To the Editor:
At the time of this report, more than 20 million people have been infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Disease pathophysiology suggests the virus initially enters the nasal cavities (1) and then infects the ciliated airway epithelium (2). Often, there is an excessive inflammatory response to the virus mediated by overexpressed TNF-α (tumor necrosis factor-α), IL-6, and IL-1β (3), which leads to significant damage to the integrity and function of the lung parenchyma, causing death in the most vulnerable populations (4). To date, additional treatments against SARS-CoV-2 infections remain needed.
An interesting drug candidate against SARS-CoV-2 is azithromycin, a drug with recognized antiinflammatory (5) and epithelial repair effects (6) already being used in the treatment of chronic obstructive pulmonary disease and cystic fibrosis (7). However, its role in the regulation of TMPRSS2 (transmembrane serine protease 2), ACE2 (angiotensin converting enzyme 2), and TMPRSS11D (transmembrane serine protease 11D) genes, coding for proteins necessary for SARS-CoV-2 activation, infection, and transmission, respectively (2), remains to be further investigated.
Methods
Briefly, three previously enrolled patients who were part of a larger descriptive study were asked to participate in this pilot study. These patients had a diagnosis of chronic rhinosinusitis according to the published American Association of Otorhinolaryngology - Head and Neck Surgery guidelines and were scheduled for endoscopic sinus surgery. A nasal biopsy at the level of the anterior ethmoid bulla was taken at the time of surgery. Three male patients of age 41, 49, and 53 years with no significant comorbidities other than chronic obstructive pulmonary disease in the latter were the sources of the nasal biopsies. No patient had received oral corticosteroids or topical or systemic antibiotic therapy in the preceding 30 days. All subjects had ceased topical intranasal corticosteroids 14 days before surgery.
Primary airway nasal epithelial cells were isolated from biopsies of the anterior ethmoid bulla and cultured according to a modified protocol from Maillé and colleagues (8). Through immunohistochemistry, the freshly isolated cell suspension was characterized to be composed of basal (cytokeratine 13–positive cells), ciliated (βIV-tubulin–positive cells), and goblet (MUC5AC-positive cells) nasal epithelial cells (Figure E1 in the data supplement). These cell types have all been described as expressing ACE2 and harbor the potential of sustaining a SARS-CoV-2 infection (9). To obtain a uniform and consistent cell population during our experiments with azithromycin treatment, this cell suspension was then expanded for 5–7 days, leading to a homogenous cell culture, predominantly composed from progenitor basal cells.
Based on previous azithromycin toxicity studies on human bronchial airway epithelial cells, the plate was treated with 10 μg/ml of azithromycin diluted in DMSO (Sigma-Aldrich) or a mock.
RNA was extracted from these cultures treated with azithromycin or mock. Then, samples for microarray studies were prepared using the Illumina RNA Amplification TotalPrep kit from Ambion (Life Technologies) and collected with the Illumina Bead Array Reader (IIlumina). Raw gene expression data was preprocessed, and pathway analysis was performed using the gene set enrichment analysis. Differential Gene Expression was then performed using the LIMMA package from Bioconductor (10). For a more detailed Methods section, refer to the data supplement.
Results
Pathway analysis using gene set enrichment analysis showed that cultures treated with 10 μg of azithromycin demonstrated a significant downregulation in serine hydrolase activity pathway (normalized enrichment score [NES] = −1.8720, P = 0.0020) together with endocytosis (NES = −1.6866, P = 0.0020) and receptor-mediated endocytosis pathway (NES = −1.5139, P = 0.0124). This is particularly interesting because the strongest associated genes included TMPRSS2 and TMPRSS11D.
Azithromycin’s antiinflammatory properties were also demonstrated by a significant downregulation of Hallmark and Gene Ontology canonical inflammatory response pathways (NES = −2.0729, P = 0.0005 and NES = −2.0569, P = 0.0020, respectively) together with IFN-γ and IFN-α pathways (NES = −2.1717, P = 0.0005 and NES = −2.1484, P = 0.0005, respectively). Moreover, downregulation of key IL signaling pathways, including IL-2, IL-6, and IL-8, was also seen.
Interestingly, Gene Ontology’s sterol biosynthetic process and Hallmark’s cholesterol homeostasis were upregulated (NES = 3.0991, P = 0.0020 and NES = 3.0543, P = 0.0005, respectively). Selected significant pathways are presented in Figure 1A and summarized in Table E1. A full table of all significantly modulated canonical pathways are presented in Table E3.
Figure 1.
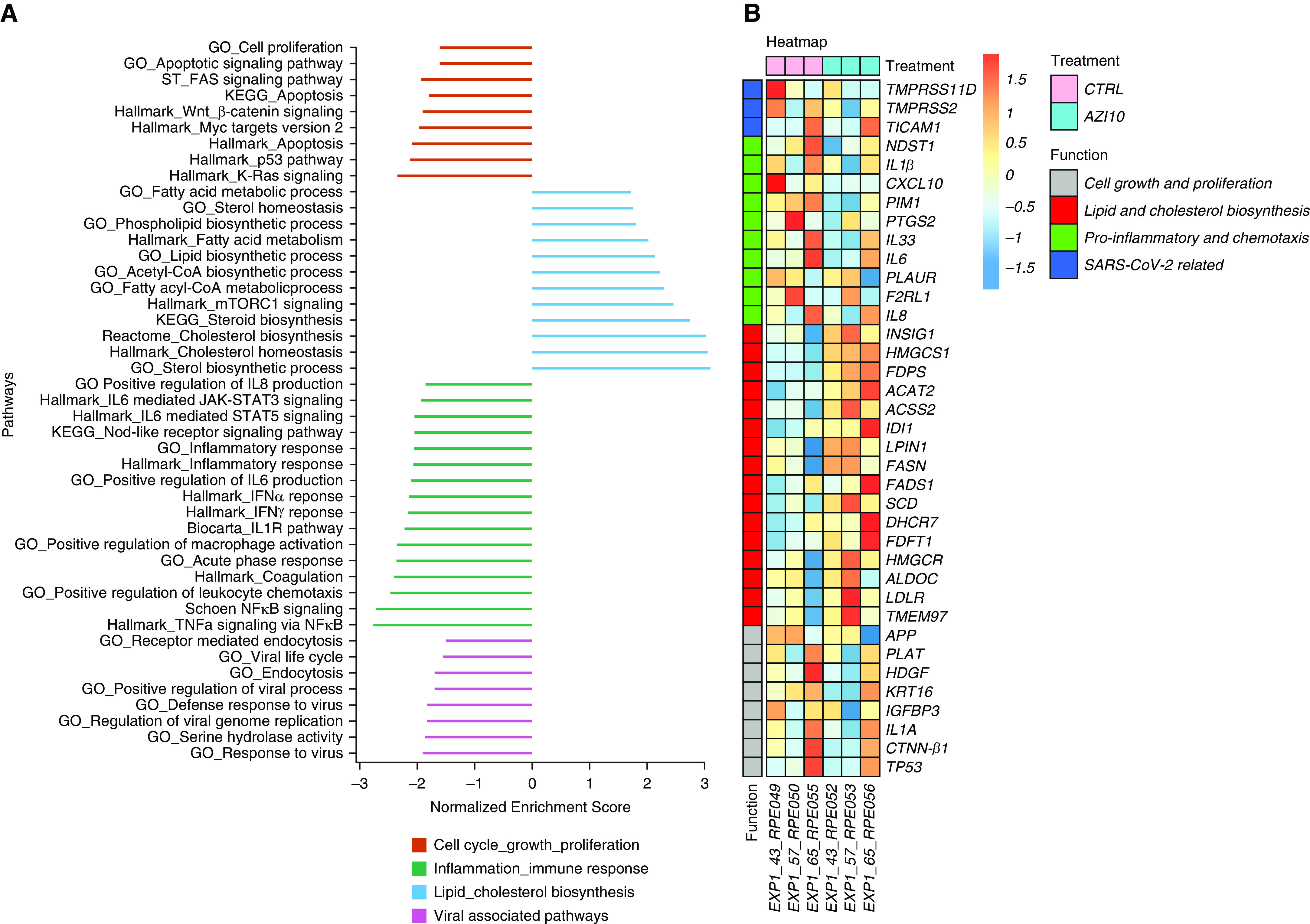
Host transcriptional response to azithromycin in basal nasal epithelial cells. (A) Gene set expression analysis comparing differential expression of a custom selection of major pathways of biological interest from MsigDB, Hallmark, C2, and C5 gene set collections. All pathways present have a false discovery rate < 0.05. Data is presented as normalized enrichment scores in which values > 0 represent upregulation and values <0 represent downregulation when comparing azithromycin-treated cell culture with mock-treated cell cultures. (B) Heatmap of a custom selection of differentially expressed genes between cell cultures treated with azithromycin and mock-treated cell cultures. Selected genes were based on biological relevance. ACAT2 = acetyl-CoA acetyltransferase 2; ACSS2 = acyl-CoA synthetase short chain family member 2; ALDOC = aldolase, fructose-bisphosphate C; APP = amyloid beta precursor protein; CTNN-β1 = catenin β1; CTRL = control; CXCL10 = C-X-C motif chemokine ligand 10; DHCR7 = 7-dehydrocholesterol reductase; F2RLI = F2R like trypsin receptor 1; FADS1 = fatty acid desaturase 1; FASN = fatty acid synthase; FDFT1 = farnesyl-diphosphate farnesyltransferase 1; FDPS = farnesyl diphosphate synthase; GO = Gene Ontology; HDGF = heparin binding growth factor; HMGCR = 3-hydroxy-3-methylglutaryl-CoA reductase; HMGCS1 = 3-hydroxy-3-methylglutaryl-CoA synthase 1; IDI-1 = isopentenyl-diphosphate Δ isomerase 1; IGFBP3 = insulin like growth factor binding protein 3; INSIG-1 = insulin induced gene 1; KEGG = Kyoto Encyclopedia of Genes and Genomes; KRT-16 = keratin 16; LDLR = low density lipoprotein receptor; LPIN-1 = lipin 1; MYC = MYC proto-oncogene; NDST1 = N-deacetylase and N-sulfotransferase 1; PIM-1 = Pim-1 proto-oncogene, serine/threonine kinase; PLAT = plasminogen activator, tissue type; PLAUR = plasminogen activator, urokinase receptor; PTGS2 = prostaglandin-endoperoxide synthase 2; SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2; SCD = stearoyl-CoA desaturase; TICAM1 = Toll-like receptor adaptor molecule 1; TMEM97 = transmembrane protein 97; TMPRSS2 = transmembrane serine protease 2; TMPRSS11D = transmembrane serine protease 11D; TP53 = tumour protein 53.
Differential Gene Expression of cultures treated with 10 μg of azithromycin demonstrated a significant downregulation of IL-1β (fold change = −1.411, P = 0.0094) and NDST-1 (fold change = −1.345, P = 0.0276).
Interestingly, within the lipid and cholesterol biosynthesis pathways, most of its individual genes were significantly upregulated. A display of selected genes is found in Figure 1B and Table E2. A full table of all tested genes are presented in Table E4.
With this study, we provide some evidence that azithromycin downregulates key pathways involving genes TMPRSS2 and TMPRSS11D, which code for two serine proteases required by SARS-CoV-2 for its activation (2) and cell-to-cell transmission (11), respectively.
Furthermore, downregulating IL-1β and NDST-1 (12) together with associated inflammation and leukocyte recruitment pathways may help reduce the characteristic excessive respiratory epithelial inflammation, a key feature of SARS-CoV-2 infection.
Finally, the unexpected upregulation of multiple genes involved in cholesterol biosynthesis is believed to be a process known as drug-induced phospholipidosis, which may decrease cholesterol in cell membrane lipid rafts (5). This may hinder SARS-CoV-2 infection, as in vitro studies demonstrated that depletion of cholesterol in the cell membrane resulted in decreased SARS-CoV-1 cell infection (13, 14). Moreover, our data are in line with a previous in vitro study in which azithromycin upregulated lipid and cholesterol pathways while decreasing important proinflammatory cytokines in differentiated human bronchial epithelial cell cultures (15).
Our study should, however, be interpreted with caution because it is limited by its small sample size, the inclusion of only a male population, and the lack of experiments validating that the observed changes in gene expression had an impact on protein levels. Nevertheless, our findings harbor significant information to better orient larger in vivo or clinical studies on future treatments against SARS-CoV-2 infections.
Supplementary Material
Footnotes
Author Contributions: A.E.R.: conception of study, data analysis, literature review, and writing of manuscript. L.M.E.: study design, data collection, patient recruitment, and in vitro experiments. D.A.: immunohistochemistry data collection and results. A.F.-M.: gene and pathway expression data processing and analysis. A.M.: conception of study. S.R.: conception of study and study design. E.B.: conception of study and study design. S.G.: study design, patient recruitment, and in vitro experiments. M.D.: conception of study and study design. All authors contributed to the correction and revision of the manuscript.
This letter has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2020-0285LE on August 28, 2020
Author disclosures are available with the text of this letter at www.atsjournals.org.
References
- 1.Lechien JR, Chiesa-Estomba CM, De Siati DR, Horoi M, Le Bon SD, Rodriguez A, et al. Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID-19): a multicenter European study. Eur Arch Otorhinolaryngol. 2020;277:2251–2261. doi: 10.1007/s00405-020-05965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280, e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simões e Silva AC, Silveira KD, Ferreira AJ, Teixeira MM. ACE2, angiotensin-(1-7) and Mas receptor axis in inflammation and fibrosis. Br J Pharmacol. 2013;169:477–492. doi: 10.1111/bph.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grasselli G, Zangrillo A, Zanella A, Antonelli M, Cabrini L, Castelli A, et al. COVID-19 Lombardy ICU Network. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy region, Italy. JAMA. 2020;323:1574–1581. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parnham MJ, Erakovic Haber V, Giamarellos-Bourboulis EJ, Perletti G, Verleden GM, Vos R. Azithromycin: mechanisms of action and their relevance for clinical applications. Pharmacol Ther. 2014;143:225–245. doi: 10.1016/j.pharmthera.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Haydar D, Cory TJ, Birket SE, Murphy BS, Pennypacker KR, Sinai AP, et al. Azithromycin polarizes macrophages to an M2 phenotype via inhibition of the STAT1 and NF-κB signaling pathways. J Immunol. 2019;203:1021–1030. doi: 10.4049/jimmunol.1801228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cigana C, Nicolis E, Pasetto M, Assael BM, Melotti P. Anti-inflammatory effects of azithromycin in cystic fibrosis airway epithelial cells. Biochem Biophys Res Commun. 2006;350:977–982. doi: 10.1016/j.bbrc.2006.09.132. [DOI] [PubMed] [Google Scholar]
- 8.Maillé E, Trinh NTN, Privé A, Bilodeau C, Bissonnette E, Grandvaux N, et al. Regulation of normal and cystic fibrosis airway epithelial repair processes by TNF-α after injury. Am J Physiol Lung Cell Mol Physiol. 2011;301:L945–L955. doi: 10.1152/ajplung.00149.2011. [DOI] [PubMed] [Google Scholar]
- 9.Sungnak W, Huang N, Bécavin C, Berg M, Queen R, Litvinukova M, et al. HCA Lung Biological Network. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, et al. Orchestrating high-throughput genomic analysis with bioconductor. Nat Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertram S, Glowacka I, Müller MA, Lavender H, Gnirss K, Nehlmeier I, et al. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. J Virol. 2011;85:13363–13372. doi: 10.1128/JVI.05300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuberi RI, Ge XN, Jiang S, Bahaie NS, Kang BN, Hosseinkhani RM, et al. Deficiency of endothelial heparan sulfates attenuates allergic airway inflammation. J Immunol. 2009;183:3971–3979. doi: 10.4049/jimmunol.0901604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baglivo M, Baronio M, Natalini G, Beccari T, Chiurazzi P, Fulcheri E, et al. Natural small molecules as inhibitors of coronavirus lipid-dependent attachment to host cells: a possible strategy for reducing SARS-COV-2 infectivity? Acta Biomed. 2020;91:161–164. doi: 10.23750/abm.v91i1.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Yang P, Liu K, Guo F, Zhang Y, Zhang G, et al. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ribeiro CMP, Hurd H, Wu Y, Martino ME, Jones L, Brighton B, et al. Azithromycin treatment alters gene expression in inflammatory, lipid metabolism, and cell cycle pathways in well-differentiated human airway epithelia. PLoS One. 2009;4:e5806. doi: 10.1371/journal.pone.0005806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


