Significance
There are pressing needs for synthetic molecular recognition techniques to augment and expand those derived from natural sources, which are widely employed for biomedical research. Corona phase molecular recognition (CoPhMoRe), whereby the adsorbed phase around a nanoparticle exhibits selective binding to an analyte, can address this need, but the underlying mechanisms remain an active scientific inquiry. Herein, we synthesize a series of corona phases around single-walled carbon nanotubes (SWNTs) and investigate the selective binding of poly(methacrylic acid-styrene) phases to an inhibitor of phosphodiesterase. A systematic study reveals that synthetic CoPhMoRe mimics the H loop of native enzyme in its activity, resulting from the corona configuration and supramolecular interactions. This work uncovers molecular mechanisms of synthetic recognition that guide future discovery.
Keywords: artificial molecular recognition, single-walled carbon nanotubes, corona phase molecular recognition
Abstract
Molecular recognition binding sites that specifically identify a target molecule are essential for life science research, clinical diagnoses, and therapeutic development. Corona phase molecular recognition is a technique introduced to generate synthetic recognition at the surface of a nanoparticle corona, but it remains an important question whether such entities can achieve the specificity of natural enzymes and receptors. In this work, we generate and screen a library of 24 amphiphilic polymers, preselected for molecular recognition and based on functional monomers including methacrylic acid, acrylic acid, and styrene, iterating upon a poly(methacrylic acid-co-styrene) motif. When complexed to a single-walled carbon nanotube, some of the resulting corona phases demonstrate binding specificity remarkably similar to that of phosphodiesterase type 5 (PDE5), an enzyme that catalyzes the hydrolysis of secondary messenger. The corona phase binds selectively to a PDE5 inhibitor, Vardenafil, as well as its molecular variant, but not to other potential off-target inhibitors. Our work herein examines the specificity and sensitivity of polymer “mutations” to the corona phase, as well as direct competitions with the native binding PDE5. Using structure perturbation, corona surface characterization, and molecular dynamics simulations, we show that the molecular recognition is associated with the unique three-dimensional configuration of the corona phase formed at the nanotube surface. This work conclusively shows that corona phase molecular recognition can mimic key aspects of biological recognition sites and drug targets, opening up possibilities for pharmaceutical and biological applications.
Molecular recognition is the property of a construct to bind to one target molecule or a class of molecules with high specificity (1). It is indispensable for biological research, due to their roles in understanding protein correlation in signaling cascades (2, 3). It is also widely used in clinical assays for identifying disease biomarkers in biofluids (4–6). Molecular recognition is also central to therapeutic efficacy of pharmaceutics in targeting abnormal bioactivities (7–9). The most commonly used molecular recognition sites are produced by immune cells: the antibodies (10). Antibodies, however, suffer from a high tendency of hydrolyzation and degradation, potential loss of activities when immobilized, low tunability of binding affinity, and limited availability for a wide range of molecules, such as posttranslational modifications (9–17). All these factors impair antibodies’ recognition functions. To develop alternatives to natural recognition molecules, extensive efforts have explored various synthetic analogues to achieve similar recognition function (1, 6, 8, 18–28).
Creating artificial recognition sites, however, remains a substantial objective and a grand challenge for synthetic and supramolecular chemistry. One approach is to utilize macrocyclic molecules, such as cucurbituril and crown ether, so that their hydrophobic cavity interacts with small guest molecules, often with limited specificity (8, 18–22). Another approach, molecular imprinting, carries out polymerization in the presence of analyte as the template, so that after extracting the template, the resulting polymer conformation has a cavity with a favorable shape and functional residues (6, 23–25). The method has been applied to various small molecules, but often suffers from incomplete template removal and low binding capacity (6, 23, 25). In addition to synthetic methods, combinatorial screening of sequence-defined oligomers/polymers is the main strategy for generating recognition sites for large biomolecules (1, 26–28). For example, random single-stranded DNAs are screened against a target protein, to generate DNA aptamers (4, 5, 7, 28, 29). Similar methods using other polymers, like peptides and nonnatural foldamers, have also been developed (1, 26, 27, 30–32). This screening approach is powerful in generating recognition sites for both small molecules and proteins, but often requires a large structure library to start with (1, 5, 27–31).
To introduce rational design in creating artificial recognition sites, our strategy, as described in previous work (2, 33–36), is to use a relatively rigid nanoscaffold to direct the folding of flexible linear polymers, to manipulate the resulting three-dimensional configuration. More specifically, we use single-walled carbon nanotubes (SWNTs)—a high aspect ratio nanostructure—to guide polymer folding through supramolecular interactions. The resulting corona phase molecular recognition (CoPhMoRe) produces a unique configuration for recognizing a target molecule found through screening. In addition to the structural function, SWNTs also serve as a signal transducer in identifying recognition interaction. More specifically, semiconducting SWNTs exhibit near infrared (nIR) bandgap fluorescence (37, 38). Because all atoms in SWNTs are surface atoms, the fluorescence is very sensitive to the surrounding dielectric and molecular environment (39, 40). A recognition interaction with the target analyte can modulate the effective dielectric constant or provide extra relaxation modes, both of which will modulate the nIR emission, serving as a direct readout of the molecular-level interaction (33–35, 39, 41). Therefore, our approach combines molecular recognition and signal transduction into one platform. The strategy has been applied to analytes including nitric oxide (35), hydrogen peroxide (2), dopamine (36), and fibrinogen (33), but has not been examined for its specificity in comparison with a naturally existing binding site, which is the topic of this work.
The natural binding site we focus on is phosphodiesterase type 5 (PDE5), a cyclic nucleotide enzyme that catalyzes the hydrolysis of secondary messenger cGMP (cyclic guanosine monophosphate) (42–44). By regulating the cGMP concentration, the enzyme controls cellular signaling in many physiological and pathological functions (42–44). PDE5 inhibitors have successfully treated erectile dysfunction, lower urinary symptoms, and pulmonary arterial hypertension and are in examination for other conditions, including cardiovascular diseases (43, 44). Considering these pharmaceutical applications of PDE5 inhibitors, synthetic constructs that specifically recognize PDE5 inhibitors could enable analytic methods to improve drug synthesis and production, as well as to detect adulterants in illegal products (45).
In this work, we identify and demonstrate an artificial molecular recognition site that mimics the interaction between PDE5 and its inhibitor Vardenafil. The corona phase around the SWNT is shown to possess functional similarities to the H-loop subunit of PDE5, suggesting that synthetic CoPhMoRe can realize properties of natural recognition systems. A library of 24 amphiphilic polymers wraps SWNT surfaces during ultrasonic processing and forms corona phases, one of which is shown to selectively interact with Vardenafil. For this recognition interaction, we examine its affinity and specificity, its sensitivity to corona composition, and its comparison with the natural PDE5 enzyme. Together with molecular dynamics simulations, we show that the interaction with Vardenafil results from the unique corona phase configuration, leading to a similar specificity to that of the PDE5 enzyme. We conclude that CoPhMoRe can mimic key aspects of biological recognition sites and provide a strategy for pharmaceutical applications.
Results and Discussion
Corona Phase Recognition Targeting Vardenafil.
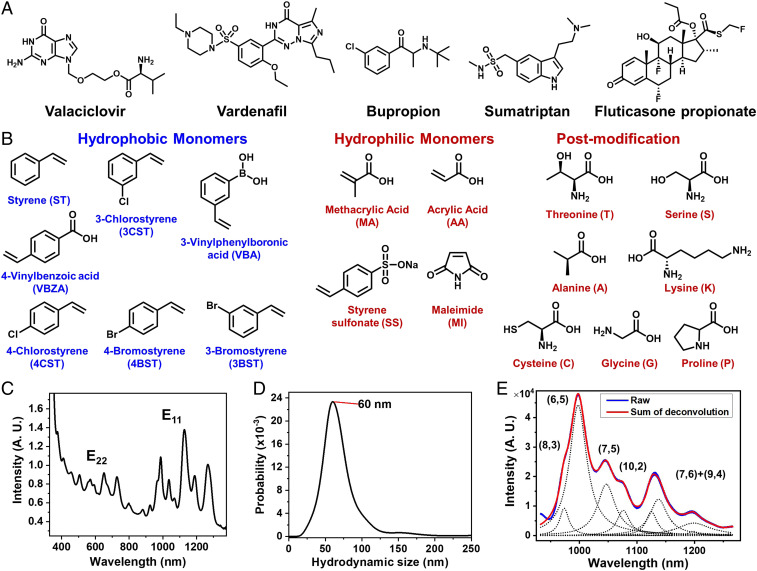
The project to build artificial molecular recognition sites for a panel of biologically active, small molecules starts with a designed library of 24 amphiphilic polymers capable of forming corona phases with SWNTs (Fig. 1 A and B). The design element focuses on the composition and the relative weighting of the hydrophobic and hydrophilic segments. The hydrophobic portion uses aromatic domains like styrene and its derivatives for their strong stacking on the nanotube surface, serving as anchors for overall polymer adsorption; the hydrophilic portion composes functional monomers such as methacrylic acid, styrene sulfonate, and maleimide, responsible for supramolecular interactions with analytes, as well as aqueous stability. Postpolymerization modifications with amino acids are introduced to increase the structural and conformational diversity (Fig. 1B). Based on the monomer structures, the library of amphiphilic polymers is preselected considering the synthetic feasibility, the aqueous stability, and whether the polymers can form stable corona phases around SWNTs. The 24 selected polymers are synthesized using reversible addition-fragmentation chain transfer (RAFT) polymerization for narrow polydispersity. The abbreviations for polymer names are assigned using their monomers’ acronym (Fig. 1B), followed by the composition percentage of the first monomer (for example, the polymer poly(methacrylic acid-co-styrene) [MA-ST-90] is made of methacrylic acid and styrene at a 90:10 ratio). (See SI Appendix for polymer characterizations.) High-pressure carbon monoxide SWNTs are used to screen various chiral species. SWNTs complexed with various polymer corona were synthesized by ultrasonic processing in aqueous polymer solution, where amphiphilic polymers adsorb and adopt different configurations at the nanotube surface (see SI Appendix for polymer–nanotube suspension preparation). Residual SWNT aggregates were removed by ultracentrifugation (105,000 relative centrifugal force for 4 h). The resulting colloidal solutions are characterized by ultraviolet-visible–near-infrared (UV-vis-nIR) absorption spectroscopy and dark-field scattering microscopy. Fig. 1C shows the UV-vis-nIR absorption spectrum for the MA-ST-90 corona phase, where the distinct and sharp peaks of E11 and E22 transitions indicate the successful isolation and suspension of individual SWNTs. Fig. 1D shows the distribution of the nanotubes’ hydrodynamic size, collected by tracking the Brownian motion of individual nanotubes through dark-field scattering microscopy. The result shows a single population of isolated nanoparticles, with a hydrodynamic size around 60 nm.
Fig. 1.
Library of target analyte molecules, monomer constituent components, and SWNT colloidal solution characterizations. (A) Chemical structures of five therapeutics as CoPhMoRe screening substrates. (B) Chemical structures of hydrophobic and hydrophilic monomers used for polymer library construction and postpolymerization modifications using amino acids. (C) The UV-vis-nIR absorption spectrum of SWNTs with the MA-ST-90 polymer corona, showing distinct and sharp peaks of E11 and E22 transitions, indicating the successful isolation of individual SWNTs. (D) Dark-field scattering microscopy is used to analyze the hydrodynamic size of individual nanotubes based on their Brownian motion, showing a single population with MA-ST-90 corona, with an average hydrodynamic size about 60 nm. (E) The near infrared emission spectrum of nanotubes under 785 nm excitation is deconvoluted to quantify the contribution of each chirality. Changes in emission spectra are used to quantify the interaction between analytes and corona phases.
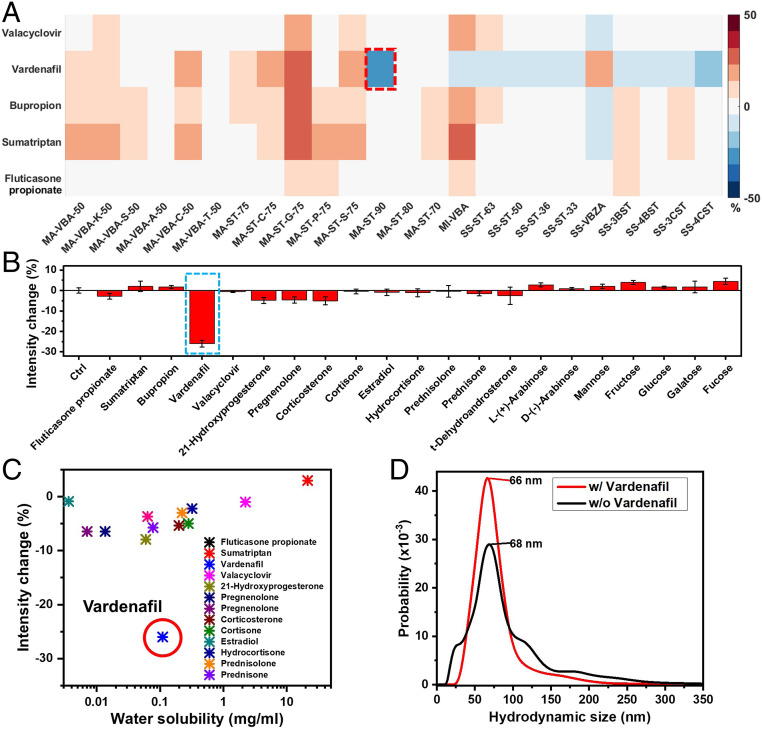
By interacting the resulting corona phases and therapeutic molecules, we have identified a corona that specifically recognizes Vardenafil. More specifically, therapeutic molecules are introduced into SWNT colloidal solutions, where the corresponding nIR spectral change of nanotubes is examined. The spectral response of each nanotube chirality is quantified by deconvoluting the emission spectra (Fig. 1E). The chirality of carbon nanotubes is indexed by a pair of integers (n,m) (37). If we imagine single-walled carbon nanotubes are rolled-up graphene sheets, the integers (n,m) define the length and the chiral angle of the roll-up vector. The diameter is proportional to the square root of . For SWNTs of (6,5) and (8,3) chirality, most of corona phases show little intensity change in response to analytes (Fig. 2A). Most of these changes are less than 15% and are possibly due to concentration variations among triplicates. [These types of small variations are commonly observed in the spectra of nanotubes (34, 36, 46).] For polymer coronas that exhibit strong spectral change, some respond to multiple analytes, suggesting nonspecific interactions. For example, Vardenafil, Bupropion, and Sumatriptan all trigger more than 20% intensity increase in MA-ST-G-75 wrapped SWNTs. In contrast, MA-ST-90, composed of methacrylic acid and styrene, demonstrates a 26% intensity reduction, exclusively in response to Vardenafil. In addition, among all of the corona phases, Vardenafil exhibits a strong emission reduction only with this corona phase. Moreover, considering that almost all intensity changes in methacrylic acid polymers are turn-on responses, it is especially unique to have a strong turn-off response in MA-ST-90 corona. All these features suggest that it is a molecule-specific recognition of Vardenafil by this polymer corona phase.
Fig. 2.
Fluorescence intensity change of SWNTs in response to a panel of drug analytes. (A) A library of amphiphilic polymers was preselected and synthesized to form corona phases on SWNTs. The nanotubes’ near infrared emission change is examined when therapeutic molecules are introduced. The colormap plots the relative intensity change of nanotubes of (6,5) and (8,3) chirality in response to analyte molecules. With the MA-ST-90 corona, nanotubes exhibit strong quenching in response to Vardenafil, in contrast to other corona phases that do not show strong specific change. MI-VBA: poly(maleimide-co-3-vinylphenylboronic acid); SS-ST: poly(styrene sulfonate-co-styrene); SS-VBZA: poly(styrene sulfonate-co-4-vinylbenzoic acid). (B) When the analyte library is expanded to 22 small molecules, Vardenafil remains as the only analyte that causes strong nIR emission change in MA-ST-90 corona. (C) No correlation is observed between the emission intensity change and the hydrophobicity of analyte, indicating that the fluorescent quenching associated with Vardenafil is not due to nonspecific hydrophobic interaction. (D) The hydrodynamic size of nanotubes with MA-ST-90 corona phase is not influenced by Vardenafil binding (66 nm vs. 68 nm), falsifying the hypothesis that the quenching is a result of nonspecific particle aggregation caused by Vardenafil.
By expanding the analyte library to 22 small molecules of different structures and hydrophobicity, we have confirmed that the recognition is not due to physical adsorption and is specific. This analyte library is chosen to examine whether nonspecific adsorption is driving the interaction observed between Vardenaifl and the MA-ST-90 corona phase. Because hydrophobicity plays an essential role in nonspecific adsorption on carbon nanotubes, the analyte molecules cover a broad spectrum of hydrophobicity. Based on the nIR spectral change (Fig. 2B), the MA-ST-90 corona phase does not appear to show strong interactions with any other analyte. In addition, we did not observe any correlation between the hydrophobicity of analytes and the emission modulation (Fig. 2C). Since hydrophobicity is essential in nonspecific adsorption on nanotubes, the lack of such correlation indicates that the recognition of Vardenafil by corona MA-ST-90 is not caused by nonspecific adsorption, but by a specific interaction.
Besides the analyte specificity, we have also confirmed that the emission intensity reduction of SWNTs is not a result of nanotube aggregation. The hypothesis is that if Vardenafil nonspecifically disrupts the electrostatic repulsion between SWNTs, it will cause bundling of nanotubes, which will reduce the nIR emission intensity. By using a dark-field scattering microscope to track an individual nanoparticle’s Brownian motion, we have quantified the hydrodynamic size of SWNTs. As shown in Fig. 2D, the distribution of nanotube hydrodynamic size is consistent regardless of the presence of Vardenafil (66 nm vs. 68 nm), falsifying the bundling hypothesis. Thus, the nIR spectral change observed in MA-ST-90 corona phase is not a result of nanoparticle aggregation.
These investigations demonstrate that the recognition of Vardenafil by the MA-ST-90 corona is not attributable to physical properties of analyte or nonspecific interactions among particles, supporting the specific recognition mechanism. The following section examines the nature of this recognition interaction.
Recognition Mechanism.
Structural properties of poly(methacrylic acid-co-styrene).
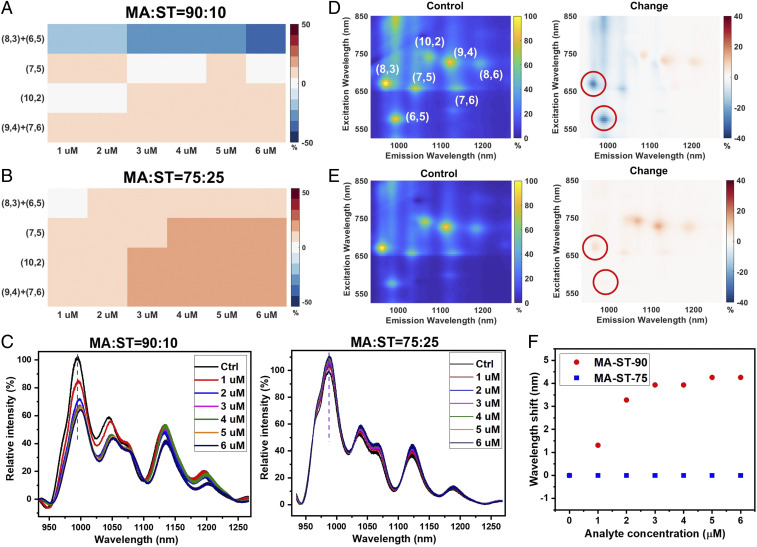
The recognition interaction appears highly sensitive to the hydrophilic–hydrophobic composition of the corona phase. Polymer MA-ST-70, made of the same monomers as in MA-ST-90, does not show a strong interaction with Vardenafil in the initial screening (Fig. 2A). MA-ST-90 has a hydrophilic–hydrophobic ratio of 90:10, in comparison to 70:30 in MA-ST-70. Moreover, as the Vardenafil concentration increases from 1 to 6 M, the emission of (6,5) and (8,3) nanotubes with the MA-ST-90 corona exhibits a 15 to 35% reduction (Fig. 3 A and C). In contrast, nanotubes with the MA-ST-75 corona, which has a hydrophilic–hydrophobic ratio of 75:25, show a much smaller intensity modulation, and the modulation is not concentration dependent (Fig. 3 B and C). In addition to the intensity change, for the MA-ST-90 corona, the wavelength of (6,5) emission redshifts, which increases with the analyte concentration, reaching about 4 nm at 5 M, whereas the MA-ST-75 emission wavelength does not change (Fig. 3F). Besides the emission spectra, we have also scanned the excitation wavelengths using a supercontinuum laser and collected the excitation–emission spectra to examine the resonant emissions of nanotubes. Similar intensity and wavelength changes are observed for MA-ST-90 as in the emission spectra (Fig. 3D and SI Appendix). No major change is observed for MA-ST-75 (Fig. 3E and SI Appendix). Results from both spectroscopic methods demonstrate that the high hydrophilic–hydrophobic ratio is essential for the recognition of Vardenafil. Because a higher MA:ST ratio is associated with fewer hydrophobic “anchors,” such a necessity on the composition ratio indicates that the recognition interaction requires the corona to be relatively flexible. Flexibility ensures that when Vardenafil approaches, the polymer corona can adapt its configuration to enable a strong interaction with the analyte.
Fig. 3.
The hydrophilic–hydrophobic ratio of the polymer corona influences the recognition interaction. (A) The colormap shows the emission intensity modulation of nanotubes when Vardenafil of different concentrations is introduced to the MA-ST-90 corona. Nanotubes of (6,5) and (8,3) chirality show a gradual emission reduction as the concentration of Vardenafil increases. (B) The emission modulations with the MA-ST-75 corona, which has a lower hydrophilic–hydrophobic ratio, are much smaller and do not change with the analyte concentration. (C) When Vardenafil of different concentrations is introduced (1 to 6 M), the emission spectra of nanotubes show a strong intensity decrease and a wavelength shift, with the MA-ST-90 corona, but not with the MA-ST-75 corona. (D) The excitation–emission spectrum presents resonant emission peaks of SWNTs, where each peak corresponds to one chirality of carbon nanotubes, labeled with a pair of integers. The spectrum of the control is shown at Left (intensity normalized), and the relative change of the spectrum is shown at Right. Upon the introduction of Vardenafil, the emission of (6,5) and (8,3) nanotubes with MA-ST-90 corona exhibits strong reductions, similar to the trend in the emission spectra. (See SI Appendix for quantified wavelength shift.) (E) In contrast, the excitation–emission spectrum of nanotubes with MA-ST-75 corona does not show major intensity or wavelength change. (F) In the emission spectra, the peak of (6,5) nanotubes with MA-ST-90 corona redshifts as a result of Vardenafil binding (red circles). The magnitude of redshifts increases with the analyte concentration. However, spectra of MA-ST-75 corona do not present a peak shift, regardless of the analyte concentration (blue squares). Color bars in heat maps are all in units of percentage.
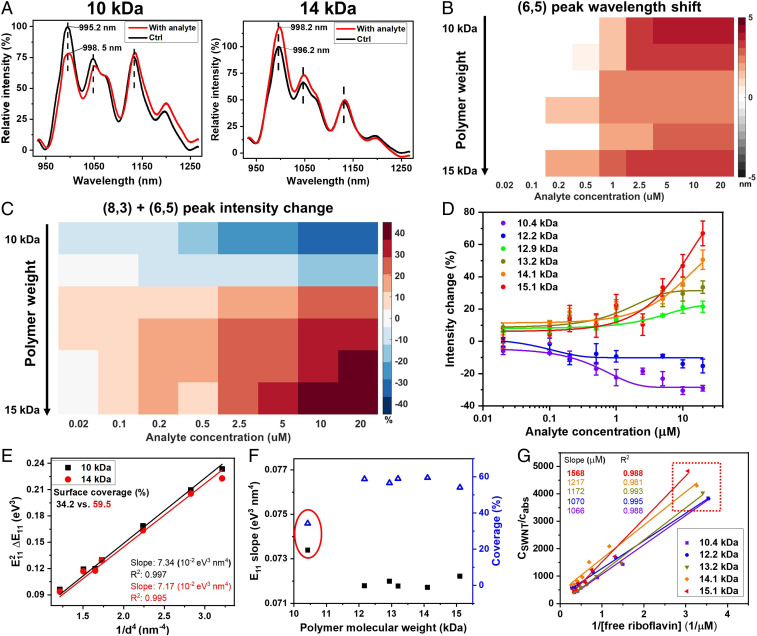
Changing polymer length can also modulate the response of corona phases to the analyte. Interestingly, when MA-ST-90 lengthens from 10 to 14 kDa (Mp), the emission intensity of (8,3) + (6,5) SWNTs changes from a reduction to an increase in response to Vardenafil (Fig. 4A; see SI Appendix for gel permeation chromatography results). Both systems show redshifts of the emission wavelengths. The trend is consistent when more polymers are examined and when different analyte concentrations are used. Six polymers with molecular weight from 10.4 to 15.1 kDa are used to prepare corona phases. As the polymer lengthens, the emission intensity modulation gradually changes from a turn-off to a turn-on response, regardless of the analyte concentration (Fig. 4C). For each polymer length, by calibrating the concentration-dependent emission intensity change, we extract the affinity between Vardenafil and the corona phase (Fig. 4D; the R-squared values for fittings are 0.97, 0.78, 0.96, 0.97, 0.88, and 0.93, from low to high polymer weight). For polymers of all lengths, the dissociation constant () values are on the order of micromolar, with the lowest being 0.5 M. The results demonstrate that the binding affinity between Vardenafil and our corona phase is in the range of antibody affinity (47, 48) and can be modulated by changing polymer length. The limits of detection range from 0.02 to 0.2 M. On the other hand, the wavelength shifts of (6,5) SWNTs are similar among all polymer lengths: all toward lower energy by 2 to 4 nm (Fig. 4B). The consistent wavelength change suggests that regardless of the polymer length, the binding of Vardenafil to the corona exerts a similar change on the dielectric environment surrounding SWNTs.
Fig. 4.
Polymer length modulates the corona phase’s response to the analyte. (A) The nIR emission spectra show that in response to 2 M Vardenafil, nanotubes wrapped with different lengths of poly(methacrylic acid-co-styrene) exhibit opposite emission intensity change. The 10-kDa polymer corona shows a turn-off response, while the 14-kDa corona shows a turn-on response. Both systems show redshifts of the emission wavelengths. (B) The colormap depicts the (6,5) peak shift in nanotubes’ emission spectra when polymer length varies. For each analyte concentration, corona phases made of MA-ST-90 of different lengths, from 10 to 15 kDa, show similar redshifts of the (6,5) peak. (C) For (6,5) + (8,3) SWNTs, when the polymer lengthens from 10 to 15 kDa, the emission intensity modulation gradually changes from a turn-off to a turn-on response, consistent over different Vardenafil concentrations. (D) The concentration-dependent intensity responses are calibrated to calculate the affinity between Vardenafil and the corona phase. The lowest dissociation constant () is 0.5 M, in the range of antibody affinity (47, 48), indicating a strong recognition interaction. (E) Solvatochromic plots are analyzed to extract the effective dielectric constant around SWNTs, for characterizing the polymer coverage on the nanotube surface (Eqs. 1–3). The calculated surface coverage by the 10-kDa polymer is 34.2% (black squares), much smaller than that of the 14-kDa polymer, which is 59.5% (red circles). (F) Based on the slopes of solvatochromic plots (black solid squares), polymers that are longer than 12 kDa have a greater coverage on nanotubes (about 60%, blue open triangles) than the shorter polymers. (G) In the probe adsorption method, the concentration ratio between SWNTs and adsorbed riboflavin () is plotted against the inverse of free riboflavin concentrations, where the linear slope is inversely proportional to the number of vacant sites on nanotubes. The linear slope increases as the polymer lengthens, indicating a decrease in the vacant sites on SWNTs. In addition, when coronas of different polymer lengths are mixed with the same amount of riboflavin (0.5 M), MA-ST-90 coronas of longer polymers correlate to higher concentrations of free dyes (red box), suggesting that these SWNTs have smaller exposed surface areas. This adsorption analysis reaches the same conclusion as the solvatochromic method: Longer polymers have higher surface coverage on SWNTs.
The different emission intensity modulations associated with polymers of different lengths result from different surface coverage of corona on the SWNTs. We used two methods to characterize the surface coverage of nanotubes: 1) an analysis of the solvatochromic shifts of SWNT emissions and 2) a technique based on the titration and adsorption of a probe molecule. The former method was introduced previously (33, 40) and characterizes solvatochromic shifts of resonant emissions for each chirality of nanotubes. The resonant emissions are extracted from excitation–emission spectra of SWNTs (Fig. 3D). Based on a semiempirical estimation (33, 40), when compared with pristine nanotubes in air or vacuum, the transition energy shift of the corona-wrapped SWNTs () is correlated to the effective dielectric constant () around nanotubes, which is approximated as a linear combination of the dielectric constants of water and polymer (Eqs. 1–3). The ratio of this linear combination () is determined by the surface coverage of SWNTs (see SI Appendix for details). In general, in the solvatochromic plot of vs. , where is the diameter of nanotubes, the bigger the slope is, the bigger the effective dielectric constant (Eqs. 1 and 2) and the smaller the surface coverage (Eq. 3). As shown in Fig. 4E, compared to the 10-kDa polymer, the 14-kDa polymer has a smaller slope in the solvatochromic plot, correlating to a smaller dielectric constant and a larger surface coverage (59.5%, in comparison to 34.2%). When comparing corona phases of variable polymer lengths (Fig. 4F), there is a decrease of slope in the solvatochromic plot and an increase of nanotube surface coverage, from less than 40 to 60%, as the polymer lengthens:
| [1] |
| [2] |
| [3] |
where E11 is the transition energy of corona-wrapped SWNTs in colloidal suspension, is the transition energy shift compared with pristine SWNTs in air or vacuum, is a fluctuation factor, is a scaling constant of the SWNT polarizability, is the nanotube’s diameter, is the slope in the linear fitting in Eq. 1, is the dielectric constant, is the reflective index, and is the percentage of coverage of nanotube surface.
We arrived at similar conclusions about the surface coverage by quantifying the hydrophobic dye adsorption to the exposed surface of SWNTs, using a technique recently developed in our laboratory (49). More specifically, as riboflavin is titrated with a SWNT colloidal solution, it adsorbs onto the exposed SWNT surface as a result of hydrophobic interactions, quenching the fluorescence of riboflavin. The adsorption amount is proportional to the exposed SWNT surface area, and can be quantified by calibrating the emission intensity of riboflavin. The linear plot in Fig. 4G is governed by the equation: , where is the concentration of SWNTs, is the concentration of adsorbed riboflavin, is the concentration of free riboflavin, is the dissociation constant of riboflavin adsorption, and is the number of vacant sites per carbon atom (see SI Appendix for detailed derivation). Because it is a physical adsorption process, it is reasonable to assume that is a constant among different coronas; therefore, the slope of the linear fitting, , is inversely proportional to the number of vacant sites ().
As highlighted in the red box in Fig. 4G, where the same amount of riboflavin (0.5 M) is introduced to SWNT solutions with different corona phases, the free riboflavin concentration increases as the corona polymer lengthens, indicating a reduction of nanotube surface exposure. In addition, as shown in Fig. 4G, the slopes of the linear fittings () increase from 1,066 to 1,568 M as the polymer lengthens, suggesting that the number of vacant sites () decreases. Thus, the exposed hydrophobic surface area decreases as the polymer lengthens, which is consistent with the results from solvatochromic shifts. Although the two methods have different mechanisms and assumptions and are not expected to directly correlate, they have demonstrated the same trend: the longer polymers have higher surface coverage on SWNTs. Overall, since shorter polymers have less coverage on SWNTs, Vardenafil’s binding to the corona phase is likely to interfere with the SWNTs’ bandgap and cause nIR emission quenching. When the polymers are longer, the surface coverage is higher and the interference with the bandgap is not efficient; thus the nIR fluorescence increases as a result of the modulation of corona configuration.
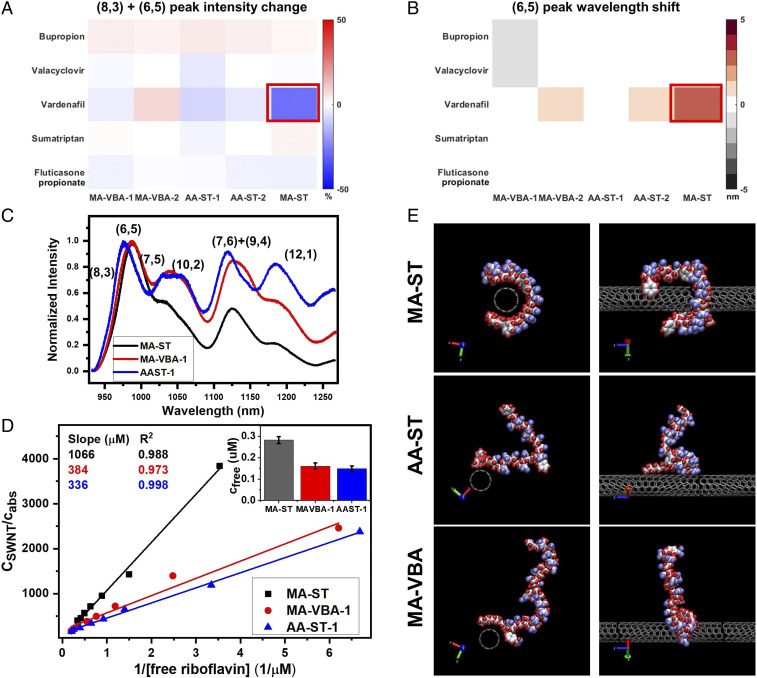
We find that methacrylic acid and styrene in the polymer composition are both essential for the corona phase to recognize Vardenafil. Two control polymers, poly(acrylic acid-co-styrene (AA-ST) and poly(methacrylic acid-co-vinylphenylboronic acid) (MA-VBA), are synthesized, replacing either the methacrylic acid or the styrene. Both polymers are made of a 90:10 hydrophilic–hydrophobic ratio like in MA-ST-90, with the polymer length around 10 kDa (AA-ST-1 and MA-VBA-1) or 15 kDa (AA-ST-2 and MA-VBA-2). None of these control polymers was able to generate a corona phase that recognizes Vardenafil, demonstrated by the lack of nIR spectral change of SWNTs when Vardenafil was introduced (Fig. 5 A and B). When analyzing the differences among these coronas, we found that both the AA-ST and MA-VBA corona phases exhibit similar emission intensity among different chiralities, whereas the MA-ST corona phase has a dominant (6,5) + (8,3) emission (Fig. 5C). Considering that the (6,5) + (8,3) nanotubes have the strongest interaction with Vardenafil (Figs. 2A and 3A), such a preference of small diameter nanotubes by the MA-ST corona appears critical for the recognition. On the other hand, when compared with AA-ST and MA-VBA, the MA-ST corona has a much higher surface coverage of SWNTs. As shown in Fig. 5 D, Inset, when 0.5 M riboflavin is mixed in SWNT solutions, less than 30% of the dyes are free when the corona is composed of control polymers, compared to more than 50% of dyes are free when the corona is made of MA-ST. This trend is consistent among different concentrations of riboflavin (Fig. 5D). In the adsorption plots, the slope of the MA-ST corona () is three times larger than those of MA-VBA and AA-ST coronas, indicating it has much smaller vacant sites on SWNTs and confirming that the MA-ST corona has a much higher surface coverage.
Fig. 5.
The monomer units, methacrylic acid and styrene, are essential in the polymer composition for the recognition of Vardenafil. (A and B) The emission intensity (A) and peak wavelength (B) modulations in nIR spectra when analytes are introduced to SWNT solutions with different corona phases. When methacrylic acid is replaced with acrylic acid, or when styrene is replaced with 3-vinylphenolboronic acid, the resulting corona phase AA-ST and MA-VBA do not show a strong interaction with Vardenafil. Neither the intensity (A) nor the wavelength (B) in emission spectra exhibits a substantial change. (C) The emission spectra of the corona phases reveal that MA-ST has a preference for small diameter nanotubes—dominant (8,3) and (6,5) peaks—while the other two coronas do not. (D) By titrating the riboflavin adsorption, we find that SWNTs with MA-ST coronas have a greater slope in the linear fitting, indicating a higher surface coverage of SWNTs compared to the MA-VBA and AA-ST coronas. When 0.5 M of riboflavin is introduced into different SWNT solutions, more than half of the dye molecules are free with the MA-ST corona, whereas less than 30% are free with the AA-ST or MA-VBA corona (Inset). (E) Molecular dynamics simulations reveal the corona configurations at thermal and density equilibrium: MA-ST has a higher surface coverage and forms a binding pocket on nanotubes, whereas AA-ST and MA-VBA do not. The shown configurations are representable of the configurations in the last 10 ns of the simulations, based on the rmsd analysis (see SI Appendix for details). Water molecules have been omitted from the simulation snapshots for clarity. Atom colors: C in nanotubes, gray; C in polymer, silver; O, blue; H, red; B, light gray.
In addition to experimental results, molecular dynamics simulations based on three starting polymer configurations reveal a “binding-pocket” configuration of the MA-ST corona on the SWNT surface, but not of the AA-ST or the MA-VBA corona. The configurations are simulated in the presence of water for 70 ns to reach thermal and density equilibrium in the NPT ensemble (T = 300 K, P = 1 bar), using the GROMACS package. A 2,200-Da segment for each polymer and one unit cell length of (6,5) SWNT are used in each simulation. Instead of starting with the polymer placed far off from the nanotube and simulating a very long molecular dynamics trajectory (50), in the initial configurations, the polymer was kept within 3 nm of the nanotube surface to facilitate quick binding. (For simulation details, please refer to SI Appendix.) In reaching equilibrium, the MA-ST polymer is shown to gradually wrap around the SWNT, reducing the solvent-accessible surface area from 15.03 to 13.43 0.17 nm2 and the polymer–nanotube distance to 1.88 nm. The resulting configuration forms a binding pocket enclosed by the nanotube and polymer surfaces (Fig. 5E). In contrast, the folding of AA-ST on the SWNT surface reduces the solvent access surface area from 15.03 to 14.59 0.10 nm2, a smaller reduction, and does not form a stable binding configuration. For MA-VBA, the solvent access surface area reduces from 15.03 to 14.54 0.23 nm2, without a binding configuration. All three polymers were extended into water in their initial configurations, and only MA-ST forms a binding-pocket configuration at the end. The higher surface coverage by the MA-ST corona is in agreement with the experimental results. Both the experimental and simulation results support that the specific recognition of Vardenafil results from the unique corona structure of MA-ST on nanotubes. Future simulation work could explore the free energetics of adsorption of Vardenafil onto the corona phase.
MA-ST corona phase in mimicking PDE5 for Vardenafil recognition.
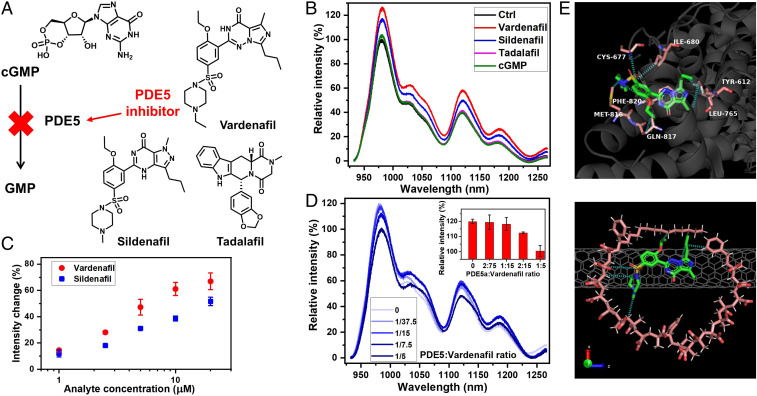
In comparison with the natural recognition site PDE5, MA-ST corona phase selectively binds to two inhibitors, Vardenafil and Sildenafil, and not to inhibitor Tadalafil and the substrate. As shown in Fig. 6A, the biochemical role of Vardenafil is to interrupt the enzymatic function of PDE5 in converting cGMP to GMP. Besides Vardenafil, Sildenafil and Tadalafil are also PED5 inhibitors. When these analytes are introduced into solutions of MA-ST-90 (15 kDa) wrapped nanotubes, only Vardenafil and Sildenafil cause strong spectral responses, whereas cGMP and Tadalafil did not (Fig. 6B), suggesting selective recognition of Vardenafil and Sildenafil. This observation aligns with their different interactions with PDE5: Sildenafil has a very similar chemical structure to Vardenafil and interacts with the same residues in the H loop of the catalytic site in PDE5 (43); Tadalafil, on the other hand, has a different structure and interacts mainly with the M loop of the catalytic site (43, 51). As the substrate, cGMP binds to both the catalytic site and the allosteric site, whereas the inhibitors do not, indicating a different binding mechanism of the substrate (52–54). The observed selectivity suggests that the MA-ST corona phase may be structurally similar, or have similar chemical interactions, to the H loop in PDE5. A further look at the affinity shows that Sildenafil consistently causes smaller emission modulations of SWNTs than Vardenafil (Fig. 6C), likely linked to a smaller binding affinity to the corona phase. This trend is comparable to the binding between PDE5 and Vardenafil being 10- to 40-fold stronger than the binding with Sildenafil (43), supporting the idea that the H loop and the MA-ST corona share structural or chemical similarities.
Fig. 6.
The MA-ST corona phase mimics PDE5 in its Vardenafil recognition. (A) The biochemical role of Vardenafil is to bind PDE5 and to interrupt its function in converting cGMP to GMP. The structures of cGMP, Vardenafil, and two other PDE5 inhibitors—Sildenafil and Tadalafil—are shown. All these four molecules interact with PDE5 directly. (B) The emission spectra show the nIR responses of the MA-ST corona phase, when PDE5 substrate (cGMP) and inhibitors are introduced. Only Vardenafil and Sildenafil exhibit strong interactions with the MA-ST corona. (C) The emission intensity modulations increase with the concentrations of Sildenafil and Vardenafil. The magnitude is notably smaller for Sildenafil. The error bars are shown for all data points. (D) The nIR spectra of MA-ST SWNTs in response to Vardenafil, when different concentrations of PDE5a are present. The PDE5a enzyme interferes with the interaction between Vardenafil and the MA-ST corona phase, gradually eliminating the spectral response as the enzyme concentration increases. Inset shows the relative intensities of the (6,5) + (3,8) emissions at different PDE5a concentrations. (E) Thermal and density equilibrated molecular dynamics simulation snapshot demonstrates the docking of Vardenafil in the binding pocket of MA-ST corona phase (Bottom). It shares similar intermolecular interactions (blue dashed lines) as when Vardenafil binds to PDE5a in the crystal structure (Top) (43). The shown configuration is representable of the configurations in the last 10 ns of the simulation, based on the rmsd analysis (see SI Appendix for details). Water molecules have been omitted from the simulation snapshots for clarity. Atom colors in Vardenafil: C, green; N, blue; O, red; S, yellow. Atom colors in polymer: C, light red; O, red; H, white. Atom colors in PDE5a residues: C, light red; N, blue; O, red; S, yellow. C atoms in nanotubes are in gray.
In addition to the selectivity, in the presence of enzyme interference, the interaction between the MA-ST corona and Vardenafil is outcompeted. When Vardenafil was introduced into a colloidal solution of MA-ST SWNTs along with PDE5a (a subset of PDE5), Vardenafil’s binding with the corona phase gradually weakens as the PDE5a concentration increases (Fig. 6D). When the molar ratio between PDE5a and Vardenafil reaches 1:5, no nIR spectral change of nanotubes is observed, suggesting that PDE5a outcompetes the corona phase. This result underscores that PDE5a has a higher binding affinity with Vardenafil than that of the corona phase, likely due to the high flexibility of the catalytic H loop and its strong intermolecular force with Vardenafil (43). This disruption of recognition, in the presence of a competitor of higher binding affinity, further illustrates that the MA-ST corona phase has a specific interaction with Vardenafil.
Molecular dynamics simulations reveal that Vardenafil docks into an identified binding pocket of the MA-ST corona phase, resulting in the recognition (Fig. 6 E, Bottom). The initial polymer configuration is the simulated configuration shown in Fig. 5E. The recognition shares similar intermolecular interactions to those when Vardenafil binds to PDE5a (Fig. 6 E, Top) (43). The propyl-imidazotriazinone of Vardenafil stacks on aromatic rings in SWNTs, similar to their interaction with residue Phe820 in the enzyme. It also feels the van der Waals force from the polymer backbone as with residue Leu765. The ethoxy group orients to a hydrophobic pocket formed by the nanotube surface and the polymer backbone. The phenyl group experiences stacking with SWNTs. The sulfonamide group interacts with the carboxylic acid group in the MA-ST polymer as with residue Cys677 in the enzyme. The ethylpiperazine group orients toward the polymer. Through the recognition process, the underlying SWNT gains more coverage (reducing the solvent accessible surface area from 13.43 0.17 nm to 12.98 0.40 nm2), and the distance between Vardenafil and SWNT is reduced from 1.90 to 1.22 nm. The rmsd of atomic positions of Vardenafil in these two environments is small (0.25 nm), indicating an extent of similarity between Vardenafil binding to the MA-ST corona phase and to the PDE5a enzyme. We also observe that the corona adopts a slightly different configuration compared to that before the binding, supporting our experimental observation that some flexibility is needed in the polymer composition to facilitate the recognition. Similar configurational changes are known to occur when Vardenafil binds to the H loop in PDE5 (55), further illustrating the binding similarity of our corona phase and the natural enzyme.
Such a synthetic mimic of enzyme binding enabled by corona phase recognition will be invaluable for both fundamental research and biomedical applications. The more immediate application is to use the nanostructure as an analytical method to detect and analyze target therapeutics, in fundamental research and in drug production. Its sensitivity, speed, potential for miniature devices, and capacity for label-free examination are features desired for sensors in these applications. In the long term, corona phase recognition may serve as a platform to study endogenous biomarkers, such as examining their transient interactions in vitro/in vivo and studying cell surface receptors. Given the complexity of therapeutic modalities, analytical methods like the one presented here will enable capacities to study unmet biomedical needs and will be in strong demand to tackle the challenges.
Summary
We have generated a synthetic mimic of PDE5 to bind its inhibitor Vardenafil, using single-walled carbon nanotube templated corona phase recognition. Among a library of amphiphilic polymers, the corona phase made of poly(methacrylic acid-co-styrene) stands out for its specific binding to Vardenafil. The interaction is proved to be highly sensitive to the polymer structure: The change of polymer composition sabotages the recognition; and the change of polymer length modulates the spectral response. In comparison with the natural binding enzyme, the MA-ST corona phase recognizes only Vardenafil and a structure very similar to it, but not the other inhibitor or substrate, indicating that the corona shares similar properties to the H loop of PDE5. The corona phase has a smaller binding affinity than the enzyme, as the interaction is disrupted in the presence of PDE5a. Molecular dynamics simulations based on three starting polymer configurations demonstrate that the recognition is associated with a binding pocket in the MA-ST corona on nanotubes, which is responsible for the intermolecular interactions with Vardenafil. This unique nanostructure presents a stable, sensitive artificial recognition site with tunable affinity for inhibitors and will open up possibilities for pharmaceutical and biological applications.
Materials and Methods
Please see SI Appendix for materials, characterization methods, polymer synthesis and characterization, polymer–SWNT suspension, fluorescence spectrum collection, molecular dynamics simulations methods, dye adsorption quantification, and supplementary plots.
Supplementary Material
Acknowledgments
This work is supported by GlaxoSmithKline plc. We have used topology files from Automated Topology Builder and Repository and are grateful for their support.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920352117/-/DCSupplemental.
Data Availability.
All data are available in the main text and SI Appendix.
References
- 1.Mahon C. S., Fulton D. A., Mimicking nature with synthetic macromolecules capable of recognition. Nat. Chem. 6, 665–672 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Jin H., et al. , Detection of single-molecule H2O2 signaling from epidermal growth factor receptor using fluorescent single-walled carbon nanotubes. Nat. Nanotechnol. 5, 302–309 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voskuil J. L., The challenges with the validation of research antibodies. F1000Res 6, 161 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zuo X., Xiao Y., Plaxco K. W., High specificity, electrochemical sandwich assays based on single aptamer sequences and suitable for the direct detection of small-molecule targets in blood and other complex matrices. J. Am. Chem. Soc. 131, 6944–6945 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiang Y., Lu Y., Using personal glucose meters and functional DNA sensors to quantify a variety of analytical targets. Nat. Chem. 3, 697–703 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wackerlig J., Lieberzeit P. A., Molecularly imprinted polymer nanoparticles in chemical sensing–Synthesis, characterization and application. Sensor. Actuator. B Chem. 207, 144–157 (2015). [Google Scholar]
- 7.Zhu G., et al. , Self-assembled, aptamer-tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics. Proc. Natl. Acad. Sci. U.S.A. 110, 7998–8003 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chinai J. M., et al. , Molecular recognition of insulin by a synthetic receptor. J. Am. Chem. Soc. 133, 8810–8813 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chames P., Van Regenmortel M., Weiss E., Baty D., Therapeutic antibodies: Successes, limitations and hopes for the future. Br. J. Pharmacol. 157, 220–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beck A., Wurch T., Bailly C., Corvaia N., Strategies and challenges for the next generation of therapeutic antibodies. Nat. Rev. Immunol. 10, 345–352 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Uhlen M., et al. , A proposal for validation of antibodies. Nat. Methods 13, 823–827 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sydow J. F., et al. , Structure-based prediction of asparagine and aspartate degradation sites in antibody variable regions. PLoS One 9, e100736 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haywood J., Mozziconacci O., Allegre K. M., Kerwin B. A., Schöneich C., Light-induced conversion of trp to gly and gly hydroperoxide in IgG1. Mol. Pharm. 10, 1146–1150 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vlasak J., Ionescu R., Fragmentation of monoclonal antibodies. MAbs 3, 253–263 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen M., Rusling J., Dixit C. K., Site-selective orientated immobilization of antibodies and conjugates for immunodiagnostics development. Methods 116, 95–111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neeli I., Radic M., Current challenges and limitations in antibody-based detection of citrullinated histones. Front. Immunol. 7, 528 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McEnaney P. J., et al. , Chemically synthesized molecules with the targeting and effector functions of antibodies. J. Am. Chem. Soc. 136, 18034–18043 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z., Mohan Nalluri S. K., Fraser Stoddart J., Surveying macrocyclic chemistry: From flexible crown ethers to rigid cyclophanes. Chem. Soc. Rev. 46, 2459–2478 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Langton M. J., Beer P. D., Rotaxane and catenane host structures for sensing charged guest species. Acc. Chem. Res. 47, 1935–1949 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Han Y., Meng Z., Ma Y. X., Chen C. F., Iptycene-derived crown ether hosts for molecular recognition and self-assembly. Acc. Chem. Res. 47, 2026–2040 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Späth A., König B., Molecular recognition of organic ammonium ions in solution using synthetic receptors. Beilstein J. Org. Chem. 6, 32 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shetty D., Khedkar J. K., Park K. M., Kim K., Can we beat the biotin–avidin pair?: Cucurbit[7]uril-based ultrahigh affinity host–guest complexes and their applications. Chem. Soc. Rev. 44, 8747–8761 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Chen L., Wang X., Lu W., Wu X., Li J., Molecular imprinting: Perspectives and applications. Chem. Soc. Rev. 45, 2137–2211 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Cieplak M., Kutner W., Artificial biosensors: How can molecular imprinting mimic biorecognition? Trends Biotechnol. 34, 922–941 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Pan J., Chen W., Ma Y., Pan G., Molecularly imprinted polymers as receptor mimics for selective cell recognition. Chem. Soc. Rev. 47, 5574–5587 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Robertson E. J., et al. , Design, synthesis, assembly, and engineering of peptoid nanosheets. Acc. Chem. Res. 49, 379–389 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Kodadek T., McEnaney P. J., Toward vast libraries of scaffold-diverse, conformationally constrained oligomers. Chem. Commun. 52, 6038–6059 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gotrik M. R., Feagin T. A., Csordas A. T., Nakamoto M. A., Soh H. T., Advancements in aptamer discovery technologies. Acc. Chem. Res. 49, 1903–1910 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Willner I., Shlyahovsky B., Zayats M., Willner B., DNAzymes for sensing, nanobiotechnology and logic gate applications. Chem. Soc. Rev. 37, 1153–1165 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Das S., et al. , A general synthetic approach for designing epitope targeted macrocyclic peptide ligands. Angew. Chem. Int. Ed. 54, 13219–13224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gray B. P., Brown K. C., Combinatorial peptide libraries: Mining for cell-binding peptides. Chem. Rev. 114, 1020–1081 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadowsky J. D., et al. , (/+)-peptide antagonists of BH3 domain/bcl-xL recognition: Toward general strategies for foldamer-based inhibition of protein-protein interactions. J. Am. Chem. Soc. 129, 139–154 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Bisker G., et al. , Protein-targeted corona phase molecular recognition. Nat. Commun. 7, 10241 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J., et al. , Molecular recognition using corona phase complexes made of synthetic polymers adsorbed on carbon nanotubes. Nat. Nanotechnol. 8, 959–968 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iverson N. M., et al. , In vivo biosensing via tissue-localizable near-infrared-fluorescent single-walled carbon nanotubes. Nat. Nanotechnol. 8, 873–880 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kruss S., et al. , Neurotransmitter detection using corona phase molecular recognition on fluorescent single-walled carbon nanotube sensors. J. Am. Chem. Soc. 136, 713–724 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Bachilo S. M., Structure-assigned optical spectra of single-walled carbon nanotubes. Science 298, 2361–2366 (2002). [DOI] [PubMed] [Google Scholar]
- 38.O’Connell M. J., et al. , Band gap fluorescence from individual single-walled carbon nanotubes. Science 297, 593–596 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Kruss S., et al. , Carbon nanotubes as optical biomedical sensors. Adv. Drug Deliv. Rev. 65, 1933–1950 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Choi J. H., Strano M. S., Solvatochromism in single-walled carbon nanotubes. Appl. Phys. Lett. 90, 223114 (2007). [Google Scholar]
- 41.Dong J., Salem D. P., Sun J. H., Strano M. S., Analysis of multiplexed nanosensor arrays based on near-infrared fluorescent single-walled carbon nanotubes. ACS Nano 12, 3769–3779 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Rotella D. P., Phosphodiesterase 5 inhibitors: Current status and potential applications. Nat. Rev. Drug Discov. 1, 674–682 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Wang H., Ye M., Robinson H., Francis S. H., Ke H., Conformational variations of both phosphodiesterase-5 and inhibitors provide the structural basis for the physiological effects of Vardenafil and sildenafil. Mol. Pharmacol. 73, 104–110 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andersson K. E., PDE5 inhibitors–Pharmacology and clinical applications 20 years after sildenafil discovery. Br. J. Pharmacol. 175, 2554–2565 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patel D. N., et al. , Screening of synthetic PDE-5 inhibitors and their analogues as adulterants: Analytical techniques and challenges. J. Pharma. Biomed. Anal. 87, 176–190 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Budhathoki-Uprety J., et al. , Synthetic molecular recognition nanosensor paint for microalbuminuria. Nat. Commun. 10, 3605 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishikori S., et al. , Broad ranges of affinity and specificity of anti-histone antibodies revealed by a quantitative peptide immunoprecipitation assay. J. Mol. Biol. 424, 391–399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barderas R., Desmet J., Timmerman P., Meloen R., Casal J. I., Affinity maturation of antibodies assisted by in silico modeling. Proc. Natl. Acad. Sci. U.S.A. 105, 9029–9034 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park M., et al. , Measuring the accessible surface area within the nanoparticle corona using molecular probe adsorption. Nano Lett. 19, 7712–7724 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shan Y., et al. , How does a drug molecule find its target binding site?. J. Am. Chem. Soc. 133, 9181–9183 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cahill K. B., Quade J. H., Carleton K. L., Cote R. H., Identification of amino acid residues responsible for the selectivity of Tadalafil binding to two closely related phosphodiesterases, PDE5 and PDE6. J. Biol. Chem. 287, 41406–41416 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rybalkin SD., Rybalkina I. G., Shimizu-Albergine M., Tang X. B., Beavo J. A., PDE5 is converted to an activated state upon cGMP binding to the GAF A domain. EMBO J. 22, 469–478 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okada D., Asakawa S., Allosteric activation of cGMP-specific, cGMP-binding phosphodiesterase (PDE5) by cGMP. Biochemistry 41, 9672–9679 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Turko I. V., Francis S. H., Corbin J. D., Binding of cGMP to both allosteric sites of cGMP-binding cGMP-specific phosphodiesterase (PDE5) is required for its phosphorylation. Biochem. J. 329, 505–510 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang H., et al. , Multiple conformations of phosphodiesterase-5: Implications for enzyme function and drug development. J. Biol. Chem. 281, 21469–21479 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text and SI Appendix.