Abstract
Ginseng polysaccharide (GPS) is known for its efficacy in cancer therapy; however, its regulatory mechanism in breast cancer (BC) remains unclear. To analyze the effect of GPS on BC cell proliferation, cell proliferation rate calculations, western blotting, plasmid transfections, electrophoretic mobility shift assays and chromatin immunoprecipitation assays were performed. GPS treatment in the culture cell medium inhibited cell proliferation in the BC cell line MDA-MB-231. In addition, the E-cadherin level was enhanced while the vimentin level was suppressed following GPS treatment (both P<0.05). Furthermore, the levels of apoptotic markers, including cleaved-Caspase-3 and p53, and inflammatory response markers, including plasminogen activator inhibitor and TNF-α, were induced by GPS treatment in MDA-MB-231 cells (all P<0.05). These results indicated that GPS supplementation activated the inflammatory response and apoptosis in BC cells. GPS treatment activated the phosphorylation levels of c-Jun N-terminal kinase, Akt and NF-κB. In MDA-MB-231 cells, GPS resulted in the accumulation of the NF-κB components p65, p50 and Ikaros family zing finger protein 1 (IKZF1; all, P<0.05). Chromatin immunoprecipitation and electrophoretic mobility shift assays indicated that p65 bound to the IKZF1 promoter. The overexpression of IKZF1 or p65 inhibited MDA-MB-231 cell proliferation (P<0.05), indicating that GPS treatment may inhibit BC cell proliferation by the activation of IKZF1. Taken together, these results suggested that GPS significantly inhibited BC cell proliferation via the control of the biological processes, including the activation of p65-IKZF1 signaling and apoptosis. The data indicated a novel mechanism for further understanding of cancer cell proliferation.
Keywords: breast cancer, ginseng polysaccharide, Ikaros family zing finger protein 1, inflammation, p65
Introduction
Breast cancer (BC) is one of the most malignant and common types of cancer among women worldwide and the sixth leading cause of cancer-associated mortality among Chinese women (1). China had 12.2% of global cases and 9.6% of cancer-related deaths of BC in 2012 (1,2). Solid tumors can occur in the lactiferous ducts, lobules of the mammary glands and in interstitial tissues of patients with BC (3). Worldwide, cancer of the lactiferous ducts and lobules accounted for 90% of all BC in 2018 (Korea Breast Cancer Society; www.kbcs.or.kr). On the molecular level, BC can be divided into four subtypes: Hormone receptor-positive BC, human epidermal growth factor receptor 2 (HER2)-positive BC, triple-negative BC (TNBC) and basal-like BC (4). Among these types of cancer, the luminal estrogen receptor (ER)-positive, HER2-negative subtype accounted for ~70% of patients with BC in China in 2012 (1,2).
Endocrine therapies, including aromatase inhibitors, selective ER downregulators, gonadotropin-releasing hormone and selective ER modulators, are effective therapeutic strategies targeting ER and HER2 in the clinical treatment of patients with BC (5). Although endocrine therapy is successful in clinical practice, patients can develop resistance to these therapies. Several important molecular signaling pathways have been identified to contribute to resistance, including estrogen-independent activation of the ER and cell-cycle regulation by cyclin D-CDK4/6, epigenetic pathways, heat shock protein 90 and an immunogenic pathway including cytotoxic T-lymphocyte associated protein 4 and programmed cell death 1 ligand 1/2- programmed cell death 1(6). Notably, the PI3K/Akt/ mTOR pathway is the most common oncogenic pathway with a crucial role in growth, survival, proliferation and differentiation of cancer cells (7,8). The PTEN signaling pathway serves as an upstream regulator of mTOR, an antagonist of PI3K/Akt signaling and a modulator of numerous cellular processes (9-11). The Akt, tuberous sclerosis complex 2 and live kinase B1 signaling pathways have been reported to inhibit mTOR activation in cancer cells (12-15). Therefore, BC could be controlled by inhibiting the mTOR pathway.
Ubiquitin-specific peptidases (USPs) regulate cell proliferation and apoptosis by ubiquitination and deubiquitination (16,17). USPs determine the accumulation levels of proteins through post-translational modifications in cells by controlling the conjugation and removal of ubiquitin (18). Additionally, the dysfunction of USPs leads to oncogenic progression in cells (19,20). USP10 regulates signaling factors associated with cell proliferation, apoptosis and cancer metabolism. Several studies have reported that UPS10 regulates the PTEN signaling pathway to inhibit cancer cell growth and invasion, and c-Myc transcription to suppress cancer formation and to affect cellular sensitivity to DNA damage (21-23). A recent study reported that USP10 expression was decreased in hepatocellular carcinoma, leading to poor prognosis (24). Consequently, it may be hypothesized that the regulation of USP10 expression inhibits the mTOR signaling pathway and the treatment and prognosis of patients.
Inflammation, a biological response to adverse physical or chemical stimuli, serves a role in cancer development and metastasis via the release of proinflammatory cytokines, including TNF-α, IL-6 and IL-1β, to activate the key transcription factor NF-κB (25). Plasminogen activator inhibitor (PAI-1) is a member of the serine protease inhibitor protein family (26). PAI-1 knockout mice exhibited low levels of inflammation compared with wild-type mice (27), indicating that this protein may serve a role in the inflammatory response. Furthermore, a previous study indicated that the PAI-1 level was closely associated with BC metastasis (28). Previous studies supported the hypothesis that chronic inflammation promoted cancer development (29,30). Certain evidence has indicated that inflammatory factors such as cyclooxygenase-2 (COX2) and lipoxygenase (LOX) were upregulated in BC (31-33) and COX2 in ER-negative BC and TNBC was associated with poor prognosis (34). COX and LOX metabolic products serve a role in BC (35). In addition, apoptosis is important for cancer cell therapy. Previous studies reported that numerous signaling pathways and molecules regulated BC cell apoptosis, including osteocyte signaling (36), miRNAs (37,38) and TNF-related apoptosis-inducing ligand (TRAIL) signaling (39), indicating that the control of inflammation and apoptosis was important for BC treatment. Clostridium difficile toxin B was reported to inhibit the inflammatory response by suppressing the COX2 level and to activate apoptosis in BC (40). In addition, ursolic acid inhibits BC development by activating apoptosis and suppressing the inflammatory response (41), indicating that inflammation is negatively associated with BC cell growth (42). However, NF-κB, a key inflammatory signaling transcription factor, activates anti- and pro-apoptotic genes (42), indicating that the regulation of inflammation and apoptosis is complex.
Panax ginseng has been used as a medicinal plant in China for thousands of years (43). The ginseng extract is composed of ginsenoside (the ginseng saponin), acanthosides, senticosides, triterpene saponins, flavonoid, vitamins, minerals and polysaccharides (44,45). Ginsenosides are the main ingredients reported to inhibit tumor metastasis in cells (46). Additionally, ginseng-derived polysaccharides were previously believed to exhibit antitumor effects and were isolated as the antitumor fraction for the first time in 1994(47). Ginseng polysaccharide (GPS) has been demonstrated to exhibit low toxicity (47). Furthermore, GPS stimulates nonspecific immune cells and activates natural killer cells and macrophages to protect the host against foreign antigens and tumor growth (48-50). Furthermore, GPS activates macrophages and inhibits tumor angiogenesis and metastasis (51-53). Therefore, the current study was performed to investigate the function of GPS in BC cell proliferation. The inflammatory response and apoptosis were analyzed. Additionally, the regulatory effect of signaling pathways associated with the inflammatory response on GPS-mediated inhibition of BC cell proliferation was assessed. The findings indicated a novel mechanism by which GPS may inhibit BC cell proliferation.
Materials and methods
Cell culture and transfection assays
The human breast adenocarcinoma cell line MDA-MB-231 obtained from the American Type Culture Collection was cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) with glutamine (Sigma-Aldrich; Merck KGaA) and supplemented with 10% FBS and 100 µg/ml penicillin and streptomycin (Gibco; Thermo Fisher Scientific, Inc.) at 37˚C. Ikaros family zing finger protein 1 (IKZF1) or p65 cDNAs were synthesized by Sangon Biotech Co., Ltd. and pcDNA3.1 (+) (Invitrogen; Thermo Fisher Scientific, Inc.) was used for constructing overexpression (OX) vectors. Subsequently, 2 µg of pcDNA3.1 (+) empty vector, IKZF1 OX and p65 OX plasmids were transfected (seeding density, 1x106 cells) on day 0 using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) and Opti-MEM I Reduced Serum Medium (Gibco; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. On day 1, 24 h post-transfection, the cells were confluent and the IKZF1 or p65 OX solutions were replaced with DMEM with glutamine supplemented with 10% FBS and 100 µg/ml penicillin and streptomycin. These transfected MDA-MB-231 cells were used for the subsequent experiments.
Western blotting
MDA-MB-231 cells treated with 100 µM or 200 µM GPS dissolved in ddH2O (Xi'an Virgin Biotechnology Co., Ltd.) for 24 h at 37˚C or transfected with IKZF1 or p65 OX plasmids for 24 h following GPS treatment were harvested in an ice-cold lysis solution (7M urea, 2M thiourea, 2% CHAPS, 40 mM Tris base, 40 mM dithiothreitol and 1% protease inhibitor) to obtain whole-cell extracts. Cells in the control group were treated with equal amounts of ddH2O. The protein concentration was measured by using BCA protein assay kit (Cell Signaling Technology, Inc.). Total proteins from each sample (20 µg/lane) were separated using SDS-PAGE (10% gel) and transferred onto Immobilon-P transfer membranes (Merck KGaA). The membranes were incubated in 1X TBS containing 5% skim milk and 0.05% Tween-20 for 1-2 h at room temperature and subsequently incubated with primary antibodies at 4˚C overnight. The following primary antibodies were used: Anti-vimentin (cat. no. ab193555; 1:1,000; Abcam), anti-E-cadherin (cat. no. ab194982; 1:1,000; Abcam), anti-p53 (cat. no. ab32389; 1:2,000; Abcam), anti-cleaved (c)-Caspase-3 (cat. no. ab2302; 1:2,000; Abcam), anti-Caspase-3 (cat. no. ab13847; 1:2,000; Abcam), anti-PAI-1 (cat. no. ab66705; 1:2,000; Abcam), anti-TNF-α (cat. no. ab1793; 1:2,000; Abcam), anti-phosphorylated p-JNK (Thr183/Tyr185; cat. no. 4668; 1:1,000; Cell Signaling Technology, Inc.), anti-JNK1 + JNK2 + JNK3 (cat. no. ab179461; 1:1,000; Abcam), anti-p-Akt (Ser473; cat. no. 4060; 1:2,000; Cell Signaling Technology, Inc.), anti-Akt (cat. no. 4691; 1:1,000; Cell Signaling Technology, Inc.), IKZF1 (cat. no. H-100; 1:2,000; Santa Cruz Biotechnology, Inc.), IκBα (cat. no. ab32518; 1:2,000, Abcam), anti-p-IκBα (cat. no. sc-8404; 1:500; Santa Cruz Biotechnology, Inc.), anti-NF-κB p50 (cat. no. ab109752; 1:2,000; Abcam), anti-NF-κB p65 (cat. no. ab16502; 1:2,000; Abcam) and anti-GAPDH (cat. no. ab8245; 1:2,000; Abcam). The membranes were washed twice with 1X PBS and incubated with anti-mouse or anti-rabbit horseradish peroxidase-conjugated secondary antibodies (cat. nos. 7074 and 7076; 1:2,000, Cell Signaling Technology) for the corresponding primary antibodies synthesized from mouse or rabbit secondary antibodies for 1 h at room temperature. Reaction products were visualized using an ECL Western Blotting Detection system (GE Healthcare). Quantification of relative band densities was performed by scanning densitometry using ImageJ software (version 2; National Institute of Health).
Cell proliferation analysis
MDA-MB-231 cells were plated in a volume of 150 µl at a density of 2x103 cells/well into 96-well plates to determine the cell proliferation rate. Cell proliferation ability was analyzed following treatment with 50 or 100 µM GPS dissolved in ddH2O (Xi'an Virgin Biotechnology Co., Ltd.) for 24, 48 and 72 h at 37˚C. Control cells were treated with equal volumes of ddH2O. Cell proliferation in the IKZF1 OX and p65 OX-transfected cells was analyzed using a Cell Counting Kit-8 (Dojindo Molecular Technologies, Inc.), following a previously published method (54).
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed on MDA-MB-231 cells using a chromatin immunoprecipitation assay kit (cat. no. 17-295; EMD Millipore), according to the manufacturer's protocol. An anti-NF-κB p65 antibody (~1 µg; cat. no. ab16502; 1:2,000; Abcam) was used for immunoprecipitation and no-antibody IP was used as the negative control. Following immunoprecipitation, a wash buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.1) and 150 mM NaCl] was used to wash the precipitates. The DNA immunoprecipitated using the antibodies was compared with the DNA precipitated without the addition of antibodies using quantitative PCR (qPCR). A SYBR-Green Master Mix (Bio-Rad Laboratories, Inc.) was used to perform the qPCR on an Illumina Eco 3.0 (Illumina, Inc.). The following thermocycling conditions were used: An initial denaturation at 95˚C for 3 min; 40 cycles of denaturation for 30 sec at 95˚C, annealing for 30 sec at 58˚C and extension at 72˚C for 30 sec; followed by a final extension at 72˚C for 5 min. The Ct value of each ChIP DNA fraction was normalized to the input DNA fraction Ct value for the same qPCR Assay to account for chromatin sample preparation differences using the 2-ΔΔCq method (50) and GAPDH was used as the negative control. The 1.5 kb of IKZF1 (NP_001207694.1) promoter sequences were downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/) and the three pairs of primers were designed. The primers used for ChIP-PCR were as follows: 1 forward, 5'-TCCTGAGTTGCTTCCCACT-3' and reverse, 5'-GGTGTGTCCCAGTAACAT-3'; 2 forward, 5'-GGGCAGAAGGAAAAGTGTCA-3' and reverse, 5'-CCAAAGGAATGTGAGCTCGT-3'; 3 forward 5'-GACCACCCCTCACATTCAAC-3' and reverse, 5'-TGGCAGTTGAGAATCAGTGG-3'; and GAPDH forward, 5'-GACCTGCCGTCTAGAAAAAC-3' and reverse, 5'-CTGTAGCCAAATTCGTTGTC-3'.
Electrophoretic mobility shift assay (EMSA)
p65 open reading frame sequences were synthesized and subcloned into T-easy vectors (Promega Corporation) and moved to XhoI and EcoRI restriction enzyme sites of pET28a (+) expression vectors to produce p65 recombinant proteins. The resulting pET28a-p65 plasmids were used for the transformation of Escherichia coli (E. coli) strain BL21 DE3 (Tiangen Biotech, Co., Ltd.). Recombinant His-p65 proteins were harvested following 4 h of 0.5 mM isopropyl β-D-1-thiogalactopyranoside (Sigma-Aldrich; Merck KGaA) treatment at 30˚C by centrifugation 12,000 rpm at 4˚C for 30 min. E. coli cells expressing His-p65 were suspended in 1x PBS solution (Tiangen Biotech, Co., Ltd.) and lysed by sonication (130-150 V; amplitude 40 µM) at 4˚C for 5 min. Crude extracts were purified by adding 1 ml of Mag-Beads His-Tag (Sangon Biotech, Co., Ltd.). The beads and crude extract were incubated overnight in a rotating instrument (BaoDragon, Inc.) at 4˚C and the beads washed with 1x PBS solution 5 times at 4˚C. Proteins were eluted by adding 100 mM imidazole (Sigma-Aldrich; Merck KGaA) and protein concentrations were measured using a BCA kit (cat. no. BCA1; Merck KGaA), according to the manufacturer's protocol. Protein were further dialyzed by removing imidazole in 1x PBS solution using dialysis tubing (Sangon Biotech, Co., Ltd.). The recombinant protein was dissolved in 1x PBS (Tiangen Biotech, Co., Ltd.). For EMSA, a standard binding reaction was performed in a total volume of 20 µl by incubating 1 µg of purified protein with 40,000 cpm of a 32P-labeled DNA probe (PCR fragments amplified in the aforementioned ChIP assay) and 1 µg of poly dI-dC, which blocked the nonspecific binding of the protein to probe DNA in the reaction buffer [25 mM HEPES-KOH (pH 7.5), 100 mM KCl, 0.1 mM EDTA, 10% (v/v) glycerol and 1mM DTT] at room temperature for 30 min. The binding reaction products were resolved on an 8% polyacrylamide gel run in 0.5X TBE buffer and bands were detected using and x-ray film (Sangon Biotech, Co., Ltd.) by detecting the 32P radiation signal.
Statistical analysis
Statistical analysis was performed with a Prism software package (version no. 5.0; GraphPad Software, Inc.). Data are presented as mean ± standard error. Comparisons between two groups and the determination of statistical significance was performed using the Student's t-test. Comparisons between more than two groups were performed using one-way ANOVA, followed by Bonferroni's multiple comparisons test. P<0.05 was considered to indicate a statistically significant difference.
Results
GPS treatment inhibits the proliferation of MDA-MB-231 cells
A BC cell line MDA-MB-231 was used in the current study to analyze the role of GPS in BC cells. Compared with the control group, 100 and 200 µM GPS significantly inhibited MDA-MB-231 cell proliferation following 24, 48 and 72 h of treatment. The changes in the cell proliferation rate following treatment with 100 and 200 µM GPS had fold changes in OD values of 0.75 and 0.72, respectively, after 24 h; 0.699 and 0.682, respectively, after 48 h; and 0.668 and 0.661, respectively, after 72 h compared with the control group (Fig. 1A). Treatment with 100 µM GPS significantly enhanced the E-cadherin level; however, the vimentin levels was reduced compared with the control group. The percentage changes in the E-cadherin level following treatment with 100 and 200 µM GPS were ~2.993 and 3.113, respectively. The percentage changes in the vimentin level following treatment with 100 and 200 µM GPS were ~0.562 and 0.529, respectively, compared with the control group (Fig. 1B and C).
Figure 1.
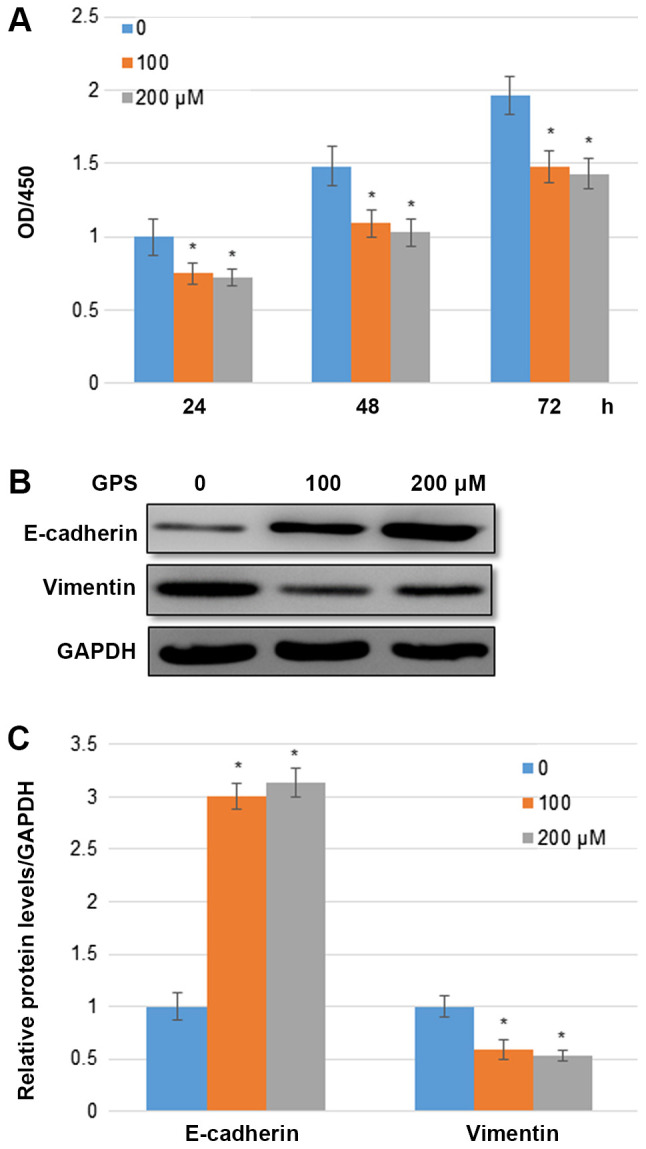
GPS treatment inhibits MDA-MB-231 BC cell proliferation. (A) Cell Counting Kit-8 assays were performed to analyze the effects of GPS on MDA-MB-231 BC cell proliferation. MDA-MB-231 cell proliferation was analyzed 0, 24 and 48 h after 100 and 200 µM GPS treatment. Experiments were repeated 20 times. (B) Western blotting was performed to determine E-cadherin and vimentin expression levels. GAPDH was used as the loading control. Experiments were performed in triplicate. (C) Relative band densities were calculated. Data are presented as mean ± standard error (n=3). *P<0.05 vs. the 0 µM group. GPS, ginseng polysaccharide; BC, breast cancer.
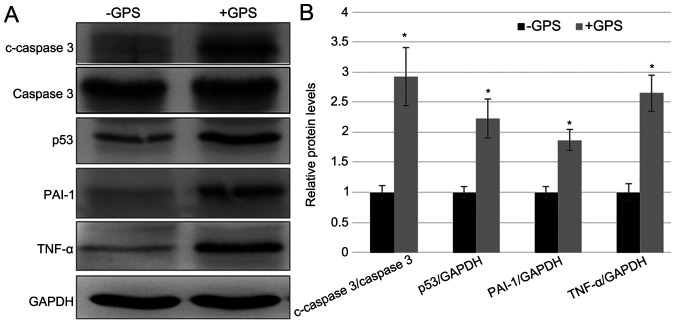
GPS treatment activates the inflammatory response and apoptosis in MDA-MB-231 cells
As discussed above, GPS treatment inhibited MDA-MB-231 cell proliferation. Therefore, the present study subsequently examined the inflammatory and apoptotic response in MDA-MB-231 cells after 100 µM GPS treatment. Apoptotic markers, including the c-Caspase-3/Caspase-3 ratio and p53 expression level, and inflammatory response markers, including PAI-1 and TNF-α, were detected in cells with and without GPS treatment. The western blotting results demonstrated that the GPS treatment enhanced the protein levels of p53, c-Caspase-3/Caspase-3, TNF-α and PAI-1 after stimulation for 24 h stimulation, while the level of Caspase-3 remained unchanged. The fold-changes in c-Caspase-3/Caspase-3, p53, PAI-1 and TNF-α after GPS treatment was 2.928, 2.194, 1.889 and 2.643, respectively, compared with the control group (Fig. 2). These results indicated that GPS treatment activated the inflammatory response and apoptosis in MDA-MB-231 cells.
Figure 2.
GPS treatment induced the expression of inflammatory and apoptosis markers in MDA-MB-231 cells. (A) Western blotting was performed to analyze the levels of apoptotic markers, including the c-Caspase-3/Caspase-3 ratio and p53, and inflammatory response markers, including PAI-1 and TNF-α. GAPDH was used as the loading control. (B) The relative band densities shown were calculated using the ratios of c-Caspase-3/Caspase-3, p53/GAPDH, PAI-1/GAPDH and TNF-α/GAPDH levels. Data are presented as mean ± standard error. Experiments were performed in triplicate. *P<0.05 vs. the -GPS group. GPS, ginseng polysaccharide; PAI-1, plasminogen activator inhibitor 1; TNF-α, tumor necrosis factor-α; c, cleaved; p, phosphorylated; +GPS, cells treated with GPS; -GPS, cells untreated with GPS.
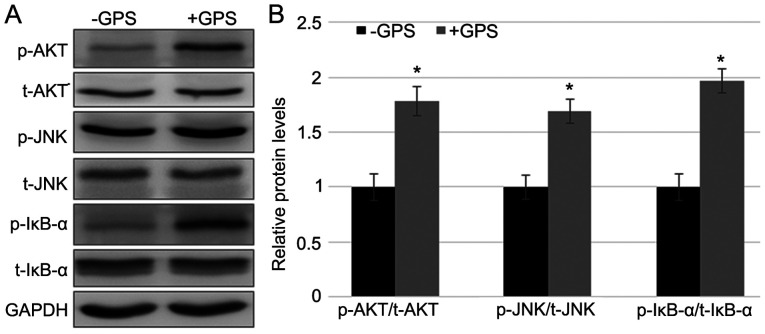
GPS treatment activates Akt, JNK and IκBα
To explore whether GPS treatment influenced the phosphorylation of Akt and JNK, western blotting was performed to evaluate the levels of total (t)-Akt and t-JNK, and p-AKT and p-JNK. The results indicated that GPS treatment enhanced p-AKT and p-JNK levels without affecting the t-AKT and t-JNK levels (Fig. 3A and B). Since, as demonstrated above, GPS treatment activated the inflammatory response by the induction of PAI-1 and TNF-α, the phosphorylation of IκBα, an NF-κB signaling regulator, was examined. Western blotting results demonstrated that GPS activated the phosphorylation of IκB-α without affecting the t-IκB-α level. The fold changes in p-Akt/t-Akt, p-JNK/t-JNK and p-IκB-α/ t-IκB-α ratios after the GPS treatment were 1.758, 1.638 and 1.953, respectively, compared with the control group (Fig. 3A and B). These results indicated that GPS supplementation activated the phosphorylation of AKT, JNK and IκB-α.
Figure 3.
GPS treatment activates Akt, JNK and IκB-α in MDA-MB-231 cells. (A) Western blotting was performed to analyze the t- and p-Akt, JNK and IκB-α levels. GAPDH was used as the loading control. (B) Relative band densities were calculated. Data are presented as mean ± standard error. Experiments were performed in triplicate. *P<0.05 vs. the -GPS group. GPS, ginseng polysaccharide; Akt, protein kinase B; JNK, c-Jun N-terminal kinase; IκB-α, inhibitor κ B-α; t, total; p, phosphorylated; +GPS, cells treated with GPS; -GPS, cells untreated with GPS.
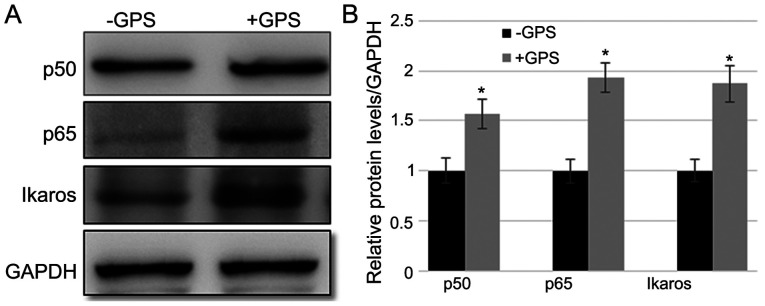
GPS treatment leads to the accumulation of p65, p50 and IKZF1 in MDA-MB-231 cells
As demonstrated above, GPS treatment increased the levels of p-IκB-α, a key NF-κB signaling regulator. Subsequently, the changes in the p65 level following GPS treatment were analyzed. Western blotting indicated that GPS treatment led to the accumulation of p65 and p50 proteins in MDA-MB-231 cells, compared with the control group (Fig. 4A and B). Furthermore, the level of IKZF1 was determined. The data indicated that GPS treatment enhanced the IKZF1 expression level compared with the control group (Fig. 4A and B). The fold changes in p50, p65 and IKZF1 after GPS treatment were 1.544, 1.909 and 1.859, respectively, compared with the control group.
Figure 4.
GPS treatment induces p50, p65 and IKZF1 expression in MDA-MB-231 cells. (A) Western blotting was performed to analyze the expression levels of p50, p65 and IKZF1. GAPDH was used as the loading control. (B) Relative band densities were calculated. Data are presented as mean ± standard error. Experiments were performed in triplicate. *P<0.05 vs. the -GPS group. GPS, ginseng polysaccharide; IKZF1, Ikaros family zing finger protein 1; +GPS, cells treated with GPS; -GPS, cells untreated with GPS.
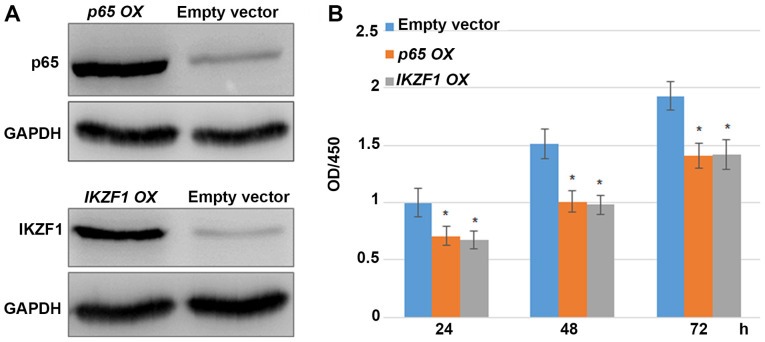
p65 or IKZF1 overexpression inhibits the proliferation of MDA-MB-231 cells
p65 and IKZF1 were overexpressed in MDA-MB-231 cells to analyze whether p65 and IKZF1 had an influence on MDA-MB-231 cell proliferation. Western blotting results indicated that p65 and IKZF1 levels were markedly higher in the OX groups compared with the empty vector control group (Fig. 5A). Furthermore, MDA-MB-231 cell proliferation was examined in p65- or IKZF1-OX groups. CCK-8 assay results indicated that the overexpression of p65 or IKZF1 inhibited cell proliferation compared with the empty vector control (Fig. 5B). The fold changes in the cell proliferation rate following the overexpression of p65 and IKZF1 were 0.712 and 0.671, respectively, after 24 h; 0.669 and 0.649, respectively, after 48 h; and 0.731 and 0.736, respectively, after 72 h compared with the control group.
Figure 5.
p65 or IKZF1 overexpression inhibits MDA-MB-231 BC cell proliferation. (A) Western blotting was performed to analyze p65 and IKZF1 levels in control and p65- or IKZF1-OX groups. GAPDH was used as the loading control. Experiments were performed in triplicate. (B) Cell Counting Kit-8 assays were performed to analyze the effects of ginseng polysaccharide on MDA-MB-231 BC cell proliferation. MDA-MB-231 cell proliferation was analyzed after 0, 24 and 48 h of transfection with p65 or IKZF1-OX plasmids. Data are presented as mean ± standard error. Experiments were repeated 20 times. *P<0.05 vs. the empty vector group. IKZF1, Ikaros family zing finger protein 1; BC, breast cancer.
p65 directly binds to the promoter of IKZF1
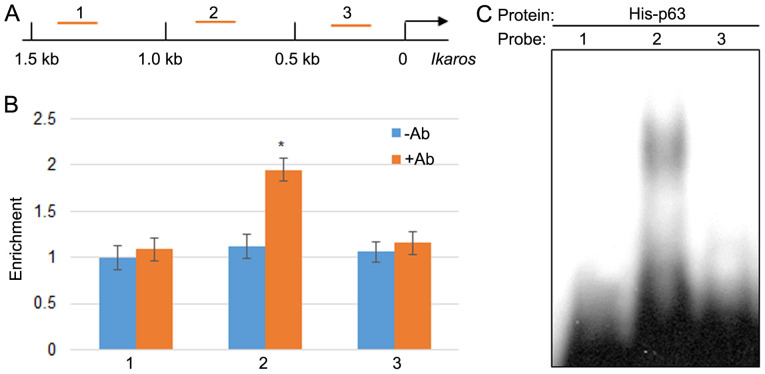
As aforementioned, the current results indicated that GPS promoted the expression of p65 and IKZF1, and inhibited MDA-MB-231 cell proliferation. Therefore, the possibility of a direct interaction between p65 and IKZF1 was explored. ChIP assay using the p65 antibody was performed to determine whether p65 bound to 1.5 kb of the IKZF1 promoter, which is 1.5 kb upstream from start codon ATG (Fig. 6A). ChIP-PCR was performed by scanning 1.5 kb of the promoter using three pairs of primer with a 0.5 kb distance for each set primer. The results indicated that p65 may bind to region 2; however, it did not bind region 1 or 3 within the IKZF1 promoter (Fig. 6B). The fold changes in the percentage of DNA content by the p65 antibody was 1.734 compared with the control group without antibody treatment. Further, EMSA was performed to test the direct binding of p65 to the IKZF1 promoter region. The EMSA results indicated that the p65 recombinant protein bound to region 2 but not regions 1 and 3 (Fig. 6C).
Figure 6.
p65 binds to the promoter of IKZF1. (A) Representation of 1.5 kb of the IKZF1 promoter region. The regions amplified in the ChIP assay and the fragments of probes for the EMSA assay were labeled as 1, 2 and 3. (B) ChIP-PCR was performed to amplify the DNA precipitated with or without an anti-p65 antibody. Experiments were performed in triplicate. *P<0.05 vs. the -Ab group. (C) EMSA was performed using His-p65 recombinant proteins and 32P-labeled DNA fragments 1, 2 and 3. Data are presented as mean ± standard error. IKZF1, Ikaros family zing finger protein 1; CHiP, chromatin immunoprecipitation; EMSA, electrophoretic mobility shift assay; -Ab, without the p65 antibody; +Ab, with the p65 antibody.
Discussion
In 2012, BC was one of the most common gynecological malignancies in China (2). Chemotherapy serves an important role in the treatment of BC; however, the side effects of chemotherapeutic drugs reduce the quality of patients' life (1). Among them, ginsenoside Rh2 and GPS have been reported to exhibit anticancer effects (46,47). The application of the Chinese medicine GPS for BC therapy may be a novel treatment approach; however, the molecular mechanism underlying the inhibition of BC cells by GPS remains largely unknown.
In the current study, the BC cell line MDA-MB-231 and GPS were used to examine the mechanism underlying the inhibition of MDA-MB-231 cell proliferation by GPS. GPS is known as an anticancer molecule (47); however, it's role in the inhibition of BC cell proliferation remains unclear. Proliferation is an important step in cancer cell metastasis in human tissues and is directly associated with disease severity (47). GPS treatment induced E-cadherin; however, vimentin levels were suppressed. These results indicated that GPS inhibited MDA-MB-231 cell viability. The inflammation and apoptosis status was evaluated by detecting the expression levels of marker proteins PAI-1 and TNF-α levels to determine the effect of GPS on the inflammatory response. Western blotting results indicated that GPS treatment activated inflammation in MDA-MB-231 cells. These results were further confirmed by examining the IκB-α phosphorylation level and the accumulation of two NF-κB components, p65 and p50. The results suggested that GPS may activate IκB-α to increase the expression of p65 in the nucleus and, subsequently, activate the expression of PAI-1 and TNF-α. GPS exhibited inhibitory activity against the p38 MAP kinase pathway, NF-κB and proinflammatory cytokines in vitro (50). In addition, ginsenoside Rg3 treatment reduced the COX2 level in mouse skin and human pro-myelocytic leukemia (HL-60) cells (55), implying a regulatory effect of ginseng extracts on inflammation in cancer cells. NF-κB is known to be the upstream regulator of COX2 and Linoleate 9S-lipoxygenase 5 (5LOX), indicating that GPS treatment may downregulate COX2 and 5LOX expression. Previous studies have demonstrated that chronic inflammation may lead to the cancerous condition (29,30). Together, these previous studies indicate that GPS may suppress COX2 and 5LOX to reduce inflammation and BC risk. However, the results of the current study reported that GPS induced the levels of inflammation marker proteins at early time points, indicating that GPS or other ginseng extract-mediated reduction of inflammation may occur in the later stages of cancer. Further studies are required to clarify this hypothesis. Furthermore, the activity of stress-responsive kinases Akt and JNK was examined by detecting the levels of t- and p-Akt and JNK. JNK functions downstream of Akt, belongs to the mitogen-activated protein kinase family and is responsive to cytokines, ultraviolet irradiation, heat and osmotic stresses (56). The results of the current study revealed that GPS activated Akt and JNK, indicating that GPS treatment led to stress in MDA-MB-231 cells. Together, the evidence revealed that GPS activated the inflammatory response by activating the NF-κB signaling in BC. In addition, apoptotic markers c-Caspase-3 and p53 were induced by GPS treatment. Furthermore, GPS treatment inhibited the proliferation of MDA-MB-231 cells, suggesting that activation of inflammation and apoptosis by GPS may be associated with the inhibition of proliferation of BC cells.
IKZF1, a negative regulator of hepatocellular carcinoma proliferation (57), was induced by GPS treatment in the current study. In mammalian cells, NF-κB1 (p50 or its precursor p105), NF-κB2 (p52 or its precursor p100), Rel (c-Rel), RelA (p65) and RelB are the five members of the NF-κB/Rel family. The NF-κB/Rel family member contains 300 amino acids in the N-terminal region termed Rel homolog domain (58). Among them, the p65-p50 heterodimer is the most abundant active form of NF-κB in numerous cell types (58). Additionally, p65 and p50 were induced by GPS treatment. Overexpression of p65 or IKZF1 was revealed to significantly inhibit MDA-MB-231 cell proliferation. IKZF1 is a transcriptional repressor while p65 is a transcriptional activator (58); therefore, the regulatory effect of p65 on the IKZF1 promoter was investigated. ChIP and EMSA data confirmed that p65 could bind to the IKZF1 promoter, indicating that the GPS-mediated induction of IKZF1 may be via p65. These data demonstrated that GPS, an anticancer molecule, inhibited breast cell proliferation possibly by activating the NF-κB signaling to induce inflammation. GPS treatment also activated the apoptotic response, which was analyzed by detecting the c-Caspase-3/Caspase-3 ratio and p53, suggesting that GPS may activate both apoptosis and inflammation to inhibit MDA-MB-231 BC cell proliferation. Numerous studies have indicated that apoptosis was important for the control of BC cell growth (36-39). The role of GPS in the activation of apoptosis in BC cells requires further investigation.
Apoptosis is one of the pathways of cell death; however, cancer cells survival is promoted by the induction of an apoptosis resistance mechanism (59). A previous study reported that aldehyde dehydrogenase family 1 member A3 (ALDH1A3) and sex determining region Y box 2 (Sox-2) regulated the mechanism of apoptosis resistance in BC cells (60); however, GPS-mediated regulation of ALDH1A3 and Sox-2 has not been reported. TNF-α is a mediator of inflammation and is a member of a family of >20 related proteins including lymphotoxin-a, CD30 ligand, CD40 ligand, Fas ligand and TRAIL (61). TRAIL is known to induce apoptosis in several cell line models; however, TRAIL-resistant tumors have also been reported and represent a challenge in cancer therapy (62,63). Furthermore, microRNA-519a-3p was reported to regulate apoptosis resistance in a TRAIL-dependent or independent manner in BC (37); however, the mechanism of TNF-α-mediated apoptosis resistance requires further investigation in the context of BC therapy.
Inflammatory factors are upregulated in BC (31-33). The results of the present study contradicted those of previous reports, since GPS treatment induced the expression of inflammatory markers. Considering the present study used only one BC cell line, MDA-MB-231, which is derived from TNBC, it is important to note that the effect of GPS on this cell line may not be applicable to all subtypes of BC. Therefore, further experiments should be conducted using different BC cell types to verify these results. In the current study, GPS promoted the expression of pro-inflammatory and pro-apoptotic markers, and inhibited the proliferation of BC cells. The results provided a novel molecular mechanism of GPS-mediated BC cell inhibition and may be used to further explore the mechanism of BC cell proliferation.
Acknowledgements
Not applicable.
Funding
The current study was supported by the Wenzhou City Public Welfare Technology Project (grant no. Y20180511).
Availability of data and materials
The datasets used and/or analyzed in the present study are available from the corresponding author on reasonable request.
Authors' contributions
HZ and RH designed the experiments. HZ, YY, XZ, TZ and JX performed the experiments. HZ, YY, BZ, TZ, JX and RH analyzed data. HZ and RH wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1. National Cancer Institute: Cancer topics: Breast cancer, 2014. Accessed 5 Jan 2015. 2015:1-1. [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Cancer incidence and mortality worldwide: IARC Cancer Base no. 10. GLOBOCAN 2008. Lyon: International Agency for Research on Cancer, 2010. [Google Scholar]
- 3.Han Z, Wei B, Zheng Y, Yin Y, Li K, Li S. Breast cancer multi-classification from histopathological images with structured deep learning model. Sci Rep. 2017;7(4172) doi: 10.1038/s41598-017-04075-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prat A, Pineda E, Adamo B, Galván P, Fernández A, Gaba L, Díez M, Viladot M, Arance A, Muñoz M. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast. 2015;24 (Suppl 2):S26–S35. doi: 10.1016/j.breast.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 5.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson S, Chia SK. Treatment algorithms for hormone receptor positive advanced breast cancer: Applying the results from recent clinical trials into daily practice-insights, limitations, and moving forward. Am Soc Clin Oncol Educ Book. 2013;33(e20) doi: 10.1200/EdBook_AM.2013.33.e20. [DOI] [PubMed] [Google Scholar]
- 7.Saxton RA, Sabatini DM. mTOR signaling in growth, 379 metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1007/s12031-016-0872-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickler MN, Tolaney SM, Rugo HS, Cortés J, Diéras V, Patt D, Wildiers H, Frenzel M, Koustenis A, Baselga J. MONARCH 1: Results from phase II study of abemaciclib, a CDK4 and CDK6 inhibitor, as monotherpay, in patients with HR+/HER2- breast cancer, after chemotherapy for advanced disease. J Clin Oncol. 2016;34(510) [Google Scholar]
- 9.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. PTEN is essential for embryonic development and tumour suppression. Nature Genetics. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Cao I, Song MS, Hobbs RM, Laurent G, Giorgi C, de Boer VC, Anastasiou D, Ito K, Sasaki AT, Rameh L, et al. Pandolfi systemic elevation of PTEN induces a tumor-suppressive metabolic state. Cell. 2012;149:49–62. doi: 10.1016/j.cell.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 12.Ma X, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 13.Huang J, Manning BD. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem Soc Trans. 2009;37:217–222. doi: 10.1042/BST0370217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signaling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 15.Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, Cantley LC. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;6:91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 16.Wilkinson KD, Hochstrasser M. Deubiquitinating enzymes. In: Ubiquitin and Biology of the Cell. Peters JM, Finley D and Harris JR (eds). Plenum Press, New York, NY, pp99-120, 1998. [Google Scholar]
- 17.Everett RD, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:566–577. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Zhang P, Wei Y, Piao HL, Wang W, Maddika S, Wang M, Chen D, Sun Y, Hung MC, et al. Deubiquitylation and stabilization of PTEN by USP13. Nat Cell Biol. 2013;15:1486–1494. doi: 10.1038/ncb2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hussain S, Zhang Y, Galardy PJ. DUBs and cancer: The role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle. 2009;8:1688–1697. doi: 10.4161/cc.8.11.8739. [DOI] [PubMed] [Google Scholar]
- 21.Zhang M, Hu C, Tong D, Xiang S, Williams K, Bai W, Li GM, Bepler G, Zhang X. Ubiquitin-specific peptidase 10 (USP10) deubiquitinates and stabilizes MutS Homolog 2 (MSH2) to regulate cellular sensitivity to DNA damage. J Biol Chem. 2016;291:10783–10791. doi: 10.1074/jbc.M115.700047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin Z, Yang H, Tan C, Li J, Liu Z, Quan Q, Kong S, Ye J, Gao B, Fang D. USP10 antagonizes c-Myc transcriptional activation through SIRT6 stabilization to suppress tumor formation. Cell Rep. 2013;5:1639–1649. doi: 10.1016/j.celrep.2013.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun J, Li T, Zhao Y, Huang L, Sun H, Wu H, Jiang X. USP10 inhibits lung cancer cell growth and invasion by upregulating PTEN. Mol Cell Biochem. 2018;41:1–7. doi: 10.1007/s11010-017-3170-2. [DOI] [PubMed] [Google Scholar]
- 24.Lu C, Ning Z, Wang A, Chen D, Liu X, Xia T, Tekcham DS, Wang W, Li T, Liu X, et al. USP10 suppresses tumor progression by inhibiting mTOR activation in hepatocellular carcinoma. Cancer Lett. 2018;436:139–148. doi: 10.1016/j.canlet.2018.07.032. [DOI] [PubMed] [Google Scholar]
- 25.Chung HY, Cesari M, Anton S, Marzetti E, Giovannini S, Seo AY, Carter C, Yu BP, Leeuwenburgh C. Molecular inflammation: Underpinnings of aging and age-related diseases. Ageing Res Rev. 2009;8:18–30. doi: 10.1016/j.arr.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Douglas EV, Rahul R, Sadiya SK, Mesut E, Asish KG. Plasminogen activator inhibitor-1 is a marker and a mediator of senescence. Arterioscler Thromb Vasc Biol. 2017;37:1446–1452. doi: 10.1161/ATVBAHA.117.309451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin SG, Koh SH, Woo CH, Lim JH. PAI-1 inhibits development of chronic otitis media and tympanosclerosis in a mouse model of otitis media. Acta Otolaryngol. 2014;134:1231–1238. doi: 10.3109/00016489.2014.940554. [DOI] [PubMed] [Google Scholar]
- 28.Wei X, Li S, He J, Du H, Liu Y, Yu W, Hu H, Han L, Wang C, Li H, et al. Tumor-secreted PAI-1 promotes breast cancer metastasis via the induction of adipocyte-derived collagen remodeling. Cell Commun Signal. 2019;17(58) doi: 10.1186/s12964-019-0373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhatelia K, Singh K, Singh R. TLRs: Linking inflammation and breast cancer. Cell Signal. 2014;26:2350–2357. doi: 10.1016/j.cellsig.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 30.Jiang X, Shapiro DJ. The immune system and inflammation in breast cancer. Mol Cell Endocrinol. 2014;382:673–682. doi: 10.1016/j.mce.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris RE, Casto BC, Harris ZM. Cyclooxygenase-2 and the inflammogenesis of breast cancer. World J Clin Oncol. 2014;5:677–692. doi: 10.5306/wjco.v5.i4.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erler JT, Bennewith KL, Nicolau M, Dornhöfer N, Kong C, Le QT, Chi JT, Jeffrey SS, Giaccia AJ. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 33.Kirschmann DA, Seftor EA, Fong SFT, Nieva DR, Sullivan CM, Edwards EM, Sommer P, Csiszar K, Hendrix MJ. A molecular role for lysyl oxidase in breast cancer invasion. Cancer Res. 2002;62:4478–4483. [PubMed] [Google Scholar]
- 34.Basudhar D, Glynn SA, Greer M, Somasundaram V, No JH, Scheiblin DA, Garrido P, Heinz WF, Ryan AE, Weiss JM, et al. Coexpression of NOS2 and COX2 accelerates tumor growth and reduces survival in estrogen receptor-negative breast cancer. Proc Natl Acad Sci USA. 2017;114:13030–13035. doi: 10.1073/pnas.1709119114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noguchi M, Rose DP, Earashi M, Miyazaki I. The role of fatty acids and eicosanoid inhibitors in breast carcinoma. Oncology. 1995;52:265–271. doi: 10.1159/000227471. [DOI] [PubMed] [Google Scholar]
- 36.Ma YV, Lam C, Dalmia S, Gao P, Young J, Middleton K, Liu C, Xu H, You L. Mechanical regulation of breast cancer migration and apoptosis via direct and indirect osteocyte signaling. J Cell Biochem. 2018;119:5665–5675. doi: 10.1002/jcb.26745. [DOI] [PubMed] [Google Scholar]
- 37.Breunig C, Pahl J, Küblbeck M, Miller M, Antonelli D, Erdem N, Wirth C, Will R, Bott A, Cerwenka A, Wiemann S. MicroRNA-519a-3p mediates apoptosis resistance in breast cancer cells and their escape from recognition by natural killer cells. Cell Death Dis. 2017;8(e2973) doi: 10.1038/cddis.2017.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu B, Gao W, Zhou H, Miao X, Chang Y, Wang L, Xu M, Ni G. Propofol induces apoptosis of breast cancer cells by downregulation of miR-24 signal pathway. Cancer Biomark. 2018;21:513–519. doi: 10.3233/CBM-170234. [DOI] [PubMed] [Google Scholar]
- 39.Yin N, Yi L, Khalid S, Ozbey U, Sabitaliyevich UY, Farooqi AA. TRAIL mediated signaling in breast cancer: Awakening guardian angel to induce apoptosis and overcome drug resistance. Adv Exp Med Biol. 2019;1152:243–252. doi: 10.1007/978-3-030-20301-6_12. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Li Y, Li H, Chen W, Liu W. Clostridium difficile toxin B recombinant protein inhibits tumor growth and induces apoptosis through inhibiting Bcl-2 expression, triggering inflammatory responses and activating C-erbB-2 and Cox-2 expression in breast cancer mouse model. Biomed Pharmacother. 2018;101:391–398. doi: 10.1016/j.biopha.2018.02.045. [DOI] [PubMed] [Google Scholar]
- 41.Luo J, Hu YL, Wang H. Ursolic acid inhibits breast cancer growth by inhibiting proliferation, inducing autophagy and apoptosis, and suppressing inflammatory responses via the PI3K/AKT and NF-κB signaling pathways in vitro. Exp Ther Med. 2017;14:3623–3631. doi: 10.3892/etm.2017.4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burstein E, Ducket CS. Dying for NF-kappaB? Control of cell death by transcriptional regulation of the apoptotic machinery. Curr Opin Cell Biol. 2003;15:732–737. doi: 10.1016/j.ceb.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 43.Helms S. Cancer prevention and therapeutics: Panax ginseng. Altern Med Rev. 2004;9:259–274. [PubMed] [Google Scholar]
- 44.Davydov M, Krikorian AD. Eleutherococcus senticosus (Rupr & Maxim.) Maxim. (Araliaceae) as an adaptogen: A closer look. J Ethnopharmacol. 2000;72:345–393. doi: 10.1016/s0378-8741(00)00181-1. [DOI] [PubMed] [Google Scholar]
- 45.Lee S, Shin DS, Oh KB, Skin KH. Antibacterial compounds from leaves of Acanthopanax senticosus. Arch Pharm Res. 2003;26:40–42. doi: 10.1007/BF03179929. [DOI] [PubMed] [Google Scholar]
- 46.Hasegawa H, Suzuki R, Nagaoka T, Tezuka Y, Kadota S, Saiki I. Prevention of growth and metastasis of murine melanoma through enhanced natural-killercytotoxicity by fatty acid conjugate of protopanaxatriol. Biol Pharm Bull. 2002;25:861–866. doi: 10.1248/bpb.25.861. [DOI] [PubMed] [Google Scholar]
- 47.Kiyohara H, Hirano M, Wen XG, Matsumoto T, Sun XB, Yamada H. Characterization of an antiulcer pectic polysaccharide from leaves of Panaxginseng C.A. Meyer. Carbohydr Res. 1994;263:89–101. doi: 10.1016/0008-6215(94)00151-0. [DOI] [PubMed] [Google Scholar]
- 48.Schepetkin IA, Quinn MT. Botanical polysaccharides: Macrophage immunomodulation and therapeutic potential. Int Immunopharmacol. 2006;6:317–333. doi: 10.1016/j.intimp.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 49.Yoon TJ, Yoo YC, Kang TB, Baek YJ, Huh CS, Song SK, Lee KH, Azuma I, Kim JB. Prophylactic effect of Korean mistletoe (Viscum album coloratum) extract on tumor metastasis is mediated by enhancement of NK cell activity. Int J Immunopharmacol. 1998;20:163–172. doi: 10.1016/s0192-0561(98)00024-1. [DOI] [PubMed] [Google Scholar]
- 50.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 51.Shin MS, Lee H, Hong HD, Shin KS. Characterization of immunostimulatory pectic polysaccharide isolated from the leaves of Diospyros kaki Tumb (Persimmon) J Funct Foods. 2016;26:319–329. [Google Scholar]
- 52.Park JY, Shin MS, Kim SN, Kim HY, Kim KH, Shin KS, Kang KS. Polysaccharides from Korean Citrus hallabong peels inhibit angiogenesis andbreast cancer cell migration. Int J Biol Macromol. 2016;85:522–529. doi: 10.1016/j.ijbiomac.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 53.Lee EH, Park HR, Shin MS, Cho SY, Choi HJ, Shin KS. Antitumor metastasis activity of pectic polysaccharide purified from the peels of Korean Citrus Hallabong. Carbohydr Polym. 2014;111:72–79. doi: 10.1016/j.carbpol.2014.04.073. [DOI] [PubMed] [Google Scholar]
- 54.Xuan YH, Huang BB, Tian HS, Chi LS, Duan YM, Wang X, Zhu ZX, Cai WH, Zhu YT, Wei TM, et al. High-glucose inhibits human fibroblast cell migration in wound healing via repression of bFGF-regulating JNK phosphorylation. PLoS One. 2014;9(e108182) doi: 10.1371/journal.pone.0108182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keum YS, Han SS, Chun KS, Park KK, Park JH, Lee SK, Surh YJ. Inhibitory effects of the ginsenoside Rg3 on phorbol ester-induced cyclooxygenase-2 expression, NF-kappaB activation and tumor promotion. Mutat Res. 2003;523-524:75–85. doi: 10.1016/s0027-5107(02)00323-8. [DOI] [PubMed] [Google Scholar]
- 56.Ip YT, Davis RJ. Signal transduction by the c-Jun N-terminal kinase (JNK)-from inflammation to development. Curr Opin Cell Biol. 1998;10:205–219. doi: 10.1016/s0955-0674(98)80143-9. [DOI] [PubMed] [Google Scholar]
- 57.Liu YY, Ge C, Tian H, Jiang JY, Zhao FY, Li H, Chen TY, Yao M, Li JJ. The transcription factor Ikaros inhibits cell proliferation by downregulating ANXA4 expression in hepatocellular carcinoma. Am J Cancer Res. 2017;7:1285–1297. [PMC free article] [PubMed] [Google Scholar]
- 58.Baeuerle PA, Baltimore D. NF-kappa B: Ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 59.Ahn JY, Choi IS, Shim JY, Yun EK, Yun YS, Jeong G, Song JY. The immunomodulator ginsan induces resistance to experimental sepsis by inhibiting Toll-like receptor-mediated inflammatory signals. Eur J Immunol. 2006;36:37–45. doi: 10.1002/eji.200535138. [DOI] [PubMed] [Google Scholar]
- 60.Kashii-Magaribuchi K, Takeuchi R, Haisa Y, Sakamoto A, Itoh A, Izawa Y, Isa M, Fukuzawa M, Murakami M, Takahashi R. Induced expression of cancer stem cell markers ALDH1A3 and Sox-2 in hierarchical reconstitution of apoptosis-resistant human breast cancer cells. Acta Histochem Cytochem. 2016;49:149–158. doi: 10.1267/ahc.16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wallach D, Varfolomeev EE, Malinin NL, Goltsev YV, Kovalenko AV, Boldin M. Tumor necrosis factor receptor and Fas signaling mechanisms. Ann Rev Immunol. 1999;17:331–367. doi: 10.1146/annurev.immunol.17.1.331. [DOI] [PubMed] [Google Scholar]
- 62.Ehrhardt H, Fulda S, Schmid I, Hiscott J, Debatin KM, Jeremias I. TRAIL induced survival and proliferation in cancer cells resistant towards TRAIL-induced apoptosis mediated by NF-kappaB. Oncogene. 2003;22:3842–3852. doi: 10.1038/sj.onc.1206520. [DOI] [PubMed] [Google Scholar]
- 63.Trauzold A, Siegmund D, Schniewind B, Sipos B, Egberts J, Zorenkov D, Emme D, Röder C, Kalthoff H, Wajant H. TRAIL promotes metastasis of human pancreatic ductal adenocarcinoma. Oncogene. 2006;25:7434–7439. doi: 10.1038/sj.onc.1209719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed in the present study are available from the corresponding author on reasonable request.