Abstract
The exquisite transcriptional control of developmental gene programs is critical for hardwiring the complex expression patterns that govern cell-fate determination and differentiation during heart development. During the past several years, studies have illuminated our understanding of a complex noncoding transcriptional landscape, primarily associated with long noncoding RNAs (lncRNAs), that is implicated in these developmental processes and has begun to reveal key functions of these transcripts. In this review, we discuss the expanding roles for lncRNAs in the earliest points of cardiac development and through differentiation and maturation of multiple cell types within the adult heart. We go on to outline the diverse mechanisms by which cardiovascular lncRNAs orchestrate these transcriptional programs, explore the challenges linked to the study of lncRNAs in developmental phenotypes, and summarize the implications for these molecules in human cardiovascular disorders.
The heart is the first organ to develop during embryogenesis, with the survival of the embryo and all subsequent developmental processes dependent on its uninterrupted and consistent function. The development of the heart is an exquisite and incredibly complex process involving the specification, determination, differentiation, migration, and integration of multiple cell lineages in the correct temporal and spatial manner to form a three-dimensional organ fully integrated with the vascular system (Evans et al. 2010; Günthel et al. 2018). Flaws in this process underpin congenital heart disease, the most common form of human birth defects, which result in a number of structural and functional abnormalities including cardiomyopathies and arrhythmias that are often fatal (Zaidi and Brueckner 2017; Moore-Morris et al. 2018). The formation and function of the cardiovascular system is precisely dictated by highly complex and integrated transcriptional programs that link upstream signaling systems with protein coding genes (PCGs) required for cardiac myogenesis, morphogenesis, and contractility (Bruneau 2013).
The gene regulatory networks (GRNs) that govern these developmental programs are under the control of the integrated activity of core lineage determining transcription factors (TFs), including HAND2, TBX5, GATA4, MEF2C, NKX2-5, and MESP1 (Bruneau 2013; Waardenberg et al. 2014). These factors combinatorially interact in a self-reinforcing manner at target cis-regulatory elements to direct specific spatial and temporal gene expression programs. Coordinated binding of these factors is tightly coupled with the dynamic remodeling of the underlying chromatin landscape, leading to global epigenomic reprogramming and reorganization of the genome’s three-dimensional nuclear architecture (Kathiriya et al. 2015; Rizki and Boyer 2015). These processes dictate PCG expression patterns that are ultimately responsible for cell-fate determination, differentiation, identity, phenotype, and behavior. Importantly, the developmental regulation of the transcriptome, the epigenome, the three-dimensional nuclear architecture, and the proteome are highly integrated to coordinate in both time and space the outputs of otherwise disparate molecular and regulatory networks (Kathiriya et al. 2015).
Until recently, the full spectrum of molecular determinants that underpin and integrate these regulatory processes was unknown. Within this context, it has emerged that our “protein-centric” view of these networks was somewhat premature (Morris and Mattick 2014). Indeed, the noncoding portion of the genome, naively called “junk DNA” and now more appropriately called genomic “dark matter” (Yue et al. 2014), encodes a vast unexplored repertoire of regulatory sequences and associated noncoding RNAs (ncRNAs) with important regulatory functions within the GRNs that dictate cardiovascular development (Devaux et al. 2015; Ounzain and Pedrazzini 2015; Frank et al. 2016). In particular, long noncoding RNAs (lncRNAs) emerged as interesting regulatory molecules that are able to integrate and couple disparate regulatory processes fundamental for heart development. Here, we focus on the surge of important recent studies that have begun to illuminate our understanding of these lncRNAs and uncovered critical roles for them in many different aspects of cardiovascular development and the myriad of cell types involved in this process.
lncRNAs: MOLECULAR GENE REGULATORY SWITCHES IN CELL STATE
The mammalian genome is predominantly composed of nonprotein coding sequences, raising the possibility for diverse regulatory functions encoded within these regions. Deep RNA-sequencing approaches have revealed that the majority of the noncoding genome is actively transcribed, generating thousands of ncRNAs (Carninci et al. 2005; Guttman et al. 2009; Cabili et al. 2011; Morris and Mattick 2014). Although GRNs have long been known to be controlled by TF proteins binding to their cognate DNA regulatory elements, emerging evidence strongly supports a similarly critical role for networks of ncRNAs (Mercer et al. 2009; Kaikkonen et al. 2011). Deep genome-wide transcriptomic profiling has identified diverse classes of ncRNA with potentially important regulatory functions. Although microRNAs (miRNAs) were the first and most comprehensively characterized class of ncRNAs, many studies in the last decade have shown the importance of lncRNAs in a variety of biological processes (Ransohoff et al. 2018). lncRNAs are operationally defined as ncRNAs that are longer than 200 nucleotides in length with minimal protein coding potential. To date, the best characterized lncRNAs are Pol II transcribed, multiexonic, alternatively spliced, and polyadenylated transcripts (Perry and Ulitsky 2016). In contrast to PCGs and miRNAs, lncRNAs show less conservation and a rapid transcriptional turnover (Johnsson et al. 2014; Ransohoff et al. 2018). This does not imply a lack of function, but instead might suggest a role for these transcripts in both increasing and scaling with the developmental complexity of the species. lncRNAs are typically expressed at a lower level than messenger RNAs (mRNAs) and often feature greater tissue- and cell-type specificity (Cabili et al. 2011; Alexanian et al. 2017). Current studies have shown that lncRNAs represent key modulators of cell state and are dysregulated in various human diseases, including cardiovascular disorders and pathological remodeling of the heart in response to stress (Han et al. 2014; Ounzain et al. 2015a; Viereck et al. 2016; Wang et al. 2016; Micheletti et al. 2017). Indeed, lncRNAs have emerged as critical regulators of nearly every aspect of GRN activity, in which they modulate gene expression transcriptionally and posttranscriptionally through a variety of mechanisms including antisense RNA base pairing, guiding chromatin and transcriptional regulators to their required genomic destination, dueling polymerase activity, and affecting chromatin structure (Fig. 1; Mercer and Mattick 2013; Marchese et al. 2017). lncRNA loci are unique in their ability to spatially amplify regulatory information encoded by their underlying DNA and to operate as a regulatory platform to fine-tune gene programs controlled by TFs and enhancers. Enhancers, known as cis-acting DNA modules that activate and sustain transcription at their target promoters, are the key information-processing units within the genome that govern cell-state-defining GRNs in specification and differentiation (Wamstad et al. 2014; Long et al. 2016). These regulatory elements are characterized by enrichment of regulatory TF-binding sites, active chromatin marks (e.g., acetylation of histone H3 lysine 27 [H3K27ac]), and binding of coactivator proteins (e.g., BRD4, MED1) (Kellis et al. 2014; Li et al. 2016). Progress in sequencing technologies have revealed that many if not all enhancers are transcribed and generate ncRNAs (Marques et al. 2013; Ounzain et al. 2014; Li et al. 2016). The majority of the enhancers give rise to nonpolyadenylated and unspliced transcripts (enhancer RNAs [eRNAs]), although a very small subset are associated with unidirectional, multiexonic, spliced, polyadenylated processed transcripts (enhancer-associated lncRNAs [elncRNAs]). Regulatory elements associated with elncRNAs show greater chromatin accessibility and increased binding of lineage-specifying TFs, with elncRNAs having recently emerged as potential regulators of cell state in both disease and development (Sigova et al. 2015; Melé and Rinn 2016; Tan et al. 2017).
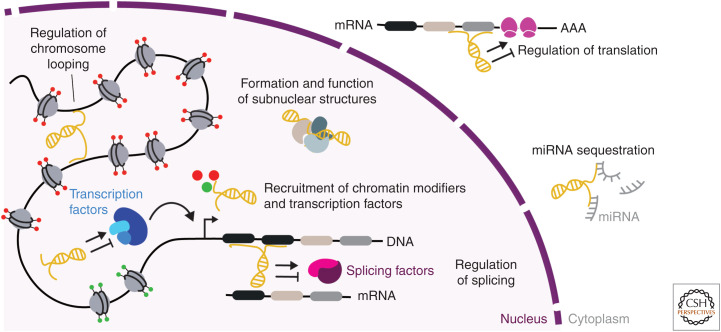
Figure 1.
Mechanisms of action of long noncoding RNAs (lncRNAs). lncRNAs modulate gene expression through a variety of mechanisms that include regulation of messenger RNA (mRNA) splicing, recruitment of chromatin modifiers and transcription factors (TFs) to specific genomic loci, formation of ribonucleoproteins, regulation of chromosome looping, regulation of translation, and microRNA (miRNA) sequestration.
DEVELOPMENTAL COMPETENCE ENCODED IN PLURIPOTENCY
A myriad of cell state transitions occur during development from the totipotent zygote to the billions of cells with highly specialized functions that comprise our tissues and organs. How the same genetic code, which is shared by all the cells in our body, is temporally and spatially regulated to ensure the generation of consistent body plans by sequential differentiation of totipotential material remains among the most fascinating questions in biology. Heart development, or that of any organ, relies on the ability of progenitor cells to appropriately respond to instructive cues—a property known as developmental competence. All heart cells share a common origin during the early events of embryogenesis; they collectively derive from a transient structure known as the primitive streak (PS) (Abu-Issa and Kirby 2007; Tam and Loebel 2007). The specification of distinct PS populations is tightly regulated in a spatial and temporal manner to give rise to both endoderm and mesoderm cells (Abu-Issa and Kirby 2007). During their in vitro differentiation into mesoderm and endoderm, embryonic stem cells (ESCs) transition through an intermediate stage called mesendoderm (ME), which is equivalent to the PS. ESCs differentiate into ME progenitors by using a defined set of regulatory elements and TFs to trigger lineage determination in response to defined instructive cues (Wang and Chen 2016). Several lines of evidence show that pluripotent stem cells possess large repertoires of regulatory elements that become heavily restricted over the course of development and differentiation irrespective of lineage (Stergachis et al. 2013). Interestingly, recent studies have shown that enhancers destined to regulate cell specification during differentiation are marked by the binding of pluripotency TFs (Kim et al. 2018). This epigenetic priming at the pluripotent stage is required for future cell-type-restricted enhancer activity in the differentiated cells. Other work has revealed establishment of a poised enhancer landscape for endodermal organ lineages in gut tube progenitor cells (Wang et al. 2015). This collection of enhancers is bound by pioneer lineage-specifying TFs such as members of the FOXA family, and are required for subsequent differentiation into specialized pancreatic endoderm. These regulatory elements confer developmental competence to endodermal intermediates and are necessary to respond appropriately to inductive signals and transition to a mature cell state. Very recent work has used a single-cell “triple-omics” approach to map chromatin accessibility, DNA methylation, and RNA expression during the exit from pluripotency and onset of gastrulation in mouse embryos (Argelaguet et al. 2019). Regulatory elements associated with each germ layer were shown to be either epigenetically primed or remodeled prior to undergoing an overt cell-fate decision during gastrulation. These data collectively support the notion that developmental competence may be defined by a subset of regulatory elements necessary for interpreting cell-fate-determining inductive signals before overt cell-fate specification.
To explore the importance of the enhancer landscape and its associated transcripts in establishing developmental competence for early cell-fate specification, we profiled the enhancer and lncRNA transcriptional landscape that defines the transition between pluripotency and the ME stage, the earliest precursor population representing the source of all endoderm and mesoderm-derived tissues, including the heart (Alexanian et al. 2017). We focused on the small subset of regulatory elements associated with the production of a fully processed elncRNA. Interestingly, we discovered that genomic loci of several ME lineage-specifying TFs such as Eomes, Sox17, and Gsc, were characterized by the close proximity of an actively transcribed enhancer at the pluripotent stage, a time point at which these TFs are either entirely absent or expressed at very low levels. To investigate the potential role of these regions in establishing developmental competence for ME-derived lineages in pluripotency, we focused on a transcribed enhancer upstream of Eomes considering its pioneer role in dictating ME specification. We named this enhancer Meteor and its associated transcript Meteor lncRNA (Alexanian et al. 2017). Meteor enhancer is active in pluripotency and repressed in EOMES-expressing cells, with Meteor lncRNA expression following a similar pattern. Deletion of the Meteor locus in ESCs completely abolishes mesendodermal competence and renders these cells unable to respond to the instructive cues for the ME fate. Instead, Meteor-deleted cells show an apparent priming for the neuroectoderm fate at the ESC stage, suggesting that Meteor controls developmental competence before the earliest commitment into the three germ layers. Importantly, Meteor-knockout (KO) cells maintain stemness features, suggesting that although this locus is specifically active in pluripotency, it is not required for sustaining the pluripotency gene circuitry linked to self-renewal. Mechanistically, the Meteor locus encodes both an enhancer and an lncRNA. Many lncRNA loci have been shown to exert their functions via processes linked to their transcription (e.g., recruitment of transcriptional coactivators or splicing factors) rather than the lncRNA transcripts themselves (Engreitz et al. 2016). Using engineered ESCs with a polyA element downstream from the Meteor lncRNA transcriptional start site (TSS) (Engreitz et al. 2016), we found that Meteor lncRNA transcription is indispensable for the ME and subsequent cardiomyocyte (CM) fates. A CRISPR-based transcriptional activation approach (CRISPR-On) (Konermann et al. 2015) to boost Meteor lncRNA transcription led to increased expression of ME-associated genes, showing that the activation of a pluripotency-specific enhancer was sufficient to drive downstream cellular fates. These results support the idea that transcribed enhancers distal to developmental genes can play a fundamental role in governing developmental competence of uncommitted cells before cell-fate specification. Importantly, depletion of the Meteor lncRNA transcript in pluripotency did not affect mesendodermal competence of ESCs, suggesting that the RNA molecule itself is dispensable for this process. Recently, the Meteor lncRNA transcript (also known as linc1405) was shown to be indispensable for proper cardiac differentiation (Guo et al. 2018). In contrast to our study, Guo and colleagues showed that Meteor/linc1405 expression is up-regulated during cardiac differentiation, with knockdown of linc1405 leading to impaired cardiac differentiation in vitro. Using an RNA immunoprecipitation (RIP) assay, these investigators show a physical interaction between Meteor/linc1405 and EOMES. Interestingly, binding of EOMES to the Mesp1 enhancer region (3.8 kb upstream of Mesp1 TSS) was disrupted on Meteor/linc1405 knockdown. Additional RIP experiments identified the histone acetyltransferase GCN5 and the trithorax group protein WDR5 as Meteor/linc1405 interactors, suggesting that Meteor/linc1405 may control cardiac mesoderm specification through interactions with known chromatin regulators. These data collectively argue that the Meteor/linc1405 locus represents an example of a transcribed regulatory element with both RNA-dependent and -independent mechanisms. Of note, a number of transcribed regulatory elements in proximity to the Haunt (Yin et al. 2015), Nanog (Blinka et al. 2016), and Hand2 (Anderson et al. 2016) loci have been shown to control cell fate via similar mechanisms, again illustrating the importance of elucidating the multifunctional roles for lncRNAs and their embedded DNA regulatory sequences in coordinating gene expression during lineage specification.
lncRNAs IN CARDIAC MESODERM SPECIFICATION
Many lncRNAs show coordinated expression with their proximal PCGs. lncRNAs that are transcribed in the opposite direction of their associated PCGs (termed divergent lncRNAs) have recently emerged as powerful regulators of lineage-specifying TFs (Luo et al. 2016). Interestingly, a disproportionate number of PCGs involved in regulating lineage specification neighbor a divergent lncRNA, suggesting that these noncoding transcripts may be important in amplifying the regulatory information contained within genomic loci critical for controlling specification and differentiation. One of the first lncRNAs implicated in heart development was Fendrr, which is transcribed divergently from the TF FOXF1 (Grote et al. 2013). Fendrr is specifically expressed in the lateral plate mesoderm of the developing embryo and interacts with both activating (TrxG/MLL) and repressive (PCR2) chromatin-modifying complexes to modulate expression of specific TFs implicated in heart development. Fendrr KO resulted in lethality at embryonic day 13.75 and affected histone modifications linked to activation and repression of transcription at key TFs regulating cardiogenic cell fate. lncRNAs are known to regulate gene expression by forming complexes with broadly expressed chromatin regulators and target their localization to defined genomic loci. Indeed, Fendrr was found to bind PRC2 and direct it to Foxf1 and Pitx2 promoters to inhibit the expression of these genes. Importantly, epigenetic signatures like those established by Fendrr persist through several stages of differentiation, thereby impacting broadly the epigenetic landscape of early development. Another example of a divergent lncRNA implicated in early ME specification is Evx1as, transcribed on the opposite strand of its nearby gene Evx1 (Luo et al. 2016). Evx1 promotes ME specification during ESC differentiation and its expression is highly correlated with Evx1as. Evx1as genomic deletion and posttranscriptional knockdown both significantly down-regulate Evx1 expression. Consistent with a cis-regulatory mechanism, increased transcription of Evx1as via a CRISPR-On system is able to boost Evx1 expression. Interestingly, tethering the Evx1as RNA to the shared promoter of Evx1as/Evx1 using a catalytically dead Cas9 (dCas9)-mediated approach showed a direct role for the Evx1as transcript in controlling Evx1 transcription through cis-regulation. Of note, down-regulation of Evx1as was associated with a more severe decrease of ME genes as compared with its proximal TF, suggesting that Evx1as may also act in trans independently of Evx1. Another divergent lncRNA transcribed in the Brachyury (T) locus was recently found to be essential for mesoderm commitment of human ESCs (Frank et al. 2019). yylncT binds the de novo DNA methyltransferase DNMT3B and is required for activation of the T locus, with yylncT depletion abolishing mesodermal commitment. Other studies have characterized the role lncRNAs transcribed in proximity of PCGs in the regulation of cardiac mesoderm commitment. The lncRNA ALIEN is proximal to the FOXA2 TF and begins to be expressed at the PS stage, showing maximal expression in the lateral plate mesoderm of E8.5 mouse embryos (Kurian et al. 2015). ALIEN loss-of-function leads to disruptions in heart and vascular formation. In another study, the same lncRNA (this time named DEANR1) was found to be a critical regulator of definitive endoderm specification through its interaction with FOXA2 and SMAD2/3 (Jiang et al. 2015). The HoxBlinc lncRNA, transcribed from the Hoxb gene locus, was also found to be a regulator of cardiac mesoderm specification through the interaction with the positive epigenetic regulators SETD1A and/or MLL1 complexes (Deng et al. 2016). Using a combination of approaches, including chromatin isolation by RNA purification (ChIRP) and chromosome conformation capture (3C), the investigators showed that the HoxBlinc transcript regulates chromatin loop at the Hoxb cluster, attracting the SETD1A and/or MLL1 complexes to target Hoxb gene promoters. Although key TFs regulating cardiogenesis are known to be extremely well conserved across species, many lncRNAs show lower interspecies conservation suggesting species-specific roles for lncRNAs. Along these lines, the lncRNA HBL1 was recently described as a human-specific regulator of CM differentiation from human-induced pluripotent stem cells (hiPSCs) (Liu et al. 2017). HBL1 is highly expressed in pluripotency and in cardiac mesoderm progenitors with its knockdown increasing CM differentiation from hiPSCs. HBL1 regulates CM differentiation by counteracting the activity of MIR1, an miRNA known to promote cardiogenic differentiation (Ivey et al. 2008; Lu et al. 2013). The HBL1 promoter is bound by SOX2, and SOX2 depletion in hiPSCs leads to decreased HBL1 expression and increased CM differentiation. Thus, the HBL1 locus is highly regulated in pluripotency and forms a regulatory network with SOX2 and MIR1 to control developmental competence of hiPSCs for subsequent cardiogenic specification.
lncRNAs IN CARDIAC PROGENITOR CELLS AND CARDIOMYOCYTES
As a first step toward identifying and characterizing lncRNAs involved in cardiac development, several genome-wide transcriptional profiling studies provided early evidence that lncRNAs are likely important components of these development-specific transcriptional networks. In an early and pioneering study investigating how chromatin structure is coupled to gene expression patterns during cardiac commitment, Wamstad et al. (2012) identified a significant number of differentially and dynamically expressed lncRNAs. These investigators explored this landscape in an in vitro model of cardiogenesis at critical transition stages, namely, ESCs, mesodermal cells, cardiac progenitor cells, and CMs. More than 200 lncRNAs were identified that were dynamically expressed in a stage-specific manner. Interestingly, many of these lncRNA expression profiles were correlated with their neighboring PCGs, suggestive of potential cis-regulatory functions during CM differentiation. A similar study using the P19 embryonal carcinoma cell line identified 40 dynamically expressed lncRNAs during CM formation (Zhu et al. 2014b).
Although these in vitro models are certainly informative, in vivo profiling and functional assessment is critical to understand the physiological functions of these transcripts. To this end, a number of studies have explored lncRNA profiles during in vivo development. For example, the transcriptomes of whole hearts from E11.5, E14.5, and E18.5 were profiled to identify dynamically expressed lncRNAs in the developing fetal mouse heart (Zhu et al. 2014a). Hundreds of lncRNAs were identified that displayed distinct and dynamic expression profiles between these embryonic time points, which coincided with key differentiation and maturation developmental steps. Supporting these observations, Ounzain and colleagues directly assessed transcripts from bona fide developmental cardiac enhancers that had previously been identified and validated in mouse embryos (Ounzain et al. 2014). A number of these dynamically active developmental enhancers were transcribed to generate putative lncRNAs. Furthermore, the expression of these lncRNAs correlated both with the activity of their embedded enhancer and with expression of their neighboring PCGs, consistent with cis-regulation. In vitro knockdown of these bona fide in vivo elncRNAs resulted in reduced expression of their putative cis-target genes. For example, activity at the enhancer named mm85 produced an associated lncRNA whose knockdown impacted the important proximal and developmental regulator Myocardin. Myocardin is a fundamental cofactor for serum response factor (SRF), a core developmental TF involved in cardiac differentiation. The investigators profiled this landscape more systematically using very deep RNA-seq on polyadenylated RNA derived from mouse ESCs and cardiac progenitor cells. De novo transcript assembly and reconstruction identified hundreds of novel, previously unannotated, multiexonic lncRNAs derived from developmental enhancers undergoing specific state transitions during cardiac specification and differentiation. Collectively, these studies established a framework and provided a plethora of uncharted lncRNAs with potential functions in cardiac specification, differentiation, growth, and homeostasis that served as an important resource for subsequent downstream functional studies.
Among the first identified and best characterized lncRNAs in cardiovascular development is Braveheart (Bhvt), which was discovered based on its unique expression pattern during CM differentiation in mouse ESCs (Klattenhoff et al. 2013). Bhvt loss-of-function in mouse ESCs resulted in perturbed differentiation and dramatically reduced the formation of CMs. Bhvt was shown to be upstream of MESP1, an important core TF that marks early cardiac precursor cells during development. It was shown that Bhvt, via its regulation of Mesp1, directed the correct temporal and spatial expression of numerous core cardiac TFs including Gata4, Gata6, Tbx2, Hand2, Hand1, and Nkx2-5. Through this fundamental regulation of the core cardiac GRN, Bhvt was necessary for the lineage transition from nascent cardiac mesoderm to the subsequent differentiation into CMs. Functionally, Bhvt acts in a trans-manner via interaction with SUZ12, a core component of the PRC2 complex. Bhvt functions as a decoy for PRC2, allowing the derepression of PRC2 target core cardiac TFs, facilitating the activation of the cardiogenic gene program. Recently, the secondary structure of Bhvt was determined using chemical probing methods and was shown to have a modular folded structure (Xue et al. 2016). The deletion of an 11-nt 5′ asymmetric G-rich internal loop (AGIL) dramatically impaired CM differentiation. This AGIL mediated an interaction with the TF CNBP/ZNF9, which subsequently drives cardiovascular lineage commitment. This seminal work, for the first time, showed that lncRNAs represent powerful regulatory molecules capable of inducing cardiac specification and differentiation. However, a Bhvt ortholog was not found in humans, suggesting other important cardiac-specifying lncRNAs are likely to exist. To this end, Ounzain et al. (2015b) investigated the orthologous and syntenic genomic locus in human cardiac precursor cells. They identified on the opposing strand a conserved lncRNA that was named cardiac mesoderm enhancer-associated ncRNA, or CARMEN (Ounzain et al. 2015b). CARMEN was identified via a transcriptomic screen for lncRNAs up-regulated during the cardiac differentiation of primary human cardiac precursor cells isolated from the fetal human heart. CARMEN was derived from an active cardiac superenhancer and was proximal to the critical cardiovascular cell-identity miRNA-143/145 cluster (Cordes et al. 2009). It was shown that CARMEN displays an RNA-dependent activity that in mouse is upstream of both Bhvt and the core cardiac mesoderm-specifying GRN. Indeed, both human and murine CARMEN knockdown abolished the ability of cardiac precursors to differentiate into CMs supporting the notion that CARMEN is an evolutionarily conserved regulator of cardiovascular cell lineage specification and differentiation. Furthermore, the same group showed that CARMEN expression is dependent on NOTCH signaling in differentiating cardiac precursor cells (Plaisance et al. 2016). Interestingly, CARMEN isoforms were identified that also dictate commitment to the smooth muscle cell (SMC) fate in a NOTCH-dependent fashion. This study found that NOTCH inhibition, via down-regulation of an SMC-specific CARMEN isoform, repressed miR-143/145 expression, thereby forcing cardiac precursor cells to adopt a CM fate.
Another important example lies in the context of Kcnq1, an important epigenetically imprinted cardiac developmental gene whose dysregulation is responsible for congenital long QT syndrome, a serious disorder that results in fatal cardiac arrhythmias (Crotti et al. 2008). Korostowski et al. (2012) showed that maternal expression of the antisense lncRNA Kcnq1ot1 has an important role in modulating Kcnq1 levels during cardiac development. This lncRNA modulates the three-dimensional chromatin structure of the Kcnq1 locus, ensuring proper cardiac-specific temporal and spatial expression of Kcnq1. Developmental regulators of CM differentiation and identity can also be identified in the stressed adult heart, as the pathological remodeling that occurs is typically associated with the reactivation of developmental and fetal gene programs. To this end, Ounzain and colleagues (2015a) comprehensively mapped the long noncoding transcriptome after myocardial infarction (MI) in a mouse model using very deep RNA sequencing followed by de novo reconstruction of the noncoding transcriptome. They identified 1500 previously unknown heart enriched lncRNAs that were primarily associated with heart-specific enhancers. Using a novel computational approach, functions were inferred for these novel lncRNAs based on chromatin dynamic state transitions associated with their genomic loci during in vitro CM differentiation. Importantly, based on this functional inference approach, the majority of these novel heart enriched lncRNAs were associated with cardiac development or structural and functional cardiac gene programs. Interestingly, one novel CM-specific lncRNA, Novlnc6, was associated with key chromatin-state transitions linked to CM differentiation and maturation gene programs. In support of this, CM loss-of-function of Novlnc6 directly impacted the expression of two fundamental CM genes, Bmp10 and the core cardiac TF Nkx2-5.
Another key TF in embryonic heart development is HAND2. The Hand2 locus encodes multiple enhancer-associated lncRNAs that have been the subject of intense scrutiny over the last several years. The first to be identified was Upperhand (Uph), a divergently transcribed elncRNA from Hand2 that shares its core promoter sequence (Anderson et al. 2016). Hand2 and Uph are coexpressed during heart development. Termination of Uph transcription in vivo, via introduction of a polyA sequence into its second exon, resulted in loss of Hand2 expression and lethal cardiac defects reminiscent of Hand2 KO embryos. Importantly, the Uph mature RNA transcript is dispensable for heart development, suggesting that the act of transcription, rather the lncRNA transcript itself, is required for proper Hand2 expression. Consistent with this notion, Uph transcription was shown to be required for proper deposition and maintenance of an active enhancer mark across the Uph-Hand2 cardiac enhancer (Anderson et al. 2016). Subsequent work confirmed the importance of the Uph locus in heart morphogenesis (Han et al. 2019). Rather surprisingly, a 1-kb deletion of the first two exons of Uph, encompassing the site of polyA knockin used to terminate Uph expression in vivo (Anderson et al. 2016), was not sufficient to produce any discernable cardiac phenotype. The full deletion of Uph/Hand2os1, however, resulted in heart defects and perinatal lethality. These data collectively suggest that the cis-regulatory elements embedded in the Uph/Hand2os1 locus, rather than its transcription, are the key regulators necessary for Hand2 expression and appropriate heart development. The discrepancy between these studies is rather unexpected as the polyA knockin approach is generally regarded as being significantly less disruptive than larger-scale promoter deletion. It is important to note that the Hand2 locus harbors a second enhancer with strong activity during cardiac development that lies downstream from the TF. A very recent publication has shown the importance of an lncRNA locus, Handsdown (Hdn), which is transcribed from this downstream regulatory element (Ritter et al. 2019). Hdn encodes multiple transcripts that are transcribed in the same direction as Hand2. Removal of the entire Hdn locus produces incomplete looping of the heart tube ultimately resulting in embryonic lethality. Interestingly, Hdn KO and Hdn transcriptional repression (by introduction of a transcriptional stop signal) were both associated with increased expression of Hand2 in an in vitro model of cardiogenic differentiation (Ritter et al. 2019). These results support a role for Hdn transcription in the cis-regulation of Hand2 and suggest that the Hdn locus is a negative regulator of Hand2. Collectively, these observations highlight the fascinating nature of this locus in which two separate regulatory elements appear to have opposing roles have evolved to fine-tune Hand2 expression during cardiac development. Unexpectedly, George and colleagues reported that a 3-kb deletion around the TSS of Hdn yielded no lethality, dramatic cardiac phenotype, or overexpression of Hand2 (George et al. 2019). Although the aforementioned studies have shown some discrepancies, they uniformly support that a functional lncRNA transcript is dispensable for appropriate cardiac development.
Appropriate temporal and spatial control of CM proliferation is fundamental during cardiac developmental stages and requires precise regulatory control to ensure appropriate three-dimensional morphogenesis of the heart. Li and colleagues (2018) recently found that silent information regulation factor 2–related enzyme (Sirt1) antisense lncRNA expression was significantly induced during heart development and correlated with CM proliferation. Gain- and loss-of-function approaches using adenovirus and LNA-GapmeRs showed that this CM-enriched lncRNA promotes CM proliferation in vitro and in vivo, whereas its suppression impeded CM proliferation. Furthermore, overexpression of Sirt1 antisense lncRNA in trans-enhanced CM proliferation, attenuated CM apoptosis, and decreased mortality in a mouse model of MI. Mechanistically, this lncRNA was found to bind the 3′-UTR of Sirt1, enhancing its stability and abundance at both the mRNA and protein levels. These increased levels of Sirt1 drove the lncRNA-mediated induction of CM proliferation.
CM maturation represents the key terminal point of cardiac development with maturation during the perinatal transition of the heart being critical for functional adaptation to the postnatal hemodynamic load and nutrient environment. Furthermore, perturbations of the maturation process are implicated in congenital heart defects. lncRNAs are also emerging as potentially important regulators of this maturation process. Touma et al. (2016) comprehensively mapped the transcriptome of the neonatal mouse left and right ventricles. They identified 196 novel lncRNAs that showed significant dynamic regulation coupled with the cardiac maturation process. In particular, a number of potential novel interactions were identified between lncRNAs and their cognate neighboring PCGs in the neonatal heart. For example, Tcap encodes a CM-specific protein involved in cardiac myogenesis, whereas Trim72 is also enriched in the heart and regulates sarcolemma responses to oxidative stress. Inverse relationships between Ppp1r1b-lncRNA and Tcap as well as between Fus-lncRNA and Trim72 in the neonatal heart support a potential role of these lncRNAs in CM maturation. Confirming this, functional studies have shown a significant impact of Ppp1r1b-lncRNA loss-of-function on maturation and sarcomere assembly by blocking myogenic differentiation. Importantly, this lncRNA is conserved and the Ppp1rb-lncRNA/Tcap expression ratio was found to be a highly sensitive molecular signature that identifies tetralogy of Fallot (TOF) and ventricular septal defect (VSD) in human infantile hearts (Touma et al. 2016).
CM-enriched lncRNAs also play important roles in the context of adult pathophysiology (Han et al. 2014; Viereck et al. 2016; Wang et al. 2016). Mhrt was the first lncRNA implicated in pathological hypertrophy (Han et al. 2014). Located in the intergenic region between murine Myh6 and Myh7, Mhrt is down-regulated in the setting of pressure overload. Transgenic overexpression of Mhrt is sufficient to protect the heart from heart failure progression in a pressure-overload model of heart failure. Mechanistically, Mhrt acts as a molecular decoy, antagonizing the activity of BRG1, a chromatin remodeling factor previously implicated in the control of pathological cardiovascular GRNs. Importantly, the human MHRT ortholog is depleted in failing human hearts, suggesting a potential conserved regulatory role of potential translational significance.
lncRNAs IN NONCARDIOMYOCYTE CELLS IN HEART DEVELOPMENT
In addition to CMs, lncRNAs are emerging as important regulators of other cell types within the heart that are fundamental for proper cardiac development, function, and homeostasis. Particularly important cells include both endothelial and SMCs. A number of lncRNAs have now emerged as important regulators of these cell types. Transcriptional profiling of human coronary artery SMCs identified many novel smooth muscle–specific lncRNAs, including a vascular enriched lncRNA named smooth muscle and endothelial cell–enriched migration/differentiation-associated lncRNA, SENCR (Bell et al. 2014). This lncRNA is transcribed on the opposing strand within the first intron of the FLI1 gene that encodes a core TF regulating blood and endothelial cell formation. FLI1 and SENCR have correlated expression profiles; however, SENCR does not simply regulate FLI1 expression in cis. Rather, SENCR is enriched in the cytoplasm and encodes primarily trans-regulatory functions. SENCR loss-of-function results in global dysregulation of the SMC contractile gene program, in particular the master SMC regulator MYOCARDIN. Furthermore, SENCR expression was altered in vascular tissue and cells derived from patients with critical limb ischemia and premature coronary artery disease. Another vascular lncRNA, smooth-muscle-induced lncRNA (SMILR) was characterized in a similar fashion (Ballantyne et al. 2016). However, in this case, increased SMILR levels were associated with vascular SMC proliferation. Mechanistically, SMILR functions via interaction with the protein HAS2, which encodes for the enzyme that catalyzes the synthesis of hyaluronic acid, a polysaccharide essential for SMC proliferation and migration. HAS2 is reduced on SMILR knockdown leading to impaired SMC proliferation. In addition to SMC proliferation, lncRNAs have also been identified that control SMC plasticity and differentiation. For example, MYOcardin-induced Smooth muscle lncRNA, Inducer of Differentiation (MYOSLID), is transactivated by Myocardin and is specifically expressed in vascular SMCs (Zhao et al. 2016). MYOSLID promotes SMC differentiation and inhibits proliferation via disrupting actin stress fiber formation and blocking the nuclear translocation of MYOCD-related TF-A (MKL1/MRTF-A). Furthermore, MYOSLID loss-of-function abrogated TGFb1-induced SMAD2 phosphorylation, leading to decreased SMC differentiation.
MALAT1 represents a highly and ubiquitously expressed lncRNA that has broadly been implicated in endothelial cell biology and behavior in the heart and the cardiovascular system (Michalik et al. 2014; Thum and Fiedler 2014). Loss of MALAT1 results in a switch from a proliferative to a migratory state for endothelial cells in vitro and significantly impairs the proliferation of endothelial cells, vessel growth, and vascularization in vivo (Michalik et al. 2014). However, MALAT1 KO mice do not display a gross developmental phenotype in vivo (Eißmann et al. 2012; Zhang et al. 2012). It is likely that MALAT1 plays important roles during cardiovascular development under stress conditions. In support of this, recent studies have implicated MALAT1 in various aspects of cardiovascular development and pathophysiology, with diverse roles in numerous cardiovascular cell types (Thum and Fiedler 2014; Huang et al. 2019). Cardiac fibroblasts represent an abundant cell population in the mammalian heart, although they have historically been overlooked in terms of functional contributions to cardiac development and adult pathophysiology. Over the past years, however, they have been recognized as key protagonists during cardiac development and disease, working together with CMs and the other cardiac cell populations through structural, paracrine, and electrical interactions (Furtado et al. 2016; Ivey and Tallquist 2016; Alexanian and Haldar 2018; Fu et al. 2018). In particular, they are sentinel regulatory effector cells of CM proliferation and growth during development, directly connected to CMs via connexins, electrically isolate various portions of the cardiac conduction system, and secrete factors to regulate CM hypertrophy (Weber et al. 2013; Prabhu and Frangogiannis 2016). Cardiac fibroblast-enriched and -specific lncRNAs have recently been identified that are fundamental for cardiac fibroblast identity, survival, and the subsequent pathological myofibroblast activation that occurs in the stressed adult heart. Micheletti and colleagues filtered for heart-enriched lncRNAs that were conserved and transcribed from human left ventricle–specific super enhancers, were induced in the setting of MI (in which there is extensive replacement and reactive fibrosis), and were highly enriched in cardiac fibroblasts versus CMs (Micheletti et al. 2017). This process led to the identification of Wisper (Wisp2 Super-Enhancer-associated RNA), which represents the first example of a cardiac fibroblast–specific regulatory lncRNA critical for fibroblast identity, proliferation, and survival. In vitro loss-of-function experiments showed that Wisper positively regulated myofibroblast gene expression and function, including proliferation, migration, survival, and extracellular matrix (ECM) deposition. Importantly, antisense-mediated depletion of Wisper in adult mice post-MI inhibited cardiac fibrosis and significantly improved cardiac structural and functional remodeling with an associated increase in survival. Mechanistically, Wisper binds to a pleiotropically acting regulator of fibrosis, TIA1 cytotoxic granule–associated RNA-binding protein-like 1 (TIAR), stimulating its nuclear translocation and promoting its processing of profibrotic target transcripts. Importantly, Wisper expression is not detected in lung or kidney fibroblasts at baseline and is not transcriptionally induced in models of kidney fibrosis, showing the tissue specificity Wisper encodes for the control of cardiac fibrosis. Furthermore, expression levels of the human Wisper ortholog correlate significantly with the severity of cardiac fibrosis in heart tissue biopsies isolated from patients with aortic stenosis, underscoring the translational relevance of these findings. Because of its indispensable role in cardiac fibroblasts identity and survival, it is also likely to have important roles during cardiac development although this awaits further exploration.
Indeed, a number of lncRNAs have been shown to play important roles in cardiovascular cell specification and differentiation (Fig. 2). Further investigation of the lncRNA transcriptional landscape during heart development has the potential to provide additional valuable insights into the molecular mechanisms governing cardiovascular development and disease.
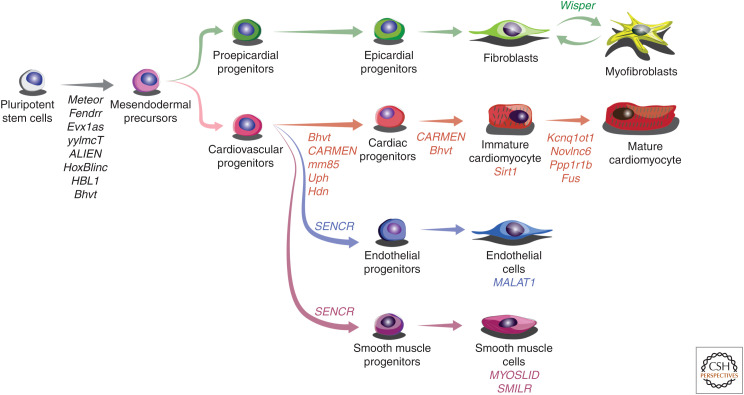
Figure 2.
Developmental cardiovascular long noncoding RNAs (lncRNAs). Schematic illustration of currently functionally characterized cardiovascular lncRNAs and the cell types they are associated with.
CONCLUSIONS, CHALLENGES, AND PERSPECTIVES
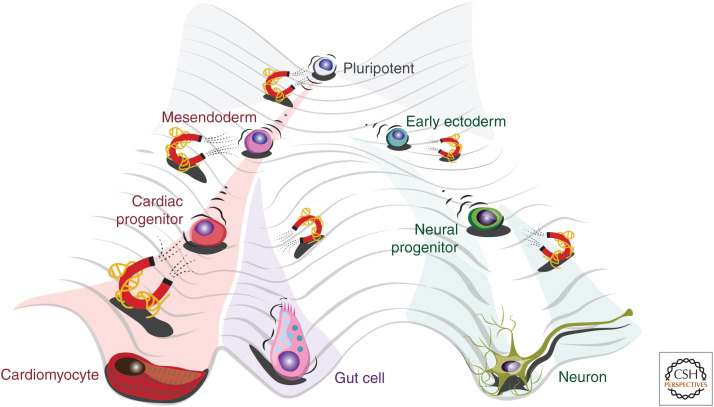
The advent of novel sequencing technologies has elucidated that much of the genome is transcribed into ncRNA molecules including lncRNAs. lncRNAs have been shown to fine-tune GRNs controlling cell fate during lineage specification. Conrad Waddington's metaphor of the “epigenetic landscape” is depicted with a ball rolling down a hill of bifurcating ridges and valleys symbolizing the possible trajectories of cell-fate specification (Waddington 2012). In this process, lncRNAs can be envisaged as “molecular magnets” that direct cell specification to a defined fate (Fig. 3).
Figure 3.
Long noncoding RNAs (lncRNAs) fine-tune the epigenetic landscape. Conrad Waddington envisioned the epigenetic landscape as a series of progressively restricted ridges and valleys a cell can traverse during its differentiation. During this process, lncRNAs (here pictured as magnets) can fine-tune cell-fate specification providing a “force” that directs cellular differentiation into a specific fate.
To date, a paucity of lncRNAs have been shown to play a causal role in cardiovascular disease, with most of those that have been identified important only in adult pathophysiology. This highlights the critical importance of expanding our understanding of the functional role of lncRNA transcripts as opposed to simply the genomic loci that encode them. The study of lncRNA function is wrought with technical challenges, largely owing to the fact that these molecules are generally transcribed from either regulatory elements or promoter regions that are shared with coding genes. Thus, classical loss-of-function experiments involving targeted disruption of the appropriate locus in the murine genome, as are regularly used for PCGs, cannot be applied in the context of lncRNAs. Instead, experimental manipulation of lncRNAs requires specific targeting of the transcript itself. The most commonly used tools for this purpose are antisense oligonucleotides or RNA-interference approaches, both of which are difficult to implement in in vivo systems, particularly during developmental transitions. This likely accounts for the bias toward in vitro systems in much of the published literature regarding lncRNA function, the limitations of which are well appreciated in the context of understanding the complexity of in vivo developmental processes. Recent evidence suggests that in vitro phenotypes associated with lncRNA loss-of-function may be less robust when evaluated in an in vivo context. For example, Meteor KO in vivo is associated with a milder phenotype than observed in vitro (Guo et al. 2018). Whether this discrepancy is disproportional for lncRNAs when compared with PCGs or if this truly represents a phenomenon of lncRNA biology remains to be addressed. Recent evaluation of nine cardiac progenitor expressed lncRNAs (George et al. 2019), all of which are strongly expressed during cardiac in vitro differentiation and in the developing heart, were not required for proper heart development. Although this cohort represents only a small fraction of the entire repertoire of cardiac lncRNAs, these results again highlight the importance of understanding how important or modest the role of this class of molecules is in organogenesis. The advent of novel tools that allow for efficient lncRNA loss-of-function in vivo will be an important step to overcoming this hurdle. Despite all these challenges, the degree of tissue- and cell-type specificity of lncRNA expression (more so than any other expressed nucleic acid or protein) raises the profile of this class of molecules as a therapeutic target for disease (Alexanian et al. 2019). Importantly, off-target toxicity frequently limits pharmacologic therapies with excellent on-target activity largely as a result of broad expression of many disease-associated pleiotropically acting proteins and pathways. The exquisite specificity of their expression when combined with their emerging role as highly specialized nodal regulators of gene expression make lncRNAs an incredibly attractive therapeutic target for human disease. Ongoing efforts to uncover the biology of this fascinating class of molecules has enormous potential to modify disease pathogenesis and improve human health.
ACKNOWLEDGMENTS
We thank Ana Catarina Silva for drawing the figures. We thank Arun Padmanabhan for the valuable comments and suggestions, which helped us to improve the quality of the manuscript. M.A. was supported by the Swiss National Science Foundation (P2LAP3_178056). S.O. is the Chief Executive Officer and Scientific Co-Founder of HAYA Therapeutics SA. S.O. is also named as an inventor on patents pertaining to the field (WO 2015/092020A2 and EP17176725.4).
Footnotes
Editors: Benoit G. Bruneau and Paul R. Riley
Additional Perspectives on Heart Development and Disease available at www.cshperspectives.org
REFERENCES
- Abu-Issa R, Kirby ML. 2007. Heart field: from mesoderm to heart tube. Annu Rev Cell Dev Biol 23: 45–68. 10.1146/annurev.cellbio.23.090506.123331 [DOI] [PubMed] [Google Scholar]
- Alexanian M, Haldar SM. 2018. The cardiac myofibroblast. Circ Res 123: 1258–1260. 10.1161/CIRCRESAHA.118.314185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexanian M, Maric D, Jenkinson SP, Mina M, Friedman CE, Ting CC, Micheletti R, Plaisance I, Nemir M, Maison D, et al. 2017. A transcribed enhancer dictates mesendoderm specification in pluripotency. Nat Commun 8: 1806 10.1038/s41467-017-01804-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexanian M, Padmanabhan A, McKinsey TA, Haldar SM. 2019. Epigenetic therapies in heart failure. J Mol Cell Cardiol 130: 197–204. 10.1016/j.yjmcc.2019.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KM, Anderson DM, McAnally JR, Shelton JM, Bassel-Duby R, Olson EN. 2016. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 539: 433–436. 10.1038/nature20128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argelaguet R, Clark SJ, Mohammed H, Stapel LC, Krueger C, Kapourani CA, Imaz-Rosshandler I, Lohoff T, Xiang Y, Hanna CW, et al. 2019. Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature 576: 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballantyne MD, Pinel K, Dakin R, Vesey AT, Diver L, Mackenzie R, Garcia R, Welsh P, Sattar N, Hamilton G, et al. 2016. Smooth muscle enriched long noncoding RNA (SMILR) regulates cell proliferation. Circulation 133: 2050–2065. 10.1161/CIRCULATIONAHA.115.021019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RD, Long X, Lin M, Bergmann JH, Nanda V, Cowan SL, Zhou Q, Han Y, Spector DL, Zheng D, et al. 2014. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler Thromb Vasc Biol 34: 1249–1259. 10.1161/ATVBAHA.114.303240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blinka S, Reimer MH Jr, Pulakanti K, Rao S. 2016. Super-enhancers at the Nanog locus differentially regulate neighboring pluripotency-associated genes. Cell Rep 17: 19–28. 10.1016/j.celrep.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau BG. 2013. Signaling and transcriptional networks in heart development and regeneration. Cold Spring Harb Perspect Biol 5: a008292 10.1101/cshperspect.a008292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. 2011. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927. 10.1101/gad.17446611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563. 10.1126/science.1112014 [DOI] [PubMed] [Google Scholar]
- Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. 2009. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 460: 705–710. 10.1038/nature08195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti L, Celano G, Dagradi F, Schwartz PJ. 2008. Congenital long QT syndrome. Orphanet J Rare Dis 3: 18 10.1186/1750-1172-3-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Li Y, Zhou L, Cho J, Patel B, Terada N, Li Y, Bungert J, Qiu Y, Huang S. 2016. HoxBlinc RNA recruits set1/MLL complexes to activate Hox gene expression patterns and mesoderm lineage development. Cell Rep 14: 103–114. 10.1016/j.celrep.2015.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux Y, Zangrando J, Schroen B, Creemers EE, Pedrazzini T, Chang CP, Dorn GW II, Thum T, Heymans S. 2015. Long noncoding RNAs in cardiac development and ageing. Nat Rev Cardiol 12: 415–425. 10.1038/nrcardio.2015.55 [DOI] [PubMed] [Google Scholar]
- Eißmann M, Gutschner T, Hämmerle M, Günther S, Caudron-Herger M, Groß M, Schirmacher P, Rippe K, Braun T, Zörnig M, et al. 2012. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol 9: 1076–1087. 10.4161/rna.21089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES. 2016. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539: 452–455. 10.1038/nature20149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SM, Yelon D, Conlon FL, Kirby ML. 2010. Myocardial lineage development. Circ Res 107: 1428–1444. 10.1161/CIRCRESAHA.110.227405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S, Aguirre A, Hescheler J, Kurian L. 2016. An lncRNA perspective into (re)building the heart. Front Cell Dev Biol 4: 128 10.3389/fcell.2016.00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S, Ahuja G, Bartsch D, Russ N, Yao W, Kuo JC, Derks JP, Akhade VS, Kargapolova Y, Georgomanolis T, et al. 2019. yylncT defines a class of divergently transcribed lncRNAs and safeguards the T-mediated mesodermal commitment of human PSCs. Cell Stem Cell 24: 318–327.e8. 10.1016/j.stem.2018.11.005 [DOI] [PubMed] [Google Scholar]
- Fu X, Khalil H, Kanisicak O, Boyer JG, Vagnozzi RJ, Maliken BD, Sargent MA, Prasad V, Valiente-Alandi I, Blaxall BC, et al. 2018. Specialized fibroblast differentiated states underlie scar formation in the infarcted mouse heart. J Clin Invest 128: 2127–2143. 10.1172/JCI98215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furtado MB, Nim HT, Boyd SE, Rosenthal NA. 2016. View from the heart: cardiac fibroblasts in development, scarring and regeneration. Development 143: 387–397. 10.1242/dev.120576 [DOI] [PubMed] [Google Scholar]
- George MR, Duan Q, Nagle A, Kathiriya IS, Huang Y, Rao K, Haldar SM, Bruneau BG. 2019. Minimal in vivo requirements for developmentally regulated cardiac long intergenic non-coding RNAs. Development (Cambridge, England) 146: dev185314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M, et al. 2013. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 24: 206–214. 10.1016/j.devcel.2012.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günthel M, Barnett P, Christoffels VM. 2018. Development, proliferation, and growth of the mammalian heart. Mol Ther 26: 1599–1609. 10.1016/j.ymthe.2018.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Xu Y, Wang Z, Wu Y, Chen J, Wang G, Lu C, Jia W, Xi J, Zhu S, et al. 2018. A Linc1405/Eomes complex promotes cardiac mesoderm specification and cardiogenesis. Cell Stem Cell 22: 893–908.e6. 10.1016/j.stem.2018.04.013 [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227. 10.1038/nature07672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P, Li W, Lin CH, Yang J, Shang C, Nuernberg ST, Jin KK, Xu W, Lin CY, Lin CJ, et al. 2014. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 514: 102–106. 10.1038/nature13596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Zhang J, Liu Y, Fan X, Ai S, Luo Y, Li X, Jin H, Luo S, Zheng H, et al. 2019. The lncRNA Hand2os1/Uph locus orchestrates heart development through regulation of precise expression of Hand2. Development 146: dev176198 10.1242/dev.176198 [DOI] [PubMed] [Google Scholar]
- Huang S, Zhang L, Song J, Wang Z, Huang X, Guo Z, Chen F, Zhao X. 2019. Long noncoding RNA MALAT1 mediates cardiac fibrosis in experimental postinfarct myocardium mice model. J Cell Physiol 234: 2997–3006. 10.1002/jcp.27117 [DOI] [PubMed] [Google Scholar]
- Ivey MJ, Tallquist MD. 2016. Defining the cardiac fibroblast. Circ J 80: 2269–2276. 10.1253/circj.CJ-16-1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE, Hsiao EC, Schwartz RJ, Conklin BR, Bernstein HS, et al. 2008. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell 2: 219–229. 10.1016/j.stem.2008.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Liu Y, Liu R, Zhang K, Zhang Y. 2015. The lncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression. Cell Rep 11: 137–148. 10.1016/j.celrep.2015.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson P, Lipovich L, Grandér D, Morris KV. 2014. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta 1840: 1063–1071. 10.1016/j.bbagen.2013.10.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikkonen MU, Lam MT, Glass CK. 2011. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res 90: 430–440. 10.1093/cvr/cvr097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiriya IS, Nora EP, Bruneau BG. 2015. Investigating the transcriptional control of cardiovascular development. Circ Res 116: 700–714. 10.1161/CIRCRESAHA.116.302832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, Marinov GK, Ward LD, Birney E, Crawford GE, Dekker J, et al. 2014. Defining functional DNA elements in the human genome. Proc Natl Acad Sci 111: 6131–6138. 10.1073/pnas.1318948111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Tan Y, Ma W, Merkurjev D, Destici E, Ma Q, Suter T, Ohgi K, Friedman M, Skowronska-Krawczyk D, et al. 2018. Pluripotency factors functionally premark cell-type-restricted enhancers in ES cells. Nature 556: 510–514. 10.1038/s41586-018-0048-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, et al. 2013. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152: 570–583. 10.1016/j.cell.2013.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. 2015. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517: 583–588. 10.1038/nature14136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostowski L, Sedlak N, Engel N. 2012. The Kcnq1ot1 long non-coding RNA affects chromatin conformation and expression of Kcnq1, but does not regulate its imprinting in the developing heart. PLoS Genet 8: e1002956 10.1371/journal.pgen.1002956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian L, Aguirre A, Sancho-Martinez I, Benner C, Hishida T, Nguyen TB, Reddy P, Nivet E, Krause MN, Nelles DA, et al. 2015. Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation 131: 1278–1290. 10.1161/CIRCULATIONAHA.114.013303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Rosenfeld MG. 2016. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet 17: 207–223. 10.1038/nrg.2016.4 [DOI] [PubMed] [Google Scholar]
- Li B, Hu Y, Li X, Jin G, Chen X, Chen G, Chen Y, Huang S, Liao W, Liao Y, et al. 2018. Sirt1 antisense long noncoding RNA promotes cardiomyocyte proliferation by enhancing the stability of Sirt1. J Am Heart Assoc 7: e009700 10.1161/JAHA.118.009700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Li Y, Lin B, Sheng Y, Yang L. 2017. HBL1 is a human long noncoding RNA that modulates cardiomyocyte development from pluripotent stem cells by counteracting MIR1. Dev Cell 42: 333–348.e5. 10.1016/j.devcel.2017.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, Prescott SL, Wysocka J. 2016. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167: 1170–1187. 10.1016/j.cell.2016.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TY, Lin B, Li Y, Arora A, Han L, Cui C, Coronnello C, Sheng Y, Benos PV, Yang L. 2013. Overexpression of microRNA-1 promotes cardiomyocyte commitment from human cardiovascular progenitors via suppressing WNT and FGF signaling pathways. J Mol Cell Cardiol 63: 146–154. 10.1016/j.yjmcc.2013.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Lu JY, Liu L, Yin Y, Chen C, Han X, Wu B, Xu R, Liu W, Yan P, et al. 2016. Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell 18: 637–652. 10.1016/j.stem.2016.01.024 [DOI] [PubMed] [Google Scholar]
- Marchese FP, Raimondi I, Huarte M. 2017. The multidimensional mechanisms of long noncoding RNA function. Genome Biol 18: 206 10.1186/s13059-017-1348-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques AC, Hughes J, Graham B, Kowalczyk MS, Higgs DR, Ponting CP. 2013. Chromatin signatures at transcriptional start sites separate two equally populated yet distinct classes of intergenic long noncoding RNAs. Genome Biol 14: R131 10.1186/gb-2013-14-11-r131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melé M, Rinn JL. 2016. “Cat's Cradling” the 3D genome by the act of lncRNA transcription. Mol Cell 62: 657–664. 10.1016/j.molcel.2016.05.011 [DOI] [PubMed] [Google Scholar]
- Mercer TR, Mattick JS. 2013. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 20: 300–307. 10.1038/nsmb.2480 [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. 2009. Long non-coding RNAs: insights into functions. Nat Rev Genet 10: 155–159. 10.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- Michalik KM, You X, Manavski Y, Doddaballapur A, Zörnig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, et al. 2014. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res 114: 1389–1397. 10.1161/CIRCRESAHA.114.303265 [DOI] [PubMed] [Google Scholar]
- Micheletti R, Plaisance I, Abraham BJ, Sarre A, Ting CC, Alexanian M, Maric D, Maison D, Nemir M, Young RA, et al. 2017. The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci Transl Med 9: aai9118 10.1126/scitranslmed.aai9118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore-Morris T, van Vliet PP, Andelfinger G, Puceat M. 2018. Role of epigenetics in cardiac development and congenital diseases. Physiol Rev 98: 2453–2475. 10.1152/physrev.00048.2017 [DOI] [PubMed] [Google Scholar]
- Morris KV, Mattick JS. 2014. The rise of regulatory RNA. Nat Rev Genet 15: 423–437. 10.1038/nrg3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ounzain S, Pedrazzini T. 2015. The promise of enhancer-associated long noncoding RNAs in cardiac regeneration. Trends Cardiovasc Med 25: 592–602. 10.1016/j.tcm.2015.01.014 [DOI] [PubMed] [Google Scholar]
- Ounzain S, Pezzuto I, Micheletti R, Burdet F, Sheta R, Nemir M, Gonzales C, Sarre A, Alexanian M, Blow MJ, et al. 2014. Functional importance of cardiac enhancer-associated noncoding RNAs in heart development and disease. J Mol Cell Cardiol 76: 55–70. 10.1016/j.yjmcc.2014.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ounzain S, Micheletti R, Beckmann T, Schroen B, Alexanian M, Pezzuto I, Crippa S, Nemir M, Sarre A, Johnson R, et al. 2015a. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur Heart J 36: 353–368a. 10.1093/eurheartj/ehu180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ounzain S, Micheletti R, Arnan C, Plaisance I, Cecchi D, Schroen B, Reverter F, Alexanian M, Gonzales C, Ng SY, et al. 2015b. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J Mol Cell Cardiol 89: 98–112. 10.1016/j.yjmcc.2015.09.016 [DOI] [PubMed] [Google Scholar]
- Perry RB, Ulitsky I. 2016. The functions of long noncoding RNAs in development and stem cells. Development 143: 3882–3894. 10.1242/dev.140962 [DOI] [PubMed] [Google Scholar]
- Plaisance I, Perruchoud S, Fernandez-Tenorio M, Gonzales C, Ounzain S, Ruchat P, Nemir M, Niggli E, Pedrazzini T. 2016. Cardiomyocyte lineage specification in adult human cardiac precursor cells via modulation of enhancer-associated long noncoding RNA expression. JACC Basic Transl Sci 1: 472–493. 10.1016/j.jacbts.2016.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu SD, Frangogiannis NG. 2016. The biological basis for cardiac repair after myocardial infarction: from inflammation to fibrosis. Circ Res 119: 91–112. 10.1161/CIRCRESAHA.116.303577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff JD, Wei Y, Khavari PA. 2018. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol 19: 143–157. 10.1038/nrm.2017.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter N, Ali T, Kopitchinski N, Schuster P, Beisaw A, Hendrix DA, Schulz MH, Müller-McNicoll M, Dimmeler S, Grote P. 2019. The lncRNA locus Handsdown regulates cardiac gene programs and is essential for early mouse development. Dev Cell 50: 644–657.e8. 10.1016/j.devcel.2019.07.013 [DOI] [PubMed] [Google Scholar]
- Rizki G, Boyer LA. 2015. Lncing epigenetic control of transcription to cardiovascular development and disease. Circ Res 117: 192–206. 10.1161/CIRCRESAHA.117.304156 [DOI] [PubMed] [Google Scholar]
- Sigova AA, Abraham BJ, Ji X, Molinie B, Hannett NM, Guo YE, Jangi M, Giallourakis CC, Sharp PA, Young RA. 2015. Transcription factor trapping by RNA in gene regulatory elements. Science 350: 978–981. 10.1126/science.aad3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergachis AB, Neph S, Reynolds A, Humbert R, Miller B, Paige SL, Vernot B, Cheng JB, Thurman RE, Sandstrom R, et al. 2013. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell 154: 888–903. 10.1016/j.cell.2013.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam PP, Loebel DA. 2007. Gene function in mouse embryogenesis: get set for gastrulation. Nat Rev Genet 8: 368–381. 10.1038/nrg2084 [DOI] [PubMed] [Google Scholar]
- Tan JY, Smith AAT, Ferreira da Silva M, Matthey-Doret C, Rueedi R, Sönmez R, Ding D, Kutalik Z, Bergmann S, Marques AC. 2017. cis-acting complex-trait-associated lincRNA expression correlates with modulation of chromosomal architecture. Cell Rep 18: 2280–2288. 10.1016/j.celrep.2017.02.009 [DOI] [PubMed] [Google Scholar]
- Thum T, Fiedler J. 2014. LINCing MALAT1 and angiogenesis. Circ Res 114: 1366–1368. 10.1161/CIRCRESAHA.114.303896 [DOI] [PubMed] [Google Scholar]
- Touma M, Kang X, Zhao Y, Cass AA, Gao F, Biniwale R, Coppola G, Xiao X, Reemtsen B, Wang Y. 2016. Decoding the long noncoding RNA during cardiac maturation: a roadmap for functional discovery. Circ Cardiovasc Genet 9: 395–407. 10.1161/CIRCGENETICS.115.001363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viereck J, Kumarswamy R, Foinquinos A, Xiao K, Avramopoulos P, Kunz M, Dittrich M, Maetzig T, Zimmer K, Remke J, et al. 2016. Long noncoding RNA Chast promotes cardiac remodeling. Sci Transl Med 8: 326ra22 10.1126/scitranslmed.aaf1475 [DOI] [PubMed] [Google Scholar]
- Waardenberg AJ, Ramialison M, Bouveret R, Harvey RP. 2014. Genetic networks governing heart development. Cold Spring Harb Perspect Med 4: a013839 10.1101/cshperspect.a013839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. 2012. The epigenotype. 1942. Int J Epidemiol 41: 10–13. 10.1093/ije/dyr184 [DOI] [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. 2012. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell 151: 206–220. 10.1016/j.cell.2012.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamstad JA, Wang X, Demuren OO, Boyer LA. 2014. Distal enhancers: new insights into heart development and disease. Trends Cell Biol 24: 294–302. 10.1016/j.tcb.2013.10.008 [DOI] [PubMed] [Google Scholar]
- Wang L, Chen YG. 2016. Signaling control of differentiation of embryonic stem cells toward mesendoderm. J Mol Biol 428: 1409–1422. 10.1016/j.jmb.2015.06.013 [DOI] [PubMed] [Google Scholar]
- Wang A, Yue F, Li Y, Xie R, Harper T, Patel NA, Muth K, Palmer J, Qiu Y, Wang J, et al. 2015. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell 16: 386–399. 10.1016/j.stem.2015.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhang XJ, Ji YX, Zhang P, Deng KQ, Gong J, Ren S, Wang X, Chen I, Wang H, et al. 2016. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat Med 22: 1131–1139. 10.1038/nm.4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber KT, Sun Y, Bhattacharya SK, Ahokas RA, Gerling IC. 2013. Myofibroblast-mediated mechanisms of pathological remodelling of the heart. Nat Rev Cardiol 10: 15–26. 10.1038/nrcardio.2012.158 [DOI] [PubMed] [Google Scholar]
- Xue Z, Hennelly S, Doyle B, Gulati AA, Novikova IV, Sanbonmatsu KY, Boyer LA. 2016. A G-rich motif in the lncRNA Braveheart interacts with a zinc-finger transcription factor to specify the cardiovascular lineage. Mol Cell 64: 37–50. 10.1016/j.molcel.2016.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Yan P, Lu J, Song G, Zhu Y, Li Z, Zhao Y, Shen B, Huang X, Zhu H, et al. 2015. Opposing roles for the lncRNA Haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell 16: 504–516. 10.1016/j.stem.2015.03.007 [DOI] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. 2014. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515: 355–364. 10.1038/nature13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S, Brueckner M. 2017. Genetics and genomics of congenital heart disease. Circ Res 120: 923–940. 10.1161/CIRCRESAHA.116.309140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, et al. 2012. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep 2: 111–123. 10.1016/j.celrep.2012.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhang W, Lin M, Wu W, Jiang P, Tou E, Xue M, Richards A, Jourd'heuil D, Asif A, et al. 2016. MYOSLID is a novel serum response factor-dependent long noncoding RNA that amplifies the vascular smooth muscle differentiation program. Arterioscler Thromb Vasc Biol 36: 2088–2099. 10.1161/ATVBAHA.116.307879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JG, Shen YH, Liu HL, Liu M, Shen YQ, Kong XQ, Song GX, Qian LM. 2014a. Long noncoding RNAs expression profile of the developing mouse heart. J Cell Biochem 115: 910–918. 10.1002/jcb.24733 [DOI] [PubMed] [Google Scholar]
- Zhu S, Hu X, Han S, Yu Z, Peng Y, Zhu J, Liu X, Qian L, Zhu C, Li M, et al. 2014b. Differential expression profile of long non-coding RNAs during differentiation of cardiomyocytes. Int J Med Sci 11: 500–507. 10.7150/ijms.7849 [DOI] [PMC free article] [PubMed] [Google Scholar]