Abstract
Bile acids (BAs) have been established as ubiquitous regulatory molecules implicated in a large variety of healthy and pathological processes. However, the scope of BA heterogeneity is often underrepresented in current literature. This is due in part to inadequate detection methods, which fail to distinguish the individual constituents of the BA pool. Thus, the primary aim of this study was to develop a method that would allow the simultaneous analysis of specific C24 BA species, and to apply that method to biological systems of interest. Herein, we describe the generation and validation of an LC-MS/MS assay for quantification of numerous BAs in a variety of cell systems and relevant biofluids and tissue. These studies included the first baseline level assessment for planar BAs, including allocholic acid, in cell lines, biofluids, and tissue in a nonhuman primate (NHP) laboratory animal, Macaca mulatta, in healthy conditions. These results indicate that immortalized cell lines make poor models for the study of BA synthesis and metabolism, whereas human primary hepatocytes represent a promising alternative model system. We also characterized the BA pool of M. mulatta in detail. Our results support the use of NHP models for the study of BA metabolism and pathology in lieu of murine models. Moreover, the method developed here can be applied to the study of common and planar C24 BA species in other systems.
Keywords: bile acid metabolism, bile salts, biosynthesis, liquid chromatography, tandem mass spectrometry, method development, high-performance liquid chromatography, quantitation
Bile acids (BAs), the amphipathic metabolites of cholesterol, have been recognized as having a multitude of regulatory properties that extend beyond the emulsification and absorption of lipids in the gut. These many functions are discussed in detail in several recent reviews (1–5). Furthermore, these roles have implicated this extremely diverse group of molecules in the progression of many disease and injury states, including, among others: hepatic and intestinal cancer (6–8), liver steatosis and associated NASH and NAFLD (9–12), diabetes (13–15), metabolic disease (16), and drug-induced liver injury (17). Understanding the particular changes in the BA pool that occur in these conditions can aid in diagnostic and prognostic assessments thereof. Moreover, a deeper awareness of the mechanisms that give rise to the perturbations in the BA pool during disease can assist in the understanding of hepatic and gastrointestinal (GI) function and pathology.
The biosynthetic pathway of BAs is highly complex, resulting in an exceptionally diverse pool of BAs mostly within the liver, GI tract, bile, plasma, urine, and feces (2, 5). BAs and their derivatives can also accumulate in other tissues, especially in illness (1, 3, 5, 18). In health, the BA pool is mainly made up of conjugated primary BAs; however, in disease and following injury, dysregulation in BA synthesis and/or homeostasis can result in the accumulation of unusual and intermediate species (1, 3, 5, 19–21). Thus, the total number of possible BA derivatives that circulate in disease is orders of magnitude more diverse than in health, demonstrating the need for a more sensitive quantification method that captures this high heterogeneity in the multiple compartments of the BA pool.
This phenomenon of distortion in the BA pool has already been demonstrated in numerous studies in human patients in the disease states mentioned above as well as in several animal models (10, 22–28). Nevertheless, there remain multiple subsets of BAs that have yet to be characterized in much detail, if at all. Further investigation is needed to determine whether the appearance of these molecules in disease is related to disease progression, either as instigators or byproducts, and whether they can be used to aid in clinical detection and prognoses. Because it is still unclear exactly why, how, and which of these BA species develop in various disease states, investigating them is critical for understanding the molecular mechanisms behind these conditions. Unfortunately, due to their high degree of structural similarity, most detection methods are insufficient to simultaneously detect and differentiate the large variety of BAs (29, 30). One such case is the planar BAs, a category that differs in their 3D structure, resulting in a “flat” or “planar” conformation contrary to the more typical “bent” or “twisted” shape of the steroid backbone (1, 25, 31). The planar BAs are of interest due to their resurgence in the circulating BA pool in several types of liver disease, but the high degree of similarity to the more abundant BAs make them challenging to detect in vivo (1, 23–25, 31). Historically, BA heterogeneity was detected with GC-MS (8, 23, 32–34). Recently, several research groups have taken advantage of the high specificity and selectivity of LC-MS/MS to reliably detect and quantify these molecules among their more typical counterparts (18, 30, 35–37). The study herein sought to build upon and extend previous methodologies and simultaneously characterize BAs in vitro and in vivo.
BA metabolism is often studied using animal models that assume similarity to human systems. Murine BA metabolism is well studied and known to have several discrepancies when compared with that of humans, but mouse and rat models are still used most often (5, 38–45). Nonhuman primate (NHP) models represent an attractive alternative because of their increased genetic resemblance to man; however, BA metabolism in NHPs has not been as well characterized (46, 47). A secondary purpose of this study was to apply the method developed herein to establish a detailed baseline profile of the BA pool in relevant tissues and biofluids in NHPs, for the comparison to injury models in this animal and to humans. Similarly, cultured cell systems of human origin are also popular in studying liver physiology and pharmacology, but these models, too, are poorly detailed with regard to BA synthesis and metabolism. Examples of primary and immortalized cell systems were examined. Herein, the development, validation, and application of a sensitive UPLC-MS/MS method for the simultaneous quantification of abundant BAs, as well as uncommon and planar mammalian BA species, is demonstrated in human cell lysates and cell culture media and in NHP liver tissue, bile, and plasma.
MATERIALS AND METHODS
Chemicals and reagents
All solvents used were of LC-MS grade or higher and purchased from Fischer Scientific (Pittsburgh, PA). Solid standards were purchased from either Sigma-Aldrich (St. Louis, MO), Toronto Research Chemicals (North York, ON, Canada), Cambridge Isotope Laboratories (Tewksbury, MA), Isosciences (Ambler, PA), Bridge Organics (Vicksburg, MI), or Steraloids (Newport, RI) (supplemental Table S1). The 3-oxo-cholic acid (3-oxo-CA) was generously provided by Dr. James E. Polli’s laboratory (University of Maryland, Baltimore, MD).
Preparation of standard solutions and calibrants
Stock solutions of each BA were prepared at 500 μg/ml by dissolving each solid standard in 100% methanol and then sonicating in a 45°C water bath. Twelve mixed standard solutions containing between 0.05 and 2,500 ng/ml of each analyte standard [CA, glycocholic acid (GCA), taurocholic acid (TCA), chenodeoxycholic acid (CDCA), glycochenodeoxycholic acid (GCDCA), taurochenodeoxycholic acid (TCDCA), ursodeoxycholic acid (UDCA), glycoursodeoxycholic acid (GUDCA), allocholic acid (ACA), isoallolithocholic acid (IALCA), lithocholic acid (LCA), glycolithocholic acid (GLCA), taurolithocholic acid (TLCA), deoxycholic acid (DCA), glycodeoxycholic acid (GDCA), taurodeoxycholic acid (TDCA), 3-oxo-CA, 3-oxo-chol-4-enic acid, and 7-α-hydroxy-3-oxo-chol-4-en-24-oic acid] and 50 ng/ml of each deuterium-labeled internal standard (IS) [cholic acid (2,2,4,4-d4) (CA-d4) and GCDCA (2,2,4,4-d4) (GCDCA-d4)] were prepared in mobile phase [1:1 acetonitrile (ACN):water with 0.01% formic acid (FA)] by serial dilution. From these, calibration curves for each analyte were constructed.
Immortalized cell lines and culture conditions
COS-1 and HepG2 cells were purchased from ATCC (Manassas, VA). HuH-7 cells were a generous gift from Dr. Hongbing Wang (University of Maryland, Baltimore, MD). COS-1, HuH-7, and HepG2 cells were cultured in DMEM supplemented with 10% FBS, penicillin (100 IU/ml), and streptomycin (100 μg/ml) (Life Technologies, Inc., Rockville, MD). For the analysis of basal BA production in immortalized cell lines, cells were plated at a density of 2.0 × 106 cells per 10 cm culture dish in DMEM containing no supplements and in hepatocyte culture medium (HCM) and maintained at 37°C with 5% CO2 for 48 h (Lonza, Basel, Switzerland). Human primary hepatocytes (HPHs) were obtained from BioIVT (Baltimore, MD). Hepatocytes at ≥90% viability were seeded at 1.5 × 106 cells/well in a 6-well collagen-coated plate in InVitroGRO CP medium (BioIVT). After overnight attachment at 37°C in a humidified atmosphere of 5% CO2, the culture medium was changed to complete Williams’ E medium. The cells were overlaid with 0.25 mg/ml Matrigel (Corning Inc., Corning, NY) for another 24 h before the medium was switched to HCM. The medium was then aspirated and stored at −20°C until further preparation for LC/MS-MS injection. Before lysis, cells were rinsed with cold PBS. If not further processed immediately, culture dishes were stored at −80°C.
Animal model
All animal procedures were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and experiments were performed with prior approval from the University of Maryland IACUC. Animals were housed and cared for in accordance with the Animal Welfare Act at the University of Maryland’s Association for Assessment and Accreditation of Laboratory Animal Care-Accredited Facility. Plasma, bile, and liver tissue were obtained from male rhesus macaques (Macaca mulatta) with a mean age of 4.5 years. These NHP samples were derived from a naïve nonirradiated (no sham treatment) group of multiple irradiation studies (48–51).
Sample preparation
HybridSPE phospholipid 96-well plates (Sigma-Aldrich), ISOLUTE PLD+ protein and phospholipid removal plates, and ISOLUTE PLD+ phospholipid depletion columns (Biotage, Uppsala, Sweden) were used to evaluate different methods of solid-phase extraction (SPE). The phospholipid depletion columns use a selective interaction with the phosphate moiety present in all phospholipids to retain and separate phospholipids from small molecule analytes, such as BAs. All SPE methods examined use solvent crash/filtration protocols to filter precipitated proteins and phospholipids from matrices with the goal of improved signal-to-noise. Sample preparation for plasma, bile, and liver utilize the ISOLUTE PLD+ phospholipid depletion columns using procedures as described below.
Cell culture sample preparation.
Cell culture medium was aspirated. One milliliter aliquots of culture medium were added to 4 ml ACN and 5 ng IS in glass culture tubes and vortexed quickly, then centrifuged at 500 g for 10 min. Four milliliters of each supernatant were transferred to fresh glass culture tubes and dried down under N2 flow. Extracts were resuspended in 100 μl 1:1 ACN:water with 0.01% FA, resulting in a final IS concentration of 50 ng/ml, and transferred to LC-MS vials and then stored at −20°C until injection. For cells, 1 ml RIPA buffer with 5 ng of each IS was added to each plate or well following a PBS wash. Culture dishes were rocked at 4°C for 1 h and then scraped. Total protein was estimated at this point using the Bradford assay. Lysates were added to 4 ml 100% ACN in glass culture tubes and vortexed quickly and then centrifuged at 2,000 rpm for 10 min. Four milliliters of each supernatant were transferred to fresh glass culture tubes and dried down under N2 flow. Extracts were resuspended in 100 μl 1:1 ACN:water with 0.01% FA, resulting in a final IS concentration of 50 ng/ml, and transferred to LC-MS vials and then stored at −20°C until injection.
Plasma and bile preparation.
ISOLUTE PLD+ phospholipid depletion columns were filled before use with 400 μl ACN with 1% FA as protein crash solvent. Five microliters of 1 ng/μl IS solution and 100 μl plasma aliquot were added to the column and vortexed. Flow-through was then collected into glass culture tubes by applying approximately 4 psi positive pressure by nitrogen gas. Eluates were further dried down by nitrogen flow and then resuspended in 100 μl 1:1 ACN:water with 0.01% FA for LC-MS/MS injection. If resuspensions were not injected that day, samples were stored at −80°C. For bile samples, the protocol was performed similarly, save that the aliquoted bile was diluted 1:9 or 1:1,000 with water and 5 μl of 1 ng/μl IS or 100 μl of 50 ng/μl IS, respectively to each dilution, were added before being applied to the ISOLUTE PLD+ column. Additionally, the bile that was diluted 1:1,000 with water preextraction was diluted 1:100 postextraction by resuspending in 100 μl 1:1 ACN:water with 0.01% FA and then diluting 10 μl into 1,000 μl of 1:1 ACN:water with 0.01% FA (resulting in a final dilution factor of 1:100,000) for the analysis of highly abundant analyte species. For all dilutions, final injections contained 50 ng/ml of each IS.
Liver sample preparation.
Approximately 50 mg sections of liver initially removed from the central right lobe of naïve rhesus macaques were preweighed and added to Fisherbrand™ prefilled bead mill tubes (2ml, 1.4mm beads) with 750 μl 50% methanol. Tissue sections were homogenized using a Precellys 24 tissue homogenizer run at 6,500 rpm for 10 s; tubes were then centrifuged at 500 g for 1 min. 600 μl of each homogenate was aspirated and spiked with IS (1 ng each IS per 10 mg tissue section weight), then transferred to a new tube with 3 ml ice-cold alkaline ACN (ACN with 5% v/v NH4OH, equal to 1.15M NH4OH) and vortexed for 5 s. Homogenates were shaken at ambient room temperature (23 ± 2°C) for 1 h, then centrifuged at 5,000 g for 10 min. 3 ml of each supernatant was applied to ISOLUTE PLD+ columns. Approximately 4 p.s.i. positive pressure was applied to collect flow-through into glass culture tubes. Flow-through fractions were dried under N2 flow and then resuspended in 1:1 ACN:water with 0.01% FA proportionally to starting tissue section weight and IS added. Final injections thus contained 0.5 mg of liver tissue per μl of resuspension. The use of a basic crash solvent in this procedure has the disadvantage of dehydrating 7-hydroxy groups in 7-hydroxy-3-oxo-4-ene BA structures, causing loss of at least one analyte of interest during sample preparation (52).
LC-MS/MS
Chromatography.
The LC-MS/MS method used was adapted from (35). LC-MS/MS analysis was performed on a Waters I-Class UPLC coupled to a Waters Xevo TQ-XS triple quadrupole mass spectrometer (Waters Corporation, Milford, MA). Separation was effected by a Waters ACQUITY BEH (ethylene bridged hybrid) C18 UPLC column (150 × 2.1 mm, 1.7 μm) using gradient elution with the following mobile phases: water with 0.01% FA (solvent A) and ACN with 0.01% FA (solvent B). The gradient was as follows: 25–40% solvent B in 12 min and then 40–75% solvent B in 14 min. Solvent B was increased to 100% over 30 s and held for 2 min to wash the column. Return to 25% B was accomplished over 30 s and allowed to reequilibrate over 4 min. The column was maintained at 55°C with a flow rate of 350 μl/min. Two microliters of sample were injected per run.
ESI-MS/MS.
Detection was performed in negative ion mode using ESI. Source conditions were as follows: capillary voltage was 2.50 kV, and cone voltage was 10 V. The desolvation temperature was set to 500°C and the source temperature to 150°C. The desolvation gas flow operated at 800 l/h, the cone gas flow at 150 l/h, and the collision gas flow at 0.15 ml/min. The Q1 low mass resolution was set to 3.06 and the high mass resolution to 14.84. The Q3 low mass resolution was set to 2.73 and high mass resolution to 15.11. Ion guide offsets were set to 3 V and 0.3 V. Both the entrance and exit potentials were set to 1.0. Dwell time for each analyte was 7.0 ms.
BA species were detected using scheduled multiple reaction monitoring wherein each BA is detected with multiple reaction monitoring according to a unique precursor to product ion m/z transition or selected ion monitoring where a single ion is detected in both Q1 and Q3 with no fragmentation (supplemental Table S3). Each BA was further identified by its chromatographic retention time (supplemental Table S3).
Data processing.
Data were processed using Waters MassLynx and TargetLynx software (version 4.1 SCN 901). Analyte responses were determined by the peak area of the BA, and analyte concentrations were determined by calibration curves constructed for each BA standard by plotting the ratio of the response for each BA to the IS response against the nominal concentration. Linearity of calibration curves was assessed through linear regression analysis using a weighting factor of 1/x. Calculated concentrations of analytes were converted from nanograms per milliliter to nanograms per milligram of starting liver tissue by dividing by a factor of 500 to reflect the theoretical 500 mg of liver tissue in every 1 ml of final prepared resuspension used for injection. Bile measurements were multiplied by their dilution factors (i.e., either ×10 or ×100,000) to reflect the initial dilution with water and postextraction dilution in mobile phase.
Method validation
Method validation studies were designed by following the FDA’s Guidance for Industry for Bioanalytical Method Validation for sensitivity, linear range, precision, accuracy, recovery, and bench-top stability. Because BAs are endogenous compounds and blank matrix is not feasible to obtain, the nonendogenous IS compounds were used as surrogates to validate the LC-MS/MS method. Intra- and inter-day precision was determined by injection of the same set of prepared plasma samples spiked with IS 4 and 24 h apart, respectively. Similarly, plasma spiked with IS was left at room temperature (23 ± 2°C) for 4 h before performing sample preparation and compared with plasma spiked with IS prepared freshly in order to assess benchtop precision. Identical validation experiments were conducted for bile, liver tissue, and cultured cells (COS-1 cells used for validation experiments) and cell media.
Statistics
Student’s t-test analyses were performed using GraphPad Prism software (version 8.3.0).
RESULTS
Method development
Sample preparation optimization.
Initially, the protocol from Han et al. (35) was duplicated for use in plasma; however, this resulted in poor extraction efficiency (5–15%) with the HybridSPE-Phospholipid 96-well plates and continuously had issues with the wells becoming clogged with biological samples. However, a similar sample preparation method provided by another vendor, i.e., the ISOLUTE PLD+ protein and phospholipid removal plate, which provided better recovery but exhibited high variability between wells. Subsequently, it was determined that using the ISOLUTE PLD+ phospholipid depletion columns yielded the most robust extraction [i.e., highest and most consistent recovery and lowest coefficient of variation values and utilized this method for the remainder of our experiments in biofluids and tissue. The protocol developed for use with these columns required less than half the time; it was also less labor- and resource-intensive than other methods (35). Biotage ISOLUTE PLD+ protein and phospholipid depletion columns were utilized for plasma, bile, and liver sample preparation. Cultured cells and media did not require SPE procedures.
LC-MS/MS.
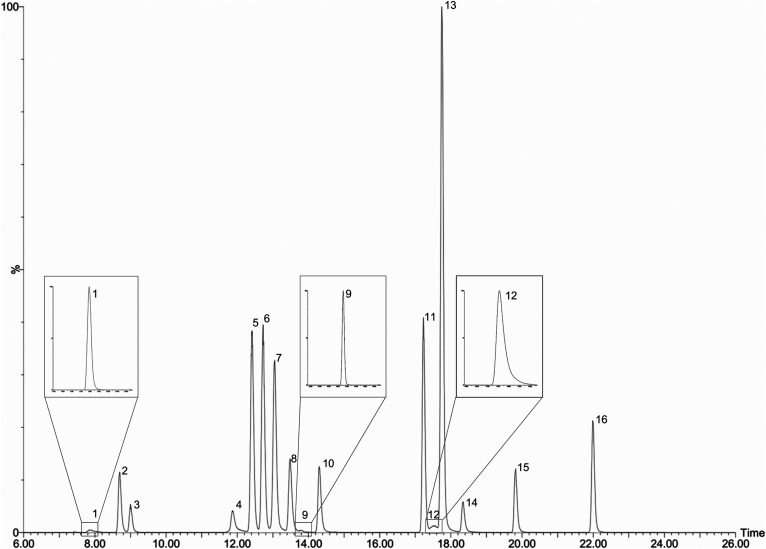
The LC-MS/MS assay included 19 BA analytes and used stable isotope-labeled ISs (supplemental Table S3). CA-d4 was used as an IS for the unconjugated BAs (LCA, CDCA, DCA, UDCA, CA, ACA, 7α-hydroxy-3-oxo-chol-4-en-24-oic acid, IALCA, 3-oxo-chol-4-enic acid, and 3-oxo-CA), and GCDCA-d4 was used as an IS for the conjugated BAs (GLCA, GDCA, GUDCA, GCA, TLCA, TDCA, TCDCA, and TCA). BAs were resolved with a reverse-phase gradient UPLC separation using a 150 mm column with a sub-2 μm particle size C18 stationary phase with an ACN/water/FA-based mobile phase. Retention times ranged from 7.9 min (TCA) to 22.0 min (LCA) (supplemental Table S3). BAs that fragmented well were detected according to an m/z transition (supplemental Table S3). Some BAs exhibited poor fragmentation efficiency and were more suitably detected by monitoring the precursor ion in both Q1 and Q3 (LCA, CDCA, DCA, UDCA, CA, ACA, and IALCA). Those BAs that shared a similar nominal mass and utilized the same selected reaction monitoring transitions were chromatographically resolved (Fig. 1).
Fig. 1.
Total ion chromatogram of included analytes inset with individual m/z transition channels for those analytes with less prominent peaks (1, 9, and 12). Authentic standards dissolved in mobile phase at 500 ng/ml each. 1: TCA; 2: GUDCA; 3: GCA; 4: TCDCA, UDCA, and 3-oxo-CA; 5: ACA; 6: CA and CA-d4; 7: TDCA; 8: GCDCA and GCDCA-d4; 9: 7α-hydroxy-3-oxo-chol-4-en-24-oic acid; 10: GDCA; 11: CDCA; 12: TLCA; 13: DCA; 14: GLCA; 15: IALCA and 3-oxo-chol-4-enic acid; 16: LCA.
Method validation
BA sensitivity and linearity were evaluated using authentic standards for each BA. Because BAs are endogenous species, ISs (CA-d4 and GCDCA-d4) were used as nonendogenous BA surrogates to evaluate intra- and inter-day precision and accuracy, apparent recovery, and benchtop stability (supplemental Table S2).
Sensitivity and linearity.
Sensitivity and linearity were determined by evaluating calibrants ranging in concentration from 0.05 to 2,500 ng/ml. The limit of detection (LOD) is defined as those points within the calibration curves with a signal-to-noise ratio (S/N) greater than 3. The lower limit of quantification (LLOQ) is defined as the lowest point on the calibration curve whose S/N is greater than 10 and whose calculated value is ≤20% of the nominal value. The LOD for BAs in this assay ranged from 0.05 to 10 ng/ml with most BAs having LODs of 0.05–0.5 ng/ml (Table 1). LLOQs for the various BAs are represented by the lower limit of the calibration curves in Table 1 and were between 0.1 and 10 ng/ml with most BAs having LLOQs having ≤0.5 ng/ml. The most sensitive LLOQ was TLCA (0.1 ng/ml), whereas the least sensitive LLOQ was for 3-oxo-chol-4-enic acid (10 ng/ml). The upper limit of quantification was 2,500 ng/ml for all BAs included in this assay. Linearity was evaluated via regression coefficients (r2) for calibration curves that exceeded 0.99, ranging from 0.990 to 0.999 (Table 1).
TABLE 1.
Summary of linear ranges and LODs and LLOQs for BAs in this assay
| Analyte | LOD (ng/ml) | Linear range (ng/ml) | r2 |
| LCA | 0.5 | 0.5–2,500 | 0.991 |
| CDCA | 0.5 | 0.5–2,500 | 0.991 |
| DCA | 0.1 | 0.5–2,500 | 0.995 |
| UDCA | 0.5 | 0.5–2,500 | 0.997 |
| CA | 0.1 | 0.5–2,500 | 0.990 |
| ACA | 0.5 | 1–2,500 | 0.991 |
| GLCA | 0.05 | 0.5–2,500 | 0.998 |
| GDCA | 0.05 | 0.5–2,500 | 0.999 |
| GUDCA | 0.05 | 0.5–2,500 | 0.999 |
| GCDCA | 0.05 | 0.5–2,500 | 0.997 |
| GCA | 0.1 | 0.5–2,500 | 0.999 |
| TLCA | 0.05 | 0.1–2,500 | 0.997 |
| TDCA | 1 | 5–2,500 | 0.995 |
| TCDCA | 0.1 | 1–2,500 | 0.997 |
| TCA | 0.5 | 1–2,500 | 0.999 |
| IALCA | 5 | 5–2,500 | 0.992 |
| 3-oxo-chol-4-enic acid | 10 | 10–2,500 | 0.990 |
| 3-oxo-CA | 0.5 | 5–2,500 | 0.990 |
| 7α-hydroxy-3-oxo-chol-4-en-24-oic acid | 0.5 | 1–2,500 | 0.996 |
These values were determined using authentic standards dissolved in mobile phase. LOD is defined as analyte peak area having S/N >3. LLOQ is represented as the lower end of the linear range and is defined as analyte peak area having S/N >10 and deviation from the nominal value less than ±20%. The r2 value reflects fit to linear regression with 1/x weighting.
Precision and accuracy.
Intra-day and inter-day instrument precision and accuracy were evaluated for plasma, bile, liver, cell lysate, and cell media. Intra-day precision for all matrices evaluated was less than 15%, ranging from 2.4% to 14.6% (supplemental Table S2). Inter-day precision was also less than 15%, ranging from 2.6% to 14.5%. (supplemental Table S2). Intra- and inter-day accuracy for all matrices ranged from 80.4% to 119.5% and from 88.2% to 113.3%, respectively.
Recovery.
Apparent recovery of CA-d4 and GCDCA-d4 in cell lysates was 74.1% and 66.0%, respectively, and in cell medium was 98.8% and 85.5%, respectively, with excellent reproducibility for each (less than ±8.4%). Bile and liver had similar recovery for CA-d4 and GCDCA-d4, 100% for both in bile and 73% for both in liver tissue. Variability in recovery was less for liver (less than ±4.5%) than in bile (less than ±28.9%). Plasma had different levels of recovery for CA-d4 and GCDCA-d4 with CA-d4 having 45.5 ± 13% recovery and GCDCA-d4 having 114.9 ± 28.6% recovery (supplemental Table S2).
Bench-top stability.
Bench-top stability was evaluated for plasma, bile, liver, cell lysate, and cell media at room temperature (23 ± 2°C) for 4 h. BA stability in each of the matrices was between 2.8% and 13.6% deviation of the nominal value, indicating acceptable stability (defined as ≤15%) over this time period at these conditions in each of these matrices (supplemental Table S2).
Application
In order to demonstrate the utility of this method, several in vitro and in vivo model systems frequently used to study hepatic function were designated for investigation. The normal BA pools of two immortalized cell lines, as well as HPHs, were determined. The BA levels in the liver tissue, plasma, and bile of healthy M. mulatta, a NHP laboratory model, were also established.
BA composition of cultured cell systems.
Hepatic cell systems commonly used include two immortalized hepatoblastoma cell lines, HepG2 and HuH-7, as well as HPHs. HepG2 and HuH-7 cells are routinely grown in DMEM, whereas HPHs require a supplemented medium such as HCM. The basal levels of BAs in the immortalized cells grown in both types of media were determined for comparison to HPHs (Tables 2, 3). Additionally, both types of medium with and without COS-1 growth were analyzed to ensure that no exogenous BAs were present. For the most part, slightly more BAs were present in HCM than in unsupplemented DMEM, though this difference was only statistically significant when examining individual BAs, not total BAs (TBAs) as a whole. This was most obvious in the cases of CDCA and its taurine-conjugate, TCDCA, and was true in both the measured medium aliquots and cell lysates (Tables 2, 3).
TABLE 2.
Concentrations of BAs found in cultured cell lysates
| BA in Cultured Cell Lysates | HuH-7 HCMa | HuH-7 DMEMa | HepG2 HCMa | HepG2 DMEMa | HPH HCMb |
| Unconjugated | 230 ± 32.3 | 162 ± 40.2 | 325 ± 101 | 339 ± 168 | 336 ± 248 |
| LCA | 0.270 | 0.185 ± 0.088 | 0.353 ± 0.187 | 0.174c | <LOD |
| CDCA | 3.40 ± 1.89c | <LOD/LLOQ | 1.09 ± 0.06 | <LOD | <LOD |
| DCA | <LOD | <LOD | <LOD | <LOD | <LOD |
| CA | 212 ± 25.2 | 150 ± 36.3 | 308 ± 97.3 | 315 ± 156 | 155 ± 27.2 |
| ACA | 15.9 ± 6.3 | 11.5 ± 4.1 | 15.5 ± 3.2 | 23.4 ± 12.8 | 36.0 ± 0.6 |
| UDCA | 0.783c | <LOD | <LLOQ | <LOD | <LOD |
| Glycine-conjugated | 0.0007 ± 0.0006 | 0.053 ± 0.031c | 0.127 ± 0.097 | 0.081 ± 0.050 | 9,270 ± 2,950 |
| GLCA | <LOD | <LOD | <LOD | <LOD | <LOD |
| GCDCA | 0.0002 ± 0.0002 | 0.031c | 0.127 ± 0.097 | 0.081 ± 0.050 | 218 ± 45.9 |
| GDCA | 0.0008c | <LOD | <LOD/LLOQ | <LOD/LLOQ | <LOD |
| GCA | <LOD/LLOQ | 0.07c | <LOD | <LLOQ | 9,060 ± 2,910 |
| GUDCA | <LOD | <LOD | <LOD | <LOD/LLOQ | <LOD |
| Taurine-conjugated | <LOD | 1.17 ± 0.90c | <LOD | 2.04 c | 161 ± 65.9 |
| TLCA | <LOD | <LOD | <LOD | <LOD | <LOD |
| TCDCA | <LOD | 1.17 ± 0.890c | <LOD | 2.04c | 42.7 ± 14.9 |
| TDCA | <LOD | <LOD | <LOD | <LOD | <LOD |
| TCA | <LOD | <LOD | <LOD | <LOD | 119 ± 51.9 |
| TBAs | 231 ± 32.3 | 162 ± 41.0 | 326 ± 100 | 340 ± 170 | 9,610 ± 3,020 |
Determined concentration is displayed as mean ± SD.
Values are presented as picograms per microgram of total protein corresponding to cells grown to confluence in n = 3 10 cm dishes over 48 h containing 10 ml total medium per plate.
Values are presented as picograms per microgram of total protein corresponding to cells grown to confluence in n = 6 wells of a 6-well plate over 48 h containing 2 ml total medium per well per 24 h.
Indicates at least one value <LLOQ and, thus, not included (n < 3 for HepG2 and HuH-7 cells).
TABLE 3.
Concentrations of BAs found in medium removed from cultured cells
| BA in Culture Medium | HuH-7 HCMa | HuH-7 DMEMa | HepG2 HCMa | HepG2 DMEMa | HPH HCMb |
| Unconjugated | <LLOQ | 31.5 ± 4.1 | 24.7 ± 10.9 | 14.3 ± 13.7 | 5.4 ± 1.6 |
| LCA | <LOD | 0.61 ± 0.12c | 9.82 ± 2.21 | 1.91c | <LOD |
| CDCA | <LOD | 31.1 ± 4.5 | 4.0c | 13.7 ± 14.3 | <LOD |
| DCA | <LOD | <LOD | <LOD | <LOD | <LOD |
| CA | <LOD | <LOD | 3.10c | <LOD | 5.40 ± 1.57 |
| ACA | <LOD | <LOD | <LOD/LLOQ | <LOD | <LOD |
| UDCA | <LOD | <LOD | 16.3 ± 13.8 | <LOD | <LOD |
| Glycine-conjugated | 121 ± 37.2 | 81.0 ± 14.3 | 192 ± 68.4 | 200 ± 89.3 | 6,750 ± 1,460 |
| GLCA | 8.03 ± 0.62 | <LOD/LLOQ | 12.20 ± 6.49 | 18.0c | 11.50 ± 3.44 |
| GCDCA | 29.2 ± 7.1 | 23.3 ± 3.1 | 45.7 ± 18.3 | 54.1 ± 28.5 | 434 ± 95.6 |
| GDCA | 41.8 ± 20.4 | 19.9 ± 6.02 | 66.7 ± 21.3 | 61.5 ± 14.4c | 35.7 ± 14.8 |
| GCA | 41.6 ± 9.1 | 34.5 ± 4.4 | 66.5 ± 24.6 | 80.3 ± 35.2 | 6,260 ± 1,360 |
| GUDCA | 0.55c | 0.58 ± 0.15 | 1.31c | 2.34 ± 2.08c | 7.45 ± 2.73 |
| Taurine-conjugated | 264 ± 68.5 | 131 ± 21.5 | 450 ± 154 | 275 ± 134 | 365 ± 34.8 |
| TLCA | 12.2 ± 4.04 | 4.36 ± 0.95 | 13.3 ± 5.69 | 6.86 ± 1.59c | 22.9 ± 2.80 |
| TCDCA | 129 ± 44.1 | 59.6 ± 6.3 | 244 ± 85.0 | 135 ± 68.7 | 130 ± 24.6 |
| TDCA | 38.2 ± 12.4 | 22.1 ± 5.2 | 62.1 ± 32.3 | 39.4 ± 24.2 | 48.1 ± 7.6 |
| TCA | 85.1 ± 16.1 | 45.0 ± 9.9 | 130 ± 38.2 | 96.7 ± 45.0 | 169 ± 21.1 |
| TBAs | 386 ± 105 | 244 ± 36.5 | 666 ± 217 | 490 ± 236 | 7,120 ± 1,490 |
Determined concentration is displayed as mean ± SD.
Values are presented as picograms per microgram of total protein corresponding to cells grown to confluence in n = 3 10 cm dishes over 48 h containing 10 ml total medium per plate.
Values are presented as picograms per microgram of total protein corresponding to cells grown to confluence in n = 6 wells of a 6-well plate over 48 h containing 2 ml total medium per well per 24 h.
Indicates at least one value <LLOQ and, thus, not included (n < 3 for HepG2 and HuH-7 cells).
HuH-7 and HepG2 cells demonstrated highly similar BA compositions both intra- and extracellularly compared with one another; however, the BA pools within the cells and in media were distinct (Tables 2 and 3, respectively). That is, the medium removed from cultured cells exhibited primarily conjugated BAs with the highest proportions being, in descending order: TCDCA (24.8–36.3% of TBAs), TCA (18.5–22.3%), GCA (9.1–16.6%), GDCA (8.2–10.7%), TDCA (7.7–9.8%), and GCDCA (6.6–10.9%) (Table 3, supplemental Fig. S1), whereas the intracellular BAs were mostly unamidated (Table 2, supplemental Fig. S1). Interestingly, the medium also revealed slightly higher concentrations of taurine-conjugated than glycine-conjugated BAs, which is contrary to the pattern found in healthy human liver (53, 54). The BA pool within the cell lysates of immortalized cells was made up nearly entirely of CA (91.7–94.5%) and ACA (4.8–7.0%) (Table 2, supplemental Fig. S1). This pattern was not reflected in HPHs, which exhibited high proportions of conjugated BAs (almost entirely GCA) both intra- and extracellularly (94.1% at an average of 9.1 ng/μg protein and 88.0% at an average of 6.3 ng/μg protein, respectively) (supplemental Fig. S2C). The remaining BA pool in HPH cell culture medium consisted of GCDCA, TCA, TCDCA, TDCA, GDCA, TLCA, GLCA, and CA; in cells, the remaining BAs were GCDCA, TCA, CA, TCDCA, and ACA (supplemental Fig. S2C).
Both the immortalized and the primary cells exhibited levels of secondary BAs (namely, UDCA, LCA, and DCA and their conjugates) to some extent, though this was much more pronounced in the HepG2 and HuH-7 cell lines and especially true for DCA and its amidated conjugates (Tables 2, 3; supplemental Fig. S1). In HPHs, these species were only detectable in conjugated forms (G- and T-amidated) in the culture medium (supplemental Fig. S2). Secondary BAs are often reported to be synthesized by bacterial species within the gut; however, it has become clear in recent years that this is not their exclusive origin (1, 3, 5, 55–57).
Additionally, there were similar intracellular and extracellular levels of TBAs produced in both lines of immortalized cells, totaling 240–670 pg of BAs per microgram of total protein in medium and 160–340 pg of BAs per microgram of total protein in cell lysates (Tables 2, 3). HPHs demonstrated much higher levels of basal BAs, approximately 30–60 times the amount of BAs per microgram of protein in cell lysates and 10–30 times the amount of BAs per microgram of protein in culture medium than immortalized cells. Specifically, HPHs had average intracellular TBAs of 9.6 ng/μg total protein and average extracellular TBAs of 7.1 ng/μg total protein (Tables 2, 3; supplemental Fig. S2B).
BA composition in plasma, bile, and liver tissue from a NHP model.
Naïve NHP plasma, bile, and liver tissue were examined to determine detailed baseline information of the BA pool for later comparison to liver and gut injury models in these animals as well as in humans. In plasma, CDCA and its taurine and glycine conjugates predominated with the unconjugated BAs as the most abundant. Hence, within plasma, unconjugated BAs were the most abundant species, totaling an estimated average of 73% of TBAs with 69% of this being CDCA (supplemental Fig. S3A). Glycine-conjugated and taurine-conjugated BAs made up averages of approximately 10% and 17% of the plasma BA pool, respectively, again being mostly conjugates of CDCA (supplemental Fig. S3A). Total circulating BAs ranged from 1.17 to 2.58 μg/ml and averaged 1.82 μg/ml (Table 4).
TABLE 4.
Concentrations of BAs discovered in NHP tissues
| BA | Plasma (n = 7) | Bile (n = 2) | Liver Tissue (n = 2) |
| Unconjugated | 1,360 ± 592a | 16.8 ± 1.36b | 477 ± 70.0c |
| LCA | 45.6 ± 46.4a | 0.8 ± 0.8b | 7.40 ± 7.6c |
| CDCA | 1,300 ± 601a | 1.5 ± 0.9b | 403 ± 110c |
| DCA | <LOD | 2.0 ± 1.4b | <LOD |
| CA | 13.0 ± 7.4a | 12.5 ± 0.1b | 57.9 ± 43.5c |
| ACA | <LOD | 0.0062 ± 0.0008b | <LOD |
| UDCA | 4.0 ± 2.6a | 0.024 ± 0.03b | 1.3d |
| 7α-hydroxy-3-oxo-chol-4-en-24-oic acid | <LOD | 0.01 ± 0.006b | <LOD |
| 3-oxo-CA | <LOD | <LLOQ | <LOD |
| Glycine-conjugated | 164 ± 118a | 133 ± 7.0e | 37.8 ± 44.2f |
| GLCA | 11.8 ± 6.05a | 5.95 ± 3.5e | 1.6 ± 2.1f |
| GCDCA | 78.1 ± 77.9a | 27.7 ± 10.6e | 8.3 ± 8.6f |
| GDCA | 22.9 ± 12.7a | 40.6 ± 14.3e | 9.0 ± 11.9f |
| GCA | 50.5 ± 33.0a | 58.5 ± 0.08e | 18.7 ± 21.5f |
| GUDCA | 0.7 ± 0.4a | 211 ± 76.6e | 0.22 ± 0.25f |
| Taurine-conjugated | 296 ± 179a | 299 ± 82.8e | 59.2 ± 66.2f |
| TLCA | 15.9 ± 4.1a | 6.9 ± 4.8e | 1.21 ± 1.54f |
| TCDCA | 234 ± 166a | 181 ± 99.9e | 17.9 ± 15.3f |
| TDCA | 23.7 ± 5.1a | 21.6 ± 15.1e | 8.13 ± 10.9f |
| TCA | 32.5 ± 16.3a | 89.4 ± 2.8e | 32.0 ± 38.5f |
| TBAs | 1.82 ± 0.59b | 431 ± 75.8e | 97.5 ± 110f |
Determined concentration is displayed as mean ± SD. Units are specified separately within the table due to the wide range of concentrations.
Values are in nanograms per milliliter.
Values are in micrograms per milliliter.
Values are picograms per milligram of tissue.
Indicates at least one value was <LLOQ and was not included.
Values are in milligrams per milliliter.
Values nanograms per milligram of tissue.
BA profiles within NHP bile and liver tissue were very similar to one another. In both, conjugated BAs, specifically TCDCA and TCA, dominated the BA pool. In bile, TCDCA occupied an average of nearly half of BA species (40.5% of TBAs) at 180.7 mg/ml (Table 4, supplemental Fig. S3B). TCDCA concurrently made up an average of approximately one-fourth of the BA pool in liver (26.5% of TBAs) at 17.9 ng/mg tissue (supplemental Fig. S3C). On average, TCA comprised 21% (89 mg/ml) of the BA pool in bile and 29.1% in liver tissue (32 ng/mg tissue). The succeeding most abundant BA species were also very similar between liver and bile. In bile, the next most prominent BA species were, in descending order: GCA (13.8% of TBAs), GDCA (9.8%), GCDCA (6.3%), and TDCA (5.4%) (supplemental Fig. S3B). In liver, the next most abundant BAs were GCA (18.7% of TBAs), GCDCA (9.8%), GDCA (6.4%), and TDCA (5.7%) (supplemental Fig. S3C). In total, conjugated BAs made up most of both the bile and liver BA pools in similar ratios, with taurine-conjugated species comprising 68.6% (bile) and 62.3% (liver) and glycine-conjugated species making up 31.4% (bile) and 36.3% (liver) of TBAs, on average. Thus, conversely to plasma, unconjugated BAs were minor constituents and totaled less than 1.5% of TBAs in liver and 0.004% of TBAs in bile, reflecting the high proficiency of the healthy hepatic conjugative enzyme systems. TBAs in bile ranged between 378 and 485 mg/ml (Table 4), an average of roughly 430 mg/ml. In liver, TBAs were highly variable and ranged between 19.5 and 175.5 ng/mg tissue, averaging 97.5 ng/mg tissue. The high variability is a consequence of this study’s small sample size and ad libitum feeding; additional samples and control of fasting/feeding status would provide a better indication of TBA concentration, as well as individual BA concentration, in healthy M. mulatta liver and bile.
DISCUSSION
LC-MS/MS methodology
A LC-MS/MS assay was developed for quantification of BA concentrations in a variety of cell systems and relevant biomatrices. These studies illustrate the validation of a simplified rapid sample preparation in comparison to recent methods. Using this method, BA isomers were reliably separated; however, additional specificity for these and similar molecules could be gained by using an instrument with higher resolution capabilities. Note that the first baseline level assessment for planar BAs, including ACA, in cell lines, biofluids, and tissue under healthy conditions is included. This single assay and simple sample preparation methods are highly adaptable and could be easily applied to a variety of experimental conditions relevant to BA homeostasis and pathology. This methodology could also be applied to human clinical samples or other animal models with the addition of species-specific BAs. Furthermore, the sensitivity of this method allows for the quantification of even low abundance BA species. A significant limitation of this method, however, is that its application for measurement of C27, sulfated, and glucuronidated BA species has not yet been assessed.
Cell culture BA quantification
Unsupplemented DMEM proved to be sufficient for the cellular production of BAs to allow measurement of the BA pool and total viability in immortalized HepG2 and HuH-7 cell lines for the time period examined (48 h); however, slightly more BAs were present per total protein in both cell lines when incubated in supplemented medium (i.e., HCM) (Tables 2, 3). In both media and cell lines, taurine conjugation was slightly more prevalent than glycine conjugation (supplemental Fig. S1), which is directly contrary to the case in the HPHs examined herein (supplemental Fig. S2C) and in humans in vivo, in which glycine conjugation predominates greatly (53, 54). This is especially interesting because neither DMEM nor HCM include taurine in their formulations, whereas glycine is included at a concentration of 30 mg/l (in DMEM) to 50 mg/l (in HCM), indicating that all three of these cell types are synthesizing taurine to some degree. Biosynthesis of taurine occurs in vivo in the liver and represents a sulfur excretion mechanism, so it is to be expected that hepatic and hepatocyte-derived cells are capable of this reaction; however, it is unexpected that the immortalized cells studied herein preferentially amidate BAs with the limited taurine instead of the more available glycine molecule that is favored in vivo (58). The altered amidation pattern in immortalized cells must be a result of modified cell metabolism, such as impedance in uptake in one or both of these amino acids or altered functionality and/or selectivity in the BA-CoA:amino acid N-acyltransferase (BAAT), the enzyme responsible for BA conjugation (1, 3, 5, 55). Indeed, qRT-PCR analysis has demonstrated that BAAT mRNA is undetectable in HepG2 and HuH-7 cell lines in comparison to HPHs (59). This is supported by this study, which demonstrated approximately equal amounts of conjugated and unconjugated BAs produced per total protein content in these immortalized cells (supplemental Fig. S1). Thus, BA levels in HepG2s and HuH-7s indicate that the cells are capable of BA production and conjugation but are deficient in comparison to HPHs and the healthy whole liver. Supporting this, HepG2 and HuH-7 cells presented impaired BA synthesis as a whole, producing only up to ∼6% of the TBAs produced by HPHs (Tables 2, 3). This is also in agreement with previous studies, which show very low or undetectable levels of mRNAs associated with BA synthetic enzymes CYP7A1, CYP7B1, and CYP8B1 in HepG2 and HuH-7 cells (59). Conversely, these cell lines express CYP27A1 mRNA, the rate-limiting enzyme of the acidic or alternative pathway of BA synthesis, at levels 130–155% those of HPHs (1, 59). This, combined with the higher proportions of CDCA and its conjugates, adds evidence to the theory that the acidic or alternative pathway of BA synthesis is more active in immortalized cells relative to primary cells; however, BA synthesis as a whole is still very depressed, which is likely due to decreased expression of the succeeding enzymes in the BA synthesis pathway (60, 61).
Of the BAs produced in immortalized cells, the amidated BAs were found almost exclusively in the culture medium, whereas the unconjugated BAs, CA and ACA, made up nearly all of the BA species within cells (supplemental Fig. S1). These distinct extracellular and intracellular profiles indicate that BA transport mechanisms selective for amidated BAs, such as BSEP, MDR2/3, and MRP, are still active within HepG2 and HuH-7 cells (1). Earlier studies demonstrated that, while BSEP and MDR2/3 mRNA levels are marginal or are undetectable in HepG2 and HuH-7 cells, MRP1 and MRP4 mRNA is nearly as abundant or many times more abundant than in HPHs (59). Hence, HepG2 and HuH-7 cells are more capable of exporting than they are at synthesizing amidated BAs, which is underpinned by both the high proportion of unconjugated BAs within the cell lysates and by the production of the secondary BAs, LCA and DCA, which are presumed to be produced in vivo by intestinal bacteria (Tables 2, 3; supplemental Fig. S1) (1, 3, 5, 55, 56). Earlier studies investigating the production of BAs by cultured cell systems did not probe for secondary BAs and enzymatically deconjugated BAs before analysis; thus, to the best of our knowledge, this study offers the most detailed characterization of BA production in HepG2 and HuH-7 cells to date (60, 62, 63). The production of secondary BAs is presumably due to the buildup of unconjugated CA and CDCA within the cells: when insufficiently conjugated and exported, these molecules are subject to dehydroxylation, which is likely performed by the same enzymes that rehydroxylate LCA and DCA in the normal liver, as this enzymatic reaction is reversible (64). The presence of secondary BAs in immortalized and primary cells calls into question the assumption that LCA and DCA are generated exclusively from the activity of intestinal bacteria in vivo. Further studies are needed to determine whether hepatic synthesis of “secondary” BAs contributes significantly to the BA pool and whether this contribution is altered during disease.
As far as we are aware, this study is the first to observe the presence of planar BAs in human-derived cultured cells, though planar BAs have been discovered in both healthy and compromised individuals (30, 65–67). A significant portion of the unconjugated BAs present in HepG2 and HuH-7 cell lysates was ACA, the 5α-epimer of CA. The planar BAs represent a subset of BAs that retain the flat 3D structure of cholesterol instead of the bent or twisted structure of typical mammalian BAs (1, 24, 25, 31). This conformation can either be due to an α-positioning of the C5 hydrogen atom, as in ACA, or a double bond between C4 and C5, as in 7α-hydroxy-3-oxo-chol-4-en-24-oic acid (1, 19, 25, 31). These molecules are abundant in pregnancy and during the first few months of life but gradually disappear after infancy and are not usually detectable in healthy adults (25, 31). In certain cases of liver or GI disease, however, these species reappear among circulating BAs (31). The mechanisms of the recurrence of planar BAs have yet to be fully elucidated, but it is thought that their synthesis is due to immature hepatic enzyme systems that can be found in the underdeveloped hepatocytes of the fetus and in the dysregulated cells of the injured liver (31). Alternatively, it is possible that these species are isomerized from their more typical counterparts, either by hepatic or microbial enzymes (31). The production of planar BAs by immortalized cells, however, substantiate the theory that proliferating hepatocytes must dedifferentiate in order to repopulate the injured liver and, therefore, express less specialized or incomplete metabolic enzymes, resulting in the production of “fetal” BA species (68). Moreover, previous studies in HepG2 cells detected high levels of additional BA precursors and intermediates for which we did not probe (62).
Though HPHs seem to produce fewer TBAs compared with the whole healthy human liver, these cells at least appear to produce similar ratios of each BA and amidate in similar patterns (supplemental Fig. S2) (69, 70). Glycine amidation predominated the BA pool greatly, but it should be noted that HCM does not provide any exogenous taurine, as stated above, which is presumably why HPHs produced such a low proportion of taurine-conjugated BAs. Also, this study examined BA production only in the 48 h following perfusion, so it should also be considered that the amount of BAs generated changes over time and does not reach maximal production until day 6 (i.e., 144 h) (60, 63). Finally, only one donor was assayed in this study; further investigations would tell whether these findings are true of all HPH cultures. Primary cells therefore offer a relatively good model of human liver BA synthesis and metabolism, as long as the above limitations are considered (60, 69, 70). Conversely, the HepG2 and HuH-7 cell lines make very poor models for the study of BA synthesis and metabolism, as they do not produce nearly as many or the same pattern of BAs as HPHs or whole human liver.
NHP BA quantification
NHP models are the most relevant and preferred model systems currently available in drug discovery and development due to the high degree of phylogeny and homogeneity to humans (71). Despite this, BA synthesis and metabolism in these animals remains poorly characterized. Herein, the BA pools of several compartments were quantified in an accepted laboratory model, the NHP.
Naïve NHP plasma BAs were dominated by unconjugated CDCA, distantly followed by TCDCA and GCDCA and then by GCA and TCA (Table 4, supplemental Fig. S3A). These results conform with previous studies indicating the prevalence of unconjugated BAs in rhesus macaque plasma; however, they disagree with previous studies regarding individual BAs, finding a higher percentage of unconjugated CDCA instead of DCA (46, 47, 72). Circulating TBAs concentration in NHP plasma proved comparable to that in human plasma as reported in scientific literature, i.e., an average of 1.82 μg/ml in NHPs compared with 1.01–2.13 μg/ml in humans (Table 4) (35, 53, 70). It is important to note, however, that the composition of the BA pool was altered in naïve NHP plasma, yielding a higher CDCA:CA ratio than is usual in human plasma. NHP plasma also presented a high proportion (∼73%) of total unconjugated BAs, specifically CDCA, whereas human plasma BAs are mostly (∼56–59%) glycine-amidated (supplemental Fig. S3A) (35, 46, 53). Yet, this has been shown to change with diet and fasting state: the glycine:taurine ratio in humans can change with diet and cooking habits, and the ratio of unconjugated to amidated BAs is increased in the fasting state compared with the nonfasting state (35, 53).
The preponderance of CDCA compared with CA that occurred in plasma was also reflected in naïve macaque bile and liver, which demonstrated similar profiles when compared with one another (supplemental Fig. S3B, C). In these tissues, the BA pools were dominated by a high proportion of taurine-conjugated primary BAs, CDCA and CA. As has been reported in human bile and liver, an overwhelming majority of BAs in naïve macaque liver and bile were amidated, with only a very small fraction escaping conjugation (supplemental Fig. S3B, C) (1, 3, 5, 19, 70, 73). Though interindividual variability in this study was relatively high due to small sample size and ad libitum feeding, it is apparent that naïve NHP liver produced comparable TBAs to healthy human liver as reported in previous studies, i.e., an average of 97.5 ng/mg of tissue versus 7.7–29.5 ng/mg of tissue in humans (Table 4) (70, 73). Although, as in plasma, NHP liver shows a notable difference in regard to the glycine/taurine ratio, preferring taurine to glycine amidation, which is converse to that in humans (supplemental Fig. S3B, C) (3, 35, 70).
In terms of TBAs, the NHP offers a high resemblance to humans in plasma, liver tissue, and bile (35, 46, 53, 54, 69, 70, 73). The individual members of the BA pool in these tissues, however, are distinct between NHPs and humans. The greatest differences observed in this study include: increased CDCA/CA ratio in the NHP BA pool, greater proportion of unconjugated compared with conjugated BAs within plasma, and the preference for taurine versus glycine amidation. Additionally, though this study did not evaluate sulfated BAs, at least one previous study has reported marked differences between the extent of this BA elimination pathway in NHPs versus humans (46, 47). Overall, however, this laboratory animal represents a highly relevant model for the study of BA synthesis, metabolism, and pharmacology with relatively minor disparities in comparison to other models.
CONCLUSIONS
The optimization, validation, and application of a sensitive and selective LC-MS/MS method for the simultaneous detection and quantification of numerous BAs in cultured cell systems and several biomatrices are described herein. The immortalized hepatoblastoma cell lines HepG2 and HuH-7 were studied for comparison with cultured HPHs and previous research examining whole liver in vivo (69, 70). Results from these studies demonstrate that these immortalized cell systems are poor indicators of healthy hepatic BA metabolism; however, HPHs are a promising alternative. Although, before using these cells as models, it should be established whether supplementing culture medium with exogenous taurine would present a similar BA profile to that produced in vivo. In addition, more HPH donors should be examined to ensure that these findings are universal.
These experiments also support the use of NHP models in favor of murine models when studying BA synthesis and metabolism, especially for comparison to human systems. Laboratory rats and mice offer the advantages of low costs, rapid growth, and straightforward handling and use, but their low homology to man makes correlation to human systems challenging and usually inaccurate (38–43, 45). In BA research, the use of murine models is additionally complicated by the utilization of a set of unique BA epimers, the muricholic acids (38, 39, 41, 45). NHPs offer a more homologous, suitable system for the study of BA synthesis and metabolism, both in physiology and in illness, that much more closely approximates that in healthy adult humans. Further characterization in these species, however, to elucidate such processes as BA elimination (i.e., renal versus biliary), sulfonation and glucuronidation, and signaling for comparison to man, should be pursued to establish physiological parameters that may be altered in relevant pathologies The continued study of NHP BA synthesis with regard to C27 BA precursors and intermediates should prove valuable in aligning this laboratory model with human conditions. Additionally, the study of conjugated planar BAs (such as TACA and GACA) both in laboratory models and in human studies would be of increased relevance due to the propensity of BAs to be amidated or otherwise conjugated in most biological systems.
In summary, these studies have proven the utility of a validated method for the quantification of common and planar C24 BAs in a variety of systems. Detailed baselines have been established herein for multiple model systems for the study of BA production and metabolism, which have implications for the selection and use of cell and animal models for the study of BA metabolism and pathology.
Data availability
All data are contained within the article.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the laboratory of Dr. James E. Polli (University of Maryland Baltimore, Baltimore, MD) for the generous donation of 3-oxo-CA.
Footnotes
Author contributions—S.J.S., P.W.S., and M.A.K. experimental design; S.J.S., P.W.S., and M.A.K. writing-original draft; A.M.F. and T.J.M. NHP care; A.M.F. and T.J.M. tissue collection; H.W. and L.L. collection, perfusion, and maintenance of human primary hepatocytes; J.W.J. initial optimization and implementation of the LC-MS/MS method; S.J.S. sample preparation, method validation, and analysis; S.J.S., J.W.J., A.M.F., T.J.M., H.W., P.W.S., and M.A.K. writing-review and editing.
This article contains supplemental data.
Funding and additional information—This work was supported by National Institutes of Health Grants R01 DK061425 (to P.W.S.) and R01 GM121550 (to H.W.) and National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services Grants HHSN272201000046C and HHSN272201500013I (to T.J.M.). Additional support was provided by University of Maryland’s Center of Excellence in Regulatory Science and Innovation (M-CERSI) Scholars Program funded by the US Food and Drug Administration (1U01FD005946) and the University of Maryland School of Pharmacy Mass Spectrometry Center (SOP1841-IQB2014). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Abbreviations—
- ACA
- allocholic acid (LMID: LMST04010092)
- ACN
- acetonitrile
- BA
- bile acid
- CA
- cholic acid (LMID: LMST04010001)
- CA-d4
- cholic acid (2,2,4,4-d4)
- CDCA
- chenodeoxycholic acid (LMID: LMST04010032)
- DCA
- deoxycholic acid (LMID: LMST04010040)
- FA
- formic acid
- GCA
- glycocholic acid (LMID: LMST05030001)
- GCDCA
- glycochenodeoxycholic acid (LMID: LMST05030008)
- GCDCA-d4
- glycochenodeoxycholic acid (2,2,4,4-d4)
- GDCA
- glycodeoxycholic acid (LMID: LMST05030006)
- GI
- gastrointestinal
- GLCA
- glycolithocholic acid (LMID: LMST05030009)
- GUDCA
- glycoursodeoxycholic acid (LMID: LMST05030016)
- HCM
- hepatocyte culture medium
- HPH
- human primary hepatocyte
- IALCA
- isoallolithocholic acid (LMID: LMST04010006)
- IS
- internal standard (CA-d4 and GCDCA-d4)
- LCA
- lithocholic acid (LMID: LMST04010003)
- LLOQ
- lower limit of quantification
- LOD
- limit of detection
- NHP
- nonhuman primate
- 3-oxo-CA
- 3-oxo-cholic acid (LMID: LMST04010443)
- S/N
- signal-to-noise ratio
- SPE
- solid-phase extraction
- TBA
- total bile acid
- TCA
- taurocholic acid (LMID: LMST05040001)
- TCDCA
- taurochenodeoxycholic acid (LMID: LMST05040005)
- TDCA
- taurodeoxycholic acid (LMID: LMST05040013)
- TLCA
- taurolithocholic acid (LMID: LMST05040003)
- UDCA
- ursodeoxycholic acid (LMID: LMST04010033)
Manuscript received February 29, 2020 and in revised form July 18, 2020. Published, JLR Papers in Press, July 22, 2020, DOI 10.1194/jlr.RD120000726.
REFERENCES
- 1.Marin J. J. G., Macias R. I. R., Briz O., Banales J. M., and Monte M. J.. 2015. Bile acids in physiology, pathology and pharmacology. Curr. Drug Metab. 17: 4–29. [DOI] [PubMed] [Google Scholar]
- 2.Kuipers F., Bloks V. W., and Groen A. K.. 2014. Beyond intestinal soap—bile acids in metabolic control. Nat. Rev. Endocrinol. 10: 488–498. [DOI] [PubMed] [Google Scholar]
- 3.Hofmann A. F., and Hagey L. R.. 2008. Bile acids: chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cell. Mol. Life Sci. 65: 2461–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrebee C. B., and Dawson P. A.. 2015. Metabolic effects of intestinal absorption and enterohepatic cycling of bile acids. Acta Pharm. Sin. B. 5: 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Aguiar Vallim T. Q., Tarling E. J., and Edwards P. A.. 2013. Pleiotropic roles of bile acids in metabolism. Cell Metab. 17: 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rees D. O., Crick P. J., Jenkins G. J., Wang Y., Griffiths W. J., Brown T. H., and Al-Sarireh B.. 2017. Comparison of the composition of bile acids in bile of patients with adenocarcinoma of the pancreas and benign disease. J. Steroid Biochem. Mol. Biol. 174: 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ressom H. W., Xiao J. F., Tuli L., Varghese R. S., Zhou B., Tsai T-H., Nezami Ranjbar M. R., Wang J., Di Poto C., Cheema A. K., et al. . 2012. Utilization of metabolomics to identify serum biomarkers for hepatocellular carcinoma in patients with liver cirrhosis. Anal. Chim. Acta. 743: 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tadano T., Kanoh M., Matsumoto M., Sakamoto K., and Kamano T.. 2006. Studies of serum and feces bile acids determination by gas chromatography-mass spectrometry. Rinsho Byori. 54: 103–110. [PubMed] [Google Scholar]
- 9.Puri P., Daita K., Joyce A., Mirshahi F., Santhekadur P. K., Cazanave S., Luketic V. A., Siddiqui M. S., Boyett S., Min H-K., et al. . 2018. The presence and severity of nonalcoholic steatohepatitis is associated with specific changes in circulating bile acids. Hepatology. 67: 534–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raselli T., Hearn T., Wyss A., Atrott K., Peter A., Frey-Wagner I., Spalinger M. R., Maggio E. M., Sailer A. W., Schmitt J., et al. . 2019. Elevated oxysterol levels in human and mouse livers reflect nonalcoholic steatohepatitis. J. Lipid Res. 60: 1270–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janssen A. W. F., Houben T., Katiraei S., Dijk W., Boutens L., van der Bolt N., Wang Z., M. Brown J., L. Hazen S., Mandard S., et al. . 2017. Modulation of the gut microbiota impacts nonalcoholic fatty liver disease: a potential role for bile acids. J. Lipid Res. 58: 1399–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiao N., Baker S. S., Chapa-Rodriguez A., Liu W., Nugent C. A., Tsompana M., Mastrandrea L., Buck M. J., Baker R. D., Genco R. J., et al. . 2018. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut. 67: 1881–1891. [DOI] [PubMed] [Google Scholar]
- 13.Ferrell J. M., and Chiang J. Y. L.. 2019. Understanding bile acid signaling in diabetes: from pathophysiology to therapeutic targets. Diabetes Metab. J. 43: 257–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maghsoodi N., Shaw N., Cross G. F., Alaghband-Zadeh J., Wierzbicki A. S., Pinkney J., Millward A., and Vincent R. P.. 2019. Bile acid metabolism is altered in those with insulin resistance after gestational diabetes mellitus. Clin. Biochem. 64: 12–17. [DOI] [PubMed] [Google Scholar]
- 15.Schonewille M., de Boer J., and Groen A.. 2016. Bile salts in control of lipid metabolism. Curr. Opin. Lipidol. 27: 295–301. [DOI] [PubMed] [Google Scholar]
- 16.Shapiro H., Kolodziejczyk A. A., Halstuch D., and Elinav E.. 2018. Bile acids in glucose metabolism in health and disease. J. Exp. Med. 215: 383–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schadt H. S., Wolf A., Pognan F., Chibout S-D., Merz M., and Kullak-Ublick G. A.. 2016. Bile acids in drug induced liver injury: key players and surrogate markers. Clin. Res. Hepatol. Gastroenterol. 40: 257–266. [DOI] [PubMed] [Google Scholar]
- 18.Griffiths W. J., Abdel-Khalik J., Yutuc E., Roman G., Warner M., Gustafsson J-Å., and Wang Y.. 2019. Concentrations of bile acid precursors in cerebrospinal fluid of Alzheimer’s disease patients. Free Radic. Biol. Med. 134: 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alnouti Y. 2009. Bile acid sulfation: a pathway of bile acid elimination and detoxification. Toxicol. Sci. 108: 225–246. [DOI] [PubMed] [Google Scholar]
- 20.Gérard P. 2013. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens. 3: 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikegami T., and Honda A.. 2018. Reciprocal interactions between bile acids and gut microbiota in human liver diseases. Hepatol. Res. 48: 15–27. [DOI] [PubMed] [Google Scholar]
- 22.Luo L., Aubrecht J., Li D., Warner R. L., Johnson K. J., Kenny J., and Colangelo J. L.. 2018. Assessment of serum bile acid profiles as biomarkers of liver injury and liver disease in humans. PLoS One. 13: e0193824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendoza M. E., Monte M. J., El-Mir M. Y., Badia M. D., and Marin J. J. G.. 2002. Changes in the pattern of bile acids in the nuclei of rat liver cells during hepatocarcinogenesis. Clin. Sci. 102: 143–150. [PubMed] [Google Scholar]
- 24.Monte M. J., Martinez-Diez M. C., El-Mir M. Y., Mendoza M. E., Bravo P., Bachs O., and Marin J. J. G.. 2002. Changes in the pool of bile acids in hepatocyte nuclei during rat liver regeneration. J. Hepatol. 36: 534–542. [DOI] [PubMed] [Google Scholar]
- 25.Stärkel P., Shindano T., Horsmans Y., Gigot J. F., Fernandez-Tagarro M., Marin J. J. G., and Monte M. J.. 2009. Foetal ‘flat’ bile acids reappear during human liver regeneration after surgery. Eur. J. Clin. Invest. 39: 58–64. [DOI] [PubMed] [Google Scholar]
- 26.Miyazaki-Anzai S., Masuda M., Kohno S., Levi M., Shiozaki Y., Keenan A. L., and Miyazaki M.. 2018. Simultaneous inhibition of FXR and TGR5 exacerbates atherosclerotic formation. J. Lipid Res. 59: 1709–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang H., Huang H., Tan P-Z., Liu Y., Nie J-H., Zhang Y-T., Zhang K-L., Diao Y., He Q., Hou B-Y., et al. . 2017. Effect of iron on cholesterol 7α-hydroxylase expression in alcohol-induced hepatic steatosis in mice. J. Lipid Res. 58: 1548–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagahashi M., Yuza K., Hirose Y., Nakajima M., Ramanathan R., Hait N. C., Hylemon P. B., Zhou H., Takabe K., and Wakai T.. 2016. The roles of bile acids and sphingosine-1-phosphate signaling in the hepatobiliary diseases. J. Lipid Res. 57: 1636–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lan K., Su M., Xie G., Ferslew B. C., Brouwer K. L. R., Rajani C., Liu C., and Jia W.. 2016. Key role for the 12-hydroxy group in the negative ion fragmentation of unconjugated C24 bile acids. Anal. Chem. 88: 7041–7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffiths W. J., Gilmore I., Yutuc E., Abdel-Khalik J., Crick P. J., Hearn T., Dickson A., Bigger B. W., Wu T. H-W., Goenka A., et al. . 2018. Identification of unusual oxysterols and bile acids with 7-oxo or 3β,5α,6β-trihydroxy functions in human plasma by charge-tagging mass spectrometry with multistage fragmentation. J. Lipid Res. 59: 1058–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shiffka S. J., Kane M. A., and Swaan P. W.. 2017. Planar bile acids in health and disease. Biochim. Biophys. Acta Biomembr. 1859: 2269–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffiths W. J., and Sjövall J.. 2010. Bile acids: analysis in biological fluids and tissues. J. Lipid Res. 51: 23–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kakiyama G., Muto A., Takei H., Nittono H., Murai T., Kurosawa T., Hofmann A. F., Pandak W. M., and Bajaj J. S.. 2014. A simple and accurate HPLC method for fecal bile acid profile in healthy and cirrhotic subjects: validation by GC-MS and LC-MS. J. Lipid Res. 55: 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee C-S., Kimura A., Wu J-F., Ni Y-H., Hsu H-Y., Chang M-H., Nittono H., and Chen H-L.. 2017. Prognostic roles of tetrahydroxy bile acids in infantile intrahepatic cholestasis. J. Lipid Res. 58: 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han J., Liu Y., Wang R., Yang J., Ling V., and Borchers C. H.. 2015. Metabolic profiling of bile acids in human and mouse blood by LC-MS/MS in combination with phospholipid-depletion solid-phase extraction. Anal. Chem. 87: 1127–1136. [DOI] [PubMed] [Google Scholar]
- 36.Fang N., Yu S., Adams S. H., Ronis M. J. J., and Badger T. M.. 2016. Profiling of urinary bile acids in piglets by a combination of enzymatic deconjugation and targeted LC-MRM-MS. J. Lipid Res. 57: 1917–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmid A., Neumann H., Karrasch T., Liebisch G., and Schäffler A.. 2016. Bile acid metabolome after an oral lipid tolerance test by liquid chromatography-tandem mass spectrometry (LC-MS/MS). PLoS One. 11: e0148869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rudling M. 2016. Understanding mouse bile acid formation: Is it time to unwind why mice and rats make unique bile acids? J. Lipid Res. 57: 2097–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takahashi S., Fukami T., Masuo Y., Brocker C. N., Xie C., Krausz K. W., Wolf C. R., Henderson C. J. and Gonzalez F. J.. 2016. Cyp2c70 is responsible for the species difference in bile acid metabolism between mice and humans. J. Lipid Res. 57: 2130–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Honda A., Miyazaki T., Iwamoto J., Hirayama T., Morishita Y., Monma T., Ueda H., Mizuno S., Sugiyama F., Takahashi S., et al. . 2020. Regulation of bile acid metabolism in mouse models with hydrophobic bile acid composition. J. Lipid Res. 61: 54–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Boer J. F., Verkade E., Mulder N. L., de Vries H. D., Huijkman N. C. A., Koehorst M., Boer T., Wolters J. C., Bloks V. W., van de Sluis B., et al. . 2019. A human-like bile acid pool induced by deletion of Cyp2c70 modulates effects of FXR activation in mice. J. Lipid Res. 61: 291–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee J. M., Ong J. R., Vergnes L., de Aguiar Vallim T. Q., Nolan J., Cantor R. M., Walters J. R. F., and Reue K. 2018. Diet1, bile acid diarrhea, and FGF15/19: mouse model and human genetic variants. J. Lipid Res. 59: 429–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wahlström A., Kovatcheva-Datchary P., Ståhlman M., Khan M-T., Bäckhed F., and Marschall H-U.. 2017. Induction of farnesoid X receptor signaling in germ-free mice colonized with a human microbiota. J. Lipid Res. 58: 412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song X., Chen Y., Valanejad L., Kaimal R., Yan B., Stoner M., and Deng R.. 2013. Mechanistic insights into isoform-dependent and species-specific regulation of bile salt export pump by farnesoid X receptor. J. Lipid Res. 54: 3030–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J., and Dawson P. A.. 2019. Animal models to study bile acid metabolism. Biochim. Biophys. Acta Mol. Basis Dis. 1865: 895–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thakare R., Alamoudi J. A., Gautam N., Rodrigues A. D., and Alnouti Y.. 2018. Species differences in bile acids I. Plasma and urine bile acid composition. J. Appl. Toxicol. 38: 1323–1335. [DOI] [PubMed] [Google Scholar]
- 47.Thakare R., Alamoudi J. A., Gautam N., Rodrigues A. D., and Alnouti Y.. 2018. Species differences in bile acids II. Bile acid metabolism. J. Appl. Toxicol. 38: 1336–1352. [DOI] [PubMed] [Google Scholar]
- 48.Cohen E. P., Hankey K. G., Farese A. M., Parker G. A., Jones J. W., Kane M. A., Bennett A., and MacVittie T. J.. 2019. Radiation nephropathy in a nonhuman primate model of partial-body irradiation with minimal bone marrow sparing—Part 1: acute and chronic kidney injury and the influence of Neupogen. Health Phys. 116: 401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parker G. A., Cohen E. P., Li N., Takayama K., Farese A. M., and MacVittie T. J.. 2019. Radiation nephropathy in a nonhuman primate model of partial-body irradiation with minimal bone marrow sparing—Part 2: histopathology, mediators, and mechanisms. Health Phys. 116: 409–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parker G. A., Li N., Takayama K., Booth C., Tudor G. L., Farese A. M., and MacVittie T. J.. 2019. Histopathological features of the development of intestine and mesenteric lymph node injury in a nonhuman primate model of partial-body irradiation with minimal bone marrow sparing. Health Phys. 116: 426–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parker G. A., Li N., Takayama K., Farese A. M., and MacVittie T. J.. 2019. Lung and heart injury in a nonhuman primate model of partial-body irradiation with minimal bone marrow sparing: histopathological evidence of lung and heart injury. Health Phys. 116: 383–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saeed A., Floris F., Andersson U., Pikuleva I., Lövgren-Sandblom A., Bjerke M., Paucar M., Wallin A., Svenningsson P., and Björkhem I.. 2014. 7α-Hydroxy-3-oxo-4-cholestenoic acid in cerebrospinal fluid reflects the integrity of the blood-brain barrier. J. Lipid Res. 55: 313–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bathena S. P. R., Mukherjee S., Olivera M., and Alnouti Y.. 2013. The profile of bile acids and their sulfate metabolites in human urine and serum. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 942–943: 53–62. [DOI] [PubMed] [Google Scholar]
- 54.Xie G., Wang Y., Wang X., Zhao A., Chen T., Ni Y., Wong L., Zhang H., Zhang J., Liu C., et al. . 2015. Profiling of serum bile acids in a healthy chinese population using UPLC-MS/MS. J. Proteome Res. 14: 850–859. [DOI] [PubMed] [Google Scholar]
- 55.Hofmann A. F., and Hagey L. R.. 2014. Key discoveries in bile acid chemistry and biology and their clinical applications: history of the last eight decades. J. Lipid Res. 55: 1553–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kriaa A., Bourgin M., Potiron A., Mkaouar H., Jablaoui A., Gérard P., Maguin E., and Rhimi M.. 2019. Microbial impact on cholesterol and bile acid metabolism: current status and future prospects. J. Lipid Res. 60: 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shoda J., Toll A., Axelson M., Pieper F., Wikvall K., and Sjövall J.. 1993. Formation of 7α- and 7β-hydroxylated bile acid precursors from 27-hydroxycholesterol in human liver microsomes and mitochondria. Hepatology. 17: 395–403. [PubMed] [Google Scholar]
- 58.Hansen S. H. 2003. Taurine homeostasis and its importance for physiological functions. In Metabolic and Therapeutic Aspects of Amino Acids in Clinical Nutrition. 2nd edition. L. A. Cynober, editor. CRC Press, Boca Raton, FL. 739–747. [Google Scholar]
- 59.Guo L., Dial S., Shi L., Branham W., Liu J., Fang J-L., Green B., Deng H., Kaput J. and Ning B.. 2011. Similarities and differences in the expression of drug metabolizing enzymes between human hepatic cell lines and primary human hepatocytes. Drug Metab. Dispos. 39: 528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Einarsson C., Ellis E., Abrahamsson A., Ericzon B-G., Bjorkhem I., and Axelson M.. 2000. Bile acid formation in primary human hepatocytes. World J. Gastroenterol. 6: 522–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi J., Wang X., Lyu L., Jiang H., and Zhu H-J.. 2018. Comparison of protein expression between human livers and the hepatic cell lines HepG2, Hep3B, and Huh7 using SWATH and MRM-HR proteomics: focusing on drug-metabolizing enzymes. Drug Metab. Pharmacokinet. 33: 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Axelson M., Mörk B., and Everson G. T.. 1991. Bile acid synthesis in cultured human hepatoblastoma cells. J. Biol. Chem. 266: 17770–17777. [PubMed] [Google Scholar]
- 63.Axelson M., Ellis E., Mörk B., Garmark K., Abrahamsson A., Björkhem I., Ericzon B. G., and Einarsson C.. 2000. Bile acid synthesis in cultured human hepatocytes: support for an alternative biosynthetic pathway to cholic acid. Hepatology. 31: 1305–1312. [DOI] [PubMed] [Google Scholar]
- 64.Penning T. M. 1997. Molecular endocrinology of hydroxysteroid dehydrogenases. Endocr. Rev. 18: 281–305. [DOI] [PubMed] [Google Scholar]
- 65.Mazzacuva F., Mills P., Mills K., Camuzeaux S., Gissen P., Nicoli E-R., Wassif C., Te Vruchte D., Porter F. D., Maekawa M., et al. . 2016. Identification of novel bile acids as biomarkers for the early diagnosis of Niemann-Pick C disease. FEBS Lett. 590: 1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maekawa M., Misawa Y., Sotoura A., Yamaguchi H., Togawa M., Ohno K., Nittono H., Kakiyama G., Iida T., Hofmann A. F., et al. . 2013. LC/ESI-MS/MS analysis of urinary 3β-sulfooxy-7β-N-acetylglucosaminyl-5-cholen-24-oic acid and its amides: New biomarkers for the detection of Niemann-Pick type C disease. Steroids. 78: 967–972. [DOI] [PubMed] [Google Scholar]
- 67.Meng L-J., Reyes H., Palma J., Hernandez I., Ribalta J., and Sjövall J.. 1997. Profiles of bile acids and progesterone metabolites in the urine and serum of women with intrahepatic cholestasis of pregnancy. J. Hepatol. 27: 346–357. [DOI] [PubMed] [Google Scholar]
- 68.Kholodenko I. V., and Yarygin K. N.. 2017. Cellular mechanisms of liver regeneration and cell-based therapies of liver diseases. BioMed Res. Int. 2017: 8910821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Setchell K. D., Rodrigues C. M., Clerici C., Solinas A., Morelli A., Gartung C., and Boyer J.. 1997. Bile acid concentrations in human and rat liver tissue and in hepatocyte nuclei. Gastroenterology. 112: 226–235. [DOI] [PubMed] [Google Scholar]
- 70.García-Cañaveras J. C., Donato M. T., Castell J. V., and Lahoz A.. 2012. Targeted profiling of circulating and hepatic bile acids in human, mouse, and rat using a UPLC-MRM-MS-validated method. J. Lipid Res. 53: 2231–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Singh V. K., and Olabisi A. O.. 2017. Nonhuman primates as models for the discovery and development of radiation countermeasures. Expert Opin. Drug Discov. 12: 695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sturman J. A., Messing J. M., Rossi S. S., Hofmann A. F., and Neuringer M.. 1991. Tissue taurine content, activity of taurine synthesis enzymes and conjugated bile acid composition of taurine-deprived and taurine-supplemented rhesus monkey infants at 6 and 12 mo of age. J. Nutr. 121: 854–862. [DOI] [PubMed] [Google Scholar]
- 73.Greim H., Czygan P., Schaffner F., and Popper H.. 1973. Determination of bile acids in needle biopsies of human liver. Biochem. Med. 8: 280–286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.