a HORMA protein, including a C-terminal seatbelt region (Fig. 1A). p31 interacted with all known subunits of the REV7-Shieldin complex (Fig. 1 B and C). Interestingly, these interactions were enhanced in the absence of TRIP13 (Fig. 1 B and C), suggesting that a p31-REV7 intermediate accumulates when TRIP13 is not present to disassemble the REV7-Shieldin complex. This is consistent with the previously observed increase in binding of p31 to MAD2-CDC20 in TRIP13−/− cells (31, 32) and to the active (closed) conformer of MAD2 (30, 33).
p31 Interacts with REV7 and MAD2 through a Common Interface.
The HORMA proteins REV7 and MAD2 are structurally very similar, despite their low amino acid sequence identity. The N-terminal ends of HORMA proteins are critical for TRIP13-mediated inactivation (26), and the cryogenic electron microscopy (cryo-EM) structure of TRIP13-p31-MAD2 explains the importance of this N-terminal sequence (SI Appendix, Fig. S1A) (25). TRIP13 binds to a conserved LTR motif in the MAD2 N terminus that feeds into its central pore (SI Appendix, Fig. S1B). Interestingly, REV7 also harbors a conserved LSR motif in its N terminus in a rare pocket of sequence conservation between the two proteins (SI Appendix, Fig. S1B).
We used the published TRIP13-p31-MAD2 structure to model REV7 onto a hypothetical TRIP13-p31-REV7 ternary complex, and detected no obvious steric clashes (SI Appendix, Fig. S1C). To validate our model, we compared it with the TRIP13-p31-MAD2 structure in order to identify residues on p31 that may be critical for its interactions with REV7, MAD2, or both binding partners (SI Appendix, Fig. S1D). Alanine replacement mutant variants of p31 were expressed in p31−/− cells and queried for their ability to interact with REV7 and MAD2 (Fig. 1D and SI Appendix, Fig. S1E). Some mutations, including F191A and Q83A, abrogated the binding of p31 to both REV7 and MAD2, supporting our model of a shared interaction surface. It is thus likely that a major function of p31 is to serve as a general adaptor between TRIP13 and HORMA substrates. Specifically, we propose that p31 works in concert with TRIP13 to unlatch the REV7 seatbelt and release its seatbelt-interacting partner SHLD3 (Fig. 1E).
Intriguingly, two of our p31 mutants—RR188, 189AA (RRAA), and Y82A—had a much stronger impact on REV7 binding than on MAD2 binding (Fig. 1D). We speculate that arginines 188 and 189 in p31 may be more important for association with REV7 through an electrostatic interaction with D138, a residue that is not conserved in MAD2 (SI Appendix, Fig. S1D). Similarly, Y82 of p31 may uniquely stabilize REV7-p31 through a hydrophobic interaction with L128 of REV7 (SI Appendix, Fig. S1D). Thus, p31-RRAA is a separation-of-function mutant that is deficient for REV7 binding but proficient for MAD2 association.
p31 Promotes End Resection and HR.
Given that TRIP13 enhances HR through REV7 inactivation (20) and that p31 interacts with REV7 (Fig. 1) (34), we asked whether p31 regulates HR. The first step of end resection was examined by quantifying DNA damage-induced RAD51 foci levels in p31−/− cell lines generated using CRISPR-Cas9 technology. As seen previously, cells lacking TRIP13 showed a marked impairment in infrared radiation (IR)-induced RAD51 focus formation compared to wild-type cells, while REV7−/− cells exhibited an enhancement (Fig. 2A) (20). Three different p31−/− clones displayed an even greater impairment in RAD51 foci levels than TRIP13−/− cells and were ∼50% of wild-type levels (Fig. 2A).
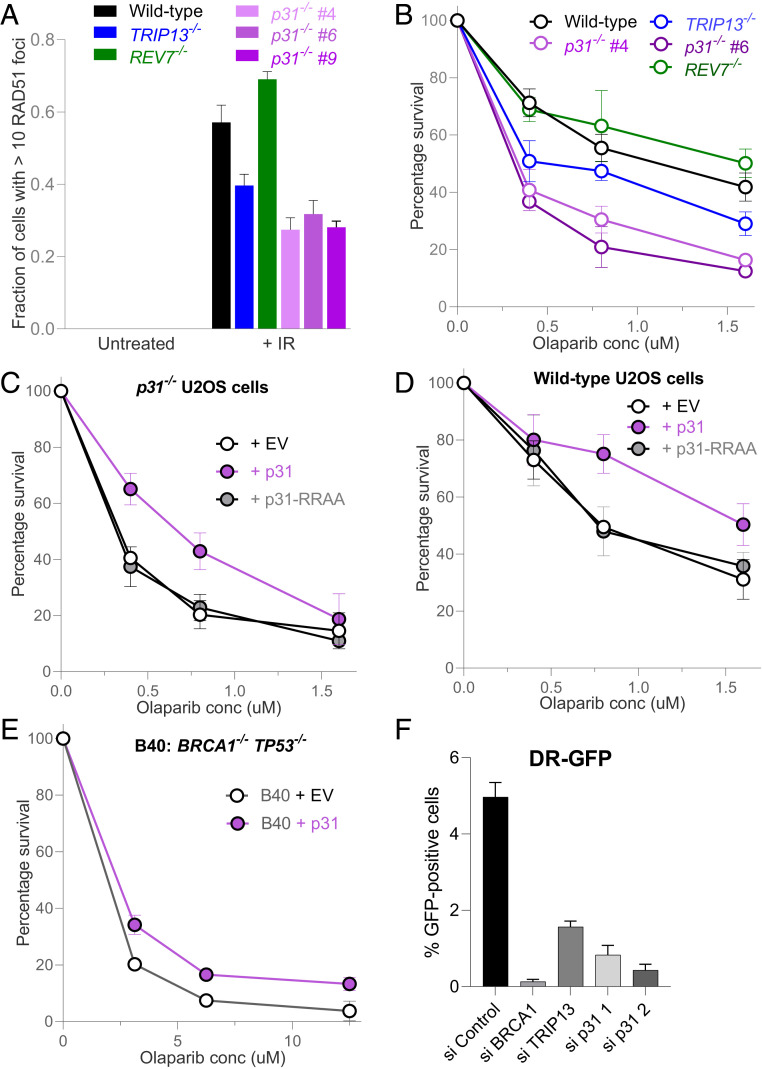
Fig. 2.
p31 promotes homologous recombination. (A) Graph showing RAD51 focus formation in wild-type, TRIP13−/−, REV7−/−, and three p31−/− clones in U2OS cells at baseline and 6 h following treatment with 5 Gy of ionizing radiation. Means and SDs of three independent experiments are depicted. (B) Clonogenic survival curve of wild type, TRIP13−/−, REV7−/−, and two p31−/− clones in U2OS cells at various doses of the PARP inhibitor, olaparib. Means and SDs of three independent experiments are depicted. (C) Clonogenic survival curve of p31−/− U2OS cells expressing empty vector (EV), wild-type p31 (WT), or RR188,189AA mutant p31 (RRAA) at various doses of olaparib. Means and SDs of three independent experiments are depicted. (D) Clonogenic survival curve of U2OS cells expressing EV or overexpressing WT or RR188,189AA (RRAA) mutant p31 at various doses of the PARP inhibitor, olaparib. Means and SDs of three independent experiments are depicted. (E) Clonogenic survival curve of BRCA1−/− TP53−/− RPE-1 cells, expressing EV or overexpressing p31. Means and SDs of two independent experiments are depicted. (F) DR-GFP assay results in U2OS cells following transfection with nontargeting or siRNAs targeting BRCA1, TRIP13, and p31. Means and SDs of two independent experiments are depicted.
As defective end resection is frequently accompanied by sensitivity to PARP inhibitors, we examined the sensitivity of p31−/− cells to olaparib in 14-d clonogenic survival assays. Consistent with our previous findings and RAD51 foci results, both p31−/− and TRIP13−/− cells were sensitive to olaparib compared to wild type, with the former exhibiting a stronger phenotype (Fig. 2B) (20). This sensitivity was also detected in a short-term 5-d Cell Titer Glo-based cytotoxicity assay (SI Appendix, Fig. S2A) and was rescued by ectopically reexpressed p31 (Fig. 2C and SI Appendix, Fig. S2 B–D), excluding off-target effects of the guide RNA or clonal artifacts. Importantly, expression of the RRAA mutant of p31, which interacts with MAD2 but not REV7, failed to rescue the olaparib sensitivity of p31−/− cells (Fig. 2C and SI Appendix, Fig. S2C), indicating that the interaction of p31 specifically with REV7 mediates olaparib resistance.
We next asked whether, conversely, overexpressing p31 can promote increased usage of the HR pathway. Consistent with this, overexpression of wild-type p31, but not the RRAA mutant, increased cellular resistance to olaparib (Fig. 2D and SI Appendix, Fig. S2 B–D). However, p31 overexpression in TRIP13−/− cells did not induce olaparib resistance (SI Appendix, Fig. S2 E and F), indicating that TRIP13 is required for HR promotion by p31.
Loss of REV7-Shieldin activity, whether through genetic inactivation or TRIP13 overexpression, also promotes HR in cells that are otherwise HR deficient due to BRCA1 deficiency (7–17). We therefore tested whether overexpression of p31 could restore HR proficiency in a human RPE-1 cell line in which BRCA1 and TP53 were knocked out with CRISPR-Cas9. Indeed, overexpression of p31 in this BRCA1-deficient cell line induced olaparib resistance (Fig. 2E and SI Appendix, Fig. S2G), consistent with a role for p31 in REV7-Shieldin complex inactivation.
A cell line-based DR-GFP reporter assay was next used to measure the efficiency of HR-dependent repair (35). Knockdown of p31 conferred a strong decrease in the GFP signal compared to control, though not to the degree observed with knockdown of the core HR factor, BRCA1 (Fig. 2F). Taken together, these data strongly implicate p31 in the promotion of HR in conjunction with TRIP13.
p31 Inhibits REV7-SHLD3 Seatbelt Association.
REV7 is recruited to DSBs by SHLD3 to counteract end resection through a seatbelt interaction (11, 13). We have previously shown that TRIP13 unlatches the REV7 seatbelt to release SHLD3, both in vitro and in cells (20). We therefore asked whether p31 has a similar effect by querying REV7-SHLD3 association levels in cells either lacking or overexpressing p31. p31−/− cells, similarly to TRIP13−/−, had higher levels of REV7-SHLD3 in complex compared to wild-type cells (Fig. 3A and SI Appendix, Fig. S3A) (20). Conversely, cells overexpressing p31 or TRIP13 had significantly less SHLD3 associated with REV7 (Fig. 3B and SI Appendix, Fig. S3A) (20). Importantly, overexpression of p31 in TRIP13−/− cells did not cause an appreciable reduction in SHLD3-REV7 association (Fig. 3C and SI Appendix, Fig. S3B). These data support a model in which p31 works in concert with TRIP13 to inactivate the REV7-Shieldin complex, specifically by releasing SHLD3 from the REV7 seatbelt (Fig. 1D).
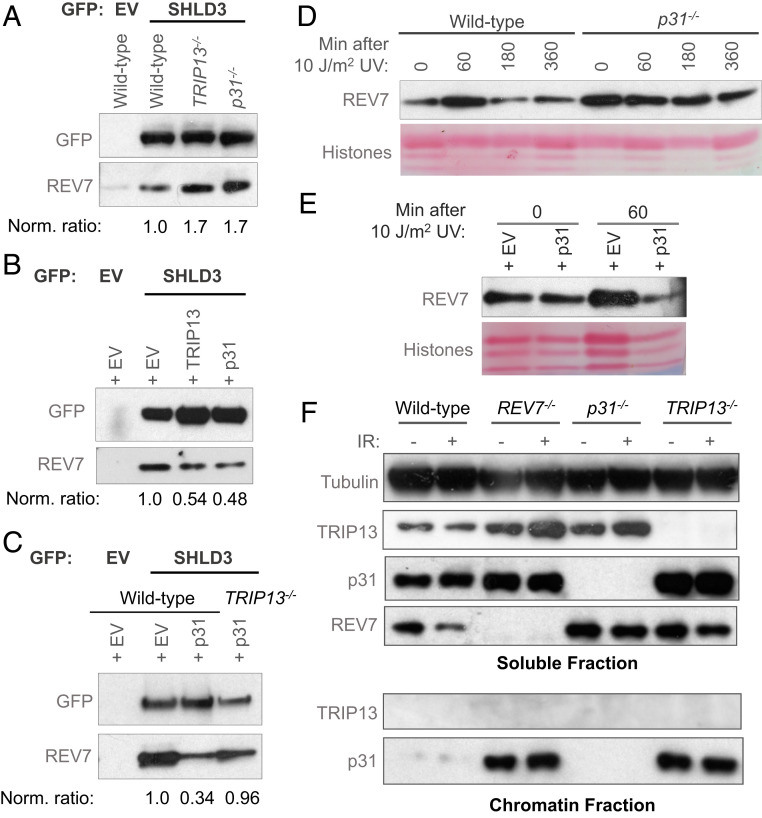
Fig. 3.
p31, like TRIP13, inhibits REV7 activity. (A) Western blot showing the GFP immunoprecipitation of GFP-empty vector (EV) or GFP-SHLD3 and the coimmunoprecipitation of endogenous REV7 in wild-type, TRIP13−/−, and p31−/− U2OS cells. (B) Western blot showing the GFP immunoprecipitation of GFP-empty vector (EV) or GFP-SHLD3 and the coimmunoprecipitation of endogenous REV7 in U2OS cells ectopically overexpressing EV or cDNAs encoding TRIP13 or p31. (C) Western blot showing the GFP immunoprecipitation of GFP- EV or GFP-SHLD3 and the coimmunoprecipitation of endogenous REV7 in wild-type or TRIP13−/− U2OS cells expressing EV or overexpressing p31. (D) Western blot of chromatin-bound REV7 from wild-type or p31−/− U2OS cells at various time points following irradiation with 10 J/m2 of UV light. (E) Western blot of chromatin-bound REV7 from U2OS cells ectopically overexpressing EV or p31 cDNA following irradiation with 10 J/m2 of UV light. (F) Western blot of soluble (Top) and chromatin-bound (Bottom) proteins from wild-type, REV7−/−, p31−/−, and TRIP13−/− U2OS cells, with and without exposure to 5 Gy of ionizing radiation. Norm. ratio is the ratio of coimmunoprecipitated REV7 to the tagged protein which was pulled down, normalized to the control condition.
p31 Promotes REV7 Chromatin Turnover.
A prediction of our model and of the literature is that hyperactive REV7-Shieldin in cells lacking p31 or TRIP13 may be enriched at DNA damage sites. Indeed, TRIP13−/− cells exhibit an increase in REV7 foci and stronger REV7 chromatin retention than wild-type cells after DNA damage (20). We determined the effect of p31 expression on damage-inducible recruitment of REV7 to chromatin. Exposure to UV light, known to induce nucleotide damage that is a substrate for Pol ζ, triggered the robust recruitment of REV7 to chromatin within 15 min, with a return to baseline by 180 min (Fig. 3D and SI Appendix, Fig. S3C). Strikingly, p31−/− cells exhibited strong REV7 chromatin binding, both at baseline and following UV irradiation (Fig. 3D and SI Appendix, Fig. S3C).
This increased chromatin retention of REV7 in p31−/− cells could be due to a loss of active extraction of REV7 by p31-TRIP13, or alternatively, it may reflect increased DNA damage in the cells. To distinguish between these possibilities, we performed the converse experiment wherein p31 was overexpressed and REV7 chromatin engagement was examined. Consistent with an active role of p31 in extracting REV7 from chromatin, overexpression of p31 appeared to decrease REV7 association with chromatin after induction of DNA damage (Fig. 3E). Thus, REV7 is likely recruited to DNA damage sites via its seatbelt interactions, whereupon it performs its functions and is subsequently released from the repair complex by the action of p31-TRIP13.
To further probe the mechanism through which p31-TRIP13 mediates chromatin extraction of REV7 complexes, we monitored the recruitment of p31 and TRIP13 to chromatin following DNA damage. Strikingly, we found p31 to be strongly chromatin associated, irrespective of DNA damage, whereas TRIP13 was primarily soluble (Fig. 3F). Furthermore, p31 accumulated on chromatin to a much greater extent in TRIP13−/− cells (Fig. 3F).
We were intrigued to notice that p31 chromatin retention was counterintuitively increased in REV7−/− cells. Importantly, we have previously shown that chronic REV7 deficiency significantly alters cell cycle kinetics, leading to an increased G2/M population (36). Given that p31 associates with the kinetochore along with MAD2 during mitosis, we speculated that this accumulation is likely unrelated to its Shieldin regulatory function. To test this, we transiently knocked down REV7 and MAD2 in HEK293T cells and monitored p31 chromatin recruitment. Unlike the genetic knockout, transient depletion of REV7 did not cause any overt cell cycle disruption, and consistent with our prediction, transient REV7 depletion did not appreciably affect p31 chromatin retention (SI Appendix, Fig. S3D). Furthermore, MAD2 depletion largely inhibited p31 chromatin association (SI Appendix, Fig. S3D), suggesting that MAD2 plays a more significant role in the recruitment of p31 to chromatin.
p31 Affects REV7 Function in Interstrand Cross-Link Repair.
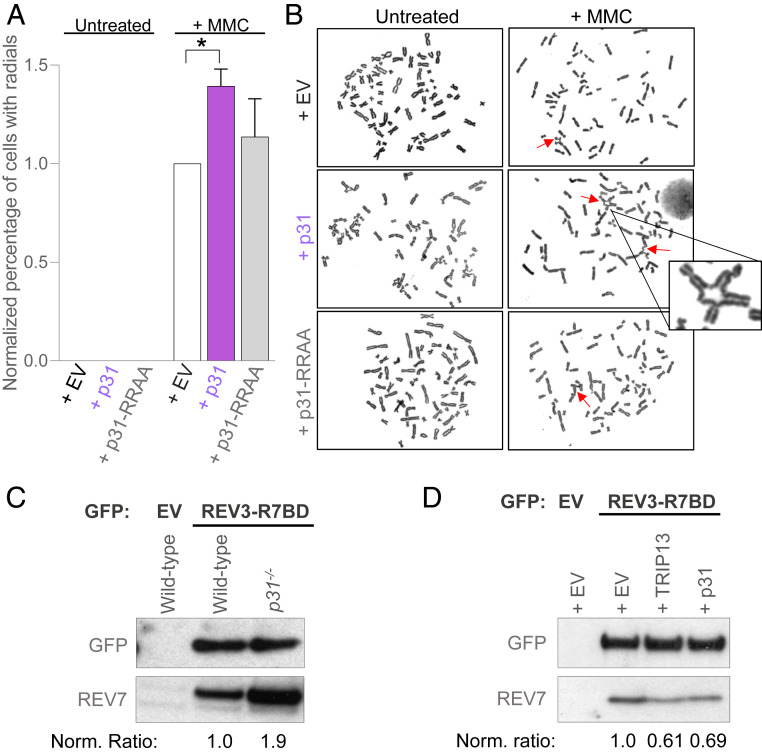
REV7 forms several seatbelt-mediated complexes, including the TLS polymerase Pol ζ. The REV7 seatbelt closes over the seatbelt-interacting region of the catalytic subunit REV3 of Pol ζ to promote ICL repair (37–39). TRIP13 disrupts both the REV7-SHLD3 and REV7-REV3 seatbelt interactions by catalyzing the closed-to-open REV7 conformational switch (20). We asked whether TRIP13 works with p31 solely for inactivation of REV7-Shieldin in HR or if p31 also regulates the Pol ζ complex in ICL repair. p31 was overexpressed and the efficiency of ICL repair was measured by the prevalence of chromosome radials after treatment with the cross-linking agent mitomycin C (MMC). Chromosome radials are formed following aberrant repair of DNA damage, especially ICLs, and are a hallmark of ICL repair deficiency. While most control cells exhibited normal metaphase spreads 48 h after exposure to MMC, p31-overexpressing cells had unresolved chromosome radials, indicative of defective ICL repair (Fig. 4 A and B). Significantly, cells overexpressing the p31-RRAA mutant that cannot interact with and inhibit REV7 showed wild-type levels of unresolved radials (Fig. 4 A and B), indicating that the p31 needs to physically interact with REV7 in order to impair ICL repair. Cells lacking p31 showed marked cohesinopathy due to compromised MAD2 function in the SAC (SI Appendix, Fig. S4 A and B), precluding their examination in this assay. Taken together, this suggests that p31-TRIP13 inhibits ICL repair by dissociating the REV7-REV3 Pol ζ complex.
Fig. 4.
p31 inhibits REV7 activity in ICL repair. (A) Graph showing the proportion of HEK293T cells showing radial chromosomes following treatment with 20 ng/mL of MMC upon expression of empty vector (EV) or overexpression of p31 wild type or RRAA mutant. Asterisk denotes a statistically significant difference. (B) Representative images of metaphase chromosome spreads quantified in A. Arrows indicate radial chromosomes. (C) Western blot showing GFP immunoprecipitation of GFP- EV or GFP-REV3-R7BD (REV7-binding domain) and the coimmunoprecipitation of endogenous REV7 in wild-type and p31−/− U2OS cells. (D) Western blot showing GFP immunoprecipitation of GFP-EV or GFP-REV3-R7BD and the coimmunoprecipitation of endogenous REV7 in U2OS cells expressing EV or overexpressing TRIP13 or p31. Norm. ratio is the ratio of coimmunoprecipitated REV7 to the tagged protein which was pulled down, normalized to the control condition.
p31 Inhibits Pol ζ Complex Formation.
We next directly tested whether p31 inactivates the Pol ζ complex in ICL repair by disengaging REV3 from REV7, similar to its disengagement of SHLD3 from REV7 in the Shieldin complex in HR (Fig. 3 A and B). As REV3 is a large protein, we expressed a minimal REV7-binding domain described previously (20). Consistent with our prediction, p31−/− cells showed greater association between REV7 and the ectopically expressed REV7-binding domain of REV3 (Fig. 4C). Conversely, overexpression of p31 decreased the interaction between REV7 and the REV7-binding domain of REV3, similarly to overexpression of TRIP13 (Fig. 4D). Thus, p31 inhibits ICL repair, mainly by promoting TRIP13-mediated disassembly of the Pol ζ complex.
High p31 Protein Levels Is Commonly Seen in Cancers and Is Associated with Poor Prognosis.
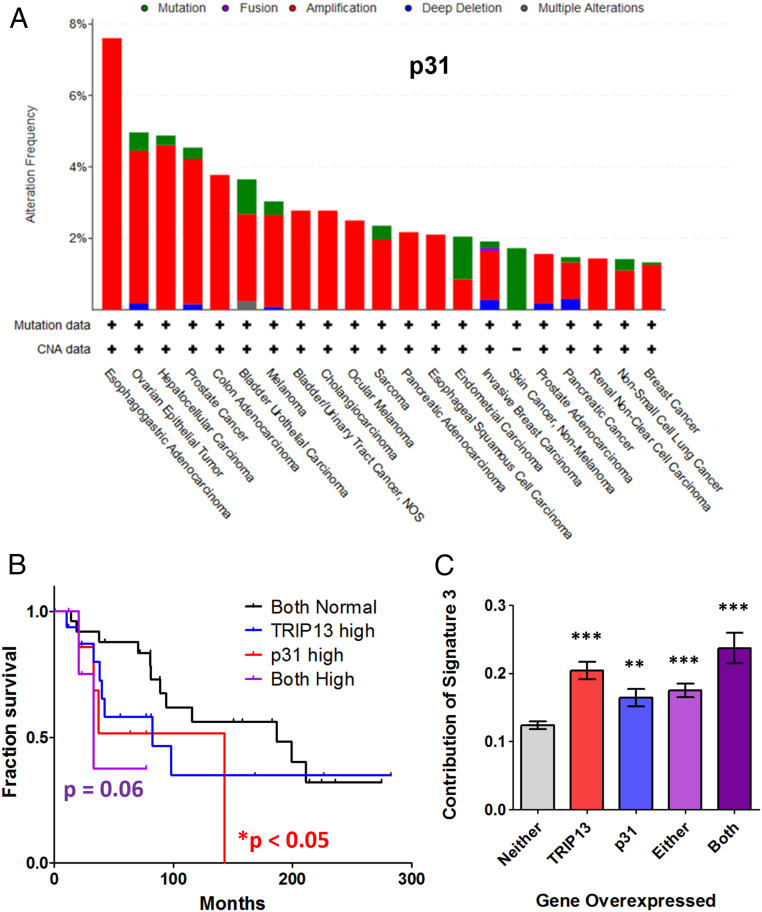
The 53BP1-RIF1-REV7-Shieldin cascade and its inactivator TRIP13 are clinically relevant. Loss of the Shieldin complex or gain of TRIP13 expression in BRCA1-deficient cells causes resistance to PARP inhibitors due to up-regulation of end resection and HR (7–17, 20). Since TRIP13 and p31 work in concert in this role in end resection regulation, it was not surprising to observe that cancer cells frequently up-regulate p31, as reported earlier for TRIP13 (Fig. 5A and SI Appendix, Fig. S5A). It is noteworthy that the cancer subtypes exhibited the same aberrant protein expression pattern for either p31 or TRIP13 (Fig. 5A and SI Appendix, Fig. S5 A and B). Intriguingly, the expression of the two proteins did not correlate in breast cancers, suggesting that they are not dependent on each other for any pro-oncogenic activity (SI Appendix, Fig. S5C).
Fig. 5.
Overexpression of p31, similar to TRIP13, is commonly observed in cancers, correlates with poor prognosis, and contributes to HR mutation signature. (A) Bar chart showing the prevalence of amplifications (red), deletions (blue), and mutations (green) of the p31 gene across an array of cancer types in TCGA. (B) Kaplan–Meier survival curve of BRCA1-deficient breast cancer patients with overexpression of TRIP13, p31, or both as compared to cells with normal expression of both genes. Normal expression vs. p31 high: P < 0.05, normal expression vs. both high: P = 0.06, Mantel–Cox log-rank test. (C) Graph showing the contribution of the HR-deficiency signature 3 in breast cancers overexpressing TRIP13 and/or p31 compared to breast cancers with normal expression of both genes. **P < 0.01, ***P < 0.001, Mann–Whitney test, two tailed.
This increased expression of p31 may be clinically important because patients with BRCA1-mutant cancers and high p31 levels had overall poorer progression-free survival, compared to those with normal levels of p31 (Fig. 5B). This holds true for TRIP13 as well (Fig. 5B) (20) and is likely because the acquired HR proficiency of the cancer cells renders them insensitive to DNA damaging treatment targeting HR deficiency. Having simultaneously high levels of both p31 and TRIP13 in a BRCA1-deficient background appears to confer a more pronounced effect on prognosis (Fig. 5B), suggesting that they act synergistically. Importantly, cancer patients are often treated with combination therapies that also include drugs targeting the mitotic spindle. As such, we cannot definitively determine which aspects of TRIP13-p31 function, which includes the regulation of HR, TLS, and the mitotic spindle assembly checkpoint, are responsible for these observed differences in patient outcome.
Cancers Overexpressing p31 and TRIP13 Have Similarly Altered Mutational Landscapes.
Usage of HR in cancer cells can be inferred by the analysis of their whole genome sequences, in order to measure the strength of the HR deficiency mutational signature (signature 3) (40). Cancer cells with high TRIP13 expression have a high signature 3 contribution, suggesting that TRIP13 up-regulation correlates with HR impairment (Fig. 5C) (20). Interestingly, cancer cells overexpressing p31 also had a higher than normal signature 3 contribution (Fig. 5C), indicative of defective HR. The presence of the HR deficiency signature, even in the presence of TRIP13 and/or p31 overexpression is informative. It implies that either p31-TRIP13 overexpression is an acquired resistance mechanism and that the mutational signature accumulated before the overexpression of these proteins, or that the HR in p31-TRIP13 overexpressing cells is unable to suppress signature 3 accumulation.
Intriguingly, the only other signature that was significantly correlated with either TRIP13 or p31 expression is mutation signature 30. We found that signature 30 was significantly higher in cells with low expression of p31 and/or TRIP13 (SI Appendix, Fig. S5D). The etiology of signature 30 is unclear, but occurs mainly in breast cancers and is elevated in the absence of the base excision repair protein NTHL1 (40, 41). An attractive hypothesis is that Pol ζ-mediated TLS across oxidized guanines, which are normally repaired by NTHL1, contributes significantly to the pattern of C > T transitions observed in signature 30. Thus, the high signature 30 in cells with low p31-TRIP13 may be reflective of hyperactive REV7 in the Pol ζ complex.
Discussion
Extensive work has highlighted the importance of DSB repair and end resection in both normal physiology and cancer progression. Aberrant or unscheduled DSB repair can lead to deleterious outcomes such as gross chromosomal rearrangements (42). Thus, cells have evolved an intricate network of proteins to regulate the crucial step of end resection. Here, we demonstrate that p31 is an essential partner of the TRIP13 ATPase, contributing to the disassembly and chromatin release of the REV7-Shieldin complex.
Upon activation, p31-TRIP13 remodels REV7 to unlatch its C-terminal seatbelt and disengage its seatbelt-binding partner SHLD3, thereby releasing the brake on end resection and switching repair from mutagenic end joining to high-fidelity templated repair (Fig. 1E). The stimuli that trigger p31-TRIP13 activation and their mechanism of activation are still unknown (43). p31 and TRIP13 activity are fully coupled, in that p31 is recruited to chromatin independently of TRIP13 and REV7 (Fig. 3F). Hence, there are potentially at least three regulated steps: the association of p31 with chromatin, the engagement of p31 with REV7-containing complexes at sites of DNA damage, and the subsequent recruitment of TRIP13. It is tempting to speculate that one or several of the physical interactors—p31, TRIP13, REV7, and SHLD3—undergo damage-induced posttranslational modification to alter the affinity of p31-TRIP13 for the substrate REV7-SHLD3. There is a precedent for this, as the mitotic phosphorylation of p31 has a functional effect on MAD2 inactivation (44–46). Elucidating the mechanism of p31 chromatin recruitment and activation is crucial.
Alternatively, the activity of TRIP13 might normally be kept in check during G1 but triggered upon DNA damage in S phase. There is no requirement for phosphorylation in vitro for the remodeling reaction (29), but the situation in cells could be different. Another possibility is that p31 or TRIP13 could undergo DNA damage-induced relocalization such that their activity is directed specifically at REV7-Shieldin engaged at DSB ends. It is therefore important to test whether p31 or TRIP13 form IR-induced foci that colocalize with REV7, paralleling how p31-TRIP13 relocalizes from the cytoplasm to unattached kinetochores during mitosis to remodel MAD2 (47–49).
p31 and SHLD2 were thought to bind to the same highly conserved region on REV7 (9, 18, 34). SHLD2 is a low abundance protein throughout the cell cycle, whereas p31 expression is increased specifically in S phase (44, 50). Cells could therefore favor end resection in S phase simply by displacement of SHLD2 from G1 phase pro-C-NHEJ REV7-SHLD2 complexes with newly minted p31 that recruits TRIP13 to chromatin to catalyze REV7 remodeling and chromatin release. However, a recent structure of REV7 bound to small fragments of SHLD2 and SHLD3 has revealed that SHLD2 binds to a different REV7 region, and that SHLD3 stabilizes a REV7 dimer through a canonical seatbelt interaction with one REV7 moiety and hydrophobic interactions with the other (51). This structure is provocative as it indicates that REV7 dimerization mediates the pro-NHEJ function of the Shieldin complex, thereby suggesting additional regulatory mechanisms impinging on REV7 homodimerization.
Other recent work has also implicated the REV7 homodimer in its TLS function, but its mechanistic significance is not understood (34). In the case of MAD2, homodimerization is required for propagating the activation signal. One closed-MAD2 moiety binds to an open MAD2 through this interface and converts it to closed MAD2, triggering a chain reaction of rapid MAD2 activation (52). p31 attenuates this amplification by competitively binding to the MAD2 dimerization region. Importantly, this function of p31 is a TRIP13-independent process (30). Is a similar mechanism at play in the case of REV7? Our observation that p31−/− cells are more sensitive to olaparib and have a stronger HR defect than TRIP13−/− cells (Fig. 2) suggests that this might be the case, although p31’s effects on REV7-SHLD3 association requires TRIP13 (Fig. 3C).
What happens to REV7 after p31-TRIP13 remodels it into the open form? No longer bound to SHLD3 at DSB ends, REV7 is likely released from chromatin to permit end resection and HR to occur (Figs. 1E and 3). Whether open REV7 remains associated with SHLD2 and SHLD1 is important to determine. Another unanswered question is how open REV7 is reactivated to form complexes closed over SHLD3 or REV3 to promote C-NHEJ and TLS, respectively. In the case of open MAD2, the seatbelt-binding activator MAD1 triggers its transition to the closed form, which then templates the closing of other open-MAD2 moieties (52). Uncovering the activation mechanism for REV7 will also have important clinical implications, because loss of that factor(s) should confer PARP inhibitor resistance.
Indeed, the REV7 regulatory axis appears to be clinically important. We show here that p31 up-regulation is commonly observed in cancers and correlates with poor prognosis in the BRCA1-deficient background (Fig. 5 A and B), similar to the up-regulation of TRIP13 (20, 53, 54). Furthermore, TRIP13 and p31 expression are significantly correlated with mutagenic processes in breast cancers, as indicated by characteristic mutation signatures (Fig. 5C). Importantly, while the loss of Shieldin pathway components is a rare event in cancer, TRIP13 and/or p31 overexpression are frequently observed, suggesting that they may represent a more clinically relevant mechanism for PARP inhibitor resistance. As PARP inhibitors are becoming widespread in the treatment of increasing types of cancers, the problem of resistance is becoming more pressing each day. Inhibition of the p31-TRIP13 axis provides a promising avenue for research into this unmet clinical need.
Materials and Methods
Cell Culture and Transfections.
Human U2OS and HEK293T cells were grown in Dulbecco's modified Eagle's medium (DMEM)/F12 + Glutamax (Invitrogen) supplemented with 10% fetal calf serum (FCS) (Invitrogen) and penicillin-streptomycin (1%) (Invitrogen). DNA transfections and siRNA knockdowns were carried out using Lipofectamine LTX (Invitrogen) and RNAiMax (Invitrogen), respectively, according to the manufacturer’s protocols.
Generation of Knockout Cell Lines with CRISPR-Cas9.
p31 guide RNA sequences were cloned into the pSpCas9(BB)-2A-GFP vector, a gift from Feng Zhang, Broad Institute, Cambridge, MA (Addgene plasmid 48138). Cells transfected with Cas9-gRNA plasmids were GFP+ selected 48 h later and seeded as single cells using a BD FACSAria II cell sorter. Single cells were cultured for 3 to 4 wk and colonies were screened for knockouts by Western blotting using the anti-p31comet antibody (Millipore-Sigma). The guide RNA target sequences used in this study were AAGGCCTCCGAAGCGTTGAG and TTAAGCTGTTCATAGGGCAG.
Structural Modeling.
All protein structures were visualized and aligned using PyMol (Schrödinger, LLC). Structural modeling of REV7 with TRIP13-p31 was done using a MAD2-p31-TRIP13 structure (PDB: 6F0X) (25) as a template. REV7 (from PDB: 3VU7) (38) was aligned with the structurally similar MAD2 protein to produce the models.
Cellular Fractionation and Immunoblot Analysis.
Cells were lysed with Nonidet P-40 buffer (1% Nonidet P-40, 300 mM NaCl, 0.1 mM EDTA, 50 mM Tris [pH 7.5]) supplemented with phosphatase and protease inhibitor mixture (Roche). Cell lysates were resolved by gel electrophoresis using precast NuPAGE Novex gels (Invitrogen). Proteins were transferred onto nitrocellulose membranes using the wet transfer method and sequentially incubated with primary and secondary antibodies and detected using chemiluminescence. For chromatin extraction, cell pellets were resuspended in hypotonic buffer (10 mM Tris⋅HCl pH 7.5, 10 mM KCl, 2.5 mM MgCl2) and passed through a 27-gauge needle 10 times to obtain nuclei. Nuclei were then washed and lysed in Nonidet P-40 buffer. Chromatin extracts were then prepared by digesting the insoluble pellet with micrococcal nuclease (Roche).
Immunoprecipitation.
Cells were lysed in 150 mM NaCl Nonidet P-40 buffer for 30 min at 4 °C with rocking. They were then incubated with antibody-bead conjugate overnight at 4 °C. Beads were washed four times with 150 mM NaCl Nonidet P-40 buffer and immunoprecipitates were eluted either using 0.5 mg/mL FLAG peptide or by boiling.
Antibodies and Chemicals.
Antibodies used in this study were: Abcam ab180579 (REV7, immunoblotting), Abcam ab128171 (TRIP13), Cell Signaling 2144 (Tubulin), Santa Cruz sc-8349 (RAD51, immunoflourescence), and Millipore-Sigma MABE451 (p31comet). MMC was purchased from Sigma and olaparib was purchased from Selleckchem.
Drug Sensitivity and Functional Cell-Based Reporter Assays.
To assay clonogenic survival, cells were seeded at 1,000 cells/well in six-well plates in triplicates. Drugs at the shown doses were added after 12 h and cells were permitted to grow for 14 d. For ionizing radiation experiments, cells were exposed to X-rays from a Rad Source RS-2000 irradiator. For UV irradiation, cells were exposed to UV-C light from a Stratagene UV Stratalinker 2400. Colony formation was scored by fixing and staining with 0.5% (wt/vol) crystal violet in 20% methanol. For short-term CellTiterGlo survival assays, cells were plated in 96-well plates at 1,000 cells/well, and treated with drugs at the indicated concentrations after 12 h. Three days later, cellular viability was measured using CellTiterGlo (Promega). Survival at each drug concentration was calculated as a percentage normalized to the corresponding untreated control, for both assays. The direct repeat homologous recombination assay using the U2OS DR-GFP reporter cell line was performed as previously described (20).
Immunofluorescence Assays.
Cells were plated on glass coverslips in 12-well plates and allowed to grow overnight. They were then either left untreated or treated at 5 Gy IR. After 6 h, they were harvested by preextraction with 0.5% Triton X-100 for 5 min, followed 4% paraformaldehyde fixation for 10 min at 4 °C. After three phosphate-buffered saline (PBS) washes, blocking was performed with 3% bovine serum albumin in PBS for 1 h at room temperature, followed by sequential primary and secondary antibody incubations overnight at 4 °C and 1 h at room temperature, respectively. The coverslips were mounted onto glass slides using DAPI-containing medium (Vector Laboratories). Images were captured using a Zeiss AX10 fluorescence microscope and Zen software, and foci were scored. At least 100 cells were counted for each sample.
Chromosomal Breakage Analysis.
HEK293T cells transfected with the indicated expression constructs were incubated for 48 h in the absence or presence of MMC at 20 ng/mL. Cells were treated with 100 ng/mL of colcemid for 2 h, followed by a hypotonic solution (0.075 M KCl) for 20 min and fixed with 3:1 methanol/acetic acid. After staining with Wright’s stain, 50 metaphase spreads were counted for aberrations. The relative number of chromosomal breaks and radials was calculated normalized to empty vector control.
The Cancer Genome Atlas (TCGA) Data Acquisition and Analysis.
TCGA mutation and expression data were obtained from cBioportal. Tumors were stratified relative to the TCGA reference level for p31 and TRIP13 expression. Mutation signature analysis used the TCGA 2012 dataset, with signature data acquired from mSignatureDB.
Supplementary Material
Acknowledgments
This work was supported by grants from the US NIH (R37HL052725, P01HL048546), the US Department of Defense (BM110181), the Breast Cancer Research Foundation, the Fanconi Anemia Research Fund, the Ludwig Center at Harvard and the Smith Family Foundation to A.D.D., and the Leukemia and Lymphoma Society (5440-16) and the Claudia Adams Barr Program in Innovative Basic Cancer Research to P.S.
Footnotes
Competing interest statement: A.D.D. is a consultant/advisory board member for Lilly Oncology, Merck-EMD Serono, Intellia Therapeutics, Sierra Oncology, Cyteir Therapeutics, Third Rock Ventures, AstraZeneca, Ideaya Inc., Cedilla Therapeutics Inc., a stockholder in Ideaya Inc., Cedilla Therapeutics Inc., and Cyteir, and reports receiving commercial research grants from Lilly Oncology and Merck-EMD Serono.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008830117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Chatterjee N., Walker G. C., Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 58, 235–263 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mirman Z., de Lange T., 53BP1: A DSB escort. Genes Dev. 34, 7–23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ceccaldi R., Rondinelli B., D’Andrea A. D., Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 26, 52–64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Symington L. S., Mechanism and regulation of DNA end resection in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 51, 195–212 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Densham R. M., Morris J. R., Moving mountains-the BRCA1 promotion of DNA resection. Front. Mol. Biosci. 6, 79 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C. C., Feng W., Lim P. X., Kass E. M., Jasin M., Homology-directed repair and the role of BRCA1, BRCA2, and related proteins in genome integrity and cancer. Annu. Rev. Cancer Biol. 2, 313–336 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmermann M., de Lange T., 53BP1: Pro choice in DNA repair. Trends Cell Biol. 24, 108–117 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu G.et al., REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature 521, 541–544 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta R.et al., DNA repair network analysis reveals shieldin as a key regulator of NHEJ and PARP inhibitor sensitivity. Cell 173, 972–988.e23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boersma V.et al., MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5′ end resection. Nature 521, 537–540 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noordermeer S. M.et al., The shieldin complex mediates 53BP1-dependent DNA repair. Nature 560, 117–121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghezraoui H.et al., 53BP1 cooperation with the REV7-shieldin complex underpins DNA structure-specific NHEJ. Nature 560, 122–127 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dev H.et al., Shieldin complex promotes DNA end-joining and counters homologous recombination in BRCA1-null cells. Nat. Cell Biol. 20, 954–965 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mirman Z.et al., 53BP1-RIF1-shieldin counteracts DSB resection through CST- and Polα-dependent fill-in. Nature 560, 112–116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Findlay S.et al., SHLD2/FAM35A co-operates with REV7 to coordinate DNA double-strand break repair pathway choice. EMBO J. 37, e100158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomida J.et al., FAM35A associates with REV7 and modulates DNA damage responses of normal and BRCA1-defective cells. EMBO J. 37, e99543 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao S.et al., An OB-fold complex controls the repair pathways for DNA double-strand breaks. Nat. Commun. 9, 3925 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Setiaputra D., Durocher D., Shieldin–The protector of DNA ends. EMBO Rep. 20, e47560 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trenner A., Sartori A. A., Harnessing DNA double-strand break repair for cancer treatment. Front. Oncol. 9, 1388 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clairmont C. S.et al., TRIP13 regulates DNA repair pathway choice through REV7 conformational change. Nat. Cell Biol. 22, 87–96 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenberg S. C., Corbett K. D., The multifaceted roles of the HORMA domain in cellular signaling. J. Cell Biol. 211, 745–755 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aravind L., Koonin E. V., The HORMA domain: A common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem. Sci. 23, 284–286 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Vader G., Pch2(TRIP13): Controlling cell division through regulation of HORMA domains. Chromosoma 124, 333–339 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Dai Y.et al., Structural basis for shieldin complex subunit 3-mediated recruitment of the checkpoint protein REV7 during DNA double-strand break repair. J. Biol. Chem. 295, 250–262 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alfieri C., Chang L., Barford D., Mechanism for remodelling of the cell cycle checkpoint protein MAD2 by the ATPase TRIP13. Nature 559, 274–278 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye Q.et al., The AAA+ ATPase TRIP13 remodels HORMA domains through N-terminal engagement and unfolding. EMBO J. 36, 2419–2434 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brulotte M. L.et al., Mechanistic insight into TRIP13-catalyzed Mad2 structural transition and spindle checkpoint silencing. Nat. Commun. 8, 1956 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miniowitz-Shemtov S., Eytan E., Kaisari S., Sitry-Shevah D., Hershko A., Mode of interaction of TRIP13 AAA-ATPase with the Mad2-binding protein p31comet and with mitotic checkpoint complexes. Proc. Natl. Acad. Sci. U.S.A. 112, 11536–11540 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye Q.et al., TRIP13 is a protein-remodeling AAA+ ATPase that catalyzes MAD2 conformation switching. eLife 4, e07367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang M.et al., p31comet blocks Mad2 activation through structural mimicry. Cell 131, 744–755 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma H. T., Poon R. Y. C., TRIP13 functions in the establishment of the spindle assembly checkpoint by replenishing O-MAD2. Cell Rep. 22, 1439–1450 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Kim D. H.et al., TRIP13 and APC15 drive mitotic exit by turnover of interphase- and unattached kinetochore-produced MCC. Nat. Commun. 9, 4354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia G.et al., Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint. EMBO J. 23, 3133–3143 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rizzo A. A.et al., Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex. Proc. Natl. Acad. Sci. U.S.A. 115, E8191–E8200 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pierce A. J., Johnson R. D., Thompson L. H., Jasin M., XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13, 2633–2638 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bluteau D.et al., Biallelic inactivation of REV7 is associated with Fanconi anemia. J. Clin. Invest. 126, 3580–3584 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makarova A. V., Burgers P. M., Eukaryotic DNA polymerase ζ. DNA Repair (Amst.) 29, 47–55 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kikuchi S., Hara K., Shimizu T., Sato M., Hashimoto H., Structural basis of recruitment of DNA polymerase ζ by interaction between REV1 and REV7 proteins. J. Biol. Chem. 287, 33847–33852 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hara K.et al., Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase zeta and REV1. J. Biol. Chem. 285, 12299–12307 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang P. J.et al., mSignatureDB: A database for deciphering mutational signatures in human cancers. Nucleic Acids Res. 46, D964–D970 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drost J.et al., Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science 358, 234–238 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bunting S. F., Nussenzweig A., End-joining, translocations and cancer. Nat. Rev. Cancer 13, 443–454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarangi P., Clairmont C. S., D’Andrea A. D., Disassembly of the shieldin complex by TRIP13. Cell Cycle 19, 1565–1575 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaisari S.et al., Role of Polo-like kinase 1 in the regulation of the action of p31comet in the disassembly of mitotic checkpoint complexes. Proc. Natl. Acad. Sci. U.S.A. 116, 11725–11730 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mo M., Arnaoutov A., Dasso M., Phosphorylation of Xenopus p31(comet) potentiates mitotic checkpoint exit. Cell Cycle 14, 3978–3985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Date D. A., Burrows A. C., Summers M. K., Phosphorylation regulates the p31Comet-mitotic arrest-deficient 2 (Mad2) interaction to promote spindle assembly checkpoint (SAC) activity. J. Biol. Chem. 289, 11367–11373 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Westhorpe F. G., Tighe A., Lara-Gonzalez P., Taylor S. S., p31comet-mediated extraction of Mad2 from the MCC promotes efficient mitotic exit. J. Cell Sci. 124, 3905–3916 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hagan R. S.et al., p31(comet) acts to ensure timely spindle checkpoint silencing subsequent to kinetochore attachment. Mol. Biol. Cell 22, 4236–4246 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson C. R., Hwang T., Chen P. H., Bhalla N., TRIP13PCH-2 promotes Mad2 localization to unattached kinetochores in the spindle checkpoint response. J. Cell Biol. 211, 503–516 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Habu T., Kim S. H., Weinstein J., Matsumoto T., Identification of a MAD2-binding protein, CMT2, and its role in mitosis. EMBO J. 21, 6419–6428 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang L.et al., Molecular basis for assembly of the shieldin complex and its implications for NHEJ. Nat. Commun. 11, 1972 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Antoni A.et al., The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 15, 214–225 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Sheng N.et al., TRIP13 promotes tumor growth and is associated with poor prognosis in colorectal cancer. Cell Death Dis. 9, 402 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Banerjee R.et al., TRIP13 promotes error-prone nonhomologous end joining and induces chemoresistance in head and neck cancer. Nat. Commun. 5, 4527 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.