Abstract
The analysis of T cell lipid raft proteome is challenging due to the highly dynamic nature of rafts and the hydrophobic character of raft-resident proteins. We explored an innovative strategy for bottom-up lipid raftomics based on suspension-trapping (S-Trap) sample preparation. Mouse T cells were prepared from splenocytes by negative immunoselection, and rafts were isolated by a detergent-free method and OptiPrep gradient ultracentrifugation. Microdomains enriched in flotillin-1, LAT, and cholesterol were subjected to proteomic analysis through an optimized protocol based on S-Trap and high pH fractionation, followed by nano-LC-MS/MS. Using this method, we identified 2,680 proteins in the raft-rich fraction and established a database of 894 T cell raft proteins. We then performed a differential analysis on the raft-rich fraction from nonstimulated versus anti-CD3/CD28 T cell receptor (TCR)-stimulated T cells. Our results revealed 42 proteins present in one condition and absent in the other. For the first time, we performed a proteomic analysis on rafts from ex vivo T cells obtained from individual mice, before and after TCR activation. This work demonstrates that the proposed method utilizing an S-Trap-based approach for sample preparation increases the specificity and sensitivity of lipid raftomics.
Keywords: immunology, lymphocytes, membranes, tandem mass spectrometry, cell signaling, detergent-free, filter-aided sample preparation, OptiPrep, raftomics
Rafts (lipid rafts or membrane rafts) are subcellular entities defined as dynamic lateral membrane microdomains (1, 2). Their formation and dynamics have been associated, by a strong body of experimental evidence, with the regulation of cellular functions. Nevertheless, for many years their existence has been questioned owing to: i) the fact that their definition is based on experimental concepts, such as detergent resistance; and ii) the limited sensitivity of the methods available for their study. A consensus official definition resulted from a Keystone Symposium back in 2006 (3). This definition establishes their size limits (10–200 nm) and composition (sterol- and sphingolipid-enriched domains), as well as their highly dynamic nature.
If almost any cell type can contain raft-like structures, the scientific community has widely accepted and integrated this concept in the context of lymphocytes. Compelling evidence indicates that T cell activation encompasses membrane reorganization events involving lipid and protein rearrangement, as well as specific protein interactions that orchestrate the clustering of cholesterol and sphingolipid-rich domains (4). It is broadly acknowledged that raft dynamics play a significant and direct role in T cell activation via T cell receptor (TCR) stimulation (5–13) and T cell migration induction via chemokine receptor stimulation (14–19).
The comprehensive characterization of the lipid raft proteome of T cells has been a challenging goal due to difficulties in the isolation of microdomains, their highly dynamic nature, and the hydrophobic character of raft-resident proteins. Nevertheless, several groups have addressed this question using state-of-the-art approaches significantly contributing to deciphering some of the mechanisms involved in T cell activation (4, 9, 17, 20–27) [reviewed in (23, 28–30)]. The numerous studies on rafts in any cell types published to date [reviewed in (31, 32)] have contributed to the generation of a curated growing mammalian lipid raft protein database (www.raftprot.org) (33, 34). This includes proteins identified by biochemical approaches, either positively by biochemical isolation or negatively by cyclodextrin-based raft-disruption experiments. Proteins identified in proteomic studies are also included, although it is specified whether their presence in rafts has been validated or not by alternative methods. This underlines the issue of purity and contamination in raft preparations.
The golden standard methods for raft isolation are based on, first, cell activity arrest at 4°C followed by differential solubilization of membrane microdomains in nonionic detergents, such as Triton X-100, Lubrol WX, and Brij 35 (35). This procedure may introduce a serious artifactual bias in that detergents are likely to alter membrane properties and, consequently, experimental resistant domains might differ significantly from those existing in physiologic conditions (36). It has been shown that detergent-based methods scramble lipid content and therefore create new artifactual detergent-resistant lipid rafts (37). Detergent-free methods have been developed to minimize this problem. They are based on membrane fragmentation by physical methods, like ultrasound treatment of cells (38) or immunoisolation targeting specific raft-resident proteins (36). These methods are considered to yield a better purity of raft fractions (38).
Detergent treatment of cell membranes is followed by ultracentrifugation on a discontinuous density gradient. Sucrose gradients are usually performed overnight, which represents a timely limitation. Shorter preparation procedures include the use of colloidal solutions, such as OptiPrep™ (36, 39, 40). The latter provides a rapid, highly reproducible, and selective isolation of raft-like microdomains, where selectivity is demonstrated by raft marker enrichment. However, OptiPrep gradients have been scarcely used as a preparation step for proteomics. The few reported cases to date correspond to proteomic analyses on extracellular vesicles (41–49), exosomes (50–53), mitosomes (54), isolated insulin secretory granules (55), synaptosomes (56), and plant organelles (57–60). To date, OptiPrep isolation has been applied to raft proteomics in a study on virus-infected algae (61). Globally, OptiPrep interferes with the LC-MS/MS analysis and needs to be eliminated thoroughly from the sample. However, efficient cleanup steps, like SDS-PAGE short separation, lead to protein loss and are not compatible with very low abundant samples, such as T cell rafts from one single mouse. Hence, there is a need for a simplified and sensitive method for proteomic analysis of T cell rafts.
In this work, we explore an innovative strategy, based on a single device suspension-trapping (S-Trap) preparation technique for proteomics on raft-like [flotillin-1-, linker for activation of T cells (LAT)-, and cholesterol-rich] microdomains. For the first time, a global proteomic analysis is performed on purified rafts from ex vivo mouse T cells, before and after activation by TCR costimulation. Our results show an increased specificity and sensitivity of the proposed method. In addition, we have created a new database of T cell raft proteins.
MATERIALS AND METHODS
Cell culture
HEK 293 cells (American Type Culture Collection) were maintained in Dulbecco’s modified Eagle’s medium containing 10% FCS. Mouse podocytes were cultured in RPMI 1640 medium containing 10% FCS, penicillin (100 U/ml), streptomycin (100 mg/ml), and interferon-g (50 U/ml) at 33°C. Differentiation was induced by maintaining stable podocyte cell lines at 37°C without interferon-g for 14 days in the presence of blasticidin and zeocin. Jurkat T cells (American Type Culture Collection) were cultured in RPMI 1640 medium supplemented with 10% FCS, penicillin (100 U/ml), and streptomycin (100 mg/ml) at 37°C.
Mouse T cell isolation and synchronization
Balb/C mice were bred following the standards established by the National Ethics Committee (COMETH) under accreditation number 29/01/13-1. Four individual mice were used as biological replicates (n = 4). After euthanization, spleens were harvested, gently minced with a scalpel, and passed through a 40 μM nylon mesh filter. T cells were isolated by negative immunoselection using the Pan T Cell isolation kit (Miltenyi Biotec GmbH, Germany). Immunoselected cells were confirmed as CD4+ and CD3+ by flow cytometry (supplemental Fig. S1). Before stimulation, T cells were synchronized at the G1 phase of the cell cycle by serum starvation in 2% FCS for 6 h. Synchronized T cells were then activated for 15 min with soluble 1 μg/ml anti-CD3 and anti-CD28 (eBiosciences, San Diego, CA) in RPMI complete medium supplemented with 10% FCS.
Lipid raft preparation using OptiPrep™ and sucrose gradients
Lipid raft-like microdomains were obtained by a detergent-free method based on the one described by McDonald and Pike (38). Between one and five million cells per sample were washed twice in cold PBS, resuspended in 800 μl of MBS/Na2CO3 buffer [25 mM MES, 150 mM NaCl, 250 mM Na2CO3 (pH 6); supplemented with 1 mM PMSF and phosphatase and protease inhibitor cocktails] and lysed by passaging 20 times through a 21 gauge needle, followed by sonication three times for 60 s in a Vibra Cell 75022 sonicator. The homogenate was mixed with two volumes of either 60% OptiPrep™ (Axis Shield) or 60% sucrose for a final volume of 2 ml of either 40% OptiPrep™ or 40% sucrose. A three-step discontinuous density gradient was made by sequentially placing 2 ml of either 30% OptiPrep™ or 30% sucrose in MBS/Na2CO3 buffer, and 1 ml of 5% OptiPrep™ or 5% sucrose sequentially on top of the homogenate. The mixture was spun in a TL-100 rotor at 268,000 g for 2 h in an Optima MAX-XP ultracentrifuge (Beckman Coulter). After spinning, one fraction of 600 μl followed by five fractions of 900 μl were collected from top to bottom. Fraction 2 containing rafts was subjected to subsequent analysis.
To analyze the distribution of flotillin-1, fractions were precipitated by addition of 10% trichloroacetic acid (final concentration), incubated overnight at −20°C, and washed three times in cold ethanol. The resulting dry protein pellets were solubilized in equal volumes of 1× Laemmli buffer and analyzed by Western blot.
Filter-aided sample preparation
Filter-aided sample preparation (FASP) was performed on OptiPrep™ raft fractions according to (62). Briefly, samples were reduced with 0.1 M DTT at 60°C for 1 h. Proteins were transferred to Microcon filter units (30 kDa cut-off) and washed twice with 200 μl of UA buffer [0.1 M Tris, 8 M urea (pH 8.9)] and concentrated by centrifugation at 14,000 g for 15 min. Proteins were alkylated with 100 μl of IAA buffer [0.05 M iodoacetamide, 0.1 M Tris (pH 8.9)] at room temperature in the dark for 20 min and centrifuged at 14,000 g for 10 min. Proteins were then washed twice by adding 100 μl of UA buffer before centrifugation at 14,000 g for 10 min, and twice with 100 μl of ABC buffer (0.05 M NH4HCO3) before centrifugation at 14,000 g for 10 min. Filter units were transferred to new collection tubes and samples were incubated with 40 μl of ABC buffer containing 1.6 μg of trypsin in a humidity chamber at 37°C for 18 h. Tubes were centrifuged at 14,000 g for 10 min, 40 μl of ABC buffer were added, and tubes were centrifuged again. Peptides were finally recovered in collection tubes.
Suspension trapping
S-Trap™ micro spin column digestion was performed on OptiPrep™ raft fractions according to the manufacturer’s protocol. Briefly, proteins were precipitated overnight using a 10% TCA final concentration and washed four times with cold ethanol. Proteins were resuspended and solubilized in 5% SDS, 50 mM triethylammonium bicarbonate (TEAB; pH 7.55), reduced with 100 mM DTT solution, and alkylated with the addition of iodoacetamide to a final concentration of 40 mM. Aqueous phosphoric acid was added to a final concentration of 1.2%. Colloidal protein particulate was formed with the addition of 231 μl of S-Trap binding buffer [90% aqueous methanol, 100 mM TEAB (pH 7.1)]. The mixture was placed on S-Trap micro 1.7 ml columns and centrifuged at 4,000 g for 10 s. Columns were washed five times with 150 μl of S-Trap binding buffer and centrifuged at 4,000 g for 10 s with 180° rotation of the columns between washes. Samples were digested with 2 μg of trypsin (Promega) at 47°C for 1 h. Peptides were eluted with 40 μl of 50 mM TEAB followed by 40 μl of 0.2% aqueous formic acid and by 35 μl of 50% acetonitrile containing 0.2% formic acid. Peptides were finally vacuum dried.
High pH fractionation
For library building, peptides were resuspended for high pH fractionation in 50 μl of 0.1% trifluoroacetic acid. Tips were homemade with one layer of Empore disk C8 and 1 mg of C18 (C18-AQ, Maisch). After washing and conditioning of C18, peptides were bound by centrifugation to C18 in acidic conditions. Peptides were sequentially eluted in eight fractions at basic pH (5, 7.5, 10, 12.5, 15, 17.5, 20, and 50% ACN in triethylamine 0.1%). Eluted peptides were concatenated pairwise to obtain four final fractions (F1F5, F2F6, F3F7, and F4F8). Samples were then vacuum dried.
Automated capillary immunoassay (WES)
Automated capillary immunoassay (Simple Western) was performed on a Western immunoassay (WES) system (Protein Simple, San Jose, CA). Akt (Cell Signaling Technologies) and Nck1/2 (Santa Cruz Biotechnology) antibodies were used at a 1:50 dilution. The analyses were performed on a 12–230 kDa separation module (SM-W004) according to the manufacturer’s instructions.
Nano-LC-MS/MS protein identification and quantification
Samples were resuspended in 35 μl of 1% ACN, 0.1% trifluoroacetic acid in HPLC-grade water. For each run, 5 μl were injected in a nanoRSLC-Q Exactive PLUS (RSLC Ultimate 3000) (Thermo Scientific, Waltham MA). Peptides were loaded onto a μ-precolumn (Acclaim PepMap 100 C18, cartridge, 300 μm ID. × 5 mm, 5 μm) (Thermo Scientific) and separated on a 50 cm reversed-phase LC column (0.075 mm ID; Acclaim PepMap 100, C18, 2 μm) (Thermo Scientific). Chromatography solvents were (A) 0.1% formic acid in water, and (B) 80% acetonitrile and 0.08% formic acid. Peptides were eluted from the column with the following gradient: 5% to 40% B (120 min), 40% to 80% (5 min). At 125 min, the gradient returned to 5% to re-equilibrate the column for 20 min before the next injection. Two blanks were run between samples to prevent sample carryover. Peptides eluting from the column were analyzed by data-dependent MS/MS, using a top-10 acquisition method. Peptides were fragmented using higher-energy collisional dissociation. Briefly, the instrument settings were as follows: resolution was set to 70,000 for MS scans and 17,500 for the data-dependent MS/MS scans in order to increase speed. The MS automatic gain control target was set to 3.106 counts with maximum injection time set to 200 ms, while MS/MS automatic gain control target was set to 1.105 with maximum injection time set to 120 ms. The MS scan range was from m/z 400 to 2,000. Dynamic exclusion was set to 30 s duration. Three separate MS runs (i.e., technical replicates) were acquired for each biological replicate under the identical mass spectrometric conditions to account for instrument-related variability and to improve accuracy of the label-free quantification.
MS data processing and bioinformatic analysis
MS data processing and bioinformatics were done as previously described with some modifications (63). Briefly, raw MS files were processed with the MaxQuant software version 1.5.2.8 and searched with the Andromeda search engine against the human UniProt database (release May 2019, 20,199 entries). To search for parent mass and fragment ions, we set the mass deviation at 4.5 and 20 ppm, respectively. The minimum peptide length was set to seven amino acids and strict specificity for trypsin cleavage was required, allowing up to two missed cleavage sites. Match between runs was allowed. Carbamidomethylation (Cys) was set as fixed modification, whereas oxidation (Met) and protein N-terminal acetylation were set as variable modifications. The false discovery rates at the protein and peptide level were set to 1%. Scores were calculated in MaxQuant as described previously (63).
Statistical and bioinformatic analyses, including heatmaps, were performed with Perseus software (version 1.5.5.3) (freely available at www.perseus-framework.org). Gene Ontology annotation was performed on Perseus software. Proteins for Gene Ontology analysis were selected if annotated with the terms membrane, mitochondrion, and nucleus in the Gene Ontology Slim Cellular Component database. RaftProt comparison was performed with the mouse database (freely available at raftprot.org).
For the T cell raft database, we used protein intensities to calculate a ratio between fraction 2 and the mean of the other fractions (fractions 3, 4, 5, and 6). Proteins with a ratio superior or equal to two were classified as “enriched in rafts,” and proteins with ratio inferior to two were classified as “non-enriched in rafts”.
For the T cell raft analysis in resting versus activating conditions, we selected proteins based on the following criteria: proteins were detected in all four samples in one condition and completely absent in the other condition. Proteins only identified by site, matching the reverse database and the potential contaminant database were filtered out. Selected proteins were identified with at least two peptides and at least five MS/MS to ensure robust identification of the proteins.
Proteome Discoverer™ software (Thermo Scientific, version 1.4) was used to evaluate the number of identified proteins for the different precipitation tests.
The MS proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD016476 and MS Viewer.
RESULTS
Membrane raft preparation
We optimized membrane raft preparation starting from T cells immunoselected from a single mouse. We aimed to perform biological experiments on single mice rather than on pooled mice in order to maintain higher statistical power. Membrane raft proteins were enriched by ultracentrifugation using an OptiPrep™ density gradient on a limited number of cells (2–10 million) after a detergent-free cell disruption.
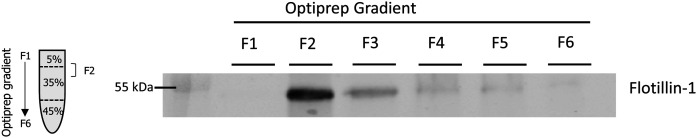
We optimized the method in order to collect raft proteins in a single fraction by adapting the volumes of fractions recovered after ultracentrifugation. As lipid rafts are enriched in flotillin-1, a protein that constitutes assembly sites for active signaling platforms (64), we first evaluated the distribution of flotillin-1 in the gradient fractions. As shown in Fig. 1, a strong flotillin-1 signal was detected in fraction 2, with lesser amounts in soluble protein-rich fractions 3–6, providing support that fraction 2 from the OptiPrep™ gradient is actually enriched in the membrane rafts.
Fig. 1.
Optimization of OptiPrep™ gradient and detection of lipid raft marker, flotillin-1, in fractions obtained from Jurkat T cells. Cells were lysed and subjected to OptiPrep density gradient separation. The diagram on the left indicates the gradient region expected to contain rafts. Six hundred microliter fractions collected from top to bottom were analyzed by Western blot with an antibody recognizing the raft marker flotillin-1. Optimization of gradient consisted of pooling fractions 2, 3, and 4 into a single combined fraction 2. The Western blot analysis of all fractions shows a strong signal for flotillin-1 in combined fraction 2.
Proteomic analysis of membrane rafts following different sample preparation methods
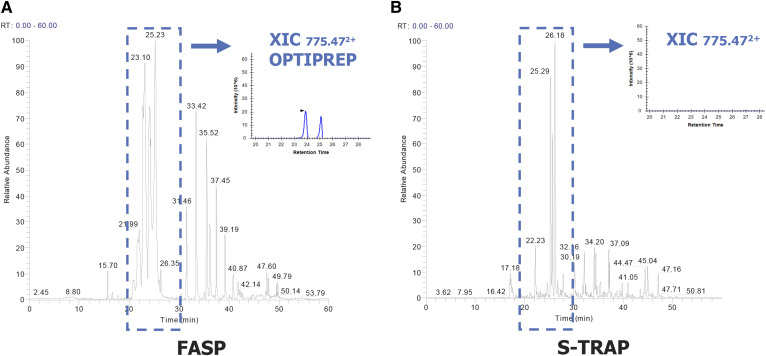
In order to identify the proteins contained in the raft-enriched fraction (F2) by LC-MS/MS analysis, we performed a classic workflow based on FASP, as we previously described (62). However, we encountered an unexpected challenge in analyzing this fraction. FASP-digested raft proteins yielded an inconsistent number of identified proteins, and flotillin-1 was not detected at all, possibly due to OptiPrep™ contamination interfering with MS analysis. Indeed, a close analysis of the base peak chromatogram of the F2 raft fraction revealed the strong presence of a doubly charged ion, m/z 775.47, corresponding to the OptiPrep™ compound iodixanol (Fig. 2A).
Fig. 2.
Comparison of FASP and S-Trap protein digestion on mouse T cell lipid rafts isolated by OptiPrep™ gradient. A: Base peak chromatogram of fraction 2 after digestion by the FASP method. The extracted ion chromatogram (in blue) shows an iodixanol ion (OptiPrep™, doubly charged ion m/z 775,47). B: Base peak chromatogram of fraction 2 digested by the S-Trap method. No iodixanol ion was detected, as shown by the extracted ion chromatogram.
To remove this persistent contamination, we combined FASP with complementary cleaning-up steps before digestion. We conducted the optimization tests on two immortalized cell lines, HEK298 cells and mouse podocytes, which constituted a nonrestricted starting material. In a first attempt we performed TCA precipitation before FASP digestion, but we could still not eliminate contamination. Also, we used an additional 3 h-long ultracentrifugation step on F2 fractions, in order to wash out contaminants and spin down membrane rafts, followed by TCA precipitation and FASP digestion. OptiPrep™ was still present when we associated these three steps together (supplemental Fig. S2). Other types of digestion were performed, including in-solution digestion and in-gel digestion, all of them leading to similar results (data not shown).
We decided to test a more recent sample preparation method, S-Trap, based on suspension-trapping filters that potentially facilitate the washing out of contaminants, such as OptiPrep™ (65). We performed S-Trap either alone or in combination with TCA precipitation and with additional SDS in the lysis step. All combinations tested led to complete and reproducible elimination of OptiPrep™ contaminants, with higher protein yields observed with the combination of TCA precipitation and S-Trap (supplemental Fig. S3). To verify the suitability of the procedure on low abundant samples, we applied this method to 2–10 million T cells immunoselected from a single mouse spleen, and we succeeded in consistently eliminating contamination (Fig. 2B). TCA precipitation allowed us to concentrate the sample and resolubilize it in 5% SDS, which contributes to solubilization of membrane proteins contained in the raft fraction. Furthermore, the S-Trap protocol proved to be faster than the other tested methods, as well as easier to perform.
Purity of raft preparation and generation of a T cell raft protein database
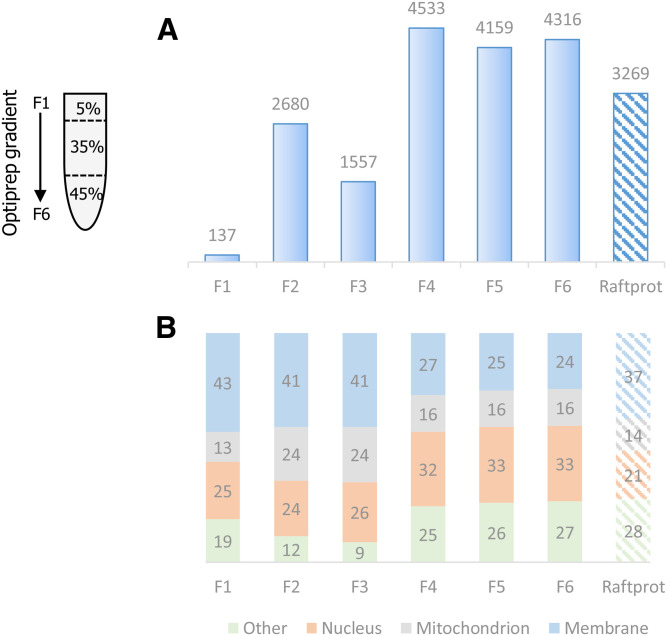
To further assess the purity of the raft-enriched fraction from nonstimulated T cells, we analyzed all OptiPrep™ gradient fractions by LC-MS/MS. We performed TCA precipitation with S-Trap digestion, and we performed high pH fractionation of the peptides for all six fractions in order to gain depth in the analysis. The number of proteins identified ranged from 137 to 4,533, with 2,680 proteins identified in fraction 2 (Fig. 3A).
Fig. 3.
Number of proteins identified and cell compartment distribution of OptiPrep gradient fractions. T cells were immunoselected from four mice and subjected to raft isolation by the simplified OptiPrep™ density gradient procedure, depicted on the left. Then, proteins were extracted separately by the S-Trap method, pooled, and analyzed by MS. A: Histogram showing the total number of proteins identified in each of the six gradient fractions in comparison with the total number of proteins present in the RaftProt database (right column). B: For each gradient fraction, the cell compartment distribution of identified proteins is indicated, as the percentage of membrane, mitochondrion, nuclear, and other localizations, according to Gene Ontology. The right column shows the global cell compartment distribution of all RaftProt database proteins according to Gene Ontology.
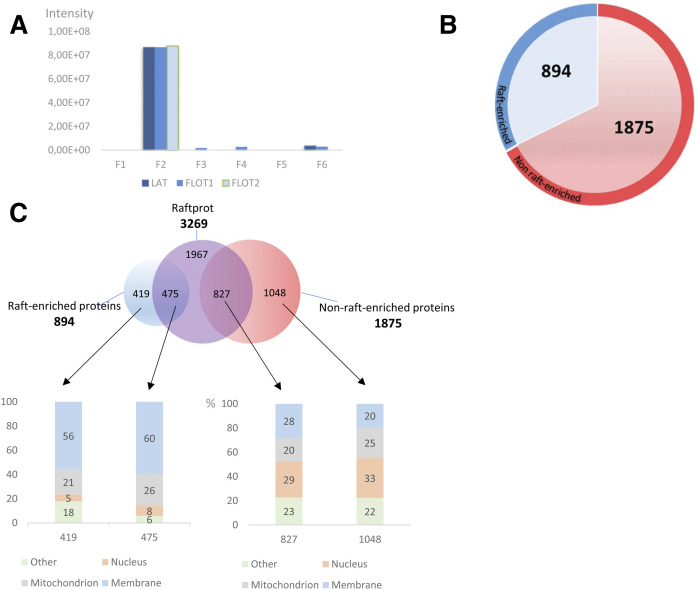
In order to determine the percentage of membrane, nuclear, and mitochondrial proteins, a Gene Ontology cellular component analysis was performed for each fraction. In fraction 2, as expected, we observed a higher percentage of membrane proteins as compared with the other fractions (Fig. 3B). We could identify with a high intensity count of 8e10 a number of proteins frequently used as raft markers: LAT, which is a raft marker specific to T cells, flotillin-1 (as confirmed above by Western blot), and flotillin-2. All of these three proteins were abundantly identified in fraction 2 and at a much lower intensity (50-fold less) in the other fractions (Fig. 4A).
Fig. 4.
Purity of raft preparation and generation of a T cell raft protein database. T cells were immunoselected from four mice and subjected to raft isolation by the simplified OptiPrep™ density gradient procedure. Then, proteins were extracted separately by the S-Trap method, pooled, and analyzed by MS. A: Intensities calculated by MaxQuant for three raft markers, namely LAT, flotillin-1 (FLOT1), and flotillin-2 (FLOT2) in all six OptiPrep™ gradient fractions. B: Representation of the generated T cell raft protein database. Proteins are divided into those enriched in rafts (abundance ratio in raft over nonraft fractions <2, red color). C: Protein comparison of generated database with RaftProt database and Gene Ontology analysis. The number of exclusive and common proteins is indicated for each case in the upper diagram. The columns in the lower side indicate the percentage of proteins corresponding to each case in the upper diagram. The columns in the lower side indicate the percentage of proteins corresponding to each cellular compartment, according to Gene Ontology.
To establish a raft protein database by following as stringent criteria as possible, we assumed that raft-residing proteins should present the same intensity profile in gradient fractions as LAT, flotillin-1, and flotillin-2, i.e., at least two times more abundant in fraction 2 compared with other fractions. Based on our LC-MS/MS data, we could establish a raft database of 894 proteins (in blue, Fig. 4B). Those proteins not falling into these criteria were classified as “nonenriched in protein rafts” (1,875 proteins in red, Fig. 3B; supplemental Table S1). We believe that these proteins can still be associated with rafts, but they are also or mainly present in cytoplasm, hence their profile in the gradient.
Our database containing 894 proteins was compared with the RaftProt database (3,269 proteins, one-star experiment). Notably, 475 proteins from murine RaftProt overlap with our raft’s T cell database, although the overlap increases to 669 when both human and murine RaftProt databases are used. Gene Ontology analysis revealed the presence of nearly 60% of membrane proteins in our database, regardless of whether they are common to RaftProt or unique (Fig. 4C, supplemental Table S2). On the contrary, the group of proteins contained in F2 and classified as nonenriched in the protein raft database corresponded to a low percentage of membrane localized proteins (28%). This Gene Ontology analysis provides support to consider the 894 proteins in F2 as true raft proteins and could be used as a reference database for nonstimulated T cells. To determine the most significantly enriched pathways and cellular location for the 894 proteins in our raft database, we performed a Fisher’s exact test using keywords and KEGG pathways. The analysis showed a clear enrichment in membrane proteins and related membrane protein pathways as compared with all the proteins identified in all fractions (supplemental Table S3).
T cell raft protein analysis in resting versus activating conditions
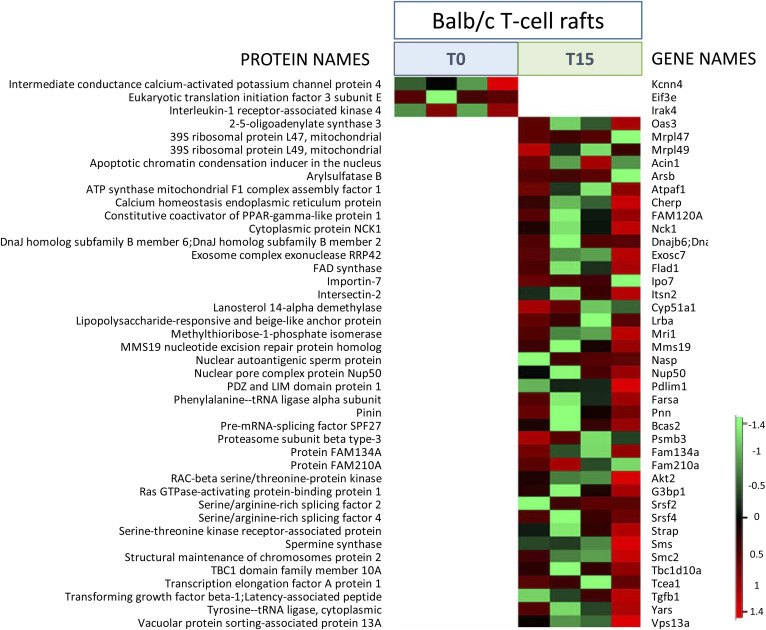
To verify the suitability of the method on low abundant samples purified by OptiPrep gradient, we used T cells from four single Balb/c mice. After negative immunoselection (yield between two and eight million cells), T cells from each mouse were split into two groups: one group (n = 4) was not stimulated (T0 minutes) and the other group (n = 4) was stimulated for 15 min (T15 minutes) with anti-CD3 and anti-CD28 antibodies. Next, we performed membrane raft preparation, TCA precipitation, and S-Trap digestion on gradient fraction 2 and analyzed proteins by LC-MS/MS. The number of identified raft proteins significantly increased in activated T cells compared with resting T cells (supplemental Fig. S4A). Likewise, the abundance of flotillin-1 was also significantly increased in activated T cells (supplemental Fig. S4B), suggesting that the abundance of rafts is increased in these cells. However, no significant differences were obtained after label-free-based quantitation between the two conditions (t-test, false discovery rate = 0.01). We therefore proceeded with a manual selection of proteins that were exclusively present in rafts either in stimulating or nonstimulating conditions. This selection was based on the detection in rafts in one condition in all four samples and nondetection in rafts in the other condition in at least four samples. A heatmap showing the selected proteins is presented in Fig. 5 (supplemental Table S4). Three proteins were found present in the raft fraction selectively at T0, while 39 proteins were selectively detected in rafts at T15. Some of these proteins are known to be involved in T cell activation at the raft level. Among them, we found as the most relevant in the context of T cell activation and raft dynamics the following: Akt2, Nck1, TgfB1, Tbc1d10, Pdlim1, and intesectin-2. The TGF-β ligand, TgfB1, is a key molecule involved in cell signaling regulating T cell activation and polarization. Akt2 is a key player in the proximal signaling events participating in TCR activation. Nck1, is an adaptor protein involved in actin cytoskeleton remodeling. Tbc1d10 is an activator of the Rab35 GTPase. Pdlim1 has a role in the regulation of actin cytoskeleton networks. Intersectin-2 (Itsn2) is known to participate in TCR internalization.
Fig. 5.
Raft protein analysis on Balb/c T cells with or without anti-CD3/CD28 TCR activation. Heatmap representation from Perseus software of proteins detected (in color) or not detected (in white) in T cell rafts from four individual mice. T cells immunoselected from each mouse were split and either nonstimulated (T0) or stimulated for 15 min (T15) with anti-CD3/anti-CD28 antibodies. The color scale denotes abundance variation (red for more abundant, green for less abundant).
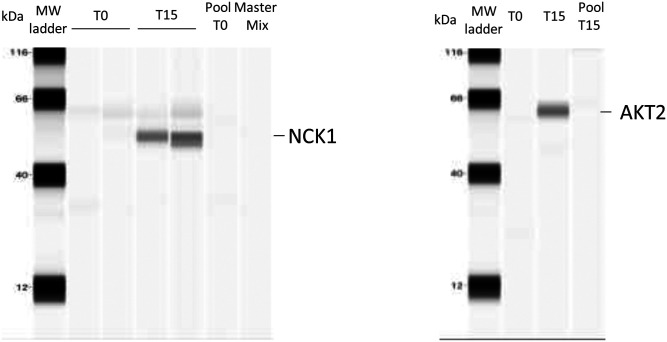
In order to validate the mass spectrometric results, Akt2 and Nck1 were selected for capillary WES, a method that allows detection of very small amounts of proteins (a few micrograms). WES analysis demonstrated the effective recruitment of Akt2 and Nck1 in rafts at T15, which is consistent with our mass spectrometric data (Fig. 6).
Fig. 6.
WES analysis of Akt2 and Nck1. Capillary Western immunoassay (WES) was used to validate some of the proteins (Akt2 and Nck1) identified as differentially recruited by MS. Two hundred microliters of raft fraction collected from the OptiPrep gradient were precipitated with 10% TCA (final concentration) and analyzed by WES. Two biological replicates for T0 and T15 were used for Nck1 detection and one biological replicate for T0 and T15 was used to detect Akt2. Nck1 and Akt2 are detected in the condition T15 only. We used a pool T0 and T15 (rabbit IgG control) and a Master Mix (primary antibodies and secondary antibodies with no protein) to rule out nonspecific binding of antibodies. No bands were detected in these lanes.
Altogether, these data demonstrate that the described method for OptiPrep™ gradient raft isolation and S-Trap protein preparation is suitable for the differential analysis of low abundant raft proteins in T cells.
DISCUSSION
The present work shows, for the first time, a global analysis of the membrane raft proteome of ex vivo T cells isolated from individual mice. Moreover, we propose a method that combines several improving features, such as the isolation capacity of OptiPrep-based density gradient, a straightforward sample preparation (S-Trap), and a label-free quantitation, globally providing a high yield of identified proteins.
The first isolation of proteins from T cells contained in triton-insoluble membranes was done back in the nineties (66). Given the early understanding that membrane rafts play a fundamental role in T cell activation (67–71), knowing the protein composition of these microdomains has represented a key objective to understand their structure, dynamics, and function. A pioneering characterization of the proteome of T cell membrane microdomains was performed on the leukemia cell line Jurkat T (20) by LC-MS/MS, while the first study of T cell proteome in activating conditions was approached by flotillin-1 immunoprecipitation (27) and MS.
Gel electrophoresis-based approaches were among the first used to address this question. For example, 1D and 2D gels with staining and MS have been used to resolve and identify proteins associated with TCR activation complexes in Jurkat cells (20). 2D fluorescence difference gel electrophoresis, based on differential fluorochrome labeling, has been applied to study lipid raft proteins after TCR costimulation (22). Bini et al. (20) succeeded in studying the variation in intensity of more than 800 spots in Jurkat cells subjected to TCR costimulation for up to 15 min using a classic 2D gel electrophoresis and MALDI-TOF setting. In the same experimental conditions, von Haller et al. (25) used a gel-free method based on LC separation.
Detergent solubility has been the landmark technique of raft (or DRM, for detergent-resistant microdomain) isolation (20, 25, 26). In our study, a detergent-free method has been used, in which mechanical disruption of membranes is performed, with a high enrichment of raft-like fractions in raft protein markers. Any study on membrane rafts must always be taken cautiously, because cellular membranes may contain a broad range of microdomains that can differ in composition, function, and dynamics (72). The experimental purification approach in all cases will determine the type of microdomain that is targeted. In our work, we have chosen a procedure that allows the purification of low-density membrane fragments highly enriched in bona fide T cell raft markers, such as flotillins, caveolins, and LAT. The high yield of RaftProt-listed proteins strongly suggests that the microdomains described in this study are close to those existing in living cells. Nevertheless, alternative methods will be needed to validate the results obtained in further functional studies on T cells.
Iodixanol, the chemical component of OptiPrep, was developed as an efficient and rapid way to isolate rat liver peroxisomes (73). Later, it replaced other density gradient procedures in the purification of subcellular structures, including rafts (36, 39, 40, 61, 74–86). The only report to date of a proteomic study following iodixanol-based raft isolation is that of Rose et al. (61), in which detergent-resistant microdomain from virus-infected plankton species Emiliania huxleyi were subjected in parallel to lipidomic and proteomic characterization. The authors reported the identification of 116 proteins from the infected cells and 86 proteins from control cells. With speed being the main advantage of iodixanol with respect to sucrose gradients, its limiting characteristic is the spectral interference with protein identification by MS, resulting in a limited and variable number of identified proteins. This drawback has limited proteomics on raft fractions isolated by this method.
Suspension trapping or S-Trap as a preparation technique for bottom-up proteomics applied to low protein amounts was recently developed for cell lysates, membrane preparations, and immunoprecipitates (65). It has been shown as more efficient than other methods, such as direct FASP and in-solution digestion, and compatible with common extraction buffers and detergents (87–89). Very recently S-Trap has been applied in the study of mouse brain microglial proteome (90) and in the search for oral squamous cell carcinoma biomarkers in saliva (91). Here, we have applied, for the first time, S-Trap to T cell proteomics and to the analysis of a submembranous fraction (rafts). Guergues et al. (90) succeeded to identify and quantify nearly 4,700 proteins in microglial cells from one single mouse, which represents a number comparable to that obtained in the current work, but from an even smaller number of cells (300,000 vs. two million). Even if the greater size of microglial cells as compared with lymphocytes could account for this difference, this suggests that our method could be further optimized to the analysis of rafts from T cell subpopulations. Due to the multiple advantages of this technique, it is likely to consolidate as a reference procedure in bottom-up proteomics in the years to come.
Interestingly, in our study comparing nonstimulated cells with cells stimulated with anti-CD3/CD28 antibodies for 15 min, label-free quantitation did not result in any statistically significant difference. It could be argued that this is due to the fact that 15 min of stimulation might be a too long lapse to find subtle protein recruitment changes in rafts. In fact, although the immunological synapse can be assembled within seconds or a few minutes, costimulation of TCR provides a more sustained response and raft redistribution is observable after at least 20 min (1). Another potential explanation is the fact that TCR stimulation induces mostly a clustering and reorganization of existing rafts, along with their resident proteins, rather than a new recruitment of proteins. Most likely, our results are due to the combination of both hypotheses. Nonetheless, several proteins were found specifically present or absent in rafts after T cell activation.
Among the 39 proteins we identified selectively recruited in rafts after 15 min of TCR stimulation, we found some that are known to play a role in T cell function and/or in raft dynamics: Akt2, Nck1, TgfB1, Tbc1d10, Pdlim1, intesectin-2, and Cherp. Akt2, a member of the PKB/Akt family, plays a key role in in the signaling events leading to activation, differentiation, and survival of Jurkat and human primary T cells after TCR activation (92–96). Akt is recruited at the plasma membrane upon TCR activation via the interaction between its pleckstrin homology domain and membrane phosphoinositides and phosphorylated by PKC at Ser473 (97). The presence of Akt2 isoform in rafts has been specifically reported in intestinal cells (98) and platelets (99).
The adaptor protein, Nck1, is also known to interact with the TCR via its SH3.1 and SH2 domains and thereby participates in T cell activation via Erk and MEK phosphorylation (100–102). Its presence in T cell rafts has not been reported to date. Conversely, it has been found in nonraft compartments in human neuroblastoma cells and embryonic cortical neurons (103).
The increased presence of the TgfB1 ligand in raft-like domains strongly suggests the presence of the cognate receptor. The TGF-β receptor has been associated with lipid rafts in previous reports (104). Most interestingly, it has been shown that TCR activation induces the recruitment of TGF-β receptor in rafts, subsequently inhibiting SMAD signaling and ultimately resulting in Th1 and Th2 differentiation (105). Intriguingly, serine-threonine kinase receptor-associated protein (Strap), known to interact with both TGF-β receptor and PDK1 and to inhibit TGF-β-dependent signaling (106), is also present within the proteins found increased in rafts in the current study. Strap has never been reported as associated with rafts or TCR activation, which evokes a complex subjacent regulatory mechanism.
Other proteins of interest include Tbc1d10, an activating protein of the GTPase Rab35. Rab35 participates in the formation of the immunological synapse by regulating TCR transport. Tbc1d10 and Rab35 colocalize with TCR in Jurkat T cells, and knockdown of the former inhibits immunological synapse relocalization of the latter (107). Pdlim1 is involved in the formation of actin networks and could play a role in the structuration of lipid raft clusters characteristic of immunological synapse. Pdlim1 expression is increased along with that of caveolin in cells treated with dexamethasone (108), while its role in T cell activation is currently unknown. Intersectin-2 is a recently described protein participating in TCR internalization, via association with Wiskott Aldrich Syndrome protein (WASp) and Cdc42, with maximum interaction with the former at 10 min post-TCR activation (109). Internalization of TCR is a key step in the regulation of T cell activation. It involves actin cytoskeleton rearrangement and clathrin-dependent endocytosis. Intriguingly, intersectin-2 is found here recruited in lipid rafts, whereas it has been reported to be involved in non-raft-dependent endocytosis (109). Finally, the calcium homeostasis endoplasmic reticulum protein (Cherp) is likely to play a role in the activation and proliferation of T cells, as its knocking down has been found to impair those processes in Jurkat cells (110). Although its localization to rafts has never been reported before, this finding is intriguing, as Ca2+ levels are known to rise rapidly following TCR engagement.
In line with the calcium homeostasis events characteristic of TCR stimulation, one of the three proteins excluded from rafts at 15 min postactivation, according to our results, was the potassium calcium-activated channel 4 (KCa3.1 or Kcnn4). This transporter, shown to be associated with rafts upon cell swelling, is critical for Ca2+ influx associated with T cell activation, and so its inhibition blocks T cell activation (111). Another raft-excluded protein, the tyrosine kinase, Irak4, has been shown to participate in TCR activation. In Jurkat cells, Irak4 was found to relocalize to rafts after stimulation with anti-CD3 antibodies, and to induce downstream signals including PKC and NF-κB activation (112). Irak4 was found in rafts in our unstimulating conditions, like in the previous report, but to our surprise, it would disappear from those fractions after stimulation instead of being increased. These unexpected and somehow contradictory results could be due either to the different cell models (mouse T cells vs. Jurkat) or to the differences in raft isolation procedures and, subsequently, in the kind of microdomains obtained. The exclusion of these two proteins from rafts 15 min after TCR stimulation warrants a closer study of their raft localization dynamics and functional implications.
In conclusion, our work shows the relevance of an S-Trap-based methodological strategy to deepen into the molecular mechanisms that govern T cell activation. We also provide a new database of T cell raft proteins. Our approach can be applied to other aspects of the biology of T cells and other cell types involving membrane rafts.
Data availability
All data are contained either within the article or as supporting information. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD016476.
Supplementary Material
Footnotes
Author contributions—C.C., S.Y.Z., V.J., D.L., and J.L. experiments; A.P. animal models; C.C., D.S., M.O., and I.C.G. experimental design; C.C., D.S., M.O., and I.C.G. writing.
This article contains supplemental data.
Author ORCIDs—Mario Ollero https://orcid.org/0000-0003-3590-706X
Funding and additional information—This work was partially supported by Fondation du Rein, Fondation Recherche Médicale Grant R13063JJ, and Fondation de l’Avenir Grant AP-RM-17020.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Abbreviations—
- FASP
- filter-aided sample preparation
- LAT
- linker for activation of T cells
- S-Trap
- suspension trapping
- TCR
- T cell receptor
- TEAB
- triethylammonium bicarbonate
- WES
- Western immunoassay
Manuscript received February 7, 2020, and in revised form August 1, 2020. Published, JLR Papers in Press, August 7, 2020, DOI 10.1194/jlr.D120000672.
REFERENCES
- 1.Lingwood D., and Simons K.. 2010. Lipid rafts as a membrane-organizing principle. Science. 327: 46–50. [DOI] [PubMed] [Google Scholar]
- 2.Simons K., and Gerl M. J.. 2010. Revitalizing membrane rafts: new tools and insights. Nat. Rev. Mol. Cell Biol. 11: 688–699. [DOI] [PubMed] [Google Scholar]
- 3.Pike L. J. 2006. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res. 47: 1597–1598. [DOI] [PubMed] [Google Scholar]
- 4.Jordan S., and Rodgers W.. 2003. T cell glycolipid-enriched membrane domains are constitutively assembled as membrane patches that translocate to immune synapses. J. Immunol. 171: 78–87. [DOI] [PubMed] [Google Scholar]
- 5.Garofalo T., Lenti L., Longo A., Misasi R., Mattei V., Pontieri G. M., Pavan A., and Sorice M.. 2002. Association of GM3 with Zap-70 induced by T cell activation in plasma membrane microdomains: GM3 as a marker of microdomains in human lymphocytes. J. Biol. Chem. 277: 11233–11238. [DOI] [PubMed] [Google Scholar]
- 6.Khoshnan A., Bae D., Tindell C. A., and Nel A. E.. 2000. The physical association of protein kinase C theta with a lipid raft-associated inhibitor of kappa B factor kinase (IKK) complex plays a role in the activation of the NF-kappa B cascade by TCR and CD28. J. Immunol. 165: 6933–6940. [DOI] [PubMed] [Google Scholar]
- 7.Langhorst M. F., Reuter A., Luxenhofer G., Boneberg E. M., Legler D. F., Plattner H., and Stuermer C. A.. 2006. Preformed reggie/flotillin caps: stable priming platforms for macrodomain assembly in T cells. FASEB J. 20: 711–713. [DOI] [PubMed] [Google Scholar]
- 8.Martin M., Schneider H., Azouz A., and Rudd C. E.. 2001. Cytotoxic T lymphocyte antigen 4 and CD28 modulate cell surface raft expression in their regulation of T cell function. J. Exp. Med. 194: 1675–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qu X., Kawauchi-Kamata K., Miah S. M., Hatani T., Yamamura H., and Sada K.. 2005. Tyrosine phosphorylation of adaptor protein 3BP2 induces T cell receptor-mediated activation of transcription factor. Biochemistry. 44: 3891–3898. [DOI] [PubMed] [Google Scholar]
- 10.Stuermer C. A., Langhorst M. F., Wiechers M. F., Legler D. F., Von Hanwehr S. H., Guse A. H., and Plattner H.. 2004. PrPc capping in T cells promotes its association with the lipid raft proteins reggie-1 and reggie-2 and leads to signal transduction. FASEB J. 18: 1731–1733. [DOI] [PubMed] [Google Scholar]
- 11.Villalba M., Bi K., Hu J., Altman Y., Bushway P., Reits E., Neefjes J., Baier G., Abraham R. T., and Altman A.. 2002. Translocation of PKC[theta] in T cells is mediated by a nonconventional, PI3-K- and Vav-dependent pathway, but does not absolutely require phospholipase C. J. Cell Biol. 157: 253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Viola A., Schroeder S., Sakakibara Y., and Lanzavecchia A.. 1999. T lymphocyte costimulation mediated by reorganization of membrane microdomains. Science. 283: 680–682. [DOI] [PubMed] [Google Scholar]
- 13.Yashiro-Ohtani Y., Zhou X. Y., Toyo-Oka K., Tai X. G., Park C. S., Hamaoka T., Abe R., Miyake K., and Fujiwara H.. 2000. Non-CD28 costimulatory molecules present in T cell rafts induce T cell costimulation by enhancing the association of TCR with rafts. J. Immunol. 164: 1251–1259. [DOI] [PubMed] [Google Scholar]
- 14.Giri B., Dixit V. D., Ghosh M. C., Collins G. D., Khan I. U., Madara K., Weeraratna A. T., and Taub D. D.. 2007. CXCL12-induced partitioning of flotillin-1 with lipid rafts plays a role in CXCR4 function. Eur. J. Immunol. 37: 2104–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gómez-Moutón C., Lacalle R. A., Mira E., Jimenez-Baranda S., Barber D. F., Carrera A. C., Martinez A. C., and Manes S.. 2004. Dynamic redistribution of raft domains as an organizing platform for signaling during cell chemotaxis. J. Cell Biol. 164: 759–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiao X., Zhang N., Xu X., Oppenheim J. J., and Jin T.. 2005. Ligand-induced partitioning of human CXCR1 chemokine receptors with lipid raft microenvironments facilitates G-protein-dependent signaling. Mol. Cell. Biol. 25: 5752–5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin S. L., Chien C. W., Han C. L., Chen E. S., Kao S. H., Chen Y. J., and Liao F.. 2010. Temporal proteomics profiling of lipid rafts in CCR6-activated T cells reveals the integration of actin cytoskeleton dynamics. J. Proteome Res. 9: 283–297. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen D. H., Espinoza J. C., and Taub D. D.. 2004. Cellular cholesterol enrichment impairs T cell activation and chemotaxis. Mech. Ageing Dev. 125: 641–650. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen D. H., Giri B., Collins G., and Taub D. D.. 2005. Dynamic reorganization of chemokine receptors, cholesterol, lipid rafts, and adhesion molecules to sites of CD4 engagement. Exp. Cell Res. 304: 559–569. [DOI] [PubMed] [Google Scholar]
- 20.Bini L., Pacini S., Liberatori S., Valensin S., Pellegrini M., Raggiaschi R., Pallini V., and Baldari C. T.. 2003. Extensive temporally regulated reorganization of the lipid raft proteome following T cell antigen receptor triggering. Biochem. J. 369: 301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta N., Wollscheid B., Watts J. D., Scheer B., Aebersold R., and DeFranco A. L.. 2006. Quantitative proteomic analysis of B cell lipid rafts reveals that ezrin regulates antigen receptor-mediated lipid raft dynamics. Nat. Immunol. 7: 625–633. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi M., Katagiri T., Kosako H., Iida N., and Hattori S.. 2007. Global analysis of dynamic changes in lipid raft proteins during T cell activation. Electrophoresis. 28: 2035–2043. [DOI] [PubMed] [Google Scholar]
- 23.Razzaq T. M., Ozegbe P., Jury E. C., Sembi P., Blackwell N. M., and Kabouridis P. S.. 2004. Regulation of T cell receptor signalling by membrane microdomains. Immunology. 113: 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomé C. H., dos Santos G. A., Ferreira G. A., Scheucher P. S., Izumi C., Leopoldino A. M., Simao A. M., Ciancaglini P., de Oliveira K. T., Chin A., et al. 2012. Linker for activation of T cell family member2 (LAT2) a lipid raft adaptor protein for AKT signaling, is an early mediator of alkylphospholipid anti-leukemic activity. Mol. Cell. Proteomics. 11: 1898–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Haller P. D., Donohoe S., Goodlett D. R., Aebersold R., and Watts J. D.. 2001. Mass spectrometric characterization of proteins extracted from Jurkat T cell detergent-resistant membrane domains. Proteomics. 1: 1010–1021. [DOI] [PubMed] [Google Scholar]
- 26.von Haller P. D., Yi E., Donohoe S., Vaughn K., Keller A., Nesvizhskii A. I., Eng J., Li X. J., Goodlett D. R., Aebersold R., et al. 2003. The application of new software tools to quantitative protein profiling via isotope-coded affinity tag (ICAT) and tandem mass spectrometry: II. Evaluation of tandem mass spectrometry methodologies for large-scale protein analysis, and the application of statistical tools for data analysis and interpretation. Mol. Cell. Proteomics. 2: 428–442. [DOI] [PubMed] [Google Scholar]
- 27.Tu X., Huang A., Bae D., Slaughter N., Whitelegge J., Crother T., Bickel P. E., and Nel A.. 2004. Proteome analysis of lipid rafts in Jurkat cells characterizes a raft subset that is involved in NF-kappaB activation. J. Proteome Res. 3: 445–454. [DOI] [PubMed] [Google Scholar]
- 28.Grant M. M., Scheel-Toellner D., and Griffiths H. R.. 2007. Contributions to our understanding of T cell physiology through unveiling the T cell proteome. Clin. Exp. Immunol. 149: 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaw A. R., and Li L.. 2003. Exploration of the functional proteome: lessons from lipid rafts. Curr. Opin. Mol. Ther. 5: 294–301. [PubMed] [Google Scholar]
- 30.Wollscheid B., von Haller P. D., Yi E., Donohoe S., Vaughn K., Keller A., Nesvizhskii A. I., Eng J., Li X. J., Goodlett D. R., et al. 2004. Lipid raft proteins and their identification in T lymphocytes. Subcell. Biochem. 37: 121–152. [DOI] [PubMed] [Google Scholar]
- 31.Mohamed A., Robinson H., Erramouspe P. J., and Hill M. M.. 2018. Advances and challenges in understanding the role of the lipid raft proteome in human health. Expert Rev. Proteomics. 15: 1053–1063. [DOI] [PubMed] [Google Scholar]
- 32.Zheng Y. Z., and Foster L. J.. 2009. Contributions of quantitative proteomics to understanding membrane microdomains. J. Lipid Res. 50: 1976–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohamed A., Shah A. D., Chen D., and Hill M. M.. 2019. RaftProt V2: understanding membrane microdomain function through lipid raft proteomes. Nucleic Acids Res. 47: D459–D463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah A., Chen D., Boda A. R., Foster L. J., Davis M. J., and Hill M. M.. 2015. RaftProt: mammalian lipid raft proteome database. Nucleic Acids Res. 43: D335–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabani V., Davani S., Gambert-Nicot S., Meneveau N., and Montange D.. 2016. Comparative lipidomics and proteomics analysis of platelet lipid rafts using different detergents. Platelets. 27: 634–641. [DOI] [PubMed] [Google Scholar]
- 36.Shah M. B., and Sehgal P. B.. 2007. Nondetergent isolation of rafts. Methods Mol. Biol. 398: 21–28. [DOI] [PubMed] [Google Scholar]
- 37.Gaus K., Rodriguez M., Ruberu K. R., Gelissen I., Sloane T. M., Kritharides L., and Jessup W.. 2005. Domain-specific lipid distribution in macrophage plasma membranes. J. Lipid Res. 46: 1526–1538. [DOI] [PubMed] [Google Scholar]
- 38.Macdonald J. L., and Pike L. J.. 2005. A simplified method for the preparation of detergent-free lipid rafts. J. Lipid Res. 46: 1061–1067. [DOI] [PubMed] [Google Scholar]
- 39.Badana A., Chintala M., Varikuti G., Pudi N., Kumari S., Kappala V. R., and Malla R. R.. 2016. Lipid raft integrity is required for survival of triple negative breast cancer cells. J. Breast Cancer. 19: 372–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu L., Zhang K., Tan L., Chen Y. H., and Cao Y. P.. 2015. Alterations in cholesterol and ganglioside GM1 content of lipid rafts in platelets from patients with Alzheimer disease. Alzheimer Dis. Assoc. Disord. 29: 63–69. [DOI] [PubMed] [Google Scholar]
- 41.Aatonen M., Valkonen S., Boing A., Yuana Y., Nieuwland R., and Siljander P.. 2017. Isolation of platelet-derived extracellular vesicles. Methods Mol. Biol. 1545: 177–188. [DOI] [PubMed] [Google Scholar]
- 42.Aatonen M. T., Ohman T., Nyman T. A., Laitinen S., Gronholm M., and Siljander P. R.. 2014. Isolation and characterization of platelet-derived extracellular vesicles. J. Extracell. Vesicles. 3: doi:10.3402/jev.v3.24692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao Z., Li C., Higginbotham J. N., Franklin J. L., Tabb D. L., Graves-Deal R., Hill S., Cheek K., Jerome W. G., Lapierre L. A., et al. 2008. Use of fluorescence-activated vesicle sorting for isolation of Naked2-associated, basolaterally targeted exocytic vesicles for proteomics analysis. Mol. Cell. Proteomics. 7: 1651–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eichenberger R. M., Talukder M. H., Field M. A., Wangchuk P., Giacomin P., Loukas A., and Sotillo J.. 2018. Characterization of Trichuris muris secreted proteins and extracellular vesicles provides new insights into host-parasite communication. J. Extracell. Vesicles. 7: 1428004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hurwitz S. N., Sun L., Cole K. Y., Ford C. R. 3rd, Olcese J. M., and Meckes D. G. Jr.. 2018. An optimized method for enrichment of whole brain-derived extracellular vesicles reveals insight into neurodegenerative processes in a mouse model of Alzheimer’s disease. J. Neurosci. Methods. 307: 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minciacchi V. R., You S., Spinelli C., Morley S., Zandian M., Aspuria P. J., Cavallini L., Ciardiello C., Reis Sobreiro M., Morello M., et al. 2015. Large oncosomes contain distinct protein cargo and represent a separate functional class of tumor-derived extracellular vesicles. Oncotarget. 6: 11327–11341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rutter B. D., and Innes R. W.. 2017. Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol. 173: 728–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sotillo J., Pearson M., Potriquet J., Becker L., Pickering D., Mulvenna J., and Loukas A.. 2016. Extracellular vesicles secreted by Schistosoma mansoni contain protein vaccine candidates. Int. J. Parasitol. 46: 1–5. [DOI] [PubMed] [Google Scholar]
- 49.Suwakulsiri W., Rai A., Xu R., Chen M., Greening D. W., and Simpson R. J.. 2019. Proteomic profiling reveals key cancer progression modulators in shed microvesicles released from isogenic human primary and metastatic colorectal cancer cell lines. Biochim. Biophys. Acta. Proteins Proteomics. 1867: 140171. [DOI] [PubMed] [Google Scholar]
- 50.Abramowicz A., Widlak P., and Pietrowska M.. 2016. Proteomic analysis of exosomal cargo: the challenge of high purity vesicle isolation. Mol. Biosyst. 12: 1407–1419. [DOI] [PubMed] [Google Scholar]
- 51.Clark D. J., Fondrie W. E., Liao Z., Hanson P. I., Fulton A., Mao L., and Yang A. J.. 2015. Redefining the breast cancer exosome proteome by tandem mass tag quantitative proteomics and multivariate cluster analysis. Anal. Chem. 87: 10462–10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kalra H., Adda C. G., Liem M., Ang C. S., Mechler A., Simpson R. J., Hulett M. D., and Mathivanan S.. 2013. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics. 13: 3354–3364.24115447 [Google Scholar]
- 53.Samuel M., Chisanga D., Liem M., Keerthikumar S., Anand S., Ang C. S., Adda C. G., Versteegen E., Jois M., and Mathivanan S.. 2017. Bovine milk-derived exosomes from colostrum are enriched with proteins implicated in immune response and growth. Sci. Rep. 7: 5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jedelský P. L., Dolezal P., Rada P., Pyrih J., Smid O., Hrdy I., Sedinova M., Marcincikova M., Voleman L., Perry A. J., et al. 2011. The minimal proteome in the reduced mitochondrion of the parasitic protist Giardia intestinalis. PLoS One. 6: e17285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li M., Du W., Zhou M., Zheng L., Song E., and Hou J.. 2018. Proteomic analysis of insulin secretory granules in INS-1 cells by protein correlation profiling. Biophys. Rep. 4: 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plum S., Helling S., Theiss C., Leite R. E. P., May C., Jacob-Filho W., Eisenacher M., Kuhlmann K., Meyer H. E., Riederer P., et al. 2013. Combined enrichment of neuromelanin granules and synaptosomes from human substantia nigra pars compacta tissue for proteomic analysis. J. Proteomics. 94: 202–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arai Y., Hayashi M., and Nishimura M.. 2008. Proteomic analysis of highly purified peroxisomes from etiolated soybean cotyledons. Plant Cell Physiol. 49: 526–539. [DOI] [PubMed] [Google Scholar]
- 58.Joseph M., Ludevid M. D., Torrent M., Rofidal V., Tauzin M., Rossignol M., and Peltier J. B.. 2012. Proteomic characterisation of endoplasmic reticulum-derived protein bodies in tobacco leaves. BMC Plant Biol. 12: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu R., Wang Y., Qin G., and Tian S.. 2016. iTRAQ-based quantitative proteomic analysis reveals the role of the tonoplast in fruit senescence. J. Proteomics. 146: 80–89. [DOI] [PubMed] [Google Scholar]
- 60.Shen H., He Z., Yan H., Xing Z., Chen Y., Xu W., and Ma M.. 2014. The fronds tonoplast quantitative proteomic analysis in arsenic hyperaccumulator Pteris vittata L. J. Proteomics. 105: 46–57. [DOI] [PubMed] [Google Scholar]
- 61.Rose S. L., Fulton J. M., Brown C. M., Natale F., Van Mooy B. A., and Bidle K. D.. 2014. Isolation and characterization of lipid rafts in Emiliania huxleyi: a role for membrane microdomains in host-virus interactions. Environ. Microbiol. 16: 1150–1166. [DOI] [PubMed] [Google Scholar]
- 62.Lipecka J., Chhuon C., Bourderioux M., Bessard M. A., van Endert P., Edelman A., and Guerrera I. C.. 2016. Sensitivity of mass spectrometry analysis depends on the shape of the filtration unit used for filter aided sample preparation (FASP). Proteomics. 16: 1852–1857. [DOI] [PubMed] [Google Scholar]
- 63.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., and Cox J.. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 13: 731–740. [DOI] [PubMed] [Google Scholar]
- 64.Otto G. P., and Nichols B. J.. 2011. The roles of flotillin microdomains–endocytosis and beyond. J. Cell Sci. 124: 3933–3940. [DOI] [PubMed] [Google Scholar]
- 65.Zougman A., Selby P. J., and Banks R. E.. 2014. Suspension trapping (STrap) sample preparation method for bottom-up proteomics analysis. Proteomics. 14: 1006-0. [DOI] [PubMed] [Google Scholar]
- 66.Caplan S., Zeliger S., Wang L., and Baniyash M.. 1995. Cell-surface-expressed T cell antigen-receptor zeta chain is associated with the cytoskeleton. Proc. Natl. Acad. Sci. USA. 92: 4768–4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Janes P. W., Ley S. C., and Magee A. I.. 1999. Aggregation of lipid rafts accompanies signaling via the T cell antigen receptor. J. Cell Biol. 147: 447–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kabouridis P. S. 2006. Lipid rafts in T cell receptor signalling. Mol. Membr. Biol. 23: 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Montixi C., Langlet C., Bernard A. M., Thimonier J., Dubois C., Wurbel M. A., Chauvin J. P., Pierres M., and He H. T.. 1998. Engagement of T cell receptor triggers its recruitment to low-density detergent-insoluble membrane domains. EMBO J. 17: 5334–5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schade A. E., and Levine A. D.. 2002. Lipid raft heterogeneity in human peripheral blood T lymphoblasts: a mechanism for regulating the initiation of TCR signal transduction. J. Immunol. 168: 2233–2239. [DOI] [PubMed] [Google Scholar]
- 71.Xavier R., Brennan T., Li Q., McCormack C., and Seed B.. 1998. Membrane compartmentation is required for efficient T cell activation. Immunity. 8: 723–732. [DOI] [PubMed] [Google Scholar]
- 72.Nicolson G. L. 2014. The fluid-mosaic model of membrane structure: still relevant to understanding the structure, function and dynamics of biological membranes after more than 40 years. Biochim. Biophys. Acta. 1838: 1451–1466. [DOI] [PubMed] [Google Scholar]
- 73.Van Veldhoven P. P., Baumgart E., and Mannaerts G. P.. 1996. Iodixanol (Optiprep), an improved density gradient medium for the iso-osmotic isolation of rat liver peroxisomes. Anal. Biochem. 237: 17–23. [DOI] [PubMed] [Google Scholar]
- 74.Annaba F., Kumar P., Dudeja A. K., Saksena S., Gill R. K., and Alrefai W. A.. 2010. Green tea catechin EGCG inhibits ileal apical sodium bile acid transporter ASBT. Am. J. Physiol. Gastrointest. Liver Physiol. 298: G467–G473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Berrou E., and Bryckaert M.. 2009. Recruitment of protein phosphatase 2A to dorsal ruffles by platelet-derived growth factor in smooth muscle cells: dephosphorylation of Hsp27. Exp. Cell Res. 315: 836–848. [DOI] [PubMed] [Google Scholar]
- 76.Gangalum R. K., Atanasov I. C., Zhou Z. H., and Bhat S. P.. 2011. AlphaB-crystallin is found in detergent-resistant membrane microdomains and is secreted via exosomes from human retinal pigment epithelial cells. J. Biol. Chem. 286: 3261–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li X., and Donowitz M.. 2008. Fractionation of subcellular membrane vesicles of epithelial and nonepithelial cells by OptiPrep density gradient ultracentrifugation. Methods Mol. Biol. 440: 97–110. [DOI] [PubMed] [Google Scholar]
- 78.Morita S. Y., Tsuda T., Horikami M., Teraoka R., Kitagawa S., and Terada T.. 2013. Bile salt-stimulated phospholipid efflux mediated by ABCB4 localized in nonraft membranes. J. Lipid Res. 54: 1221–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Norambuena A., Poblete M. I., Donoso M. V., Espinoza C. S., Gonzalez A., and Huidobro-Toro J. P.. 2008. P2Y1 receptor activation elicits its partition out of membrane rafts and its rapid internalization from human blood vessels: implications for receptor signaling. Mol. Pharmacol. 74: 1666–1677. [DOI] [PubMed] [Google Scholar]
- 80.Persaud-Sawin D. A., Lightcap S., and Harry G. J.. 2009. Isolation of rafts from mouse brain tissue by a detergent-free method. J. Lipid Res. 50: 759–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qiu Y., Wang Y., Law P. Y., Chen H. Z., and Loh H. H.. 2011. Cholesterol regulates micro-opioid receptor-induced beta-arrestin 2 translocation to membrane lipid rafts. Mol. Pharmacol. 80: 210–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rimmerman N., Bradshaw H. B., Kozela E., Levy R., Juknat A., and Vogel Z.. 2012. Compartmentalization of endocannabinoids into lipid rafts in a microglial cell line devoid of caveolin-1. Br. J. Pharmacol. 165: 2436–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Riquier A. D., Lee D. H., and McDonough A. A.. 2009. Renal NHE3 and NaPi2 partition into distinct membrane domains. Am. J. Physiol. Cell Physiol. 296: C900–C910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Siddiqi S., Sheth A., Patel F., Barnes M., and Mansbach C. M. 2nd.. 2013. Intestinal caveolin-1 is important for dietary fatty acid absorption. Biochim. Biophys. Acta. 1831: 1311–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sultan A., Luo M., Yu Q., Riederer B., Xia W., Chen M., Lissner S., Gessner J. E., Donowitz M., Yun C. C., et al. 2013. Differential association of the Na+/H+ exchanger regulatory factor (NHERF) family of adaptor proteins with the raft- and the non-raft brush border membrane fractions of NHE3. Cell. Physiol. Biochem. 32: 1386–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wani N. A., and Kaur J.. 2011. Reduced levels of folate transporters (PCFT and RFC) in membrane lipid rafts result in colonic folate malabsorption in chronic alcoholism. J. Cell. Physiol. 226: 579–587. [DOI] [PubMed] [Google Scholar]
- 87.Elinger D., Gabashvili A., and Levin Y.. 2019. Suspension trapping (S-Trap) is compatible with typical protein extraction buffers and detergents for bottom-up proteomics. J. Proteome Res. 18: 1441–1445. [DOI] [PubMed] [Google Scholar]
- 88.HaileMariam M., Eguez R. V., Singh H., Bekele S., Ameni G., Pieper R., and Yu Y.. 2018. S-Trap, an ultrafast sample-preparation approach for shotgun proteomics. J. Proteome Res. 17: 2917–2924. [DOI] [PubMed] [Google Scholar]
- 89.Ludwig K. R., Schroll M. M., and Hummon A. B.. 2018. Comparison of in-solution, FASP, and S-Trap based digestion methods for bottom-up proteomic studies. J. Proteome Res. 17: 2480–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guergues J., Zhang P., Liu B., and Stevens S. M. Jr.. 2019. Improved methodology for sensitive and rapid quantitative proteomic analysis of adult-derived mouse microglia: application to a novel in vitro mouse microglial cell model. Proteomics. 19: e1800469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lin Y. H., Eguez R. V., Torralba M. G., Singh H., Golusinski P., Golusinski W., Masternak M., Nelson K. E., Freire M., and Yu Y.. 2019. Self-assembled STrap for global proteomics and salivary biomarker discovery. J. Proteome Res. 18: 1907–1915. [DOI] [PubMed] [Google Scholar]
- 92.Bauer B., and Baier G.. 2002. Protein kinase C and AKT/protein kinase B in CD4+ T-lymphocytes: new partners in TCR/CD28 signal integration. Mol. Immunol. 38: 1087–1099. [DOI] [PubMed] [Google Scholar]
- 93.Genot E. M., Arrieumerlou C., Ku G., Burgering B. M., Weiss A., and Kramer I. M.. 2000. The T cell receptor regulates Akt (protein kinase B) via a pathway involving Rac1 and phosphatidylinositide 3-kinase. Mol. Cell. Biol. 20: 5469–5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Juntilla M. M., Wofford J. A., Birnbaum M. J., Rathmell J. C., and Koretzky G. A.. 2007. Akt1 and Akt2 are required for alphabeta thymocyte survival and differentiation. Proc. Natl. Acad. Sci. USA. 104: 12105–12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lafont V., Astoul E., Laurence A., Liautard J., and Cantrell D.. 2000. The T cell antigen receptor activates phosphatidylinositol 3-kinase-regulated serine kinases protein kinase B and ribosomal S6 kinase 1. FEBS Lett. 486: 38–42. [DOI] [PubMed] [Google Scholar]
- 96.Parry R. V., Reif K., Smith G., Sansom D. M., Hemmings B. A., and Ward S. G.. 1997. Ligation of the T cell co-stimulatory receptor CD28 activates the serine-threonine protein kinase protein kinase B. Eur. J. Immunol. 27: 2495–2501. [DOI] [PubMed] [Google Scholar]
- 97.Yang L., Qiao G., Ying H., Zhang J., and Yin F.. 2010. TCR-induced Akt serine 473 phosphorylation is regulated by protein kinase C-alpha. Biochem. Biophys. Res. Commun. 400: 16–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li X., Leu S., Cheong A., Zhang H., Baibakov B., Shih C., Birnbaum M. J., and Donowitz M.. 2004. Akt2, phosphatidylinositol 3-kinase, and PTEN are in lipid rafts of intestinal cells: role in absorption and differentiation. Gastroenterology. 126: 122–135. [DOI] [PubMed] [Google Scholar]
- 99.Ohtsuka H., Iguchi T., Hayashi M., Kaneda M., Iida K., Shimonaka M., Hara T., Arai M., Koike Y., Yamamoto N., et al. 2017. SDF-1alpha/CXCR4 Signaling in Lipid Rafts Induces Platelet Aggregation via PI3 Kinase-Dependent Akt Phosphorylation. PLoS One. 12: e0169609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ngoenkam J., Paensuwan P., Preechanukul K., Khamsri B., Yiemwattana I., Beck-Garcia E., Minguet S., Schamel W. W., and Pongcharoen S.. 2014. Non-overlapping functions of Nck1 and Nck2 adaptor proteins in T cell activation. Cell Commun. Signal. 12: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Paensuwan P., Hartl F. A., Yousefi O. S., Ngoenkam J., Wipa P., Beck-Garcia E., Dopfer E. P., Khamsri B., Sanguansermsri D., Minguet S., et al. 2016. Nck binds to the T cell antigen receptor using its SH3.1 and SH2 domains in a cooperative manner, promoting TCR functioning. J. Immunol. 196: 448–458. [DOI] [PubMed] [Google Scholar]
- 102.Paensuwan P., Ngoenkam J., Khamsri B., Preechanukul K., Sanguansermsri D., and Pongcharoen S.. 2015. Evidence for inducible recruitment of Wiskott-Aldrich syndrome protein to T cell receptor-CD3 complex in Jurkat T cells. Asian Pac. J. Allergy Immunol. 33: 189–195. [DOI] [PubMed] [Google Scholar]
- 103.Petrie R. J., Zhao B., Bedford F., and Lamarche-Vane N.. 2009. Compartmentalized DCC signalling is distinct from DCC localized to lipid rafts. Biol. Cell. 101: 77–90. [DOI] [PubMed] [Google Scholar]
- 104.Ito T., Williams J. D., Fraser D. J., and Phillips A. O.. 2004. Hyaluronan regulates transforming growth factor-beta1 receptor compartmentalization. J. Biol. Chem. 279: 25326–25332. [DOI] [PubMed] [Google Scholar]
- 105.Giroux M., Delisle J. S., O’Brien A., Hébert M. J., and Perreault C.. 2010. T cell activation leads to protein kinase C theta-dependent inhibition of TGF-beta signaling. J. Immunol. 185: 1568–1576. [DOI] [PubMed] [Google Scholar]
- 106.Seong H. A., Jung H., Choi H. S., Kim K. T., and Ha H.. 2005. Regulation of transforming growth factor-beta signaling and PDK1 kinase activity by physical interaction between PDK1 and serine-threonine kinase receptor-associated protein. J. Biol. Chem. 280: 42897–42908. [DOI] [PubMed] [Google Scholar]
- 107.Patino-Lopez G., Dong X., Ben-Aissa K., Bernot K. M., Itoh T., Fukuda M., Kruhlak M. J., Samelson L. E., and Shaw S.. 2008. Rab35 and its GAP EPI64C in T cells regulate receptor recycling and immunological synapse formation. J. Biol. Chem. 283: 18323–18330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Clark R., Nosie A., Walker T., Faralli J. A., Filla M. S., Barrett-Wilt G., and Peters D. M.. 2013. Comparative genomic and proteomic analysis of cytoskeletal changes in dexamethasone-treated trabecular meshwork cells. Mol. Cell. Proteomics. 12: 194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.McGavin M. K., Badour K., Hardy L. A., Kubiseski T. J., Zhang J., and Siminovitch K. A.. 2001. The intersectin 2 adaptor links Wiskott Aldrich Syndrome protein (WASp)-mediated actin polymerization to T cell antigen receptor endocytosis. J. Exp. Med. 194: 1777–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.O’Rourke F. A., LaPlante J. M., and Feinstein M. B.. 2003. Antisense-mediated loss of calcium homoeostasis endoplasmic reticulum protein (CHERP; ERPROT213-21) impairs Ca2+ mobilization, nuclear factor of activated T-cells (NFAT) activation and cell proliferation in Jurkat T-lymphocytes. Biochem. J. 373(Pt 1): 133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Srivastava S., Zhdanova O., Di L., Li Z., Albaqumi M., Wulff H., and Skolnik E. Y.. 2008. Protein histidine phosphatase 1 negatively regulates CD4 T cells by inhibiting the K+ channel KCa3.1. Proc. Natl. Acad. Sci. USA. 105: 14442–14446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Suzuki N., Suzuki S., Millar D. G., Unno M., Hara H., Calzascia T., Yamasaki S., Yokosuka T., Chen N-J., Elford A. R., et al. 2006. A critical role for the innate immune signaling molecule IRAK-4 in T cell activation. Science. 311: 1927–1932. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained either within the article or as supporting information. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD016476.