Abstract
Human genetic studies recently identified an association of SNPs in the 17-β hydroxysteroid dehydrogenase 13 (HSD17B13) gene with alcoholic and nonalcoholic fatty liver disease development. Mutant HSD17B13 variants devoid of enzymatic function have been demonstrated to be protective from cirrhosis and liver cancer, supporting the development of HSD17B13 as a promising therapeutic target. Previous studies have demonstrated that HSD17B13 is a lipid droplet (LD)-associated protein. However, the critical domains that drive LD targeting or determine the enzymatic activity have yet to be defined. Here we used mutagenesis to generate multiple truncated and point-mutated proteins and were able to demonstrate in vitro that the N-terminal hydrophobic domain, PAT-like domain, and a putative α-helix/β-sheet/α-helix domain in HSD17B13 are all critical for LD targeting. Similarly, we characterized the predicted catalytic, substrate-binding, and homodimer interaction sites and found them to be essential for the enzymatic activity of HSD17B13, in addition to our previous identification of amino acid P260 and cofactor binding site. In conclusion, we identified critical domains and amino acid sites that are essential for the LD localization and protein function of HSD17B13, which may facilitate understanding of its function and targeting of this protein to treat chronic liver diseases.
Keywords: nonalcoholic fatty liver disease, alcoholic liver disease, retinoids, lipid droplets, enzyme regulation, protein structure
Associated with the global epidemic of lifestyle-associated obesity and metabolic syndrome, NAFLD has become a major global health burden and one of the leading causes for end-stage liver disease, hepatocellular carcinoma, and liver transplants (1, 2). Limited treatment options are available, stimulating the search for novel molecular targets suitable for therapeutic pharmacological intervention (3). One approach to the discovery of these novel targets, based on the known heritability of NAFLD, is the use of genetic studies (4–6). Several genes related to the incidence of NAFLD and to its progressive form of NASH were identified using this approach, including PNPLA3 (7–12), TM6SF2 (13–15), MBOAT7 (16), and the recently discovered HSD17B13 (17–19). Of these, HSD17B13 (17-β hydroxysteroid dehydrogenase 13) represents a likely therapeutic target for the treatment of chronic liver disease with a potential for pharmaceutical intervention.
HSD17B13 is a hepatic lipid droplet (LD)-associated enzyme with steroid substrates, bioactive lipids (19), and retinol (17) suggested as potential enzymatic substrates. Three independent genetic variants in HSD17B13 were found to confer protection from injury in NASH (17, 19, 20), alcoholic liver disease (17, 19), and hepatocellular carcinoma (21, 22). We and others identified a splice-site SNP rs72613567 that leads to the formation of two novel splicing variants (HSD17B13-G insertion and HSD17B13-exon 6 deletion) (17, 19, 23); the nonsynonymous SNP rs62305723 encodes a proline to serine mutation at amino acid (AA) position 260 (17); and the rs143404524 SNP leads to premature truncation (24). All three protective variants generate protein products that are devoid, or predicted to be devoid of, enzymatic activity, confirming the importance of understanding the enzymatic function of HSD17B13. Collectively, these data also suggest that HSD17B13 activity can be modulated by large domain truncations and deletions, as well as by single or double critical AA mutations, implying the possibility of modulating the enzymatic activity therapeutically, either by interfering with gene expression (i.e., by using antisense oligonucleotides) or by inhibiting activity directly using small synthetic compounds.
Excess cellular lipids are esterified to neutral lipids and stored in LDs, which function as main storage reservoirs of metabolic energy and membrane lipid components (25). LD-associated proteins, which play pivotal roles in lipid metabolism regulation, are embeded in or adherent to the phospholipid monolayer, which includes PC, PE, PI, lyso-PC, and lyso-PE (26). MS proteomic analyses have identified hundreds of LD-associated proteins, including a couple of dozen proteins confirmed to be LD resident proteins (25). Two types of LD targeting signals have been proposed for directing proteins to the LD surface: amphipathic α-helices and hydrophobic hairpins (25, 27). The PAT protein family encompasses the most widely studied LD-associated proteins (perilipin, ADRP, TIP47, and S3-12) (28) that share conserved PAT domains that are essential for their LD targeting (29, 30). HSD17B13 is an LD-associated protein (17, 19, 31, 32), and we identified a PAT-like domain at its N-terminus that is essential for its stability, targeting to LD, and enzymatic function (17). However, other domains in HSD17B13 that are essential for correct trafficking to LDs have not yet been fully defined.
Pharmacological efforts to silence HSD17B13 by siRNA have been initiated by pharmaceutical companies (33). However, beyond the naturally occurring mutants and cofactor binding sites that we identified (17), other sites or domains critical for its enzymatic activity and targeting are still unknown. In the current study, we aimed to identify critical domains and AAs that are essential for LD targeting and the enzymatic activity of HSD17B13 by studying multiple truncated and point-mutated proteins. Our work may facilitate the design of small molecule inhibitors and lead to a better understanding of the protein function that is essential for HSD17B13 to be considered a therapeutic target in chronic liver disease.
MATERIALS AND METHODS
Hydropathy analysis
Hydropathy analysis of HSD17B13 was performed using the online TMHMM server (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0) (34) and ProtScale (https://web.expasy.org/protscale) (35) with default settings.
Cell culture and stock solution
HepG2 and HEK293 cells were cultured in DMEM (Corning) supplemented with 10% FBS (Sigma-Aldrich) under 5% CO2 at 37°C. Primary human hepatocytes (Gibco) were plated on a polylysine (Sigma-Aldrich) coated 4-well chamber slide in William’s E medium (Gibco) supplemented with 10% FBS. Oleate and palmitate were solubilized in a PBS (Corning) solution by heating to 55°C and 65°C, respectively. Solubilized fatty acids were conjugated with 10% fatty acid-free-BSA in culture medium to generate fatty acid stock solution. LDs were induced by adding fatty acid stock solution into culture medium for 48 h with a final concentration of 200 µM oleate and 200 µM palmitate unless otherwise indicated. Transfections of HepG2 cells stably expressing HSD17B13-GFP (17) were carried out using Lipofectamine 3000 (Thermo Fisher Scientific). To study homodimerization, cells were grown in 6-well cell culture dishes and transfected using 2 µg HSD17B13-FLAG plasmid DNA and 10 µl Lipofectamine 3000 per well. Forty-eight hours after transfection, cells were lysed in 500 µl lysis buffer [1% Triton X-100, 50 mM Tris (pH 8), 150 mM NaCl] for 10 min on ice and collected after centrifugation at 13,000 g for 30 min at 4°C.
Plasmids and mutagenesis
The full-length protein HSD17B13-GFP (RG213132) and the exon-2-deleted variant (Δ71-106, variant B) HSD17B13-variant B-GFP (RG227799) were obtained from OriGene. HSD17B13-FLAG (VB150430-10020) was designed and constructed by VectorBuilder Inc. (Cyagen Biosciences, Santa Clara, CA). The Q5® Site-Directed Mutagenesis Kit (NEB, E0554S) was used to generate mutant HSD17B13 plasmids with designed mutagenesis primers (supplemental Table S1). HSD17B13-GFP and HSD17B13-FLAG plasmids were used as mutagenesis templates for the cellular localization study and for the enzymatic assay, respectively, unless otherwise indicated. Mutant plasmids were confirmed by Sanger sequencing.
The selection of the sites for point mutation was based on a prediction of sites relevant to activity using the NCBI conserved domain search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and relying on similarity with other short-chain dehydrogenase/reductases (SDRs).
Cellular localization
Cells were seeded in a NuncTM Lab-TekTM II Chambered Coverglass (Thermo Fisher Scientific) and transfected with wild-type or mutant plasmids using Lipofectamine 3000 Reagent (Thermo Fisher Scientific) following the manufacturer’s instructions. To study LD targeting, fatty acids were added 48 h after the transfection to induce LDs. After 48 h of fatty acid treatment cells were fixed in 4% paraformaldehyde (Electron Microscopy Science) for 10 min and counterstained with 1 µg/ml Hoechst (Thermo Fisher Scientific) for nuclei and LipidTox (1:500; Thermo Fisher Scientific) for LDs.
To study the colocalization of HSD17B13 naturally occurring variant B (HSD17B13-B, Δ71-106) with the ER, cells were cotransfected with SEC61β-GFP (ER marker protein; gift from Alexandre Toulmay) and HSD17B13-B-Flag. Cells were fixed in 4% paraformaldehyde and permeabilized in 0.3% Triton-X, 3% BSA, and 10% normal goat serum (Vector Laboratories) in PBS. Immunofluorescence staining of HSD17B13-B-FLAG was performed by incubating cells with FLAG M2 antibody (F3165; Sigma-Aldrich) at room temperature for 1 h. Alexa Fluor 568 goat anti-mouse secondary antibody was used after washing. Hoechst was used to stain nuclei.
Immunofluorescence staining of apoptosis-inducing factor (AIF), a marker for mitochondria, and HSD17B13 were performed to demonstrate mitochondrial targeting of mutant HSD17B13 after transfection. Antibodies against AIF (5318; Cell Signaling Technology) and FLAG (F3165) were used for primary incubation. Alexa Fluor 647 goat anti-rabbit and Alexa Fluor 568 goat anti-mouse secondary antibodies were used to recognize rabbit anti-AIF and mouse anti-FLAG antibodies, respectively. Cellular fluorescence images were taken by confocal microscopy (Zeiss LSM 700).
Retinol dehydrogenase activity assay
Enzymatic activity was measured using the retinol dehydrogenase (RDH) activity assay as previously described (17, 27). Briefly, HEK293 cells were seeded 1 day before being transiently transfected in triplicate with HSD17B13, HSD17B13 mutant, or empty vector plasmids. All-trans-retinol (Toronto Research Chemicals) at 2 or 5 µM in ethanol, with a final ethanol concentration ≤0.5% (v/v), was added to the culture medium, and cells were incubated for 6 or 8 h. Retinoids were extracted twice by equal volume of ethanol and double volume of hexane and were separated by normal-phase HPLC with a Spherisorb S3W column (4.6 × 100 mm) (Waters Corp.). Retinaldehyde and retinoic acid levels were normalized per total protein amount and are shown relative to empty vector. Wild-type HSD17B13 was used as a positive control, with multiple constructs tested in the same round of experiments sharing the positive control. Aliquots of cell suspensions were taken for protein quantification and Western blot analysis.
Coimmunoprecipitation and Western blot
Protein G Dynabeads (Invitrogen) were incubated with anti-FLAG antibody (clone F1804) from Sigma-Aldrich or anti-turboGFP (clone OTI2H8) antibody from Origene overnight with rotation at 4°C. For each pulldown, 3 µg of antibody were incubated with 50 µl of bead slurry. Lysates of transfected HepG2 were incubated with antibody-conjugated beads with rotation for 1 h at room temperature. Bound proteins were washed and eluted with SDS containing sample buffer per the manufacturer’s guidelines. Proteins were separated in 4% to 15% precast PAGE gels (Bio-Rad) and transferred to PVDF membranes (Invitrogen). The membranes were blocked with a 5% nonfat milk in Tris-buffered saline containing 0.05% Tween-20 for 1 h. Membranes were incubated with polyclonal anti-HSD17B13 antibody (1:2000) from Origene (TA350064) in conjunction with an HRP-conjugated secondary antibody (GE Healthcare) and Pierce enhanced chemiluminescent substrate for the detection of HRP (Thermo Fisher Scientific).
Statistical analysis
Differences between wild-type HSD17B13 and empty vector or HSD17B13 mutants were tested using Student’s t-test (GraphPad Prism version 8).
RESULTS
Essential domains for lipid droplet targeting of HSD17B13
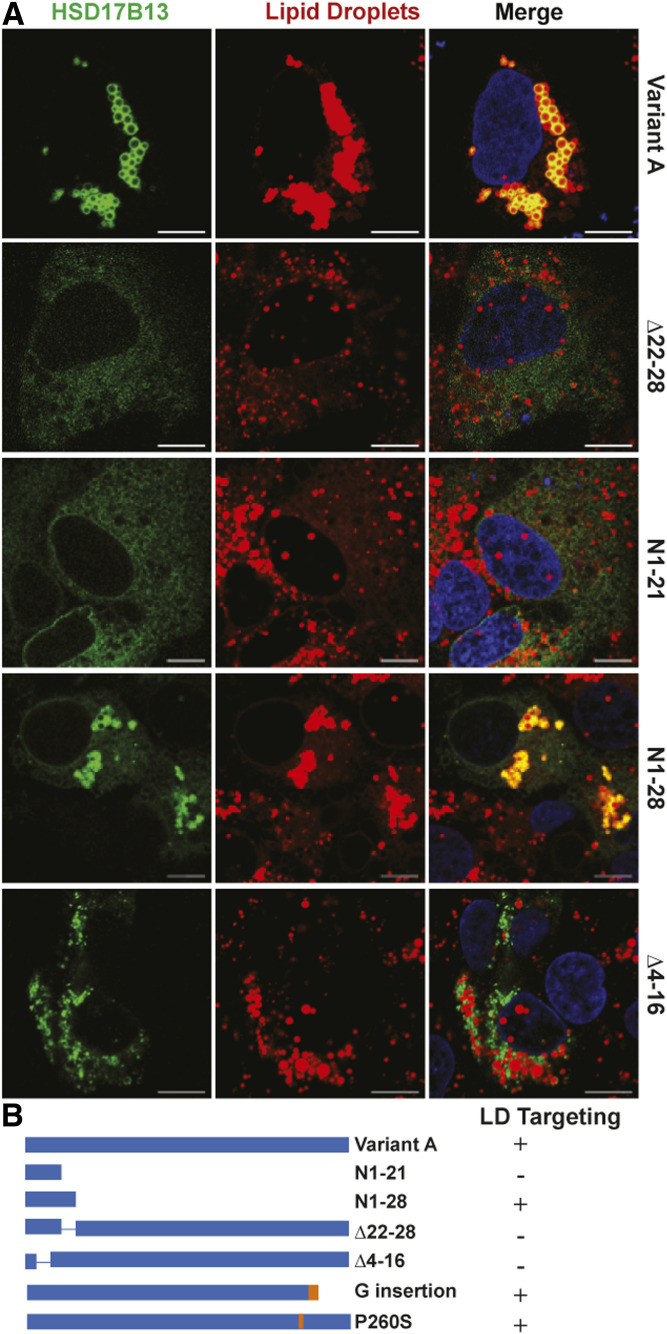
The full-length HSD17B13 protein (variant A) is targeted to LDs when cells are lipid-loaded (Fig. 1A). We previously described the reduction of protein stability and LD targeting with the loss of the PAT-like N-terminal AAs 22–28 (Δ22–28) or with the loss of AAs 71–106 (variant B, a naturally occurring variant with exon 2 skipping) (17) (Fig. 1, Fig. 2A). The N terminus of HSD17B13 is predicted by hydropathy analysis to be a putative transmembrane domain (supplemental Fig. S1) and thus could serve to anchor the protein to LDs; AAs 30–300 are likely to reside on the outside membrane surface. We thus extended our study in detail to delineate the characteristics of the N-terminal sequences that are critical for LD targeting. Not surprisingly, a fragment containing only the hydrophobic domain (N1-21) of HSD17B13 was not targeted to LDs in the absence of AAs 22–28 and AAs 71–106 (Fig. 1), suggesting that the hydrophobic domain is not sufficient to drive protein LD targeting. Interestingly, the addition of the PAT-like domain (AAs 22–28) to the hydrophobic domain generates a peptide fragment (N1-28) of HSD17B13 that localizes to LDs despite the absence of AAs 71–106 (Fig. 1). To test whether the hydrophobic domain is necessary for HSD17B13 to target LDs, we further generated an HSD17B13 devoid of the hydrophobic AAs 4–16 (Δ4–16) and, as expected, found that without this hydrophobic sequence HSD17B13 does not target to LDs (Fig. 1A). Interestingly, Δ4–16 has a nonrandom pattern of cellular distribution, and we found it to be localized in close proximity to mitochondria (supplemental Fig. S2). Thus, our data indicates that at the N terminus of HSD17B13, both AAs 4–16 and AAs 22–28 are necessary for LD targeting.
Fig. 1.
N-terminal domains for LD targeting of HSD17B13. A: HepG2 cells were transiently transfected with HSD17B13 wild-type or mutant plasmids and treated with 200 μM oleate and palmitate to induce LDs. Proteins were C-terminally tagged with GFP, which was used to determine their cellular localization (green). Nuclei were counterstained with Hoechst (blue), and LDs were stained with LipidTox (red). Images were analyzed by confocal microscopy. The bar indicates 10 μM. B: Schematic representation of full-length HSD17B13 (variant A) and mutant proteins. The two naturally occurring protective mutants found in humans (G insertion and P260S) are included for comparison. The plus sign indicates targeting to LDs; the minus sign indicates no targeting.
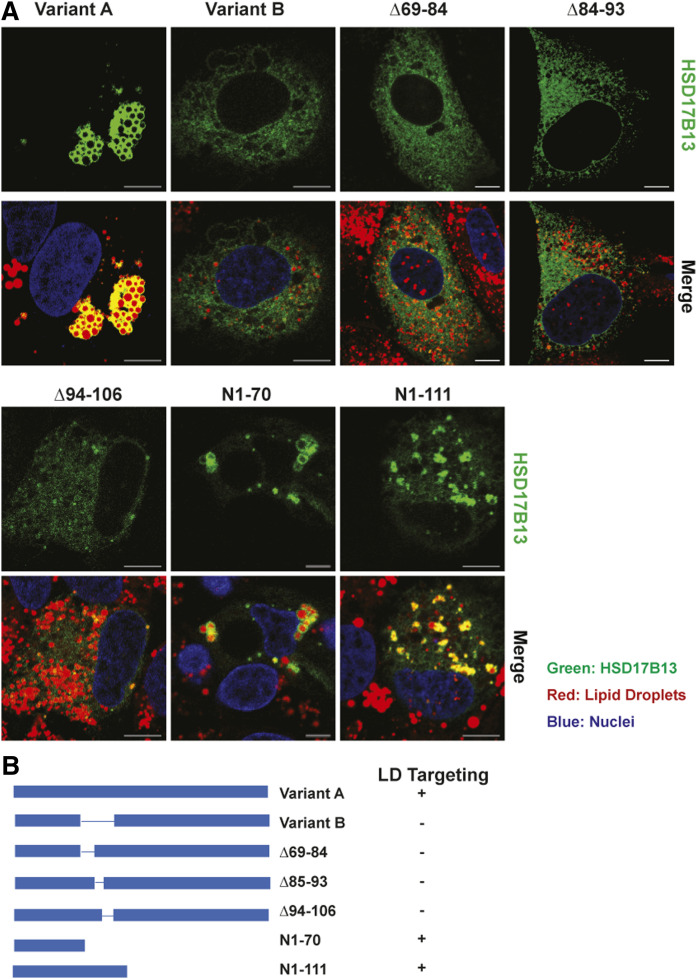
Fig. 2.
Identification of domains critical for LD targeting of HSD17B13. A: HepG2 cells were transiently transfected with HSD17B13 wild-type, the naturally occurring variant B (Δ71-106), or mutant plasmids and treated with fatty acids to induce LDs. Proteins were C-terminally tagged with GFP, which was used to determine their cellular localization (green). Nuclei were counterstained with Hoechst (blue), and LDs were stained with LipidTox (red). Images were analyzed by confocal microscopy. The bar indicates 10 μM. B: Schematic representation of full-length HSD17B13 (variant A) and mutant proteins. The plus sign indicates targeting to LDs; the minus sign indicates no targeting.
The naturally occurring variant B of HSD17B13 is a consequence of exon 2 skipping (without a frame shift), leading to the absence of AAs 71–106. Although the structure of HSD17B13 has not been resolved to date, X-ray crystallography data are available for HSD17B11 (PDB ID: 1YB1; https://www.rcsb.org/structure/1YB1), a close paralogue of HSD17B13, with 77% AA similarity. Based on the structure of HSD17B11, AAs 69–106 in HSD17B13 are predicted to form a α-helix/β-sheet/α-helix structure, which is absent in variant B (Δ71–106). As we previously identified that variant B is not targeted to LDs, despite having the hydrophobic domain (AAs 4–16) and PAT-like domain (AAs 22–28), and has no enzymatic function (17), we tested the importance of this region in more detail. To determine which part of the α-helix/β-sheet/α-helix structure is essential for LD targeting, we generated three mutant proteins of HSD17B13: Δ69–84 (first α-helix deletion), Δ85–93 (β-sheet deletion), and Δ94–106 (second α-helix deletion). The loss of any of the three domains was sufficient to impair LD localization (Fig. 2), indicating that the entire intact structure is required. Without these domains, HSD17B13-variant B is retained in the ER and colocalized with SEC61β, an ER marker protein (supplemental Fig. S3). It is possible that the deletion of the α-helix/β-sheet/α-helix structure leads to misfolding of subsequent sequences in the protein, preventing its transport from the ER to LDs. C-terminal-deleted variants, either without (N1-70) or with the α-helix/β-sheet/α-helix structure (N1-111), localized correctly to LDs (Fig. 2). Taken together, we conclude that the N-terminal hydrophobic and PAT-like domains likely promote the anchoring of of HSD17B13 to the ER/LD membrane, whereas the α-helix/β-sheet/α-helix structure is required for the transportation of the full-length protein from the ER to LDs. To extend our observations beyond cell lines, we transfected primary human hepatocytes (PHHs) with wild-type HSD17B13 or mutant proteins with critical LD-targeting domains modified. We confirmed that the same domains are needed in PHHs to drive the LD targeting of HSD17B13 (supplemental Fig. S4). To confirm whether HSD17B13 will target LD differently under oleate- or palmitate-induced conditions, we treated PHHs with oleate, palmitate, or a combination of both and found that HSD17B13 targets LD under all conditions (supplemental Fig. S5).
Essential domains for enzymatic activity of HSD17B13
Enzymes in the SDR superfamily have a conserved Rossmann-fold motif consisting of six parallel α-helices surrounding the central seven parallel β-sheets (36). This classical Rossmann-fold structure was observed in the predicted 3D structure of HSD17B13 using HSD17B11 as a template (Fig. 3). HSD17B13 is believed to affect chronic liver disease through its enzymatic activity, and enzymatically nonfunctional mutants are associated with protection from liver injury (17, 19). We therefore sought to identify the sites that are critical for its enzymatic activity. We have previously demonstrated the importance of the conserved cofactor binding site (37) Gly47/Gly49 for enzymatic activity (17). In a similar manner, we generated plasmids expressing HSD17B13 with mutations in the putative catalytic site, substrate binding site, or homodimer interaction site and tested their RDH activity.
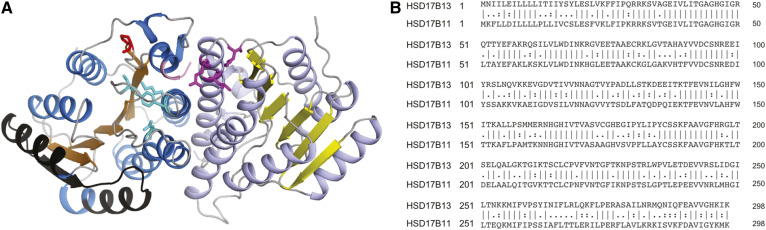
Fig. 3.
Predicted 3D structure of HSD17B13 homodimers using HSD17B11 (67% sequence identity on modeled sequence, PDB entry 1YB1) as the template. A: Structure prediction performed by SWISS-MODEL homology modeling web server. The classical Rossmann-fold structure was observed in the predicted 3D structure of HSD17B13. The β-sheets are colored in orange and yellow, α-helices in marine and light blue, loop regions in gray, amino acids in exon 2 (which are missing in HSD17B13-variant B) in black, catalytic tetrad Asn144-Ser172-Tyr185-Lys189 in cyan, and substrate binding sites Leu199, Glu202, Lys208 in magenta. The Pro260 affected by the P260S mutation is marked in red, and the truncated animo acids affected by the rs72613567-driven G-insertion (only three amino acids are shown in this model) are marked in pink. B: Sequence alignment of human HSD17B13 and HSD17B11 showing overall identity of 191/298 (64.1%) and similarity of 232/298 (77.9%) amino acids.
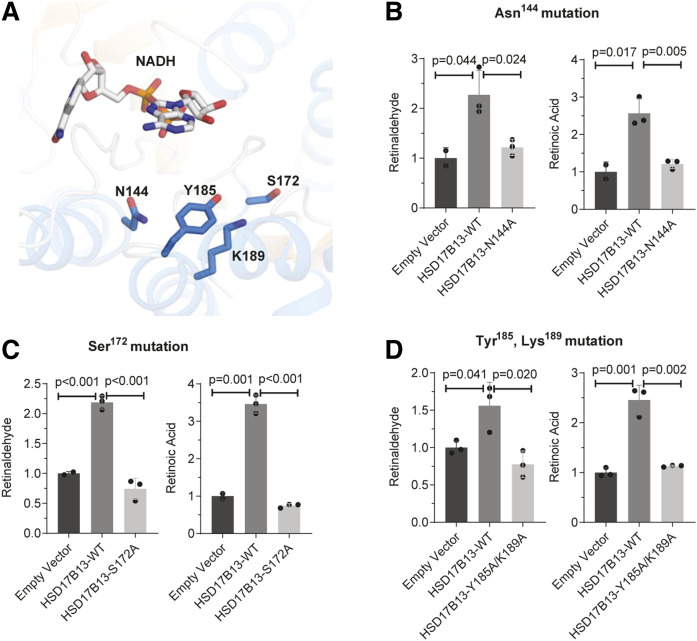
The conserved Asn144-Ser172-Tyr185-Lys189 catalytic tetrad (38) is predicted to be in close proximity to the cofactor NAD+ (Fig. 4A). A single AA mutation from asparagine to alanine at the predicted catalytic site Asn144 (HSD17B13-N144A) abolished the RDH activity of HSD17B13 (Fig. 4B), and similar results were obtained when we mutated another putative catalytic site residue at Ser172 (Fig. 4C). A double mutation of Tyr185 and Lys189 revealed additional AA residues that are critical for activity (Fig. 4D). With the exception of N144A, none of these mutations interfered with protein stability (supplemental Fig. S6) or LD targeting (supplemental Fig. S7).
Fig. 4.
The catalytic tetrad Asn144-Ser172-Tyr185-Lys189 is important for HSD17B13 enzymatic activity. A: Ribbon diagram for the structure of the catalytic tetrad of HSD17B13. NAD and essential residues are labeled and shown as sticks. The binding pose of NAD and retinol were predicted using the HSD17B13 homology model on the SwissDock online web server. B–D: Enzymatic activity of mutant HSD17B13. HEK293 cells were seeded 1 day before and transiently transfected in triplicate with HSD17B13, HSD17B13 mutant, or empty vector plasmids. All-trans-retinol was added to the culture and incubated for 6 or 8 h. Retinaldehyde and retinoic acid were separated by normal-phase HPLC and quantified by retinoid standards. Retinoid levels were normalized to protein concentrations and are shown as relative values to empty vector controls. Data are presented as means ± SEMs. WT, wild-type (variant A).
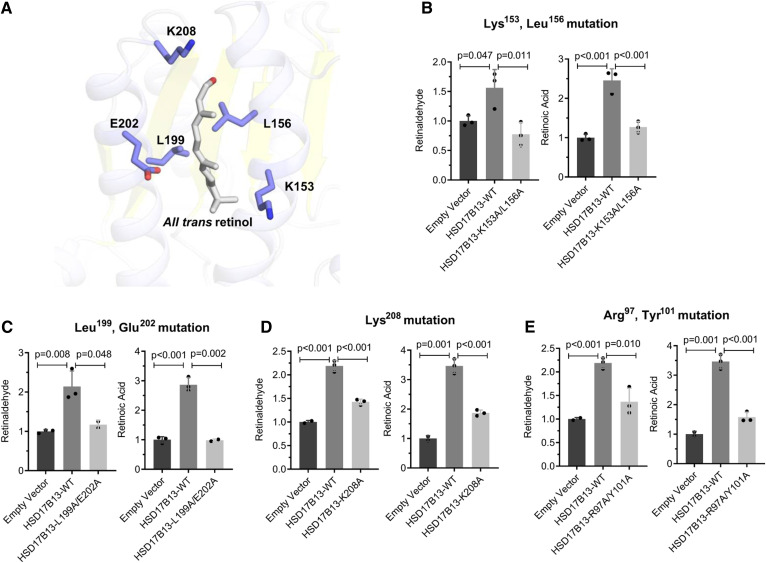
We also generated mutations in putative substrate binding sites in HSD17B13 and tested the RDH activity of the mutant proteins (Fig. 5A). A double mutant at Lys153 and Leu156 (HSD17B13-K153A/L156A) or at Leu199 and Glu202 (HSD17B13-L199A/E202A) abolished the enzymatic activity of HSD17B13 completely (Fig. 5B, C) but was also associated with lower protein levels (supplemental Fig. S6), suggesting decreased stability. A single mutation of Lys208 (HSD17B13-K208A) decreased but did not fully abolish RDH activity (Fig. 5D), indicating that both sites are critical for the enzymatic activity of HSD17B13. The close paralogue and a major retinol dehydrogenase, RDH10, functions either as a homodimer or as an oxidoreductive heterooligometric complex with retinaldehyde reductase 3 to tightly control the steady-state levels of retinoic acid (39). Given the strong structural similarity, it is plausible that HSD17B13 also requires dimerization for its activity. Indeed, we were able to confirm homodimerization of HSD17B13 in vitro by cotransfecting cells with HSD17B13-GFP and HSD17B13-FLAG and performing coimmunoprecipitation (supplemental Fig. S8). We therefore mutated a putative homodimer interaction site at R97/Y101 and found that the mutant protein significantly reduced RDH activity (Fig. 5E). A summary of essential sites for HSD17B13 enzymatic activity is shown in Table 1 and Fig. 6.
Fig. 5.
The amino acid residues in the putative substrate binding sites Lys153, Leu156, Leu199, Glu202, Lys208, and the homodimer binding sites Arg97 and Tyr101 are important for HSD17B13 enzymatic activity. A: Ribbon diagram for the predicted structure of the substrate binding sites of HSD17B13 with retinol as the substrate. Retinol and essential residues are labeled and shown as sticks. B–E: Enzymatic activity of mutant HSD17B13. HEK293 cells were seeded 1 day before and transiently transfected in triplicate with HSD17B13, HSD17B13 mutant, or empty vector plasmids. All-trans-retinol was added to the culture and incubated for 6 or 8 h. Retinaldehyde and retinoic acid were separated by normal-phase HPLC and quantified by retinoid standards. Retinoid levels were normalized to protein concentrations and are shown as relative value to empty vector controls. Data are presented as mean ± SEMs. WT, wild-type (variant A).
TABLE 1.
Critical sites for enzymatic activity of HSD17B13
| Protein Name | Enzymatic Activity | Possible Reason for Loss of Activity |
| HSD17B13 | Yes | NA |
| HSD17B13-variant B | No | Nonlipid droplet targeting/low protein stability |
| HSD17B13-Δ22-28 | No | Nonlipid droplet targeting/low protein stability |
| HSD17B13-Δexon 6 | No | Not known |
| HSD17B13-G insertion | No | Not known |
| HSD17B13-P260S | No | Not known |
| HSD17B13-G47A/G49A | No | Impaired cofactor binding |
| HSD17B13-N144A | No | Impaired catalytic site/low protein stability |
| HSD17B13-S172A | No | Impaired catalytic site |
| HSD17B13-Y185A/K189A | No | Impaired catalytic site |
| HSD17B13-L199A/E202A | No | Impaired substrate binding/low protein stability |
| HSD17B13-K208A | No | Impaired substrate binding |
| HSD17B13-K153A/L156A | No | Impaired substrate binding/low protein stability |
| HSD17B13-R97A/Y101A | No | Impaired dimer formation |
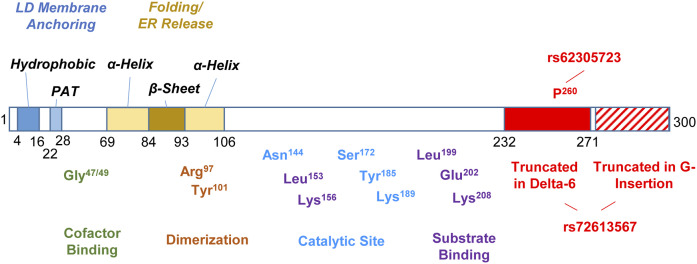
Fig. 6.
Essential domains and amino acids in HSD17B13. Domains essential for localization are marked in italics. Amino acids essential for enzymatic activity are marked with predicted site function. The consequence of naturally occurring genetic variants is marked in red. Not to scale.
DISCUSSION
Human genetic studies identified an association between HSD17B13 and NASH (17, 19, 20), alcoholic liver disease (17, 19), and hepatocellular carcinoma (21, 22). Enzymatically inactive protein mutants of HSD17B13, generated by genetic variants, confer protection from the progression of fatty liver disease (17, 19), confirming the importance of the enzymatic activity of HSD17B13, although this protection was not seen in Hsd17b13 knockout mice (40). Pharmaceutical efforts to silence HSD17B13 by siRNA and to inhibit HSD17B13 enzymatic activity are being pursued by pharmaceutical companies (https://investor.regeneron.com/news-releases/news-release-details/regeneron-and-alnylam-pharmaceuticals-announce-collaboration; https://clinicaltrials.gov/ct2/show/NCT04202354). Understanding the critical residues for its enzymatic activity is important for facilitating small molecule inhibitor design and to better explore the protein function. In this study, we focused on residues based on two premises: first, our previous finding that targeting to LDs is required for enzymatic activity (17), and second, in silico predictions based on structural similarity to other enzymes. With this approach, we identified critical domains (N-terminal hydrophobic domain, PAT-like domain, and a putative α-helix/β-sheet/α-helix domain) for LD targeting and several important residues (predicted catalytic sites, substrates binding sites, and homodimer interaction sites) for the enzymatic activity of HSD17B13.
The N-terminal sequence AAs 1–35 has been shown to be sufficient in localizing HSD17B13 to LDs (31). Here we scale down the required sequence to the N-terminal AAs 1–28 and identify it as consisting of two domains with distinct roles: an N-terminal hydrophobic domain predicted to be a transmembrane helix and an adjacent PAT-like domain (supplemental Figure S1), with both domains required for LD targeting. HSD17B13 is structurally related to two families: the HSD family with steroids, bile acids, or fatty acids as potential substrates (41) and the short-chain dehydrogenase/reductase 16C family, of which some family members have been reported to regulate retinoid metabolism by acting as retinol dehydrogenases (42). Some members of the short-chain dehydrogenase/reductase 16C family such as HSD17B11 and RDH10 have also been identified as LD-associated proteins (43, 44), and a sequence comparison can help clarify the role of targeting domains. An N-terminal-sequence hydrophobic transmembrane domain is known to be required for LD targeting of HSD17B11 and RDH10 (43, 44). The noncanonical PAT motif (PAT-like) has also been described in HSD17B11 (44, 45) but is not present in RDH10, consistent with the known ability of the latter two proteins to localize to other cellular compartments as well.
Interestingly, deletions of the specific HSD17B13 N-terminal domains resulted in different cellular distribution patterns. PAT-like domain deletion (Δ22–28) largely reduced protein stability, whereas the deletion of the transmembrane hydrophobic domain (Δ4–16) redistributed HSD17B13 from LDs to mitochondria. Small deletions leading to protein redistribution from LDs to mitochondria have been described previously for ADRP (46), HSD17B11 (44), and RDH10 mutation studies (43), suggesting similarities between LD and mitochondria targeting machinery that may play a role in the close contact and dynamic interaction of these two organelles (47). Notably, the PAT-like domain in HSD17B13 is also a recognition motif for mitochondrial surface import receptor TOM20 (http://mitf.cbrc.jp/MitoFates/cgi-bin/top.cgi) (48). With the loss of the anchoring transmembrane helix, HSD17B13-Δ4–16 may be released from the ER to the cytosol, where it can be recognized and imported by the TOM20-TOM40 complex (49). Whether this has physiological importance or is purely an artifact of laboratory deletions is unknown.
In this article we describe for the first time a unique motif determining the cellular distribution of HSD17B13 in the α-helix/β-sheet/α-helix domain (AAs 69–106), most of which (AAs 71–106) is missing in the naturally occurring variant B isoform of HSD17B13. The deletion of this domain led to the protein being retained in the ER and not being targeted to LDs. Interestingly, if the C-terminal domain is deleted, the presence of these domains is no longer critical, as evidenced by the normal LD targeting of N1-70 and N1-111. One possibility is that the loss of the α-helix/β-sheet/α-helix domain leads to protein misfolding of the C-terminal domain. Instead of exiting the ER, misfolded proteins are retained in the ER and subjected to proteasomal degradation through a process termed ER-associated degradation (50). Variant B is less stable than the full-length protein (17), hinting that the ER-associated degradation may be triggered by HSD17B13-B. Alternatively, this domain may be needed for additional membrane interaction for the full protein. A hairpin α-helix structure is crucial for membrane interaction and targeting from the ER to LDs for many proteins (25), and the α-helix/β-sheet/α-helix we report here may play a similar role. The same domain is highly conserved in HSD17B11 and is present in RDH10, although in the latter protein it is disrupted by a 24-AA insertion between the first α-helix and the β-sheet. The high degree of structural conservation suggests its importance beyond folding only. Furthermore, the conserved Cys80 residue contained in this region was found to be palmitoylated in mouse Hsd17b13 (51), increasing the hydrophobicity and suggesting important membrane interactions.
Beyond localization to LDs, modifying key residues can affect protein function by interfering with sites essential for its enzymatic activity. The genetically driven splicing variants (HSD17B13-G insertion and HSD17B13-exon 6 deletion) associated with rs72613567 generate an inactive enzyme, possibly due to protein instability (17, 19, 23). Another inactive variant we previously identified is the P260S mutant, encoded by the nonsynonymous genetic variant rs62305723 (17). Although distant from the predicted conserved substrate biding, catalytic, or cofactor binding sites, the highly conserved Pro260 is an essential residue for the enzymatic activity of HSD17B13 (17), and its mutation to serine may lead to an impaired hinge function and introduce protein conformational and subsequent functional changes (52). An additional variant (rs143404524, encoding A192Lfs) was reported to confer protection against chronic liver disease (24); given the critical role of Pro260 (17) and potential protein conformation change, we predict this frame shift protein to be inactive.
NAD(H)/NADP(H) are essential cofactors for SDRs (41), and mutations in conserved cofactor binding glycine residues abolished the enzymatic activity of RDH10 (53) and HSD17B13 (17). Despite a relatively low sequence identity, the 3D structures of SDRs display a highly conserved α/β folding pattern with a predicted catalytic tetrad of Asn-Ser-Tyr-Lys (38). Within this catalytic tetrad, TyrXXXLys is the most conserved site across the SDR family (54). We identified HSD17B13 residues Asn144, Ser172, and Tyr185/Lys189 as the key components of that tetrad and could abolish enzymatic activity by mutating them. Our findings for the HSD17B13 Y185A/L189A double mutant and the N144A mutant are thus consistent with similar studies in RDH10 and in 3 β-HSD (38, 53). Replacing the Ser172 residue in HSD17B13 has also led to a loss of function, although replacing that catalytic serine in other SDRs showed discrepant results; S142A in 17β-HSD-1 (55) and S138A in 3β-HSD (38) abolished their activity, while in RDH10 S197A the activity was retained but not in S197T, which abolished the enzymatic activity (53). These observations suggest slightly different catalytic structures for each enzyme.
We have also tested the predicted substrate binding-site residues Lys153, Leu156, Leu199, Glu202, and Lys208 found them to be essential for HSD17B13 activity. The substrate binding loop is the least conserved domain in the SDR family, which implies a variety of substrate specificities (56, 57) and possibly explains why the mutagenetic replacement of Glu282 failed to affect the activity of another enzyme, 17β-HSD-1 (55). Interestingly, some of the essential sites seem to affect protein stability (L199/E202, K153/L156, and N144), a similar phenotype to what has been described in the protective human variants (19).
In conclusion, we identified common and unique domains and AA sites that are essential for the LD targeting (N-terminal hydrophobic domain, PAT-like domain, and a putative α-helix/β-sheet/α-helix domain) and enzymatic activity (catalytic sites, substrates binding sites, and homodimer interaction sites) of HSD17B13, which could facilitate the therapeutic targeting of this protein against chronic liver diseases.
Data availability
All data are contained herein and are also available upon request from the corresponding author.
Supplementary Material
Acknowledgments
The authors thank Seung Bum Park for his technical assistance with the primary hepatocyte culture.
Footnotes
This article contains supplemental data.
Author contributions—Y.M. and Y.R. conceptualization; Y.M., S.K., P.M.B., D.D.L., M.C.P., W.Z., and O.V.B. investigation; Y.M., and Y.R. formal analysis; Y.M. and Y.R. writing-original draft; Y.M., S.K., P.M.B., D.D.L., M.C.P., W.Z., O.V.B., N.Y.K., and Y.R. writing-review and editing; Y.R. supervision.
Author ORCIDs—Natalia Y. Kedishvili https://orcid.org/0000-0001-6917-4891 Yaron Rotman https://orcid.org/0000-0002-7549-8216
Funding and additional information—This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Disease and by National Institute on Alcohol Abuse and Alcoholism Grant AA012153 (N.Y.K.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Abbreviations—
- AA
- amino acid
- AIF
- apoptosis-inducing factor
- HSD17B13
- 17-β hydroxysteroid dehydrogenase 13
- LD
- lipid droplet
- PHH
- primary human hepatocyte
- PNPLA3
- patatin-like phospholipase domain-containing protein 3
- RDH
- retinol dehydrogenase
- SDR
- short-chain dehydrogenase/reductase
Manuscript received May 13, 2020, and in revised form September 8, 2020. Published, JLR Papers in Press, September 24, 2020, DOI 10.1194/jlr.RA120000907.
REFERENCES
- 1.Younossi Z., Anstee Q. M., Marietti M., Hardy T., Henry L., Eslam M., George J., and Bugianesi E.. 2018. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 15: 11–20. [DOI] [PubMed] [Google Scholar]
- 2.Pais R., Barritt A. St., Calmus Y., Scatton O., Runge T., Lebray P., Poynard T., Ratziu V., and Conti F.. 2016. NAFLD and liver transplantation: current burden and expected challenges. J. Hepatol. 65: 1245–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rotman Y., and Sanyal A. J.. 2017. Current and upcoming pharmacotherapy for non-alcoholic fatty liver disease. Gut. 66: 180–190. [DOI] [PubMed] [Google Scholar]
- 4.Loomba R., Schork N., Chen C. H., Bettencourt R., Bhatt A., Ang B., Nguyen P., Hernandez C., Richards L., Salotti J., et al. 2015. Heritability of hepatic fibrosis and steatosis based on a prospective twin study. Gastroenterology. 149: 1784–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwimmer J. B., Celedon M. A., Lavine J. E., Salem R., Campbell N., Schork N. J., Shiehmorteza M., Yokoo T., Chavez A., Middleton M. S., et al. 2009. Heritability of nonalcoholic fatty liver disease. Gastroenterology. 136: 1585–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browning J. D., Szczepaniak L. S., Dobbins R., Nuremberg P., Horton J. D., Cohen J. C., Grundy S. M., and Hobbs H. H.. 2004. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 40: 1387–1395. [DOI] [PubMed] [Google Scholar]
- 7.Romeo S., Kozlitina J., Xing C., Pertsemlidis A., Cox D., Pennacchio L. A., Boerwinkle E., Cohen J. C., and Hobbs H. H.. 2008. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 40: 1461–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan X., Waterworth D., Perry J. R., Lim N., Song K., Chambers J. C., Zhang W., Vollenweider P., Stirnadel H., Johnson T., et al. 2008. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am. J. Hum. Genet. 83: 520–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sookoian S., Castano G. O., Burgueno A. L., Gianotti T. F., Rosselli M. S., and Pirola C. J.. 2009. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J. Lipid Res. 50: 2111–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rotman Y., Koh C., Zmuda J. M., Kleiner D. E., Liang T. J., and NASH CRN . 2010. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (PNPLA3) with histological severity of nonalcoholic fatty liver disease. Hepatology. 52: 894–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Speliotes E. K., Butler J. L., Palmer C. D., Voight B. F., GIANT Consortium, MIGen Consortium, NASH CRN, and Hirschhorn J. N.. 2010. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology. 52: 904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valenti L., Al-Serri A., Daly A. K., Galmozzi E., Rametta R., Dongiovanni P., Nobili V., Mozzi E., Roviaro G., Vanni E., et al. 2010. Homozygosity for the patatin-like phospholipase-3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology. 51: 1209–1217. [DOI] [PubMed] [Google Scholar]
- 13.Holmen O. L., Zhang H., Fan Y., Hovelson D. H., Schmidt E. M., Zhou W., Guo Y., Zhang J., Langhammer A., Løchen M-L., et al. 2014. Systematic evaluation of coding variation identifies a candidate causal variant in TM6SF2 influencing total cholesterol and myocardial infarction risk. Nat. Genet. 46: 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozlitina J., Smagris E., Stender S., Nordestgaard B. G., Zhou H. H., Tybjaerg-Hansen A., Vogt T. F., Hobbs H. H., and Cohen J. C.. 2014. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 46: 352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahdessian H., Taxiarchis A., Popov S., Silveira A., Franco-Cereceda A., Hamsten A., Eriksson P., and van’t Hooft F.. 2014. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc. Natl. Acad. Sci. USA. 111: 8913–8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buch S., Stickel F., Trepo E., Way M., Herrmann A., Nischalke H. D., Brosch M., Rosendahl J., Berg T., Ridinger M., et al. 2015. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat. Genet. 47: 1443–1448. [DOI] [PubMed] [Google Scholar]
- 17.Ma Y., Belyaeva O. V., Brown P. M., Fujita K., Valles K., Karki S., de Boer Y. S., Koh C., Chen Y., Du X., et al. 2019. 17-Beta hydroxysteroid dehydrogenase 13 is a hepatic retinol dehydrogenase associated with histological features of nonalcoholic fatty liver disease. Hepatology. 69: 1504–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chambers J. C., Zhang W., Sehmi J., Li X., Wass M. N., Van der Harst P., Holm H., Sanna S., Kavousi M., Baumeister S. E., et al. 2011. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat. Genet. 43: 1131–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abul-Husn N. S., Cheng X., Li A. H., Xin Y., Schurmann C., Stevis P., Liu Y., Kozlitina J., Stender S., Wood G. C., et al. 2018. A protein-truncating HSD17B13 variant and protection from chronic liver disease. N. Engl. J. Med. 378: 1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pirola C. J., Garaycoechea M., Flichman D., Arrese M., San Martino J., Gazzi C., Castano G. O., and Sookoian S.. 2019. Splice variant rs72613567 prevents worst histologic outcomes in patients with nonalcoholic fatty liver disease. J. Lipid Res. 60: 176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stickel F., Lutz P., Buch S., Nischalke H. D., Silva I., Rausch V., Fischer J., Weiss K. H., Gotthardt D., Rosendahl J., et al. 2020. Genetic variation in HSD17B13 reduces the risk of developing cirrhosis and hepatocellular carcinoma in alcohol misusers. Hepatology. 72: 88–102. [DOI] [PubMed] [Google Scholar]
- 22.Yang J., Trepo E., Nahon P., Cao Q., Moreno C., Letouze E., Imbeaud S., Bayard Q., Gustot T., Deviere J., et al. 2019. A 17-Beta-hydroxysteroid dehydrogenase 13 variant protects from hepatocellular carcinoma development in alcoholic liver disease. Hepatology. 70: 231–240. [DOI] [PubMed] [Google Scholar]
- 23.Ma Y., Brown P. M., and Rotman Y.. 2020. Letter to the Editor: does the HSD17B13 rs72613567 splice variant actually yield a new type of alternative splicing? Hepatology. 71: 1885–1886. [DOI] [PubMed] [Google Scholar]
- 24.Kozlitina J., Stender S., Hobbs H. H., and Cohen J. C.. 2018. HSD17B13 and chronic liver disease in Blacks and Hispanics. N. Engl. J. Med. 379: 1876–1877. [DOI] [PubMed] [Google Scholar]
- 25.Thiam A. R., Farese R. V. Jr., and Walther T. C.. 2013. The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. Cell Biol. 14: 775–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujimoto T., and Parton R. G.. 2011. Not just fat: the structure and function of the lipid droplet. Cold Spring Harb. Perspect. Biol. 3: a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kory N., Farese R. V. Jr., and Walther T. C.. 2016. Targeting fat: mechanisms of protein localization to lipid droplets. Trends Cell Biol. 26: 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bickel P. E., Tansey J. T., and Welte M. A.. 2009. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim. Biophys. Acta. 1791: 419–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miura S., Gan J. W., Brzostowski J., Parisi M. J., Schultz C. J., Londos C., Oliver B., and Kimmel A. R.. 2002. Functional conservation for lipid storage droplet association among Perilipin, ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and Dictyostelium. J. Biol. Chem. 277: 32253–32257. [DOI] [PubMed] [Google Scholar]
- 30.Ohsaki Y., Maeda T., Maeda M., Tauchi-Sato K., and Fujimoto T.. 2006. Recruitment of TIP47 to lipid droplets is controlled by the putative hydrophobic cleft. Biochem. Biophys. Res. Commun. 347: 279–287. [DOI] [PubMed] [Google Scholar]
- 31.Horiguchi Y., Araki M., and Motojima K.. 2008. 17beta-Hydroxysteroid dehydrogenase type 13 is a liver-specific lipid droplet-associated protein. Biochem. Biophys. Res. Commun. 370: 235–238. [DOI] [PubMed] [Google Scholar]
- 32.Su W., Wang Y., Jia X., Wu W., Li L., Tian X., Li S., Wang C., Xu H., Cao J., et al. 2014. Comparative proteomic study reveals 17beta-HSD13 as a pathogenic protein in nonalcoholic fatty liver disease. Proc. Natl. Acad. Sci. USA. 111: 11437–11442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedman S. L., Neuschwander-Tetri B. A., Rinella M., and Sanyal A. J.. 2018. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 24: 908–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sonnhammer E. L., von Heijne G., and Krogh A.. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6: 175–182. [PubMed] [Google Scholar]
- 35.Wilkins M. R., Gasteiger E., Bairoch A., Sanchez J. C., Williams K. L., Appel R. D., and Hochstrasser D. F.. 1999. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112: 531–552. [DOI] [PubMed] [Google Scholar]
- 36.Lukacik P., Kavanagh K. L., and Oppermann U.. 2006. Structure and function of human 17beta-hydroxysteroid dehydrogenases. Mol. Cell. Endocrinol. 248: 61–71. [DOI] [PubMed] [Google Scholar]
- 37.Duax W. L., Pletnev V., Addlagatta A., Bruenn J., and Weeks C. M.. 2003. Rational proteomics I. Fingerprint identification and cofactor specificity in the short-chain oxidoreductase (SCOR) enzyme family. Proteins. 53: 931–943. [DOI] [PubMed] [Google Scholar]
- 38.Filling C., Berndt K. D., Benach J., Knapp S., Prozorovski T., Nordling E., Ladenstein R., Jörnvall H., and Oppermann U.. 2002. Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J. Biol. Chem. 277: 25677–25684. [DOI] [PubMed] [Google Scholar]
- 39.Belyaeva O. V., Adams M. K., Wu L., and Kedishvili N. Y.. 2017. The antagonistically bifunctional retinoid oxidoreductase complex is required for maintenance of all-trans-retinoic acid homeostasis. J. Biol. Chem. 292: 5884–5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma Y., Brown P. M., Lin D. D., Ma J., Feng D., Belyaeva O. V., Podszun M. C., Roszik J., Allen J., Umarova R., et al. 2020. Hsd17b13 deficiency does not protect mice from obesogenic diet injury. Hepatology. Epub ahead of print. August 11, 2020; doi: 10.1002/hep.31517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchais-Oberwinkler S., Henn C., Moller G., Klein T., Negri M., Oster A., Spadaro A., Werth R., Wetzel M., Xu K., Frotscher M., et al. 2011. 17β-Hydroxysteroid dehydrogenases (17β-HSDs) as therapeutic targets: protein structures, functions, and recent progress in inhibitor development. J. Steroid Biochem. Mol. Biol. 125: 66–82. [DOI] [PubMed] [Google Scholar]
- 42.Belyaeva O. V., Chang C., Berlett M. C., and Kedishvili N. Y.. 2015. Evolutionary origins of retinoid active short-chain dehydrogenases/reductases of SDR16C family. Chem. Biol. Interact. 234: 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang W., and Napoli J. L.. 2013. The retinol dehydrogenase Rdh10 localizes to lipid droplets during acyl ester biosynthesis. J. Biol. Chem. 288: 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horiguchi Y., Araki M., and Motojima K.. 2008. Identification and characterization of the ER/lipid droplet-targeting sequence in 17beta-hydroxysteroid dehydrogenase type 11. Arch. Biochem. Biophys. 479: 121–130. [DOI] [PubMed] [Google Scholar]
- 45.Tsachaki M., and Odermatt A.. 2019. Subcellular localization and membrane topology of 17beta-hydroxysteroid dehydrogenases. Mol. Cell. Endocrinol. 489: 98–106. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura N., and Fujimoto T.. 2003. Adipose differentiation-related protein has two independent domains for targeting to lipid droplets. Biochem. Biophys. Res. Commun. 306: 333–338. [DOI] [PubMed] [Google Scholar]
- 47.Ingelmo-Torres M., Gonzalez-Moreno E., Kassan A., Hanzal-Bayer M., Tebar F., Herms A., Grewal T., Hancock J. F., Enrich C., Bosch M., et al. 2009. Hydrophobic and basic domains target proteins to lipid droplets. Traffic. 10: 1785–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fukasawa Y., Tsuji J., Fu S. C., Tomii K., Horton P., and Imai K.. 2015. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell. Proteomics. 14: 1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamamoto H., Itoh N., Kawano S., Yatsukawa Y., Momose T., Makio T., Matsunaga M., Yokota M., Esaki M., Shodai T., et al. 2011. Dual role of the receptor Tom20 in specificity and efficiency of protein import into mitochondria. Proc. Natl. Acad. Sci. USA. 108: 91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith M. H., Ploegh H. L., and Weissman J. S.. 2011. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 334: 1086–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen L. F., Chen Y. J., Liu K. M., Haddad A. N. S., Song I. W., Roan H. Y., Chen L. Y., Chen Y-J., Wu J-Y., Chen Y-T.. 2017. Role of S-palmitoylation by ZDHHC13 in mitochondrial function and metabolism in liver. Sci. Rep. 7: 2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park C. B., Yi K. S., Matsuzaki K., Kim M. S., and Kim S. C.. 2000. Structure-activity analysis of buforin II, a histone H2A-derived antimicrobial peptide: the proline hinge is responsible for the cell-penetrating ability of buforin II. Proc. Natl. Acad. Sci. USA. 97: 8245–8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi Y., Moiseyev G., Farjo K., and Ma J. X.. 2009. Characterization of key residues and membrane association domains in retinol dehydrogenase 10. Biochem. J. 419: 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jörnvall H., Persson B., Krook M., Atrian S., Gonzalez-Duarte R., Jeffery J., and Ghosh D.. 1995. Short-chain dehydrogenases/reductases (SDR). Biochemistry. 34: 6003–6013. [DOI] [PubMed] [Google Scholar]
- 55.Huang Y. W., Pineau I., Chang H. J., Azzi A., Bellemare V., Laberge S., and Lin S. X.. 2001. Critical residues for the specificity of cofactors and substrates in human estrogenic 17beta-hydroxysteroid dehydrogenase 1: variants designed from the three-dimensional structure of the enzyme. Mol. Endocrinol. 15: 2010–2020. [DOI] [PubMed] [Google Scholar]
- 56.Oppermann U., Filling C., Hult M., Shafqat N., Wu X., Lindh M., Shafqat J., Nordling E., Kallberg Y., Persson B., et al. 2003. Short-chain dehydrogenases/reductases (SDR): the 2002 update. Chem. Biol. Interact. 143–144: 247–253. [DOI] [PubMed] [Google Scholar]
- 57.Ghosh D., and Vihko P.. 2001. Molecular mechanisms of estrogen recognition and 17-keto reduction by human 17beta-hydroxysteroid dehydrogenase 1. Chem. Biol. Interact. 130–132: 637–650. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained herein and are also available upon request from the corresponding author.