Significance
The need for mass-spectrometric determination of protein identity severely limits the throughput of many whole-cell proteomic analyses. We have developed an approach to physically connect in vivo-expressed proteins to their encoding messenger RNAs (mRNAs). This genotype–phenotype linkage enables us to carry out a variety of whole-proteome assays and determine the identity of each protein using nucleic acid sequencing. We show that the resulting in vivo mRNA display technology enables determination of protein subcellular localization and protein–protein interactions, with high sensitivity and specificity, using simple purification followed by high-throughput sequencing. In vivo mRNA display should enable more efficient interrogation of proteome behavior in a variety of applications that are currently limited by the cost and throughput of mass-spectrometric analysis.
Keywords: proteomics, mRNA display, protein display technologies, protein–protein interactions, MS2 tagging
Abstract
Large-scale proteomic methods are essential for the functional characterization of proteins in their native cellular context. However, proteomics has lagged far behind genomic approaches in scalability, standardization, and cost. Here, we introduce in vivo mRNA display, a technology that converts a variety of proteomics applications into a DNA sequencing problem. In vivo-expressed proteins are coupled with their encoding messenger RNAs (mRNAs) via a high-affinity stem-loop RNA binding domain interaction, enabling high-throughput identification of proteins with high sensitivity and specificity by next generation DNA sequencing. We have generated a high-coverage in vivo mRNA display library of the Saccharomyces cerevisiae proteome and demonstrated its potential for characterizing subcellular localization and interactions of proteins expressed in their native cellular context. In vivo mRNA display libraries promise to circumvent the limitations of mass spectrometry-based proteomics and leverage the exponentially improving cost and throughput of DNA sequencing to systematically characterize native functional proteomes.
Cellular proteins act in concert with each other to achieve a diverse set of functions through protein–protein interactions (PPIs), regulatory interactions, posttranslational modifications, and subcellular localization. Proteomic technologies allow us to dissect the functional roles of proteins in the context of biological processes, cellular compartments, and metabolic/signaling pathways (1). A comprehensive view of this complex proteomic landscape depends on our ability to reliably identify and characterize proteins in their native physiological contexts with precision and specificity (2). Current high-throughput approaches utilize mass spectrometry coupled with affinity purification (3) or subfractionation (4) as well as reporter assays such as the yeast two hybrid (Y2H) (5–7), fluorescence resonance energy transfer (8), and protein complementation (9–11). Additionally, methods developed under the category of “spatial proteomics” can label and purify proteins within a certain radius of a chosen bait (12–17). Although such studies have provided a snapshot of the cellular proteome in many contexts (17–24), the picture is far from comprehensive due to the vast space of possible interactions, the diverse roles played by proteins in different cellular contexts, as well as the transient nature of many interactions (25–27). Meanwhile, next generation sequencing (NGS) has ushered genomics into a new age due to its low cost, precision, accuracy, and capacity for massive multiplexing. Furthermore, many biochemical assays have been adapted to take advantage of NGS by mapping functional assays to a DNA sequencing readout. Such applications include Hi-C (28), assay for transposase-accessible chromatin using sequencing (29), bisulfite sequencing (30), and others. However, functional proteomics have not yet tapped into NGS’s full potential at a similar scale and fashion (31, 32).
Here, we introduce in vivo mRNA display, a technology that enables diverse proteomics applications using NGS as the readout. A variety of existing display technologies create a link between genotype and phenotype, whereby a protein or peptide is linked to its encoding nucleic acid. For example, in phage display, the nucleic acid encoding the capsid displayed peptide is contained within the phage (33). The resulting collection of displayed peptides can be used for the in vitro characterization of protein interactions, protein engineering, and selection of human antibody fragment libraries (34–36). Alternative in vitro methods linking nucleotide information to phenotype include mRNA display (37, 38), ribosome display (39), and yeast display (40). In the past decade, many of these technologies have been coupled with NGS (41–44). More recently, a method based on affinity capture of polyribosomes (45) converts in vitro interactions of nascent polypeptides and their polyribosomes to RNA sequencing. Another approach adapted DNA sequencing chips to immobilize collections of DNA–RNA–protein complexes and carry out fluorescence-based functional assays on the chip (46). Despite their diverse utility, these existing display technologies are limited to analysis of proteins in vitro, significantly limiting their physiological relevance due to lack of appropriate cellular context, in vivo posttranslational modifications, and even proper folding states (41).
We set out to engineer a scalable display technology that functions in vivo. To this end, we co-opted the high-affinity interaction between the MS2 bacteriophage coat protein and its cognate RNA stem loop (47, 48). This interaction was previously utilized as a reporter system to track mRNA molecules in living cells in a variety of organisms (49). In these assays, tandem copies of the stem-loop sequence were inserted adjacent to the monitored gene, which enables the detection of its mRNA through the interaction of the stem loop with a fluorescent protein fused to the MS2 coat protein (MCP) (50, 51). MS2 RNA affinity purification has been also utilized to capture RNA-centric in vivo proteomics (52). Here, we modified the MS2 tagging system in order to associate a translated protein with its own mRNA. We fused the MCP to the N terminus of a target protein while we introduced the cognate RNA stem loop downstream of the gene, establishing a direct link between gene and protein (Fig. 1A). In contrast to in vitro display technologies, assayed proteins are expressed, processed, and tagged in vivo in their relevant cellular contexts. Our approach, termed in vivo mRNA display, identifies proteins in a variety of in vivo functional assays using nucleic acid sequencing as the readout.
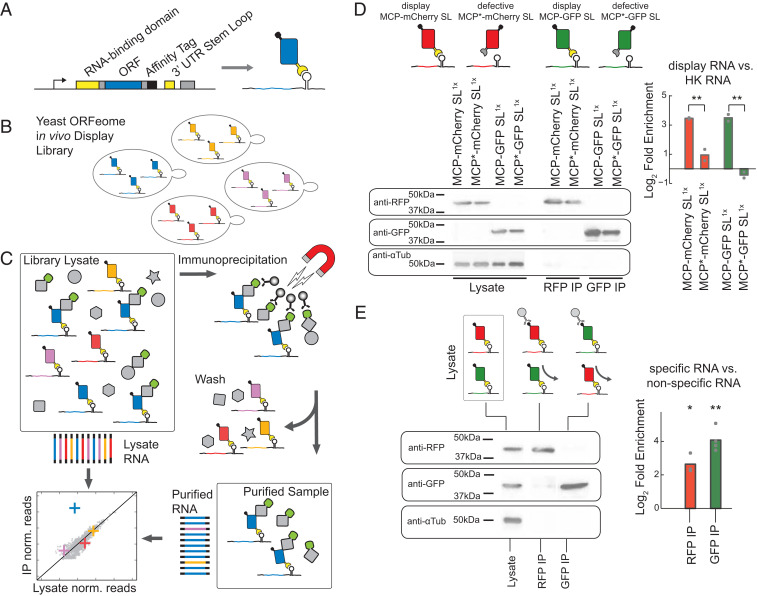
Fig. 1.
High-throughput proteomics using in vivo mRNA display and NGS. (A) N-terminal MS2 coat protein fused to a gene of interest binds the RNA stem loop present on the 3′ untranslated region (UTR) of its encoding mRNA. (B) In vivo mRNA display libraries of the yeast ORFeome consist of a mixed population of strains, each expressing a single displayed protein interacting with its native cellular environment independently of other library species. (C) A proteomic assay with RNA sequencing as the readout. Scheme for a copurification assay of a given bait from in vivo mRNA display extracts, whereby RNA is processed from both purified and input lysates. Potential interactors are detected by comparing RNA read frequencies in the two samples for each displayed mRNA. (D) Log2 fold enrichment of displayed mRNA for purified proteins compared with the lysate is calculated using qPCR for a given construct with ACT1 as a reference housekeeping (HK) gene. Two in vivo display constructs (MCP-mCherry, MCP-GFP) vs. defective coat protein constructs (MCP*-mCherry, MCP*-GFP) show significant relative enrichment (P = 0.002, P = 0.009, one-way ANOVA). (E) Log2 fold enrichment of displayed mRNA for purified proteins from a mixed construct population (MCP-mCherry and MCP-GFP) for anti-RFP and anti-GFP purifications. The mRNA species of each specific protein is enriched relative to the input lysate over the nonspecific species (RFP-IP: P = 0.016; GFP-IP: P = 0.001, t test). For all purifications, cropped western blot images against RFP, GFP, and α-Tubulin are shown to the left (full images are in SI Appendix, Fig. S2). Biological replicates are represented as gray dots; bars represent mean signal; **P < 0.01, *P < 0.05.
In Vivo mRNA Display for Protein Identification
To demonstrate in vivo mRNA display, we generated an episomally expressed inducible construct, expressing an MCP–open reading frame (ORF) fusion. This fusion includes a short polypeptide purification tag and is followed by a single copy of the 19-nt stem loop (47) such that, upon translation, the fusion product binds to its encoding mRNA (Fig. 1A). Following transformation, each strain contains a single species of the in vivo mRNA display construct corresponding to a single displayed protein, which interacts with its cellular context independently from all of the other species in the library (Fig. 1B). Induced cells can be assayed according to the desired biochemical assay (e.g., immunoprecipitation of a bait), which should preserve the RNA–protein interaction (Fig. 1C). The enrichment/depletion of each ORF sequence can be quantified by comparing their abundance in isolated RNA before and after the assay.
To demonstrate the propensity of an mRNA displayed protein to stably interact with its encoding mRNA, we constructed a set of strains expressing MCP fluorescent protein fusions. Each fusion protein was immunoprecipitated using magnetic beads that specifically recognize each construct (Fig. 1D and SI Appendix). Immunoprecipitation (IP) of the target protein copurifies its self-identifying mRNA with an enrichment of eightfold (P < 0.01) relative to the input lysate as measured by RT-PCR over native housekeeping mRNAs. In contrast, a defective coat protein construct, MCP* (N55D, K57E) (48), shows no enrichment of its respective mRNA upon purification. Similarly, deleting the downstream stem loop also removes the enrichment (SI Appendix, Fig. S1).
Next, we assessed the precision with which a displayed protein–mRNA complex can be isolated in the presence of other displayed proteins. Cotransformation of two in vivo display constructs into yeast, one expressing a GFP and the other an mCherry fusion, results in a mixed population of yeast cells each expressing one or the other. As expected, we observe a significant enrichment of the GFP mRNA compared with mCherry mRNA when purifying GFP using anti-GFP magnetic beads from the mixed population (Fig. 1E) (∼11-fold, P = 0.001) and vice versa for RFP (∼5-fold, P = 0.016). Therefore, we are able to correctly identify in vivo mRNA display proteins in one-on-one competitive assays by comparing the enrichment of mRNA levels with each other.
Discriminative Ability of In Vivo mRNA Display for High-Throughput Protein Identification
In order to systematically determine the sensitivity and specificity of in vivo mRNA display, we constructed a mix of three in vivo display libraries consisting of a few hundred distinct yeast Saccharomyces cerevisiae proteins (53). Each library carried a different C-terminal purification tag (FLAG, MYC, and HIS) and was transformed into a haploid yeast strain. The purification tags were used to specifically isolate each protein subpopulation.
In order to quantify the frequency of each displayed ORF, we designed a sequencing preparation protocol compatible with NGS (SI Appendix, Figs. S3–S5). In brief, RNA was isolated from starting and purified protein samples. The library mRNAs were processed utilizing universal sequences flanking the ORF of each construct, and Illumina adapters were added to the fragments corresponding to the 5′ and 3′ ends of each ORF, allowing us to quantify frequencies with a minimal number of reads. Frequencies of fragments in the starting sample were compared with the frequencies from the isolated protein samples and normalized to the frequencies of nonspecific functional controls (SI Appendix, Figs. S6 and S7). The nonspecific functional controls are a set of constructs that display their mRNA but are not isolated in a given assay (Dataset S1). For every ORF, we calculated a relative enrichment, termed display score (DS). Additionally, a z score and a significance value for the DS of each ORF were calculated from the distribution of the nonspecific functional controls (SI Appendix, Fig. S8).
Since unbound MCPs and stem loops are free to interact with nonspecific partners postlysis, they could compromise precision (SI Appendix, Figs. S9 and S10). Therefore, we provided an excess of coat protein in order to titrate any nonspecific interactions (SI Appendix, Fig. S11). Moreover, lysis and all purification steps were performed at 4 °C in order to minimize likelihood of partner exchanges due to possible disassociation of mRNA and MCP at higher temperatures (SI Appendix, Fig. S12).
As expected, when using anti-FLAG beads to purify the FLAG-tagged proteins from the mixed population, we observed a substantial enrichment of the ORF mRNAs from the FLAG library in the purified sample with respect to the lysate (Fig. 2A), while ORFs in the MYC- and HIS-tagged libraries were not enriched. We conducted three separate purifications for each tag from the mixed population and quantified the DSs for the ORFs in each library (Fig. 2 B and C, Dataset S2, and SI Appendix, Fig. S13). We used the DS for each ORF to classify proteins as members of the immunoprecipitated population, resulting in the receiver operating curves in Fig. 2C. The high values for the area under the curve (AUC; AUC = 0.98, 0.96, and 0.77 for FLAG, MYC, and HIS, respectively) (Fig. 2C) demonstrate that in vivo mRNA display classified proteins to the correct population while maintaining low false-positive rates. Although all three assays demonstrate relatively high discriminative ability, the FLAG and MYC purifications perform better than HIS, suggesting a higher background during isolation of histidine-tagged proteins based on immobilized metal affinity chromatography.
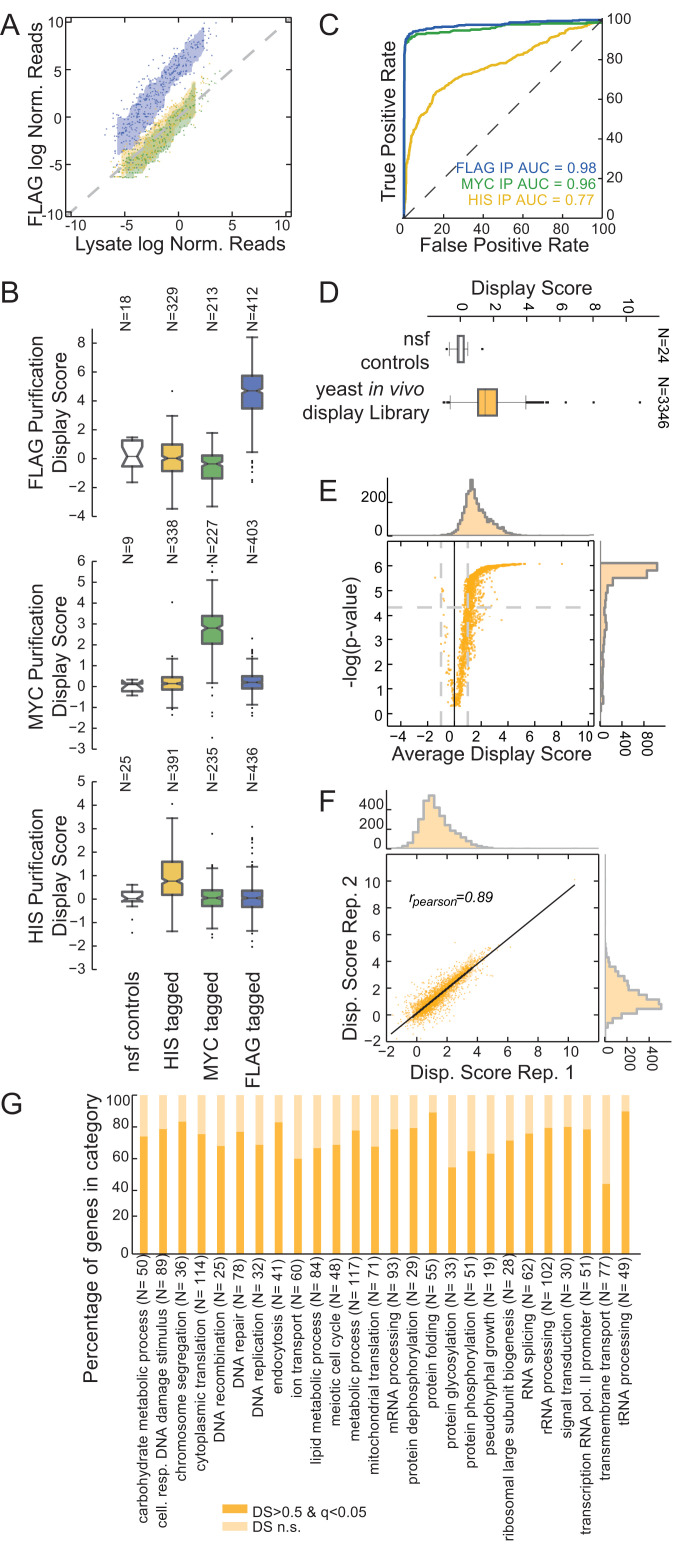
Fig. 2.
In vivo mRNA display enables precise protein identification in a complex mixture. (A) Assessment of in vivo mRNA display precision by identification of a specific protein subpopulation. Anti-FLAG immunoprecipitation from a mixed population containing HIS-tagged (yellow), MYC-tagged (green), and FLAG-tagged (blue) yeast in vivo mRNA display constructs. Scatterplot for log-normalized reads for the lysate (x axis) against the purified samples (y axis). Reads for each sample were normalized by the mean of nonspecific functional controls. For each population, the area between rolling 10th and 90th percentiles is shaded with the respective color. (B) DS box plots for anti-FLAG, anti-MYC, and anti-HIS purifications. Shown are box plots of DSs for each subpopulation and the nonspecific functional controls (nsf). The box extends from the lower to the upper quartile values, while whiskers extend 1.5 times the interquartile range from the edge of the box, and outliers are shown as individual points. (C) Receiver operating characteristic curves for the purifications in B. Members of the mixed library were classified according to their respective DSs. (D) Yeast in vivo mRNA display library purification. Average DSs were calculated per gene for over 3,300 ORFs over four biological replicates. Shown is a box plot for the ORFs in the library compared with the set of nonspecific functional controls. (E) Volcano plot for the DS of the yeast library purifications. P values were calculated with respect to the nonspecific functional controls (SI Appendix) (q values calculated using a Benjamini–Hochberg correction). (F) Scatter plot for DSs between two replicates (Pearson correlation is reported). (G) Percentage of in vivo mRNA display proteins with significant and nonsignificant (n.s.) DSs per gene ontology biological process category.
An In Vivo mRNA Display Library for Exploration of Yeast Proteomics
Next, we built an in vivo display library of the yeast ORFeome for high-throughput proteomic exploration. Starting from the plasmid ORFeome library (53) encoding ∼4,700 validated yeast proteins, we pooled the ORFs and introduced them into an in vivo mRNA display backbone using the Gateway cloning system (SI Appendix). We transformed the resulting pooled library into the BY4742 S288c Matα strain. To estimate the overall ability of every protein to display its encoding mRNA effectively, we purified the proteome from library lysate utilizing a 6xHIS tag. As with all fusion libraries, the ability to capture interactions is limited by proper protein folding, the proper positioning of any functional domains as well as for this approach, the ability of the coat protein domain to bind the stem loop efficiently. We used the 6xHIS tag for library construction in order to preserve other tags for future functional assays when more specific tags would be needed for the purification of cellular complexes. Since the histidine tag purification has a relatively poor ability to enrich for bound RNA relative to other tags (Fig. 2C), we expect this assay to underestimate display efficiency. Overall, the constructed yeast in vivo display library captured ∼3,400 proteins, which were consistently present in either the lysate or the purified samples across four replicates (Fig. 2D and Dataset S3). We sequenced each replicate for <5 million reads (SI Appendix, Fig. S14) and calculated the DS of each ORF in the purified samples against the lysate, relative to the nonspecific functional controls (SI Appendix, Fig. S15); 73% of the ORFs captured in the assay exhibit a significant display enrichment score compared with the nonspecific functional controls (average DS > 0.5; Mann–Whitney U test, Benjamini–Hochberg corrected q value < 0.05) (Fig. 2E). DSs were reproducible across replicates (rspear = 0.76 to 0.89) (Fig. 2F and SI Appendix, Fig. S16). Overall, yeast proteins that efficiently display their own mRNA span a wide range of biological processes, functions, and cellular compartments (54) (Fig. 2G and SI Appendix, Figs. S17–S19).
In Vivo mRNA Display Retains Native Organellar Localization of the Proteome
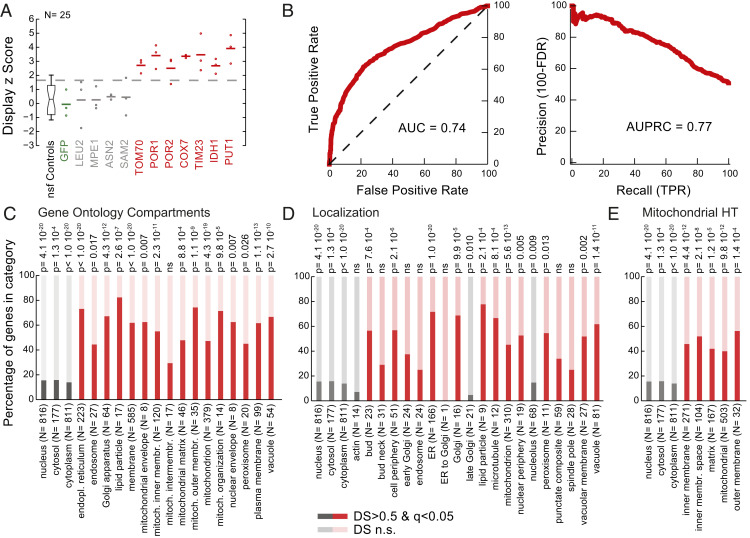
We wondered whether in vivo mRNA displayed proteins retain their native subcellular compartmentalization, despite their episomal overexpression, fusion with the coat protein, and association with their cognate mRNA. To test this, we performed a subcellular fractionation experiment in order to isolate proteins localized in specific cellular compartments. In particular, we performed a crude mitochondrial purification (55, 56), whereby induced in vivo displayed library spheroplasts were disrupted with a dounce homogenizer, and a fraction was enriched by means of differential centrifugation in triplicate (SI Appendix). This crude mitochondrial fractionation is commonly used as it is fast and does not require large amounts of starting material, even though it is known to be enriched in proteins and membranes from other organelles. We, thus, isolated and sequenced RNA from the supernatant and the pelleted samples of the final centrifugation step. We calculated a DS score comparing read frequencies for each mRNA displayed species present in our assay between the two fractions (Dataset S4 and SI Appendix, Fig. S20). For example, the mRNAs for mitochondrial outer membrane protein TOM70 and porins POR1 and POR2 are significantly enriched in the fraction compared with the nonspecific controls (z score = 4.7, 4.3, and 5.9, respectively), as well as for inner membrane proteins COX7 and TIM23 (z score = 5.8 and 6.0, respectively) and mitochondrial matrix proteins IDH1 and PUT1 (z score = 3.9 and 4.1, respectively). On the other hand, in vivo display mRNAs for cytosolic proteins LEU2 (z score = 0.4), MPE1 (z score = 0.5), ASN2 (z score = 0.8), and SAM2 (z score = 0.8) are not significantly enriched in the organelle fraction (Fig. 3A). Gene ontology (GO) term enrichment analysis showed that DSs are indicative of protein membership in the expected organelles (AUC = 0.74, AUPRC = 0.77) (Fig. 3B and SI Appendix, Fig. S21). In general, proteins known to localize to the mitochondria (57–59) were three times more likely to be significantly displayed in the pellet compared with cytosolic proteins (P < 10−18) (Fig. 3 C–E). Our analysis revealed that proteins of the mitochondrial outer membrane (×4.7; P < 10−8) and inner membrane proteins (×3.5; P < 10−10) are all significantly enriched (Fig. 3C). As expected, endoplasmic reticulum (ER)-, Golgi-, and lipid particle-associated proteins were over four times more likely to be significantly displayed in the pellet. Also, as expected, proteins known to localize to the cytoplasm and nucleus were significantly depleted (P < 10−20 and P < 10−19, respectively).
Fig. 3.
In vivo mRNA display captures native protein localization. (A) Display z scores of the enrichment for individual mRNAs in a crude mitochondrial isolation. Replicates are denoted with circles, while averages are reported as horizontal lines. z scores were calculated with respect to the nonspecific functional (nsf) controls. (B) Receiver operating characteristic and precision recall curves for the crude mitochondrial isolation. Members of the library were classified according to their respective DS and compared with the GO term compartment categories. True positive rates (TPR), false positive rates (FPR), and false discovery rates (FDR) were calculated. Area under the curve (AUC) and area under the precision recall curve (AUPRC) values are reported. (C–E) Percentage of in vivo mRNA display proteins with significant and nonsignificant (n.s.) DSs per (C) GO term compartment category, (D) localization category (58), and (E) high-throughput (HT) mitochondrial study (59). Categories specific to the crude mitochondrial enrichment are shown in red, while cytoplasmic and nuclear fractions are reported in gray. Organelle and membrane proteins are significantly enriched, while cytosolic proteins are depleted (hypergeometric test for P values, nonsignificant P values marked as ns).
In Vivo mRNA Display Enables Accurate Discovery of In Vivo PPIs
Mapping the network of PPIs has been a central challenge of postgenome biology. We aimed to determine whether in vivo mRNA display can be used to efficiently identify the in vivo interaction partners of a protein of interest. We, thus, generated libraries for systematic PPI assays by mating our haploid in vivo display MATα library with an MATa strain expressing a protein bait of interest. The protein bait was fused with a C-terminal GFP epitope tag, enabling its efficient IP. After induction and homogenization, we compared RNA reads from the lysate with a sample purified using anti-GFP magnetic beads and calculated a corresponding DS. We chose to investigate the interaction partners of two proteins: SAM2, a highly expressed S-adenosylmethionine synthetase (60), and ARC40, a member of the Arp2/3 complex that is an actin nucleation center playing a critical role in the motility and integrity of actin patches (61, 62). We generated two libraries for each of the SAM2- and ARC40-GFP baits, one with the fusion protein integrated into the genome and driven by the native promoter (58) and another episomally expressed and inducible. In addition, we generated two control libraries containing either an inducible GFP, not fused to any other peptides, or a null library containing no bait. We tested each of the described libraries in duplicate (Datasets S5–S7 and SI Appendix, Figs. S22–S24).
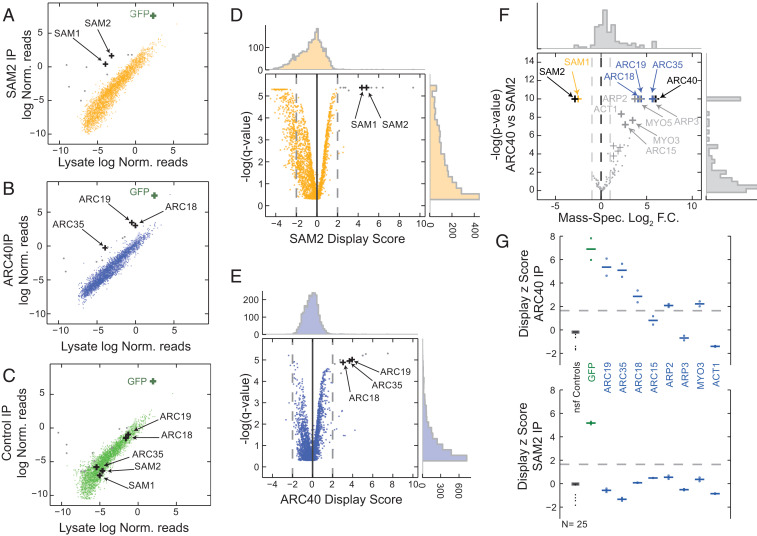
For a given bait (SAM2 or ARC40), we considered a library protein to be a PPI hit if the mRNA of the corresponding ORF was enriched in the corresponding samples (average DS > 2, q value < 0.001) (Fig. 4 A–C) compared with the lysate but not enriched in the control samples (q value > 0.05). For SAM2, we find two hits: SAM2 itself (DS = 4.8, q value = 6 × 10−4) and its paralog SAM1 (DS = 4.4, q value = 6 × 10−4) (Fig. 4 A and D). Indeed, SAM2 has been reported to interact with its paralog in traditional affinity capture–mass spectrometry (MS) studies (21, 22), and it has also been predicted to interact with itself by Y2H (6). On the other hand, the hits for ARC40 are members of the same complex (62): ARC19 (DS = 3.9, q value = 1 × 10−5), ARC35 (DS = 3.7, q value = 1.1 × 10−5), and ARC18 (DS = 3.3, q value = 1.3 × 10−5) (Fig. 4 B and E). ARC40 forms a seven-subunit complex along with ARC19, ARC35, ARC18, ARC15, ARP2, and ARP3 (62). ARP2 is only moderately enriched in our assay (DS = 0.55, q value = 0.006), while ARP3 is not enriched in the purified ARC40 samples. ARC15 is not present in our library and therefore, could not be assessed.
Fig. 4.
In vivo mRNA display enables high-throughput protein interaction assays. (A–C) Copurification using anti-GFP magnetic beads from SAM2-GFP (A), ARC40-GFP (B), or control GFP (C). Experiments were performed in biological quadruplicates. Scatterplots are shown for the log-normalized reads for the lysate (x axis) against the purified samples (y axis). The area between rolling 10th and 90th percentiles is shaded with the respective color. GFP mRNA is a positive control for the assay and is enriched in all three purifications. Hits for SAM2 and ARC40 are noted as black crosses. Gray dots denote nonspecific ARC40 and SAM2 hits that are also significantly enriched in the GFP samples (common background). (D and E) Volcano plots for the DS for SAM2 (D) and ARC40 (E). P values were calculated with respect to the nonspecific functional controls (SI Appendix). (F) Volcano plot for mass spectrometry of purified SAM2 and ARC40 samples (black crosses). The common hits for both MS and in vivo mRNA display for the two purified proteins are shown in yellow and blue, respectively. The remainder MS hits are denoted in gray. Log2 fold change (F.C.) is plotted against -log P values. (G) Display z scores for individual ARC40 interactors in a low-throughput purification of ARC40 (Upper) and SAM2 (Lower). Replicates are denoted with circles, while averages are reported as horizontal lines; z scores were calculated with respect to the nonspecific functional (nsf) controls (shown in black).
We performed affinity capture followed by liquid chromatography with tandem MS to validate our results using samples processed identically to our in vivo display assays. We confirmed that SAM1 was copurified with SAM2, while ARC40 samples were enriched in ARP2/3 complex subunits as expected (Fig. 4F). Additionally, we find that actin-related proteins MYO3, MYO5, and ACT1 were enriched in the ARC40 samples. MYO3 was not a member of our pooled library, while MYO5 was not included in the yeast ORFeome set. MS cannot discriminate between self-interaction and presence as a bait, and hence, the identified targets SAM2 and ARC40 (Fig. 4F) are due to the purified bait itself. On the other hand, in vivo mRNA display is able to capture such self-interactions as demonstrated by the enrichment of SAM2 reads in the SAM2 purified samples.
The lack of strong enrichment for the known ARC40 interactors ARP2 and ARP3 may be due to multiple factors. These include the inability of the MCP fusions to fold properly or to bind their respective mRNAs efficiently, the interference of the fused domains with the interaction under study, or even library construction biases. In order to probe the sensitivity of in vivo mRNA display further, we designed a low-throughput display experiment that included all of the possible targets of ARC40 from the mass spectrometry assay. We cloned the respective ORFs into our construct one at a time and validated their sequences. In addition to ARC35, ARC18, and ARC19, we observed that ARP2 (DS = 1.8, q value = 0.002) and MYO3 (DS = 1.9, q value = 0.0015) are significantly enriched when ARC40 is purified, while they are not enriched in SAM2 samples (Fig. 4G). On the other hand, ARC15, ARP3, and ACT1 are not enriched, showcasing possible limitations of our approach. While ARC15 was not present in the high-throughput library, ARP3 and ACT1 did not significantly display their mRNA in the whole-library purification assay (Dataset S3), which explains the lack of enrichment in the copurification assay.
Discussion
Here, we have shown that the MS2–MCP interaction enables stable noncovalent linking of proteins to their encoding mRNAs in vivo. We have demonstrated that this feature can be exploited to convert a variety of standard proteomics-based assays to a sequencing readout. We have shown that the in vivo mRNA displayed proteins maintain their organellar distributions in a manner that can be utilized for sequencing-based protein cartography. We have also shown that in vivo mRNA display can be used for high-specificity detection of in vivo PPIs. However, our demonstration of this technology has some limitations. As expected, not all library proteins are able to display their mRNAs effectively. Future studies could determine whether a C-terminal coat protein fusion library will complement the current N-terminal library and extend our proteome coverage. A C-terminal design may alleviate potential pitfalls with the proper processing of N-terminal transit signals (SI Appendix, Fig. S25) and assist with the correct function of membrane protein constructs with N termini on the outer side of the cellular membrane. Additionally, using the mRNA of each protein for display purposes allows for potential length and RNA stability biases, which if ignored, could partly mask biological phenotypes under study. One possible remedy would be to decouple the display protein from the displayed mRNA utilizing a library of same-length bar codes. In addition, any possible construction biases could be remedied in an automated ORF by ORF library construction (vs. our simple pooled approach).
Overall, in vivo mRNA display enables high-throughput proteomics, leveraging the ease, cost, and capacity for massive parallelization of NGS. While a medium-throughput mass spectrometry experiment can cost over $1,000, the same samples can be processed for ∼1/10th of the cost with in vivo mRNA display. As with all display technologies, such as Y2H and phage display, our approach depends on library construction, which requires some initial labor and cost, but this initial investment would pay off in the longer-term benefits of this resource for diverse applications across the community. Moreover, in vivo mRNA display interrogates proteins in their native cellular context, including posttranslational modifications, the presence of cofactors, and subcellular localization, making it compatible with affinity capture assays, which are the gold standard for proteomics.
NGS has revolutionized genomics, and we envision that in vivo mRNA display has the potential of similarly improving the throughput, labor, and cost of a variety of proteomics applications. In vivo mRNA display may enable studies of in vivo enzymatic and regulatory activity of proteins and characterize their biochemistry by means of NGS. For example, our approach has the potential to capture the dynamic nature of phosphorylation by modulating the activity of a kinase or phosphatase and determining the full spectrum of its substrates using phosphor-specific antibodies. Similarly, DNA- and RNA-centric regulatory interactions may be explored utilizing the purely sequence-based approach outlined here.
Protein functional studies are critical in studying basic biology but also in better understanding the molecular etiology of disease and development of novel therapeutics. As the MS2 tagging system has already been utilized in many different cellular contexts, we anticipate that in vivo mRNA display can be a powerful tool for proteomic studies in mammalian systems. Furthermore, much as other display technologies such as phage display have enabled in vitro protein optimization in industrial and biomedical applications (63, 64), we envision that in vivo mRNA display will enable similar optimization of peptides and proteins for therapeutic benefit within physiologically relevant contexts in vivo.
Materials and Methods
A summary of key methods is given below. Detailed methods for all experiments and analyses are included in SI Appendix.
Plasmids.
All in vivo mRNA display plasmids and respective controls were based on the plasmid pSH100 (URA3 selection marker; MET25 promoter) (51). MCP was PCR amplified from pSH100, while stem-loop sequences were ordered as blocks from IDT based on pDZ415 (51); defective MCP (MCP*) mutations were introduced via overlap PCR. Destination vectors were constructed to allow for Gateway cloning of ORFs into the display constructs (Dataset S1A).
Yeast Strains.
The BY4742 S288c MATα laboratory deletion strain was used as the starting strain for all strains harboring in vivo mRNA display constructs. Plasmids were transformed using the LiAc-PEG-ssDNA method (65) and selected in appropriate 2% glucose dropout media (Dataset S1).
In Vivo mRNA Display Library Generation.
Plasmid was extracted from the pooled yeast ORFeome plasmid collection. Yeast ORFs were PCR amplified using flanking sequences, and a two-step Gateway recombination reaction was used to transfer the sequences into the in vivo mRNA display vector. Colonies were selected in semiliquid soft agarose gel Luria Bertani media with the appropriate antibiotics. The final in vivo mRNA display library was transformed into BY4742 using the LiAc-PEG-ssDNA method and selected in 2% glucose synthetic complete (SC) media lacking uracil semiliquid soft agarose gel.
Yeast Cell Culture.
S. cerevisiae strains were cultured in the appropriate SC dropout media supplemented with 2% glucose at 30 °C and shaken at 220 rpm. Overnight cultures were induced by seeding 0.1 optical density (OD600) per milliliter into a new liquid culture with a similar SC dropout media additionally lacking methionine. Strains were outgrown for 6 to 8 h to 0.6 to 0.8 OD600 per milliliter, collected by centrifugation, washed twice with ultrapure water, split in aliquots, flash frozen, and stored at −80 °C until further processing.
Excess Coat Protein.
In order to titrate any nonspecific interactions, we mixed into each sample an excess culture of yeast cells that express an MCP fusion that does not display its own mRNA, unless otherwise noted. The excess MCP is not isolated specifically in any given protein assay, and its mRNA is not processed during first-strand and second-strand synthesis (Dataset S1). Upon induction, the equivalent of 30 OD600 units of strains expressing excess coat protein was mixed with 10 OD600 of in vivo mRNA display library cells immediately prior to or immediately after freezing.
Nonspecific Functional Controls for In Vivo mRNA Display.
We included a set of in vivo mRNA display constructs in every library that function as internal negative and positive controls for a given protein purification assay. Their mRNA frequencies provide a background with respect to which we normalize the frequencies of each ORF (SI Appendix, Figs. S7 and S8). The controls consist of a small set of reporter genes and peptides that should not participate in any biological interactions inside the cell (Dataset S1). Overall, the controls represented 2 to 5% of the total processed cell culture.
Whole-Cell Lysate Preparation.
Frozen cell pellets were resuspended in 750 μL of ice-cold Lysis Buffer (20 mM Hepes, pH 7.5, 140 mM KCl, 1.5 mM MgCl2, 1% Triton X-100, 1× Complete Mini Protease Inhibitor EDTA-free, 0.2 U/µL SUPERase RNase Inhibitor) and homogenized on a bead mill at 4 °C. Whole-cell lysate was cleared by a 1-min centrifugation at 7,000 × g and a 30-s spin at 11,000 × g.
In Vivo mRNA Display Protein Bait Purification.
We used appropriate magnetic beads for all tagged protein purifications. Purifications were performed at 4 °C. All beads were washed three times before use with 200 μL of Wash Buffer (50 mM sodium phosphate, pH 8, 300 mM NaCl, 0.01% Tween-20, 0.02 U/µL SUPERase RNase Inhibitor). Whole-cell lysate was incubated on a roller with the magnetic beads, and the bound beads were washed four times with 300 μL Wash Buffer and resuspended in 100 μL of Storage Buffer.
Crude Mitochondrial Isolation.
We processed frozen library pellets equivalent to 20 OD600 units of cells per replicate with no excess coat protein culture added. We performed a crude mitochondrial isolation by means of differential centrifugation using a commercially available kit from Sigma (MITOISO3).
RNA Extraction and Complementary DNA (cDNA) Synthesis.
RNA was extracted from all protein samples using TRIzol in combination with a spin column. RNA was treated with double-strand specific DNAse and reverse transcribed using a construct-specific primer binding downstream of the in vivo mRNA display construct ORF (SI Appendix, Fig. S3). For second-strand synthesis, we performed a PCR amplification using construct-specific primers upstream and downstream of the in vivo mRNA display ORF.
qPCR.
We assessed extracted RNA for quality and in vivo mRNA display efficiency using qPCR. We determined the relative abundance of mCherry and GFP transcripts in each sample with respect to each other or to ACT1 as a reference gene. Library protein purification experiments were designed such that either GFP or mCherry is copurified in the experiment (specific positive control) and the other is washed away (nonspecific reference). We calculated a ΔCt value for each sample and a −ΔΔCt between purified sample and input lysate. The log2 fold enrichment is
In Vivo mRNA Display Library Sequencing Preparation.
To prepare libraries for NGS, double-stranded cDNA from each sample was digested with a mixture of restriction enzymes (HinP1I and AciI or MspI and HpyCH4IV) followed by a Y-Linker ligation. Adapters for multiplexing and Illumina sequencing were added by means of PCR amplification.
In Vivo mRNA Display Sequencing Data Analysis.
Reads were mapped to the 5′ and 3′ ends of all yeast ORFs. The average log frequency of the 5′ fragments and the 3′ fragments was calculated for each ORF. The log frequencies were normalized by the average frequency of the nonspecific functional control constructs within each sample. The DS (DSi) for each ORF i is calculated as the difference of the log-normalized frequencies between two matched samples (SI Appendix, Fig. S8). The DS represents an enrichment (DSi > 0) or depletion (DSi < 0) of the reads of ORF i in the purified sample compared with the lysate with respect to the nonspecific functional controls. The distribution of the nonspecific functional controls was used to calculate a z score for the DS of each ORF.
Supplementary Material
Acknowledgments
We thank R. Singer (Albert Einstein College of Medicine) for the generous sharing of key plasmids, Rajesh Soni (Columbia University Medical Campus) for his assistance with mass spectrometry, and L. Symington for sharing yeast protein extraction protocols and equipment as well as the members of the Barasch laboratory for sharing protocols and equipment for western blotting (Columbia University). We thank the laboratory of S.T. for insightful advice on the conceptualization and application of the technology. This work was supported by NIH Grant R01HG009065 (to S.T.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002650117/-/DCSupplemental.
Data Availability.
All data generated or analyzed during this study are included in Datasets S1–S7 (SI Appendix has details). Sequencing data are available in NCBI's Gene Expression Omnibus (accession no. GSE154643). Plasmids are available on Addgene (https://www.addgene.org/Saeed_Tavazoie/). Scripts and support files for processing of in vivo mRNA display sequencing data are available in Github at https://github.com/tavalab/in-vivo-mRNA-Display.
References
- 1.Altelaar A. F. M., Munoz J., Heck A. J. R., Next-generation proteomics: Towards an integrative view of proteome dynamics. Nat. Rev. Genet. 14, 35–48 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Snider J.et al., Fundamentals of protein interaction network mapping. Mol. Syst. Biol. 11, 848 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morris J. H.et al., Affinity purification-mass spectrometry and network analysis to understand protein-protein interactions. Nat. Protoc. 9, 2539–2554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Havugimana P. C.et al., A census of human soluble protein complexes. Cell 150, 1068–1081 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fields S., Song O., A novel genetic system to detect protein-protein interactions. Nature 340, 245–246 (1989). [DOI] [PubMed] [Google Scholar]
- 6.Yu H.et al., High-quality binary protein interaction map of the yeast interactome network. Science 322, 104–110 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snider J.et al., Detecting interactions with membrane proteins using a membrane two-hybrid assay in yeast. Nat. Protoc. 5, 1281–1293 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Kenworthy A. K., Imaging protein-protein interactions using fluorescence resonance energy transfer microscopy. Methods 24, 289–296 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Remy I., Michnick S. W., Clonal selection and in vivo quantitation of protein interactions with protein-fragment complementation assays. Proc. Natl. Acad. Sci. U.S.A. 96, 5394–5399 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tarassov K.et al., An in vivo map of the yeast protein interactome. Science 320, 1465–1470 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Weill U.et al., Genome-wide SWAp-Tag yeast libraries for proteome exploration. Nat. Methods 15, 617–622 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Branon T. C.et al., Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roux K. J., Kim D. I., Raida M., Burke B., A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernández-Suárez M., Chen T. S., Ting A. Y., Protein-protein interaction detection in vitro and in cells by proximity biotinylation. J. Am. Chem. Soc. 130, 9251–9253 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhee H.-W.et al., Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hung V.et al., Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. eLife 6, e24463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lobingier B. T.et al., An approach to spatiotemporally resolve protein interaction networks in living cells. Cell 169, 350–360.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hein M. Y.et al., A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Huttlin E. L.et al., Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan C.et al., Panorama of ancient metazoan macromolecular complexes. Nature 525, 339–344 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gavin A.-C.et al., Proteome survey reveals modularity of the yeast cell machinery. Nature 440, 631–636 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Krogan N. J.et al., Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Rolland T.et al., A proteome-scale map of the human interactome network. Cell 159, 1212–1226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luck K., Sheynkman G. M., Zhang I., Vidal M., Proteome-scale human interactomics. Trends Biochem. Sci. 42, 342–354 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vidal M., Cusick M. E., Barabási A.-L., Interactome networks and human disease. Cell 144, 986–998 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menche J.et al., Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science 347, 1257601 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovács I. A.et al., Network-based prediction of protein interactions. Nat. Commun. 10, 1240 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lieberman-Aiden E.et al., Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krueger F., Kreck B., Franke A., Andrews S. R., DNA methylome analysis using short bisulfite sequencing data. Nat. Methods 9, 145–151 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Sidoli S., Kulej K., Garcia B. A., Why proteomics is not the new genomics and the future of mass spectrometry in cell biology. J. Cell Biol. 216, 21–24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Low T. Y., Mohtar M. A., Ang M. Y., Jamal R., Connecting proteomics to next-generation sequencing: Proteogenomics and its current applications in biology. Proteomics 19, e1800235 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Smith G. P., Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 228, 1315–1317 (1985). [DOI] [PubMed] [Google Scholar]
- 34.McCafferty J., Griffiths A. D., Winter G., Chiswell D. J., Phage antibodies: Filamentous phage displaying antibody variable domains. Nature 348, 552–554 (1990). [DOI] [PubMed] [Google Scholar]
- 35.Smith G. P., Petrenko V. A., Phage display. Chem. Rev. 97, 391–410 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Sidhu S. S., Koide S., Phage display for engineering and analyzing protein interaction interfaces. Curr. Opin. Struct. Biol. 17, 481–487 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Nemoto N., Miyamoto-Sato E., Husimi Y., Yanagawa H., In vitro virus: Bonding of mRNA bearing puromycin at the 3′-terminal end to the C-terminal end of its encoded protein on the ribosome in vitro. FEBS Lett. 414, 405–408 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Roberts R. W., Szostak J. W., RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl. Acad. Sci. U.S.A. 94, 12297–12302 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanes J., Plückthun A., In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl. Acad. Sci. U.S.A. 94, 4937–4942 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boder E. T., Wittrup K. D., Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15, 553–557 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Gu L.et al., Multiplex single-molecule interaction profiling of DNA-barcoded proteins. Nature 515, 554–557 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larman H. B., Liang A. C., Elledge S. J., Zhu J., Discovery of protein interactions using parallel analysis of translated ORFs (PLATO). Nat. Protoc. 9, 90–103 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Younger D., Berger S., Baker D., Klavins E., High-throughput characterization of protein-protein interactions by reprogramming yeast mating. Proc. Natl. Acad. Sci. U.S.A. 114, 12166–12171 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trigg S. A.et al., CrY2H-seq: A massively multiplexed assay for deep-coverage interactome mapping. Nat. Methods 14, 819–825 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng X.et al., Affinity capture of polyribosomes followed by RNAseq (ACAPseq), a discovery platform for protein-protein interactions. eLife 7, e40982 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Layton C. J., McMahon P. L., Greenleaf W. J., Large-scale, quantitative protein assays on a high-throughput DNA sequencing chip. Mol. Cell 73, 1075–1082.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johansson H. E., Liljas L., Uhlenbeck O. C., RNA recognition by the MS2 phage coat protein. Semin. Virol. 8, 176–185 (1997). [Google Scholar]
- 48.Peabody D. S., The RNA binding site of bacteriophage MS2 coat protein. EMBO J. 12, 595–600 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tyagi S., Imaging intracellular RNA distribution and dynamics in living cells. Nat. Methods 6, 331–338 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Bertrand E.et al., Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2, 437–445 (1998). [DOI] [PubMed] [Google Scholar]
- 51.Hocine S., Raymond P., Zenklusen D., Chao J. A., Singer R. H., Single-molecule analysis of gene expression using two-color RNA labeling in live yeast. Nat. Methods 10, 119–121 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsai B. P., Wang X., Huang L., Waterman M. L., Quantitative profiling of in vivo-assembled RNA-protein complexes using a novel integrated proteomic approach. Mol. Cell. Proteomics 10, M110.007385 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gelperin D. M.et al., Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 19, 2816–2826 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.The Gene Ontology Consortium , The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao P.-C., Boldogh I. R., Siegmund S. E., Freyberg Z., Pon L. A., Isolation of mitochondria from Saccharomyces cerevisiae using magnetic bead affinity purification. PLoS One 13, e0196632 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daum G., Böhni P. C., Schatz G., Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 257, 13028–13033 (1982). [PubMed] [Google Scholar]
- 57.Ashburner M.et al.; The Gene Ontology Consortium , Gene ontology: Tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huh W.-K.et al., Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Morgenstern M.et al., Definition of a high-confidence mitochondrial proteome at quantitative scale. Cell Rep. 19, 2836–2852 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thomas D., Rothstein R., Rosenberg N., Surdin-Kerjan Y., SAM2 encodes the second methionine S-adenosyl transferase in Saccharomyces cerevisiae: Physiology and regulation of both enzymes. Mol. Cell. Biol. 8, 5132–5139 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winter D. C., Choe E. Y., Li R., Genetic dissection of the budding yeast Arp2/3 complex: A comparison of the in vivo and structural roles of individual subunits. Proc. Natl. Acad. Sci. U.S.A. 96, 7288–7293 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson R. C.et al., Crystal structure of Arp2/3 complex. Science 294, 1679–1684 (2001). [DOI] [PubMed] [Google Scholar]
- 63.Hoogenboom H. R., “Overview of antibody phage-display technology and its applications” in Antibody Phage Display: Methods and Protocols, O’Brien P. M., Aitken R., Eds. (Methods in Molecular Biology 178, Humana Press, Totowa, NJ, 2002), pp. 1–37. [DOI] [PubMed] [Google Scholar]
- 64.Clackson T., Hoogenboom H. R., Griffiths A. D., Winter G., Making antibody fragments using phage display libraries. Nature 352, 624–628 (1991). [DOI] [PubMed] [Google Scholar]
- 65.Gietz R. D., Woods R. A., “Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method” in Methods in Enzymology, Guthrie C., Fink G. R., Eds. (Elsevier, Amsterdam, the Netherlands, 2002), vol. 350, pp. 87–96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in Datasets S1–S7 (SI Appendix has details). Sequencing data are available in NCBI's Gene Expression Omnibus (accession no. GSE154643). Plasmids are available on Addgene (https://www.addgene.org/Saeed_Tavazoie/). Scripts and support files for processing of in vivo mRNA display sequencing data are available in Github at https://github.com/tavalab/in-vivo-mRNA-Display.