Abstract
Bile acids, which are synthesized from cholesterol by the liver, are chemically transformed along the intestinal tract by the gut microbiota, and the products of these transformations signal through host receptors, affecting overall host health. These transformations include bile acid deconjugation, oxidation, and 7α-dehydroxylation. An understanding of the biogeography of bile acid transformations in the gut is critical because deconjugation is a prerequisite for 7α-dehydroxylation and because most gut microorganisms harbor bile acid transformation capacity. Here, we used a coupled metabolomic and metaproteomic approach to probe in vivo activity of the gut microbial community in a gnotobiotic mouse model. Results revealed the involvement of Clostridium scindens in 7α-dehydroxylation, of the genera Muribaculum and Bacteroides in deconjugation, and of six additional organisms in oxidation (the genera Clostridium, Muribaculum, Bacteroides, Bifidobacterium, Acutalibacter, and Akkermansia). Furthermore, the bile acid profile in mice with a more complex microbiota, a dysbiosed microbiota, or no microbiota was considered. For instance, conventional mice harbor a large diversity of bile acids, but treatment with an antibiotic such as clindamycin results in the complete inhibition of 7α-dehydroxylation, underscoring the strong inhibition of organisms that are capable of carrying out this process by this compound. Finally, a comparison of the hepatic bile acid pool size as a function of microbiota revealed that a reduced microbiota affects host signaling but not necessarily bile acid synthesis. In this study, bile acid transformations were mapped to the associated active microorganisms, offering a systematic characterization of the relationship between microbiota and bile acid composition.
Keywords: biosynthesis, metabolome, microbiome, proteomics, Oligo-MM12, fibroblast growth factor 15, Cyp7a1, Sult2a8, farnesoid X receptor
The gut is a complex ecosystem hosting a wide diversity of microorganisms. The gut microbiome is sometimes referred to as the forgotten endocrine organ because of its profound influence on host physiology. Gut microbes convert dietary and other exogenous molecules into signaling metabolites to communicate with the host (1). Bile acids (BAs) are one of these microbiota-derived signaling metabolites.
Primary BAs are synthesized in the liver from cholesterol (dietary or endogenous). In the hepatocytes, they are conjugated to glycine or taurine and stored continuously in the gall bladder as the main component of bile. The human liver synthesizes only two primary BAs [cholic acid (CA) and chenodeoxycholic acid (CDCA)], whereas the rodent liver synthesizes five [CA, CDCA, two muricholic acids (αMCA and βMCA), and ursodeoxycholic acid, the 7β epimer of CDCA (2)]. After food intake, the presence of fat molecules in the duodenum stimulates the release of a hormone, cholecystokinin, which triggers the contraction of the gall bladder and the relaxation of the Oddi sphincter, leading to the release of bile into the small intestine (3). Along the intestinal tract, primary BAs undergo several chemical transformations that are catalyzed by gut microorganisms, leading to the formation of secondary BAs. Thus, the activity of the gut microbiota increases the diversity of the BA pool. The microbial transformation of the BA pool is essential, as it modifies their detergent properties (i.e., their cytotoxicity) and their affinity for host BA receptors (2). For instance, the secondary BAs deoxycholic acid (DCA) and lithocholic acid (LCA), resulting from microbial 7α-dehydroxylation of CA and CDCA, have a higher affinity for the membrane receptor Takeda G-protein receptor 5 (TGR5) compared with the host liver-derived primary BAs (2, 4).
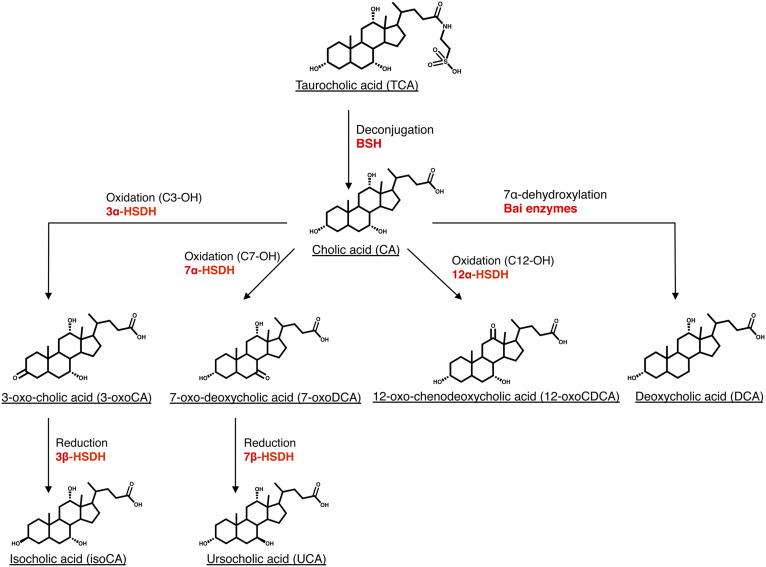
Gut microorganisms act on the side chain, the hydroxyl groups, and the cyclohexane rings in the BA structure. Thus, microbial BA transformations include deconjugation or hydrolysis (i.e., the removal of the taurine/glycine moiety on the side chain); oxidation and epimerization of hydroxyl groups (at the C3, C7, and C12 positions) and of the C5 hydrogen; reduction of ketone groups; dehydroxylation at C7 and C12 (5); desulfation (6); esterification of hydroxyl groups (7); and the oxidation of a steroid ring to form unsaturated BAs (8, 9) (Fig. 1). A new microbial BA transformation was recently identified in both humans and mice: reconjugation. After being deconjugated by bacteria carrying bile salt hydrolase (BSH), the unconjugated BA can be reconjugated by bacteria with amino acids such as tyrosine, phenylalanine, leucine forming tyrosocholic acid, phenylalanocholic acid, and leucocholic acid (10).
Fig. 1.
Microbial BA transformations. Deconjugation (bile salt hydrolysis), oxidation of hydroxyl groups, reduction of ketone group, and BA 7α-dehydroxylation are major BA transformations occurring in the intestinal tract and are those on which we focused in this study. The enzymes catalyzing each transformation are indicated in red. Other microbial BA transformations [e.g., desulfation (65), reconjugation (10), the oxidation of C12-hydroxyl group on taurocholic acid by 12α-HSDH (13, 37)] are not presented in this figure for the sake of clarity.
The abundance and diversity of bacteria carrying out the various BA transformations vary enormously. Deconjugating bacteria, synthesizing BSH, are reported to be abundant in the gut and widespread across bacterial phyla (11, 12). Similarly, oxidation of the hydroxyl groups in BAs is also a common activity in gut microorganisms and is catalyzed by hydroxysteroid dehydrogenases (HSDHs) (13). In contrast, 7α-dehydroxylating bacteria are considered as rare organisms in the gut and so far only belong to the order Clostridiales (14). The 7α-dehydroxylation reaction is catalyzed by a series of proteins (Bai proteins) encoded by genes belonging to the bai operon (15). However, because deconjugation is a prerequisite for BA 7α-dehydroxylation, it is important to consider the relative localization of these two BA transformations (16, 17).
Connecting the spatial organization of the gut microorganisms to biological function remains challenging because of the high diversity of the gut microbial community (18). For BA transformations, this difficulty is compounded by the large number of BA chemical species that can be generated by the gut microbiota. The original study of microbial BA transformation biogeography dates back to 1968, when Midtvedt and Norman sampled luminal content along the intestinal tract of three rats and probed for in vitro BA transformations in each sample (19). First, they noticed variability in the distribution of microbial BA transformations among the three rats. For instance, one rat had very little deconjugating activity in the small intestine (ileum) compared with the two other rats. Two rats showed 7α-HSDH activity (oxidation of the 7α-hydroxyl) in the ileum, while the third rat did not. Since then, other studies have investigated BA profiles longitudinally (20, 21). However, probing for the presence/absence of specific BAs may not accurately reflect the in vivo microbial BA transformation activity or the presence/absence of the organisms responsible for the synthesis of that particular BA. For instance, it was recently observed that BA 7α-dehydroxylation activity was absent in the ileum of gnotobiotic mice despite colonization of the 7α-dehydroxylating organism Clostridium scindens (22). It was hypothesized that the high level of tauro-conjugated BAs in the ileum likely precluded BA 7α-dehydroxylation (22). Furthermore, the ability to transform BAs in vitro does not necessarily reflect the in vivo activity. For instance, Narushima et al. reported that the 7α-dehydroxylating bacterium C. hiranonis (formerly Clostridium strain TO-931), known to deconjugate TCA to CA and to 7α-dehydroxylate cholic acid in vitro, did not show any activity when amended to germ-free (GF) mice (23). Thus, in vivo studies are needed to identify organisms active in BA metabolism within the microbial community of the gut.
In this study, we explored the longitudinal distribution of BA transformations in gnotobiotic mouse models and identified the microorganisms responsible for these transformations using a combined metabolomic and metaproteomic approach. BA 7α-dehydroxylation was a transformation of particular interest because of its connection to TGR5 signaling. We contrasted gnotobiotic mice lacking BA 7α-dehydroxylating activity with gnotobiotic mice containing the same base microbiota but complemented with the 7α-dehydroxylating organism C. scindens ATCC 35704. Finally, for comparison, we considered the BA profile in mice with either a complex microbiota [conventional, specific pathogen-free (SPF) mice], a reduced microbiota (antibiotic-treated SPF mice), or no microbiota (GF mice).
MATERIALS AND METHODS
Animal experiments
Groups of age-matched C57BL/6 mice (6–12 weeks old) were used. GF and gnotobiotic sDMDMm2 mice were established and maintained at the University of Bern Clean Mouse Facility. sDMDMm2 mice were colonized with a mouse-intestine-derived 12-species mouse microbiota [Oligo-MM12] (supplemental Table S1) (24). All Oligo-MM12 strains are available at http://www.dsmz.de/miBC. In-depth descriptions of the Oligo-MM12 consortium species and novel taxa are provided elsewhere (25–27). SPF mice were purchased from Envigo and acclimatized to the local animal facility for 7 days before the start of experiments. SPF mice were pretreated with the antibiotic clindamycin (100 µl by i.p. injection, 2 mg/ml in PBS) or streptomycin (100 µl by gavage, 200 mg/ml in PBS) 24 h before being euthanized. sDMDMm2 mice were inoculated with C. scindens ATCC 35704 by gavage of 107 to 109 colony-forming units of C. scindens and colonized for 7 days before being euthanized. C. scindens precolonization was performed in flexible film isolators. These mice are denoted sDMDMm2+Cs.
All animals were euthanized inside a sterile laminar flow hood, and an aseptic technique was used to collect the specific tissues and contents (liver, terminal ileum tissue, distal duodenum content, ileum content, cecum content, and colon content). The collected samples were flash-frozen in liquid nitrogen and kept at −80°C until subsequent analyses. All animal experiments were performed in accordance with the Swiss Federal and the Bernese Cantonal regulations and were approved by the Bernese Cantonal ethical committee for animal experiments under license numbers BE 82/13 and BE 111/16.
Quantitative real-time PCR
The liver (30 mg of the caudate lobe) and terminal ileum tissue were lysed and homogenized in tubes filled with 2.8 mm zirconium oxide beads using Precellys 24 Tissue Homogenizer (Bertin Instruments) at 6,500 rpm for 3 × 10 s. Total RNA was isolated with the RNeasy Plus Mini Kit (QIAGEN). The QuantiTect Reverse Transcription Kit (QIAGEN) was used to synthesize 20 µl cDNA templates from 1 µg purified RNA. cDNA templates were diluted 10× before use in subsequent reactions. SensiFAST SYBR No-ROX Kit (Bioline) was used for quantitative real-time PCR with a final reaction volume of 10 µl (7.5 µl mix and 2.5 µl diluted cDNA template). Gene-specific primers (400 nM) were used in each reaction, and all results were normalized to the ribosomal protein L32 mRNA (primer sequences can be found in supplemental Table S2). PCR product specificity was verified by performance of a melting curve for each primer set. Assays were performed using Mic qPCR Cycler (Bio Molecular Systems) with a three-step program (2 min at 95°C followed by 40 cycles of 95°C for 5 s, 60°C for 10 s, and 72°C for 10 s). Four replicates of each sample for each primer set were performed, as well as negative controls (no reverse transcription and no template controls). Relative quantification analysis using the REST method was performed with the Mic qPCR analysis software (Bio Molecular Systems).
Preparation of BA standard solutions
Stock solutions (10 mM) of each BA standard were prepared in methanol (supplemental Table S3). Individual standard solutions (100 µM) were mixed together and diluted with a 50:50 (v/v) mix of ammonium acetate (5 mM) and methanol to construct external standard curves between 1 and 10,000 nM. Deuterated CDCA and DCA were used as recovery standards.
BA extraction
Approximately 10 mg of a freeze-dried intestinal sample was homogenized with 150 µl H2O and spiked with 40 µl recovery standard (100 μM) in 2 ml tubes filled with 2.8 mm zirconium oxide beads. Homogenization was performed using an automated Precellys 24 Tissue Homogenizer (Bertin Instruments) at 5,000 rpm for 20 s. Mixed samples were equilibrated on ice for 1 h. A volume of 500 µl ice-cold alkaline acetonitrile (5% ammonia in acetonitrile) was added to the homogenates, which were then vigorously vortexed and continuously shaken for 1 h at room temperature. The mixtures were centrifuged at 16,000 g for 10 min, and the supernatants were collected. The pellets were extracted with another 500 µl of ice-cold alkaline acetonitrile. Supernatants from the two extraction steps were pooled and lyophilized in a rotational vacuum concentrator (Christ) before reconstitution in 100 µl of a 50:50 (v/v) mix of 5 mM ammonium acetate and methanol (pH 6) and stored at −20°C. Supernatants were diluted according to the intestinal compartment (4,000-fold for the small intestine and liver samples and 100-fold for the cecum and colon samples) before LC/MS injection. The extraction recovery ratios ranged between 75% and 85%.
UHPLC/MS analyses
Both qualitative and quantitative analyses were conducted on an Agilent 6530 Accurate-Mass Q-TOF LC/MS mass spectrometer coupled to an Agilent 1290 series UHPLC system. The separation was achieved using a Zorbax Eclipse-Plus C18 column (2.1 × 100 mm, 1.8 μm; Agilent) heated at 50°C. A binary gradient system consisted of 5 mM ammonium acetate (pH 6) in water as eluent A and acetonitrile as eluent B. The sample separation was carried out at 0.4 ml/min over a 22 min total run time using the following program: 0–5.5 min, isocratic 21.5% B; 5.5–6 min, 21.5–24.5% B; 6–10 min, 24.5–25% B; 10–10.5 min, 25–29% B; 10.5–14.5 min, isocratic 29% B; 14.5–15 min, 29–40% B; 15–18 min, 40–45% B; 18–20.5 min, 45–95% B; 20.5–22 min, isocratic 95%.
The system was reequilibrated to initial conditions for 3 min. The sample manager system temperature was maintained at 4°C, and the injection volume was 5 μl. Mass spectrometer detection was operated in the negative ionization mode using the Dual AJS Jet stream ESI Assembly. The QTOF instrument was operated in the 4 GHz high-resolution mode [typical resolution 17,000 (full width at half maximum) at m/z 1,000] in profile mode and calibrated in negative full-scan mode using ESI-L solution (Agilent). Internal calibration was performed during acquisition via continuous infusion of a reference mass solution [5 mM purine, 1 mM HP-921 (Agilent reference mass kit) in 95% methanol acidified with 0.1% formic acid] and allowed to permanently achieve a mass accuracy better than 5 ppm. High-resolution mass spectra were acquired over the range of m/z 300–700 at an acquisition rate of three spectra. AJS settings were as follows: drying gas flow, 8 l/min; drying gas temperature, 300°C; nebulizer pressure, 35 psi; capillary voltage, 3,500 V; nozzle voltage, 1,000 V; fragmentor voltage, 175 V; skimmer voltage, 65 V; octopole 1 RF voltage, 750 V. Data were processed using the MassHunter Workstation (Agilent). According to this method, 36 BAs (supplemental Table S3) were quantified by external calibration curves. Extracted ion chromatograms were based on a retention-time window of ±0.5 min with mass-extraction windows of ±30 ppm centered on the m/ztheor of each BA.
BA-metabolizing enzyme database
Whole genomes for the 12 Oligo-MM12 species (27) and C. scindens ATCC 35704 (16) were obtained from the NCBI and EBI (accession numbers available in supplemental Table S4). Sequences were processed by Prodigal (28) to produce the gene sequences forming the BLASTP search database. Reference protein sequences for BSH, 3α-HSDH, 3β-HSDH, 7α-HSDH, 7β-HSDH, and 12α-HSDH proteins were obtained from NCBI via the accession numbers presented in supplemental Table S5. These sequences were subsequently blasted against the produced protein database. The top five matches, as identified by bit score, were selected for manual review. The final sequences with the highest scores were selected and are presented in supplemental Table S6.
Metaproteome analysis
Proteins were extracted from the small intestine, cecum, and colon content samples of sDMDMm2 (n = 3) and sDMDMm2+Cs mice (n = 3) at the Oak Ridge National Laboratory. Protein extraction and digestion followed by LC/MS/MS analysis were performed as described previously (29) with slight modifications. Briefly, intestinal content samples were suspended in SDS lysis buffer (4% SDS, 100 mM Tris-HCl, 10 mM dithiothreitol, pH 8.0) and subjected to bead beating. The proteins were isolated by chloroform-methanol extraction and resuspended in 4% sodium deoxycholate (SDC) in 100 mM ammonium bicarbonate buffer. The crude protein concentration was estimated using a Nanodrop (Thermo Fisher Scientific). Protein samples (250 μg) were then transferred to a 10 kDa MWCO spin filter (Vivaspin 500; Sartorius), washed with ammonium bicarbonate buffer, and digested with sequencing-grade trypsin.
After proteolytic digestion, the tryptic peptide solution was adjusted to 1% formic acid to precipitate the remaining SDC. The precipitated SDC was removed using water-saturated ethyl acetate. For each sample, 9 μg of peptides were analyzed by automated 2D LC/MS/MS using a Vanquish UHPLC system with the autosampler plumbed directly in-line with a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) across three successive salt cuts of ammonium acetate (35, 100, and 500 mM), each followed by a 155-min organic gradient. Eluting peptides were measured and sequenced by data-dependent acquisition on the Q Exactive Plus as described previously (30).
Peptide identification and protein inference were performed as follows: MS raw data files were searched against the protein database of 12 bacterial species genomes published by Garzetti et al. (27) and the C. scindens ATCC 35704 genome published by Devendran et al. (16) (supplemental Table S4) along with common contaminates (e.g., trypsin, human keratin) appended with a decoy database consisting of reverse protein sequences to control the false-discovery rate (FDR) to 1% at the spectral level (31).
MS/MS spectra data analysis (searching and filtering) was done by the Crux MS analysis toolkit (version 3.2-f7929ba) (32). Tide-search (33) was used for searching. For Tide-search, default settings were used except for the following parameters: allowed clip N-terminal methionine, allowed four missed cleavages, a precursor mass tolerance of 10 ppm, a static modification on cysteines (iodoacetamide; +57.0214 Da), dynamic modifications on methionine (oxidation; +15.9949), and a fragment ion mass tolerance of 0.02 Da. Trypsin was used as the digestion enzyme that cleaves the C terminus to arginine and lysine residues with an exception of proline in the C terminus to lysine and arginine. The obtained search result was processed by Percolator (34) to estimate q values using default parameters. MS1-level precursor intensities were derived from moFF (35) using default parameters along with match between run features from biological replicates. An FDR cutoff of 1% was used as the peptide level. Protein intensities were calculated by summing together FDR-filtered quantified peptides and normalized by performing LOESS and median central tendency procedures on log2-transformed values by InfernoRDN (36). All proteins that had at least one unique peptide sequence were retained for biological interpretation. The list of proteins obtained along with their intensities is available in supplemental Table S7.
RESULTS AND DISCUSSION
Mapping BA transformations in gnotobiotic mice
The first goal of this study was to explore, using a combined metabolomic and metaproteomic approach, the longitudinal distribution of BA transformations in gnotobiotic mouse models (sDMDMm2 and sDMDMm2 + C. scindens) and to identify the microorganisms responsible for these transformations.
Bile salt deconjugation is an active transformation in the large intestine.
Bile salt deconjugation (hydrolysis) constitutes the first step in the microbial BA metabolism (Fig. 1) (12). For instance, 7α-dehydroxylation requires unconjugated BAs (17), while other processes such as dehydrogenation (oxidation) have been reported to occur with conjugated BA (13, 37).
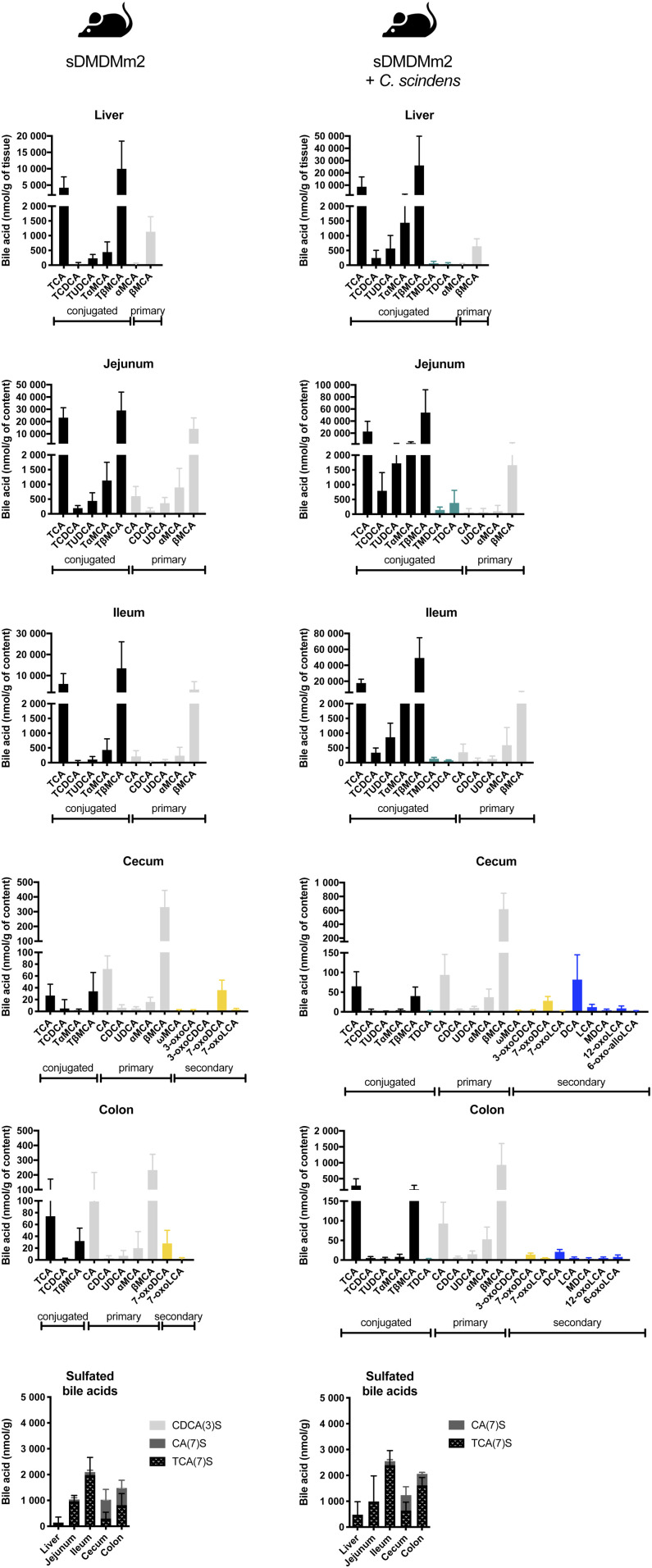
In the small intestine, the amendment of C. scindens to the Oligo-MM12 microbial community led to a decrease in the deconjugating activity. Indeed, in the jejunum of C. scindens-colonized gnotobiotic mice, not only was the concentration of the dominant unconjugated primary BA (βMCA) 8-fold lower than that in the sDMDMm2 cohort in absolute terms, but the ratio of βMCA to its conjugated counterpart (TβMCA) was also 16-fold lower than that in the microbiota lacking C. scindens (Fig. 2). Accordingly, BSH peptides were not detected in the small intestine of C. scindens-colonized gnotobiotic mice (Fig. 3). In contrast, BSH expression was detected in all compartments in the sDMDMm2 cohort. Together, the BA profile and the BSH protein expression data suggest decreased BSH activity in the small intestine in the presence of C. scindens (Figs. 2, 3). This could be due either to differences in microbiota colonization of the gut or, more directly, to changes in the expression of the BSH enzymes. Notable differences were observed in the protein abundance of some strains in the small intestine between the two gnotobiotic mouse models (e.g., Blautia coccoides YL58 and C. clostridioforme YL32; Fig. 4). However, protein abundance of the small intestine BSH-producer Muribaculum intestinale YL27 was comparable between C. scindens-colonized mice and the control group (sDMDMm2 mice) (Fig. 4). Thus, we hypothesize that the expression of BSH enzymes was diminished in the C. scindens-colonized mice relative to the control.
Fig. 2.
BA composition along the intestinal tract of sDMDMm2 and C. scindens-colonized sDMDMm2 gnotobiotic mice based on LC/MS/MS analysis. sDMDMm2 mice (n = 5) and sDMDMm2 mice precolonized 7 days with C. scindens (n = 5) were euthanized, and intestinal content samples were harvested and processed for BA LC/MS/MS measurements. Values indicate measured BA concentrations in nmol/g tissue or intestinal content. Black bars indicate primary conjugated BAs, teal bars indicate secondary conjugated BAs, gray bars indicate primary deconjugated BAs, yellow bars indicate secondary BAs (all but 7-dehydroxylated), and blue bars indicate 7-dehydroxylated BAs.
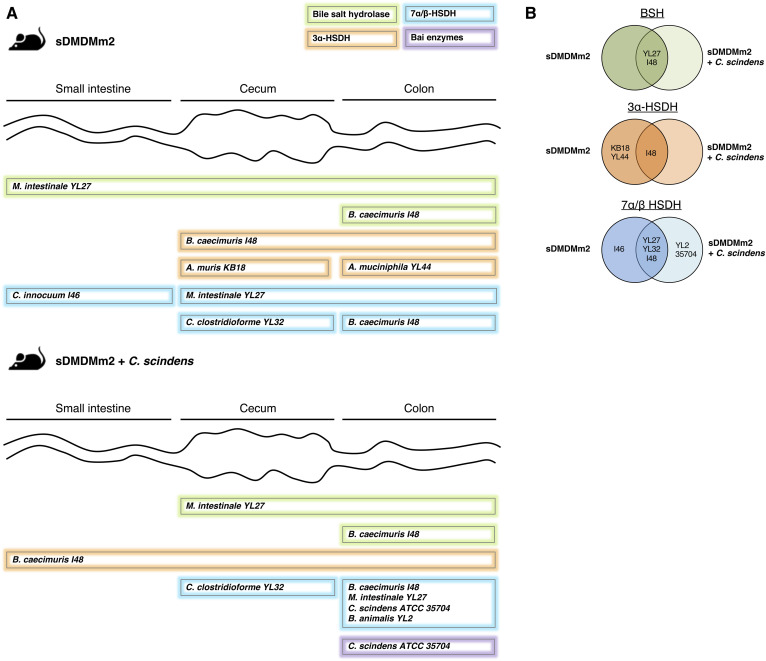
Fig. 3.
Biogeography of specific BA transformation enzymes and the associated microorganisms based on metaproteomic analysis. sDMDMm2 mice (n = 3) and sDMDMm2 mice precolonized 7 days with C. scindens (n = 3) were killed, and intestinal content samples were harvested and processed for metaproteomic analysis. Metaproteomes were mined to find homologous sequences of known BSH, 3α/β-HSDH, 7α/β-HSDH, and 12α-HSDH as well as C. scindens bai genes. A: Scheme illustrating where BSHs, 3α-, 7α-, and 7β-HSDH, and Bai enzymes are expressed in the gut and which bacterial species produce these enzymes. 12α-HSDH and 3β-HSDH were not detected in the metaproteome and therefore are not presented on this figure. Almost all enzymes indicated in panel A were found in the three biological triplicates (supplemental Table S8). The only one that was found in only two mice out of three was the 3α-HSDH from B. caecimuris I48 in the small intestine (supplemental Table S8). B: Venn diagrams summarizing the BA-metabolizing enzymes detected in sDMDMm2 mice and C. scindens-colonized sDMDMm2 mice metaproteomes. For each enzyme detected, the strain producing the enzyme is indicated on the diagram (on the left or right circle if the enzyme was detected in sDMDMm2 mice or C. scindens-colonized sDMDMm2 mice, respectively). If an enzyme was found in both sDMDMm2 and C. scindens colonized-sDMDMm2 mice, the producing strain is indicated in the overlapping part of the two circles.
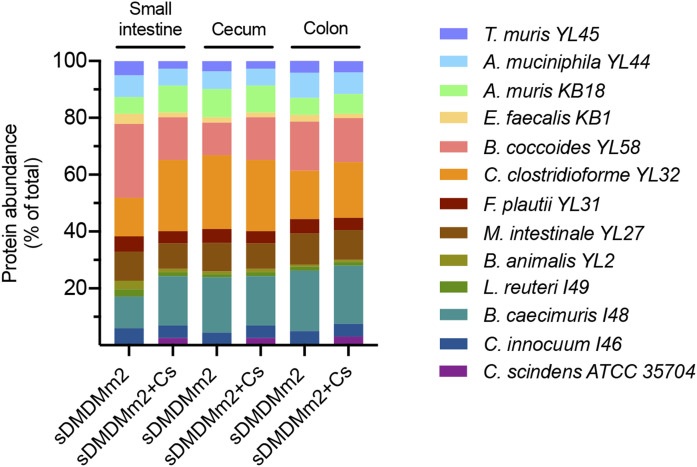
Fig. 4.
Protein abundance along the intestinal tract of sDMDMm2 and C. scindens-colonized sDMDMm2 gnotobiotic mice based on the metaproteomic analysis of intestinal content. sDMDMm2 mice (n = 3) and sDMDMm2 mice precolonized 7 days with C. scindens (n = 3) were sacrificed, and intestinal content samples were harvested and processed for metaproteomic analysis. Of note, proteins (2–4% of total) in the control mice were assigned to C. scindens ATCC 35704 and are not depicted in this figure. Contamination of control mice with C. scindens was evaluated experimentally by plating and confirmed to be negative. C. scindens protein misassignment is likely due to genome relatedness, which is a current challenge in the field of metaproteomic. Using a more stringent approach (matched metaproteome database), we resolved this issue of genome relatedness and did not detect C. scindens proteins in control mice (supplemental Fig. S3).
In the large intestine of the two gnotobiotic mouse models, deconjugated BAs predominated and BSHs were detected, suggesting active bile salt hydrolysis in this compartment (Figs. 2, 3). In contrast to the small intestine, the protein abundance composition was comparable between the two gnotobiotic mouse models in the cecum and colon for all 12 strains (Fig. 4).
While 7 of the 12 organisms in the Oligo-MM12 microbiota harbor BSH-encoding genes (supplemental Table S6), BSHs from only two organisms, M. intestinale YL27 and Bacteroides caecimuris I48, were detected in the metaproteome (Fig. 3, supplemental Table S8). Thus, we propose that these two organisms are the main bile salt-hydrolyzing strains in this mouse model. The BSH of M. intestinale YL27 was highly expressed along the entire intestinal tract of sDMDMm2 mice, while the BSH of B. caecimuris I48 was detectable only in the colon in both types of gnotobiotic mice (Fig. 3).
Thus, based on the BA profile and the metaproteomic data of gnotobiotic mice (both sDMDMm2 and C. scindens-colonized sDMDMm2), we show that bile salt hydrolysis activity was mainly extant in the large intestine (Figs. 2, 3).
BA oxidation is the most widespread transformation in the intestinal tract.
Another common microbial BA transformation is oxidation (dehydrogenation), producing BAs such as 3-oxoCA or 7-oxoDCA (Fig. 1). It has been hypothesized that BA oxidation is a detoxification process. It increases the hydrophilicity of the BA pool and consequently reduces its toxicity for the host and the microbiota (38).
HSDHs catalyze the oxidation and epimerization of hydroxyl groups in the BA structure (Fig. 1). Thirty-two predicted HSDHs were found in the genomes of the Oligo-MM12 species and C. scindens. They included 3α-HSDH (oxidation of 3α-OH and reduction of 3-oxo group), 3β-HSDH (oxidation of 3β-OH and reduction of 3-oxo group), 7α-HSDH (oxidation of 7α-OH and reduction of 7-oxo group), 7β-HSDH (oxidation of 7β-OH and reduction of 7-oxo group), and 12α-HSDH (oxidation of 12α-OH and reduction of 12-oxo group) (Fig. 1, supplemental Table S6). Only 9 of 32 predicted HSDHs were detected with the metaproteomic analysis (Fig. 3; supplemental Tables S6, S8) and corresponded exclusively to 3α-HSDH, 7α-HSDH, and 7β-HSDH (thus missing 3β-HSDH and 12α-HSDH). Furthermore, the BA profiles of gnotobiotic mice suggested that microbial oxidation of BAs only takes place in the large intestine (Fig. 2).
Indeed, oxo-BAs were not detected in the small intestine (Fig. 2) despite the presence of 7β-HSDH from C. innocuum I46 in sDMDMm2 mice and of 3α-HSDH from B. caecimuris I48 in C. scindens-colonized sDMDMm2 mice (Fig. 3, supplemental Table S8). We could speculate that the oxygen gradient (39) along the intestinal tract may have impacted the activity of the HSDH in vivo, but further research should be carried out to test this hypothesis. Hirano and Masuda have reported that oxygen inhibited the activity of the 7α-HSDH of Eubacterium lentum (currently known as Eggerthella lenta) (40). Thus, if the C. innocuum 7α-HSDH is oxygen-sensitive, the higher oxygen level in the small intestine might inactivate it, precluding the formation of 7-oxo BAs. However, oxygen sensitivity has never been reported for 3α-HSDH.
The metaproteomic analysis revealed that seven species of the Oligo-MM12 as well as C. scindens were producers of 3α-HSDH and/or 7α/β-HSDH in the intestinal tract of the gnotobiotic mice (Fig. 3, supplemental Table S8). Interestingly, the amendment of C. scindens clearly decreased the expression of 3α-HSDH in vivo. In the original gnotobiotic mice, 3α-HSDH from Acutalibacter muris KB18, Akkermansia muciniphila YL44, and B. caecimuris I48 were present in the large intestine, while in the mice amended with C. scindens only the one from B. caecimuris I48 was detected (Fig. 3).
In contrast, the amendment of C. scindens enhanced the expression of 7α/β-HSDHs. The 7β-HSDH from Bifidobacterium animalis YL2 and the 7α-HSDH from C. scindens were observed only in the metaproteome of C. scindens-colonized animals. Additionally, in both cohorts, the 7α-HSDHs from M. intestinale YL27 and B. caecimuris I48, as well as the 7β-HSDH from C. clostridioforme YL32, were encountered. These findings demonstrate that a single strain can profoundly alter the BA composition and expression of BA-metabolizing enzymes within a given microbial community. The remaining question is whether C. scindens affected the expression of enzymes directly or impacted the abundance of certain species. Previously, Studer et al. have observed only minor changes in the fecal Oligo-MM12 microbiota composition following the addition of C. scindens (41). These minor changes included a decrease in abundance of A. muris KB18 that could explain the absence of 3α-HSDH from this species in the C. scindens-colonized cohort (Fig. 3). However, the protein abundance for the 12 Oligo-MM12 species was comparable in the large intestine of the two-gnotobiotic mouse model, suggesting that the amendment of C. scindens had little impact on the large intestine microbial community (Fig. 4).
Furthermore, Devendran and colleagues have reported a downregulation, by the secondary BA DCA, of many genes encoding BA-transforming enzymes in C. scindens (16). Among them, baiA2 (HDCHBGLK_01433) encodes a 3α-HSDH (42). We hypothesize that C. scindens-specific metabolites, such as 7-dehydroxylated secondary BAs (e.g., DCA), may affect the expression of BA-metabolizing enzymes by other bacteria of the Oligo-MM12 consortium. The nondetection of 3α-HSDH associated with A. muris KB18 and A. muciniphila YL44 in gnotobiotic mice colonized with C. scindens (Fig. 3) supports this hypothesis. Interestingly, 3β-HSDHs were entirely absent in the metaproteome. Consistent with this result, iso- BAs were not detected in the gnotobiotic animals (Figs. 1, 2). 12α-HSDHs were also lacking in the metaproteome despite the presence of 12-oxoLCA in C. scindens-colonized mice (Fig. 2). C. scindens is known to carry a gene encoding a 12α-HSDH and could be an active producer of 12-oxoLCA in vivo. Marion et al. have reported the formation of 12-oxoLCA in vitro and proposed it as a transient intermediate of CA 7α-dehydroxylation by C. scindens (22). Here, we hypothesize that 12α-HSDH from C. scindens is present in too low abundance to be detected with metaproteomic analysis. In line with this hypothesis, the other enzymes of the 7α-dehydroxylation pathway were absent in the metaproteome despite the clear presence of 7α-dehydroxylated secondary BAs (Fig. 2).
BA 7α-dehydroxylation is restricted to the large intestine.
The bile acid profile in the large intestine was very similar for mice models with and without C. scindens, with the exception of the presence of 7α-dehydroxylated species in the model including C. scindens (Fig. 2). The 7α-dehydroxylated species detected included LCA, DCA, and murideoxycholic acid (MDCA), the conjugated form of the latter two (TDCA and TMDCA), as well as 7α-dehydroxylated and oxidized species (6-oxoLCA, 6-oxo-alloLCA, 12-oxoLCA).
Among enzymes of the 7α-dehydroxylation pathway, only the 7α-hydroxy-3-oxo-Δ4-cholenoic acid oxidoreductase (43) was detected in the colon of the C. scindens-colonized cohort. None of the Bai enzymes were detected in the small intestine of these mice (Fig. 3, supplemental Table S8).
Based on the metaproteomic and metabolomic data, we show that BA 7α-dehydroxylation occurs only in the large intestine of mice colonized with C. scindens (Figs. 2, 3). This observation supports an earlier study in which the large intestine was demonstrated to be the ecological niche of C. scindens in the gnotobiotic sDMDMm2 mouse model (22).
While all 12 microorganisms within the Oligo-MM12 microbiota harbor BA-modifying enzymes (supplemental Table S6), peptides from only seven of them were detected that mapped onto BA-related enzymes (Fig. 3, supplemental Table S8). Because the metaproteomic approach is limited in measurement dynamic range, we cannot rule out the activity of the five other strains but can conclude that these seven are the most active in BA transformations.
The second goal of the study was to assess the impact of the microbiota on the BA pool. To tackle this, we compared the BA pools of gnotobiotic mice to ones of mouse models with various gut microbiota complexities: a very complex microbiota (SPF), a reduced microbiota (antibiotic-treated SPF), or the absence of microbiota (GF).
The microbiota shapes the BA pool composition
GF mice are devoid of microbial BA transformations. Thus, their BA pool was exclusively composed of liver-derived tauro-conjugated primary BAs (Fig. 5, supplemental Fig. S2). TCA and TβMCA dominated the BA pool in all compartments (i.e., they represented ∼94% of the liver BA pool). Smaller contributions from TCDCA, TUDCA, and TαMCA were also observed. In the liver, a small fraction of unconjugated βMCA (836 ± 544 nmol/g tissue) was also noted along with very minor contributions (<10 nmol/g content) of the αMCA and βMCA in some intestinal compartments (ileum, cecum, and colon). These observations support an earlier study from Sayin et al. (20), who have also observed minor fractions of unconjugated MCAs in GF mice. We hypothesize that the high BA synthesis activity in the liver does not allow for conjugating enzymes to act on all of the newly synthesized BAs, resulting in the release into the intestine of a fraction of primary BAs that have escaped conjugation in the liver. As expected, secondary BAs were altogether absent in the GF mouse intestine due to the dearth of microorganisms.
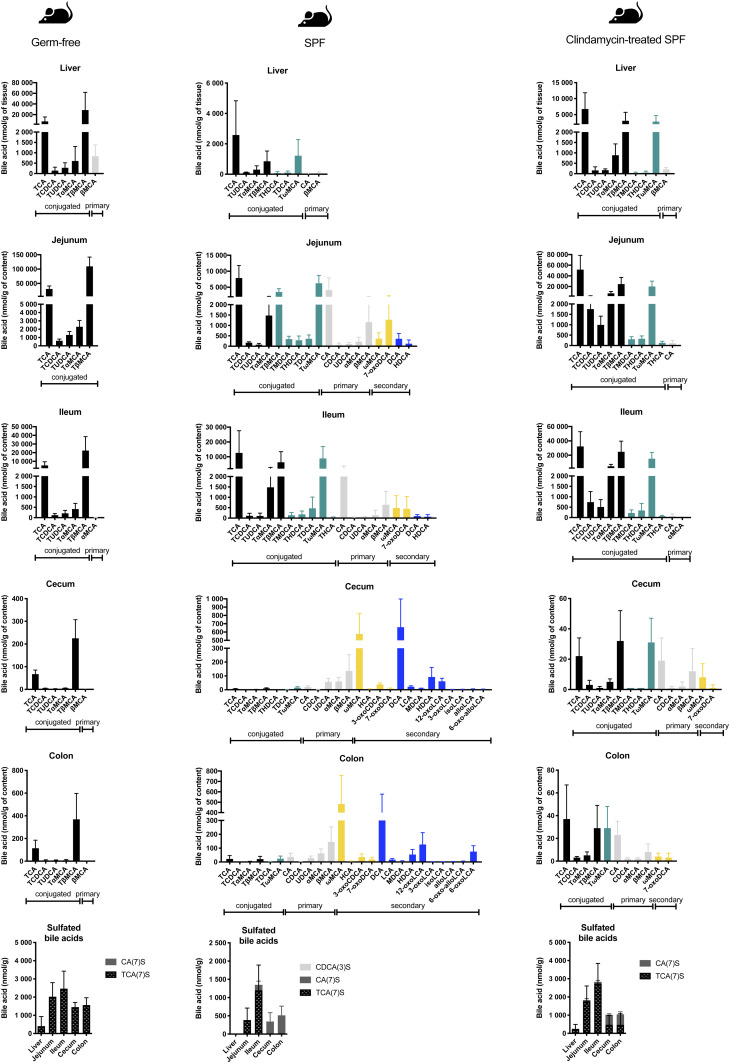
Fig. 5.
BA composition along the intestinal tract in GF, SPF, and clindamycin-treated SPF mice based on LC/MS/MS analysis. SPF mice were pretreated with clindamycin 24 h before being euthanized. Intestinal content samples were harvested from GF mice (n = 5), SPF mice (n = 5), and clindamycin-treated SPF mice (n = 5). Samples were processed for BA LC/MS/MS measurements. Values indicate measured BA concentrations in nmol/g tissue or intestinal content. Black bars indicate primary conjugated BAs, teal bars indicate secondary conjugated BAs, gray bars indicate primary deconjugated BAs, yellow bars indicate secondary BAs (all but 7-dehydroxylated), and blue bars indicate 7-dehydroxylated BAs.
For comparison, we also considered SPF mice with or without antibiotic treatment. As expected, SPF mice exhibited the highest BA diversity (Fig. 5). Secondary BAs in SPF mice were present in the small intestine, while in C. scindens-bearing gnotobiotic animals, they were detected only in the large intestine (Figs. 2, 5). As the mouse liver is able to reconjugate deconjugated BAs, tauro-conjugated 7α-dehydroxylated BAs (TMDCA, THDCA, TDCA, TωMCA) were observed in the upper intestinal tract. Thus, it is not possible to determine whether the 7α-dehydroxylated BAs DCA and HDCA in the small intestine come from 7α-dehydroxylation of CA and hyocholic acid (HCA; the 6α epimer of αMCA) or from the deconjugation of TDCA and THDCA in that compartment (Fig. 5).
In contrast to the other models, secondary BAs represented more than half of the BA pool in the large intestine of SPF mice, and the conjugated BAs were present at low levels (supplemental Fig. S2). Additionally, 7α-dehydroxylated secondary BAs, comprising the 7-dehydroxylated form of the primary BAs (DCA, LCA, MDCA, HDCA), the oxidized species (12-oxoLCA, 3-oxoLCA, 6-oxoLCA, 6-oxo-alloLCA), and the epimerized species (isoLCA, alloLCA), represented 57% of the pool of secondary BAs (Fig. 5, supplemental Fig. S2). Finally, SPF mice were the only mice to produce iso-BAs (3β epimers), allo-BAs (5α epimers), and 6α epimers (e.g., HCA) in the large intestine (Fig. 5).
Antibiotic treatment had a tremendous impact on BA transformation throughout the intestinal tract. In the small intestine, BA diversity was extremely reduced compared with that in untreated SPF mice (Fig. 5). The most salient impacts were the suppression of BA deconjugation in the small intestine and the complete abolition of 7α-dehydroxylation by the clindamycin treatment (Fig. 5). Streptomycin-treated animals exhibited decreased amounts of 7α-dehydroxylated species but this antibiotic was not as efficient as clindamycin at inhibiting 7α-dehydroxylation, suggesting the survival of 7α-dehydroxylating bacteria to streptomycin treatment (supplemental Fig. S1). Interestingly, tauroconjugated 7α-dehydroxylated species (TMDCA and THDCA) were detected in the small intestine (Fig. 5). We attribute the presence of those species to the 7-dehydroxylation of MCAs (αMCA, βMCA, ωMCA) and HCA prior to the antibiotic treatment, followed by recycling via the enterohepatic circulation and reconjugation in the liver before release into the small intestine.
7α-Dehydroxylated BAs have been linked to protection against Clostridioides difficile infections (CDIs) through the inhibitory role of 7-dehydroxylated BAs in C. difficile spore germination and outgrowth (44, 45). Based on the BA profile of clindamycin- and streptomycin-treated mice (Figs. 5, supplemental Fig. S1), it would be predicted that treatment with streptomycin is less of a risk factor for CDI than treatment with clindamycin and that 7α-dehydroxylating organisms are sensitive to clindamycin but not streptomycin. In accordance with these hypotheses, Theriot et al. have shown that cecal content from mice treated with clindamycin triggers spore germination and cell outgrowth of C. difficile (46).
Furthermore, Slimings and Riley have reported no association between the use of aminoglycosides (an antibiotic class that includes streptomycin) and CDIs, while clindamycin was the antibiotic most frequently associated with CDIs (both hospital-acquired and community-acquired) in patient meta-analyses (47–49). Additionally, two of the major BA 7α-dehydroxylating organisms, C. scindens and C. hylemonae, were sensitive to clindamycin (supplemental Table S9). Furthermore, two studies have observed that restricting the use of clindamycin led to a sustained decrease in CDIs (50, 51). Buffie et al. have shown that the cecal microbiota of SPF mice treated with clindamycin underwent profound losses of Lachnospiraceae, a family that includes several 7α-dehydroxylating strains (e.g., C. scindens and C. hylemonae) (52). These observations are all consistent with clindamycin effectively inhibiting BA 7-dehydroxylation and thus creating conditions propitious for the germination and outgrowth of C. difficile spores. These data support previous research that reported drastic changes in the fecal BA pool following antibiotic treatment (46, 53). The novelty of the present study is the extensive BA profile for the liver and four intestinal compartments (duodenum, ileum, cecum, and colon). Other studies have focused on fewer sample types [liver and plasma (53), gallbladder and serum (20), or ileum and cecum (46)]. Furthermore, the number of BAs screened in this study is greater compared with previous studies (20, 46, 53), offering more in-depth reporting of the impact of antibiotic treatment on BA composition. This study also reports for the first time the antibiotic susceptibility profiles of three major BA 7-dehydroxylating species: C. scindens, C. hylemonae, and C. hiranonis (supplemental Table S9).
Microbiota-host interactions regulate the BA pool size
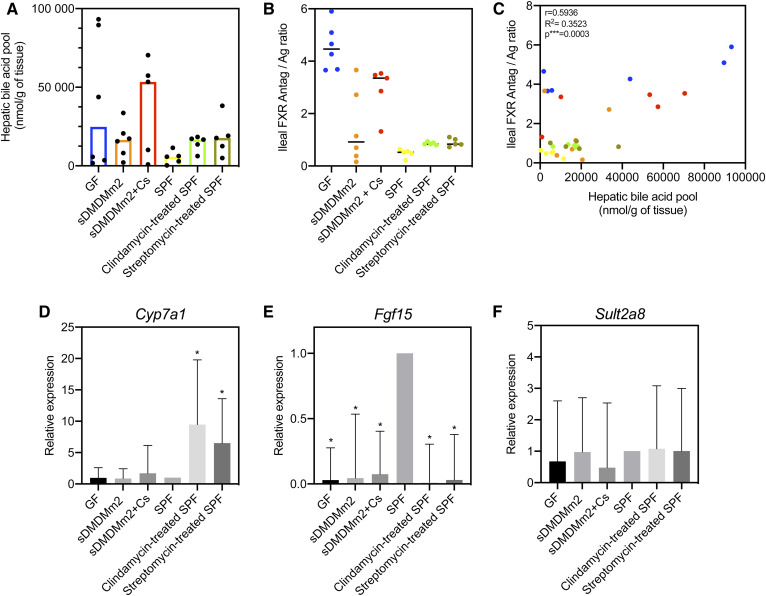
Supporting earlier research (20), our data showed that GF mice harbor an abundant BA pool (Fig. 5, supplemental Fig. S2). Here, we also observed that mouse models with a reduced microbiota (gnotobiotic and antibiotic-treated mice) have an increased hepatic BA pool compared with conventional mice (SPF) harboring a more diverse microbiota (Fig. 6A). Furthermore, we noted a large interindividual variability of the hepatic BA pool in GF and gnotobiotic mice cohorts compared with that in SPF and antibiotic-treated SPF mice (Fig. 6A). This clearly indicates that BA synthesis is more tightly regulated in hosts with more complex microbial communities (e.g., SPF). In mammals, the nuclear receptor farnesoid X receptor (FXR), highly expressed in the ileum (54), controls hepatic BA synthesis through a negative feedback loop (55). The activation of FXR by BAs induces the release of fibroblast growth factor 15 (Fgf15) in the intestine, which reaches the liver via portal vein circulation and inhibits the expression of the hepatic enzyme Cyp7a1 (cholesterol 7α-hydroxylase), which is responsible for catalyzing the rate-limiting step in BA synthesis (20).
Fig. 6.
Probing the influence of the gut microbiota on BA synthesis and detoxification. A: Hepatic BA pool. Each dot corresponds to an individual mouse. Histograms show the median of the hepatic BA pool for the mouse models considered. B: Ileal FXR antagonist/agonist ratio. TαMCA and TβMCA were considered to be FXR antagonists, and CDCA, CA, DCA, LCA, TCDCA, TCA, TDCA, and TLCA were considered to be FXR agonists, according to Parks et al. (57). Each dot corresponds to an individual mouse. The horizontal bar in the scatterplot indicates the median for the mouse models considered. C: Correlation between the hepatic BA pool and the ileal FXR antagonist/agonist ratio. Each dot corresponds to an individual mouse. Pearson correlation analysis was computed with GraphPad Prism. D–F: Gene expression of Cyp7a1, Fgf15, and Sult2a8 in each mouse model considered relative to SPF mice (n = 5 mice for each mouse model). A value of 1 is attributed to SPF mice. *P < 0.05.
The tauro-conjugated murine primary BAs are strong antagonists of FXR (56), while CDCA and the 7α-dehydroxylated species DCA and LCA are potent FXR agonists (57), thus acting as repressors of BA synthesis. Gut microbes are important actors in FXR-mediated regulation of BA synthesis because they decrease the concentration of FXR antagonists (e.g., TβMCA, TαMCA) and produce unconjugated BAs and secondary BAs that activate FXR (20). The role of the deconjugating microbial community is particularly important, as deconjugation contributes to a decrease in the concentration of the FXR antagonists TMCAs (58) and allows further transformation such as 7α-dehydroxylation (17). GF mice, devoid of microbial BA transformations, had the largest BA pool size compared with that of other mice models harboring a microbial community (Fig. 5, supplemental Fig. S2). As has previously been demonstrated (20), we observed that the concentration of TβMCA was high in GF mice, resulting from the absence of microbial deconjugation (Fig. 5). Consequently, the FXR-dependent negative feedback loop was inhibited, resulting in increased BA synthesis (20) and hepatic BA pool (Fig. 6A).
The ileal ratio of FXR antagonists (TMCAs) over FXR agonists (57) (CDCA + CA + TCDCA + TCA + DCA + LCA + TDCA + TLCA) was a good indicator of the size of the hepatic BA pool (Fig. 6B). Indeed, this relationship offers an explanation as to why C. scindens-colonized sDMDMm2 mice have a higher BA pool size compared with sDMDMm2 mice (Fig. 6A). This higher pool size was due to the increased FXR antagonist/agonist ratio in those mice relative to mice lacking C. scindens (Fig. 6B), explaining the higher BA synthesis. The increased ratio was likely due to reduced BSH activity in the small intestine, as was reflected in the metaproteomic data, with no BSH activity detected in the small intestine (Fig. 3). The reduced BSH activity led to an increased concentration of FXR antagonists (particularly TMCAs) in the ileum. Indeed, the TβMCA level was 3-fold greater in the ileum of the C. scindens-colonized cohort compared with that in the sDMDMm2 mice (Fig. 2). Similar to the hepatic BA pool, there was a large interindividual variability in the ileal FXR antagonist/agonist ratio in GF and gnotobiotic cohorts compared with that in SPF and antibiotic-treated SPF mice (Fig. 6B). This ileal ratio is relevant only for mouse studies, as it takes into account the murine-specific conjugated primary BA (TMCA) as FXR antagonists, which are absent in humans. In humans, very little is known about endogenous FXR antagonists. Only LCA has been reported to have FXR antagonist activity in addition to partial agonist activity in cells (59). Further research is needed to investigate the ligand potency of human primary, secondary, and tertiary BAs toward the FXR receptor.
Antibiotic treatment also affected BA-deconjugating activity, and most likely, FXR signaling, leading to increased BA synthesis (Fig. 6). To better understand the FXR-mediated regulation of BA synthesis by the gut microbiota, we measured the ileal expression of Fgf15 and hepatic expression of Cyp7a1 in our mice models. Mice models with no or a reduced microbiota (GF, gnotobiotic, and antibiotic-treated SPF) showed a significantly reduced ileal expression of Fgf15 (more than 10-fold) relative to SPF mice (Fig. 6E).
The expression of Cyp7a1 was significantly increased in antibiotic-treated mice compared with untreated ones (Fig. 6D). In contrast, we did not detect a significant difference in Cyp7a1 expression between GF and SPF mice, as previously reported by Sayin et al. (20). However, the increase reported by Sayin et al. is only 2-fold and could be considered minor when discussing gene expression. In line with our observation, a recent study have reported no difference in Cyp7a1 mRNA expression between GF and SPF mice with a chow diet (60). Although not statistically significant, C. scindens-colonized mice showed an increased Cyp7a1 expression compared with sDMDMm2 and GF mice, which is consistent with the higher BA pool (Fig. 6D).
Altogether, these data show that having a reduced microbiota impacts FXR signaling (via the downregulation of Fgf15). BA synthesis was mostly impacted by the antibiotic-mediated dysbiosis. There was a large interindividual variation in the hepatic BA pool and ileal FXR antagonist/agonist ratio in GF and gnotobiotic mice (Fig. 6A–C) that was not observed in SPF and antibiotic-treated mice. This suggests that the BA synthesis was more tightly regulated in mice harboring a more diverse microbial community.
BA detoxification
Intestinal sulfated BAs were measured in this study. BA sulfation is a major host metabolic pathway for detoxifying and eliminating BAs. Sulfotransferases (e.g., Sult2a1) catalyze the sulfation (or sulfonation) of BAs in the liver. Sulfation of BAs increases their solubility, reduces the intestinal absorption, and consequently fosters their urinary and fecal elimination. In humans, about 70% of the BA pool in the urine is sulfated, but under pathological conditions (e.g., cholestatic diseases) the fraction of sulfated BA increases (61).
Sulfation in SPF mice was low compared with the GF, gnotobiotic, and antibiotic-treated mouse models (Figs. 2, 5). In SPF mice, sulfated BAs were not detected in the liver, and the amount of sulfated BAs in the intestine was 2-fold smaller than that of the mouse models with a reduced microbial community (Figs. 2, 5). Miyata et al. have reported that the hepatic sulfotransferase Sult2a1 is negatively regulated through FXR in mice and humans (62). Sult2a8 was recently identified as a major BA-sulfonating enzyme in mice (63). Here, we observed that the expression of Sult2a8 was comparable among the various mouse models, suggesting that Sult2a8 is neither regulated through the FXR nor influenced by the gut microbiota (Fig. 6F).
CONCLUSION
BAs exert crucial functions in the host that go beyond their role as lipid detergents, and gut microbes play a central role in BA metabolism. By modifying the structure of the BA, microorganisms influence their affinity to host BA receptors. In this study, we characterized the biogeography of BA transformation in gnotobiotic mice. We identified the main organisms involved in BA metabolism in a gnotobiotic mouse model and the localization of their activity in the gut. Furthermore, by comparing the BA profiles of mouse models with either reduced or more complex microbiota, we emphasized the profound influence of the gut microbial community on the regulation of the BA pool size and composition. Finally, by measuring the expression of Fgf15, Cyp7a1, and Sult2a8, we demonstrated that a reduced microbiota affects FXR signaling but not necessarily BA synthesis. The latter was mainly impacted by antibiotic-mediated dysbiosis. Finally, we report that BA sulfonation by Sult2a8, the major BA sulfotransferase in mice, does not seem to be mediated by the FXR or influenced by the gut microbiota.
Data availability
All raw mass spectra for the proteome measurements have been deposited into the ProteomeXchange repository with the following accession numbers: MassIVE accession: MSV000084484; ProteomeXchange: PXD015971; FTP link to files: ftp://MSV000084484@massive.ucsd.edu. All other data are either contained in the article or available upon request from the corresponding author.
Supplementary Material
Acknowledgments
The authors thank Gilbert Greub and Guy Prod’Hom for their help with the antimicrobial susceptibility testing; the academic and technical staff at the University of Bern Clean Mouse Facility for their services and support; Alessia Perino for her assistance with the quantitative real-time PCR; and Léonard Jequier for his assistance with the bioinformatic analysis.
Footnotes
This article contains supplemental data.
Author contributions—S.M., L.D., N.S., S.H., and R.B.L. conceived the experiments; S.M., N.S., Y.D., and M.N. performed in vivo experiments; L.D., L.M., and S.M. performed bile acid measurements and analysis; S.P. and R.H. performed the metaproteomic analysis; S.M. performed RT-qPCR analysis; A.J. performed bioinformatic analysis; S.M. and R.B.L. wrote the paper. All authors read and commented on the final manuscript.
Author ORCIDs—Solenne Marion https://orcid.org/0000-0003-1662-7898; Rizlan Bernier-Latmani https://orcid.org/0000-0001-6547-722X
Funding and additional information—This work was supported by Swiss National Science Foundation Sinergia Grants CRSII3_147603 and CRSII5_180317.
Conflict of interest—The authors declare that they have no conflict of interest with the contents of this article.
Abbreviations—
- BA
- bile acid
- BSH
- bile salt hydrolase
- CA
- cholic acid
- CDCA
- chenodeoxycholic acid
- CDI
- Clostridioides difficile infection
- DCA
- deoxycholic acid
- FDR
- false-discovery rate
- Fgf15
- fibroblast growth factor 15
- FXR
- farnesoid X receptor
- GF
- germ-free
- HCA
- hyocholic acid
- HDCA
- hyodeoxycholic acid
- HSDH
- hydroxysteroid dehydrogenase
- LCA
- lithocholic acid
- MCA
- muricholic acid
- MDCA
- murideoxycholic acid
- SDC
- sodium deoxycholate
- SPF
- specific pathogen-free
Manuscript received July 9, 2020. Published, JLR Papers in Press, July 13, 2020, DOI 10.1194/jlr.RA120001021.
REFERENCES
- 1.Schroeder B. O., and Bäckhed F.. 2016. Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 22: 1079–1089. [DOI] [PubMed] [Google Scholar]
- 2.Wahlström A., Sayin S. I., Marschall H. U., and Bäckhed F.. 2016. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 24: 41–50. [DOI] [PubMed] [Google Scholar]
- 3.Hofmann A. F. 1999. The continuing importance of bile acids in liver and intestinal disease. Arch. Intern. Med. 159: 2647–2658. [DOI] [PubMed] [Google Scholar]
- 4.Ridlon J. M., Kang D. J. D., Hylemon P. B. P., and Bajaj J. J. S.. 2014. Bile acids and the gut microbiome. Curr. Opin. Gastroenterol. 30: 332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edenharder R. 1984. Dehydroxylation of cholic acid at C12 and epimerization at C5 and C7 by Bacteroides species. J. Steroid Biochem. 21: 413–420. [DOI] [PubMed] [Google Scholar]
- 6.Huijghebaert S., Parmentier G., and Eyssen H.. 1984. Specificity of bile salt sulfatase activity in man, mouse and rat intestinal microflora. J. Steroid Biochem. 20: 907–912. [DOI] [PubMed] [Google Scholar]
- 7.Kelsey M. I., Molina J. E., Huang S. K., and Hwang K. K.. 1980. The identification of microbial metabolites of sulfolithocholic acid. J. Lipid Res. 21: 751–759. [PubMed] [Google Scholar]
- 8.Macdonald I. A., Bokkenheuser V. D., Winter J., McLernon A. M., and Mosbach E. H.. 1983. Degradation of steroids in the human gut. J. Lipid Res. 24: 675–700. [PubMed] [Google Scholar]
- 9.Prabha V., and Ohri M.. 2006. Review: bacterial transformations of bile acids. World J. Microbiol. Biotechnol. 22: 191–196. [Google Scholar]
- 10.Quinn R. A., Vrbanac A., Melnik A. V., Patras K. A., Christy M., Nelson A. T., Aksenov A., Tripathi A., Humphrey G., da Silva R., et al. . 2020. Global chemical effects of the microbiome include new bile-acid conjugations. Nature 579: 123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urdaneta V., and Casadesús J.. 2017. Interactions between bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front. Med. (Lausanne). 4: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foley M. H., O’Flaherty S., Barrangou R., and Theriot C. M.. 2019. Bile salt hydrolases: gatekeepers of bile acid metabolism and host-microbiome crosstalk in the gastrointestinal tract. PLoS Pathog. 15: e1007581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doden H., Sallam L. A., Devendran S., Ly L., Doden G., Daniel S. L., Alves J. M. P., and Ridlon J. M.. 2018. Metabolism of oxo-bile acids and characterization of recombinant 12α-hydroxysteroid dehydrogenases from bile acid 7α-dehydroxylating human gut bacteria. Appl. Environ. Microbiol. 84: e00235-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ridlon J. M., Kang D. J., and Hylemon P. B.. 2006. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47: 241–259. [DOI] [PubMed] [Google Scholar]
- 15.Ridlon J. M., Harris S. C., Bhowmik S., Kang D-J. J., and Hylemon P. B.. 2016. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes. 7: 22–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devendran S., Shrestha R., Alves J. M. P., Wolf P. G., Ly L., Hernandez A. G., Méndez-García C., Inboden A., Wiley J., Paul O., et al. . 2019. Clostridium scindens ATCC 35704: integration of nutritional requirements, the complete genome sequence, and global transcriptional responses to bile acids. Appl. Environ. Microbiol. 85: e00052-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Batta A. K., Salen G., Arora R., Shefer S., Batta M., and Person A.. 1990. Side chain conjugation prevents bacterial 7-dehydroxylation of bile acids. J. Biol. Chem. 265: 10925–10928. [PubMed] [Google Scholar]
- 18.Tropini C., Earle K. A., Huang K. C., and Sonnenburg J. L.. 2017. The gut microbiome: connecting spatial organization to function. Cell Host Microbe. 21: 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Midtvedt T., and Norman A.. 1968. Anaerobic, bile acid transforming microorganisms in rat intestinal content. Acta Pathol. Microbiol. Scand. 72: 337–344. [DOI] [PubMed] [Google Scholar]
- 20.Sayin S. I., Wahlström A., Felin J., Jäntti S., Marschall H-U., Bamberg K., Angelin B., Hyötyläinen T., Orešič M., and Bäckhed F.. 2013. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17: 225–235. [DOI] [PubMed] [Google Scholar]
- 21.Selwyn F. P., Csanaky I. L., Zhang Y., and Klaassen C. D.. 2015. Importance of large intestine in regulating bile acids and glucagon-like peptide-1 in germ-free mice. Drug Metab. Dispos. 43: 1544–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marion S., Studer N., Desharnais L., Menin L., Escrig S., Meibom A., Hapfelmeier S., and Bernier-Latmani R.. 2019. In vitro and in vivo characterization of Clostridium scindens bile acid transformations. Gut Microbes. 10: 481–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narushima S., Itoh K., Takamine F., and Uchida K.. 1999. Absence of cecal secondary bile acids in gnotobiotic mice associated with two human intestinal bacteria with the ability to dehydroxylate bile acids in vitro. Microbiol. Immunol. 43: 893–897. [DOI] [PubMed] [Google Scholar]
- 24.Eberl C., Ring D., Münch P. C., Beutler M., Basic M., Slack E. C., Schwarzer M., Srutkova D., Lange A., Frick J. S., et al. . 2020. Reproducible colonization of germ-free mice with the oligo-mouse-microbiota in different animal facilities. Front. Microbiol. 10: 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagkouvardos I., Pukall R., Abt B., Foesel B. U., Meier-Kolthoff J. P., Kumar N., Bresciani A., Martínez I., Just S., Ziegler C., et al. . 2016. The Mouse Intestinal Bacterial Collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nat. Microbiol. 1: 16131. [DOI] [PubMed] [Google Scholar]
- 26.Brugiroux S., Beutler M., Pfann C., Garzetti D., Ruscheweyh H-J., Ring D., Diehl M., Herp S., Lötscher Y., Hussain S., et al. . 2016. Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium. Nat. Microbiol. 2: 16215. [DOI] [PubMed] [Google Scholar]
- 27.Garzetti D., Brugiroux S., Bunk B., Pukall R., McCoy K. D., Macpherson A. J., and Stecher B.. 2017. High-quality whole-genome sequences of the oligo-mouse-microbiota bacterial community. Genome Announc. 5: e00758-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hyatt D., Brugiroux S., Bunk B., Pukall R., McCoy K. D., Macpherson A. J., and Stecher B.. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 11: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patnode M. L., Beller Z. W., Han N. D., Cheng J., Peters S. L., Terrapon N., Henrissat B., Le Gall S., Saulnier L., Hayashi D. K., et al. . 2019. Interspecies competition impacts targeted manipulation of human gut bacteria by fiber-derived glycans. Cell. 179: 59–73.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clarkson S. M., Giannone R. J., Kridelbaugh D. M., Elkins J. G., Guss A. M., and Michener J. K.. 2017. Construction and optimization of a heterologous pathway for protocatechuate catabolism in Escherichia coli enables bioconversion of model aromatic compounds. Appl. Environ. Microbiol. 83: e01313-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elias J. E., and Gygi S. P.. 2007. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 4: 207–214. [DOI] [PubMed] [Google Scholar]
- 32.Park C. Y., Klammer A. A., Käli L., MacCoss M. J., and Noble W. S.. 2008. Rapid and accurate peptide identification from tandem mass spectra. J. Proteome Res. 7: 3022–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diament B. J., and Noble W. S.. 2011. Faster SEQUEST searching for peptide identification from tandem mass spectra. J. Proteome Res. 10: 3871–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Käll L., Canterbury J. D., Weston J., Noble W. S., and MacCoss M. J.. 2007. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 4: 923–925. [DOI] [PubMed] [Google Scholar]
- 35.Argentini A., Goeminne L. J. E., Verheggen K., Hulstaert N., Staes A., Clement L., and Martens L.. 2016. moFF: a robust and automated approach to extract peptide ion intensities. Nat. Methods. 13: 964–966. [DOI] [PubMed] [Google Scholar]
- 36.Polpitiya A. D., Qian W-J., Jaitly N., Petyuk V. A., Adkins J. N., Camp D. G., Anderson G. A., and Smith R. D.. 2008. DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics. 24: 1556–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris S. C., Devendran S., Méndez-García C., Mythen S. M., Wright C. L., Fields C. J., Hernandez A. G., Cann I., Hylemon P. B., and Ridlon J. M.. 2018. Bile acid oxidation by Eggerthella lenta strains C592 and DSM 2243 T. Gut Microbes. 9: 523–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe M., Fukiya S., and Yokota A.. 2017. Comprehensive evaluation of the bactericidal activities of free bile acids in the large intestine of humans and rodents. J. Lipid Res. 58: 1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donaldson G. P., Lee S. M., and Mazmanian S. K.. 2016. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14: 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirano S., and Masuda N.. 1981. Transformation of bile acids by Eubacterium lentum. Appl. Environ. Microbiol. 42: 912–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Studer N., Desharnais L., Beutler M., Brugiroux S., Terrazos M. A., Menin L., Schürch C. M., McCoy K. D., Kuehne S. A., Minton N. P., et al. . 2016. Functional intestinal bile acid 7α-dehydroxylation by Clostridium scindens associated with protection from Clostridium difficile infection in a gnotobiotic mouse model. Front. Cell. Infect. Microbiol. 6: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhowmik S., Jones D. H., Chiu H-P., Park I-H., Chiu H-J., Axelrod H. L., Farr C. L., Tien H. J., Agarwalla S., and Lesley S. A.. 2014. Structural and functional characterization of BaiA, an enzyme involved in secondary bile acid synthesis in human gut microbe. Proteins. 82: 216–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kang D-J., Ridlon J. M., Moore D. R., Barnes S., and Hylemon P. B.. 2008. Clostridium scindens baiCD and baiH genes encode stereo-specific 7α/7β-hydroxy-3-oxo-Δ4-cholenoic acid oxidoreductases. Biochim. Biophys. Acta. 1781: 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen A. 2015. A gut odyssey: the impact of the microbiota on Clostridium difficile spore formation and germination. PLoS Pathog. 11: e1005157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buffie C. G., Bucci V., Stein R. R., McKenney P. T., Ling L., Gobourne A., No D., Liu H., Kinnebrew M., Viale A., et al. . 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 517: 205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Theriot C. M., Bowman A. A., and Young V. B.. 2016. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. MSphere. 1: e00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deshpande A., Pasupuleti V., Thota P., Pant C., Rolston D. D. K., Sferra T. J., Hernandez A. V., and Donskey C. J.. 2013. Community-associated Clostridium difficile infection antibiotics: a meta-analysis. J. Antimicrob. Chemother. 68: 1951–1961. [DOI] [PubMed] [Google Scholar]
- 48.Brown K. A., Khanafer N., Daneman N., and Fisman D. N.. 2013. Meta-analysis of antibiotics and the risk of community-associated Clostridium difficile infection. Antimicrob. Agents Chemother. 57: 2326–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Slimings C., and Riley T. V.. 2014. Antibiotics and hospital-acquired Clostridium difficile infection: update of systematic review and meta-analysis. J. Antimicrob. Chemother. 69: 881–891. [DOI] [PubMed] [Google Scholar]
- 50.Pear S. M., Williamson T. H., Bettin K. M., Gerding D. N., and Galgiani J. N.. 1994. Decrease in nosocomial Clostridium difficile-associated diarrhea by restricting clindamycin use. Ann. Intern. Med. 120: 272–277. [DOI] [PubMed] [Google Scholar]
- 51.Climo M. W., Israel D. S., Wong E. S., Williams D., Coudron P., and Markowitz S. M.. 1998. Hospital-wide restriction of clindamycin: effect on the incidence of Clostridium difficile-associated diarrhea and cost. Ann. Intern. Med. 128: 989–995. [DOI] [PubMed] [Google Scholar]
- 52.Buffie C. G., Jarchum I., Equinda M., Lipuma L., Gobourne A., Viale A., Ubeda C., Xavier J., and Pamer E. G.. 2012. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect. Immun. 80: 62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swann J. R., Want E. J., Geier F. M., Spagou K., Wilson I. D., Sidaway J. E., Nicholson J. K., and Holmes E.. 2011. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc. Natl. Acad. Sci. USA. 108: 4523–4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Houten S. M., Volle D. H., Cummins C. L., Mangelsdorf D. J., and Auwerx J.. 2007. In vivo imaging of farnesoid X receptor activity reveals the ileum as the primary bile acid signaling tissue. Mol. Endocrinol. 21: 1312–1323. [DOI] [PubMed] [Google Scholar]
- 55.Chiang J. Y. L. 2009. Bile acids: regulation of synthesis. J. Lipid Res. 50: 1955–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang H., Chen J., Hollister K., Sowers L. C., and Forman B. M.. 1999. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell. 3: 543–553. [DOI] [PubMed] [Google Scholar]
- 57.Parks D. J., Blanchard S. G., Bledsoe R. K., Chandra G., Consler T. G., Kliewer S. A., Stimmel J. B., Willson T. M., Zavacki A. M., Moore D. D., et al. . 1999. Bile acids: natural ligands for an orphan nuclear receptor. Science. 284: 1365–1368. [DOI] [PubMed] [Google Scholar]
- 58.Li F., Jiang C., Krausz K. W., Li Y., Albert I., Hao H., Fabre K. M., Mitchell J. B., Patterson A. D., and Gonzalez F. J.. 2013. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat. Commun. 4: 2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu J., Lo J-L., Huang L., Zhao A., Metzger E., Adams A., Meinke P. T., Wright S. D., and Cui J.. 2002. Lithocholic acid decreases expression of bile salt export pump through farnesoid X receptor antagonist activity. J. Biol. Chem. 277: 31441–31447. [DOI] [PubMed] [Google Scholar]
- 60.Just S. 2017. Impact of the Interplay between Bile Acids, Lipids, Intestinal Coriobacteriaceae and Diet on Host Metabolism. PhD Dissertation. Technical University of Munich, Munich, Germany. [Google Scholar]
- 61.Alnouti Y. 2009. Bile acid sulfation: a pathway of bile acid elimination and detoxification. Toxicol. Sci. 108: 225–246. [DOI] [PubMed] [Google Scholar]
- 62.Miyata M., Matsuda Y., Tsuchiya H., Kitada H., Akase T., Shimada M., Nagata K., Gonzalez F. J., and Yamazoe Y.. 2006. Chenodeoxycholic acid-mediated activation of the farnesoid X receptor negatively regulates hydroxysteroid sulfotransferase. Drug Metab. Pharmacokinet. 21: 315–323. [DOI] [PubMed] [Google Scholar]
- 63.Dawson P. A., and Setchell K. D. R.. 2017. Will the real bile acid sulfotransferase please stand up? Identification of Sult2a8 as a major hepatic bile acid sulfonating enzyme in mice. J. Lipid Res. 58: 1033–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huijghebaert S. M., Mertens J. A., and Eyssen H. J.. 1982. Isolation of a bile salt sulfatase-producing Clostridium strain from rat intestinal microflora. Appl. Environ. Microbiol. 43: 185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw mass spectra for the proteome measurements have been deposited into the ProteomeXchange repository with the following accession numbers: MassIVE accession: MSV000084484; ProteomeXchange: PXD015971; FTP link to files: ftp://MSV000084484@massive.ucsd.edu. All other data are either contained in the article or available upon request from the corresponding author.