Abstract
Interactions between multiple functional groups are key to catalysis. Previously we reported synergistic interactions in catalytic amyloids formed by mixtures of heptameric peptides which lead to significant improvements in esterase activity. Here we describe the in-depth investigation of synergistic interactions within a family of amyloid fibrils, exploring the results of functional group interactions, the effects of chirality and the use of mixed enantiomers within fibrils. Remarkably, we find that synergistic interactions (either positive or negative) are found in the vast majority of binary mixtures of catalytic amyloid-forming peptides. The productive arrangements of functionalities rapidly identified by mixing different peptides will undoubtedly lead to development of more active catalysts for a variety of different transformations.
Keywords: self-assembly, catalysis, amyloids, synergistic interactions, peptides
Graphical Abstract
Synergistic interactions are prevalent in catalytic amyloids providing insight how early prebiotic peptides might have found productive combinations of functional groups to become precursors of modern day enzymes. @IvanKSyracuse @KorendovychLab @mailsharllam
The emergence of enzymes from the primordial soup is a topic of much discussion.[1] The observation of the spontaneous abiotic formation of short peptides that subsequently assemble into amyloid-like structures has led to an “amyloid world” hypothesis, which posits that the spontaneous assembly of short peptide chains led to functional supramolecules that could evolve into ancestors for modern enzymes.[2]
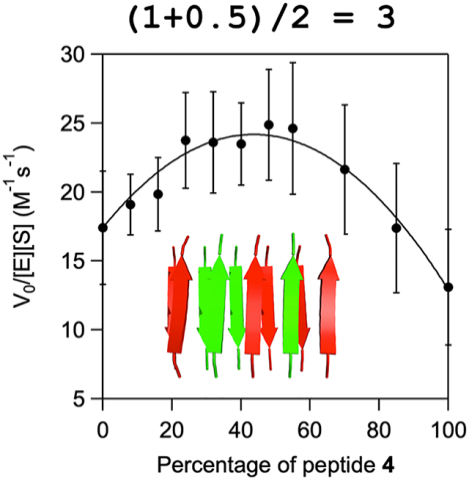
We and others have previously demonstrated that amphiphatic peptides self-assemble into amyloid-like fibrils which form active esterases in the presence of metal ions.[3] Metals simultaneously stabilize the fibrils while also acting as cofactors to promote hydrolysis of p-nitrophenyl acetate (pNPA), a commonly used substrate for testing hydrolytic catalysts.[4] These assemblies show high activity for pNPA hydrolysis, reaching the levels by weight as those shown by some enzymes in the same reaction. During these experiments, we observed substantial positive synergy when two peptides with different sequences were mixed (Figure 1).[3a] We proposed that this would allow us to perform combinatorial screening of functional groups within a single fibril, without the need to prepare each amino acid sequence of every combination, a fundamentally impossible task. However, the question remained whether the synergistic relationship between Arg and Tyr-containing peptides observed in our previous work is only an isolated example, or if it is representative of typical behavior in catalytic amyloids. A broader investigation of the mixing behavior in catalytic amyloids would allow us to determine the general applicability of short catalytic amyloids to form synergistic mixtures, something which has been observed in other model catalytic systems,[5] and has been shown to substantially increase reactivity in other de novo hydrolytic systems.[6] In addition to the fundamental importance of this question for the elucidation of the Origins of Life, the combinatorial investigation of short model complexes can help refine strategies for peptide and protein design.[7]
Figure 1.
The activity profile in pNPA hydrolysis of mixtures of preformed fibrils formed by peptides 4 and 7 shows a linear dependence (top), whereas fibrils formed by mixing of 4 and 7 in monomeric states prior to fibrillation demonstrates a synergistic behaviour (bottom). Kinetic plots reproduced from Rufo et al.[3a]
In this work we set out to study the binary interactions between all the originally reported peptides that utilized the leucine core (Table 1) using our standard mixing protocols.[8]
Table 1.
Peptides used in the study
| Identifier | Sequence | Catalytic efficiency, kcat/KM (M−1 s−1) |
|---|---|---|
| 1 | Ac-LHLHLDL-NH2 | 0.2 ± 0.1† |
| 2 | Ac-LHLHLEL-NH2 | <0.2† |
| 3 | Ac-LHLHLQL-NH2 | 30 ± 3† |
| 4 | Ac-LHLHLYL-NH2 | 13 ± 5† |
| 5 | Ac-LHLHLHL-NH2 | 0.6 ± 0.08† |
| 6 | Ac-LHLHLKL-NH2 | 12 ± 2† |
| 7 | Ac-LHLHLRL-NH2 | 18 ± 4† |
| 7F | Ac-LRLHLHL-NH2 | 28 ± 3.5 |
Taken from ref.[3a]
Remarkably, synergistic interactions were observed in nearly all the binary mixtures tested (Figures 2, S4–S8, Supporting Information). The analysis of the synergistic pattern is quite instructive (Figure 2a). Peptides with positively charged residues show positive synergy when mixed with the ones containing Glu or Asp (importantly the latter are inactive on their own) as well as the neutral residues Gln and Tyr, in the case of arginine-containing peptide 7. This synergy was preserved over time course experiments, with optimal mixtures showing a nearly a two-fold increase in catalytic activity of the mixture of 4 and 7 based on a binary fit (Supporting Information) relative to the corresponding mixture of preformed homomeric fibrils with the same composition. Productive interactions between the two most individually active peptides in the screen, 3 and 7, increased the activity by nearly two-fold when the two are mixed in a 1:1 ratio (Table S2, Supporting Information).
Figure 2.
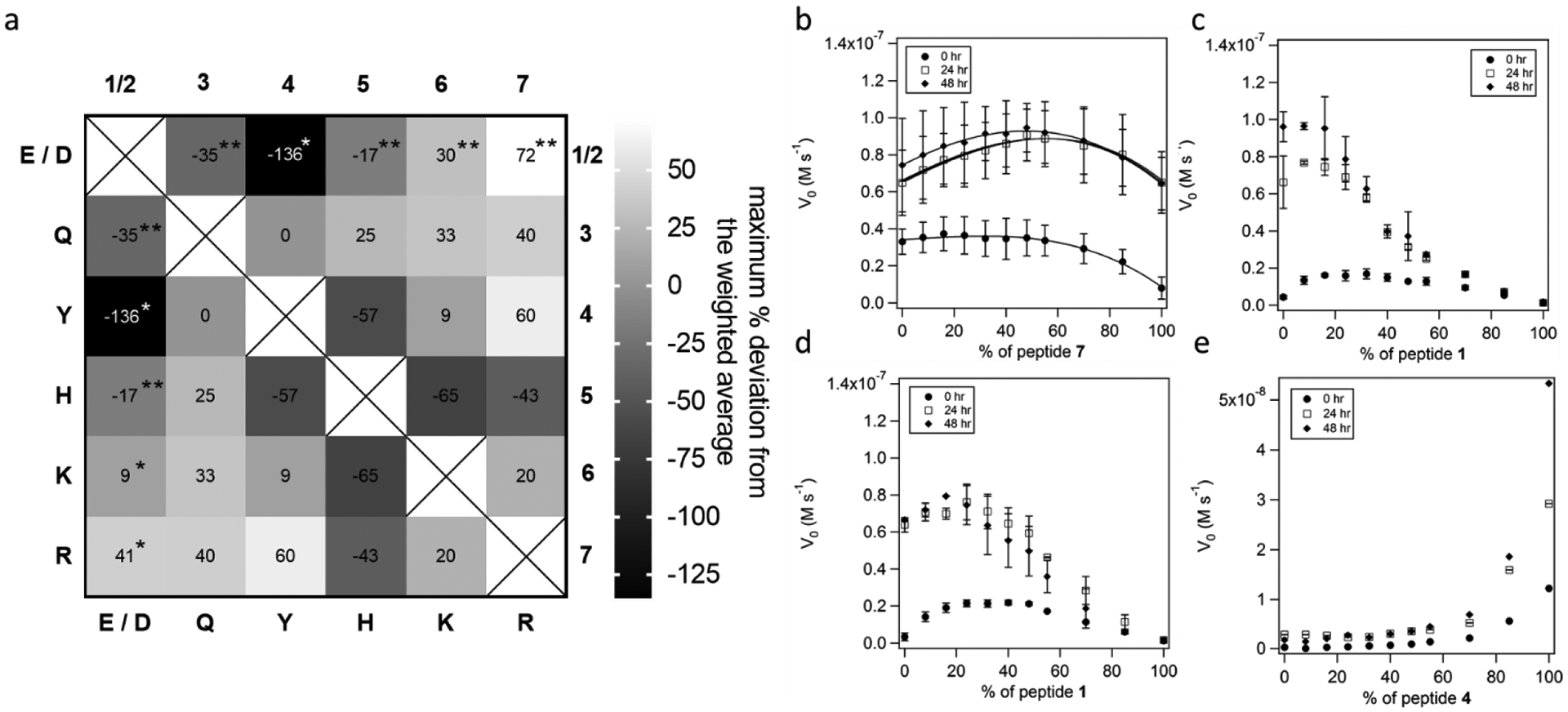
a) A graphic representation of all binary interactions between the peptides studied in this work. The scale indicates the maximum percent deviation from a weighted average of individual activities predicted if no synergy were present. *indicates mixing with peptide 1, ** with 2. (b-e) Hydrolytic activity of fibrils as a function of mixture compositions for b) 7 and 3 c) 6 and 1 d) 7 and 1 e) 4 and 1. Experimental conditions: 25 mM Tris, pH 8.0, 25 μM total peptide, 0.1 mM Zn2+, 195 μM pNPA.
We then turned our attention to the interactions of 6, a fibril with another positively charged residue (Lys) with 1 or 2 (Figure 2c). Interestingly, while we still observed positive synergy, it was not nearly as pronounced as it was in the case of 7. Moreover, the ratio representing the maximum activity shifted, so the resulting curve could not be described using the model that implies stochastic distribution of dimeric species. We assumed that lysine and arginine synergistic interactions would possess significant overlap, and they do share the ability to interact positively with the negatively charged amino acids as well as 3 or 4, but the percentage increase in activity is lower for lysine containing peptides. 6 and 7 even showed mild positive synergy when mixed. These findings indicate that while charge complementarity was important to the formation of synergistic interactions, even subtle modifications of peptide sequence and structure (i.e. the introduction of a more flexible and longer side chain) can lead to drastic changes in catalytic activity.
To study the factors that lead to increased activity when a catalytically inactive fibril is added, and to gain a deeper understanding of the factors that determine positive synergy between 7 and the negatively charged 1 and 2, we performed in-depth characterization studies of the mixtures. The activity profile showed that esterase activity of peptide 7 increased when mixed with the catalytically inactive peptide 1 or 2 (Figure 2d, Figure S4). Heterogeneous peptide mixtures showed faster fibrillation and slightly increased magnitudes of the minima at ~218 nm in circular dichroism studies (Figure 3, S1, S2, Supporting Information). A 24:76 mixture of peptides 1 and 7 demonstrated in a 15% increase in signal at 218 nm in the CD spectrum when compared to 7 alone. At the same time transmission electron micrographs for the peptide mixtures showed fibrils of a similar morphology to those reported for 7.
Figure 3.
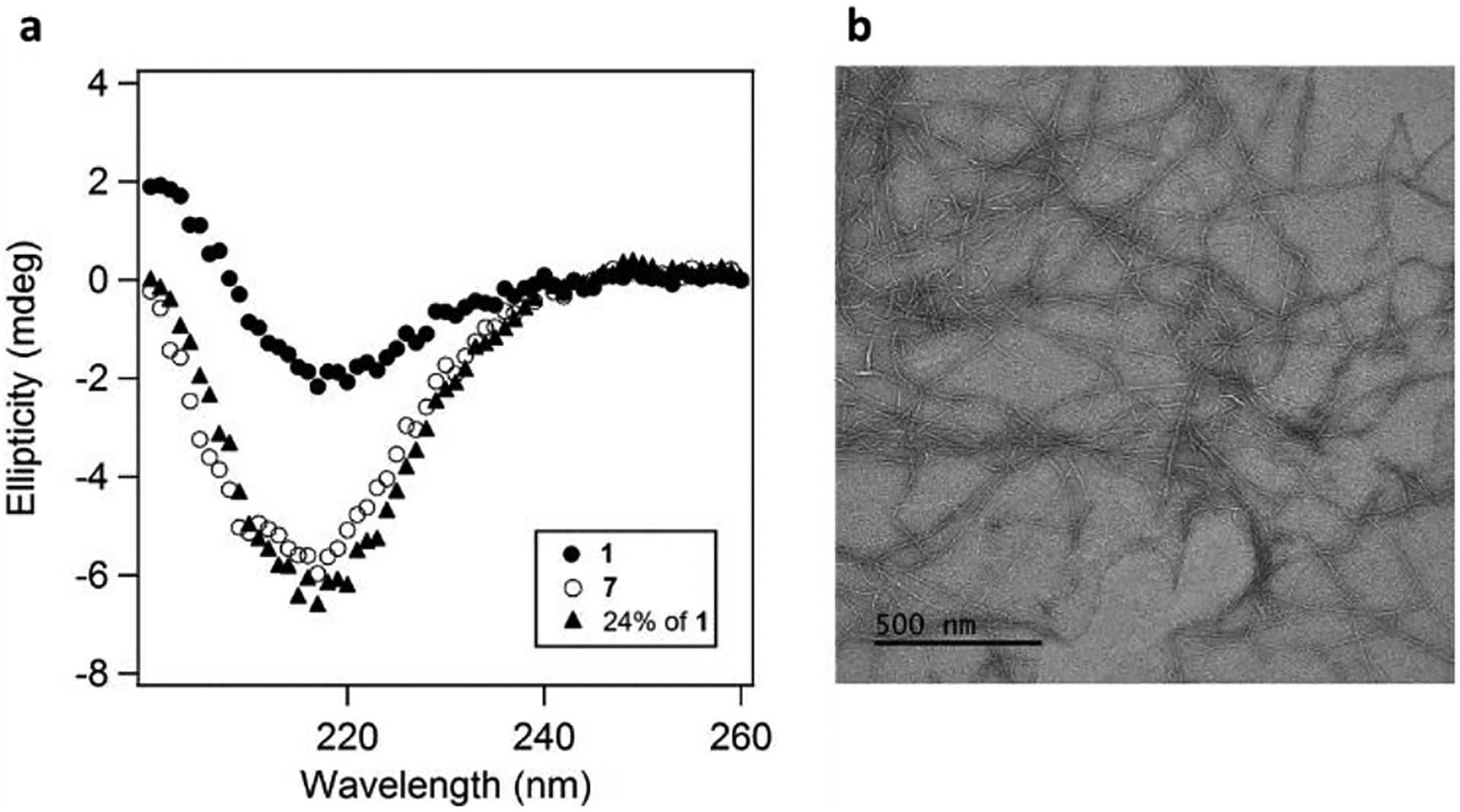
a) Circular dichroism spectra of 1, 7, and the mixture (24:76) of 1 and 7; b) TEM image of the fibrils formed by a mixture (24:76) of 1 and 7. Experimental conditions: 25 mM Tris, pH 8.0, 25 μM peptide, 0.1 mM ZnCl2, the samples were incubated for 24 h prior to measurements.
The kinetic parameters of two of the peptide mixtures were examined more closely, to further explore how the interactions of functional groups in peptide mixtures affect catalytic activity (Figure 4). We used a mixture of 4 and 1 as a representative example of negative synergy (Figure 2e).
Figure 4.
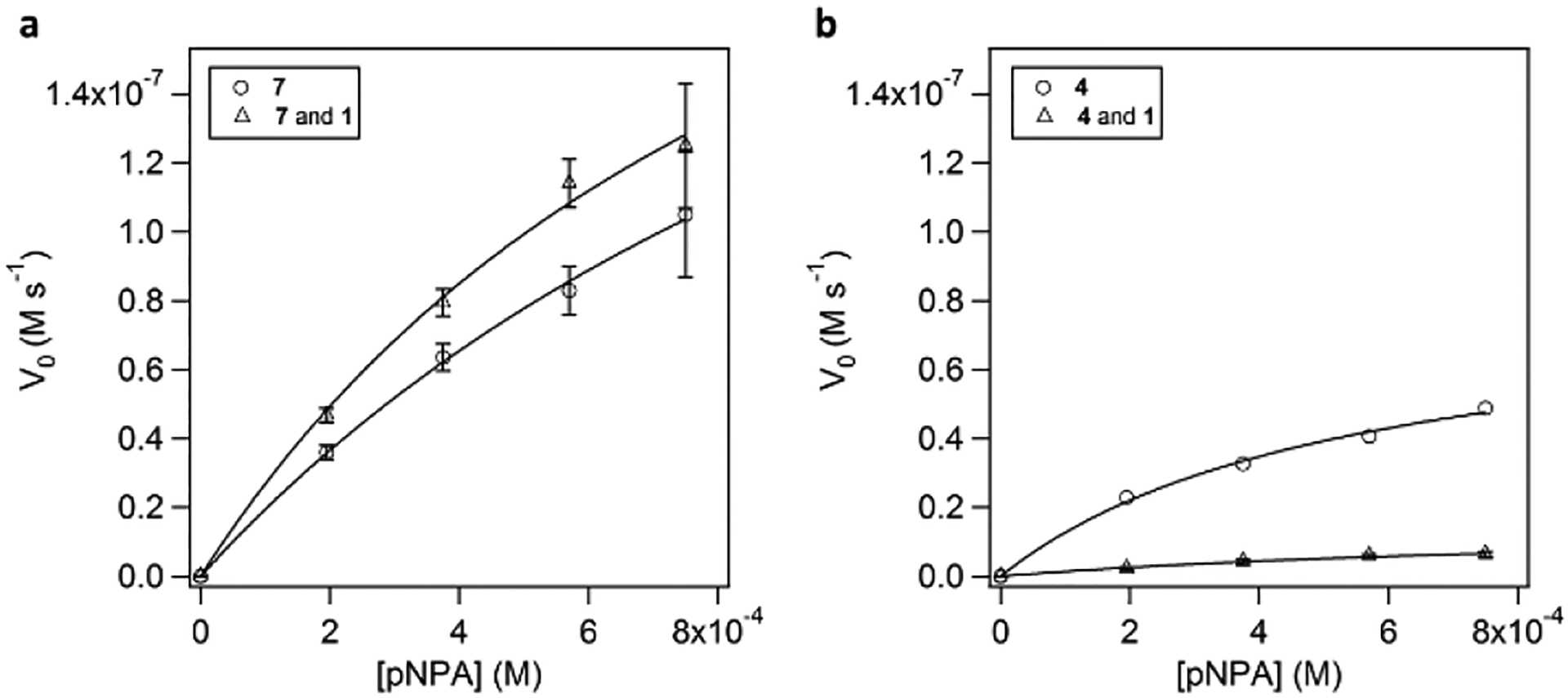
Michaelis-Menten plots of pNPA hydrolysis promoted by a) 7 and the mixture (24:76) of 1 and 7; and b) 4 and the mixture (50:50) of 1 and 4. Experimental conditions: 25 mM Tris, pH 8.0, 25 μM total peptide, 0.1 mM Zn2+. 25 μM peptide, 6.25 μM active 2–7 species, the samples were incubated for 24 h prior to measurements.
The fibrils containing both 7 and 1 displayed 50% higher catalytic efficiencies predominantly derived from increased kcat, with KM remaining largely unchanged (Figure 4a). This finding indicated that the observed improvement in activity likely comes from concerted action of multiple functional groups and not the improved substrate binding affinity. In the case of a mixtures showing strong negative synergy, such as 4 and 1, the catalytic activity was diminished due to both decreased kcat and increased KM (Figure 4b).
Homochirality is a key feature of all ribosomally produced proteins, but the origin of homochirality in natural systems has been frequently debated.[9] All previous examples of catalytic amyloids have employed the natural L-enantiomers and we wondered what the effects of mixed chirality on catalysis in amyloid structures might be. As expected 7D, the enantiomer of 7 made of all D-amino acids, self-assembled in the presence of zinc to produce catalytic amyloids with activity identical to that of 7. Pauling’s proposal that a mixture of L- and D- β-strands would result in racemic rippled sheets has been demonstrated by both Nilsson and Schneider.[10] The most relevant to this work is an excellent study by Nilsson and co-workers on octapeptides which showed that co-assemblies displayed an enthalpic advantage over those of homochirality.[11] We were curious to investigate the effects this kind of assembly would have on the catalytic efficiency, and so measured the activity of the mixtures of 7 and 7D in different proportions. The result of this experiment proved to be quite striking: unlike the fairly smooth negative synergy observed in studies described above we observed a sharp drop in activity in all mixtures (Figure 5a). Addition of even 10% of the opposite enantiomer leads to nearly complete loss of catalytic activity. TEM characterization of the mixtures shows amyloid formation in all cases with morphology similar to that of 7 (Figure S9). The strong increase in thioflavin T fluorescence upon mixing of the species indicates the formation of amyloid-like species (Figure 5b). The magnitude of fluorescence increase for the mixtures over that of the pure enantiomer is consistent with a molecular level change of peptide arrangements, possible representing a pleated β-sheet arrangement. The same behavior (although with a slightly less pronounced drop) was observed for 4 (Figure S8, Supporting Information). This feature was not unique to enantiomers of the same fibril. We mixed L-Ac-LHLHLQL-NH2 and D-Ac-LHLHLRL-NH2, which was shown above to result in strong positive synergy when carried out with the corresponding L-peptides. However, the mixture of 3 and 7D again showed a profound disruption of catalytic activity in the heteromeric assemblies (Figure S9, Supporting Information), suggesting the general inability of a rippled β-sheet to promote catalysis with the current arrangements of functional groups.
Figure 5.
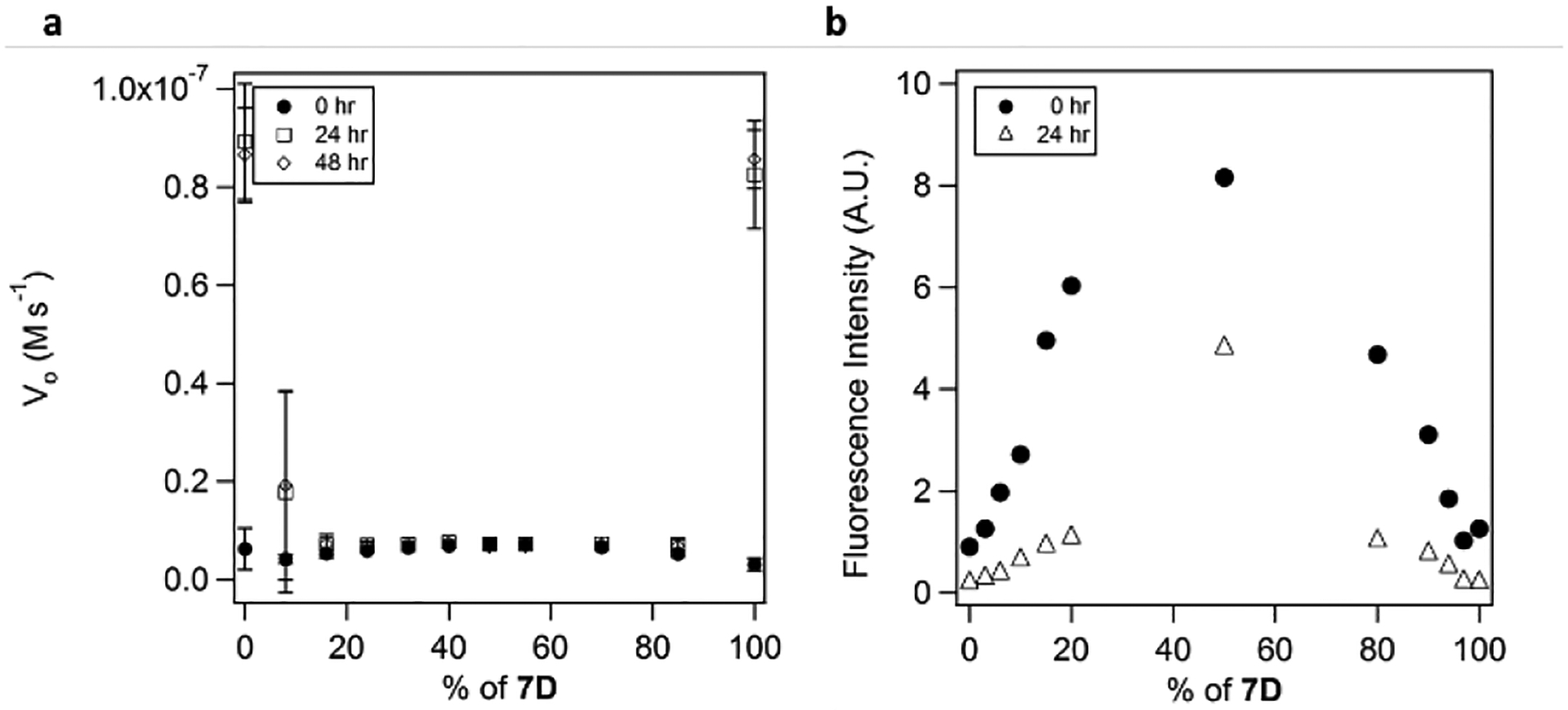
a) The dependence of catalytic efficiency in pNPA hydrolysis on the composition of the fibrils made by mixing 7 and 7D. Experimental conditions: 25 mM Tris, pH 8.0, 25 μM total peptide, 0.1 mM Zn2+, 195 μM pNPA. b) ThT fluorescence of the fibrils formed by mixing 7D with 7 as a function of fibril composition.
To determine whether an alternative arrangement of functional groups in the rippled sheet geometry might render the peptides active we mixed 7F with 7D. 7F is a “flipped” version of peptide 7, where we swapped the position of the metal-binding site: Ac-LRLHLHL-NH2 (7F). Just like 7, 7F self-assembled in the presence of zinc to produce an efficient esterase with catalytic efficiency of 28 ± 3.5 M−1s−1 (Figure S11, Supporting Information). Similar to 7-7D mixtures, a sharp activity drop was observed when 7D and 7F were mixed in essentially any proportions (Figure S8, Supporting Information). The sharp drop in activity observed upon addition of just a relatively small amount of the opposite enantiomer regardless of the exact sequence is very remarkable. While the mechanistic origins of this finding require additional structural and modeling studies, it is reasonable to assume that even a relatively small amount of the opposite enantiomer can break an extended array of strands in the proper configuration required for the catalytic activity. Future studies will be also necessary to elucidate the effect of fibril morphology on catalytic activity, which may affect substrate access and/or fibril aggregation.
In conclusion, we have demonstrated that catalytic amyloids possess a high potential for synergistic interactions: nearly all binary peptide mixtures in a limited peptide set shows non-linear behavior. This observation is important from several standpoints. First, it can help explain how a small subset of short peptides can provide a robust library for rapid identification of catalytically productive arrangements of functional groups in a primordial soup. Second, peptide mixtures can rapidly provide very basic structural and functional information about the fibrils themselves, serving as a straightforward and inexpensive characterization tool. While the structural and mechanistic origins of the synergistic effect will require further studies, the productive arrangements identified in these rapid screens will undoubtedly lead to development of more active catalysts for a variety of different transformations, including redox chemistry.[12]
Supplementary Material
Acknowledgements
This work was supported by the NIH (grant GM119634), the CRDF (grant OISE 18-63891-0), and the Alexander von Humboldt Foundation.
References
- [1].a) Lazcano A, Miller SL, Cell 1996, 85, 793–798; [DOI] [PubMed] [Google Scholar]; b Paecht-Horowitz M, Berger J, Katchalsky A, Nature 1970, 228, 636–639. [DOI] [PubMed] [Google Scholar]
- [2].a) Maury CPJ, Origins Life Evol. B 2009, 39, 141–150; [DOI] [PubMed] [Google Scholar]; b) Maury CPJ, Cell Mol. Life Sci 2018, 75, 1499–1507; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Rout SK, Friedmann MP, Riek R, Greenwald J, Nat. Commun 2018, 9, 234–234; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Greenwald J, Friedmann MP, Riek R, Angew. Chem. Int. Ed 2016, 55, 11609–11613; [DOI] [PubMed] [Google Scholar]; e) Tena-Solsona M, Nanda J, Diaz-Oltra S, Chotera A, Ashkenasy G, Escuder B, Chem. Eur. J 2016, 22, 6687–6694; [DOI] [PubMed] [Google Scholar]; f) Vaidya N, Walker Sara I., Lehman N, Chem. Biol 2013, 20, 241–252; [DOI] [PubMed] [Google Scholar]; g) Friedmann MP, Torbeev V, Zelenay V, Sobol A, Greenwald J, Riek R, PLoS One 2015, 10, e0143948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].a) Rufo CM, Moroz YS, Moroz OV, Stöhr J, Smith TA, Hu X, DeGrado WF, Korendovych IV, Nat. Chem 2014, 6, 303–309; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Makhlynets OV, Gosavi PM, Korendovych IV, Angew. Chem. Int. Ed 2016, 55, 9017–9020; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Lengyel Z, Rufo CM, Moroz YS, Makhlynets OV, Korendovych IV, ACS Catal. 2018, 8, 59–62; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Al-Garawi ZS, McIntosh BA, Neill-Hall D, Hatimy AA, Sweet SM, Bagley MC, Serpell LC, Nanoscale 2017, 9, 10773–10783; [DOI] [PubMed] [Google Scholar]; e) Monasterio O, Nova E, Diaz-Espinoza R, Biochem. Biophys. Res. Commun 2017, 482, 1194–1200. [DOI] [PubMed] [Google Scholar]
- [4].a) Lee M, Wang T, Makhlynets OV, Wu Y, Polizzi NF, Wu H, Gosavi PM, Stöhr J, Korendovych IV, DeGrado WF, Hong M, Proc. Natl. Acad. Sci. U. S. A 2017, 114, 6191–6196; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Song R, Wu X, Xue B, Yang Y, Huang W, Zeng G, Wang J, Li W, Cao Y, Wang W, Lu J, Dong H, J. Am. Chem. Soc 2019, 141, 223–231. [DOI] [PubMed] [Google Scholar]
- [5].a) Fichman G, Guterman T, Adler-Abramovich L, Gazit E, CrystEngComm 2015, 17, 8105–8112; [Google Scholar]; b) Wang M, Lv Y, Liu X, Qi W, Su R, He Z, ACS Appl. Mater. Interfaces 2016, 8, 14133–14141; [DOI] [PubMed] [Google Scholar]; c) Pan T, Liu Y, Si C, Bai Y, Qiao S, Zhao L, Xu J, Dong Z, Luo Q, Liu J, ACS Catal. 2017, 7, 1875–1879; [Google Scholar]; d) Gulseren G, Khalily MA, Tekinay AB, Guler MO, J. Mater. Chem. B 2016, 4, 4605–4611; [DOI] [PubMed] [Google Scholar]; e) Marshall LR, Zozulia O, Lengyel-Zhand Z, Korendovych IV, ACS Catal. 2019, 9, 9265–9275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tolbert AE, Ervin CS, Ruckthong L, Paul TJ, Jayasinghe-Arachchige VM, Neupane KP, Stuckey JA, Prabhakar R, Pecoraro VL, Nat. Chem 2020, 12, 405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].a) Maeda Y, Javid N, Duncan K, Birchall L, Gibson KF, Cannon D, Kanetsuki Y, Knapp C, Tuttle T, Ulijn RV, Matsui H, J. Am. Chem. Soc 2014, 136, 15893–15896; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Schoonen L, van Esterik KS, Zhang C, Ulijn RV, Nolte RJM, Hest J. C. M. v., Sci. Rep 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lengyel Z, Rufo CM, Korendovych IV, Methods Mol. Bio 2018, 1777, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].a) Blackmond DG, Cold Spring Harb Perspect. Biol 2010, 2, a002147–a002147; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Breslow R, Tetrahedron Lett. 2011, 52, 4228–4232. [Google Scholar]
- [10].a) Pauling L, Corey RB, Proc. Natl. Acad. Sci. U. S. A 1953, 39, 253; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Swanekamp RJ, Welch JJ, Nilsson BL, ChemComm. 2014, 50, 10133–10136; [DOI] [PubMed] [Google Scholar]; c) Nagy KJ, Giano MC, Jin A, Pochan DJ, Schneider JP, J. Am. Chem. Soc 2011, 133, 14975–14977; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Nagy-Smith K, Beltramo PJ, Moore E, Tycko R, Furst EM, Schneider JP, ACS Cent. Sci 2017, 3, 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].a) Swanekamp RJ, DiMaio JT, Bowerman CJ, Nilsson BL, J. Am. Chem. Soc 2012, 134, 5556–5559; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Urban JM, Ho J, Piester G, Fu R, Nilsson BL, Molecules 2019, 24, 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].a) Solomon LA, Kronenberg JB, Fry HC, J. Am. Chem. Soc 2017, 139, 8497–8507; [DOI] [PubMed] [Google Scholar]; b) Zozulia O, Korendovych IV, Angew. Chem. Int. Ed 2020, doi i 10.1002/anie.201916712; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Dolan MA, Basa PN, Zozulia O, Lengyel Z, Lebl R, Kohn EM, Bhattacharya S, Korendovych IV, ACS Nano 2019, 13, 9292–9297; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Moffet DA, Certain LK, Smith AJ, Kessel AJ, Beckwith KA, Hecht MH, J. Am. Chem. Soc 2000, 122, 7612–7613. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


