Abstract
RanBPM is a multimodular scaffold protein that interacts with a great variety of molecules including nuclear, cytoplasmic, and membrane proteins. By building multiprotein complexes, RanBPM is thought to regulate various signaling pathways, especially in the immune and nervous system. However, the diversity of these interactions does not facilitate the identification of its precise mechanism of action, and therefore the physiological role of RanBPM still remains unclear. Recently, RanBPM has been shown to be critical for the fertility of both genders in mouse. Although mechanistically it is still unclear how RanBPM affects gametogenesis, the data collected so far suggest that it is a key player in this process. Here, we examine the RanBPM sterility phenotype in the context of other genetic mutations affecting mouse gametogenesis to investigate whether this scaffold protein affects the function of other known proteins whose deficiency results in similar sterility phenotypes.
1. INTRODUCTION
Multimodular scaffold proteins are crucial regulators of a great variety of physiological functions. They coordinate the physical assembly of proteins, regulating signal transduction cascades, and shaping signaling responses. RanBPM, also named RanBP9, is a scaffold protein that belongs to the Ran-binding protein family. Proteins of this family were initially identified by yeast two-hybrid as binding partners of the small Ras-like GTPase Ran. However, RanBPM is structurally and functionally unrelated to the other members of this family, as it lacks the consensus Ran-binding domain and seems devoid of any role in nuclear trafficking (Beddow, Richards, Orem, & Macara, 1995). Instead, RanBPM has been described as an adaptor protein. Its structure does not contain any known catalytic domain but includes several protein-binding domains that participate in the formation of large protein complexes. Indeed, RanBPM has been reported to interact so far with more than 45 proteins. These include structural and adhesion proteins, cytosolic kinases, cell surface tyrosine kinase receptors, nuclear receptors, and a number of proteins in the nervous system (Suresh, Ramakrishna, & Baek, 2012). RanBPM has been reported to function in the immune and nervous system (Murrin & Talbot, 2007). Yet, the in vivo significance of these activities needs further studies. Moreover, it is still unclear how RanBPM orchestrates the activities of such a broad spectrum of proteins that are functionally unrelated. RanBPM has a multimodular structure with “sticky” properties that makes it possible for it to associate nonspecifically with many other proteins in vitro. Indeed, many of its binding partners have been identified by yeast-two-hybrid screening and coimmunoprecipitation experiments in overexpression systems, suggesting that additional functional assays may be needed to validate the existing data. In an attempt to study the in vivo function of RanBPM, we recently generated and characterized mice lacking RanBPM (Puverel, Barrick, Dolci, Coppola, & Tessarollo, 2011). This loss-of-function study revealed a crucial role of RanBPM in both spermatogenesis and oogenesis. However, we still do not know how it orchestrates the signal transduction machinery required for the execution of these biological processes. In this chapter, we discuss the RanBPM-deficient phenotypes, the possible mechanism of RanBPM action in relation to its structure, spatiotemporal pattern of expression, and how it relates to other mutations affecting gametogenesis.
2. PROTEIN STRUCTURE
RanBPM comprises five main domains. These include an N-terminal proline-rich domain, a consensus SPRY protein–protein interaction domain originally identified in the SplA kinase and the ryanodine receptor (Ponting, Schultz, & Bork, 1997), a lissencephaly type-1-like homology (LisH) motif described to act as a dimerization domain (Gerlitz, Darhin, Giorgio, Franco, & Reiner, 2005; Kim et al., 2004; Mateja, Cierpicki, Paduch, Derewenda, & Otlewski, 2006), a CTLH (carboxy-terminal to LisH) domain of unknown function, and a C-terminal CRA (CT11-RanBPM) motif, reported to interact with the fragile X mental retardation protein FMRP (Menon, Gibson, & Pastore, 2004). The CRA domain has been reported to contain a nuclear localization signal (Lakshmana et al., 2010). The LisH-CTLH domain is mostly described in proteins involved in the regulation of microtubule dynamics and cell migration and is present in several proteins which associate with RanBPM to form a large protein complex (Emes & Ponting, 2001; Kobayashi et al., 2007; Nishitani et al., 2001). Thus, all these domains provide potential protein-binding sites for a great variety of molecules (Fig. 13.1).
Figure 13.1.
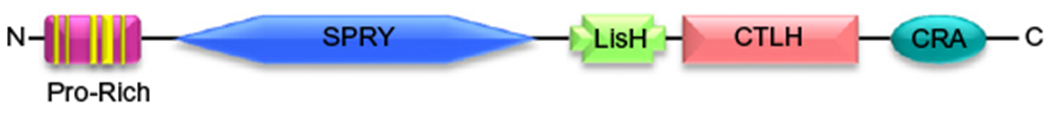
Structure of RanBPM protein. RanBPM is composed of five protein–protein interaction domains, including a proline-rich domain (Pro-Rich), a SPRY domain, the functionally related LisH and CTLH domains, and a C-terminal CRA domain. Adapted from Suresh et al. (2012).
In an initial study, RanBPM was described as a centrosomal 55-kDa protein implicated in the regulation of microtubule nucleation (Nakamura et al., 1998). Later work by the same group revealed that this was instead an N-terminal truncated protein and that the full-length RanBPM cDNA encodes a 90-kDa protein which does not localize to the microtubule-organizing center (Nishitani et al., 2001). However, it is interesting to note that in a recent study, some authors reported the occurrence of a 60-kDa-processed form (C-terminal truncation) with an enhanced stability compared to the full-length RanBPM. While the full-length protein is localized in both the nucleus and cytoplasm, the truncated form is mostly localized in the cytoplasm (Lakshmana et al., 2010). Yet, this fragment retains the capacity to form self-interacting multimeric complexes.
More recently, another protein named RanBP10 has been described as the closest homolog of RanBPM. This protein shares significant sequence similarities and, except for lacking the N-terminal proline-rich domain, it possesses all other domains and these are well conserved. So far, RanBP10 has been reported to play a role in regulating the platelet shape and function in the adult mouse (Kunert et al., 2009). The close structural similarities between RanBPM and RanBP10 suggest the possibility of functional redundancy during the analysis of mutant mice lacking one of these proteins.
3. FUNCTION OUTSIDE THE REPRODUCTIVE SYSTEM
RanBPM is evolutionarily conserved and is ubiquitously expressed; to our knowledge, it has been found in all tissues and cell lines examined so far. It can undergo phosphorylation and ubiquitination, but the precise role of these posttranslational modifications still needs to be evaluated (Denti et al., 2004; Ideguchi et al., 2002). At the subcellular level, RanBPM has been found in the cytoplasm, nucleus, as well as associated with the cell membrane (Denti et al., 2004; Suresh et al., 2012; Valiyaveettil et al., 2008).
Among the best-known partners of RanBPM are the Met tyrosine kinase receptor for hepatocyte growth factor (HGF), whose Ras–Erk pathway is facilitated by RanBPM, and the kelch-repeat protein muskelin which acts on the regulation of cell morphology (Valiyaveettil et al., 2008; Wang, Li, Messing, & Wu, 2002). In addition to its involvement in these general cellular processes, RanBPM is known to play a role in the immune response by interacting with the β2-integrin lymphocyte function-associated antigen 1 (Denti et al., 2004). Moreover, a growing number of studies are reporting a function for RanBPM in the nervous system. For example, it has been involved in the modulation of axonal and neurite outgrowth by cooperating with the Plexin-A receptors and inhibiting the neural cell adhesion molecule L1 signaling (Cheng, Lemmon, & Lemmon, 2005; Togashi, Schmidt, & Strittmatter, 2006). Interestingly, RanBPM binds to both the low- and the high-affinity neurotrophin receptors p75NTR, TrkA, and TrkB (Bai, Chen, & Huang, 2003; Yin et al., 2010; Yuan et al., 2006), interacts with the fragile X mental retardation protein, possibly modulating its RNA-binding properties, and binds to metabotropic glutamate receptors (Menon et al., 2004; Seebahn, Rose, & Enz, 2008). RanBPM has also been found to associate with citron kinase (CITK), regulating the production of pyramidal neurons in the embryonic neocortex (Chang et al., 2010). Finally, RanBPM has been involved in Alzheimer’s disease, affecting amyloid precursor protein processing and amyloid β generation (Lakshmana et al., 2009; Woo et al., 2012).
4. PHENOTYPE OF RanBPM-DEFICIENT MICE
While RanBPM has been involved in a number of biological processes, the recent generation of RanBPM-deficient mice has provided a tool that will allow investigators to test their physiological significance in vivo. Homozygous mutant mice for RanBPM develop to term, suggesting that the gene is not crucial for embryonic development. However, a little more than half of the mutant animals die postnatally, while the rest survive to adulthood. Surviving RanBPM−/− mice have a normal life span but are significantly smaller than their control littermates. The major organs, including brain, heart, liver, lung, kidney, thymus, spleen, and intestine, are proportional to the body size and do not show any obvious abnormality. By contrast, mutant gonads are severely atrophic leading to sterility of both genders (Puverel et al., 2011) (Fig. 13.2). In the adult mutant testis, seminiferous tubules are decreased in size and completely devoid of germ cells, and the epididymal duct lumen does not contain any sperm. In the mutant ovary, only a few follicles develop in young mice, but disappear in older mice.
Figure 13.2.
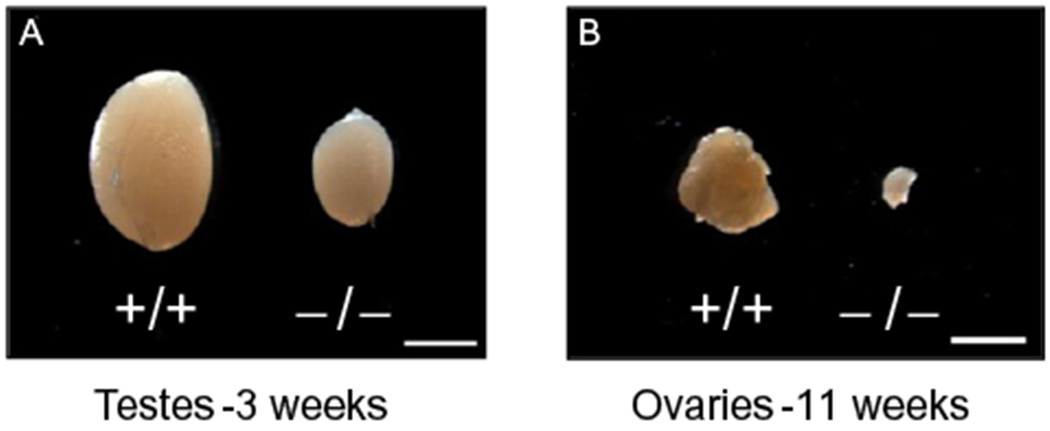
Representative pictures showing the gonad atrophy in RanBPM homozygous mutant mice. Mutant and wild-type mouse testes (A) and ovaries (B) from 3-week-old males and 11-week-old females. Scale bars: 2 mm.
5. EXPRESSION IN THE REPRODUCTIVE SYSTEM
RanBPM has been reported to be highly expressed in the mouse testis, especially in the maturating spermatocytes (Shibata et al., 2004). Another study has shown that the RanBPM protein is produced in spermatogonia and primary spermatocytes in the human testis, whereas in the rat testis, it appears to be expressed only in round and elongated spermatids (Tang et al., 2004). Moreover, gene expression profiling of mouse male meiotic germ cells and northern blot analysis have found RanBPM transcripts in spermatogonia and primary spermatocytes, as well as in spermatids (Rossi et al., 2004).
In order to clarify the RanBPM pattern of expression, we decided to evaluate the activity of this gene during testis development by taking advantage of a LacZ reporter gene inserted in the targeted RanBPM locus (Puverel et al., 2011). Since the LacZ gene product, β-galactosidase, is expressed under the control of the endogenous RanBPM promoter, X-gal staining constitutes a sensitive and accurate method to study the expression pattern of the RanBPM gene. We found that in males, RanBPM mRNA expression starts during embryogenesis; it is first detected in the testis of heterozygous animals at embryonic day 17.5 in clustered mouse vasa homolog (MVH)-positive germ cells. In newborns, expression is still mainly restricted to the gonocytes, while 5 days later, interstitial cells begin expressing RanBPM. At 2 weeks, both spermatogonia and Sertoli cells, the supporting cells of the spermatogenic process, are strongly labeled. RanBPM transcript is thus sequentially expressed by distinct cell populations during testis postnatal development, yet its expression is always present in the germ cell lineage. Interestingly, RanBPM mRNA expression is cyclically regulated during adult steady state spermatogenesis. In the adult testis, expression is detected in spermatogonia residing at the periphery of the seminiferous tubules and maintained in both primary and secondary spermatocytes. Remarkably, the highest expression is observed in primary spermatocytes undergoing the first meiotic division, although it seems to stop at the stage of differentiated tailed spermatids. RanBPM expression is thus dynamically regulated during the spermatogenic wave. In addition to RanBPM transcriptional activity, we were able to show that the protein is also expressed in the testis. Between birth and 2 weeks, RanBPM protein level is fairly constant but it increases significantly between 2 and 3 weeks, corresponding to the time when the spermatocytes of the first wave of spermatogenesis undergo the first meiotic division. By immunohistochemistry, we found that primary spermatocytes are the cells with the highest RanBPM protein expression.
The first study focusing on RanBPM function in the female reproductive system was conducted in Drosophila where there are two proteins (a short and a long) coded by transcripts produced by alternative splicing. The short RanBPM isoform appears to be expressed in all somatic and germline cells of the ovary, while the long isoform is specifically enriched in the germline stem cell niche where it is required for the proper arrangement of niche cells and niche cell size (Dansereau & Lasko, 2008). In mice, the RanBPM gene is active in both the developing and adult ovary. Interestingly, RanBPM expression starts at the birth and is restricted to the germinal cell lineage, that is, oocytes reaching the dictyate arrest (Puverel et al., 2011). In the adult ovaries, RanBPM is present at a high level in oocytes and is maintained during follicle development, from primary to Graafian follicles. As in males, RanBPM transcripts are also expressed in the somatic cell lineage in the ovary since expression is detected in the theca cell layers, while granulosa cells are negative. However, whether RanBPM transcripts are translated into proteins in every cell type in which they have been detected is still unknown and, as suggested by the results obtained in males, RanBPM mRNA expression may be broader than that of the protein (Puverel et al., 2011).
6. RanBPM IS DISPENSABLE FOR PRIMORDIAL GERM CELL MIGRATION
The data available so far suggest that RanBPM is not involved in primordial germ cell (PGC) migration (Puverel et al., 2011). PGCs are derived from the epiblast from where they migrate to colonize the genital ridges by 11.5 days of embryogenesis (Bendel-Stenzel, Anderson, Heasman, & Wylie, 1998; Tres, Rosselot, & Kierszenbaum, 2004). Upon entry into the developing gonads, PGCs coalesce with somatic cells that have initiated sexual differentiation. PGCs proliferate during migration and for a short time after colonization of the gonads. Male germ cells then enter mitotic arrest while female germ cells begin to enter the first meiotic division. Several factors have been involved in this process, including the chemokine stromal cell-derived factor 1 and its receptor CXCR4, as well as stem cell factor (SCF), and the c-Kit tyrosine kinase receptor (Ara et al., 2003; Buehr, McLaren, Bartley, & Darling, 1993; Mahakali Zama, Hudson, & Bedell, 2005; Molyneaux et al., 2003; Runyan et al., 2006). X-gal staining failed to detect any RanBPM expression in E13.5 embryos, as RanBPM expression starts during late embryogenesis in males and at birth in females (Puverel et al., 2011). In addition, the lack of RanBPM does not seem to affect germ cells during embryogenesis, as gonocytes are present in the seminiferous tubules of newborn males, and the number of meiotic oocytes present in the developing ovary of mutants at day E17.5 is comparable with that of WT embryos. These observations suggest that the migration of PGCs occurs normally in mutant mice and that RanBPM does not play a role in the germ cell migration and proliferation events occurring during embryonic development.
7. FUNCTION OF RanBPM DURING POSTNATAL TESTIS DEVELOPMENT
In males, the second week of postnatal life is marked by the initiation of the spermatogenesis process with massive proliferation of spermatogonia. Macroscopically, this transition is reflected by a rapid increase in testis volume as germ cells start filling the seminiferous tubules. RanBPM mutant testes do not show any overt phenotype during the first postnatal week. However, at 2 weeks, contrary to WT testes which undergo a significant growth, they only slightly increase in size. Histological examination at 2 weeks revealed a clear decrease in the diameter of seminiferous tubules, compared to WT (Puverel et al., 2011). These tubules contain a dramatically decreased number of both spermatogonia and spermatocytes. However, the levels of apoptosis were comparable between WT and mutant at this stage, suggesting that the lack of germ cells is not the consequence of cell death events. By contrast, and detectable as early as P8, there was a clear defect in proliferation which was revealed by a dramatic decrease in the number of BrdU-positive cells, as well as an altered organization of the proliferating cells, when compared to WT mouse testes in which these cells are typically forming a continuous ring delineating the seminiferous tubules (Puverel et al., 2011). Altogether, these data demonstrate that RanBPM is a crucial factor for spermatogonia production. Whether RanBPM acts on the differentiation of spermatogonia or on their proliferation remains to be determined.
One of the factors reported to act on spermatogonia proliferation is the tyrosine kinase receptor c-Kit. While the downregulation of its ligand, SCF affects PGCs migration, proliferation, and survival (Mahakali Zama et al., 2005; Runyan et al., 2006), c-Kit also plays a role in spermatogonia proliferation (Blume-Jensen et al., 2000; Kissel et al., 2000). RanBPM has been reported to interact with several tyrosine kinase receptors, including the HGF receptor Met and both the TrkA and TrkB neurotrophin receptors, and to possibly influence their signaling pathways. Considering that c-Kit function is critical for normal spermatogenesis, it is tempting to speculate that RanBPM may interact with this receptor and affect its signaling (Wang et al., 2002; Yin et al., 2010; Yuan et al., 2006). However, c-Kit acts on spermatogonia cell survival and entry into meiosis, and the phenotype of c-Kit mutants with a defect in the PI3-kinase pathway differs from RanBPM mutant mice in that spermatogonia are depleted by apoptosis and meiotic cells are rare or absent (Blume-Jensen et al., 2000; Kissel et al., 2000; Puverel et al., 2011). Outside the reproductive system, some studies reported a role for RanBPM in cell division. CITK is a protein that localizes to the surface of the lateral ventricles during embryonic cortex development and is essential in neurogenic mitoses (Di Cunto et al., 2000). A biochemical interaction between CITK and RanBPM has been reported recently, and the use of in utero RNAi to decrease RanBPM expression leads to an increased number of cells in mitosis, and a concomitant decrease in the number of cells in cytokinesis (Chang et al., 2010). Both RanBPM and RanBP10 have also been described to interact with YPEL5, a protein located at different mitosis-related subcellular structures and involved in cell cycle progression (Hosono et al., 2010). Taken together, these reports suggest that RanBPM could play a role in the proliferation of different cell types. This hypothesis is also supported by the fact that RanBPM is ubiquitously expressed and that its deletion causes partial neonatal lethality and growth retardation for reasons that are still unknown (Puverel et al., 2011).
Androgens are essential hormones for the initiation and maintenance of spermatogenesis. The androgen receptor (AR) is a nuclear receptor acting as a ligand-inducible transcription factor to modulate the expression of a number of target genes. RanBPM has been reported to associate with AR and to enhance its transcriptional activity in a ligand-dependent manner when overexpressed in prostate cancer cell lines (Rao et al., 2002). AR has been reported to be expressed in all cell types of the testis. Knockout mice for AR exhibit a severe phenotype, with an abdominal localization of the testes and an absence of epididymis formation (Wang, Yeh, Tzeng, & Chang, 2009). If RanBPM mediates AR activity in the testis, its effect is thus likely limited to specific cell types. Testicular cell-specific AR knockout mice have been described in the literature and provide a better understanding of the role of this receptor in spermatogenesis (Wang et al., 2009). Interestingly, Sertoli cell-specific and Leydig cell-specific AR mutants exhibit a decrease in testis size and seminiferous tubules development as well as an arrest in germ cell maturation, mainly at the diplotene and round spermatid stages, respectively. By contrast, germ cell-AR-deficient mice are comparable to controls, with a normal range of spermatogenesis. If the defect in postnatal testis development observed in RanBPM-deficient mice was due to an effect of RanBPM on AR activity, one could speculate that this regulation occurs either in Sertoli or Leydig cells. However, such AR deregulation is accompanied by changes in serum hormone levels. In AR-knock-out mice and Sertoli- and Leydig-specific mutants, the levels of testosterone are decreased while LH levels are increased, whereas in RanBPM mutant mice, these hormone levels are not significantly different from those of control littermates (Puverel et al., 2011; Wang et al., 2009). In addition, RanBPM protein expression has been detected in primary spermatocytes, and we have found that RanBPM is acting in germ cells in a cell-autonomous way (see Section 10). Finally, the meiotic arrest observed in AR-deficient mice is unlikely to be due to the same defect occurring in RanBPM mice, since mutant females for AR do not exhibit any arrest in differentiation during the prophase of the first meiotic division, in contrast to RanBPM mutant females (Hu et al., 2004; Shiina et al., 2006) (Section 8).
8. MEIOTIC ARREST IN MICE DEFICIENT FOR RanBPM
Despite the reduction in spermatogonia proliferation, primary spermatocytes are still present, yet in reduced numbers, in the seminiferous tubules of RanBPM−/− male mice during the first wave of spermatogenesis. By contrast, postmeiotic cells are totally absent and no sperm is produced at any developmental stage. While somatic Sertoli and Leydig cells appear normal in the young mutant animal, spermatogonia, pachytene, and only a few diplotene spermatocytes are the most mature germ cells present in the tubules. Thus, RanBPM−/− males exhibit a stage-specific arrest of spermatogenesis, as generally observed in mutant mice in which male sterility is due to a germinal cell primary defect. The fact that mutant mice are affected at as early as 3 weeks of age shows that the first wave of germ line differentiation does not take place. In mice, the first wave of meiotic division is synchronous throughout the testis and occurs around 3 weeks, when the secondary spermatocytes and round spermatids first appear in the seminiferous tubules (Bellve et al., 1977). At this stage, RanBPM mutant testes display a high level of cell death and primary spermatocytes undergo apoptotic death instead of further developing into spermatids (Puverel et al., 2011). Meiosis is a critical step of gametogenesis. It allows genetic exchange between the paternal and maternal genomes, which occurs by recombination during the prophase of the first meiotic division. Prophase I is tightly regulated and can be divided into different stages (see also Fig. 13.3). The leptotene stage is marked by the compaction of chromatin and the programmed induction of localized double-strand breaks (DSBs) which allows the genetic exchange. Chromosome pairing and synapsis are initiated during zygotene, with full synapsis being achieved at the pachytene stage. During zygotene and pachytene stages, the DSBs are repaired by interaction with the homologs. The subsequent disjunction of homologous chromosomes is initiated at the diplotene stage; chromosomes start to separate but remain attached in the regions where crossing over has occurred (Cohen & Pollard, 2001; Zickler & Kleckner, 1999). The proper alignment and synapsis of homologous chromosomes are achieved by the formation of a large zipper-like tripartite protein complex called the synaptonemal complex, which connects the chromosomes along their entire length (Page & Hawley, 2004; Yang & Wang, 2009).
Figure 13.3.
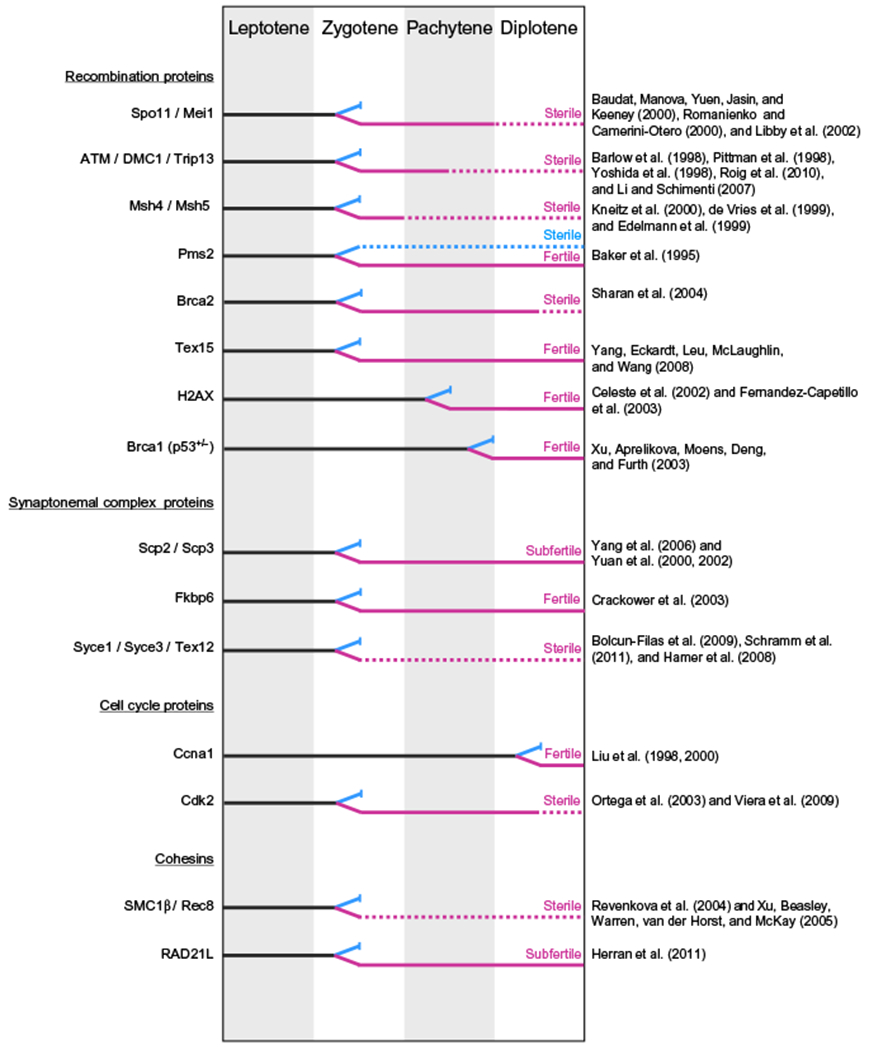
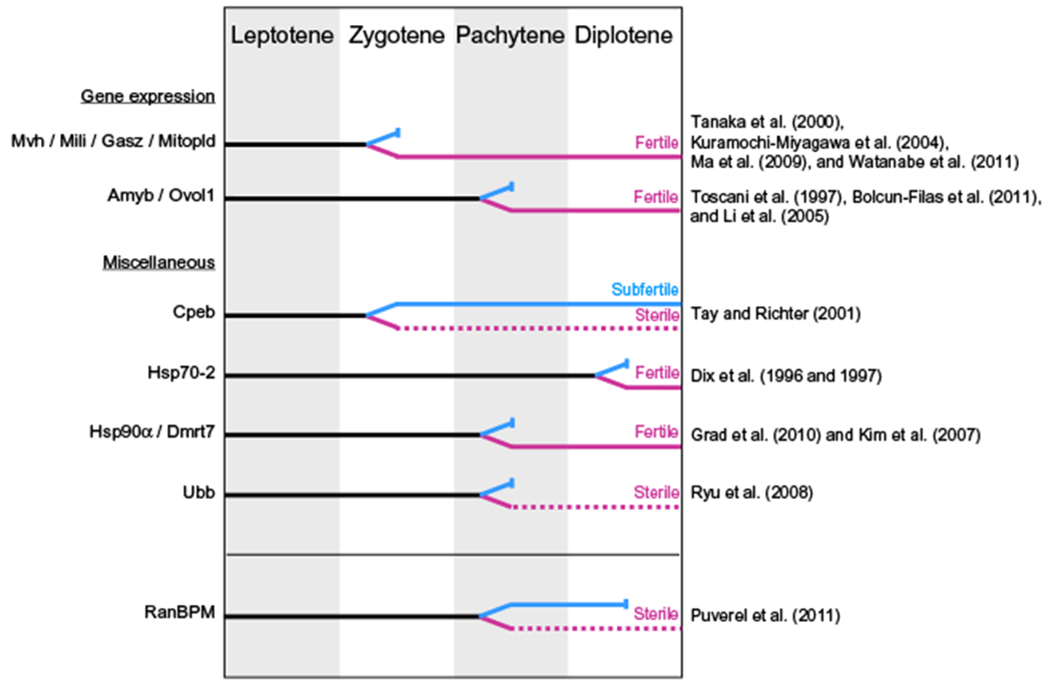
Diagram comparing known mouse mutants exhibiting an arrest during the prophase of the first meiotic division. A black solid line indicates stages during which male and female meioses are similar. When meiotic progression is different between the genders, a blue line indicates male progression and a purple line female progression. A meiotic arrest is represented by a solid vertical bar and dotted lines indicate that only a proportion of germ cells persists. Only genes for which information is available for both genders have been included. Adapted from Hunt and Hassold (2002).
This protein complex mediates the DSBs resolution events leading to recombination. In males, the X and Y chromosomes synapse only at their pseudoautosomal region forming what is called the sex body, located in the nuclear periphery. Instead, in females, the XX chromosomes synapse along their entire length to undergo homologous recombination. In humans, incorrect assembly of the synaptonemal complex causes infertility by triggering germ cell elimination in males and the formation of aneuploid oocytes in females (Hassold & Hunt, 2001;Judis, Chan, Schwartz, Seftel, & Hassold, 2004; Miyamoto et al., 2003). The high level of apoptosis observed in RanBPM−/− seminiferous tubules is correlated to a decrease in the expression of genes that are specifically expressed at the end of Prophase I, such as Calmegin and Hox1.4 (Puverel et al., 2011; Rubin, Toth, Patel, D’Eustachio, & Nguyen-Huu, 1986; Watanabe et al., 1994). Expression of CyclinA1, which has been shown to rise dramatically in late pachytene spermatocytes and to reach a maximum in diplotene cells, is even more affected in RanBPM mutant mice (Sweeney et al., 1996). By contrast, genes expressed earlier in the differentiation process, such as A-myb, Dmc1, and Hsp70.2, are not affected (Mettus et al., 1994; Rosario, Perkins, O’Brien, Allen, & Eddy, 1992; Yoshida et al., 1998; Zakeri, Wolgemuth, & Hunt, 1988). In addition, the genes encoding the synaptonemal complex proteins SCP1 and SCP3 are expressed at normal levels in the mutant, indicative of the presence of pachytene spermatocytes (Klink, Lee, & Cooke, 1997; Meuwissen et al., 1992).
Female germ cells enter meiosis during embryonic development, around E13, and subsequently arrest at the end of Prophase I, around birth (dictyate arrest) (Speed, 1982). Oocytes remain arrested until puberty, when a pool of oocytes is recruited and completes the first meiotic division. The second meiotic round occurs only when fertilization takes place (Edson, Nagaraja, & Matzuk, 2009). Ovaries of adult RanBPM−/− females exhibit a severe dysgenesis resulting from an absence of follicles. Premature ovarian failure has many possible causes: for example, it can result from a defect in PGC migration, meiosis, and formation, survival or activation of primordial follicles (Jagarlamudi, Reddy, Adhikari, & Liu, 2010). Analysis of RanBPM−/− fetal ovaries at E17.5 revealed that the number of oocytes is comparable to that of controls. However, by postnatal day 0, there is a drastic decrease in their number (Puverel et al., 2011). Interestingly, RanBPM is first expressed at birth in the ovaries suggesting that it is essential from the very earliest stages of its expression since most germ cells are absent immediately after birth. Oocyte loss thus occurs between late embryogenesis and early postnatal development in RanBPM−/− females, and the vast majority of the oocytes fail to progress to the dictyate arrest. Early studies of meiosis in mouse fetal ovaries suggest that entry into the pachytene stage starts at E16 (Speed, 1982). Since the majority of the oocytes are between late pachytene and diplotene at the time of birth, the phenotype of RanBPM-deficient mice suggests that oocytes reach the pachytene stage unaffected but display the same meiotic arrest as observed in males. Importantly, this arrest, occurring at the same meiotic stage in both genders, suggests that RanBPM deletion affects mechanisms common to both male and female meiosis. This is even more interesting considering that the timing and synchrony of meiosis show striking differences between the two genders (Hunt & Hassold, 2002; Kolas et al., 2005). In females, the entire germ cell pool synchronously initiates meiosis in utero around E13.5 and meiosis transiently arrests at birth. By contrast, male meiosis is not initiated until puberty and spermatocyte development in the adult testis is continuous and proceeds without the arrest periods occurring in females (Nebel, Amarose, & Hacket, 1961). In addition, a growing number of mouse mutants for genes involved in meiotic progression display sexual dimorphism in fertility (Fig. 13.3). These studies revealed clear differences in the stringency of the meiotic events between males and females (Cohen, Pollack, & Pollard, 2006; Handel & Eppig, 1998; Matzuk & Lamb, 2002; Morelli & Cohen, 2005). Female meiosis is more “forgiving” to errors; oocytes with meiotic defects often reach a more advanced stage than spermatocytes and can give rise to aneuploid gametes. Thus, females derived from gene-targeting experiments of genes that affect meiosis display a variety of phenotypes, ranging from subfertility with production of aneuploid embryos to complete infertility. The same meiotic defect in spermatocytes almost always leads to a complete meiotic failure, absence of progression past the zygotene- or pachytene-like stage, and apoptosis of germ cell leading to infertility (Hunt & Hassold, 2002) (see also Fig. 13.3). Although the cellular mechanisms that regulate the Prophase I events, such as homologous recombination and synapsis, appear to be largely conserved between the two genders, these processes are subjects to a different level of control (commonly defined as “checkpoints”), which is more stringent in males than in females.
In order to gain insight into the mechanism of the meiotic defect caused by RanBPM deletion, here, we compare the RanBPM−/− mouse meiotic phenotype with those of other mutants, including some of the most recently characterized (Fig. 13.3). Most mouse models lacking proteins involved in the recombination process such as Spo1, known to initiate the DSBs possibly with the cooperation of Mei1, proteins involved in DNA repair such as ATM, Dmc1, the mismatch proteins Msh4 and Msh5, and the recently identified protein Trip13, exhibit a meiotic arrest in spermatogenesis during Prophase I (Baudat et al., 2000; de Vries et al., 1999; Kneitz et al., 2000; Libby et al., 2002; Pittman et al., 1998; Roig et al., 2010; Yoshida et al., 1998). This is caused by a failure of chromosomes to synapse properly, impairing the progression of spermatocytes beyond the zygotene stage. Since, as mentioned above, the mechanisms monitoring meiosis are less stringent in females, meiosis can progress further in females but most oocytes are eliminated soon after birth as the meiotic defects become apparent during the checkpoint process. By contrast, mutant females for Tex15, a gene encoding a protein recently identified as essential for DSBs repair and synapsis, retain normal fertility (Yang et al., 2008). In mice deficient for the synaptonemal complex proteins, spermatocytes also arrest at the zygotene–pachytene transition, with mutant females either retaining fertility (Scp3, Fkbp6) or being sterile (Syce1, Tex12) (Bolcun-Filas et al., 2009; Crackower et al., 2003; Hamer et al., 2008; Yuan et al., 2002). Throughout the first meiotic prophase, the sister chromatids are held together by cohesin complex proteins. These proteins, such as SMC1β and Rec8, are essential for proper recombination, completion of synapsis, and chiasmata formation, and mutants for these genes exhibit a similar arrest at the zygotene–pachytene transition (Revenkova et al., 2004; Xu et al., 2005). By contrast, in RanBPM mutant mice, meiosis progresses apparently normally up to the pachytene stage. At this stage, there is no obvious defect in chromosome synapsis, since SCP3 immunolabeling in spermatocytes is comparable between mutants and controls (Puverel et al., 2011). The distribution of phosphorylated histone γH2AX, which marks sites of DSBs at the leptotene stage and subsequently converges around the sex chromosomes to form the sex body, also appeared normal in RanBPM−/− spermatocytes. Thus, in RanBPM homozygous mutant males, meiosis still progresses slightly further than in the mouse mutants mentioned above, and the spermatocytes reach the pachytene stage with no obvious synapsis abnormalities. These data suggest that RanBPM-deficient spermatocytes are able to go through the first checkpoint, which ensures the complete synapsis of homologous chromosomes prior to entry into the pachytene stage, but may be stopped at a later checkpoint (Morelli & Cohen, 2005; Odorisio, Rodriguez, Evans, Clarke, & Burgoyne, 1998). Since recombination is required for the proper synapsis process in mice, we can also suggest that recombination is initiated properly in RanBPM−/− male mice. The mechanisms of synapsis and recombination in RanBPM−/− oocytes have not yet been elucidated, but the timing of meiotic progression in the embryo suggests that the defect is similar to that of males, making the RanBPM−/− mouse phenotype quite unique (see Fig. 13.3). A more detailed analysis of spermatocytes and oocytes using several markers of synapsis and recombination may help determine the exact nature of this defect apparently common to both genders. Nevertheless, we should note that, at this point in time, we still need to investigate whether the phenotype observed in these mutants is uniquely caused by the lack of RanBPM or whether possible compensatory effects by its homolog RanBP10 can influence its penetrance on gametogenesis.
RanBPM has been reported to interact with the MVH protein, a member of the DEAD-box family of genes encoding an ATP-dependent RNA helicase (Fujiwara et al., 1994; Shibata et al., 2004). MVH is part of a group of male-specific regulators that include Mili and Gasz; mutants for these genes display similar phenotypes, marked by a meiotic arrest at the zygotene–pachytene transition (Kuramochi-Miyagawa et al., 2004; Ma et al., 2009; Tanaka et al., 2000). These genes seem to be involved in posttranscriptional regulation during spermatogenesis via RNA processing. However, contrary to RanBPM−/− mice where both males and females are arrested at the pachytene stage, in these mutants, females are completely fertile. It is therefore difficult to link RanBPM and MVH by the phenotypes caused by the deletion of these genes.
One mutation that appears to be the closest to the RanBPM mutant phenotype in terms of timing of meiotic arrest in both genders is the one for the polyubiquitin gene, Ubb. Ubb−/− germ cells progress to the pachytene stage and form apparently normal synaptonemal complexes in both genders (Ryu et al., 2008). However, in males, spermatocytes do not mature beyond the pachytene stage and undergo cell death, while in females, although some oocytes reach the dictyate arrest, they are not able to complete meiosis I. Therefore, it would be of interest to determine whether the Ubb−/− phenotype can be correlated with the defects observed in RanBPM−/− mice. However, it is possible that the phenotypes caused by either Ubb or RanBPM loss have in common only the fact that they affect multiple pathways: RanBPM, by interacting with a variety of proteins through its scaffolding activity, and Ubb, in addition to its role in directing protein degradation, by affecting a great variety of other functions including membrane trafficking and transcriptional regulation.
9. ROLE OF RanBPM IN THE ADULT GONAD
As described above, in RanBPM−/− pubertal males, germ cell differentiation is arrested at Prophase I. Histological examination of mutant testes at 6 weeks of age revealed totally empty seminiferous tubules (Sertoli cell only), which is followed by tubular degeneration at later stages (Puverel et al., 2011) (Fig. 13.4). This severe phenotype is not typical of mutants with exclusive meiotic arrest. In fact, in mutant mice displaying an arrest in spermatocyte differentiation, the production of germ cells is usually maintained for a certain period despite the elimination of primary spermatocytes. For example, in mice lacking the cyclin-dependent kinase 2 gene, the alteration of chromosome synapsis leads to the elimination of spermatocytes at prophase I, yet some spermatocytes are still produced in 3-month-old mouse testes (Berthet, Aleem, Coppola, Tessarollo, & Kaldis, 2003; Ortega et al., 2003).
Figure 13.4.
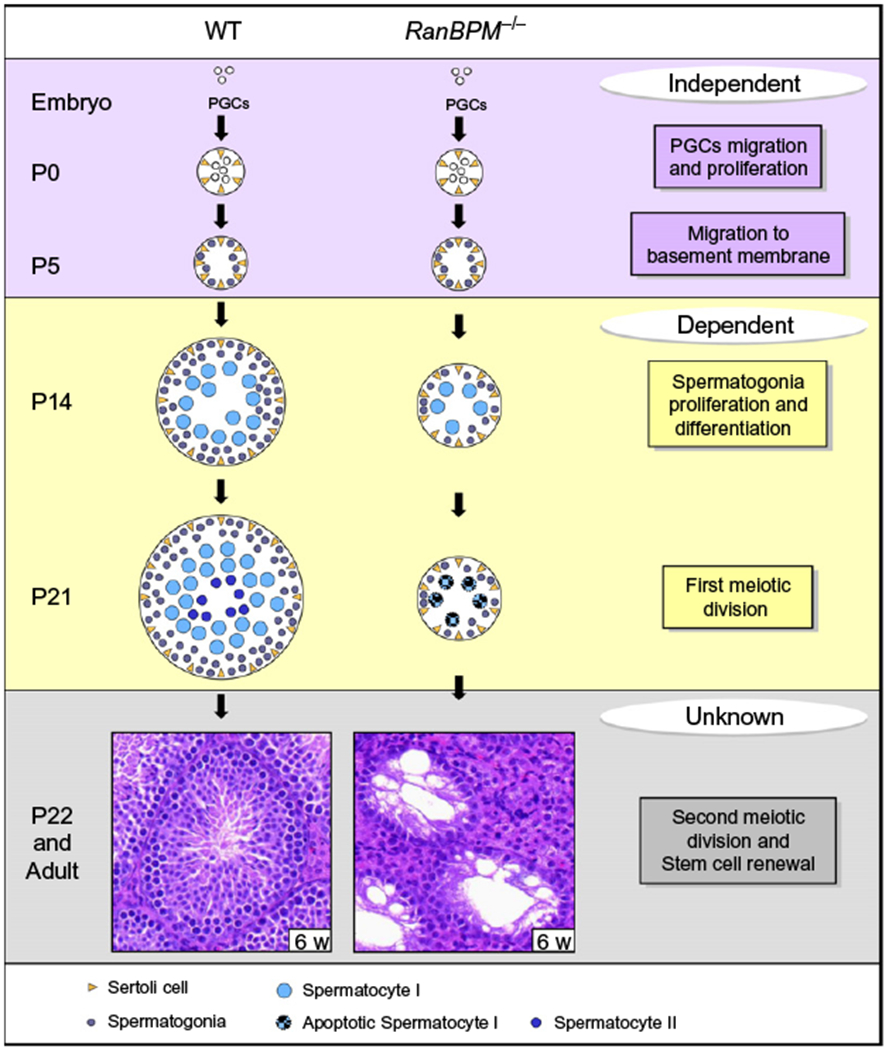
Model illustrating the role of RanBPM in spermatogenesis. Schematic representing the stages at which RanBPM plays a role in spermatogenesis. RanBPM is not required for primordial germ cell migration and proliferation and gonocyte migration to the basement membrane of developing seminiferous tubules in the newborn (purple box). However, it is required in the pubertal male at the crucial step of spermatogonia production and for the meiotic progression of primary spermatocytes (yellow box). Whether RanBPM is involved in the second meiotic division and the maintenance of adult spermatogenesis is still unknown, since the elimination of all germ cells in the young mutant does not allow the study of any time point past the meiotic arrest occurring during the prophase of the first meiotic division (gray box). P: postnatal day.
Instead, mice with defects in spermatogonial stem cells (SSCs) display a phenotype that more closely resembles the one observed in the RanBPM-deficient mice. SSCs provide differentiating cells by a self-renewal mechanism, thus keeping the spermatogenesis going (Oatley & Brinster, 2008). Some authors have suggested that SSCs are not involved in the first round of mouse spermatogenesis since the first wave of germline differentiation initiates directly from gonocytes (Yoshida et al., 2006). By using a transgenic system, these authors provided evidence that germ cells originating from gonocytes specifically contribute to the leading edge of pubertal spermatogenesis, generating the first spermatozoa that are released at around P35. Several factors have been shown to be involved in the stem cell renewal process, including the Glial cell line-derived neurotrophic factor Gdnf (Meng et al., 2000), the transcription factor Plzf (Buaas et al., 2004; Costoya et al., 2004), the ERM protein, produced by Sertoli cells (Chen et al., 2005), and the RNA-binding protein NANOS2 (Sada, Suzuki, Suzuki, & Saga, 2009). For example, null mice for GDNF and ERM show an arrest of germ cell production between 4 and 6 weeks (Chen et al., 2005; Meng et al., 2000). Therefore, the similarity of this phenotype with the one displayed by RanBPM−/− mutant animals, which are severely depleted of all germ cell types including transit-amplifying spermatogonia as early as 1 month of age, suggests that RanBPM may cause a defect in the spermatogonia self-renewal process as well (Puverel et al., 2011). However, the lack of germ cell production could also be related to the defect in spermatogonia proliferation observed in juvenile animals (Section 7). Taken together, the mutant phenotype displayed by RanBPM−/− males shows distinct features, suggesting that this scaffold protein may be critical in more than one mechanism in the control of spermatogenesis.
As outlined in females, RanBPM is crucial for normal oocyte development. Mutant females display a premature ovarian failure and only young adults still retain some follicles in their ovaries (Fig. 13.5). The measure of vaginal impedance in RanBPM−/− pubertal females indicates normal estrus cycles, suggesting that follicle and stromal cell functions are intact (Puverel et al., 2011). By contrast, mutant females older than 5 months are not cycling anymore, in accordance with the absence of follicles at this age. Thus, although mutant ovaries contain decreased numbers of follicles, these are sufficient to support normal estrus cycles. The finding that pubertal mutant females are cycling strongly suggests that the sterility of these mice is not caused by hormonal deficiencies and argues in favor of a cell-autonomous role of RanBPM in the oocyte. Oocytes orchestrate and coordinate the development of the mammalian ovarian follicles (Eppig, Wigglesworth, & Pendola, 2002). Following the superovulation treatment (PMSG, pregnantmare serumgonadotropin injection), RanBPM−/− females developed morphologically normal antral follicles, suggesting that follicles are able to mature in absence of RanBPM (Puverel et al., 2011). Whether RanBPM−/− females are able to ovulate and if oocytes are meiotically competent to give rise to normal embryos is still an open question.
Figure 13.5.
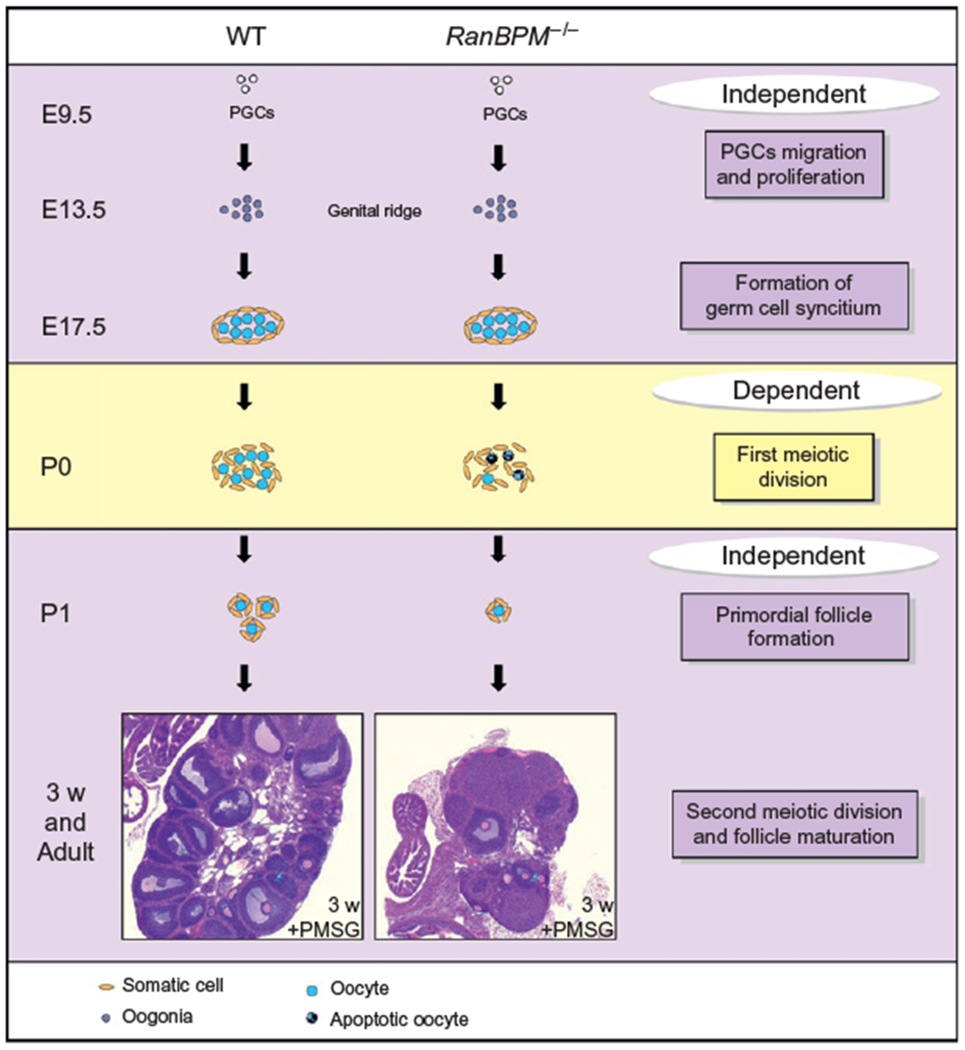
Model illustrating the role of RanBPM in oogenesis. Schematic representing the stages at which RanBPM plays a role in oogenesis. As in males, RanBPM is not essential for primordial germ cell proliferation and migration toward the developing gonad (top purple box). However, RanBPM is crucial for the meiotic progression of oocytes, since germ cells exhibit a meiotic arrest during the prophase of the first meiotic division, as in males (yellow box). The observation that some oocytes survive to form primordial follicles and that these follicles can mature upon PMSG stimulation suggests that RanBPM is not required for the maturation of germ cells in pubertal and adult mice, although the ability of these oocytes to produce embryos has not been assessed (bottom purple box). E: embryonic day and P: postnatal day.
10. CELL-AUTONOMOUS FUNCTION OF RanBPM
RanBPM transcripts are present in both the somatic and germinal cell lineages of the testis (Section 5), suggesting that the spermatogenesis defect displayed by RanBPM−/− mutant males could be either due to a defect in the Sertoli cell nursing function or to a defect intrinsic to the germ cells. We were able to address this issue by generating chimeric mice with homozygous mutant ES cells to study the extent of the ES cell contribution to the chimeras (Puverel et al., 2011). These experiments showed that while control cells could contribute to all cell types in the male testis, mutant ES cells failed to produce any germ cell while producing other cell types. Importantly, mice with testicular supporting cells produced by mutant ES cells had normal spermatogenesis. Together, these data suggest that RanBPM functions in a cell-autonomous fashion in male germ cells and that it is dispensable from supporting cells for spermatogenesis completion.
11. CONCLUSION
Since its first characterization, RanBPM has been the object of a growing interest from the scientific community. Devoid of any known functional domain, it has been suggested that RanBPM is a scaffolding protein important to assemble high molecular weight protein complexes that shape the activity of various signal transduction pathways. Functionally, RanBPM has been implicated in diverse biological roles, especially in the immune and nervous systems. However, with the recent generation and characterization of a mouse lacking this scaffolding protein, it has become apparent that RanBPM plays a crucial role in spermatogenesis and oogenesis. The main features of this mutant are (1) a defect in postnatal testis development suggesting that RanBPM is essential for spermatogonia production in the young male and (2) a meiotic arrest in both genders during the prophase of the first meiotic division, showing that RanBPM is required for normal progression of meiosis. Even though the exact mechanism of action of RanBPM in the gonad remains to be determined, the results from chimera experiments strongly suggest a cell-autonomous function of RanBPM in germ cells. Importantly, the unique phenotype of RanBPM mutant mice compared to mice lacking other genes critical for normal mouse reproduction (Fig. 13.3) suggests that this scaffold protein may influence the activity of multiple pathways or interact with other, yet unknown, proteins. Regardless, the characterization of RanBPM function in mouse gonad development provides new insight into some key events regulating mammalian gametogenesis.
REFERENCES
- Ara T, Nakamura Y, Egawa T, Sugiyama T, Abe K, Kishimoto T, et al. (2003). Impaired colonization of the gonads by primordial germ cells in mice lacking a chemokine, stromal cell-derived factor-1 (SDF-1). Proceedings of the National Academy of Sciences of the United States of America, 100, 5319–5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai D, Chen H, & Huang BR (2003). RanBPM is a novel binding protein for p75NTR. Biochemical and Biophysical Research Communications, 309, 552–557. [DOI] [PubMed] [Google Scholar]
- Baker SM, Bronner CE, Zhang L, Plug AW, Robatzek M, Warren G, et al. (1995). Male mice defective in the DNA mismatch repair gene PMS2 exhibit abnormal chromosome synapsis in meiosis. Cell, 82, 309–319. [DOI] [PubMed] [Google Scholar]
- Barlow C, Liyanage M, Moens PB, Tarsounas M, Nagashima K, Brown K, et al. (1998). Atm deficiency results in severe meiotic disruption as early as leptonema of prophase I. Development, 125, 4007–4017. [DOI] [PubMed] [Google Scholar]
- Baudat F, Manova K, Yuen JP, Jasin M, & Keeney S (2000). Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Molecular Cell, 6, 989–998. [DOI] [PubMed] [Google Scholar]
- Beddow AL, Richards SA, Orem NR, & Macara IG (1995). The Ran/TC4 GTPase-binding domain: Identification by expression cloning and characterization of a conserved sequence motif. Proceedings of the National Academy of Sciences of the United States of America, 92, 3328–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellve AR, Cavicchia JC,Millette CF, O’Brien DA, Bhatnagar YM, & Dym M. (1977). Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. The Journal of Cell Biology, 74, 68–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendel-Stenzel M, Anderson R, Heasman J, & Wylie C (1998). The origin and migration of primordial germ cells in the mouse. Seminars in Cell & Developmental Biology, 9, 393–400. [DOI] [PubMed] [Google Scholar]
- Berthet C, Aleem E, Coppola V, Tessarollo L, & Kaldis P (2003). Cdk2 knockout mice are viable. Current Biology, 13, 1775–1785. [DOI] [PubMed] [Google Scholar]
- Blume-Jensen P, Jiang G, Hyman R, Lee KF, O’Gorman S, & Hunter T (2000). Kit/stem cell factor receptor-induced activation of phosphatidylinositol 3’-kinase is essential for male fertility. Nature Genetics, 24, 157–162. [DOI] [PubMed] [Google Scholar]
- Bolcun-Filas E, Hall E, Speed R, Taggart M, Grey C, de Massy B, et al. (2009). Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair. PLoS Genetics, 5, e1000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolcun-Filas E, Bannister LA, Barash A, Schimenti KJ, Hartford SA, Eppig JJ, et al. (2011). A-MYB (MYBL1) transcription factor is a master regulator of male meiosis. Development, 138, 3319–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, et al. (2004). Plzf is required in adult male germ cells for stem cell self-renewal. Nature Genetics, 36, 647–652. [DOI] [PubMed] [Google Scholar]
- Buehr M, McLaren A, Bartley A, & Darling S (1993). Proliferation and migration of primordial germ cells in We/We mouse embryos. Developmental Dynamics, 198, 182–189. [DOI] [PubMed] [Google Scholar]
- Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, et al. (2002). Genomic instability in mice lacking histone H2AX. Science, 296, 922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Paramasivam M, Girgenti MJ, Walikonis RS, Bianchi E, & LoTurco JJ (2010). RanBPM regulates the progression of neuronal precursors through M-phase at the surface of the neocortical ventricular zone. Developmental Neurobiology, 70, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ouyang W, Grigura V, Zhou Q, Carnes K, Lim H, et al. (2005). ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature, 436, 1030–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Lemmon S, & Lemmon V (2005). RanBPM is an L1-interacting protein that regulates L1-mediated mitogen-activated protein kinase activation. Journal of Neurochemistry, 94, 1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen PE, Pollack SE, & Pollard JW (2006). Genetic analysis of chromosome pairing, recombination, and cell cycle control during first meiotic prophase in mammals. Endocrine Reviews, 27, 398–426. [DOI] [PubMed] [Google Scholar]
- Cohen PE, & Pollard JW (2001). Regulation of meiotic recombination and prophase I progression in mammals. BioEssays, 23, 996–1009. [DOI] [PubMed] [Google Scholar]
- Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, et al. (2004). Essential role of Plzf in maintenance of spermatogonial stem cells. Nature Genetics, 36, 653–659. [DOI] [PubMed] [Google Scholar]
- Crackower MA, Kolas NK, Noguchi J, Sarao R, Kikuchi K, Kaneko H, et al. (2003). Essential role of Fkbp6 in male fertility and homologous chromosome pairing in meiosis. Science, 300, 1291–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansereau DA, & Lasko P (2008). RanBPM regulates cell shape, arrangement, and capacity of the female germline stem cell niche in Drosophila melanogaster. The Journal of Cell Biology, 182, 963–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries SS, Baart EB, Dekker M, Siezen A, de Rooij DG, de Boer P, et al. (1999). Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes & Development, 13, 523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denti S, Sirri A, Cheli A, Rogge L, Innamorati G, Putignano S, et al. (2004). RanBPM is a phosphoprotein that associates with the plasma membrane and interacts with the integrin LFA-1. The Journal of Biological Chemistry, 279, 13027–13034. [DOI] [PubMed] [Google Scholar]
- Di Cunto F, Imarisio S, Hirsch E, Broccoli V, Bulfone A, Migheli A, et al. (2000). Defective neurogenesis in citron kinase knockout mice by altered cytokinesis and massive apoptosis. Neuron, 28, 115–127. [DOI] [PubMed] [Google Scholar]
- Dix DJ, Allen JW, Collins BW, Mori C, Nakamura N, Poorman-Allen P, et al. (1996). Targeted gene disruption of Hsp70-2 results in failed meiosis, germ cell apoptosis, and male infertility. Proceedings of the National Academy of Sciences of the United States of America, 93, 3264–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix DJ, Allen JW, Collins BW, Poorman-Allen P, Mori C, Blizard DR, et al. (1997). HSP70-2 is required for desynapsis of synaptonemal complexes during meiotic prophase in juvenile and adult mouse spermatocytes. Development, 124, 4595–4603. [DOI] [PubMed] [Google Scholar]
- Edelmann W, Cohen PE, Kneitz B, Winand N, Lia M, Heyer J, et al. (1999). Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nature Genetics, 21, 123–127. [DOI] [PubMed] [Google Scholar]
- Edson MA, Nagaraja AK, & Matzuk MM (2009). The mammalian ovary from genesis to revelation. Endocrine Reviews, 30, 624–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emes RD, & Ponting CP (2001). A new sequence motif linking lissencephaly, Treacher Collins and oral-facial-digital type 1 syndromes, microtubule dynamics and cell migration. Human Molecular Genetics, 10, 2813–2820. [DOI] [PubMed] [Google Scholar]
- Eppig JJ, Wigglesworth K, & Pendola FL (2002). The mammalian oocyte orchestrates the rate of ovarian follicular development. Proceedings of the National Academy of Sciences of the United States of America, 99, 2890–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, Romanienko PJ, Camerini-Otero RD, Bonner WM, et al. (2003). H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Developmental Cell, 4, 497–508. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Komiya T, Kawabata H, Sato M, Fujimoto H, Furusawa M, et al. (1994). Isolation of a DEAD-family protein gene that encodes a murine homolog of Drosophila vasa and its specific expression in germ cell lineage. Proceedings of the National Academy of Sciences of the United States of America, 91, 12258–12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlitz G, Darhin E, Giorgio G, Franco B, & Reiner O (2005). Novel functional features of the Lis-H domain: Role in protein dimerization, half-life and cellular localization. Cell Cycle, 4, 1632–1640. [DOI] [PubMed] [Google Scholar]
- Grad I, Cederroth CR, Walicki J, Grey C, Barluenga S, Winssinger N, et al. (2010). The molecular chaperone Hsp90alpha is required for meiotic progression of spermatocytes beyond pachytene in the mouse. PloS One, 5, e15770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer G, Wang H, Bolcun-Filas E, Cooke HJ, Benavente R, & Hoog C (2008). Progression of meiotic recombination requires structural maturation of the central element of the synaptonemal complex. Journal of Cell Science, 121, 2445–2451. [DOI] [PubMed] [Google Scholar]
- Handel MA, & Eppig JJ (1998). Sexual dimorphism in the regulation of mammalian meiosis. Current Topics in Developmental Biology, 37, 333–358. [DOI] [PubMed] [Google Scholar]
- Hassold T, & Hunt P (2001). To err (meiotically) is human: The genesis of human aneuploidy. Nature Reviews. Genetics, 2, 280–291. [DOI] [PubMed] [Google Scholar]
- Herran Y, Gutierrez-Caballero C, Sanchez-Martin M, Hernandez T, Viera A, Barbero JL, et al. (2011). The cohesin subunit RAD21L functions in meiotic synapsis and exhibits sexual dimorphism in fertility. The EMBO Journal, 30, 3091–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosono K, Noda S, Shimizu A, Nakanishi N, Ohtsubo M, Shimizu N, et al. (2010). YPEL5 protein of the YPEL gene family is involved in the cell cycle progression by interacting with two distinct proteins RanBPM and RanBP10. Genomics, 96, 102–111. [DOI] [PubMed] [Google Scholar]
- Hu YC, Wang PH, Yeh S, Wang RS, Xie C, Xu Q, et al. (2004). Subfertility and defective folliculogenesis in female mice lacking androgen receptor. Proceedings of the National Academy of Sciences of the United States of America, 101, 11209–11214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt PA, & Hassold TJ (2002). Sex matters in meiosis. Science, 296, 2181–2183. [DOI] [PubMed] [Google Scholar]
- Ideguchi H, Ueda A, Tanaka M, Yang J, Tsuji T, Ohno S, et al. (2002). Structural and functional characterization of the USP11 deubiquitinating enzyme, which interacts with the RanGTP-associated protein RanBPM. The Biochemical Journal, 367, 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagarlamudi K, Reddy P, Adhikari D, & Liu K (2010). Genetically modified mouse models for premature ovarian failure (POF). Molecular and Cellular Endocrinology, 315, 1–10. [DOI] [PubMed] [Google Scholar]
- Judis L, Chan ER, Schwartz S, Seftel A, & Hassold T (2004). Meiosis I arrest and azoospermia in an infertile male explained by failure of formation of a component of the synaptonemal complex. Fertility and Sterility, 81, 205–209. [DOI] [PubMed] [Google Scholar]
- Kim MH, Cooper DR, Oleksy A, Devedjiev Y, Derewenda U, Reiner O, et al. (2004). The structure of the N-terminal domain of the product of the lissencephaly gene Lis1 and its functional implications. Structure, 12, 987–998. [DOI] [PubMed] [Google Scholar]
- Kim S, Namekawa SH, Niswander LM, Ward JO, Lee JT, Bardwell VJ, et al. (2007). A mammal-specific Doublesex homolog associates with male sex chromatin and is required for male meiosis. PLoS Genetics, 3, e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissel H, Timokhina I, Hardy MP, Rothschild G, Tajima Y, Soares V, et al. (2000). Point mutation in kit receptor tyrosine kinase reveals essential roles for kit signaling in spermatogenesis and oogenesis without affecting other kit responses. The EMBO Journal, 19, 1312–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink A, Lee M, & Cooke HJ (1997). The mouse synaptosomal complex protein gene Sycp3 maps to band C of chromosome 10. Mammalian Genome, 8, 376–377. [DOI] [PubMed] [Google Scholar]
- Kneitz B, Cohen PE, Avdievich E, Zhu L, Kane MF, Hou H Jr., et al. (2000). MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes & Development, 14, 1085–1097. [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N, Yang J, Ueda A, Suzuki T, Tomaru K, Takeno M, et al. (2007). RanBPM, Muskelin, p48EMLP, p44CTLH, and the armadillo-repeat proteins ARMC8alpha and ARMC8beta are components of the CTLH complex. Gene, 396, 236–247. [DOI] [PubMed] [Google Scholar]
- Kolas NK, Marcon E, Crackower MA, Hoog C, Penninger JM, Spyropoulos B, et al. (2005). Mutant meiotic chromosome core components in mice can cause apparent sexual dimorphic endpoints at prophase or X-Y defective male-specific sterility. Chromosoma, 114, 92–102. [DOI] [PubMed] [Google Scholar]
- Kunert S, Meyer I, Fleischhauer S, Wannack M, Fiedler J, Shivdasani RA, et al. (2009). The microtubule modulator RanBP10 plays a critical role in regulation of platelet discoid shape and degranulation. Blood, 114, 5532–5540. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, et al. (2004). Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development, 131, 839–849. [DOI] [PubMed] [Google Scholar]
- Lakshmana MK, Chung JY, Wickramarachchi S, Tak E, Bianchi E, Koo EH, et al. (2010). A fragment of the scaffolding protein RanBP9 is increased in Alzheimer’s disease brains and strongly potentiates amyloid-beta peptide generation. The FASEB Journal, 24, 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmana MK, Yoon IS, Chen E, Bianchi E, Koo EH, & Kang DE (2009). Novel role of RanBP9 in BACE1 processing of amyloid precursor protein and amyloid beta peptide generation. The Journal of Biological Chemistry, 284, 11863–11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Nair M, Mackay DR, Bilanchone V, Hu M, Fallahi M, et al. (2005). Ovol1 regulates meiotic pachytene progression during spermatogenesis by repressing Id2 expression. Development, 132, 1463–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XC, & Schimenti JC (2007). Mouse pachytene checkpoint 2 (trip13) is required for completing meiotic recombination but not synapsis. PLoS Genetics, 3, e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby BJ, De La Fuente R, O’Brien MJ, Wigglesworth K, Cobb J, Inselman A, et al. (2002). The mouse meiotic mutation mei1 disrupts chromosome synapsis with sexually dimorphic consequences for meiotic progression. Developmental Biology, 242, 174–187. [DOI] [PubMed] [Google Scholar]
- Liu D, Liao C, & Wolgemuth DJ (2000). A role for cyclin A1 in the activation of MPF and G2-M transition during meiosis of male germ cells in mice. Developmental Biology, 224, 388–400. [DOI] [PubMed] [Google Scholar]
- Liu D, Matzuk MM, Sung WK, Guo Q, Wang P, & Wolgemuth DJ (1998). Cyclin A1 is required for meiosis in the male mouse. Nature Genetics, 20, 377–380. [DOI] [PubMed] [Google Scholar]
- Ma L, Buchold GM, Greenbaum MP, Roy A, Burns KH, Zhu H, et al. (2009). GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genetics, 5, e1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahakali Zama A, Hudson FP 3rd, & Bedell MA (2005). Analysis of hypomorphic KitlSl mutants suggests different requirements for KITL in proliferation and migration of mouse primordial germ cells. Biology of Reproduction, 73, 639–647. [DOI] [PubMed] [Google Scholar]
- Mateja A, Cierpicki T, Paduch M, Derewenda ZS, & Otlewski J (2006). The dimerization mechanism of LIS1 and its implication for proteins containing the LisH motif. Journal of Molecular Biology, 357, 621–631. [DOI] [PubMed] [Google Scholar]
- Matzuk MM, & Lamb DJ (2002). Genetic dissection of mammalian fertility pathways. Nature Cell Biology, 4(Suppl.), s41–s49. [DOI] [PubMed] [Google Scholar]
- Meng X, Lindahl M, Hyvonen ME, Parvinen M, de Rooij DG, Hess MW, et al. (2000). Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science, 287, 1489–1493. [DOI] [PubMed] [Google Scholar]
- Menon RP, Gibson TJ, & Pastore A (2004). The C terminus of fragile X mental retardation protein interacts with the multi-domain Ran-binding protein in the microtubule-organising centre. Journal of Molecular Biology, 343, 43–53. [DOI] [PubMed] [Google Scholar]
- Mettus RV, Litvin J, Wali A, Toscani A, Latham K, Hatton K, et al. (1994). Murine A-myb: Evidence for differential splicing and tissue-specific expression. Oncogene, 9, 3077–3086. [PubMed] [Google Scholar]
- Meuwissen RL, Offenberg HH, Dietrich AJ, Riesewijk A, van Iersel M, & Heyting C (1992). A coiled-coil related protein specific for synapsed regions of meiotic prophase chromosomes. The EMBO Journal, 11, 5091–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T, Hasuike S, Yogev L, Maduro MR, Ishikawa M, Westphal H, et al. (2003). Azoospermia in patients heterozygous for a mutation in SYCP3. Lancet, 362, 1714–1719. [DOI] [PubMed] [Google Scholar]
- Molyneaux KA, Zinszner H, Kunwar PS, Schaible K, Stebler J, Sunshine MJ, et al. (2003). The chemokine SDF1/CXCL12 and its receptor CXCR4 regulate mouse germ cell migration and survival. Development, 130, 4279–4286. [DOI] [PubMed] [Google Scholar]
- Morelli MA, & Cohen PE (2005). Not all germ cells are created equal: Aspects of sexual dimorphism in mammalian meiosis. Reproduction, 130, 761–781. [DOI] [PubMed] [Google Scholar]
- Murrin LC, & Talbot JN (2007). RanBPM, a scaffolding protein in the immune and nervous systems. Journal of Neuroimmune Pharmacology, 2, 290–295. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Masuda H, Horii J, Kuma K, Yokoyama N, Ohba T, et al. (1998). When overexpressed, a novel centrosomal protein, RanBPM, causes ectopic microtubule nucleation similar to gamma-tubulin. The Journal of Cell Biology, 143, 1041–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebel BR, Amarose AP, & Hacket EM (1961). Calendar of gametogenic development in the prepuberal male mouse. Science, 134, 832–833. [DOI] [PubMed] [Google Scholar]
- Nishitani H, Hirose E, Uchimura Y, Nakamura M, Umeda M, Nishii K, et al. (2001). Full-sized RanBPM cDNA encodes a protein possessing a long stretch of proline and glutamine within the N-terminal region, comprising a large protein complex. Gene, 272, 25–33. [DOI] [PubMed] [Google Scholar]
- Oatley JM, & Brinster RL (2008). Regulation of spermatogonial stem cell self-renewal in mammals. Annual Review of Cell and Developmental Biology, 24, 263–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorisio T, Rodriguez TA, Evans EP, Clarke AR, & Burgoyne PS. (1998). The meiotic checkpoint monitoring synapsis eliminates spermatocytes via p53-independent apoptosis. Nature Genetics, 18, 257–261. [DOI] [PubMed] [Google Scholar]
- Ortega S, Prieto I, Odajima J, Martin A, Dubus P, Sotillo R, et al. (2003). Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nature Genetics, 35, 25–31. [DOI] [PubMed] [Google Scholar]
- Page SL, & Hawley RS (2004). The genetics and molecular biology of the synaptonemal complex. Annual Review of Cell and Developmental Biology, 20, 525–558. [DOI] [PubMed] [Google Scholar]
- Pittman DL, Cobb J, Schimenti KJ, Wilson LA, Cooper DM, Brignull E, et al. (1998). Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Molecular Cell, 1, 697–705. [DOI] [PubMed] [Google Scholar]
- Ponting C, Schultz J, & Bork P (1997). SPRY domains in ryanodine receptors (Ca(2+)-release channels). Trends in Biochemical Sciences, 22, 193–194. [DOI] [PubMed] [Google Scholar]
- Puverel S, Barrick C, Dolci S, Coppola V, & Tessarollo L (2011). RanBPM is essential for mouse spermatogenesis and oogenesis. Development, 138, 2511–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MA, Cheng H, Quayle AN, Nishitani H, Nelson CC, & Rennie PS (2002). RanBPM, a nuclear protein that interacts with and regulates transcriptional activity of androgen receptor and glucocorticoid receptor. The Journal of Biological Chemistry, 277, 48020–48027. [DOI] [PubMed] [Google Scholar]
- Revenkova E, Eijpe M, Heyting C, Hodges CA, Hunt PA, Liebe B, et al. (2004). Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nature Cell Biology, 6, 555–562. [DOI] [PubMed] [Google Scholar]
- Roig I, Dowdle JA, Toth A, de Rooij DG, Jasin M, & Keeney S (2010). Mouse TRIP13/PCH2 is required for recombination and normal higher-order chromosome structure during meiosis. PLoS Genetics, 6, e1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanienko PJ, & Camerini-Otero RD (2000). The mouse Spo11 gene is required for meiotic chromosome synapsis. Molecular Cell, 6, 975–987. [DOI] [PubMed] [Google Scholar]
- Rosario MO, Perkins SL, O’Brien DA, Allen RL, & Eddy EM (1992). Identification of the gene for the developmentally expressed 70 kDa heat-shock protein (P70) of mouse spermatogenic cells. Developmental Biology, 150, 1–11. [DOI] [PubMed] [Google Scholar]
- Rossi P, Dolci S, Sette C, Capolunghi F, Pellegrini M, Loiarro M, et al. (2004). Analysis of the gene expression profile of mouse male meiotic germ cells. Gene Expression Patterns, 4, 267–281. [DOI] [PubMed] [Google Scholar]
- Rubin MR, Toth LE, Patel MD, D’Eustachio P, & Nguyen-Huu MC. (1986). A mouse homeo box gene is expressed in spermatocytes and embryos. Science, 233, 663–667. [DOI] [PubMed] [Google Scholar]
- Runyan C, Schaible K, Molyneaux K, Wang Z, Levin L, & Wylie C (2006). Steel factor controls midline cell death of primordial germ cells and is essential for their normal proliferation and migration. Development, 133, 4861–4869. [DOI] [PubMed] [Google Scholar]
- Ryu KY, Sinnar SA, Reinholdt LG, Vaccari S, Hall S, Garcia MA, et al. (2008). The mouse polyubiquitin gene Ubb is essential for meiotic progression. Molecular and Cellular Biology, 28, 1136–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sada A, Suzuki A, Suzuki H, & Saga Y (2009). The RNA-binding protein NANOS2 is required to maintain murine spermatogonial stem cells. Science, 325, 1394–1398. [DOI] [PubMed] [Google Scholar]
- Schramm S, Fraune J, Naumann R, Hernandez-Hernandez A, Hoog C, Cooke HJ, et al. (2011). A novel mouse synaptonemal complex protein is essential for loading of central element proteins, recombination, and fertility. PLoS Genetics, 7, e1002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seebahn A, Rose M, & Enz R (2008). RanBPM is expressed in synaptic layers of the mammalian retina and binds to metabotropic glutamate receptors. FEBS Letters, 582, 2453–2457. [DOI] [PubMed] [Google Scholar]
- Sharan SK, Pyle A, Coppola V, Babus J, Swaminathan S, Benedict J, et al. (2004). BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development, 131, 131–142. [DOI] [PubMed] [Google Scholar]
- Shibata N, Tsunekawa N, Okamoto-Ito S, Akasu R, Tokumasu A, & Noce T (2004). Mouse RanBPM is a partner gene to a germline specific RNA helicase, mouse vasa homolog protein. Molecular Reproduction and Development, 67, 1–7. [DOI] [PubMed] [Google Scholar]
- Shiina H, Matsumoto T, Sato T, Igarashi K, Miyamoto J, Takemasa S, et al. (2006). Premature ovarian failure in androgen receptor-deficient mice. Proceedings of the National Academy of Sciences of the United States of America, 103, 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed RM (1982). Meiosis in the foetal mouse ovary. I. An analysis at the light microscope level using surface-spreading. Chromosoma, 85, 427–437. [DOI] [PubMed] [Google Scholar]
- Suresh B, Ramakrishna S, & Baek KH (2012). Diverse roles of the scaffolding protein RanBPM. Drug Discovery Today, 17, 379–387. [DOI] [PubMed] [Google Scholar]
- Sweeney C, Murphy M, Kubelka M, Ravnik SE, Hawkins CF, Wolgemuth DJ, et al. (1996). A distinct cyclin A is expressed in germ cells in the mouse. Development, 122, 53–64. [DOI] [PubMed] [Google Scholar]
- Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, et al. (2000). The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes & Development, 14, 841–853. [PMC free article] [PubMed] [Google Scholar]
- Tang X, Zhang J, Cai Y, Miao S, Zong S, Koide SS, et al. (2004). Sperm membrane protein (hSMP-1) and RanBPM complex in the microtubule-organizing centre. Journal of Molecular Medicine, 82, 383–388. [DOI] [PubMed] [Google Scholar]
- Tay J, & Richter JD (2001). Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Developmental Cell, 1, 201–213. [DOI] [PubMed] [Google Scholar]
- Togashi H, Schmidt EF, & Strittmatter SM (2006). RanBPM contributes to Semaphorin3A signaling through plexin-A receptors. The Journal of Neuroscience, 26, 4961–4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toscani A, Mettus RV, Coupland R, Simpkins H, Litvin J, Orth J, et al. (1997). Arrest of spermatogenesis and defective breast development in mice lacking A-myb. Nature, 386, 713–717. [DOI] [PubMed] [Google Scholar]
- Tres LL, Rosselot C, & Kierszenbaum AL (2004). Primordial germ cells: What does it take to be alive? Molecular Reproduction and Development, 68, 1–4. [DOI] [PubMed] [Google Scholar]
- Valiyaveettil M, Bentley AA, Gursahaney P, Hussien R, Chakravarti R, Kureishy N, et al. (2008). Novel role of the muskelin-RanBP9 complex as a nucleocytoplasmic mediator of cell morphology regulation. The Journal of Cell Biology, 182, 727–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viera A, Rufas JS, Martinez I, Barbero JL, Ortega S, & Suja JA (2009). CDK2 is required for proper homologous pairing, recombination and sex-body formation during male mouse meiosis. Journal of Cell Science, 122, 2149–2159. [DOI] [PubMed] [Google Scholar]
- Wang D, Li Z, Messing EM, & Wu G (2002). Activation of Ras/Erk pathway by a novel MET-interacting protein RanBPM. The Journal of Biological Chemistry, 277, 36216–36222. [DOI] [PubMed] [Google Scholar]
- Wang RS, Yeh S, Tzeng CR, & Chang C (2009). Androgen receptor roles in spermatogenesis and fertility: Lessons from testicular cell-specific androgen receptor knockout mice. Endocrine Reviews, 30, 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe D, Yamada K, Nishina Y, Tajima Y, Koshimizu U, Nagata A, et al. (1994). Molecular cloning of a novel Ca(2+)-binding protein (calmegin) specifically expressed during male meiotic germ cell development. The Journal of Biological Chemistry, 269, 7744–7749. [PubMed] [Google Scholar]
- Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, et al. (2011). MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Developmental Cell, 20, 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo JA, Jung AR, Lakshmana MK, Bedrossian A, Lim Y, Bu JH, et al. (2012). Pivotal role of the RanBP9-cofilin pathway in Abeta-induced apoptosis and neurodegeneration. Cell Death and Differentiation, 19, 1413–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Aprelikova O, Moens P, Deng CX, & Furth PA (2003). Impaired meiotic DNA-damage repair and lack of crossing-over during spermatogenesis in BRCA1 full-length isoform deficient mice. Development, 130, 2001–2012. [DOI] [PubMed] [Google Scholar]
- Xu H, Beasley MD, Warren WD, van der Horst GT, & McKay MJ (2005). Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Developmental Cell, 8, 949–961. [DOI] [PubMed] [Google Scholar]
- Yang F, De La Fuente R, Leu NA, Baumann C, McLaughlin KJ, & Wang PJ (2006). Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. The Journal of Cell Biology, 173, 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Eckardt S, Leu NA, McLaughlin KJ, & Wang PJ. (2008). Mouse TEX15 is essential for DNA double-strand break repair and chromosomal synapsis during male meiosis. The Journal of Cell Biology, 180, 673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, & Wang PJ (2009). The Mammalian synaptonemal complex: A scaffold and beyond. Genome Dynamics, 5, 69–80. [DOI] [PubMed] [Google Scholar]
- Yin YX, Sun ZP, Huang SH, Zhao L, Geng Z, & Chen ZY (2010). RanBPM contributes to TrkB signaling and regulates brain-derived neurotrophic factor-induced neuronal morphogenesis and survival. Journal of Neurochemistry, 114, 110–121. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Kondoh G, Matsuda Y, Habu T, Nishimune Y, & Morita T. (1998). The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Molecular Cell, 1, 707–718. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Sukeno M, Nakagawa T, Ohbo K, Nagamatsu G, Suda T, et al. (2006). The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development, 133, 1495–1505. [DOI] [PubMed] [Google Scholar]
- Yuan L, Liu JG, Zhao J, Brundell E, Daneholt B, & Hoog C (2000). The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Molecular Cell, 5, 73–83. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Fu C, Chen H, Wang X, Deng W, & Huang BR. (2006). The Ran binding protein RanBPM interacts with TrkA receptor. Neuroscience Letters, 407, 26–31. [DOI] [PubMed] [Google Scholar]
- Yuan L, Liu JG, Hoja MR, Wilbertz J, Nordqvist K, & Hoog C (2002). Female germ cell aneuploidy and embryo death in mice lacking the meiosis-specific protein SCP3. Science, 296, 1115–1118. [DOI] [PubMed] [Google Scholar]
- Zakeri ZF, Wolgemuth DJ, & Hunt CR (1988). Identification and sequence analysis of a new member of the mouse HSP70 gene family and characterization of its unique cellular and developmental pattern of expression in the male germ line. Molecular and Cellular Biology, 8, 2925–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, & Kleckner N (1999). Meiotic chromosomes: Integrating structure and function. Annual Review of Genetics, 33, 603–754. [DOI] [PubMed] [Google Scholar]


