Abstract
BACKGROUND
In the majority of cases, the cause of stillbirth remains unknown despite detailed clinical and laboratory evaluation. Approximately 10 to 20% of stillbirths are attributed to chromosomal abnormalities. However, the causal nature of single-nucleotide variants and small insertions and deletions in exomes has been understudied.
METHODS
We generated exome sequencing data for 246 stillborn cases and followed established guidelines to identify causal variants in disease-associated genes. These genes included those that have been associated with stillbirth and strong candidate genes. We also evaluated the contribution of 18,653 genes in case–control analyses stratified according to the degree of depletion of functional variation (described here as “intolerance” to variation).
RESULTS
We identified molecular diagnoses in 15 of 246 cases of stillbirth (6.1%) involving seven genes that have been implicated in stillbirth and six disease genes that are good candidates for phenotypic expansion. Among the cases we evaluated, we also found an enrichment of loss-of-function variants in genes that are intolerant to such variation in the human population (odds ratio, 2.15; 95% confidence interval [CI], 1.46 to 3.06). Loss-of-function variants in intolerant genes were concentrated in genes that have not been associated with human disease (odds ratio, 2.22; 95% CI, 1.41 to 3.34), findings that differ from those in two postnatal clinical populations that were also evaluated in this study.
CONCLUSIONS
Our findings establish the diagnostic utility of clinical exome sequencing to evaluate the role of small genomic changes in stillbirth. The strength of the novel risk signal (as generated through the stratified analysis) was similar to that in known disease genes, which indicates that the genetic cause of stillbirth remains largely unknown. (Funded by the Institute for Genomic Medicine.)
An Illustrated Glossary is available at NEJM.org
STILLBIRTH (DEFINED AS FETAL DEATH IN utero at ≥20 weeks of gestation) accounts for 60% of all perinatal deaths and is unexplained in 25 to 60% of cases.1–3 Data are limited regarding the contribution of known mendelian diseases to stillbirth. In many clinical disciplines, it is now standard practice to perform exome sequencing to test for pathogenic single-nucleotide variants and small insertions and deletions to determine the cause of disease.4–12 Clinical exome sequencing has been particularly useful in diagnosing otherwise unexplained childhood disorders (in 20 to 30% of such cases) and fetal structural anomalies (in 10 to 20%).11,13,14
Given the incidence of stillbirth and the presumption of a strong genetic contribution, it is unfortunate that clinical exome sequencing has not been applied more rigorously in this context. Previous studies have been small and have concentrated on predetermined causes, included data regarding early miscarriage, or had a high prevalence of recurrent cases or cases with structural anomalies.15,16 The identification of monogenic disorders that are responsible for stillbirth may facilitate closure and bereavement for families, inform recurrence risk and management in subsequent pregnancies, allow for the avoidance of ineffective and potentially harmful therapies and interventions, and identify novel targets for risk stratification and therapy. Thus, we evaluated the diagnostic utility of clinical exome sequencing in a cohort of 246 stillbirth cases in a multicenter case–control study involving a geographically and demographically diverse population in the United States.
METHODS
STUDY DESIGN AND PARTICIPANTS
The Stillbirth Collaborative Research Network (SCRN) of the Eunice Kennedy Shriver National Institute of Child Health and Human Development conducted the study from March 2006 through September 2008.2 The initial cytogenetic studies of this cohort were completed in 2012, and remaining tissue samples became available from the SCRN for exome sequencing in 2017; the complete data set became available in May 2018. The study was approved by the institutional review board of the data coordinating center at each clinical site. All the participants provided written informed consent.
The study included fetuses with a gestation of 20 weeks or more; in cases in which the dating criteria were questionable, that window was expanded to include fetuses with a gestation of 18 weeks 0 days to 19 weeks 6 days. All case reviews included a standardized maternal interview and a detailed medical-chart abstraction.
The study series, which has been described previously,2,3 consisted of 953 potential participants; of these women, 663 enrolled, and 639 consented to participate in the genetic study. Of these women, 560 provided consent for fetuses to undergo partial or complete postmortem examination (Fig. S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org).
In 512 cases for which data were available, the investigators used the Initial Causes of Fetal Death (INCODE) algorithmic classification tool17 to determine the probable cause of death in 312 fetuses (60.9%; 95% confidence interval [CI], 56.5 to 65.2) and the possible or probable cause of death in 390 fetuses (76.2%; 95% CI, 72.2 to 79.8).2 The most common causes were obstetrical conditions (e.g., preterm labor, placental abruption, cervical insufficiency, and preterm premature rupture of membranes) in 150 (29.3%; 95% CI, 25.4 to 33.5), placental abnormalities in 121 (23.6%; 95% CI, 20.1 to 27.6), fetal structural abnormalities in 70 (13.7%; 95% CI, 10.9 to 17.0), infection in 66 (12.9%; 95% CI, 10.2 to 16.2), umbilical cord abnormalities in 53 (10.4%; 95% CI, 7.9 to 13.4), hypertensive disorders in 47 (9.2%; 95% CI, 6.9 to 12.1), and other maternal medical conditions in 40 (7.8%; 95% CI, 5.7 to 10.6), with more than one cause identified in some cases.2
DNA PREPARATION AND SEQUENCING
DNA was extracted with the use of established methods (Puregene, Qiagen Systems). (Details are provided in the Methods section in the Supplementary Appendix.) Exome sequencing of all samples was performed at the Institute for Genomic Medicine at Columbia University, which also provided control samples that had been obtained from healthy relatives of probands of mixed ancestry in a collection of studies. Exomes were captured and sequenced according to standard protocols and processed with the use of an in-house bioinformatics pipeline.18 The analysis was limited to 18,653 protein-coding genes that had a consensus coding sequence (release 20), which included two-base intronic extensions to accommodate canonical splice variants. To limit confounding caused by differential coverage, we used a previously described site-based pruning strategy.19 Sites of consensus coding sequences were excluded from the analysis if the absolute difference in the percentage of cases as compared with controls with adequate (10×) coverage of the site differed by more than 10 percentage points.19 This process resulted in the pruning of 7% of bases of consensus coding sequence. All testing of case–control genes to determine the presence of enrichment of genetic variants in cases was performed on the pruned consensus coding sequence.
MOLECULAR DIAGNOSTIC EVALUATION
We screened 246 stillbirth cases for molecular diagnoses using the guidelines of the American College of Medical Genetics and Genomics (ACMG).20 We prioritized variants with characteristics designed to enrich for pathogenicity in mendelian disease genes from the Online Mendelian Inheritance in Man (OMIM) database (Fig. S2).21 Given the lack of data regarding parental genotypes, we considered compound heterozygous variants (allele frequency, <1%) as possible causes of stillbirth only if the variants were confirmed through read-based phasing in the Integrative Genomics Viewer.22
We identified two sets of molecular diagnoses in 221 genes that have been previously described in stillbirth (Table S1) and in genes that have not been previously described but that are strong candidates for phenotype expansion. Laboratory-based molecular diagnoses do not always match precisely with the expected phenotype and may indicate phenotypic expansion, in which the spectrum of phenotypes that are known to be caused by pathogenic variants in a specific gene is broadened. In this study, phenotype expansions were determined through case-based literature searches and considered for genes associated with infant and adult disorders with a plausible lethal disease mechanism. Molecular diagnoses were classified as either “pathogenic” or “likely pathogenic” according to the ACMG criteria or “suggestive” by a multidisciplinary clinical and genetics team. (Details regarding this classification are provided in the Methods section in the Supplementary Appendix.)
STATISTICAL ANALYSIS
We performed a standard gene-based collapsing analysis in which variants in a specific gene in the cases were compared with variants in the same gene in the controls. A two-tailed Fisher’s exact test was performed across 18,653 protein-coding genes with a consensus coding sequence for three models (Fig. S5). (Such genes are consistently annotated and are of high quality in human and mouse genomes.) We implemented standard procedures to reduce bias caused by relatedness and population stratification (Figs. S3 and S4).19,23,24
We also performed the same collapsing analysis to compare variants in the cases with those in controls across genes in specific categories of “intolerance to variation” (Fig. 1). (Genes that harbor few functional variants in healthy persons are known to be more likely to cause disease than genes that carry many functional variants. A gene is said to be “intolerant” if it has relatively less functional genetic variation in the general population than is expected. Expected values can be calculated differently on the basis of the size of the gene and frequency of mutation, the amount of putatively neutral variation, or the overall number of observed variants.) Specifically, we sought to evaluate the contribution of genes intolerant to loss-of-function variants by testing whether stillborn fetuses were more likely than unaffected persons to carry loss-of-function variants in such genes. Genic intolerance to variation was stratified on the basis of the loss-of-function observed-to-expected upper boundary fraction (LOEUF) value (i.e., the upper boundary of a Poisson-derived confidence interval of the observed-to-expected ratio) based on the Genome Aggregation Database, version 2.1.1.29 We used Loss-of-Function Transcript Effect Estimator (LOFTEE) annotations,29 along with low-complexity regions, common structural variants, and segmental duplications,35 to filter for “high-confidence” loss-of-function variants in cases and controls. We then tested for a case or control enrichment of these loss-of-function variants at each LOEUF threshold in the data set. We used a logistic-regression model to test for an association between the case–control status and the presence or absence of a loss-of-function variant in each gene grouping. The logistic model accounted for background genetic variation in the gene grouping by controlling for presumed neutral ultra-rare synonymous variants.19 We then assessed the significance of the observed minimum P value across all thresholds by means of permutation (number of permutations, 10,000). (Details are provided in the Methods section in the Supplementary Appendix.)
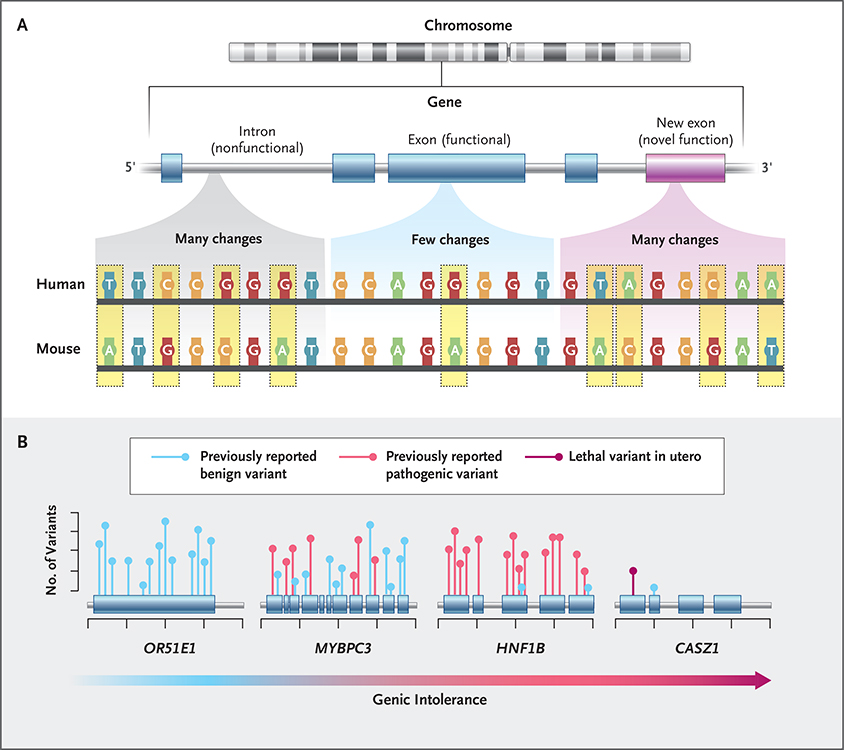
Figure 1. Intolerance to Genetic Variation.
Regions of the human genome that are under the strongest natural selection are the most likely to cause disease when they are variant. Until 2013, the primary approach that was used to identify such regions relied on the genetic similarities of different species.25 Genomic regions under selection show fewer DNA changes (e.g., nucleotide substitutions, deletions, or insertions) across species and are likely to be functionally important (Panel A). Pathogenic variants that cause human diseases have long been shown to fall preferentially within these “constrained” regions. Although this approach is useful, it cannot identify genomic regions of particular importance in humans, as might happen because of the evolution of a novel function. In 2013, a new framework was developed to address this limitation: variation, solely within the human population, was used to identify genes with less functional variation than expected according to genomewide averages. Genes with a depletion of human variation are termed “intolerant”26 and reflect parts of the genome under strong selection specifically in humans. Since the introduction of intolerance scoring, there have been a number of important elaborations focused on regions of genes, specific types of variants, and regulatory regions.27‑30 Intolerance scores have now been shown to provide independent information about where in the human genome pathogenic variants are found (Panel B).24,29,31‑34 For example, the gene encoding olfactory receptor 51E1 (OR51E1) is tolerant to variation and does not cause disease, whereas MYBPC3 and HNF1B are intolerant and are known to cause cardiac and kidney disease, respectively. CASZ1 is highly intolerant to variation in the healthy population, but no pathogenic variants have been reported in the literature regarding postnatal disease, which indicates that it may result in lethality when variant in utero.
We then compared enrichment of loss-of-function variation in the stillbirth cohort with that in two additional clinical cohorts: live-born infants with fetal structural anomalies and a large cohort of patients with postnatal diseases. The patients with postnatal disease were ascertained for a mixture of phenotypes with variable ages of onset and severity. All additional cases were sequenced and processed at the Institute for Genomic Medicine. All the sequencing data underwent the same quality-control filtering and pruning procedures to ensure parity between cases and controls. We then partitioned a loss-of-function risk signal in intolerant genes according to OMIM disease-association status in a binary fashion according to whether the genes had previously been implicated in disease.
RESULTS
STUDY POPULATION
Among the 560 women who provided consent to participate in the study and for partial or complete postmortem fetal examination, DNA was available for exome sequencing in 392 stillbirths. Of these samples, sequencing provided data in 337 cases, and the quality was adequate23,33 in 296 cases (Fig. S1). A total of 50 cases were excluded from downstream analyses because of a well-defined cause of stillbirth (i.e., multiple gestation, infection, maternal hypertension, maternal medical complications, or previously identified pathological karyotype or microarray).3,4 Among the remaining 246 cases, we included those with probable placental disease, umbilical cord abnormalities, obstetrical complications, or fetal structural abnormalities because the genetic underpinnings of such cases are uncertain. In the majority of cases, both ultrasonography and autopsy (full or limited) were performed (Table S2). Of the 246 women in this study, 105 (42.7%) were non-Hispanic and of European ancestry. The percentage of women in this ancestry group was larger than the percentage among those who enrolled but were not included in the final analysis (129 of 417 [30.9%]) (Table S3). There was no significant difference in the reported characteristics between the 639 women who consented for genetic evaluation and those who were included in this study (Table S4).
The sources of the control samples that were used in studies at the Institute for Genomic Medicine are provided in Table S5. The average coverage rate of pruned bases of consensus coding sequence (at 10×) was 98.8% for cases and 98.6% for controls. The average coverage rate of pruned sites (at 30×) was 95.6% for cases and 95.6% for controls.
DIAGNOSTIC YIELD
We identified molecular diagnoses in 9 of 246 stillborn cases (3.7%) across seven genes that have previously been described in stillbirth (interactive Table 1, available at NEJM.org). We further identified 6 cases (2.4%) with molecular diagnoses in six disease genes that are strong candidates for phenotype expansion (interactive Table 2). We report a cumulative diagnostic yield of 6.1%, with 15 of 246 stillborn cases receiving a molecular diagnosis (according to ACMG criteria) in a known disease gene. An additional 6 cases (2.4%) had a “suggestive” genotype in either a known stillbirth gene or a gene candidate for phenotype expansion (Table S6). With the inclusion of suggestive variants in the diagnostic yield, 21 of 246 of cases (8.5%) received a probable molecular diagnosis.
Of the 15 cases with a secure molecular diagnosis, 6 (40.0%) had a multisystem developmental disorder and 5 (33.3%) had an isolated cardiac disorder. The disease genes that were identified were enriched for several cardiac-related gene-ontology biologic processes (Table S7). The remaining cases had a disorder primarily affecting the kidney (in 2 [13.3%]) and either the brain or bone (in 1 each [6.7%]).
The 13 cases with a structural anomaly were more likely to receive a molecular diagnosis than cases without a structural anomaly (odds ratio, 8.80; 95% CI, 1.7 to 38.4). However, we found no significant difference in the frequency of diagnosis between cases with a probable INCODE cause of death and cases with an unexplained cause (odds ratio, 1.38; 95% CI, 0.39 to 4.54).
CASE-LEVEL DIAGNOSTIC FINDINGS
A recurrent molecular diagnosis occurred in PTPN11, which is associated with Noonan syndrome. We identified three previously reported ultra-rare missense variants in the region of PTPN11 encoding the interacting surfaces of the N-terminal src-homology 2 (N-SH2) and protein tyrosine phosphatase (PTP) domains. Most pathogenic missense variants that cause Noonan syndrome are known to cluster in these regions.36 One of the identified missense variants in PTPN11, which predicted the substitution of alanine with threonine at position 72 (Ala72Thr), is a hot spot for isolated juvenile myelomonocytic leukemia (JMML). With few exceptions, germline variants causing Noonan syndrome do not occur as causative somatic variants in JMML.37 Somatic variants in PTPN11 that cause JMML may have stronger gain-of-function activity than those associated with Noonan syndrome and have been speculated to result in embryonic death if the variants are germline.
We found some cases with loss-of-function variants in genes in which missense alleles are known to cause postnatal disease in infants. We identified an ultra-rare frameshift variant in SMC3, a gene in which missense variants have been reported to cause a mild form of Cornelia de Lange syndrome,31,38 in a stillbirth case with severe fetal growth restriction (birth weight, 0.06 percentile). Similarly, we found a novel frameshift variant in FBN2, in which missense variants are known to cause congenital contractural arachnodactyly.39 Given the strong selection against loss-of-function variants in these genes and the lack of loss-of-function variation in the postnatal disease population, we suspect that loss-of-function variants in these genes caused the stillbirth.
POPULATION-BASED CASE–CONTROL ANALYSES
No individual gene reached study-wide significance in any of the three gene-based collapsing models. Under the model of known pathogenic variants (i.e., those that have previously been implicated in causing disease40,41), PTPN11 was the top-ranked gene, and HNF1B and RYR2 were among the genes in the top 20 (Table S8). Owing to the genetic heterogeneity of the stillbirth phenotype, larger sample sizes will probably be needed to determine significant associations between variation in individual genes and death in utero.
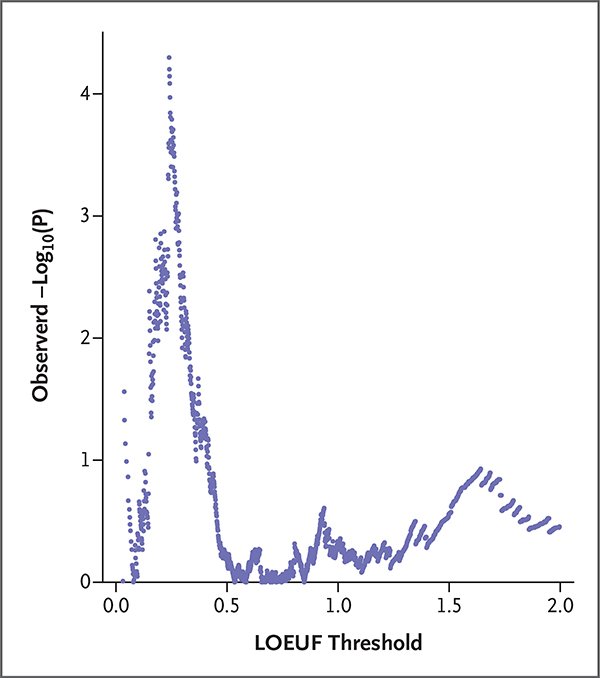
Separately, we explored enrichment of loss-of-function variation at every possible LOEUF threshold for intolerance in our data set and assessed the significance of the observed minimum P value by permutation (number of permutations, 10,000). The 8081 genes harboring a loss-of-function variant in 241 case samples and 7239 control samples reflected 1825 unique LOEUF divisions among all the genes that were sequenced. The minimum observed P value was at position 61 of 10,000 permutations, which reflects a significant enrichment of loss-of-function variants at a threshold for high intolerance (LOEUF, ≤0.239) (Fig. 2). In this gene grouping, there was at least one loss-of-function variant in 35 of 241 cases (14.5%) and in 531 of 7239 controls (7.3%) (Table S9). In accounting for the control carrier frequency, we estimated that loss-of-function variants in intolerant genes contributed to the risk of stillbirth in approximately 7.2% of cases. We found no significant difference in the burden of loss-of-function variants between cases with a probable INCODE cause of death and those with an unexplained cause in this gene grouping (Table S10).
Figure 2. Enrichment of Loss-of-Function Variants in Stillbirth Cases, as Compared with Controls.
To explore which genetic variants may be associated with stillbirth, we assigned an LOEUF (loss-of-function observed-to-expected upper boundary fraction) value to all 8081 genes with a loss-of-function variant in 241 case samples and 7239 control samples. The process resulted in 1825 unique LOEUF values. The LOEUF value is the upper boundary of a Poisson-derived confidence interval of the observed-to-expected ratio. A low LOEUF score indicates a depletion of loss-of-function variation (also referred to as “intolerance” to loss-of-function variation). The enrichment of loss-of-function variants was then assessed by comparing the count of variants in cases and controls in each of these 1825-gene groupings. Enrichment was evaluated with the use of a logistic-regression model that evaluated case–control status regarding the presence or absence of a loss-of-function variant in each gene grouping. Enrichment of presumed neutral synonymous variants at each intolerance threshold was used as a covariate to control for differences between case and control genomes that are unrelated to disease risk. The negative log of the enrichment statistic (unadjusted P value) at each LOEUF threshold is shown on the y axis. The x axis reflects the LOEUF threshold for each gene grouping. The minimum observed P value, which indicates the most significant enrichment across all LOEUF thresholds, occurs at a LOEUF threshold of 0.239 or less. The significance of the observed minimum P value was then assessed by permutation (number of permutations, 10,000).
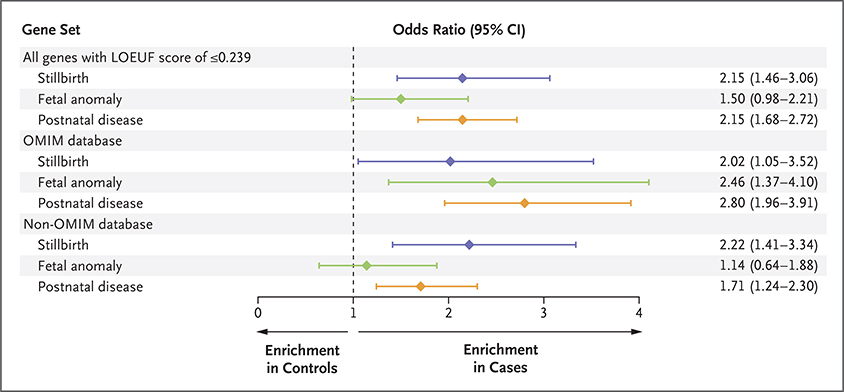
We then partitioned the loss-of-function risk signal on the basis of an association with OMIM disease. We found that enrichment of loss-of-function variants in intolerant genes in the stillbirth cohort was concentrated in genes without an existing OMIM disease association (odds ratio, 2.22; 95% CI, 1.41 to 3.34) (Fig. 3). This enrichment of loss-of-function variants in genes that are not currently associated with any known diseases (non-OMIM genes) is similar to enrichment in genes already known to cause a human disease before or after birth (OMIM genes) (odds ratio, 2.02; 95% CI, 1.05 to 3.52). Among the intolerant genes (LOEUF, ≤0.239), at least one loss-of-function variant in a non-OMIM gene was found in 25 of 241 cases (10.4%) and in 358 of 7239 controls (4.9%) (interactive Table 3). Accordingly, we estimate that 5.5% of stillbirth cases are potentially explained by loss-of-function variants in these candidate novel disease genes. Of these candidate genes, 44% have an orthologue in the mouse that causes a lethal phenotype when the gene is knocked out, according to the Mouse Genome Informatics database. The corresponding percentage for genes carrying a loss-of-function mutation in controls was 25.5%. The candidate disease genes were enriched for several biologic processes listed in the Gene Ontology database (Table S11).
Figure 3. Loss-of-Function Signal in Intolerant Genes in Three Clinical Cohorts.
In this study, the LOEUF threshold that had the strongest enrichment of loss-of-function variants (≤0.239) in the 241 cases in the stillbirth cohort was used to make point comparisons with the loss-of-function signal in a group of 251 live-birth cases with fetal anomalies and in 589 cases with postnatal disease. The odds ratio indicates the enrichment of loss-of-function variants in the gene set in cases as compared with a large group of controls. The gene sets are shown in three categories: all the genes below the LOEUF threshold (≤0.239), those that had an existing disease association in the Online Mendelian Inheritance in Man (OMIM) database, and those that did not have such an association (non-OMIM).
We also found an enrichment of loss-of-function variants at the same intolerance threshold as in the stillbirth cohort in a postnatal disease cohort of 589 cases (odds ratio, 2.15; 95% CI, 1.68 to 2.72) and in a live-birth fetal-anomaly cohort of 251 cases (odds ratio, 1.50; 95% CI, 0.98 to 2.21). Unlike in the stillbirth cohort, however, loss-of-function signal was driven predominantly by known disease genes in the postnatal disease cohort (odds ratio, 2.80; 95% CI, 1.96 to 3.91) and in the live-birth fetal-anomaly cohort (odds ratio, 2.46; 95% CI, 1.37 to 4.10) (Fig. 3).
DISCUSSION
On the basis of exome sequencing analysis of singleton stillbirths, we found that 8.5% of those with a normal chromosomal microarray and without probable maternal or obstetrical causes were probably attributable to mendelian disorders. When we combined these data with the previously published results of a cytogenetic analysis performed in this cohort, in which 6.9% of stillbirths were aneuploid and 2.6% harbored a pathogenic copy number, we determined that a total of 18% of stillbirths had a known genetic cause.40
Our yield from clinical exome sequencing was lower than that for cohorts with severe early-onset diseases.11,13,14 The yield was probably reduced by the absence of genotypic data from the parents, since we could not identify compound heterozygous variants that are potentially identifiable with knowledge of parental genotypes. Of the 21 possibly compound heterozygous variants that we identified but could not confirm through read-based phasing, 4 were in genes known to cause stillbirth (ACE and INVS in 1 case each and NEB in 2 cases). The interpretation of novel missense variants is also compromised by the absence of inheritance information. For this reason, such variants were excluded in our diagnostic pipeline. To evaluate the effect of these exclusions, we estimated the potential added contribution of de novo novel missense variants by pulling all ultra-rare damaging novel missense variants in our cohort. We identified an additional 7 ultra-rare variants in 3 genes known to cause stillbirth (COL2A1 in 3 cases and PEIZO1 and HNF1B in 2 cases each). With parental genotype information, a fraction of the identified novel missense variants would probably be confirmed as de novo and possibly compound heterozygous variants confirmed in trans (i.e., on separate alleles). We can therefore estimate that the diagnostic yield in known stillbirth genes lies between 3.7 and 8.1% (in 20 of 246 cases). This percentage is consistent with values in other studies that have reported a doubling of the diagnostic yield in family trio sequencing as compared with singleton sequencing and suggests that parental samples should be included in the analysis of stillbirths in clinical care.12,14,42,43
In assessing how intolerant genes may contribute to stillbirth, we took an approach that makes no prior assumption regarding which human genes may have enrichment of loss-of-function variants. Because loss-of-function variants in the genes with less variation than expected are known to have a high likelihood of causing disease on the basis of research carried out over the past decade,29,43 our unbiased approach was highly conservative. Our approach did not assume in advance that any particular set of genes carried contributing variants. Rather, we used a statistical framework that led to the discovery of the highest concentration of causal variants and corrected appropriately for all the comparisons that were evaluated. We think that this approach constitutes a more robust statistical method for thresholding in enrichment analyses of this kind.
Previous sequencing studies that have used a similar approach to prioritizing variants in postnatal disease cohorts have reported a preponderance of variants in genes that are associated with established diseases.43 In this study, we observed in stillbirths an enrichment of loss-of-function variants in genes that are not currently known to cause human disease, whereas in two postnatal clinical populations (live-born infants with fetal anomalies and patients with undiagnosed postnatal disease) enrichment of loss-of-function variants was concentrated in known disease genes. To our knowledge, stillbirth may be the only condition that has been studied in which a novel risk signal (in non-OMIM genes) is of the same order of magnitude as that in known OMIM disease genes.
Almost half the candidate genes for an association with stillbirth are essential for life in the mouse, which supports the relevance of these genes to survival of the human fetus. Moreover, we find some consistency between the mouse and human phenotypes. For instance, we found a novel stop–gain variant in CASZ1 (encoding zinc-finger transcription factor) in a stillborn infant with hydrops and intrauterine growth restriction. Similarly, in one study, Casz1−/− mice were found to have edema and morphologic anomalies of the heart before embryonic death at day 17.5.44 A limited autopsy was performed in the stillborn infant, and additional abnormalities in the heart may have gone undetected. We also identified a frameshift variant in NUP98, encoding part of the nuclear pore complex, in a stillborn infant with severe intrauterine growth restriction (5th percentile for birth weight at 25 weeks of gestation). Nup98−/− mice die early in utero with severe growth delay,45 and deletions in NUP98 have been identified in biopsy samples of placental tissue from women with recurrent pregnancy loss.46
In the majority of stillbirth cases in our study, structural anomalies were not revealed on ultrasonography or autopsy (Table S3). However, ion channelopathies and cardiomyopathies could still be responsible for a proportion of stillbirths without structural anomalies. Previous studies have shown pathogenic mutations in cardiac-associated genes in cases of stillbirth without structural anomalies.10,16,47,48 In particular, several forms of cardiomyopathy (including hypertrophic, dilated, and arrhythmogenic) may be accompanied by very minor or even no structural abnormalities. The presence of such disorders has been suggested to underlie a portion of sudden infant deaths and stillbirths.49
Our findings suggest that a portion of stillbirth cases are caused by pathogenic variants in genes known to cause disorders in infants and adults, whereas a similar number of cases are caused by loss-of-function variants in genes that are critical to in utero survival but that are not currently known to be associated with stillbirth or postnatal disease. Exome sequencing analysis of larger, independent stillbirth cohorts would test the validity of our results and almost certainly uncover additional genes that are associated with stillbirth. We anticipate that the identification of specific genes and variants will further improve our understanding of the pathways leading to stillbirth and improve the counseling of affected families. Currently, no sequencing data from a sufficiently large cohort exist to perform this analysis. Our results show that exome sequencing in stillbirth can identify variants of genes that are incompatible with survival in utero.
Supplementary Material
Acknowledgments
The views expressed in this article are those of the authors and do not necessarily reflect the views of the National Institutes of Health or the Eunice Kennedy Shriver National Institute of Child Health and Development (NICHD).
Supported by the Institute for Genomic Medicine with grants (HD45925, HD45944, HD45952, HD45953, HD45954, and HD45955) from the NICHD.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
We thank the patients, physicians, study coordinators, and research nurses who participated in the Stillbirth Collaborative Research Network.
APPENDIX
The authors’ full names and academic degrees are as follows: Kate E. Stanley, B.A., Jessica Giordano, M.S., C.G.C., Vanessa Thorsten, M.P.H., Christie Buchovecky, Ph.D., Amanda Thomas, Ph.D., Mythily Ganapathi, Ph.D., Jun Liao, Ph.D., Avinash V. Dharmadhikari, Ph.D., Anya Revah-Politi, M.S., C.G.C., Michelle Ernst, M.S., C.G.C., Natalie Lippa, M.S., C.G.C., Halie Holmes, M.S., C.G.C., Gundula Povysil, M.D., Ph.D., Joseph Hostyk, B.S., Corette B. Parker, Dr.P.H., Robert Goldenberg, M.D., George R. Saade, M.D., Donald J. Dudley, M.D., Halit Pinar, M.D., Carol Hogue, Ph.D., M.P.H., Uma M. Reddy, M.D., M.P.H., Robert M. Silver, M.D., Vimla Aggarwal, M.B., B.S., Andrew S. Allen, Ph.D., Ronald J. Wapner, M.D., and David B. Goldstein, Ph.D.
Contributor Information
K.E. Stanley, Institute for Genomic Medicine at Vagelos College of Physicians and Surgeons, Columbia University Irving Medical Center
J. Giordano, Institute for Genomic Medicine at Vagelos College of Physicians and Surgeons, Columbia University Irving Medical Center Department of Obstetrics and Gynecology, Columbia University Medical Center, New York.
V. Thorsten, RTI International, Research Triangle Park, North Carolina
C. Buchovecky, Department of Pathology and Cell Biology, Columbia University Medical Center, New York RTI International, Research Triangle Park, North Carolina.
A. Thomas, Department of Pathology and Cell Biology, Columbia University Medical Center, New York
M. Ganapathi, Department of Pathology and Cell Biology, Columbia University Medical Center, New York
J. Liao, Department of Pathology and Cell Biology, Columbia University Medical Center, New York
A.V. Dharmadhikari, Department of Pathology and Cell Biology, Columbia University Medical Center, New York
A. Revah‑Politi, Institute for Genomic Medicine at Vagelos College of Physicians and Surgeons, Columbia University Irving Medical Center
M. Ernst, Institute for Genomic Medicine at Vagelos College of Physicians and Surgeons, Columbia University Irving Medical Center
N. Lippa, Institute for Genomic Medicine at Vagelos College of Physicians and Surgeons, Columbia University Irving Medical Center
H. Holmes, Institute for Genomic Medicine at Vagelos College of Physicians and Surgeons, Columbia University Irving Medical Center
G. Povysil, Institute for Genomic Medicine at Vagelos College of Physicians and Surgeons, Columbia University Irving Medical Center
J. Hostyk, Institute for Genomic Medicine at Vagelos College of Physicians and Surgeons, Columbia University Irving Medical Center
C.B. Parker, RTI International, Research Triangle Park, North Carolina
R. Goldenberg, Department of Obstetrics and Gynecology, Columbia University Medical Center, New York
G.R. Saade, Department of Obstetrics and Gynecology and Cell Biology, University of Texas Medical Branch, Galveston
D.J. Dudley, Division of Maternal–Fetal Medicine, Department of Obstetrics and Gynecology, University of Virginia School of Medicine, Charlottesville
H. Pinar, Division of Perinatal and Pediatric Pathology, Women and Infants Hospital, Warren Alpert School of Medicine of Brown University, Providence, RI
C. Hogue, Rollins School of Public Health, Emory University, Atlanta
U.M. Reddy, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Pregnancy and Perinatology Branch, Bethesda, MD
R.M. Silver, University of Utah and Intermountain Healthcare, Salt Lake City
V. Aggarwal, Institute for Genomic Medicine at Vagelos College of Physicians and Surgeons, Columbia University Irving Medical Center Department of Pathology and Cell Biology, Columbia University Medical Center, New York.
A.S. Allen, Department of Biostatistics and Bioinformatics, Duke University, Durham, North Carolina
R.J. Wapner, Institute for Genomic Medicine at Vagelos College of Physicians and Surgeons, Columbia University Irving Medical Center Department of Obstetrics and Gynecology, Columbia University Medical Center, New York.
D.B. Goldstein, Institute for Genomic Medicine at Vagelos College of Physicians and Surgeons, Columbia University Irving Medical Center
References
- 1.Wapner RJ, Lewis D. Genetics and metabolic causes of stillbirth. Semin Perinatol 2002;26:70–4. [DOI] [PubMed] [Google Scholar]
- 2.Stillbirth Collaborative Research Network Writing Group. Causes of death among stillbirths. JAMA 2011;306:2459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reddy UM, Page GP, Saade GR, et al. Karyotype versus microarray testing for genetic abnormalities after stillbirth. N Engl J Med 2012;367:2185–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee H, Deignan JL, Dorrani N, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA 2014;312:1880–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med 2013;369:1502–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groopman EE, Marasa M, Cameron-Christie S, et al. Diagnostic utility of exome sequencing for kidney disease. N Engl J Med 2019;380:142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Splinter K, Adams DR, Bacino CA, et al. Effect of genetic diagnosis on patients with previously undiagnosed disease. N Engl J Med 2018;379:2131–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bagnall RD, Crompton DE, Petrovski S, et al. Exome-based analysis of cardiac arrhythmia, respiratory control, and epilepsy genes in sudden unexpected death in epilepsy. Ann Neurol 2016;79:522–34. [DOI] [PubMed] [Google Scholar]
- 9.Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 2014;312:1870–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neubauer J, Lecca MR, Russo G, et al. Post-mortem whole-exome analysis in a large sudden infant death syndrome cohort with a focus on cardiovascular and metabolic genetic diseases. Eur J Hum Genet 2017;25:404–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lord J, McMullan DJ, Eberhardt RY, et al. Prenatal exome sequencing analysis in fetal structural anomalies detected by ultrasonography (PAGE): a cohort study. Lancet 2019;393:747–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meng L, Pammi M, Saronwala A, et al. Use of exome sequencing for infants in intensive care units: ascertainment of severe single-gene disorders and effect on medical management. JAMA Pediatr 2017; 171(12):e173438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrovski S, Aggarwal V, Giordano JL, et al. Whole-exome sequencing in the evaluation of fetal structural anomalies: a prospective cohort study. Lancet 2019;393: 758–67. [DOI] [PubMed] [Google Scholar]
- 14.Wright CF, Fitzgerald TW, Jones WD, et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet 2015;385:1305–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shamseldin HE, Kurdi W, Almusafri F, et al. Molecular autopsy in maternal-fetal medicine. Genet Med 2018;20: 420–7. [DOI] [PubMed] [Google Scholar]
- 16.Sahlin E, Gréen A, Gustavsson P, et al. Identification of putative pathogenic single nucleotide variants (SNVs) in genes associated with heart disease in 290 cases of stillbirth. PLoS One 2019;14(1):e0210017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dudley DJ, Goldenberg R, Conway D, et al. A new system for determining the causes of stillbirth. Obstet Gynecol 2010; 116:254–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Analysis tool for annotated variants— a comprehensive platform for population-scale genomic analyses. 2016. (https://www.biorxiv.org/content/10.1101/2020.06.08.136507v1.full). [DOI] [PMC free article] [PubMed]
- 19.Epi4K Consortium, Epilepsy Phenome/Genome Project. Ultra-rare genetic variation in common epilepsies: a case-control sequencing study. Lancet Neurol 2017;16: 135–43. [DOI] [PubMed] [Google Scholar]
- 20.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamosh A, Scott AF, Amberger J, Valle D, McKusick VA. Online Mendelian Inheritance in Man (OMIM). Hum Mutat 2000;15:57–61. [DOI] [PubMed] [Google Scholar]
- 22.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 2013;14:178–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrovski S, Todd JL, Durheim MT, et al. An exome sequencing study to assess the role of rare genetic variation in pulmonary fibrosis. Am J Respir Crit Care Med 2017;196:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu X, Padmanabhan R, Copeland B, et al. A case-control collapsing analysis identifies epilepsy genes implicated in trio sequencing studies focused on de novo mutations. PLoS Genet 2017;13(11):e1007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol 2010; 6(12):e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet 2013;9(8):e1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samocha KE, Robinson EB, Sanders SJ, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet 2014;46:944–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuller ZL, Berg JJ, Mostafavi H, Sella G, Przeworski M. Measuring intolerance to mutation in human genetics. Nat Genet 2019;51:772–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karczewski KJ, Francioli LC, Tiao G, et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. January 30, 2019. (https://www.biorxiv.org/content/10.1101/531210v2). preprint.
- 30.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eilbeck K, Quinlan A, Yandell M. Settling the score: variant prioritization and Mendelian disease. Nat Rev Genet 2017; 18:599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartha I, di Iulio J, Venter JC, Telenti A. Human gene essentiality. Nat Rev Genet 2018;19:51–62. [DOI] [PubMed] [Google Scholar]
- 33.Gelfman S, Dugger S, de Araujo Martins Moreno C, et al. A new approach for rare variation collapsing on functional protein domains implicates specific genic regions in ALS. Genome Res 2019;29:809–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kosmicki JA, Samocha KE, Howrigan DP, et al. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat Genet 2017;49: 504–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Bloom JM, Farjoun Y, et al. A synthetic-diploid benchmark for accurate variant-calling evaluation. Nat Methods 2018;15:595–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tartaglia M, Kalidas K, Shaw A, et al. PTPN11 mutations in Noonan syndrome: molecular spectrum, genotype-phenotype correlation, and phenotypic heterogeneity. Am J Hum Genet 2002;70:1555–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mason-Suares H, Toledo D, Gekas J, et al. Juvenile myelomonocytic leukemia-associated variants are associated with neo-natal lethal Noonan syndrome. Eur J Hum Genet 2017;25:509–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gil-Rodríguez MC, Deardorff MA, Ansari M, et al. De novo heterozygous mutations in SMC3 cause a range of Cornelia de Lange syndrome-overlapping phenotypes. Hum Mutat 2015;36:454–62. [DOI] [PubMed] [Google Scholar]
- 39.Park ES, Putnam EA, Chitayat D, Child A, Milewicz DM. Clustering of FBN2 mutations in patients with congenital contractural arachnodactyly indicates an important role of the domains encoded by exons 24 through 34 during human development. Am J Med Genet 1998;78:350–5. [PubMed] [Google Scholar]
- 40.Landrum MJ, Lee JM, Riley GR, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 2014;42: D980–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stenson PD, Mort M, Ball EV, et al. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet 2017;136:665–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yates CL, Monaghan KG, Copenheaver D, et al. Whole-exome sequencing on deceased fetuses with ultrasound anomalies: expanding our knowledge of genetic disease during fetal development. Genet Med 2017;19:1171–8. [DOI] [PubMed] [Google Scholar]
- 43.Zhu X, Petrovski S, Xie P, et al. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet Med 2015;17:774–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Z, Li W, Ma X, et al. Essential role of the zinc finger transcription factor Casz1 for mammalian cardiac morphogenesis and development. J Biol Chem 2014;289:29801–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu X, Kasper LH, Mantcheva RT, Mantchev GT, Springett MJ, van Deursen JM. Disruption of the FG nucleoporin NUP98 causes selective changes in nuclear pore complex stoichiometry and function. Proc Natl Acad Sci U S A 2001;98: 3191–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kasak L, Rull K, Sõber S, Laan M. Copy number variation profile in the placental and parental genomes of recurrent pregnancy loss families. Sci Rep 2017;7: 45327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brownstein CA, Poduri A, Goldstein RD, Holm IA. The genetics of sudden infant death syndrome In: Duncan JR, Byard RW, eds. SIDS sudden infant and early childhood death: the past, the present and the future. Adelaide, SA, Australia: University of Adelaide Press, 2018. [PubMed] [Google Scholar]
- 48.Crotti L, Tester DJ, White WM, et al. Long QT syndrome-associated mutations in intrauterine fetal death. JAMA 2013; 309:1473–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narula N, Tester DJ, Paulmichl A, Maleszewski JJ, Ackerman MJ. Post-mortem whole exome sequencing with gene-specific analysis for autopsy-negative sudden unexplained death in the young: a case series. Pediatr Cardiol 2015;36:768–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.