Significance
Some intracellular bacteria develop inside a vacuole, which expands during the infection process. We show, here, that the protein ATG16L1 restricts the expansion of the Chlamydia trachomatis vacuole. ATG16L1 is well known for its role in autophagy, a process that contributes to the elimination of intracellular microbes. However, the restriction exerted by ATG16L1 on vacuole expansion relies on a different ATG16L1 function. We demonstrate that the bacteria secrete an effector protein that prevents ATG16L1 binding to TMEM59 and allows rerouting of vesicular traffic to the vacuole. The discovery that one bacterial effector evolved to target ATG16L1’s engagement in intracellular traffic emphasizes the importance this secondary activity of ATG16L1 for maintaining host cell homeostasis.
Keywords: host-pathogen interactions, autophagy, ATG16L1, intracellular traffic, Chlamydia trachomatis
Abstract
The obligate intracellular bacteria Chlamydia trachomatis, the causative agent of trachoma and sexually transmitted diseases, multiply in a vacuolar compartment, the inclusion. From this niche, they secrete “effector” proteins, that modify cellular activities to enable bacterial survival and proliferation. Here, we show that the host autophagy-related protein 16-1 (ATG16L1) restricts inclusion growth and that this effect is counteracted by the secretion of the bacterial effector CT622/TaiP (translocated ATG16L1 interacting protein). ATG16L1 is mostly known for its role in the lipidation of the human homologs of ATG8 (i.e., LC3 and homologs) on double membranes during autophagy as well as on single membranes during LC3-associated phagocytosis and other LC3-lipidation events. Unexpectedly, the LC3-lipidation-related functions of ATG16L1 are not required for restricting inclusion development. We show that the carboxyl-terminal domain of TaiP exposes a mimic of an eukaryotic ATG16L1-binding motif that binds to ATG16L1’s WD40 domain. By doing so, TaiP prevents ATG16L1 interaction with the integral membrane protein TMEM59 and allows the rerouting of Rab6-positive compartments toward the inclusion. The discovery that one bacterial effector evolved to target ATG16L1’s engagement in intracellular traffic rather than in LC3 lipidation brings this “secondary” activity of ATG16L1 in full light and emphasizes its importance for maintaining host cell homeostasis.
Chlamydia trachomatis is an obligate intracellular pathogen responsible for the most common sexually transmitted bacterial infection (1). The bacteria reside within a vacuolar compartment, called the inclusion, which expands throughout the developmental cycle. The host and the bacteria contribute collectively to the making of this compartment. In particular, host lipids are diverted to the inclusion membrane both through vesicular and through nonvesicular traffic (2). The nature of the intercepted vesicles is not fully understood, and the presence of many different Rab GTPases at the inclusion membrane suggests that several trafficking pathways are involved (3). Key players in this rerouting of host-derived vesicles are the bacterial Inc proteins, that are inserted into the inclusion membrane, and that interact with regulators of intracellular traffic (4). However, Inc proteins are confined to the inclusion membrane, which limits their range of action. We recently observed that the loss of expression of the soluble effector CT622 in a CtrΔct622 strain resulted in several deficiencies, including a defect in inclusion growth, supporting the hypothesis that this soluble effector might contribute to the diversion of host-derived material toward the inclusion (5). In the present study, we identify the host protein ATG16L1 as a target of CT622. ATG16L1 is best known for its role as part of the ATG12-ATG5-ATG16L1 complex, which catalyzes the lipidation of the human homologs of ATG8 (i.e., LC3 and homologs) on double membranes during autophagy as well as on single membranes during LC3-associated phagocytosis and other LC3-lipidation events (6–9). ATG16L1 also plays an important role in the control of inflammation through its ability to bind NOD1 and NOD2 (10). Very unexpectedly, we show here that the ATG16L1-driven function that is targeted by CT622 is not related to its LC3-lipidation capacity nor to its ability to bind NODs but to its involvement in regulating intracellular traffic by interacting with the transmembrane protein TMEM59. We show that CT622 inhibits the formation of the ATG16L1/TMEM59 complex, allowing the rerouting of vesicular traffic to the inclusion thereby rescuing inclusion growth in the CtrΔCT622 strain.
Results
CT622 Binds to ATG16L1 through Its Carboxyl-Terminal (C-Terminal) Domain.
The observation that loss of CT622 impaired inclusion growth suggested that this soluble effector might contribute to the diversion of host-derived material toward the inclusion. To identify the targets of CT622 in the host cytoplasm, we performed two independent pull-down (PD) experiments. We identified 33 proteins that were significantly enriched in the GST-CT622 PD fraction compared to GST, three of which being recovered in the two independent experiments (SI Appendix, Table S1). The autophagy-related proteins ATG16L1 and ATG5 were recovered in the two experiments with the highest total peptide counts. To test their ability to interact with CT622, we first performed co-immunoprecipitation (IP) experiments in cells transfected with plasmids expressing Flag-CT622, GFP-ATG5, and/or GFP-ATG16L1. After cell lysis, we immunoprecipitated Flag-CT622, separated the proteins by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE), and probed this fraction with antibodies against the green fluorescent protein (GFP) by Western blot. GFP-ATG16L1 co-immunoprecipitated with Flag-CT622 while GFP-ATG5 did not (Fig. 1A). This result suggests that CT622 interacts with ATG16L1. The recovery of ATG5 in the PD but not in the co-IP suggests that when all protein were expressed at the endogenous level, ATG5 cofractionated with CT622 via its ability to bind ATG16L1. By immunofluorescence, we observed that coexpression of GFP-ATG16L1 with Flag-CT622 led to the relocation of Flag-CT622 to GFP-ATG16L1 puncta, further supporting the hypothesis that the two proteins interact (SI Appendix, Fig. S1A).
Fig. 1.
TaiP binds directly to ATG16L1 through its C-terminal domain to promote C. trachomatis infection. (A) HeLa cells were cotransfected with Flag-CT622 and the indicated GFP-tagged construct for 24 h, lysed, and IP was performed with anti-Flag coupled beads. Proteins were separated on SDS/PAGE, transferred on a poly(vinylidene difluoride) (PVDF) membrane, and probed with the indicated antibody ([IB]: immunoblot). An aliquot of each cell lysate was loaded on a separate gel to visualize the expression of Flag-CT622 and of each of the GFP-tagged proteins (input, Left). (B) Same as in A, using full-length CT622 or constructs corresponding to the N-terminal (CT622Nterm, amino acids 1–345) or C-terminal (CT622Cterm, amino acids 346–647) domains. (C) HeLa cells were infected for 35 h with CtrΔct622+CT622-Flag bacteria, lysed, and IP was performed with anti-Flag coupled beads. Proteins were separated on SDS/PAGE, transferred on a PVDF membrane and probed with anti-Flag and anti-ATG16L1 antibodies. (D) Recombinant ATG16L1 (100 nM) was incubated with recombinant GST-CT622 or GST-CT622Nterm (100 nM) for 60 min at 4 °C before performing GST-PD using glutathione beads. PD fractions were analyzed by Western blot as in A. GST-CT622Nterm used here as a negative control shows the level of nonspecific ATG16L1 binding to the beads. ATG16L1 was pulled-down together with TaiP, demonstrating that the interaction is direct. The experiment shown is representative of three independent experiments. (E) Quantification of the effect of KO atg16l1 on inclusion size. WT or atg16l1 KO cells seeded on coverslips were infected with CtrWT, CtrΔtaiP, or CtrΔtaiP+TaiP-Flag for 20 h at multiplicity of infection (MOI) = 0.2. After fixation, infected cells were permeabilized, and the inclusion membrane was stained with antibodies against the inclusion protein Cap1. The area of inclusion was measured using imageJ software. The dot plot shows the median ± SD of three independent experiments (n > 50 in total) and displays the P values of the Student’s t tests. The Right shows the absence of ATG16L1 in atg16l1 KO whole cell lysates probed by Western blot with anti-ATG16L1 antibodies. ACTIN IB serves as a loading control. (F) Quantification of the effect of TaiP expression on inclusion size. Cells were transfected with constructs for Flag-CymR or Flag-TaiP for 24 h before being infected with Ctr or CtΔtaiP. The cells were fixed 20-h postinfection, permeabilized, and the inclusion membrane was stained with antibodies against the inclusion protein Cap1. The area of inclusion was measured using imageJ software. The dot plot displays the median ± SD of three independent experiments (n > 50 in total) and displays the P values of the Student’s t test.
CT622 exhibits a highly conserved C-terminal domain (CT622Cterm) and a somewhat less conserved amino-terminal (N-terminal) domain (CT622Nterm) (5). Co-IP experiments with each of these domains expressed individually revealed that the interaction with ATG16L1 occurred via CT622Cterm (Fig. 1B). To confirm this interaction in the context of an infection and in the absence of protein overexpression we used the CtrΔct622 strain complemented with ct622 with a C-terminal Flag tag (CtrΔct622+CT622-Flag), which we had characterized previously (5). Cells were infected for 35 h, lysed, and anti-Flag antibodies were used to immunoprecipitate CT622-Flag. We detected endogenous ATG16L1 in the IP fraction, supporting the hypothesis that CT622 interacted with ATG16L1 in infection (Fig. 1C). Finally, to determine whether the interaction was direct, we incubated purified ATG16L1 with either recombinant GST-CT622 or recombinant GST-CT622Nterm as a negative control. We pulled-down GST using glutathione-bound resin and measured the levels of ATG16L1 that copurified in these fractions by quantifying band intensities in Western blot. ATG16L1 copurified with GST-CT622, demonstrating that the interaction between the two proteins was direct (Fig. 1D). Based on these data, we propose to rename CT622 translocated ATG16L1 interacting protein (TaiP), which is consistent with the nomenclature for other soluble chlamydial effectors (e.g., TarP, TepP, and TmeA).
ATG16L1 Restricts C. trachomatis Development and the Restriction Is Exacerbated in the Absence of TaiP.
To study the role of ATG16L1 in C. trachomatis infection, we generated atg16l1 knockout (KO) HeLa cells (Fig. 1E) in which we verified that LC3B lipidation was fully impaired (SI Appendix, Fig. S1B). We compared the ability of wild type (WT) bacteria (CtrWT), CtrΔtaiP, and CtrΔtaiP+TaiP-Flag to establish an infection in this cellular background compared to the parental HeLa cells. The loss of ATG16L1 resulted in a 20% increase in the size of the inclusions of CtrWT and CtrΔtaiP+TaiP-Flag (Fig. 1E), indicating that ATG16L1 restricts the development of C. trachomatis. Strikingly, the increase in inclusion diameter was much more pronounced (∼70% increase) for the CtrΔtaiP strain. As a result, the CtrΔtaiP inclusions in the atg16l1 background reached the average size for CtrWT in control cells (Fig. 1E). The recovery in inclusion size was observed in four independent atg16l1 KO clones and is, therefore, not a clonal effect (SI Appendix, Fig. S1C). The same observations were made when using small interfering RNA (siRNA) targeting ATG16L1 in HeLa cells as well as in the endometrial epithelial cell line HEC-1-B (SI Appendix, Fig. S1 D and E). These data indicate that, at least, part of the decrease in inclusion growth observed in the CtrΔtaiP strain is due to its inability to counteract an ATG16L1-driven restriction on inclusion development. In support of this, we observed that the transfection of Flag-CT622 prior to infection resulted in a 50% increase in inclusion size for the CtrΔtaiP strain and a 40% increase for the CtrWT strain (Fig. 1E). A construct expressing an irrelevant Flag-tagged protein (CymR) expressed at similar levels as Flag-TaiP was used as a negative control. Altogether, we concluded from these series of experiments that ATG16L1 hinders inclusion development and that one of the roles of the effector TaiP is to counteract this brake by binding to ATG16L1. Importantly, the absence of ATG16L1 resulted in an approximately twofold increase in progeny for the CtrΔtaiP strain (SI Appendix, Fig. S1F). This is modest compared to the 25-fold difference of infectivity that exists between the CtrΔtaiP and the CtrWT strains (5), implicating that while the inclusions recovered a normal size, the other phenotype associated with TaiP loss, i.e., defect of infectivity of EBs, remained. This is not surprising, considering that the loss of progeny in the CtrΔtaiP strain is largely due to the formation of nonfunctional EBs (e.g., defects in TarP secretion, for instance), which is likely disconnected from the defect on the inclusion size. The absence of ATG16L1 did not significantly affect the progeny of the CtrWT or CtrΔtaiP+TaiP-Flag strains (SI Appendix, Fig. S1F).
ATG16L1 Restricts Inclusion Growth through Its WD40 Domain.
The ATG16L1 N-terminal part is structurally and functionally conserved in yeast and is responsible for its role in LC3 lipidation. Its C-terminal part is made of an unstructured region followed with seven WD40 repeats (amino acids 320–607) and is absent in the yeast ortholog (Fig. 2A). The seven WD40 repeats form part of a β-propeller domain which recruits several ATG16L1 effector proteins including NOD1, NOD2, and TLR2 (11–13). Notably, while the WD40 domain is dispensable for LC3 lipidation on double membrane autophagosomes during classical autophagy, it was shown to be required for ATG12-ATG5-ATG16 complex-mediated LC3 lipidation at single membranes (7, 9). To investigate the mechanism by which ATG16L1 restricts inclusion development, we expressed in WT or atg16l1 KO HeLa GFP, full length GFP-ATG16L1 (GFP-ATG16L1FL), a truncated form of ATG16L1 lacking the WD40 domain (GFP-ATG16L11–319), or a truncated form of ATG16L1 lacking the ATG5-binding and coiled-coil domains (GFP-ATG16L1266–607). As expected, expression of GFP-ATG16L1FL and of GFP-ATG16L11–319 in atg16l1 KO cells rescued LC3B lipidation, and GFP-ATG16L1266–607 did not (SI Appendix, Fig. S2A). These cells were infected with CtrΔtaiP, and the median size of inclusions in the GFP-positive cells was measured (SI Appendix, Fig. S2B). We observed that the expression of GFP-ATG16L1FL and GFP-ATG16L1266–607 decreased the size of the CtrΔtaiP inclusions compared to GFP expressing cells, whereas the expression of GFP-ATG16L11–319 did not (Fig. 2B). These experiments demonstrate that the E3-ligase activity of ATG16L1 is dispensable for the restriction this protein exerts on C. trachomatis development. Interestingly, the presence of LC3B at the inclusion periphery had been reported in a previous study, and the authors had concluded that this observation did not depend on a functional autophagy machinery (14). In agreement with that report, we observed an enrichment of LC3B around the inclusion, labeled with an antibody against the inclusion protein Cap1 (Fig. 2C). Strikingly, the presence of LC3B around the inclusion was independent of the LC3B lipidation by the ATG12-ATG5-ATG16 complex since it was also observed in atg16l1 KO HeLa cells (Fig. 2C), or in atg16l1 KO or atg3 KO HEK293 cells (SI Appendix, Fig. S2B). It was also independent of the expression of TaiP as CtrΔtaiP inclusions were also decorated with LC3B (Fig. 2C). Thus, while the presence of LC3B at the inclusion membrane is an intriguing observation, we concluded from these experiments that it is not related to the TaiP/ATG16L1 interaction.
Fig. 2.
TaiP targets ATG16L1’s WD40 domain. (A) Schematic of the ATG16L1 structure including the coiled-coil (C.C.) domain and the WD40 domain (WD40). The binding sites to ATG5, WIPI2B, and FIP200 are highlighted, along with the serine phosphorylated by ULK1. WD40 binding partners are indicated on the right. (B) WT or atg16l1 KO HeLa cells seeded on coverslips were transfected with the indicated constructs for 24 h. Cells were then infected with CtrΔtaiP for 20 h at MOI = 0.2 before being fixed, permeabilized, and the inclusion membrane was stained with antibodies against the inclusion protein Cap1. The graph displays the median ± SD of three independent experiments (n > 50 cells in total) and the P values of the Student’s t test. (C) WT and HeLa cells were infected with either CtrWT or CtrΔtaiP for 20 h before fixation, permeabilization, and immunostaining with antibodies against LC3B and the bacterial inclusion protein Cap1. LC3B is localized at the inclusion membrane independent of TaiP or ATG16L1. (Scale bar: 10 µm.) The dot plot displays the median intensity of LC3B staining at the inclusion membrane (n > 10 cells from three independent experiments). Student’s t tests showed no significant difference. (D) Lysates from atg16l1 KO cells transfected with either GFP-ATG16L1 or GFP-ATG16L11–319 were incubated with 100 pmol of recombinant GST-TaiP for 1 h at 4 °C, before performing a GST-PD using glutathione beads. Fractions were analyzed, such as in Fig. 1A. ATG16L1 was no longer pulled down withTaiP when its WD40 domain was deleted. (E) atg16l1 KO HeLa cells were cotransfected with Flag-TaiP and either GFP-ATG16L1 or GFP-ATG16L11–319. The cells were fixed 24 h later, permeabilized, and stained with anti-Flag antibodies. In the absence of the WD40 domain, Flag-TaiP was no longer recruited to ATG16L1 puncta. (Scale bar: 10 µm.)
Since the expression of the WD40 domain was sufficient to restrict the growth of CtrΔtaiP inclusions, we tested whether this domain was implicated in the interaction between ATG16L1 and TaiP. Purified GST-CT622 was incubated with lysates from atg16l1 KO cells expressing either GFP-ATG16L11–319 or GFP-ATG16L1FL before purifying GST-TaiP together with associated proteins (Fig. 2D). We worked in an atg16l1 KO background to avoid possible dimerization of the expressed constructs with endogenous ATG16L1. ATG16L1FL copurified with GST-TaiP while ATG16L11–319 did not, indicating that the WD40 is necessary for the formation of the TaiP/ATG16L1 complex (Fig. 2D). Consistent with this result, we observed that Flag-TaiP no longer relocalized to GFP-ATG16L1 puncta in the absence of the WD40 (Fig. 2E). Altogether, we concluded from these experiments that TaiP targets the WD40 domain of ATG16L1. This interaction results in a gain in inclusion growth through a pathway that does not require the LC3-lipidation capacity of ATG16L1.
TaiP Promotes Inclusion Growth by Disrupting ATG16L1/TMEM59 Interaction.
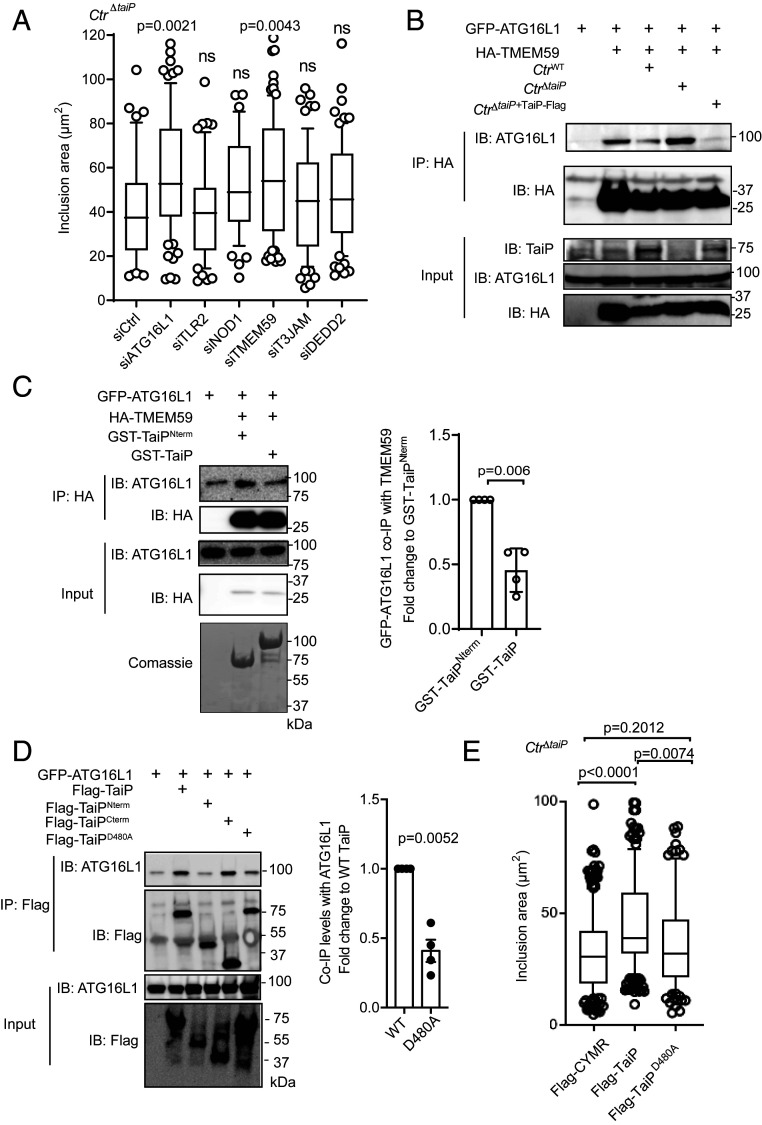
To identify the ATG16L1-related complex or pathway that is targeted by TaiP, we reasoned that silencing the expression of proteins involved in this process should result in the same phenotype as ATG16L1 silencing, i.e., a rescue of the growth of the CtrΔtaiP strain. We, thus, transfected cells with siRNA against the five best characterized binding partners of the WD40 domain of ATG16L1, i.e., TLR2, NOD1, TMEM59, T3JAM, and DEDD2 (12). NOD2 was not tested as it is not expressed in HeLa cells. siRNA against ATG16L1 was included in the screen as a positive control, and the efficiency of the silencing was verified by qRT-PCR (SI Appendix, Fig. S3A). Thirty h later, the cells were infected with CtrΔtaiP. The cells were fixed 20 h after infection and processed for measuring the size of the inclusions by immunofluorescence. We observed that silencing the expression of the protein TMEM59 phenocopied the phenotype observed with ATG16L1 silencing, i.e., an increase in the size of CtrΔtaiP inclusions (Fig. 3A), a result we confirmed in the HEC-1-B epithelial cell line (SI Appendix, Fig. S3B). To avoid the possible sampling bias inherent to microscopy quantification, we measured the percentage of infected cells detected by flow cytometry. When working at low MOI, this parameter is directly linked to bacterial load because only cells bearing sufficient bacteria are recorded as infected (15). We observed that silencing the expression of the protein TMEM59, but not of the other proteins tested, significantly increased the percentage of infected cells, confirming the results obtained on inclusion size measurement (SI Appendix, Fig. S3C). These results indicate that the restriction ATG16L1 exerts on the growth of C. trachomatis inclusions depends on its ability to interact with TMEM59. To strengthen this hypothesis, we tested whether TaiP interfered with the formation of ATG16L1/TMEM59 complexes. We first confirmed, using HeLa cells expressing GFP-ATG16L1 and hemagglutinin (HA)-TMEM59, that the two proteins co-immunoprecipitated (Fig. 3B). When the IP was performed on a cell infected with CtrWT or CtrΔtaiP+TaiP-Flag strains, we observed a decrease in the amount of ATG16L1 that co-immunoprecipitated with TMEM59. In contrast, infection with CtrΔtaiP did not prevent the interaction of ATG16L1 with TMEM59 (Fig. 3B). This result strongly supports an inhibitory role of TaiP on the formation of an ATG16L1/TMEM59 complex. To confirm these data, we expressed separately HA-TMEM59 and GFP-ATG16L1 in HeLa cells by transfection, then, mixed cell lysates in the presence of purified GST-TaiP. GST-TaiPNterm was used as a negative control in this assay since it does not bind to ATG16L1 (Fig. 1B). After incubation, HA-TMEM59 was immunoprecipitated, and we analyzed, by Western blot, the levels of GFP-ATG16L1 in this fraction. In the presence of GST-TaiPNterm GFP-ATG16L1 immunoprecipitated together with HA-TMEM59. However, in the presence of GST-TaiP, the quantity of GFP-ATG16L1 that coprecipitated with HA-TMEM59 amounted to the signal observed in the absence of expression of HA-TMEM59 and, thus, corresponded to nonspecific GFP-ATG16L1 binding to the beads (Fig. 3C). Interestingly, addition of GST-TaiP did not compromise the ability for ATG16L1 to interact with NOD1, NOD2, nor TLR2 (SI Appendix, Fig. S3D). Thus, TaiP blocks specifically TMEM59/ATG16L1 interaction in epithelial cells.
Fig. 3.
TaiP blocks the TMEM59/ATG16L1 complex to promote C. trachomatis inclusion expansion. (A) Cells transfected with the indicated siRNA were infected with CtΔtaiP the following day at MOI = 0.2. Cells were fixed 20 h later, permeabilized, and the inclusion membrane was stained with anti-Cap1 antibodies. Inclusion areas were measured using imageJ software. The box and whiskers plots represent the median area and 90th percentile of intracellular inclusions (n > 50 cells from three independent experiments). Statistical analysis was performed using a one-way ANOVA test with a Dunnett’s multiple comparison test to small interfering control (siCtrl). (B) Cells transfected with the indicated plasmids were infected or not 12 h later with CtWT, CtΔtaiP, or CtrΔtaiP+TaiP-Flag bacteria at MOI = 1. Cells were lysed 30 h later, and IP was performed with anti-HA coupled beads. Proteins were separated on SDS/PAGE, transferred on a PVDF membrane, and probed with the indicated antibodies. (C) Lysates from cells expressing HA-TMEM59 or GFP-ATG16L1 were mixed in the presence of 100 pmol of GST-TaiP or GST-TaiPNterm (for negative control) for 90 min at 4 °C in a final volume of 1.5 mL. TMEM59 was IP with anti-HA coupled beads and the levels of GFP-ATG16L1 in the IP fraction was analyzed by Western blot. IBs of the input fractions show the expression of each individual protein and represents 1.5% of the total reaction and comassie staining shows purified GST-TaiP and GST-TaiPNterm. The histogram on the Right displays the mean ± SD of three independent experiments and the P value of the Student’s t test. Addition of GST-TaiP decreased the amount of GFP-ATG16L1 that co-IP with HA-TMEM59 by about 50%. (D) HeLa cells were cotransfected with Flag-TaiP, Flag-TaiPD480A, and GFP-ATG16L1 constructs for 24 h, lysed, and IP was performed with anti-Flag antibody. Proteins were separated on SDS/PAGE, transferred on a PVDF membrane, and probed with the indicated antibody. An aliquot of each cell lysate was loaded on separate gels to visualize the expression of the tagged proteins (input). Quantification on the Right shows the decrease in the amount of ATG16L1 that co-immunoprecipitates with Flag-TaiPD480A compared to Flag-TaiP. Statistical analysis was performed using Student’s unpaired t test, n = 4. (E) Quantification of the average inclusion size in cells expressing Flag-CymR, Flag-TaiP, and Flag-TaiPD480A. Cells were transfected for 24 h before they were infected for 20 h with CtrΔtaiP at MOI = 0.2. After fixation and permeabilization, inclusions were stained using antibodies against Cap1. Inclusion areas were measured using imageJ software. The dot plot shows the median ± SD of three independent experiments (n > 50 cells in each experiment) and displays the P values of the Student’s t test.
TaiP Mimics a Eukaryotic Domain for Binding to the ATG16L1 WD40 Domain, and D480 Is a Critical Residue for TaiP/ATG16L1 Interaction.
Our results indicate that TaiP targets the ATG16L1/TMEM59 complex. TMEM59 is a type I transmembrane protein whose role is poorly understood. Initially described as residing in the Golgi apparatus, it was later described as a player in endocytic trafficking from late endosomes to lysosomes with a clear colocalization with lysosomal markers (12, 16). Overexpression of TMEM59 induced LC3 lipidation of the compartment in which the protein resides, through its ability to attract ATG16L1 (12). The Pimentel-Muiños’ Laboratory identified, in its short cytoplasmic tail, a WD40-binding motif defined as [YW]-X3-[ED]-X4-[YWF]-X2-L (12). This motif was found in several other proteins that bind to ATG16L1, such as TLR2 and the CARD domain of NOD2. We analyzed the TaiP sequence and found a single matching motif Y474AAALSD480GYSAY485KTL488, that corresponds to the sixth helix of TaiPCter, which is well exposed at the surface of the protein (5). To test whether this motif was implicated in ATG16L1/TaiP interaction, we mutated aspartate 480 into an alanine (D480A). Co-IP experiments showed that the introduction of this point mutation reduced the ability for TaiP to interact with ATG16L1 by about 50% (Fig. 3D). To confirm the engagement of this motif in ATG16L1/TaiP interaction, we next measured the gain in inclusion size in HeLa cells transfected with either TaiP or TaiPD480A and infected with CtrΔtaiP. Transfection of an irrelevant Flag-tagged construct, Flag-CymR, was used as a negative control. As previously observed, expression of TaiP resulted in increased CtrΔtaiP inclusion size. However, the mutated form of TaiP was unable to rescue inclusion growth (Fig. 3E). We concluded from these experiments that TaiPCterm sixth helix mimics the ATG16L1-binding motif [YW]-X3-[ED]-X4-[YWF]-X2-L thereby allowing the bacterial effector to disrupt TMEM59/ATG16L1 interaction and favor inclusion growth.
TaiP Diverts Rab6- and ATG16L1-Dependent Vesicular Traffic toward the Inclusion.
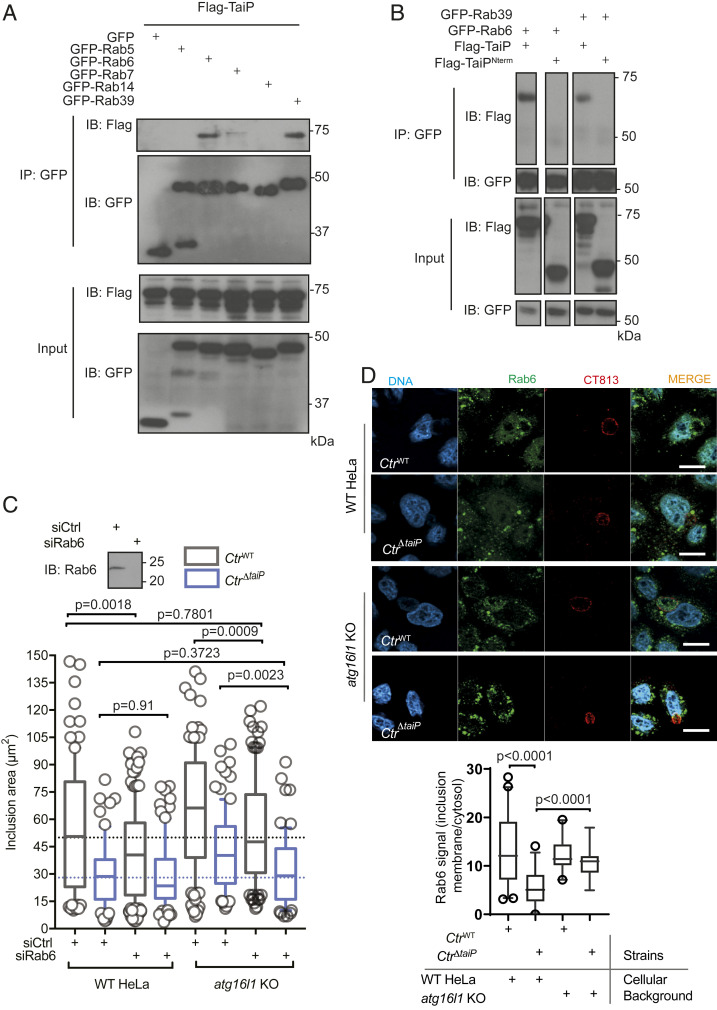
We next analyzed TMEM59 localization in CtrWT and CtrΔtaiP infected HeLa cells. We observed TMEM59 in punctate structures with no enrichment at the inclusion membrane, neither in cells infected with WT nor with CtrΔtaiP bacteria (SI Appendix, Fig. S4). Our data show that silencing ATG16L1 or TMEM59 converge to a similar phenotype, i.e., a rescue of the growth of CtrΔtaiP inclusions. One possible explanation for this phenomenon is that, in the absence of those players (or in the presence of TaiP, that disrupts their interaction), a pool of vesicles becomes available for inclusion growth. To strengthen this scenario, we looked for a molecular marker associated with such a pool of vesicles. Among the proteins that were significantly enriched in the GST-TaiP PD compared to GST alone (SI Appendix, Table S1), we had identified several small Rab GTPases: Rab5, Rab7, and small GTPases that could not be identified because the peptide recovered was common to several Rab proteins (Rab6, 27, 34, 39, 41, and 44). Out of these potential candidates, Rab 6 and Rab39 are recruited to the chlamydial inclusion (17, 18). Furthermore, silencing Rab6 reduces the delivery of the lipid ceramide to the inclusion (19). To test if TaiP was able to interact with Rab proteins as indicated by the mass spectrometry data, we performed co-IP experiments in cells cotransfected with flag-tagged TaiP and GFP-tagged Rab proteins. We tested several of the Rab proteins that the PD assay had hinted to as potential interactors (Rab 5, 6, 7, and 39) as well as Rab14 because it is one of the Rab proteins recruited to the inclusion membrane (20). TaiP co-immunoprecipitated with Rab6a and Rab39a and with none of the other Rab proteins tested (Fig. 4A). Deletion of the C-terminal domain of TaiP abolished these interactions, indicating that this domain is involved (Fig. 4B). To determine if the growth defect of the CtrΔtaiP strain was due to an inability to mobilize Rab6 or Rab39-regulated vesicular traffic, we tested the effect of silencing Rab6 or Rab39 on bacterial development. Silencing Rab39 had no effect on the ability for the bacteria to infect and grow in HeLa cell, and this small GTPase was not investigated further. Silencing of Rab6 resulted in a 25% decrease in the inclusion size of WT bacteria, confirming its role in feeding Chlamydia-inclusion growth (19) (Fig. 4C). In contrast, depleting Rab6 had no significant impact on the size of CtrΔtaiP inclusions. This result suggests that the TaiP/ATG16L1-dependent vesicular traffic that contributes to the growth of the inclusion in the WT strain requires Rab6. To further link Rab6 to the TaiP/ATG16L1-dependent growth of Chlamydia inclusions, we looked at the consequence of silencing Rab6 in the WT versus the atg16l1 KO background. We observed that the benefice for the CtrΔtaiP strain of knocking out ATG16L1 expression was lost when Rab6 was silenced (Fig. 4C). This observation indicates that the source of membrane that allows faster growth of the CtrWT strain compared to the CtrΔtaiP strain is Rab6 dependent. Rab6 is highly enriched in the Golgi apparatus, which is localized close to the inclusion (SI Appendix, Fig. S4). To facilitate the quantification of vesicular Rab6 enrichment at the inclusion periphery, we applied a short treatment with nocodazole, an inhibitor of microtubule polymerization, that results in the dispersion of Golgi stacks (21). Infected cells were fixed and stained for endogenous Rab6 and for the inclusion protein CT813 (Fig. 4D). We observed a stronger enrichment in Rab6 at the periphery of CtrWT inclusions compared to CtrΔtaiP, an observation fully consistent with the hypothesis that TaiP is required for efficient recruitment of Rab6 positive vesicles to the inclusion. Furthermore, the recruitment of Rab6 to CtrΔtaiP inclusions but not to CtrWT inclusions, increased significantly in the atg16l1 KO background, indicating that ATG16L1 restricts Rab6 traffic toward the inclusion in the absence of TaiP (Fig. 4D). Altogether, our data converge to establish that one the functions of the chlamydial effector TaiP is to disrupt the ATG16L1/TMEM59 interaction through mimicry of the WD40-binding motif. This unleashes access to a Rab6-dependent supply of membrane that feeds inclusion growth (Fig. 5A). Our model predicts that Rab6-positive vesicles normally feed TMEM59-positive compartments and that this pathway requires ATG16L1/TMEM59 interaction. The large overlap of Rab6- and TMEM59-positive compartments observed by immunofluorescence is consistent with this prediction (SI Appendix, Fig. S4). To test it further, we coexpressed GFP-Rab6 and HA-TMEM59 in WT and in the atg16l1 KO background. We observed that, indeed, Rab6 co-immunoprecipitated with TMEM59 in the WT HeLa but not in the absence of ATG16L1 (Fig. 5B). Thus, our study has uncovered a trafficking pathway, controlled by ATG16L1, that feeds TMEM59-positive compartments with Rab6-positive material.
Fig. 4.
TaiP redirects Rab6-positive vesicular traffic to the inclusion. (A) HeLa cells were cotransfected with Flag-TaiP and the indicated GFP-tagged Rab constructs for 24 h. Cells were lysed, and IP was performed with the anti-GFP antibody. Proteins were separated on SDS/PAGE, transferred to a PVDF membrane, and probed with the indicated antibody. An aliquot of each cell lysate was loaded on a separate gel to visualize the expression of Flag-TaiP and of each of the GFP-tagged proteins (input panels). (B) HeLa cells were cotransfected with the indicated constructs for 24 h and analyzed as in B. (C) WT and atg16l1 KO cells were transfected as indicated and infected with CtrWT or CtrΔtaiP the following day at MOI = 0.2. Cells were fixed 20-h postinfection, permeabilized, and the inclusion membrane was stained with anti-Cap1 antibodies. Inclusion areas were measured using imageJ software. The box and whiskers plots display the median area of intracellular inclusions (n > 50 cells from three independent experiments). Statistical analysis was performed using a one-way ANOVA with a Dunnett’s multiple comparison test to siCtrl in WT HeLa. The Inset shows the efficacy of the siRNA against Rab6 at silencing Rab6 expression probed by Western blot. (D) WT and atg16l1 KO cells were infected with CtrWT or CtrΔtaiP the following day at MOI = 0.2. Some 20 h after infection, the cells were treated with 10 nM nocodazole and incubated further for 60 min before fixation. After permeabilization, the cells were stained with rabbit antibodies against endogenous Rab6 and mouse antibodies against the inclusion membrane protein CT813. Representative images are shown. (Scale bar: 10 µm.) The box and whiskers plot displays the median intensity of the Rab6 staining at the inclusion periphery, relative to its cytosolic intensity (n > 50 cells from three independent experiments, see Methods for details). Statistical analysis was performed using Student’s t test.
Fig. 5.
TaiP disrupts the ATG16L1-controlled traffic of Rab6-positive vesicles toward TMEM59-positive compartments. (A) Model a. In unifected cells, ATG16L1’s binding to TMEM59 facilitates the supply of Rab6-positive veiscles to TMEM59-positive compartments. b. In cells infected with CtrWT, TaiP secretion by the bacteria prevents ATG16L1/TMEM59 interaction, and Rab6-positive vesicles are hijacked by the inclusion. c. In the absence of TaiP, the bacteria lose access to this pool of vesicles. CtrΔtaiP inclusions are smaller as they only rely on alternative membrane sources. d. Silencing ATG16L1 or TMEM59 expression reverts this phenotype. (B) WT and atg16l1 KO HeLa cells were cotransfected with the indicated plasmids. One day later, the cells were lysed, and IP was performed with anti-HA coupled beads. Proteins were separated on SDS/PAGE, transferred to a PVDF membrane, and probed with the indicated antibody. GFP-Rab6 co-IP with TMEM59 only in cells expressing ATG16L1. The use of cells that do not express HA-TMEM59 allows to measure the level of nonspecific association of GFP-Rab6 to the beads.
Discussion
The observation that the absence of expression of TaiP resulted in several developmental defects had led us to hypothesize that this effector may contribute to multiple key events in infection (5). In this report, we show that the C-terminal part of TaiP is engaged in the regulation of membrane supply to the inclusion, accounting for the defect in inclusion growth of the CtrΔtaiP strain. At the molecular level, we demonstrated that TaiP interacted with the host protein ATG16L1 through its C-terminal domain. Moreover, we found that TaiP competitively inhibited the interaction of TMEM59 and ATG16L1 in vitro and in the infectious context. Remarkably, silencing either TMEM59 or ATG16L1 expression allows to revert the inclusion growth defect of the CtrΔtaiP strain.
Most work on ATG16L1 has focused on its role in ATG8 lipidation as part of the ATG12-ATG5-ATG16 complex. We observed that silencing atg16l1 or disrupting its expression enhanced Chlamydia growth, even for the WT strain. This observation led us to hypothesize that ATG16L1 might restrict Chlamydia growth through an autophagy-related mechanism. However, we demonstrated that it was not the case since the N-terminal part of ATG16L1 required and sufficient for ATG8 lipidation was not able to restrict Chlamydia growth. Conversely, expression of the C-terminal part of ATG16L1, that contains the WD40, was sufficient for that effect. Therefore, the restriction of bacterial growth exerted by ATG16L1 is not due to its ability to mediate ATG8 lipidation but is provided by a separate function of the WD40 domain.
ATG16L1’s WD40 domain is proposed to interact with a variety of proteins (13, 22). A motif common to several ATG16L1-binding proteins had been identified as [YW]-X3-[ED]-X4-[YWF]-X2-L (12). TaiPCterm contains a single sequence that matches this definition: Y474AAALSDGYSAYKTL488, and we showed that the D480A mutation impaired TaiP binding to ATG16L1. Thus, our data confirm the privileged interaction between the [YW]-X3-[ED]-X4-[YWF]-X2-L motif and the ATG16L1 WD40. Interestingly, the interaction among ATG16L1 and TLR2 or NOD2, which also carry the [YW]-X3-[ED]-X4-[YWF]-X2-L motif, appear insensitive to the addition of TaiP. This might be explained by the difference in affinities and/or by the limitation of our readout (co-IP), that might not be sensitive enough to reveal competition between these ATG16L1 binders. Also, other surfaces are likely implicated in the interactions between these different molecules and ATG16L1 as exemplified by NOD1, which binds ATG16L1 but does not present the motif (12). These additional binding surfaces could compensate for the competition exerted by helix 6 of TaiP.
We observed that exogenous expression of TaiP (by transfection) partially reverted the inclusion size defect of the CtrΔct622 strain. Importantly, the TaiP D480A point mutation abolished both the beneficial effect of TaiP expression on inclusion growth and the disruptive effect of TaiP on ATG16L1/TMEM59 complexes, strongly supporting the hypothesis that TaiP supports C. trachomatis growth by disrupting ATG16L1/TMEM59 interaction. In favor of this scenario, we showed that TMEM59 silencing led to a similar phenotype as ATG16L1 silencing, i.e., a recovery of CtrΔtaiP inclusion size. TMEM59 is a transmembrane protein associated with several compartments, including the Golgi apparatus and late endocytic compartments, but its role in membrane traffic remains unclear. Our results converge to the hypothesis that ATG16L1/TMEM59 interaction limits bacterial access to host vesicular traffic (Fig. 5A). Translocation of TaiP disrupts ATG16L1/TMEM59 interaction, unleashing access to a membrane pool that feeds inclusion growth. TaiP was detected by immunofluorescence in the bacteria throughout the developmental cycle and in the cytosol late in the cycle probably due to a detection threshold (5). Our data are consistent with a role of TaiP throughout the inclusion growth phase and, in particular, in the first half of the developmental cycle when the difference in inclusion sizes compared to WT inclusions is the strongest. We showed that the ability of this membrane pool to support inclusion growth was dependent on the expression of the small GTPase Rab6. Rab6 is associated with several exocytic pathways emerging from the Golgi apparatus (23). Our data confirm the role of Rab6 in feeding Chlamydia-inclusion growth (19) and revealed the control by ATG16L1 of a Rab6-positive membrane flow toward TMEM59 compartments.
ATG16L1 has attracted a lot of attention since the identification of an amino acid substitution (T300A) that sensitizes the protein to caspase-3 processing and that is associated with diminished autophagy and increased risk of developing Crohn's disease (24, 25). The molecular links between this variant and the susceptibility to Crohn's disease are still unclear (26) and could involve impaired trafficking events (8, 27). The finding that the ATG16L1T300A protein functions as a dominant negative raises the possibility that the cleavage products have deleterious activity of their own (28). The cleavage site liberates the WD40 domain that we show here to be implicated in the control of the traffic of, at least, a subset of Rab6 positive vesicles. The fact that evolution shaped a bacterial effector that targets this WD40 domain in order to redirect intracellular traffic to the bacterial compartment indicates that the role played by WD40 in this process is very central. Future work will, thus, need to consider the possibility that impaired ATG16L1-controlled trafficking events could play a major role in the susceptibility to Crohn's disease and other pathologies in which ATG16L1 has been implicated.
Methods
Cells and Bacteria.
HeLa (ATCC), HEC-1-B (ATCC), and HEK293 cells (Invitrogen) were grown in Dulbecco’s modified Eagle’s medium (DMEM) with Glutamax (DMEM, Invitrogen), supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum. HEK293 atg3 KO and atg16l1 KO cells used for SI Appendix, Fig. S2 were described in ref. 9. The C. trachomatis serovar LGV L2 strain 434 (CtrWT) and the taiP genetic disruption CtrΔtaiP mutant were propagated on HeLa cells as described (5).
Generation of atg16l1 KO HeLa Cells.
The atg16l1 KO HeLa cells were generated as described in ref. 29, inserting single guide CACCGCTGCAGAGACAGGCGTTCG (forward) and AAACCGAACGCCTGTCTCTGCAGC (reverse) in the pSpCas9(BB)-puro. After transfection, the cells were treated with 0.5 µg/mL puromycin for 24 h before individual clone selection. After monoclonal expansion, the loss of ATG16L1 expression was verified by Western blot. All experiments used cells passaged four times or less after freezing.
PD of GST-TaiP Partners in Infected and Noninfected Cells and Mass Spectrometry.
HeLa cells (about 107 cells per point), infected or not for 24 h with C. trachomatis, were lysed in Nonidet P-40 lysis buffer with gentle rocking at 4 °C (150 mM NaCl, 50 mM 2-amino-2-hydroxymethyl-1,3-propanediol [Tris]⋅HCl pH 7.5, 1 mM [ethylenedinitrilo]tetraacetic acid [EDTA], 1 mM ethylene glycol bis[β-aminoethyl ether]-N,N,N′,N′-tetraacetic acid [EGTA], 10 mM NaF, and 5% glycerol supplemented with 0.5% Nonidet P-40 [vol/vol] for the first 20 min of lysis in a 0.1-mL volume, diluted 1:10 to reach 0.05% Nonidet P-40 for the last 20 min of lysis, and the rest of the procedure). Lysates were centrifuged at 17,000 × g for 15 min at 4 °C and precleaned with glutathione agarose beads with 20 µg of GST for 90 min at 4 °C in a rocking platform. Equal amounts of precleaned supernatants were incubated with 50 µL of a 50% slurry of glutathione sepharose 4B beads and 30 µg of purified glutathione S-transferase (GST) or GST-TaiP for 90 min at 4 °C on a rocking platform. After a brief centrifugation, beads were washed five times with cold GST lysis buffer. The bound proteins were eluted with urea buffer (8 M urea, 1% SDS, 150 mM NaCl, and 30 mM Tris⋅HCl, pH 8.0), then, identified by mass spectrometry by the Proteopole of the Institut Pasteur as described in ref. 5.
Recombinant Protein Purification.
GST-TaiP (GST-CT622) purification was described in ref. 5. The same protocol was used to produce GST-TaiPNterm. ATG16L1 complementary DNA (cDNA) was cloned into pCoofy 29 vector via sequence- and ligation-independent cloning. The ATG16L1 protein was expressed in H5 insect cells grown in EX-CELL 420 serum-free medium (Sigma-Aldrich). Baculovirus-infected insect cells were added in a ratio of 1:1,000. Cultures were shaken for 72 h at 25 °C and 85 rpm. Insect cells were harvested by centrifugation at 2,000 × g for 15 min, washed with Dulbecco’s phosphate-buffered saline (PBS) (Gibco), and resuspended in lysis buffer (0.1 M Tris⋅HCl pH 8.0, 0.5 M NaCl, 20 mM imidazole, pH 8.0, 10% glycerol, 5 mM β-mercaptoethanol, and 0.5% protease inhibitor mixture, Sigma-Aldrich]). Cells were lysed using a dounce homogenizer. ATG16L1 was purified from cell lysates using a His-trap Ni-NTA agarose column followed by gel filtration in a Superdex 200 column. The His tag was removed by preScission protease, and the released protein was subjected to size exclusion chromatography. Fractions containing ATG16L1 were pooled and concentrated using Vivaspin cellulose centrifugation filters (Sartorius Stedim). The protein was flash frozen in liquid nitrogen and stored at −80 °C.
In Vitro Assay of ATG16L1/TaiP Interaction and IBs.
GST-TaiP and GST-TaiPNterm at 100 µM were incubated with ATG16L1 at 33 µM, in the binding buffer (0.4 M NaCl and 25 mM Tris⋅HCl, pH 7.4) and rocked on an Eppendorf tube roller for 2 h at 4 °C. Prewashed 30 µL slurry glutathione sepharose-4B beads were, then, added to the protein mix and further incubated for 40 min at 4 °C. The beads were centrifuged at 1,500 × g for 5 min, and more than 90% of the supernatant was removed. The sedimented beads were washed twice in binding buffer supplemented with 0.1% Triton X-100 and once in binding buffer. The samples were eluted by boiling in SDS sample buffer and analyzed by immunoblotting. IBs were analyzed using horseradish peroxidase secondary antibodies, and chemiluminescence was analyzed on a Syngene Pxi4 imaging system. For the quantification, the background signal was subtracted from all bands before calculating the fold enrichment to the indicated control.
LC3B-PE Turnover Measurement.
Some 1.5 × 105 WT and atg16l1 KO HeLa cells were seeded in duplicate 12-well plates for each condition. The following day they were treated with vehicle (dimethyl sulfoxide-D2650, Sigma-Aldrich) or 10 nM bafilomycinA1 (Sigma-Aldrich B1793). Three hours later, the cells were washed with PBS before lysis with Laemmli buffer supplemented with 2% β-mercaptoethanol and boiled for 5 min. The lysates were separated by electrophoresis on SDS/PAGE at 15% acrylamide concentration before being transferred to a PVDF membrane. LC3B and LC3B-PE were revealed using the antibody NB100-2,220 from Novus Biologicals.
Plasmids and Transfections.
Genomic DNA from C. trachomatis D/UW-3/CX was prepared from bacteria using the RapidPrep Micro Genomic DNA isolation kit (Amersham Pharmacia Biotech). attB-containing primers (SI Appendix, Table S2, Gateway, Life Technologies) were used to amplify and clone ct622 into a destination vector derived from the mammalian expression vector pCiNeo, providing a N-terminal 3xflag tag and into pDEST15 (Gateway) for production of GST-tagged proteins. All constructs were verified by sequencing. GFP-conjugated ATG16L1β constructs (WT and GFP-ATG16L1266–607) were from E. Morel, Institut Necker Enfants Malades (INEM), France, TMEM59 was from S. Lichtenthaler, Technical University of Munich (TUM), Germany, Flag NOD1/2 constructs were from L. Boyer, University of Nice, France, TLR2 was from P. Cossart, Institut Pasteur, France, GFP-Rab6 was from B. Goud, Institut Curie, France, GFP-Rab5 was obtained from A. Echard, Institut Pasteur, France, and GFP-Rab7, GFP-Rab14, and GFP-Rab39 were kindly provided by M. T. Damiani, University of Cuyo, Mendoza, Argentina. Single point mutagenesis was performed to generate Flag-TaiPD480A and GFP-ATG16L11–319 using the Quickchange Lightning Site-Directed Mutagenesis Kit from Agilent following the manufacturer’s protocol and primers reported in SI Appendix, Table S2.
DNA transfection was performed using Jet prime and following the manufacturer’s protocol. For siRNA transfection, we used RNAi Max as recommended by the manufacturer (see SI Appendix, Table S2 for sequences).
Immunofluorescence.
Some 1.5 × 105 HeLa cells were seeded on glass coverslips in 12-well plates before transfection (for expression of Flag-tagged proteins or for silencing) for 24 h. Transfected cells were then infected for 20 h at a MOI = 0.2, before fixation with 4% paraformaldehyde in PBS for 20 min, followed with 10 min quenching with 50 mM NH4Cl in PBS. The cells were washed with PBS, permeabilized with 0.05% saponin, 5 mg/mL BSA in PBS (permeabilization buffer) for 20 min, and immunolabeled for 60 min with primary antibodies diluted in permeabilization buffer. Rabbit antibodies against the bacterial inclusion protein Cap1 are described in ref. 30, mouse antibodies against CT813 were kindly provided by G. Zhong, University of Texas, San Antonio, TX, mouse antibodies against LC3B were from MBL (no. M152-3), antibodies against Flag(M2) were from Sigma-Aldrich, and rat antibodies against TMEM59 (clone 4E5) were generously provided by S. Lichtenthaler (16). Coverslips were then washed three times with PBS before incubating for 60 min in fluorochrome-coupled secondary antibodies diluted in permeabilization buffer. DNA was stained using 0.5 µg mL−1 of Hoechst 33342 (Thermo Fisher Scientific) added in the secondary antibody solution. Images were acquired on an Axio observer Z1 microscope equipped with an ApoTome module (Zeiss, Germany) and a 63× Apochromat lens. Images were taken with an ORCAflash4.OLT camera (Hamamatsu, Japan) using the software Zen. Quantification of LC3B and Rab6 at the inclusion periphery was performed using the brush tool of ImageJ set, at 9 pixels, and following the inclusion membrane marker (Cap1 and CT813, respectively). To measure Rab6 enrichment at the inclusion periphery, the mean green fluorescence (Rab6) along the this line was normalized to the mean value of green fluorescence in two randomly selected ∼1 μm2 areas in the cytosol. Since the LC3B level in the cytosol was hardly above background, this normalization was not applied for LC3B measurements, and the data in Fig. 2C display the mean green (LC3B) fluorescence along the inclusion membrane.
Inclusion Size Measurements.
Some 1.5 × 105 HeLa cells were seeded in 12-well plates before transfection with either DNA or siRNA for 24 h. The transfected cells were infected in triplicates at MOI = 0.2 for 20 h before being fixed and permeabilized for immunofluorescence. The inclusion membrane was stained using an antibody against Cap1, and 5–10 random pictures were taken for each coverslips as described in the immunofluorescence methods. Using the imageJ software the scale was set from pixels to micrometers and the area of individual inclusions were measured. Each condition was analyzed in a blind fashion from three individual coverslips with a minimum of 50 inclusions per simplicate. For Figs. 1D, 3C, and 4D and SI Appendix, Fig. S1C, the size of all inclusions was analyzed, whereas in Figs. 1E and 3B, the inclusions of only cells positive for Flag were analyzed. For Fig. 2B, only inclusions in GFP positive cells were taken into account and were analyzed. Note that we consistently observed that the inclusions grew slower in transfected cells compared to nontransfected cells (compare, for instance, the average diameter of inclusions for CtrWT in nontransfected cells in Fig. 1D and in cells transfected with GFP in Fig. 2B).
Progeny Assay and Flow Cytometry.
For progeny assays displayed in SI Appendix, Fig. S1F WT or atg16l1 KO HeLa cells infected for 40 h with the indicated strains were detached, lysed using glass beads, and the supernatant was used to infect fresh HeLa cells plated the day before (100,000 cells/well in a 24-well plate), in serial dilution. The next day, three wells per condition with an infection lower than 30% (checked by microscopy) were detached, fixed in 70% ethanol and stained with a home-made rabbit antibody against GroEL followed with Alexa488-coupled secondary antibodies. Acquisition was performed using a CytoFLEX S (Beckman Coulter), and 10,000 events per sample were acquired and, then, analyzed using FlowJo (version 10.0.7) to determine the bacterial titer as described in ref. 15. For determining the consequence of infection on bacterial load in SI Appendix, Fig. S3C, only the primary infection was analyzed by flow cytometry after staining the bacteria with anti-GroEL antibodies.
IP.
Some 5 × 106 HeLa cells were seeded in 10-cm2 dishes. On the following day, cells were transfected with 5 µg of plasmid. One day later, cells were lysed in 250 µL of lysis buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.5, 5% glycerol, and 0.5% Nonidet P-40) supplemented with a protease mixture inhibitor (Roche Complete, EDTA free). The lysates were incubated on a rotating wheel at 4 °C for 20 min before adding 1 mL of dilution buffer (150 mM NaCl and 50 mM Tris-HCl pH 7.5 supplemented with Protease mixture inhibitor) thereby reducing the glycerol to 1% and Nonidet P-40 to a final concentration of 0,1%. The lysates were then centrifuged at 10,000 × g for 10 min, and the insoluble pellet was discarded. For GFP IP, the lysates were then incubated with 2 µg of antibody (Invitrogen, no. A11122) at 4 °C for 2 h before adding 20 µL of slurry protein G beads (Sigma-Aldrich Fast Flow Protein G sepharose) for 20 min. For HA and Flag IPs, antibody-coupled beads were used (Sigma-Aldrich). The beads were then washed before adding 20 µL of Laemmli buffer supplemented with 2% β-mercaptoethanol and boiled for 5 min. For Fig. 1 A and B, the plasmids were cotransfected, whereas for Fig. 3C and SI Appendix, Fig. S3 A–C, the plasmids were transfected individually before mixing 0.75 mL of each diluted lysate in the presence or absence of 100 pmol of GST-TaiPNterm or GST-TaiP. For the IP in infected cells shown in Fig. 1C, cell lysis was performed in RIPA buffer (150 mM sodium chloride, 0.5% Nonidet P-40, 0.5 Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris, pH 8.0). Antibodies used for probing the membranes are described in the Immunofluorescence section, in addition, antibodies against GFP (no. NB600-308), ATG16L1 (no. PM040), and HA (clone 12CA5) were purchased from Novus, MBL and Sigma-Aldrich, respectively.
PD Assays.
Some 2.5 × 106 HeLa cells were seeded in a 6-cm2 dish the day before transfection with 2.5 µg of plasmid. On the following day, the cells were lysed in 250 µL of lysis buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 5% glycerol, and 0.5% Nonidet P-40) supplemented with a protease mixture inhibitor (Roche Complete, EDTA free). The lysates were incubated on a rotating wheel at 4 °C for 20 min before reducing the glycerol to 1% and Nonidet P-40 to a final concentration of 0.1% as above. The lysates were then incubated with 100 pmol of GST-TaiP for 60 min on a spinning wheel at 4 °C before adding 30 µL of slurry glutathione sepharose beads for 20 min. The beads were then washed three times using lysis buffer before adding 20 µL of Laemmli buffer supplemented with 2% β-mercaptoethanol and boiled for 5 min.
RT-qPCR.
Total RNAs were isolated 35 h after transfection with the RNeasy Mini Kit (Qiagen) with DNase treatment (DNase I, Roche). Reverse transcription was performed on 500 ng of total RNA using the high capacity cDNA Reverse transcription kit (Applied Biosystems) according to manufacturer’s instructions. cDNA were diluted five times, and qPCR was performed on 1 µL of cDNA with the LightCycler 480 system using LightCycler 480 SYBR Green Master I (Roche). Data were analyzed using the ΔΔCt method with the 36B4 gene as a control gene (31).
Supplementary Material
Acknowledgments
We thank Dr. A. Simonsen for discussions, Dr. S. Lichtenthaler (TUM Munich, Germany) for the gift of anti-TMEM59 antibodies and TMEM59-HA plasmid, Dr. B. Goud (Institut Curie, France) and Dr. M. T. Damiani (Mendoza, Argentina) for supplying Rab constructs and anti-Rab6 antibody, Dr. G. Zhong (San Antonio, Texas) for anti-CT813 antibodies. We also thank Dr. E. Morel (INEM, France) for providing the GFP-ATG16L1 constructs, Dr. Laurent Boyer (Nice, France) for the NOD constructs, and Dr. P. Cossart (Institut Pasteur, France) for the TLR2 vector. We thank Thibault Chaze (Proteomics platform Institut Pasteur) for the analysis of the PD experiments. This work was supported by a European Research Council (ERC) Starting Grant (NUChLEAR 282046), and by a Grant from the Pasteur-Weizmann Collaborative Research Funds. D.H. was funded by the ERC and by the Fondation pour la Recherche Médicale (Grant SPF20170938695).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005389117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Brunham R. C., Rey-Ladino J., Immunology of Chlamydia infection: Implications for a Chlamydia trachomatis vaccine. Nat. Rev. Immunol. 5, 149–161 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Triboulet S., Subtil A., Make it a sweet home: Responses of Chlamydia trachomatis to the challenges of an intravacuolar lifestyle. Microbiol. Spectr. 7, BAI-0005-2019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Damiani M. T., Gambarte Tudela J., Capmany A., Targeting eukaryotic Rab proteins: A smart strategy for chlamydial survival and replication. Cell. Microbiol. 16, 1329–1338 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Elwell C., Mirrashidi K., Engel J., Chlamydia cell biology and pathogenesis. Nat. Rev. Microbiol. 14, 385–400 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cossé M. M.et al., The loss of expression of a single type 3 effector (CT622) strongly reduces Chlamydia trachomatis infectivity and growth. Front. Cell. Infect. Microbiol. 8, 145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohsumi Y., Historical landmarks of autophagy research. Cell Res. 24, 9–23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fletcher K.et al., The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. EMBO J. 37, e97840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cadwell K., Debnath J., Beyond self-eating: The control of nonautophagic functions and signaling pathways by autophagy-related proteins. J. Cell Biol. 217, 813–822 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lystad A. H.et al., Distinct functions of ATG16L1 isoforms in membrane binding and LC3B lipidation in autophagy-related processes. Nat. Cell Biol. 21, 372–383 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sorbara M. T.et al., The protein ATG16L1 suppresses inflammatory cytokines induced by the intracellular sensors Nod1 and Nod2 in an autophagy-independent manner. Immunity 39, 858–873 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Bajagic M., Archna A., Büsing P., Scrima A., Structure of the WD40-domain of human ATG16L1. Protein Sci. 26, 1828–1837 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boada-Romero E.et al., TMEM59 defines a novel ATG16L1-binding motif that promotes local activation of LC3. EMBO J. 32, 566–582 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slowicka K.et al., Physical and functional interaction between A20 and ATG16L1-WD40 domain in the control of intestinal homeostasis. Nat. Commun. 10, 1834 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Younes H. M.et al., Autophagy-independent function of MAP-LC3 during intracellular propagation of Chlamydia trachomatis. Autophagy 7, 814–828 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Vromman F., Laverrière M., Perrinet S., Dufour A., Subtil A., Quantitative monitoring of the Chlamydia trachomatis developmental cycle using GFP-expressing bacteria, microscopy and flow cytometry. PLoS One 9, e99197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ullrich S.et al., The novel membrane protein TMEM59 modulates complex glycosylation, cell surface expression, and secretion of the amyloid precursor protein. J. Biol. Chem. 285, 20664–20674 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rzomp K. A., Scholtes L. D., Briggs B. J., Whittaker G. R., Scidmore M. A., Rab GTPases are recruited to chlamydial inclusions in both a species-dependent and species-independent manner. Infect. Immun. 71, 5855–5870 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gambarte Tudela J.et al., The late endocytic Rab39a GTPase regulates the interaction between multivesicular bodies and chlamydial inclusions. J. Cell Sci. 128, 3068–3081 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Rejman Lipinski A.et al., Rab6 and Rab11 regulate Chlamydia trachomatis development and golgin-84-dependent Golgi fragmentation. PLoS Pathog. 5, e1000615 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capmany A., Damiani M. T., Chlamydia trachomatis intercepts Golgi-derived sphingolipids through a Rab14-mediated transport required for bacterial development and replication. PLoS One 5, e14084 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogalski A. A., Singer S. J., Associations of elements of the Golgi apparatus with microtubules. J. Cell Biol. 99, 1092–1100 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jain B. P., Pandey S., WD40 repeat proteins: Signalling scaffold with diverse functions. Protein J. 37, 391–406 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Boncompain G., Weigel A. V., Transport and sorting in the Golgi complex: Multiple mechanisms sort diverse cargo. Curr. Opin. Cell Biol. 50, 94–101 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Hampe J.et al., A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 39, 207–211 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Murthy A.et al., A Crohn’s disease variant in Atg16l1 enhances its degradation by caspase 3. Nature 506, 456–462 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Matsuzawa-Ishimoto Y., Hwang S., Cadwell K., Autophagy and inflammation. Annu. Rev. Immunol. 36, 73–101 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Cadwell K.et al., A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao P.et al., The inflammatory bowel disease-associated autophagy gene Atg16L1T300A acts as a dominant negative variant in mice. J. Immunol. 198, 2457–2467 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Ran F. A.et al., Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gehre L.et al., Sequestration of host metabolism by an intracellular pathogen. eLife 5, e12552 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmittgen T. D., Livak K. J., Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.