Serratia marcescens is an opportunistic pathogen that can use chitin-enriched nutrients as an alternative energy source. We identified a chitoporin (ChiP), termed SmChiP, from the outer membrane of S. marcescens and report its crystallization without and with chitohexaose. Initial assessment of the complex structure revealed electron density for the sugar ligand.
Keywords: chitin, chitoporin, outer membrane proteins, Serratia marcescens, sugar transport, chitoligosaccharides, sugar-specific porins
Abstract
Serratia marcescens is an opportunistic pathogen that commonly causes hospital-acquired infections and can utilize chitin-enriched nutrients as an alternative energy source. This study reports the identification of a chitoporin (ChiP), termed SmChiP, from the outer membrane of S. marcescens. Sequence alignment with genetically characterized ChiPs suggests that SmChiP is more closely related to the monomeric EcChiP from Escherichia coli than to the trimeric VhChiP from Vibrio campbellii. A single crystal of SmChiP grown under the condition 22%(w/v) PEG 8000, 0.1 M calcium acetate, 0.1 M MES pH 6.0 diffracted X-ray synchrotron radiation to 1.85 Å resolution. SmChiP co-crystallized with chitohexaose under the condition 19%(w/v) PEG 1500, 2 M ammonium phosphate monobasic, 0.1 M HEPES pH 7.0 diffracted X-rays to 2.70 Å resolution. Preliminary crystallographic analysis shows that both SmChiP crystal forms contain one molecule per asymmetric unit and that they belong to the tetragonal space groups P42212 and P41212, respectively. The SmChiP crystal has unit-cell parameters a = 82.97, b = 82.97, c = 189.53 Å, α = β = γ = 90°, while the crystal of SmChiP in complex with chitohexaose has unit-cell parameters a = 73.24, b = 73.24, c = 213.46 Å, α = β = γ = 90°. Initial assessment of the complex structure clearly revealed electron density for the sugar ligand. Structure determination of SmChiP in the absence and presence of chitohexaose should reveal the molecular basis of chitin utilization by S. marcescens.
1. Introduction
Serratia marcescens is a species of Gram-negative, facultative bacteria that belongs to the family Enterobacteriaceae (Golemi-Kotra, 2008 ▸; Hejazi & Falkiner, 1997 ▸). It has a widespread occurrence in different settings, such as in soil, water, plants and the digestive tracts of animals and humans (Hejazi & Falkiner, 1997 ▸; Mahlen, 2011 ▸). Several strains of S. marcescens have been identified as opportunistic pathogens that cause urinary-tract, bloodstream and respiratory-tract infections in hospitalized adults, and in the gastrointestinal systems of children (Golemi-Kotra, 2008 ▸; Gupta et al., 2014 ▸; Hejazi & Falkiner, 1997 ▸; Mahlen, 2011 ▸). S. marcescens species usually grow on various organic materials and starchy surfaces by secreting several hydrolytic enzymes that degrade polymeric biomolecules, such as DNases, gelatinases, lipases and proteases. They also secrete several chitinolytic enzymes, such as ChiA, ChiB, ChiC, ChiD (Cabib, 1988 ▸; Młynarczyk et al., 2007 ▸; Monreal & Reese, 1969 ▸; Vaaje-Kolstad et al., 2013 ▸) and chitin-binding protein (CBP21; Vaaje-Kolstad et al., 2005 ▸), which allow them to utilize chitinous materials as alternative sources of carbon and nitrogen. Despite the expression of these well characterized chitin-degrading enzymes and chitin-binding protein, there is only scarce evidence on how the chitin-degradation products (chitooligosaccharides) are taken up through the outer membrane (OM) of S. marcescens. A chitoligoosaccharide-specific porin (ChiP) was first described in a marine Vibrio species (Keyhani et al., 2000 ▸). The wild-type V. furnissii expressing ChiP could grow on minimal medium containing (GlcNAc)2,3 and could take up these chitooligosaccharides, while the ChiP-null mutant showed a drastic decrease in the rate of cell growth and chitooligosaccharide uptake.
Detailed functional characterization of VhChiP from V. campbellii (formerly V. harveyi) type strain BAA-1116 has been reported, including pore-forming properties (Suginta, Chumjan, Mahendran, Janning et al., 2013 ▸), sugar specificity (Suginta, Chumjan, Mahendran, Schulte et al., 2013 ▸) and statistical analysis of chitooligosaccharide transport (Suginta, Khunkaewla et al., 2013 ▸). When reconstituted into artificial phospholipid bilayers, VhChiP facilitates ion flow with an average conductance of 1.8 nS and is highly responsive to exposure to long-chain chitooligosaccharides, with chitohexaose being the most preferred substrate (Suginta, Chumjan, Mahendran, Janning et al., 2013 ▸). The crystal structures of OM-expressed (PDB entry 5mdq) and in vitro folded VhChiP (PDB entry 5mdo) have been reported at resolutions of 1.95 and 2.5 Å, respectively (Aunkham et al., 2018 ▸). The VhChiP structure is a trimeric assembly, with each monomeric subunit containing a 16-stranded β-barrel and an N-terminal periplasmic sequence (the so-called N-plug) that is believed to regulate the open and closed states of the channel. Structures of VhChiP in complex with chitotetraose (PDB entry 5mds) and chitohexaose (PDB entry 5mdr) were also solved at resolutions of 2.6 and 1.9 Å, respectively. The sugar ligand was contained inside the protein lumen; the pore-lining residues formed a large hydrogen-bond network with the sugar side chains, while some surface-exposed aromatic residues formed hydrophobic interactions with the pyranose rings of the bound sugar. Stochastic analysis of ion currents in the presence of chitohexaose suggested that sugar translocation is likely to be achieved through correlated trapping/untrapping dynamics, causing transient movements along the sugar passage inside the long, narrow channel (Suginta & Smith, 2013 ▸; Suginta et al., 2016 ▸).
A different class of ChiPs has been also documented. We previously identified a quiescent chiP gene encoding EcChiP from the genome of Escherichia coli strain K-12 substrain MG1655 (Soysa & Suginta, 2016 ▸). The chiP gene was cloned into pET-23d(+) expression vector, which could be expressed in the Omp-deficient E. coli BL21 (Omp8) strain. Unlike VhChiP, which is an OmpC-like channel that consists of three identical subunits (Suginta, Chumjan, Mahendran, Janning et al., 2013 ▸) and has exceptional specificity towards long-chain chitooligosaccharides (Suginta, Chumjan, Mahendran, Schulte et al., 2013 ▸), the recombinant EcChiP was characterized as a monomeric OprD-like porin with a molecular weight of ∼52 kDa and showed only 12% sequence identity to VhChiP. EcChiP could form a stable channel and could fully conduct ion flows with an average single-channel conductance of 0.5 nS (Soysa et al., 2017 ▸, 2018 ▸). Nevertheless, it exhibited a significantly lower affinity towards chitooligosaccharides, with the binding constant towards the most preferred substrate being tenfold lower than that of VhChiP (Soysa et al., 2017 ▸). In further relevant studies, the chiP gene encoding SmChiP from S. marcescens 2170 was identified to be part of the ybfMN-ctp cluster (Toratani et al., 2012 ▸). This gene cluster has subsequently been referred to as the chiPQ-ctb cluster (Takanao et al., 2014 ▸). Cell growth has been reported to be affected in S. marcescens mutants with deletions of the chiP, chiX and chiQ genes. In particular, the ΔchiP mutant showed a significantly lower ability to grow on media supplemented with colloidal chitin and chitobiose, and showed no growth on media containing chitotriose. Subsequent studies demonstrated that the chiX small RNA controls the expression of chitin-degrading enzymes (chitinases and chitobiase), a chitin-binding protein (CBP21) and a chitooligosaccharide transporter (ChiP) (Toratani et al., 2012 ▸; Suzuki et al., 2016 ▸). Nevertheless, no detailed functional and structural characterization of chitin acquisition by SmChiP has been performed to date. This short communication describes the gene identification, gene synthesis, recombinant expression, purification and crystallization of SmChiP expressed in E. coli. Crystallization of SmChiP in the absence and presence of a chitooligosaccharide substrate was successful, allowing high-quality crystallographic data for SmChiP and SmChiP in complex with chitooligosaccharide to be obtained. An initial sequence analysis suggested that SmChiP is similar to EcChiP, but differs from VhChiP. Therefore, structure determination will reveal the exclusive properties of SmChiP with respect to how it conducts ion currents and acquires sugar substrate.
2. Materials and methods
2.1. Recombinant expression, protein extraction and purification
AR325_08275, an ORF corresponding to a chiP gene in the genome of S. marcescens, was identified from the UniProtNK database (https://www.uniprot.org/uniprot/A0A0P0QBS3). The Serratia chiP gene fragment containing 1416 base pairs was synthesized by GenScript and cloned into pET-23a(+) using BamHI/XhoI restriction sites. The gene construct, termed pET-23a(+)_Serratia chiP, was transformed into Omp-deficient E. coli BL21 (Omp8) Rosetta host cells and the overnight culture of transformed cells was transferred to Luria–Bertani broth containing 100 µg ml−1 ampicillin and 25 µg ml−1 kanamycin and grown at 300 K. During the exponential growth phase (OD600 of ∼0.6–0.8), SmChiP expression was induced with 0.5 mM (final concentration) isopropyl β-d-1-thiogalactopyranoside. After 6 h of further incubation at 300 K, the cell pellet was harvested by centrifugation at 2590g for 30 min at 277 K.
To extract and purify SmChiP, the cell pellet was resuspended in lysis buffer (20 mM Tris–HCl pH 8.0, 2.5 mM MgCl2, 0.1 mM CaCl2) containing 0.1%(v/v) Triton X-100, 10 µg ml−1 RNase A and 10 µg ml−1 DNase I. Cells were disrupted on ice with a high-speed ultrasonic processor (Cole-Parmer, Vernon Hills, Illinois, USA) for 10 min; 2%(w/v) SDS was then added to the cell suspension, which was further incubated at 323 K for 60 min using a Thermomixer Comfort (Eppendorf AG, Hamburg, Germany) with a thermoblock in 4 × 50 ml Falcon tubes to ensure complete lysis. Cell-wall components were removed by ultracentrifugation at 35 000g at 277 K for 45 min. The pale pellet containing the membrane fraction and recombinant SmChiP was extracted twice with 2.5%(v/v) n-octylpolyoxyethylene (octyl-POE; Alexis Biochemicals, Lausanne, Switzerland) in 10 mM HEPES buffer pH 7.5, 100 mM NaCl using a homogenizer. The suspension was incubated at 300 K with gentle shaking for 1 h using the Thermomixer and the supernatant containing solubilized SmChiP was then collected by ultracentrifugation at 115 600g for 1 h. The supernatant was concentrated, and the detergent was exchanged to 0.2%(v/v) lauryldimethylamine oxide (LDAO; Sigma–Aldrich, Singapore) using an Amicon concentrator (10 kDa molecular-weight cutoff; Merck Millipore, Tullagreen, Cork, Ireland). SmChiP was further purified to homogeneity by size-exclusion chromatography using a Sephacryl S-200 HR 26/100 column connected to an ÄKTA pure FPLC (GE Healthcare Bio-Sciences, Uppsala, Sweden), with a flow rate of 1.0 ml min−1. The purity of the SmChiP peak from the size-exclusion step was verified by SDS–PAGE, the fractions containing highly purified SmChiP were pooled and the buffer was exchanged to 10 mM HEPES pH 7.4 containing 100 mM LiCl, 0.4%(w/v) tetraethylene glycol monooctyl ether (C8E4; Anatrace, Maumee, Ohio, USA) in an Amicon concentrator as described above. The protein concentration was estimated from A 280 using a molar extinction coefficient of 126 170 M −1 cm−1. The freshly prepared sample (15.0 mg ml−1) was used to set up crystallization screens, while the remaining sample was aliquoted into small volumes, snap-frozen in liquid nitrogen and immediately stored at 193 K for further use. Detergent extraction and purification of SmChiP for the purposes of co-crystallization were carried out separately following the protocol described above, except that the protein was buffer-exchanged into 10 mM HEPES pH 7.4 containing 100 mM LiCl and 0.4%(v/v) C8E4 using an Amicon concentrator for crystallization trials. The molecular weight of SmChiP was confirmed by electrospray mass-spectrometry analysis (First BASE Laboratories, Selangor Darul Ehsan, Malaysia).
2.2. Protein crystallization and data collection
Sitting-drop crystallization screening was carried out in Swissci 96-Well 2-Drop MRC Crystallization Plates (Molecular Dimensions, Sheffield, UK) using a Mosquito crystallization robot (TTP Labtech, Melbourn, UK). For crystallization trials of SmChiP, the freshly prepared protein (9.0 mg ml−1) was screened using the MemGold and MemGold2 screening kits (Molecular Dimensions) with protein solution:mother liquor ratios of 200 nl:200 nl in the top wells and 200 nl:150 nl in the bottom wells. The screening plates were sealed immediately and incubated at 293 K. The appearance of single crystals was observed daily under a stereo microscope. Conditions that yielded single crystals were further optimized in 24-well plates using the hanging-drop vapour-diffusion method. In each well, two drops with different protein:precipitant volume ratios, 1.0 µl:1.0 µl and 1.5 µl:1.0 µl, were set up on an 18 mm cover slip. The crystallization plates were incubated at 293 K. Single crystals obtained from the optimized condition were harvested with nylon loops and plunged into liquid nitrogen using 20%(w/v) PEG 400 added to the mother liquor as a cryoprotectant and were tested for X-ray diffraction. For SmChiP in complex with chitooligosaccharide, the freshly prepared protein (15 mg ml−1) dissolved in 10 mM HEPES pH 7.4 containing 100 mM LiCl and 0.4%(v/v) C8E4 was mixed with 10 mM chitohexaose prior to crystallization. The protein mixed with chitohexaose was incubated for 1 h before setting up crystallization screening. Initial screening and optimization were carried out using the MemTrans and MemChannel screening kits (Molecular Dimensions), with protein solution:mother liquor ratios of 150 nl:150 nl in the top well and 150 nl:200 nl in the bottom well, as described for SmChiP. The screening plates were incubated at 292 K. A hit condition with a large number of single crystals was optimized in 24-well plates using the hanging-drop method as for SmChiP. At this point, the protein:precipitant volume ratios were changed to 0.5 µl:0.5 µl and 0.5 µl:1.0 µl and the drops were incubated at 292 K. The single crystals from the optimized condition were harvested under a liquid-nitrogen steam as for SmChiP.
2.3. X-ray data collection and processing
X-ray diffraction data were collected from the SmChiP crystals on beamline I04-1 at Diamond Light Source (DLS), Didcot, UK. The data set was processed with DIALS and scaled with AIMLESS (Evans & Murshudov, 2013 ▸; Winter et al., 2018 ▸). X-ray diffraction data from SmChiP in complex with chitohexaose were collected on beamline TPS 05A at the National Synchrotron Radiation Research Centre (NSRRC), Hsinchu, Taiwan. One data set was collected with 94.9% completeness and was indexed by iMosflm (Battye et al., 2011 ▸). X-ray data scaling was performed by AIMLESS, which is available in the CCP4 package (Winn et al., 2011 ▸).
3. Results and discussion
3.1. Identification of the chiP gene, gene synthesis and sequence analysis
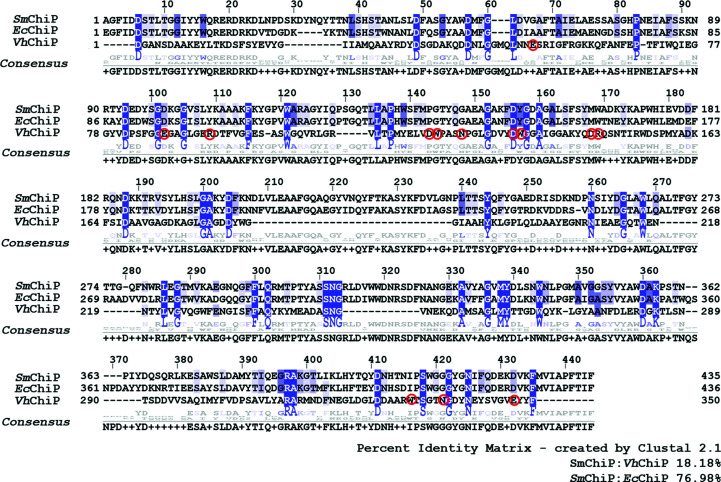
The full-length chiP gene was identified from the UniProtKB Protein Database (UniProtKB entry A0A0P0QBS3). The Serratia_chiP gene construct encodes SmChiP that includes two extra amino acids GS from the BamHI cloning site and the 27-amino-acid signal sequence, followed by the mature SmChiP polypeptide of 435 amino acids. The predicted molecular weight of mature SmChiP was 48 987 Da and the pI was 5.07. Multiple sequence alignment of SmChiP with two other functionally characterized ChiPs, EcChiP from E. coli (Soysa & Suginta, 2016 ▸; Soysa et al., 2017 ▸, 2018 ▸) and VhChiP from V. campbellii (formerly V. harveyi; Suginta, Chumjan, Mahendran, Janning et al., 2013 ▸), gave 77% and 18% sequence identity to EcChiP and VhChiP, respectively (Fig. 1 ▸). This result indicated that SmChiP is likely to be closely related, both structurally and functionally, to EcChiP.
Figure 1.
Multiple sequence alignment of SmChiP from S. marcescens (UniProtKB entry A0A0P0QBS3, this study) with EcChiP from E. coli (UniProtKB entry P75733) and VhChiP from V. campbellii (UniProtKB entry L0RVU0). The sequence alignment was generated by Clustal 2.1 and displayed by Jalview version 2.11.1.0. Blue shades indicate strictly conserved amino acids and red circles the amino-acid residues of VhChiP that bind to chitohexaose (Aunkham et al., 2018 ▸).
The amino-acid sequences of SmChiP and EcChiP are relatively different from that of VhChiP, with 45 amino-acid residues (highlighted in blue) that are completely conserved and distributed throughout the entire sequences. These conserved residues are presumed to participate in critical architectural features of the three porins but are not involved in sugar transport. It is notable that except for Asp135, none of the amino-acid residues (indicated with red circles) that interact with the substrate in VhChiP are found in EcChiP or SmChiP. However, these residues are identical in the latter two channels. Such differences within the interior of the channel may give rise to a different binding mode and substrate specificity of the monomeric SmChiP/EcChiP versus the trimeric VhChiP.
3.2. Protein expression, purification and mass determination
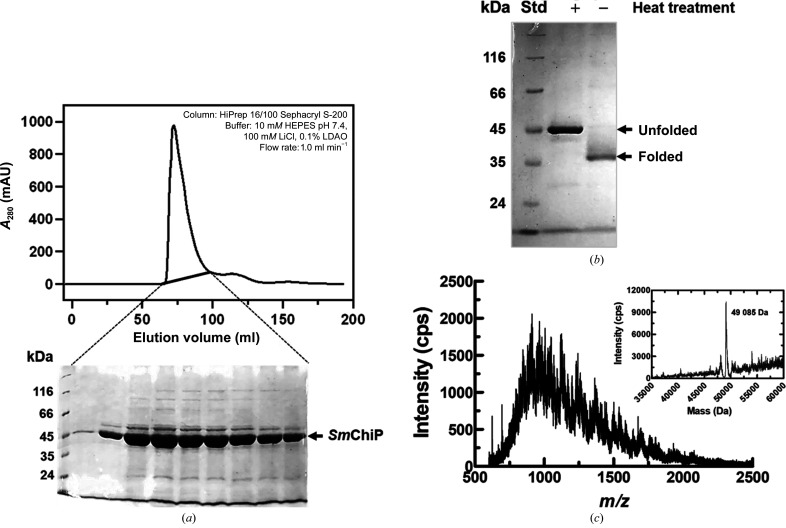
Recombinant SmChiP containing its own signal peptide was successfully expressed in the OM of the Omp-deficient E. coli (Omp8) Rosetta host. After cell lysis, the SmChiP-containing OM fraction was prepared by extraction with SDS followed by octyl-POE. At this stage, SmChiP with 70–80% purity was usually obtained. The protein was then purified to homogeneity by size-exclusion chromatography for crystallization trials. Fig. 2 ▸(a) shows a representative elution profile of SmChiP purified by HiPrep 16/100 Sephacryl S-200 HR column chromatography (upper panel), yielding a single peak in A 280 with no peak corresponding to aggregated protein. The peak contained a major protein that migrated with an apparent molecular weight of approximately 45 kDa on SDS–PAGE (Fig. 2 ▸ a, lower panel). We then performed thermal denaturation to identify the monomeric/oligomeric state of SmChiP. It is known that outer membrane porins are structurally resistant to SDS treatment at room temperature, but unfold at high temperature (Reid et al., 1988 ▸; Noinaj et al., 2015 ▸). Fig. 2 ▸(b) shows the thermal dissociation of SmChiP. The protein was found to migrate at a position of about 45 kDa on SDS–PAGE after heating at 373 K for 10 min, while the unheated protein, corresponding to the folded protein, migrated at a position of about 35 kDa (Fig. 2 ▸ b). The folded protein moved faster on SDS–PAGE, owing to its compact barrel structure, compared with the unfolded SmChiP. Similar results were observed with the monomeric E. coli chitoporin (EcChiP), which consists of one subunit of molecular weight 50 kDa (Soysa & Suginta, 2016 ▸). This is in contrast to the previous report on VhChiP from V. campbellii (formerly V. harveyi), where the folded protein migrated at a higher molecular weight under nondenatured conditions, while the unfolded protein appeared three times lower and was reported to be a trimeric protein (Soysa & Suginta, 2016 ▸; Aunkham et al., 2018 ▸). Thus, the results obtained from thermal denaturation support the notion from multiple sequence alignment that SmChiP is more closely related to EcChiP than to VhChiP. The monomeric state of the purified protein was confirmed by intact mass analysis by ESI-LC mass spectrometry, which allows determination of the molecular weight with high precision. ESI-MS analysis of SmChiP is shown as individual spectra in Fig. 2 ▸(c), with the monoisotopic peaks in the ESI mass spectrum representing intact molecular species with variable charges. The vertical axis shows the relative abundances (intensities) of multiply charged species of SmChiP, and the horizontal axis represents the mass to charge ratio (m/z) of the multiply charged analyte. The intact mass of SmChiP was then determined by automated charge-state deconvolution to be 49 085 Da (inset in Fig. 2 ▸ c), which is consistent within 0.2% standard error with the predicted theoretical mass of mature SmChiP (48 987 Da) as described earlier.
Figure 2.
Preparation of SmChiP expressed in E. coli. (a) A representative elution profile of SmChiP eluted from a HiPrep 16/100 Sephacryl S-200 HR column with 10 mM HEPES pH 7.4, 100 mM LiCl, 0.1% LDAO (upper panel). SDS–PAGE showed that a single protein peak with an elution volume of 65–100 ml contained SmChiP as the major component (lower panel). (b) SDS–polyacrylamide gel showing migration of the purified SmChiP after heating at 373 K for 10 min (lane +) and without heat treatment (lane −). Lane Std contains molecular-weight markers (labelled in kDa). (c) Isotopic ionization pattern of SmChiP as determined by electrospray mass spectrometry. The inset shows the peak deconvolution.
3.3. Crystallization and initial X-ray crystallographic analysis
Small single crystals of SmChiP were observed after one week in four conditions: conditions A8 [30%(w/v) PEG 400, 0.1 M CdCl2, 0.1 M LiCl, 0.1 M sodium acetate pH 4.5] and D6 [26%(v/v) PEG 350 MME, 0.2 M CaCl2 dihydrate, 0.1 M MES pH 6.5] from MemGold (designated MG1_A8 and MG1_D6, respectively) and conditions C10 [22%(w/v) PEG 8000, 0.1 M calcium acetate, 0.1 M MES pH 6.0] and D12 [24%(w/v) PEG 400, 0.2 M calcium acetate, 0.1 M HEPES pH 7.0] from MemGold2 (designated MG2_C10 and MG2_D12, respectively). During co-crystallization, single crystals were observed within one week from conditions F12 [38%(w/v) PEG 550 MME, 0.1 M MgCl2 hexahydrate, 0.1 M Tris–HCl pH 8.7] in both the top and bottom wells (1:1 and 1:1.75 protein:mother liquor ratios) and H12 [20%(w/v) PEG 1500, 0.2 M ammonium phosphate monobasic, 0.1 M HEPES pH 7.0] from MemTrans (designated MT_F12 and MT_H12, respectively). The initial hits were further optimized as described above and a summary of the crystallization conditions for SmChiP and co-crystallized SmChiP is given in Table 1 ▸.
Table 1. Crystallization conditions for SmChiP.
| SmChiP | SmChiP co-crystallized with chitohexaose | |
|---|---|---|
| Screening | ||
| Method | Sitting drop | Sitting drop |
| Temperature (K) | 293 | 292 |
| Protein concentration (mg ml−1) | 9 | 15 |
| Molar ratio of protein:ligand | — | 0.3 mM:10 mM |
| Buffer composition | 10 mM HEPES pH 7.5, 100 mM NaCl, 0.4%(v/v) C8E4 | 10 mM HEPES pH 7.4, 100 mM LiCl, 0.4%(v/v) C8E4 |
| Condition composition | MG1_A8: 30%(w/v) PEG 400, 0.1 M CdCl2, 0.1 M LiCl, 0.1 M sodium acetate pH 4.5 | MT_F12: 38%(w/v) PEG 550 MME, 0.1 M MgCl2·6H2O, 0.1 M Tris–HCl pH 8.7 |
| MG1_D6: 26%(w/v) PEG 350 MME, 0.2 M CaCl2 dihydrate, 0.1 M MES pH 6.5 | MT_H12: 20%(w/v) PEG 1500, 0.2 M ammonium phosphate monobasic, 0.1 M HEPES pH 7.0 | |
| MG2_C10: 22%(w/v) PEG 8000, 0.1 M calcium acetate, 0.1 M MES pH 6.0 | ||
| MG2_D12: 24%(w/v) PEG 400, 0.2 M calcium acetate, 0.1 M HEPES pH 7.0 | ||
| MG2_D12: 24%(w/v) PEG 400, 0.2 M calcium acetate, 0.1 M HEPES pH 7.0 | ||
| Volume and ratio of drop | 400 nl, 1:1; 350 nl, 1.75:1 | 300 nl, 1:1; 350 nl, 1:1.75 |
| Volume of reservoir (µl) | 80 | 80 |
| Optimization | ||
| Method | Hanging drop | Hanging drop |
| Temperature (K) | 293 | 292 |
| Protein concentration (mg ml−1) | 9 | 15 |
| Molar ratio of SmChiP:chitohexaose | — | 0.3 mM:10 mM |
| Buffer composition of protein solution | 10 mM HEPES pH 7.5, 100 mM NaCl, 0.4%(v/v) C8E4 | 10 mM HEPES pH 7.4, 100 mM LiCl, 0.4%(v/v) C8E4 |
| Condition composition | 22%(w/v) PEG 8000, 0.1 M calcium acetate, 0.1 M MES pH 6.0 (from MG_C10) | 19%(w/v) PEG 1500, 0.2 M ammonium phosphate monobasic 0.1 M HEPES pH 7.0 (from MT_H12) |
| Volume and ratio of drop | 2 µl, 1:1; 2.5 µl, 1:1.15 | 1 µl, 1:1; 1.5 µl, 1:1.5 |
| Volume of reservoir (µl) | 300 | 300 |
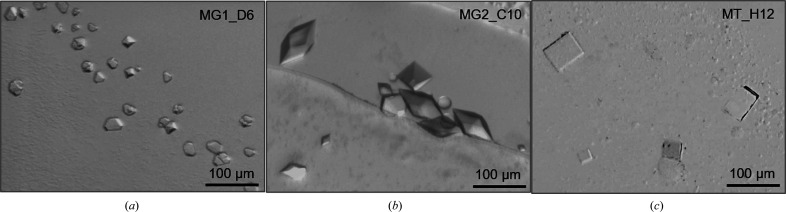
Although several single hexagonal crystals of SmChiP were obtained from MG1_D6, the crystal dimensions were relatively small (approximately 20 × 30 µm; Fig. 3 ▸ a), so this condition was not selected for further optimization and the crystals were not tested for X-ray diffraction. After hanging-drop optimization, good-sized and well defined hexagonal bipyramidal crystals (dimensions of about 50 × 100 µm) were observed after one day of incubation at 293 K in the optimized condition MG2_C10 (Fig. 3 ▸ b). Crystals of SmChiP in complex with chitohexaose were successfully grown under the condition 19%(w/v) PEG 1500, 0.2 M ammonium phosphate monobasic, 0.1 M HEPES pH 7.0. Thin plate tetragonal crystals appeared after two days of incubation at 292 K, with dimensions of approximately 30 × 50 µm (Fig. 3 ▸ c).
Figure 3.
Single crystals of SmChiP. (a) Crystals of SmChiP obtained from the initial screen condition MG1_D6. (b) Crystals of SmChiP obtained from the optimized condition 22%(w/v) PEG 8000, 0.1 M calcium acetate, 0.1 M MES pH 6.0. (c) Crystals of SmChiP co-crystallized with chitohexaose from the optimized condition 19%(w/v) PEG 1500, 0.2 M ammonium phosphate monobasic, 0.1 M HEPES pH 7.0.
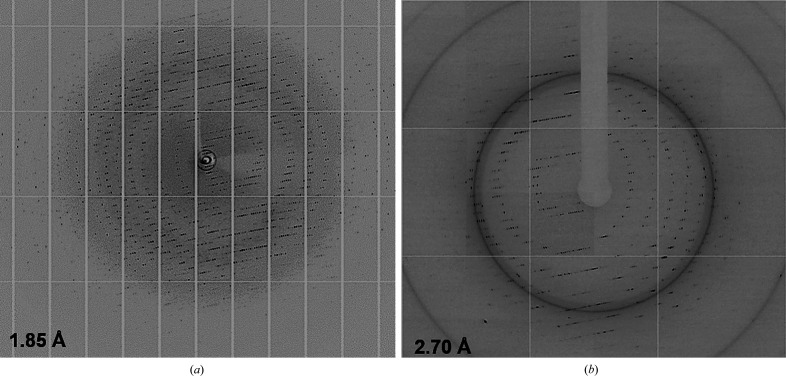
X-ray diffraction data were collected from the SmChiP crystals obtained under the optimized condition on beamline I04-1 at Diamond Light Source. The best data set was collected with 100% completeness to a resolution of 1.85 Å (Fig. 4 ▸ a). The L-test estimated a twinning fraction of 0.063, indicating no twinning problems. The crystal diffraction showed substantially low mosaicity and a single lattice, indicating the overall high quality of this data set. POINTLESS predicted P42212 to be the most likely space group, with a space-group confidence of 0.91. The unit-cell parameters are a = 82.97, b = 82.97, c = 189.53 Å, α = β = γ = 90°. The estimated Matthews coefficient of 3.33 Å3 Da−1 suggested the presence of one molecule in the asymmetric unit and a solvent content of 63.1%.
Figure 4.
High-resolution images of X-ray data from (a) SmChiP and (b) SmChiP in complex with chitohexaose.
The X-ray diffraction data for SmChiP in complex with chitohexaose were collected on beamline TPS 05A at the National Synchrotron Radiation Research Center (NSRRC), Taiwan. Data from a crystal grown under the condition 19%(w/v) PEG 1500, 0.2 M ammonium phosphate monobasic, 0.1 M HEPES pH 7.0 were collected to a resolution of 2.38 Å. Using the half-set correlation coefficient (CC1/2) and I/σ(I) criteria (Winn et al., 2011 ▸) with the accepted values of CC1/2 = 0.3 and I/σ(I) = 1.0, the data were scaled to a resolution cutoff of 2.70 Å (Fig. 4 ▸ b) with 94.9% completeness. The processed data have unit-cell parameters a = 73.24, b = 73.24, c = 213.46 Å, α = β = γ = 90°, and a Matthews coefficient of 2.89 Å3 Da−1 indicates one molecule per asymmetric unit (solvent content of 57%). The indexing statistics were compatible with the tetragonal space group P41212. Data-quality assessment of the final MTZ file by phenix.xtriage (Liebschner et al., 2019 ▸; Otwinowski & Minor, 1997 ▸) estimated the mean Z-score of ice rings to be 2.05, indicating that no ice rings were detected (Morris et al., 2004 ▸). The distribution of |L| values indicates a twin fraction of 0.0, suggesting no twinning problems (Padilla & Yeates, 2003 ▸). The preliminary crystallographic parameters of both SmChiP crystal forms are summarized in Table 2 ▸.
Table 2. Data-collection and processing statistics.
Values in parentheses are for the outer resolution shell.
| Apo SmChiP | SmChiP–chitohexaose | |
|---|---|---|
| Resolution cutoff (Å) | 1.85 | 2.70 |
| Beamline | I04-1, DLS | TPS 05A, NSRRC |
| Wavelength (Å) | 0.97886 | 0.99984 |
| Resolution range (Å) | 94.76–1.85 (1.89–1.85) | 71.15–2.70 (2.83–2.70) |
| Space group | P42212 | P41212 |
| a, b, c (Å) | 82.9, 82.97, 189.53 | 73.24, 73.24, 213.46 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Total No. of reflections | 800417 | 100353 |
| No. of unique reflections | 57273 | 15892 |
| Completeness (%) | 100 (94.5) | 99.0 (94.9) |
| Multiplicity | 11.7 (9.8) | 6.3 (6.5) |
| Mean I/σ(I)† | 24.8 (2.5) | 8.7 (3.0) |
| R merge ‡ | 0.101 (0.903) | 0.164 (0.922) |
| CC1/2 | 0.993 (0.647) | 0.966 (0.569) |
Mean I/σ(I) = 〈〈I h〉/σ′(〈I h〉)〉.
R
merge = 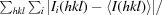
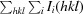 , where Ii(hkl) is the intensity of the ith measurement of an equivalent reflection with indices hkl.
, where Ii(hkl) is the intensity of the ith measurement of an equivalent reflection with indices hkl.
3.4. Future prospects for structure determination
Since the X-ray data set for SmChiP without ligand was the first to be collected, we attempted to determine it independently of the crystal complex. The model of SmChiP was built by molecular replacement (MR) using the crystal structure of unliganded EcChiP from E. coli (PDB entry not yet publicly available) as a template. Although the final model of SmChiP has still to be refined, the electron density was of sufficient quality for all 435 amino-acid residues to be visible in the 2F o − F c map contoured at a σ level of 1.0. The electron density for SmChiP in complex with chitohexaose gave an OMIT map with reasonable electron density, allowing all of the amino acids to be visualized. We observed clear electron density for four GlcNAc rings of chitohexaose in the middle of the protein pore, while partial electron densities for the GlcNAc rings at the termini of the sugar chain were seen owing to the high mobility of the sugar rings at the reducing and nonreducing ends of the sugar backbone. Refinement of both SmChiP models and structure validation are ongoing. Future research will examine the structural similarities and discrepancies between the monomeric SmChiP and the trimeric VhChiP in order to understand the molecular basis of sugar-channel interactions and chitooligosaccharide uptake by S. marcescens.
Acknowledgments
The authors declare no conflicts of interest. We thank the experimental facility and the technical services provided by the Synchrotron Radiation Protein Crystallography Facility of the National Core Facility Program for Biotechnology, Ministry of Science and Technology and the National Synchrotron Radiation Research Centre, a national user facility supported by the Ministry of Science and Technology, Taiwan, and Diamond Light Source, UK. We would like to thank Dr Yoshihito Kitaoku for assisting with data collection and processing, and Professor Takeshi Watanabe, Department of Applied Biological Chemistry, Niigata University, Japan for introducing the chiP gene from the S. marcescens system for functional characterization. We would like to thank Dr David Apps of the University of Edinburgh for critical reading and English improvement of the manuscript.
Funding Statement
This work was funded by Vidyasirimedhi Institute of Science and Technology grant BRG610008 to Wipa Suginta. Thailand Research Fund grant BRG610008.
References
- Aunkham, A., Zahn, M., Kesireddy, A., Pothula, K. R., Schulte, A., Baslé, A., Kleinekathöfer, U., Suginta, W. & van den Berg, B. (2018). Nat. Commun. 9, 220. [DOI] [PMC free article] [PubMed]
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Cabib, E. (1988). Methods Enzymol. 161, 460–462.
- Evans, P. R. & Murshudov, G. N. (2013). Acta Cryst. D69, 1204–1214. [DOI] [PMC free article] [PubMed]
- Golemi-Kotra, D. (2008). xPharm: The Comprehensive Pharmacology Reference, edited by S. J. Enna & D. B. Bylund, pp. 1–6. New York: Elsevier.
- Gupta, N., Hocevar, S. N., Moulton-Meissner, H. A., Stevens, K. M., McIntyre, M. G., Jensen, B., Kuhar, D. T., Noble-Wang, J. A., Schnatz, R. G., Becker, S. C., Kastango, E. S., Shehab, N. & Kallen, A. J. (2014). Clin. Infect. Dis. 59, 1–8. [DOI] [PMC free article] [PubMed]
- Hejazi, A. & Falkiner, F. R. (1997). J. Med. Microbiol. 46, 903–912. [DOI] [PubMed]
- Keyhani, N. O., Li, X.-B. & Roseman, S. (2000). J. Biol. Chem. 275, 33068–33076. [DOI] [PubMed]
- Liebschner, D., Afonine, P. V., Baker, M. L., Bunkóczi, G., Chen, V. B., Croll, T. I., Hintze, B., Hung, L.-W., Jain, S., McCoy, A. J., Moriarty, N. W., Oeffner, R. D., Poon, B. K., Prisant, M. G., Read, R. J., Richardson, J. S., Richardson, D. C., Sammito, M. D., Sobolev, O. V., Stockwell, D. H., Terwilliger, T. C., Urzhumtsev, A. G., Videau, L. L., Williams, C. J. & Adams, P. D. (2019). Acta Cryst. D75, 861–877.
- Mahlen, S. D. (2011). Clin. Microbiol. Rev. 24, 755–791. [DOI] [PMC free article] [PubMed]
- Młynarczyk, A., Młynarczyk, G., Pupek, J., Bilewska, A., Kawecki, D., Łuczak, M., Gozdowska, J., Durlik, M., Pączek, L., Chmura, A. & Rowińnski, W. (2007). Transplant. Proc. 39, 2879–2882. [DOI] [PubMed]
- Monreal, J. & Reese, E. (1969). Can. J. Microbiol. 15, 689–696. [DOI] [PubMed]
- Morris, R. J., Zwart, P. H., Cohen, S., Fernandez, F. J., Kakaris, M., Kirillova, O., Vonrhein, C., Perrakis, A. & Lamzin, V. S. (2004). J. Synchrotron Rad. 11, 56–59. [DOI] [PubMed]
- Noinaj, N., Kuszak, A. J. & Buchanan, S. K. (2015). Methods Mol. Biol. 1329, 51–56. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol 276, 307–326. [DOI] [PubMed]
- Padilla, J. E. & Yeates, T. O. (2003). Acta Cryst. D59, 1124–1130. [DOI] [PubMed]
- Reid, J., Fung, H., Gehring, K., Klebba, P. E. & Nikaido, H. (1988). J. Biol. Chem. 263, 7753–7759. [PubMed]
- Soysa, H. S. M. & Suginta, W. (2016). J. Biol. Chem. 291, 13622–13633. [DOI] [PMC free article] [PubMed]
- Soysa, H. S. M., Schulte, A. & Suginta, W. (2017). J. Biol. Chem. 292, 19328–19337. [DOI] [PMC free article] [PubMed]
- Soysa, H. S. M., Suginta, W., Moonsap, W. & Smith, M. F. (2018). Phys. Rev. E, 97, 052417. [DOI] [PubMed]
- Suginta, W., Chumjan, W., Mahendran, K. R., Janning, P., Schulte, A. & Winterhalter, M. (2013). PLoS One, 8, e55126. [DOI] [PMC free article] [PubMed]
- Suginta, W., Chumjan, W., Mahendran, K. R., Schulte, A. & Winterhalter, M. (2013). J. Biol. Chem. 288, 11038–11046. [DOI] [PMC free article] [PubMed]
- Suginta, W., Khunkaewla, P. & Schulte, A. (2013). Chem. Rev. 113, 5458–5479. [DOI] [PubMed]
- Suginta, W. & Smith, M. F. (2013). Phys. Rev. Lett. 110, 238102. [DOI] [PubMed]
- Suginta, W., Winterhalter, M. & Smith, M. F. (2016). Biochim. Biophys. Acta, 1858, 3032–3040. [DOI] [PubMed]
- Suzuki, K., Shimizu, M., Sasaki, N., Ogawa, C., Minami, H., Sugimoto, H. & Watanabe, T. (2016). Biosci. Biotechnol. Biochem. 80, 376–385. [DOI] [PubMed]
- Takanao, S., Honma, S., Miura, T., Ogawa, C., Sugimoto, H., Suzuki, K. & Watanabe, T. (2014). Biosci. Biotechnol. Biochem. 78, 524–532. [DOI] [PubMed]
- Toratani, T., Suzuki, K., Shimizu, M., Sugimoto, H. & Watanabe, T. (2012). Biosci. Biotechnol. Biochem. 76, 1920–1924. [DOI] [PubMed]
- Vaaje-Kolstad, G., Horn, S. J., Sørlie, M. & Eijsink, V. G. (2013). FEBS J. 280, 3028–3049. [DOI] [PubMed]
- Vaaje-Kolstad, G., Houston, D., Riemen, A., Eijsink, V. & van Aalten, D. (2005). J. Biol. Chem. 280, 11313–11319. [DOI] [PubMed]
- Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., Keegan, R. M., Krissinel, E. B., Leslie, A. G. W., McCoy, A., McNicholas, S. J., Murshudov, G. N., Pannu, N. S., Potterton, E. A., Powell, H. R., Read, R. J., Vagin, A. & Wilson, K. S. (2011). Acta Cryst. D67, 235–242. [DOI] [PMC free article] [PubMed]
- Winter, G., Waterman, D. G., Parkhurst, J. M., Brewster, A. S., Gildea, R. J., Gerstel, M., Fuentes-Montero, L., Vollmar, M., Michels-Clark, T., Young, I. D., Sauter, N. K. & Evans, G. (2018). Acta Cryst. D74, 85–97. [DOI] [PMC free article] [PubMed]