Abstract
Metastasis is a highly dynamic process during which cancer and microenvironmental cells undergo a cascade of events required for efficient dissemination throughout the body. During the metastatic cascade, tumor cells can change their state and behavior, a phenomenon commonly defined as cellular plasticity. To monitor cellular plasticity during metastasis, high-resolution intravital microscopy (IVM) techniques have been developed and allow us to visualize individual cells by repeated imaging in animal models. In this review, we summarize the latest technological advancements in the field of IVM and how they have been applied to monitor metastatic events. In particular, we highlight how longitudinal imaging in native tissues can provide new insights into the plastic physiological and developmental processes that are hijacked by cancer cells during metastasis.
Metastasis, the dispersion of tumor cells throughout the body, represents the largest cause of cancer-related death. Various multifaceted events occur during the escape of metastatic cells from the primary tumor and dissemination to secondary sites (Chaffer et al. 2016). A detailed understanding of these events is required for the development of efficient antimetastatic strategies (Steeg 2016). Throughout the different steps of the metastatic cascade, tumor cells can change their state, a phenomenon commonly defined as “cellular plasticity,” and this plasticity complicates the identification of therapeutic targets. Investigating these plastic events is rather limited when using conventional techniques, such as biochemical and histology assays, that draw static pictures. In contrast, development of microscopy techniques for in vivo imaging, often referred to as intravital microscopy (IVM), has provided unprecedented insights into the dynamics of cancer progression and metastatic spread (Beerling et al. 2011; Entenberg et al. 2013; Conway et al. 2014; Nobis et al. 2018). Imaging tissues in living animals was first reported in the 19th century, however due to the limited properties of the optics and the lack of contrast, IVM was mainly used to visualize tissue vasculature (Wagner 1839). In 1958, metastatic cells in the rabbit ear were imaged for the first time using an imaging chamber (Wood 1958). Since the 21st century, the development of genetic rodent models that express fluorescent proteins, and technical improvements in the field of microscopy have enabled researchers to visualize biological events deeply into live tissues, with enhanced resolution and timescale. In this review, we describe the latest technical developments for high-resolution fluorescence IVM as well as infrequently used high-end techniques that could be further used to image metastatic spread. Next, we discuss the latest biological findings provided by IVM regarding the plastic behaviors of disseminating cancer cells and stromal compartments during metastasis.
RECENTLY DEVELOPED IVM TECHNIQUES
Here, we provide an update on the most recently developed IVM technologies, for more detailed reviews on the historical development of IVM techniques and the tools now commonly used for IVM, we refer to (Beerling et al. 2011; Pittet and Weissleder 2011; Conway et al. 2014; Ellenbroek and van Rheenen 2014; Nobis et al. 2018).
Novel Technologies to Identify Cancer Cells and Their Microenvironment
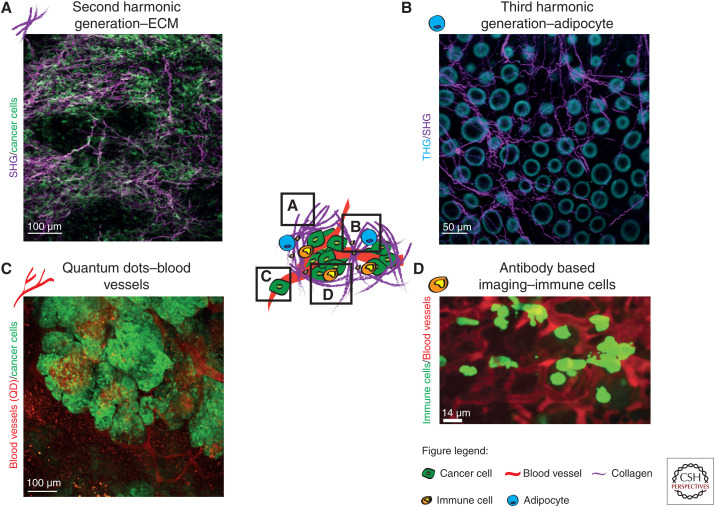
The ability to simultaneously acquire signals from several channels using IVM enables the combination of multiple fluorescent probes in a single IVM experiment. While mice genetically engineered to endogenously express fluorescent labels are commonly used to study specific biological processes (Abe and Fujimori 2013; Conway et al. 2014; Ellenbroek and van Rheenen 2014; Suijkerbuijk and van Rheenen 2017; Nobis et al. 2018), they can be combined with different strategies to visualize multiple cancer compartments. For instance, infrared light is often used to excite fluorophores deep inside tissues, but this light can also produce label-free signals from structures within the tumor microenvironment (TME) (Weissleder 2001). For example, noncentrosymmetric structures such as coiled-coil and polymeric proteins can be visualized using second harmonic generation (SHG) imaging (Cicchi and Pavone 2017). Similarly, third-harmonic generation (THG) imaging is a label-free strategy that relies on the light scattering properties and mismatches of refractive indexes of water–lipid and water–protein interfaces (Yildirim et al. 2015). During SHG and THG, two or three photons are combined into one photon with a wavelength of half or a third of the excitation wavelength. SHG has enabled the visualization of extracellular matrix (ECM) remodeling during cancer progression and response to therapy (Fig. 1A; Weigelin et al. 2012; Nobis et al. 2013a; Burke and Brown 2014; Lawson et al. 2015; Vennin et al. 2017) and THG can be used to visualize elastin, erythrocytes, adipocytes, endothelial cells, and leukocytes (Fig. 1B; You et al. 2018; van Huizen et al. 2019). SHG and THG imaging were recently combined to localize implanted biomaterials in mice and to study immunological responses upon surgery (Dondossola et al. 2016). Similarly, two- and three-photon imaging can be used to monitor metabolism via for instance label-free detection of FAD and NADH (Blacker and Duchen 2016). Recently, You et al. (2018) developed a multiharmonic technique relying on a single-excitation source nonlinear imaging platform for concurrent imaging of FAD/NADH along with SHG and THG signals, to map the microenvironment of breast tumors. A recent labeling strategy relies on quantum dots (QD), which are nanoparticles with excellent optical properties for in vivo imaging (Matea et al. 2017). QD are frequently used for real-time imaging of blood vessels (Ornes 2016) and have also been used to monitor cell extravasation, angiogenesis, tumor vessel leakiness and to label rare cell populations such as hematopoietic stem cells (HSCs) (Fig. 1C; Nobis et al. 2013a; Gallego-Ortega et al. 2015; Bruns et al. 2017; Vennin et al. 2017; Wang et al. 2018). In addition, QD can be conjugated with antibodies to visualize specific cell subtypes and to quantify the amount of targeted cell population using in vivo cytometry (Lidke et al. 2007). Similarly, injecting fluorescently labeled dextran or antibodies can mark tumor-associated vasculature or specific cell populations such as immune cells (Fig. 1D; Progatzky et al. 2013; Freise and Wu 2015; Thanabalasuriar et al. 2016). Last, near-infrared (NIR) fluorescent probes have increased the possibilities for multichannel imaging (Shcherbakova et al. 2016). Because of the low absorption of NIR emission light by tissues and due to the low autofluorescence of NIR wavelengths, deeper structures can be imaged. However, the low sensitivity of most detectors at infrared light often lowers the signal-to-background ratio (Luo et al. 2011). NIR probes were recently used for IVM imaging of senescent cells, protein–protein interactions, cell cycle status, RNA detection, signaling cascades, and lipoprotein turnover (Shcherbakova et al. 2016; Bruns et al. 2017). Interestingly, NIR probes such as indocyanine green are used in patients to visualize blood vessels (Carr et al. 2018). The combination of this large range of probes for IVM represents powerful avenues to simultaneously study multiple metastatic events.
Figure 1.
Novel developments to label cancer cells and their microenvironment. (A) Live SHG imaging (purple) of the ECM in a pancreatic cancer patient-derived xenograft with high ECM content. [Panel A adapted from Vennin et al. (2017) with permission from The American Association for the Advancement of Science © 2017.] (B) THG imaging of adipocytes (blue) and SHG imaging of the ECM (magenta) in the healthy mammary fat pad. (C) Quantum dot labeling of tumor-associated vasculature in breast tumors. [Panel C reprinted from Gallego-Ortega et al. (2015) under the terms of the Creative Attribution License.] (D) Visualization of natural killer cells residing in the lungs of a healthy mouse using conjugated-antibodies and injected in vivo. [Panel D based on data in Thanabalasuriar et al. (2016)].
IVM of Fluorescent Properties to Reveal Molecular Events
Fluorescent biosensors developed for IVM can report subcellular events, such as transient protein–protein interactions or protein activity (Nobis et al. 2013b, 2018; Conway et al. 2014). Genetically encoded FRET (Förster resonance energy transfer) biosensors rely on two fluorescent proteins (a donor and an acceptor), where excited donors transfer their energy to acceptors, triggering a loss of donor emission and a gain in acceptor emission. FRET depends on the proximity between the donor and the acceptor, their spectral overlap and their alignment (Rajoria et al. 2014). Intravital measurements of FRET are achieved by either ratiometric measurements of donor and acceptor signals, by measuring the sensitized emission of the acceptor, or by measuring the donor's fluorescence lifetime (FLIM) (Piston and Kremers 2007; Jalink 2013; Conway et al. 2014; Martin et al. 2018). Cell motility can be studied using for example FRET-reporters for RhoA (Nobis et al. 2017), Rac (Johnsson et al. 2014; Yukinaga et al. 2014), Src (Nobis et al. 2013a; Vennin et al. 2017), SWAP70 (Kriplani et al. 2019), Arf6 (Hall et al. 2008), or FAK (Seong et al. 2013) activity. Similarly, FRET biosensors have shed light on the dynamics of tyrosine kinase signaling transduction (Akt, Erk reporters) (Aoki et al. 2017; Conway et al. 2018; Muta et al. 2018), membrane receptor activity (PDGFR, GPCR biosensors) (Seong et al. 2017; Halls 2019), cell response to tissue tension (MMP-9, Src, FAK, or Eph4 biosensors) (Wang et al. 2005; Ouyang et al. 2010; Seong et al. 2011; Stawarski et al. 2014; Pan et al. 2019), immune cell activity (Fyn, ZAP-70 biosensors) (Li et al. 2016; Ouyang et al. 2019), and cancer cell metabolism (ATP, cAMP biosensors) (Imamura et al. 2009; Klarenbeek et al. 2015; Annamdevula et al. 2018; Husada et al. 2018). Last, monitoring cell cycle progression and response to treatments can be assessed using CDK1 (Gavet and Pines 2010a,b; Vennin et al. 2017), PLK1 (Gheghiani et al. 2017) or caspase-3 FRET reporters (van der Wal et al. 2001; Janssen et al. 2013). A caveat of FRET sensors is their rather low dynamic range, with a typical 5%–10% change in FRET upon modification of molecule activity or interactions in vitro (Lam et al. 2001). This limitation is further increased in vivo, where the signal-to-noise level is low. In addition, emission at shorter wavelengths suffers from more scattering when imaging in deep tissues. This leads to differences in detection efficiency of donor and acceptor signals and impairs FRET measurements that depend on ratiometric and sensitized emission (Jares-Erijman and Jovin 2003). In addition, FLIM-FRET imaging requires long acquisition times of the signal, and physiological motions in vivo can impair the measurements. This can be resolved by software developed for the correction of sample motion blurring, which offer new opportunities for FLIM-FRET quantification of in vivo processes (Lowe 2004; Kaifosh et al. 2014; Warren et al. 2018).
Fluorescence recovery after photobleaching (FRAP) also allows to study molecular processes. Here, a fluorescently labeled molecule is photobleached in a targeted subcellular region and fluorescence recovery in this region is recorded by time-lapse microscopy. The speed and extent of fluorescence recovery is used as a readout of the mobility and turnover of the bleached molecule (Fritzsche and Charras 2015). FRAP has increased our understanding of processes driving the mobility of membrane proteins, protein–protein interactions, cell–cell junction turnover, cell motility and invasion, signal transduction, DNA damage, force transmission, and ECM features (Dunham 2004; Goehring et al. 2010; Erami et al. 2016; Khait et al. 2016; Kumar et al. 2016; Steurer et al. 2019). FRAP can be complemented with fluorescence loss in photobleaching (FLIP), where loss of fluorescence in areas adjacent to the bleached region is measured, and thereby strengthens the observed changes in molecule mobility obtained by FRAP (Erami et al. 2016). Moreover, optogenetic probes represent powerful tools for manipulating and monitoring cell behavior with enhanced spatiotemporal resolution. Optogenetic probes are light-responsive proteins that can be used to selectively manipulate molecular processes (Shemesh et al. 2017). Such sensors were initially engineered to probe neural activity, however recently optogenetic tools were used to dissect molecular events occurring during cell motility, epithelial-to-mesenchymal transition (EMT), transcription, mechanotransduction, and immune modulation (Tan et al. 2017; Valon et al. 2017; Fielmich et al. 2018; O'Neill et al. 2018; Qin et al. 2018; Zago et al. 2018; Zhou et al. 2018).
Considering the intricate interdependence of signaling pathways, combined imaging and analysis of multiple fluorescent biosensors in the same cell could in the future increase our understanding of the complexity of biological processes driving metastasis.
Recently Developed Optical Windows
Most sites where solid tumors metastasize to, such as the lungs and the liver, have mainly been imaged upon surgical exposure, which limits the visualization of the metastatic process to up to 72 h (Ewald et al. 2011; Entenberg et al. 2015). To overcome this, optical imaging windows allow the longitudinal monitoring of tumor progression in native tissues (Alieva et al. 2014; Conway et al. 2014; Ellenbroek and van Rheenen 2014). Historically, the first windows were engineered to image tumors transplanted in the brain or skin (Brown et al. 2003; Kienast et al. 2010). More recently, the development of lung and abdominal windows and technologies for IVM in the bone and bone marrow allow us to study metastatic spread in secondary sites. For instance, we previously developed an abdominal imaging window (AIW) (Ritsma et al. 2012), consisting of a titanium ring with a coverslip on top, which can be tightly implanted to the skin and abdominal wall using a purse-string suture. The AIW can be maintained in mice for up to 9 wk without significant alterations of animal behavior. We used the AIW to track metastatic cells and revealed that in the early stages of liver colonization, migration is required for efficient establishment of metastasis (Supplemental Movie S1; Ritsma et al. 2012). Similarly, Entenberg et al. developed a new window for lung imaging, a metastatic site for which the success of treatments remains moderate. The authors engineered a steel, passivated ring which can be inserted into mice between the ribs (Entenberg et al. 2018). While previous approaches for lung IVM required mechanical ventilation of the animal and were therefore terminal procedures (Looney et al. 2011; Entenberg et al. 2015), this novel lung window was used daily for 2 wk and was combined with microcartography to study multiple steps of lung metastasis (Entenberg et al. 2018). Other common metastatic sites are the bone and bone marrow, which are optically dense and highly scattering tissues, and therefore also present difficulties for IVM. While bone metastasis is a critical burden for patients with advanced cancer (Croucher et al. 2016), our knowledge of this disease is derived mostly from static approaches. Repeated surgical exposure of bones can be used to longitudinally study metastasis. For instance, monitoring dormant cancer cells in the endosteal niche was achieved via surgical exposure of the same bone area and using fiducial marks to localize single cells over time (Lawson et al. 2015). Implantation of miniaturized tissue-engineered bone constructs under a skin-fold window was also used to image bone-like tissues (Dondossola et al. 2018; Khosravi et al. 2018; Stiers et al. 2018). For instance, Dondossola et al. (2018), engineered an ossicle that recapitulates important mechanical and biological features of the native bone, while also displaying high optical accessibility. Combining a skin window with this implanted engineered bone constructs allowed visualization of tumor-stroma crosstalk during metastatic spread, as well as the remodeling of the bone niche during therapy (Dondossola et al. 2018). Similarly, endothelial cells in bone marrow vessels can be specifically visualized in GFP-Flk1 mice, and this enabled the direct in vivo monitoring of HSCs homing in the bone marrow, as well as the measurement of blood cell flow dynamics (Bixel et al. 2017). Last, intracranial windows have been optimized to image the dynamics of brain colonization (Kienast et al. 2010; Goldey et al. 2014; Alieva et al. 2017, 2019). Following craniotomy of the parietal bone and removal of the meninges to expose the brain cortex, a glass window is glued along with an adaptor ring for fixation onto the microscope stage. Such strategies have enabled long-term monitoring of the different steps of metastatic spread, cell invasion and changes of the TME in the brain (Kienast et al. 2010; Erapaneedi et al. 2016; Alieva et al. 2017, 2019).
Combining IVM with Multiple Techniques to Enhance Our Understanding of Metastasis
While each microscopy technique comes with its benefits and caveats, combining IVM with complimentary approaches can enhance our understanding of the complexity of metastasis. For instance, we previously developed cryosection labeling and intravital imaging (CLIM) to integrate IVM with histochemistry analyses (Ritsma et al. 2013). Here, an area imaged by IVM can be “tattooed” using light to subsequently relocate this specific area in histological sections (Ritsma et al. 2013). We used CLIM to interrogate how microenvironmental events influence tumor cell invasion, and we found that interactions with T cells lead to enhance cancer cell motility (Ritsma et al. 2013). Similarly, IVM can be combined with superresolution imaging. For instance, correlative light and electron microscopy (CLEM) combines IVM data with electron microscopy (EM) to obtain a complete view at the nanoscale level of tissues (Karreman et al. 2016a; Hyenne et al. 2019). The main bottleneck of CLEM is the difficulty to precisely locate by EM the region imaged previously by IVM, however this can be facilitated using fiducial marks such as blood vessels, nuclei, or myelinated axons (Karreman et al. 2016a; Goetz 2018; Luckner et al. 2018). CLEM has provided new high-resolution information into the subcellular events driving tumor cell invasion, extravasation of circulating tumor cells (CTCs) and uptake of cancer cell-derived extracellular vesicles (EVs) (Karreman et al. 2014, 2016b; Follain et al. 2018; Hyenne et al. 2019). Lastly, single-cell sequencing has also been used to characterize cells identified using IVM (Beerling et al. 2016; Scheele et al. 2017). This enables researchers to link dynamic behaviors visualized via IVM with molecular signatures obtained using single-cell sequencing.
Imaging Plasticity of Cell Lineages
Progression through the metastatic cascade can occur over periods of weeks, months, or even years. During this period, tumor cells can change their state and fate (Chaffer et al. 2016). Fluorescent-based reporters are used together with IVM to track the same cells and their progeny over time and enable linking cell status to cell fate. For instance, photoconvertible fluorophores such as Dendra (Kedrin et al. 2008; Fumagalli et al. 2017), PSmOrange (Subach et al. 2011), EosFP (McKinney et al. 2009), or Kaede (Ando et al. 2002; Torcellan et al. 2017) can change color upon laser exposure and are used to track cells in time and space and during the lifetime of the photoconverted fluorophore.
While photoconvertible probes allow transient marking of targeted cells, lineage tracing tools label cells with an inheritable mark. A common lineage tracing strategy relies on the expression of unbiased or lineage-specific Cre-recombinase to permanently activate a fluorescent reporter (Livet et al. 2007; Snippert et al. 2010; Scheele et al. 2017). Lineage tracing probes are used to study plasticity of tumor cell identity, hierarchy, and clonality. For example, static and IVM-based lineage tracing recently showed that the cellular hierarchy of nontransformed tissues is maintained in a number of tumors (Driessens et al. 2012; Schepers et al. 2012; Zomer et al. 2013). Using lineage tracing, these studies identified a small population of cancer stem cells (CSCs) in skin, colorectal, and breast tumors that have self-renewal capacity and give rise to more differentiated cancer cells (Driessens et al. 2012; Schepers et al. 2012; Zomer et al. 2013). Moreover, IVM of lineage tracing tools revealed that cancer stemness is a plastic state that can be lost or gained over time, rather than a fixed cell property (Zomer et al. 2013). In addition to differences in stem cell properties, cancer cells can acquire different growth potentials, and this could be due to accumulation of mutations or alterations of the microenvironment. For instance, static lineage tracing revealed that a strong selection pressure can occur during progression from benign lesions to carcinoma stages, and this results in the dominance of one single clone (Reeves et al. 2018). Similarly, IVM revealed that precursor lesions in pancreatic cancer are polyclonal, however, this diversity decreases during cancer progression while a significant fraction of metastatic lesions are polyclonal (Maddipati and Stanger 2015). Similarly in breast cancer, metastatic cells homing in distant sites were found to be polyclonal (Cheung et al. 2016). Last, IVM showed that stem cells harboring oncogenic activation of the Wnt/β-catenin pathway in mouse skin epithelium can form tumor lesions (Brown et al. 2017). This study further showed that wild-type cells can actively eliminate transformed stem cells. This leads to tumor regression and even complete disappearance of the tumor, thereby reestablishing normal homeostasis (Brown et al. 2017). Lineage tracing can also be used to label cells that take up material released by other cells. For example, packaging of Cre-recombinase into EVs released by donor cells enables the specific labeling of recipient cells expressing a Cre-dependent reporter (Zomer et al. 2015; Steenbeek et al. 2018). This method of cell labeling, combined with IVM, revealed for instance that aggressive breast tumor cells can transmit their invasive behavior to recipient cells through the exchange of biomolecules carried by EVs (Supplemental Movie S2; Ridder et al. 2014, 2015; Zomer et al. 2015; Steenbeek et al. 2018). Recently, new recombinase-based systems were engineered to label cells in a more controllable manner. For instance, a photoactivatable Cre-recombinase (PA-Cre) was developed to target cells with enhanced spatiotemporal precision (Schindler et al. 2015). The PA-Cre can be activated in native tissues using fiber optic illumination, enabling the modification of gene expression while also permanently labeling native cells (Schindler et al. 2015). In addition, dual-recombinase-activated lineage tracing (DeaLT) was recently developed to trace multiple lineages while also reducing off-target labeling that can occur using the Cre-mediated lineage tracing (He et al. 2018). DeaLT integrates the Cre-LoxP system with a second system, the Dre-rox system, for DNA targeting with enhanced specificity while also enabling the tracing of multiple lineages (He et al. 2018). DeaLT was recently used to specifically label and trace different lineages of cardiac stem cells expressing c-Kit (He et al. 2018). Similarly, a split Cre system, which relies on concurrent expression of two markers for activation of functional Cre was recently developed (Hirrlinger et al. 2009; Buczacki et al. 2013). This system was used to identify quiescent cells in the intestine that are precursors of mature and differentiated secretory cells (Buczacki et al. 2013). In the future, combining these novel Cre-based reporters with IVM may further enhance our understanding of plastic switches undergone by cancer cells during metastasis.
LATEST BIOLOGICAL INSIGHTS INTO CELL PLASTICITY DURING METASTASIS UNCOVERED BY IVM
In the next section, we discuss the latest biological insights into plastic metastatic events as assessed by IVM.
Monitoring Cancer Plasticity during Invasion
In order to be able to leave the primary tumor, cancer cells are thought to adopt distinct modes of invasion (Friedl and Alexander 2011), which have been studied extensively using static approaches. Recently, IVM has facilitated the study of these heterogeneous migratory behaviors. As such, IVM identified single cells escaping from the primary tumor (Supplemental Movie S3; Beerling et al. 2016), as well as multicellular streaming entities, and cells switching from cohesive cell migration to single-cell migration (Giampieri et al. 2009). Interestingly, IVM demonstrated that the switch from collective to single-cell invasion can be regulated by TGFβ1 signaling (Giampieri et al. 2009). Moreover, in vivo measurements of FRET- and FRAP-reporters of cell invasion in primary pancreatic tumors and during the early steps of liver colonization (Johnsson et al. 2014; Erami et al. 2016; Warren et al. 2018) have provided insights into how invasive cancer cells adapt their invasive behavior upon genetic modification or during treatment with anti-invasive drugs (Johnsson et al. 2014; Erami et al. 2016; Warren et al. 2018).
It has been extensively debated whether disseminating epithelial cancer cells acquire migratory and stem cells properties by hijacking the developmental process EMT (Del Pozo Martin et al. 2015; Fischer et al. 2015; Zheng et al. 2015; Zhao et al. 2016; Brabletz et al. 2018). For example, the lack of evidence for EMT in histological sections of human primary tumors that have metastasized, as well as in metastases, is often raised to question the very existence of EMT during metastasis (Bukholm et al. 2000; Kowalski et al. 2003; Zheng et al. 2015). Recently, IVM-studies have shed light on the plastic nature of epithelial and mesenchymal states by demonstrating the ability of cancer cells to switch between the two states. For instance, we used IVM to monitor cancer cells that change color upon EMT. We showed that migratory and disseminating cells undergo EMT but revert to an epithelial state at the metastatic site, a process referred to as mesenchymal-to-epithelial transition (MET) (Fig. 2A; Beerling et al. 2016). Importantly, MET is believed to be necessary for efficient metastatic colonization (Kalluri and Weinberg 2009). Similarly, IVM identified a population of cancer cells in the mammary gland that undergo EMT, are highly motile, commonly reside close to blood vessels (Zhao et al. 2016) and which can revert back to an epithelial state when colonizing secondary sites (Beerling et al. 2016; Harper et al. 2016). Moreover, IVM in melanoma revealed that invasive cells that intravasate into the tumor vasculature at the primary site are poorly differentiated (Pinner et al. 2009). However, at secondary sites, this undifferentiated, highly motile phenotype is not maintained, suggesting that metastatic cells can switch from one state to another (Pinner et al. 2009).
Figure 2.
Imaging plasticity during the metastatic cascade. (A) IVM of migrating tumor cells in primary and secondary sites. Time-lapse microscopy of migrating breast tumor cells (yellow and blue for cancer cells expressing E-cadherin) leaving the primary tumor. [Panel A is reprinted from Beerling et al. (2016) under the terms of the Creative Commons Attribution-NonCommercial-No Derivatives License.] Scale bar, 50 µm. (B) IVM of tumor cells homing to and colonizing secondary sites. (i) IVM through a cranial window of extravasation of cancer cells and formation of micrometastases (green) in close proximity to blood vessels (red). [Panel B(i) is reprinted from Follain et al. (2018) with permission from Elsevier © 2018.] Scale bar, 100 µm. (ii) Longitudinal IVM tracking through a cranial window of metastatic outgrowth of lung cancer cells (red) in the brain adjacent to blood vessels (green). [Panel B(ii) is reprinted from Kienast et al. (2010) with permission from Springer Nature © 2009.] Scale bar, left 100 µm, right 20 µm. (C) IVM microscopy of dormant leukemic cells in the bone marrow and attached to osteopontin (red), labeled with DiR (green), which is retained in noncycling cells. [Panel C reprinted from Boyerinas et al. (2013) with permission from the American Society of Hematology © 2013.] Estimated scale bar, 20 µm. (D) IVM monitoring of the plastic interactions between cancer cells and the immune system. (i) IVM of patrolling monocytes (green) taking up material derived from lung CTCs (red) in the lungs. [Panel D(i) reprinted from Hanna et al. (2015) with permission from The American Association for the Advancement of Science © 2015.] Scale bar, 100 µm. (ii) Tissue resident T cells (green) in primary tumors interacting with melanoma cells (red). [Panel D(ii) reprinted from Park et al. (2019) with permission from Springer Nature © 2019.] Estimated scale bar, 10 µm.
Tumor cell invasion is also dependent on microenviromental cues, such as cell density, collagen fiber crosslinking, proximity to blood vessels and interactions with immune cells (Gligorijevic et al. 2014). For instance, IVM tracking of the early steps of liver colonization by metastatic pancreatic tumor cells revealed that the activity of Src, a key regulator of cancer cell invasion, was reduced upon TME editing with the Rho-kinase inhibitor Fasudil (Warren et al. 2018). Moreover, photoswitchable proteins were used to label invasive mammary tumor cells and to dissect how the TME influences cell invasion (Kedrin et al. 2008; Gligorijevic et al. 2014). Using this approach, cancer cells in close proximity to blood vessels were found to be more migratory than tumor cells located in areas devoid of vessels (Kedrin et al. 2008; Gligorijevic et al. 2014). Following intravasation, CTCs are subjected to hemodynamic forces, and recently CLEM was used to characterize how these forces influence the arrest and extravasation of disseminated tumor cells in the brain (Fig. 2B(i); Follain et al. 2018). In addition, IVM through a cranial window enabled the monitoring of metastatic growth in proximity to blood vessels following extravasation (Fig. 2B(ii); Kienast et al. 2010). IVM also revealed that tumor-associated macrophages (TAMs) can promote tumor cell invasion and metastasis. For instance, TAMs were found to migrate together with breast cancer cells upon stimulation with either epidermal growth factor or colony-stimulating factor 1 (Wyckoff et al. 2007). Similarly, paracrine signaling between TAMs and cancer cells can lead to enhanced expression of the actin-binding protein MenaINV in the cancer cell compartment, and this can increase invadopodia formation, cancer cell motility, and intravasation (Philippar et al. 2008; Roussos et al. 2011; Gligorijevic et al. 2014). Last, IVM monitoring of breast cancer cells targeted with chemotherapy demonstrated that macrophages with high levels of Tie2 expression (Tie2Hi) accumulate at tumor-microenvironment of metastasis (TMEM) and can facilitate local transient permeability of the endothelium and tumor cell intravasation (Harney et al. 2015; Karagiannis et al. 2017). This suggests that inhibiting Tie2Hi macrophages in combination with chemotherapy may be beneficial in the neoadjuvant setting (Harney et al. 2015; Karagiannis et al. 2017).
Plasticity of Dormancy and Senescence
Dormant and senescent (D/S) cancer cells are quiescent cancer cells that survive within primary and secondary tumors until cell intrinsic or microenvironmental cues reactivate their proliferative state. In addition, because they do not proliferate, dormant cells are thought to be resistant to chemotherapy and may therefore drive tumor recurrence (Aguirre-Ghiso 2006; Goss and Chambers 2010). While senescence has long been considered to be irreversible, senescent cells were recently suggested to maintain some plasticity and to regain the ability to proliferate (Milanovic et al. 2018). Fluorescent probes have been engineered to monitor the plastic behavior of D/S cells in native tissues. For instance, lipophilic fluorescent dyes are gradually diluted upon cell proliferation but are maintained by nonproliferating cells (Boyerinas et al. 2013; Lawson et al. 2015). In addition, the fluorescence ubiquitination cell cycle (FUCCI) and Ki67-reporters can be used to identify nonproliferating cells in live tissue (Newman and Zhang 2008; Sugiyama et al. 2009; Basak et al. 2014; Yano et al. 2014; Chou et al. 2018). Similarly, a p16Ink4a trimodality reporter (p16-3MR), consisting of a fusion protein of synthetic Renilla luciferase, mRFP and Herpes simplex virus thymidine kinase allows visualization and specific killing of cells expressing p16Ink4a, an important driver of dormancy and senescence (Demaria et al. 2014, 2017). Moreover, labeling β-galactosidase+ senescent cells can be achieved using beads loaded with a fluorophore and coated with galacto-liposaccharides (Muñoz-Espín et al. 2018).
IVM of D/S cancer cells has led to the identification of microenvironmental mechanisms driving the plastic switch toward proliferation. For instance, in acute lymphoblastic leukemia (ALL), osteopontin (OPN) can induce dormancy by promoting anchorage of leukemic cells in the endosteal niche (Boyerinas et al. 2013). IVM of label-retaining cells in the calvarial bone marrow demonstrated that blocking OPN triggers dormant cells to switch back to proliferation and renders them more sensitive to chemotherapy (Fig. 2C; Boyerinas et al. 2013). Similarly, repeated imaging of multiple myeloma cells in the endosteal niche identified cancer cells entering dormancy for long time-periods before resuming proliferation (Lawson et al. 2015). Moreover, distinct components of the endosteal niche were found to regulate the dormancy-to-proliferation switch, where interactions with osteoblasts maintained dormancy, while the osteoclast-driven remodeling of the niche reactivated cancer cell proliferation (Lawson et al. 2015). Last, longitudinal tracking of metastatic melanoma and lung cancer cells through an intracranial window revealed that dormant cells reside in close proximity to the perivascular niche and that blocking angiogenesis can induce dormancy (Kienast et al. 2010). Surprisingly, dormant metastatic melanoma cells were also found to be highly migratory in the brain, while metastatic lung cancer cells were mostly static at this secondary site, suggesting that this phenotype is not driven by the brain microenvironment (Kienast et al. 2010).
Recently, non-IVM based studies have demonstrated that cancer cell dormancy can also be regulated by local inflammation and autophagy (Albrengues et al. 2018; Vera-Ramirez et al. 2018), and that tumor cells that exit senescence are reprogramed into a stem-like state to fuel cancer growth (Milanovic et al. 2018). In addition, dormant cells that are mesenchymal can undergo MET during the switch toward proliferation and metastatic growth (Yang et al. 2018). Future IVM assessments of dormant cell plasticity may enhance our understanding of these newly discovered phenomena and can help identify new targets to tackle therapy resistance and tumor recurrence.
Plasticity of the Immune Landscape
While the identity and fate of cancer cells continuously evolve during metastatic spread, the TME is also plastic and can be coopted by cancer cells to support their dissemination (Vennin et al. 2016). For instance, IVM of immune cells during cancer progression and metastatic spread has provided novel insights into the plasticity of both antitumor and protumor behavior of immune cells in response to cancer-related stimuli (Vennin et al. 2016; Suijkerbuijk and van Rheenen 2017; Torcellan et al. 2017; Nobis et al. 2018; Wellenstein and de Visser 2018). For instance, in melanoma, IVM recently identified a subset of dendritic cells (DCs) that present antigens to CD4+ T cells but fail to promote their differentiation toward an antitumor phenotype (Binnewies et al. 2019). Interestingly, blocking regulatory T cells (Tregs) removed the brake on DCs and reestablished CD4+ T cell antitumor functions (Binnewies et al. 2019). Similarly, DCs were found to cluster with natural killer cells, and this correlated with enhanced responses to immunotherapy (Barry et al. 2018). In addition, monocytes are classically considered to promote metastatic spread, however recently patrolling monocytes were shown to reduce metastatic colonization of the lungs (Hanna et al. 2015). Here, IVM of Nra1-GFP mice to specifically monitor patrolling monocytes revealed that upon intravenous injection of lung, melanoma, or breast cancer cells, patrolling monocytes are recruited to the lungs and prevent tumor cell invasion (Fig. 2D(i); Hanna et al. 2015). Similarly, live imaging in primary breast tumors demonstrated that monocytes that enter tumor tissues differentiate into highly migratory TAMs, which guide cancer cells toward blood vessels (Arwert et al. 2018). These migratory TAMs can undergo a unidirectional switch toward a metastatic-assisting perivascular phenotype (Arwert et al. 2018). In addition, a population of tumor-experienced T cells were recently identified using photolabeling strategies. These T cells can readily egress from the primary tumor and migrate to metastatic sites, where they may mediate tumor-immunity (Torcellan et al. 2017). The interactions between cancer and immune cells can vary over time and space. For instance, in clinically dormant melanoma lesions, resident memory T cells accumulate in stable and persistent melanoma lesions, and can either retain prolonged contact with cancer cells or display a migratory phenotype (Fig. 2D(ii); Park et al. 2019). Similarly, interactions between CTCs and immune cells residing in secondary sites were shown to influence the fate of disseminated cancer cells (Headley et al. 2016). Here, using fluorescent lineage reporters of myeloid cell populations, Headley et al. (2016) visualized waves of myeloid cell populations that take up microparticles released by CTCs and that subsequently accumulate in the lungs to promote metastatic growth. Successful establishment of metastasis can also be driven by interactions of cancer cells with neutrophils. For instance, CTCs that bind to neutrophil extracellular traps (NET) or that extravasated in close proximity to neutrophils in the liver have a higher ability to seed in this tissue (Spicer et al. 2012; Cools-Lartigue et al. 2013).
Plasticity of the ECM and the Vasculature
Remodeling of the ECM provides cancer cells with biochemical and mechanical cues that regulate their growth, invasion, and response to therapies (Pickup et al. 2014). For instance, increased ECM deposition is thought to impair drug delivery (Pickup et al. 2014; Vennin et al. 2018). IVM FRAP combined with histology and transmission EM demonstrated that the diffusion coefficient of macromolecules in patient-derived cranial and skin tumors is reduced in areas with a dense ECM (Netti et al. 2000). In line with this, IVM of mouse tumors and patient-derived xenografts revealed that reducing ECM crosslinking can enhance chemotherapy efficacy (Vennin et al. 2017). Cancer cell invasion and liver metastases can also be reduced upon ECM manipulation, suggesting that reestablishing normal ECM homeostasis could be used as an antimetastatic strategy (Vennin et al. 2017). Cancer-associated fibroblasts (CAFs) are thought to be the main drivers of ECM remodeling, and IVM demonstrated that CAFs can also influence cancer cell metastasis. For instance, CAFs and cancer cells can establish physical interactions that are mechanically active and promote collective migration of cancer cells (Labernadie et al. 2017). Similarly, CAFs can travel along with cancer cells from a primary tumor to secondary sites, and depletion of CAFs was shown to reduce metastatic burden and extended mouse survival (Duda et al. 2010).
Tumor cells can also coopt the vasculature. IVM has shown that vasculature remodeling occurs during the early stages of malignant transformation and that angiogenesis appears when tumors progress from hyperplasia to carcinoma (Wyckoff et al. 2007; Kedrin et al. 2008; Vennin et al. 2016). This results in a leaky and dysfunctional blood vessel network (Hagendoorn et al. 2006; Vennin et al. 2017), which provides a route for metastatic cells toward secondary sites (Kedrin et al. 2008). In addition, IVM of breast tumors revealed that cancer cells migrate toward macrophages in the perivascular niche (PVN), and this correlated with the intravasation of tumor cells (Wyckoff et al. 2007). In line with this, IVM through a cranial window demonstrated that dormant metastatic cells reside in close proximity to the PVN (Kienast et al. 2010) and that the switch from dormancy to growth can either be driven by angiogenesis or by vessel cooption (Kienast et al. 2010). Because of the dysfunctional vasculature, oxygen is not evenly distributed within the tumor and this can lead to the establishment of hypoxia (Wilson and Hay 2011). IVM mapping of oxygen levels in pancreatic tumors recently showed that hypoxic areas move within tumor tissues over time (Conway et al. 2018). In addition, an Akt FRET-reporter was used to reveal that hypoxia induces resistance to the mTORC1/2 inhibitor AZD2014. This resistance was overcome by combining AZD2014 with cytotoxic-prodrugs specifically activated in hypoxia (Conway et al. 2018). Last, an intraoperative epifluorescent platform was recently used to image the tumor vasculature of melanoma patients during surgical removal of the tumor. This suggests that in the future, IVM in a human setting could help monitoring drug delivery and could facilitate the development of new treatments (Fisher et al. 2016).
Plastic Responses to Therapies
IVM has shed new light on cell intrinsic and microenvironmental mechanisms of resistance to therapy. For instance, in glioma, where chemotherapy efficacy is commonly low, IVM identified tumor microtubes, which are long-lived, ultralong and thick membrane extensions that interconnect tumor cells via gap junctions (Osswald et al. 2015). The density of these tumor microtubes increased upon chemotherapy and prevent cancer cell apoptosis (Fig. 3A; Osswald et al. 2015; Weil et al. 2017). Moreover, in T cell acute lymphoblastic leukemia (T-ALL), cells that survive treatments adopt a more motile phenotype and colonize secondary sites (Fig. 3B), in line with recent findings suggesting that cytotoxic treatments may induce metastatic spread (Hawkins et al. 2016; Karagiannis et al. 2017).
Figure 3.
Plastic responses to therapies. Cancer cells display plastic responses to therapies, which can be cell intrinsic or a result of microenvironmental protection. Here, we depict some examples of both types of mechanisms of resistance: (A) A network of tumor microtubes protects glioma cells from dying. After chemotherapy, the number of connections increases and this correlates with reduced response to treatment. White arrows point to microtubes. [Panel A is reprinted from Osswald et al. (2015) with permission from Springer Nature © 2015.] (B) T-ALL cells become more migratory and colonize the bone marrow after chemotherapy. [Panel B reprinted from Hawkins et al. (2016) et al. with permission from Springer Nature © 2016.] (C) CAF activation upon treatments with BRAF inhibitor leads to ECM remodeling and reduced tumor cell response to treatment. [Panel C reprinted from Hirata et al. (2015) under the terms of the Creative Commons Attribution-NonCommercial-No Derivatives License (CC BY NC ND).] (D) Peritumoral Tregs prevent infiltration of adoptive cytotoxic T cells (CTLs) into melanoma lesions, reducing immunotherapy efficacy. [Panel D reprinted from Qi et al. (2016) under the terms of the Creative Commons Attribution License (CC-BY).] (E) Biopsy induces inflammation and tumor cell migration, demonstrated by the change in the red area postbiopsy. [Panel E reprinted from Alieva et al. (2017) under the terms of the Creative Commons Attribution License (CC BY).]
Cancer cells can also modify their niche upon treatment and alterations of the TME can influence response to therapy (Binnewies et al. 2018). For instance, in T-ALL, treatment can increase the ability of tumor cells to trigger apoptosis of osteoblasts, thereby misbalancing normal hematopoiesis and conferring an advantage for T-ALL cells to repopulate the bone marrow (Hawkins et al. 2016). Similarly, in melanoma, tumor cells subjected to MEK and BRAF inhibitors can activate neighboring CAFs, leading to ECM remodeling and reduced cancer cell response to treatment (Fig. 3C; Hirata et al. 2015). While therapy resistance can be driven by ECM stiffness (Hirata et al. 2015), long-term treatment with MEK and BRAF inhibitors can also push cancer cells toward an epithelial phenotype, and this correlated with increased resistance to treatment (Brighton et al. 2018). In addition, IVM was used to show that chemotherapy in acute myeloid leukemia (AML) induces apoptosis of endothelial cells, which subsequently leads to the establishment of hypoxia and reduces drug delivery to the tumor cells (Passaro et al. 2017). Similarly, IVM revealed that AML cells can disrupt the bone marrow vasculature after treatment with cytarabine and doxorubicin, leading to transendothelial migration of HSCs, loss of osteoblasts and increased survival of leukemic cells (Passaro et al. 2017). Restoring normal tumor vasculature can improve treatment efficacy (Vennin et al. 2018). For instance, encaging chemotherapies into transferrin-functionalized nanoparticles that specifically target tumor-associated blood vessels can overcome treatment resistance in glioma (Lam et al. 2018). Immune cells can also undergo dynamic changes upon treatments, and this in turn influences cancer cell responses to therapies. For example, IVM demonstrated that anti-VEGF therapy switched the TME toward an immunosuppressive state, with enhanced extravasation of neutrophils into colorectal tumors and increased the production of immunosuppressive IL-10 (Jung et al. 2017). In addition, in melanoma, peritumoral Tregs can prevent tumor infiltration of adoptive CTLs and thereby reduce immunotherapy efficacy (Fig. 3D; Qi et al. 2016). This was reverted by administering cyclophosphamide, which eliminated Tregs and increased migration of adoptive CTLs into tumors (Qi et al. 2016). In addition, IVM revealed that CCR2+ macrophages are recruited to breast tumors upon chemotherapy, and this correlates with tumor relapse (Lam et al. 2018). Similarly, we recently used IVM to demonstrate that biopsies, which are routinely performed in the clinic, can trigger the recruitment of macrophages to the biopsy site and subsequently induce cancer cell invasion and proliferation (Fig. 3E; Supplemental Movie S4; Alieva et al. 2017). Importantly, blocking the recruitment of macrophages prevents this plastic switch and reduces cancer cell invasion (Alieva et al. 2017). IVM can also facilitate the development of novel immune-based therapies. For instance, fluorescently labeled bisphosphonates, which are thought to solely target bone cells, were shown to accumulate in TAMs infiltrating breast tumors, suggesting that bisphosphonates may be used in treatment of breast cancer patients (Junankar et al. 2015). Similarly, Yu et al. (2019) developed α-melittin nanoparticles, which are specifically taken up by liver sinusoidal endothelial cells, triggering a switch in the cytokine milieu to recruit innate and adaptive immune cells to the liver, thereby reducing metastatic. Lastly, interactions between a subset of DCs and T cells were recently shown to be required for efficient anti-PD1 treatment in colorectal tumors (Garris et al. 2018). Here, IVM of cytokine production demonstrated that DCs sense IFNγ produced by T cells during immunotherapy, and in turn release IL-2 to further stimulate antitumor immunity (Garris et al. 2018).
CONCLUDING REMARKS
IVM comprises a range of powerful tools, which over the last decade have shed light on the dynamics of metastasis. In this review, we provide a glance of how IVM enables the monitoring of plastic events followed by cancer cells during metastasis. Discoveries uncovered using IVM may in the future facilitate the development of novel antimetastatic therapeutics. In addition, in order to expand our knowledge of metastasis, IVM should be combined with high-throughput approaches for profiling of cancer cells as well as with new cancer models such as patient-derived models and humanized mouse models.
ACKNOWLEDGMENTS
The authors regret not being able to discuss additional publications related to our topic, due to space limitations. The authors thank members of the van Rheenen group for critical reading of the manuscript. We also thank the authors of the papers highlighted in our figures for sharing with us their IVM images. J.v.R. is supported by CancerGenomics.nl, a European Research Council Grant CANCER-RECURRENCE 648804, the Doctor Josef Steiner Foundation and the European Union's Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement No. 642866. C.V. is the recipient of a HFSP fellowship. A.S.M. is supported by the GABBA PhD program (FCT fellowship PD/BD/105748/2014).
Footnotes
Editors: Jeffrey W. Pollard and Yibin Kang
Additional Perspectives on Metastasis: Mechanism to Therapy available at www.perspectivesinmedicine.org
REFERENCES
- Abe T, Fujimori T. 2013. Reporter mouse lines for fluorescence imaging. Dev Growth Differ 55: 390–405. 10.1111/dgd.12062 [DOI] [PubMed] [Google Scholar]
- Aguirre-Ghiso JA. 2006. The problem of cancer dormancy: understanding the basic mechanisms and identifying therapeutic opportunities. Cell Cycle 5: 1740–1743. 10.4161/cc.5.16.3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrengues J, Shields MA, Ng D, Park CG, Ambrico A, Poindexter ME, Upadhyay P, Uyeminami DL, Pommier A, Küttner V, et al. 2018. Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science 361: eaao4227 10.1126/science.aao4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alieva M, Ritsma L, Giedt RJ, Weissleder R, van Rheenen J. 2014. Imaging windows for long-term intravital imaging: general overview and technical insights. Intravital 3: e29917 10.4161/intv.29917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alieva M, Margarido AS, Wieles T, Abels ER, Colak B, Boquetale C, Jan Noordmans H, Snijders TJ, Broekman ML, van Rheenen J. 2017. Preventing inflammation inhibits biopsy-mediated changes in tumor cell behavior. Sci Rep 7: 7529 10.1038/s41598-017-07660-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alieva M, Leidgens V, Riemenschneider MJ, Klein CA, Hau P, van Rheenen J. 2019. Intravital imaging of glioma border morphology reveals distinctive cellular dynamics and contribution to tumor cell invasion. Sci Rep 9: 2054 10.1038/s41598-019-38625-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. 2002. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci 99: 12651–12656. 10.1073/pnas.202320599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annamdevula NS, Sweat R, Griswold JR, Trinh K, Hoffman C, West S, Deal J, Britain AL, Jalink K, Rich TC, et al. 2018. Spectral imaging of FRET-based sensors reveals sustained cAMP gradients in three spatial dimensions. Cytometry A 93: 1029–1038. 10.1002/cyto.a.23572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Kondo Y, Naoki H, Hiratsuka T, Itoh RE, Matsuda M. 2017. Propagating wave of ERK activation orients collective cell migration. Dev Cell 43: 305–317.e5. 10.1016/j.devcel.2017.10.016 [DOI] [PubMed] [Google Scholar]
- Arwert E, Harney AS, Entenberg D, Wang Y, Sahai E, Pollard JW, Condeelis JS. 2018. A unidirectional transition from migratory to perivascular macrophage is required for tumor cell intravasation. Cell Rep 23: 1239–1248. 10.1016/j.celrep.2018.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry KC, Hsu J, Broz ML, Cueto FJ, Binnewies M, Combes AJ, Nelson AE, Loo K, Kumar R, Rosenblum MD, et al. 2018. A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments. Nat Med 24: 1178–1191. 10.1038/s41591-018-0085-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak O, van de Born M, Korving J, Beumer J, van der Elst S, van Es JH, Clevers H. 2014. Mapping early fate determination in Lgr5+ crypt stem cells using a novel Ki67-RFP allele. EMBO J 33: 2057–2068. 10.15252/embj.201488017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerling E, Ritsma L, Vrisekoop N, Derksen PW, van Rheenen J. 2011. Intravital microscopy: new insights into metastasis of tumors. J Cell Sci 124: 299–310. 10.1242/jcs.072728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerling E, Seinstra D, de Wit E, Kester L, van der Velden D, Maynard C, Schäfer R, van Diest P, Voest E, van Oudenaarden A, et al. 2016. Plasticity between epithelial and mesenchymal states unlinks EMT from metastasis-enhancing stem cell capacity. Cell Rep 14: 2281–2288. 10.1016/j.celrep.2016.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI, Ostrand-Rosenberg S, Hedrick CC, et al. 2018. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 24: 541–550. 10.1038/s41591-018-0014-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnewies M, Mujal AM, Pollack JL, Combes AJ, Hardison EA, Barry KC, Tsui J, Ruhland MK, Kersten K, Abushawish MA, et al. 2019. Unleashing type-2 dendritic cells to drive protective antitumor CD4+ T cell immunity. Cell 177: 556–571.e16. 10.1016/j.cell.2019.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bixel MG, Kusumbe AP, Ramasamy SK, Sivaraj KK, Butz S, Vestweber D, Adams RH. 2017. Flow dynamics and HSPC homing in bone marrow microvessels. Cell Rep 18: 1804–1816. 10.1016/j.celrep.2017.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacker TS, Duchen MR. 2016. Investigating mitochondrial redox state using NADH and NADPH autofluorescence. Free Radic Biol Med 100: 53–65. 10.1016/j.freeradbiomed.2016.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyerinas B, Zafrir M, Yesilkanal AE, Price TT, Hyjek EM, Sipkins DA. 2013. Adhesion to osteopontin in the bone marrow niche regulates lymphoblastic leukemia cell dormancy. Blood 121: 4821–4831. 10.1182/blood-2012-12-475483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabletz T, Kalluri R, Neito MA, Weinberg RA. 2018. EMT in cancer. Nat Rev Cancer 18: 128–134. 10.1038/nrc.2017.118 [DOI] [PubMed] [Google Scholar]
- Brighton HE, Angus SP, Bo T, Roques J, Tagliatela AC, Darr DB, Karagoz K, Sciaky N, Gatza ML, Sharpless NE, et al. 2018. New mechanisms of resistance to MEK inhibitors in melanoma revealed by intravital imaging. Cancer Res 78: 542–557. 10.1158/0008-5472.CAN-17-1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E, McKee T, diTomaso E, Pluen A, Seed B, Boucher Y, Jain RK. 2003. Dynamic imaging of collagen and its modulation in tumors in vivo using second-harmonic generation. Nat Med 9: 796–800. 10.1038/nm879 [DOI] [PubMed] [Google Scholar]
- Brown S, Pineda CM, Xin T, Boucher J, Suozzi KC, Park S, Matte-Martone C, Gonzalez DG, Rytlewski J, Beronja S, et al. 2017. Correction of aberrant growth preserves tissue homeostasis. Nature 548: 334–337. 10.1038/nature23304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns OT, Bischof TS, Harris DK, Franke D, Shi Y, Riedemann L, Bartelt A, Jaworski FB, Carr JA, Rowlands CJ, et al. 2017. Next-generation in vivo optical imaging with short-wave infrared quantum dots. Nat Biomed Eng 1: 0056 10.1038/s41551-017-0056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buczacki SJ, Zecchini HI, Nicholson AM, Russell R, Vermeulen L, Kemp R, Winton DJ. 2013. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature 495: 65–69. 10.1038/nature11965 [DOI] [PubMed] [Google Scholar]
- Bukholm IK, Nesland JM, Børresen-Dale AL. 2000. Re-expression of E-cadherin, α-catenin and β-catenin, but not of γ-catenin, in metastatic tissue from breast cancer patients [seecomments]. J Pathol 190: 15–19.<15::AID-ATH489>3.0.CO;2-L [DOI] [PubMed] [Google Scholar]
- Burke K, Brown E. 2014. The use of second harmonic generation to image the extracellular matrix during tumor progression. Intravital 3: e984509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr JA, Franke D, Caram JR, Perkinson CF, Saif M, Askoxylakis V, Datta M, Fukumura D, Jain RK, Bawendi MG, et al. 2018. Shortwave infrared fluorescence imaging with the clinically approved near-infrared dye indocyanine green. Proc Natl Acad Sci 115: 4465–4470. 10.1073/pnas.1718917115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffer CL, San Juan BP, Lim E, Weinberg RA. 2016. EMT, cell plasticity and metastasis. Cancer Metastasis Rev 35: 645–654. 10.1007/s10555-016-9648-7 [DOI] [PubMed] [Google Scholar]
- Chou A, Froio D, Nagrial AM, Parkin A, Murphy KJ, Chin VT, Wohl D, Steinmann A, Stark R, Drury A, et al. 2018. Tailored first-line and second-line CDK4-targeting treatment combinations in mouse models of pancreatic cancer. Gut 67: 2142–2155. 10.1136/gutjnl-2017-315144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchi R, Pavone FS. 2017. Probing collagen organization: practical guide for second-harmonic generation (SHG) imaging. Methods Mol Biol 1627: 409–425. 10.1007/978-1-4939-7113-8_27 [DOI] [PubMed] [Google Scholar]
- Conway J, Carragher N, Timpson P. 2014. Developments in preclinical cancer imaging: innovating the discovery of therapeutics. Nat Rev Cancer 14: 314–328. 10.1038/nrc3724 [DOI] [PubMed] [Google Scholar]
- Conway JRW, Warren SC, Herrmann D, Murphy KJ, Cazet AS, Vennin C, Shearer RF, Killen MJ, Magenau A, Mélénec P, et al. 2018. Intravital imaging to monitor therapeutic response in moving hypoxic regions resistant to PI3K pathway targeting in pancreatic cancer. Cell Rep 23: 3312–3326. 10.1016/j.celrep.2018.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools-Lartigue J, Spicer J, McDonald B, Gowing S, Chow S, Giannias B, Bourdeau F, Kubes P, Ferri L. 2013. Neutrophil extracellular traps sequester circulating tumor cells and promote metastasis. J Clin Invest 123: 3446–3458 10.1172/JCI67484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher PI, McDonald MM, Martin TJ. 2016. Bone metastasis: the importance of the neighbourhood. Nat Rev Cancer 16: 373–386. 10.1038/nrc.2016.44 [DOI] [PubMed] [Google Scholar]
- del Pozo Martin Y, Park D, Ramachandran A, Ombrato L, Calvo F, Chakravarty P, Spencer-Dene B, Derzsi S, Hill CS, Sahai E, et al. 2015. Mesenchymal cancer cell-stroma crosstalk promotes niche activation, epithelial reversion, and metastatic colonization. Cell Rep 13: 2456–2469. 10.1016/j.celrep.2015.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaria M, Ohtani N, Youssef SA, Rodier F, Toussaint W, Mitchell JR, Laberge RM, Vijg J, Van Steeg H, Dollé ME, et al. 2014. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev Cell 31: 722–733. 10.1016/j.devcel.2014.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaria M, O'Leary MN, Chang J, Shao L, Liu S, Alimirah F, Koenig K, Le C, Mitin N, Deal AM, et al. 2017. Cellular senescence promotes adverse effects of chemotherapy and cancer relapse. Cancer Discov 7: 165–176. 10.1158/2159-8290.CD-16-0241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondossola E, Holzapfel BM, Alexander S, Filippini S, Hutmacher DW, Friedl P. 2016. Examination of the foreign body response to biomaterials by nonlinear intravital microscopy. Nat Biomed Eng 1: 0007 10.1038/s41551-016-0007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondossola E, Alexander S, Holzapfel BM, Filippini S, Starbuck MW, Hoffman RM, Navone N, De-Juan-Pardo EM, Logothetis CJ, Hutmacher DW, et al. 2018. Intravital microscopy of osteolytic progression and therapy response of cancer lesions in the bone. Sci Transl Med 10: eaao5726 10.1126/scitranslmed.aao5726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. 2012. Defining the mode of tumour growth by clonal analysis. Nature 488: 527–530. 10.1038/nature11344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte D, Hawkins ED, Akinduro O, Ang H, De Filippo K, Kong IY, Haltalli M, Ruivo N, Straszkowski L, Vervoort SJ, et al. 2018. Inhibition of endosteal vascular niche remodeling rescues hematopoietic stem cell loss in AML. Cell Stem Cell 22: 64–77.e6. 10.1016/j.stem.2017.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda AM, Duyverman AM, Kohno M, Snuderl M, Steller EJ, Fukumura D, Jain RK. 2010. Malignant cells facilitate lung metastasis by bringing their own soil. Proc Natl Acad Sci 50: 21677–21682. 10.1073/pnas.1016234107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham SM, Pudavar HE, Prasad PN, Stachowiak MK. 2004. Cellular signaling and protein–protein interactions studied using fluorescence recovery after photobleaching. J Phys Chem B 108: 10540–10546. 10.1021/jp0400972 [DOI] [Google Scholar]
- Ellenbroek SI, van Rheenen J. 2014. Imaging hallmarks of cancer in living mice. Nat Rev Cancer 14: 406–418. 10.1038/nrc3742 [DOI] [PubMed] [Google Scholar]
- Entenberg D, Kedrin D, Wyckoff J, Sahai E, Condeelis J, Segall JE. 2013. Imaging tumor cell movement in vivo. Curr Protoc Cell Biol 19: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entenberg D, Rodriguez-Tirado C, Kato Y, Kitamura T, Pollard JW, Condeelis J. 2015. In vivo subcellular resolution optical imaging in the lung reveals early metastatic proliferation and motility. Intravital 4: 1–11. 10.1080/21659087.2015.1086613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entenberg D, Voiculescu S, Guo P, Borriello L, Wang Y, Karagiannis GS, Jones J, Baccay F, Oktay M, Condeelis J. 2018. A permanent window for the murine lung enables high-resolution imaging of cancer metastasis. Nat Methods 15: 73–80. 10.1038/nmeth.4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erami Z, Herrmann D, Warren SC, Nobis M, McGhee EJ, Lucas MC, Leung W, Reischmann N, Mrowinska A, Schwarz JP, et al. 2016. Intravital FRAP imaging using an E-cadherin-GFP mouse reveals disease- and drug-dependent dynamic regulation of cell-cell junctions in live tissue. Cell Rep 14: 152–167. 10.1016/j.celrep.2015.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erapaneedi R, Belousov VV, Schäfers M, Kiefer F. 2016. A novel family of fluorescent hypoxia sensors reveal strong heterogeneity in tumor hypoxia at the cellular level. EMBO J 35: 102–113. 10.15252/embj.201592775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald AJ, Werb Z, Egeblad M. 2011. Monitoring of vital signs for long-term survival of mice under anesthesia. Cold Spring Harb Protoc 2011: pdb prot5563 10.1101/pdb.prot5563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielmich LE, Schmidt R, Dickinson DJ, Goldstein B, Akhmanova A, van den Heuvel S. 2018. Optogenetic dissection of mitotic spindle positioning in vivo. Elife 7: e38198 10.7554/eLife.38198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J, et al. 2015. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527: 472–476. 10.1038/nature15748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DT, Muhitch JB, Kim M, Doyen KC, Bogner PN, Evans SS, Skitzki JJ. 2016. Intraoperative intravital microscopy permits the study of human tumour vessels. Nat Commun 7: 10684 10.1038/ncomms10684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follain G, Osmani N, Azevedo AS, Allio G, Mercier L, Karreman MA, Solecki G, Garcia Leòn MJ, Lefebvre O, Fekonja N, et al. 2018. Hemodynamic forces tune the arrest, adhesion, and extravasation of circulating tumor cells. Dev Cell 45: 33–52.e12. 10.1016/j.devcel.2018.02.015 [DOI] [PubMed] [Google Scholar]
- Freise A, Wu A. 2015. In vivo imaging with antibodies and engineered fragments. Mol Immunol 67: 142–152. 10.1016/j.molimm.2015.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Alexander S. 2011. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell 147: 992–1009. 10.1016/j.cell.2011.11.016 [DOI] [PubMed] [Google Scholar]
- Fritzsche M, Charras G. 2015. Dissecting protein reaction dynamics in living cells by fluorescence recovery after photobleaching. Nat Protoc 10: 660–680. 10.1038/nprot.2015.042 [DOI] [PubMed] [Google Scholar]
- Fumagalli A, Drost J, Suijkerbuijk SJ, van Boxtel R, de Ligt J, Offerhaus GJ, Begthel H, Beerling E, Tan EH, Sansom OJ, et al. 2017. Genetic dissection of colorectal cancer progression by orthotopic transplantation of engineered cancer organoids. Proc Natl Acad Sci 114: E2357–E2364. 10.1073/pnas.1701219114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Ortega D, Ledger A, Roden DL, Law AM, Magenau A, Kikhtyak Z, Cho C, Allerdice SL, Lee HJ, Valdes-Mora F, et al. 2015. ELF5 drives lung metastasis in luminal breast cancer through recruitment of Gr1+ CD11b+ myeloid-derived suppressor cells. PLoS Biol 13: e1002330 10.1371/journal.pbio.1002330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garris CS, Arlauckas SP, Kohler RH, Trefny MP, Garren S, Piot C, Engblom C, Pfirschke C, Siwicki M, Gungabeesoon J, et al. 2018. Successful anti-PD-1 cancer immunotherapy requires T cell-dendritic cell crosstalk involving the cytokines IFN-γ and IL-12. Immunity 49: 1148–1161.e7. 10.1016/j.immuni.2018.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavet O, Pines J. 2010a. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev Cell 18: 533–543. 10.1016/j.devcel.2010.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavet O, Pines J. 2010b. Activation of cyclin B1–Cdk1 synchronizes events in the nucleus and the cytoplasm at mitosis. J Cell Biol 189: 247–259. 10.1083/jcb.200909144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheghiani L, Loew D, Lombard B, Mansfield J, Gavet O. 2017. PLK1 activation in late G2 sets up commitment to mitosis. Cell Rep 19: 2060–2073. 10.1016/j.celrep.2017.05.031 [DOI] [PubMed] [Google Scholar]
- Giampieri S, Manning C, Hooper S, Jones L, Hill CS, Sahai E. 2009. Localized and reversible TGFβ signalling switches breast cancer cells from cohesive to single cell motility. Nat Cell Biol 11: 1287–1296. 10.1038/ncb1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gligorijevic B, Bergman A, Condeelis J. 2014. Multiparametric classification links tumor microenvironments with tumor cell phenotype. PLoS Biol 12: e1001995 10.1371/journal.pbio.1001995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring NW, Chowdhury D, Hyman AA, Grill SW. 2010. FRAP analysis of membrane-associated proteins: lateral diffusion and membrane-cytoplasmic exchange. Biophys J 99: 2443–2452. 10.1016/j.bpj.2010.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz JG. 2018. Exploiting anatomical landmarks for efficient in vivo CLEM. Trends Biochem Sci 43: 744–747. 10.1016/j.tibs.2018.08.003 [DOI] [PubMed] [Google Scholar]
- Goldey GJ, Roumis DK, Glickfeld LL, Kerlin AM, Bonin V, Schafer DP, Andermann ML. 2014. Removable cranial windows for long-term imaging in awake mice. Nat Protoc 9: 2515–2538. 10.1038/nprot.2014.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss PE, Chambers AF. 2010. Does tumour dormancy offer a therapeutic target? Nat Rev Cancer 10: 871–877. 10.1038/nrc2933 [DOI] [PubMed] [Google Scholar]
- Hagendoorn J, Tong R, Fukumura D, Lin Q, Lobo J, Padera TP, Xu L, Kucherlapati R, Jain RK. 2006. Onset of abnormal blood and lymphatic vessel function and interstitial hypertension in early stages of carcinogenesis. Cancer Res 66: 3360–3364. 10.1158/0008-5472.CAN-05-2655 [DOI] [PubMed] [Google Scholar]
- Hall B, McLean MA, Davis K, Casanova JE, Sligar SG, Schwartz MA. 2008. A fluorescence resonance energy transfer activation sensor for Arf6. Anal Biochem 374: 243–249. 10.1016/j.ab.2007.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halls M. 2019. Localised GPCR signalling as revealed by FRET biosensors. Curr Opin Cell Biol 57: 48–56. 10.1016/j.ceb.2018.11.001 [DOI] [PubMed] [Google Scholar]
- Hanna RN, Cekic C, Sag D, Tacke R, Thomas GD, Nowyhed H, Herrley E, Rasquinha N, McArdle S, Wu R, et al. 2015. Patrolling monocytes control tumor metastasis to the lung. Science 350: 985–990. 10.1126/science.aac9407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harney AS, Arwert EN, Entenberg D, Wang Y, Guo P, Qian BZ, Oktay MH, Pollard JW, Jones JG, Condeelis JS. 2015. Real-time imaging reveals local, transient vascular permeability, and tumor cell intravasation stimulated by TIE2Hi macrophage-derived VEGFA. Cancer Discov 5: 932–943. 10.1158/2159-8290.CD-15-0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper KL, Sosa MS, Entenberg D, Hosseini H, Cheung JF, Nobre R, Avivar-Valderas A, Nagi C, Girnius N, Davis RJ, et al. 2016. Mechanism of early dissemination and metastasis in Her2+ mammary cancer. Nature 540: 588–592. 10.1038/nature20609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins ED, Duarte D, Akinduro O, Khorshed RA, Passaro D, Nowicka M, Straszkowski L, Scott MK, Rothery S, Ruivo N, et al. 2016. T-cell acute leukaemia exhibits dynamic interactions with bone marrow microenvironments. Nature 538: 518–522. 10.1038/nature19801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Li Y, Huang X, Li Y, Pu W, Tian X, Cai D, Huang H, Lui KO, Zhou B. 2018. Genetic lineage tracing of resident stem cells by DeaLT. Nat Protoc 13: 2217–2246. 10.1038/s41596-018-0034-5 [DOI] [PubMed] [Google Scholar]
- Headley MB, Bins A, Nip A, Roberst EW, Looney MR, Gerard A, Krummel MF. 2016. Visualization of immediate immune responses to pioneer metastatic cells in the lung. Nature 531: 513–517. 10.1038/nature16985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata E, Girotti MR, Viros A, Hooper S, Spencer-Dene B, Matsuda M, Larkin J, Marais R, Sahai E. 2015. Intravital imaging reveals how BRAF inhibition generates drug-tolerant microenvironments with high integrin β1/FAK signaling. Cancer Cell 13: 574–588. 10.1016/j.ccell.2015.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirrlinger J, Scheller A, Hirrlinger PG, Kellert B, Tang W, Wehr MC, Goebbels S, Reichenbach A, Sprengel R, Rossner MJ, et al. 2009. Split-Cre complementation indicates coincident activity of different genes in vivo. PLoS One 4: e4286 10.1371/journal.pone.0004286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husada F, Bountra K, Tassis K, de Boer M, Romano M, Rebuffat S, Beis K, Cordes T. 2018. Conformational dynamics of the ABC transporter McjD seen by single-molecule FRET. EMBO J 37: e100056 10.15252/embj.2018100056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyenne V, Ghoroghi S, Collot M, Bons J, Follain G, Harlepp S, Mary B, Bauer J, Mercier L, Busnelli I, et al. 2019. Studying the fate of tumor extracellular vesicles at high spatiotemporal resolution using the zebrafish embryo. Dev Cell 48: 554–572.e7. 10.1016/j.devcel.2019.01.014 [DOI] [PubMed] [Google Scholar]
- Imamura H, Nhat KP, Togawa H, Saito K, Lino R, Kato-Yamada Y, Nagai T, Noji H. 2009. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc Natl Acad Sci 106: 15651–15656. 10.1073/pnas.0904764106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalink K. 2013. hiFRET: some tailwind for FRET resolves weak protein interactions. Nat Methods 10: 947–948. 10.1038/nmeth.2652 [DOI] [PubMed] [Google Scholar]
- Janssen A, Beerling E, Medema R, van Rheenen J. 2013. Intravital FRET imaging of tumor cell viability and mitosis during chemotherapy. PLoS One 8: e64029 10.1371/journal.pone.0064029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jares-Erijman EA, Jovin TM. 2003. FRET imaging. Nat Biotechnol 21: 1387–1395. 10.1038/nbt896 [DOI] [PubMed] [Google Scholar]
- Johnsson AE, Dai Y, Nobis M, Baker MJ, McGhee EJ, Walker S, Schwarz JP, Kadir S, Morton JP, Myant KB, et al. 2014. The Rac-FRET mouse reveals tight spatiotemporal control of Rac activity in primary cells and tissues. Cell Rep 6: 1153–1164. 10.1016/j.celrep.2014.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junankar S, Shay G, Jurczyluk J, Ali N, Down J, Pocock N, Parker A, Nguyen A, Sun S, Kashemirov B, et al. 2015. Real-time intravital imaging establishes tumor-associated macrophages as the extraskeletal target of bisphosphonate action in cancer. Cancer Discov 5: 35–42. 10.1158/2159-8290.CD-14-0621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K, Heishi T, Khan OF, Kowalski PS, Incio J, Rahbari NN, Chung E, Clark JW, Willett CG, Luster AD, et al. 2017. Ly6Clo monocytes drive immunosuppression and confer resistance to anti-VEGFR2 cancer therapy. J Clin Invest 127: 3039–3051. 10.1172/JCI93182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaifosh P, Zaremba JD, Danielson NB, Losonczy A. 2014. SIMA: Python software for analysis of dynamic fluorescence imaging data. Front Neuroinform 8: 80 10.3389/fninf.2014.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. 2009. The basics of epithelial-mesenchymal transition. J Clin Invest 119: 1420–1428. 10.1172/JCI39104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagiannis GS, Pastoriza JM, Wang Y, Harney AS, Entenberg D, Pignatelli J, Sharma VP, Xue EA, Cheng E, D'Alfonso TM, et al. 2017. Neoadjuvant chemotherapy induces breast cancer metastasis through a TMEM-mediated mechanism. Sci Transl Med 9: eaan0026 10.1126/scitranslmed.aan0026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreman MA, Mercier L, Schieber NL, Shibue T, Schwab Y, Goetz JG. 2014. Correlating intravital multi-photon microscopy to 3D electron microscopy of invading tumor cells using anatomical reference points. PLoS One 9: e114448 10.1371/journal.pone.0114448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreman M, Hyenne V, Schwab Y, Goetz JG. 2016a. Intravital correlative microscopy: imaging life at the nanoscale. Trends Cell Biol 26: 848–863. 10.1016/j.tcb.2016.07.003 [DOI] [PubMed] [Google Scholar]
- Karreman MA, Mercier L, Schieber NL, Solecki G, Allio G, Winkler F, Ruthensteiner B, Goetz JG, Schwab Y. 2016b. Fast and precise targeting of single tumor cells in vivo by multimodal correlative microscopy. J Cell Sci 129: 444–456. 10.1242/jcs.181842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedrin D, Gligorijevic B, Wyckoff J, Verkhusha VV, Condeelis J, Segall JE, van Rheenen J. 2008. Intravital imaging of metastatic behavior through a mammary imaging window. Nat Methods 5: 1019–1021. 10.1038/nmeth.1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khait I, Orsher Y, Golan O, Binshtok U, Gordon-Bar N, Amir-Zilberstein L, Sprinzak D. 2016. Quantitative analysis of delta-like 1 membrane dynamics elucidates the role of contact geometry on notch signaling. Cell Rep 14: 225–233. 10.1016/j.celrep.2015.12.040 [DOI] [PubMed] [Google Scholar]
- Khosravi N, Mendes VC, Nirmal G, Majeed S, DaCosta RS, Davies JE. 2018. Intravital imaging for tracking of angiogenesis and cellular events around surgical bone implants. Tissue Eng Part C Methods 24: 617–627. 10.1089/ten.tec.2018.0252 [DOI] [PubMed] [Google Scholar]
- Kienast Y, von Baumgarten L, Fuhrmann M, Klinkert WE, Goldbrunner R, Herms J, Winkler F. 2010. Real-time imaging reveals the single steps of brain metastasis formation. Nat Med 16: 116–122. 10.1038/nm.2072 [DOI] [PubMed] [Google Scholar]
- Klarenbeek J, Goedhart J, van Batenburg A, Groenewald D, Jalink K. 2015. Fourth-generation epac-based FRET sensors for cAMP feature exceptional brightness, photostability and dynamic range: characterization of dedicated sensors for FLIM, for ratiometry and with high affinity. PLoS One 10: e0122513 10.1371/journal.pone.0122513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski PJ, Rubin MA, Kleer CG. 2003. E-cadherin expression in primary carcinomas of the breast and its distant metastases. Breast Cancer Res 5: R217–R222. 10.1186/bcr651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriplani N, Duncan R, Leslie N. 2019. SWAP70 undergoes dynamic conformational regulation at the leading edge of migrating cells. FEBS Lett 593: 395–405. 10.1002/1873-3468.13326 [DOI] [PubMed] [Google Scholar]
- Kumar A, Ouyang M, Van den Dries K, McGhee EJ, Tanaka K, Anderson MD, Groisman A, Goult BT, Anderson KI, Schwartz MA, et al. 2016. Talin tension sensor reveals novel features of focal adhesion force transmission and mechanosensitivity. J Cell Biol 213: 371–383. 10.1083/jcb.201510012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labernadie A, Kato T, Brugués A, Serra-Picamal X, Derzsi S, Arwert E, Weston A, González-Tarragó V, Elosegui-Artola A, Albertazzi L, et al. 2017. A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nat Cell Biol 19: 224–237. 10.1038/ncb3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam AJ, St-Pierre F, Gong Y, Marshall JD, Cranfill PJ, Baird MA, McKeown MR, Wiedenmann J, Davidson MW, Schnitzer MJ, et al. 2001. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat Methods 9: 1005–1012. 10.1038/nmeth.2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam FC, Morton SW, Wyckoff J, Vu Han TL, Hwang MK, Maffa A, Balkanska-Sinclair E, Yaffe MB, Floyd SR, Hammond PT. 2018. Enhanced efficacy of combined temozolomide and bromodomain inhibitor therapy for gliomas using targeted nanoparticles. Nat Commun 9: 1991 10.1038/s41467-018-04315-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson MA, McDonald MM, Kovacic N, Hua Khoo W, Terry RL, Down J, Kaplan W, Paton-Hough J, Fellows C, Pettitt JA, et al. 2015. Osteoclasts control reactivation of dormant myeloma cells by remodelling the endosteal niche. Nat Commun 6: 8983 10.1038/ncomms9983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Xiang X, Sun J, He HT, Wu J, Wang Y, Zhu C. 2016. Imaging spatiotemporal activities of ZAP-70 in live T cells using a FRET-based biosensor. Ann Biomed Eng 44: 3510–3521. 10.1007/s10439-016-1683-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidke D, Nagy P, Arndt-Jovin DJ. 2007. In vivo imaging using quantum-dot-conjugated probes. Curr Protoc Cell Biol. 10.1002/0471143030.cb2501s36 [DOI] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. 2007. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450: 56–62. 10.1038/nature06293 [DOI] [PubMed] [Google Scholar]
- Looney MR, Thornton EE, Sen D, Lamm WJ, Glenny RW, Krummel MF. 2011. Stabilized imaging of immune surveillance in the mouse lung. Nat Methods 8: 91–96. 10.1038/nmeth.1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe DG. 2004. Distinctive image features from scale-invariant keypoints. Int J Comput Vis 60: 91–110. 10.1023/B:VISI.0000029664.99615.94 [DOI] [Google Scholar]
- Luckner M, Burgold S, Filser S, Scheungrab M, Niyaz Y, Hummel E, Wanner G, Herms J. 2018. Label-free 3D-CLEM using endogenous tissue landmarks. iScience 6: 92–101. 10.1016/j.isci.2018.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Zhang E, Su Y, Cheng T, Shi C. 2011. A review of NIR dyes in cancer targeting and imaging. Biomaterials 32: 7127–7138. 10.1016/j.biomaterials.2011.06.024 [DOI] [PubMed] [Google Scholar]
- Maddipati R, Stanger BZ. 2015. Pancreatic cancer metastases harbor evidence of polyclonality. Cancer Discov 5: 1086–1097. 10.1158/2159-8290.CD-15-0120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KJ, McGhee EJ, Schwarz JP, Drysdale M, Brachmann SM, Stucke V, Sansom OJ, Anderson KI. 2018. Accepting from the best donor; analysis of long-lifetime donor fluorescent protein pairings to optimise dynamic FLIM-based FRET experiments. PLoS ONE 13: e0183585 10.1371/journal.pone.0183585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matea CT, Mocan T, Tabaran F, Pop T, Mosteanu O, Puia C, Iancu C, Mocan L. 2017. Quantum dots in imaging, drug delivery and sensor applications. Int J Nanomedicine 12: 5421–5431. 10.2147/IJN.S138624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. 2009. A bright and photostable photoconvertible fluorescent protein. Nat Methods 6: 131–133. 10.1038/nmeth.1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milanovic M, Fan DNY, Belenki D, Däbritz JHM, Zhao Z, Yu Y, Dörr JR, Dimitrova L, Lenze D, Monteiro Barbosa IA, et al. 2018. Senescence-associated reprogramming promotes cancer stemness. Nature 553: 96–100. 10.1038/nature25167 [DOI] [PubMed] [Google Scholar]
- Muñoz-Espín D, Rovira M, Galiana I, Giménez C, Lozano-Torres B, Paez-Ribes M, Llanos S, Chaib S, Muñoz-Martín M, Ucero AC, et al. 2018. A versatile drug delivery system targeting senescent cells. EMBO Mol Med 10: e9355 10.15252/emmm.201809355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muta Y, Fujita Y, Sumiyama K, Sakurai A, Taketo MM, Chiba T, Seno H, Aoki K, Matsuda M, Imajo M. 2018. Composite regulation of ERK activity dynamics underlying tumour-specific traits in the intestine. Nat Commun 9: 2174 10.1038/s41467-018-04527-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netti PA, Berk DA, Swartz MA, Grodzinsky AJ, Jain RK. 2000. Role of extracellular matrix assembly in interstitial transport in solid tumors. Cancer Res 60: 2497–2503. [PubMed] [Google Scholar]
- Newman RH, Zhang J. 2008. Fucci: street lights on the road to mitosis. Chem Biol 15: 97–98. 10.1016/j.chembiol.2008.02.003 [DOI] [PubMed] [Google Scholar]
- Nobis M, McGhee EJ, Morton JP, Schwarz JP, Karim SA, Quinn J, Edward M, Campbell AD, McGarry LC, Evans TR, et al. 2013a. Intravital FLIM-FRET imaging reveals dasatinib-induced spatial control of src in pancreatic cancer. Cancer Res 73: 4674–4686. 10.1158/0008-5472.CAN-12-4545 [DOI] [PubMed] [Google Scholar]
- Nobis M, Carragher NO, McGhee EJ, Morton JP, Sansom OJ, Anderson KI, Timpson P. 2013b. Advanced intravital subcellular imaging reveals vital three-dimensional signalling events driving cancer cell behaviour and drug responses in live tissue. FEBS J 280: 5177–5197. 10.1111/febs.12348 [DOI] [PubMed] [Google Scholar]
- Nobis M, Herrmann D, Warren SC, Kadir S, Leung W, Killen M, Magenau A, Stevenson D, Lucas MC, Reischmann N, et al. 2017. A RhoA-FRET biosensor mouse for intravital imaging in normal tissue homeostasis and disease contexts. Cell Rep 21: 274–288. 10.1016/j.celrep.2017.09.022 [DOI] [PubMed] [Google Scholar]
- Nobis M, Warren SC, Lucas MC, Murphy KJ, Hermann D, Timpson P. 2018. Molecular mobility and activity in an intravital imaging setting—implications for cancer progression and targeting. J Cell Sci 131: jcs206995 10.1242/jcs.206995 [DOI] [PubMed] [Google Scholar]
- O'Neill PR, Castillo-Badillo JA, Meshik X, Kalyanaraman V, Melgarejo K, Gautam N. 2018. Membrane flow drives an adhesion-independent amoeboid cell migration mode. Dev Cell 46: 9–22.e4. 10.1016/j.devcel.2018.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornes S. 2016. Core concept: quantum dots. Proc Natl Acad Sci 113: 2796–2797. 10.1073/pnas.1601852113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osswald M, Jung E, Sahm F, Solecki G, Venkataramani V, Blaes J, Weil S, Horstmann H, Wiestler B, Syed M, et al. 2015. Brain tumour cells interconnect to a functional and resistant network. Nature 528: 93–98. 10.1038/nature16071 [DOI] [PubMed] [Google Scholar]
- Ouyang M, Huang H, Shaner NC, Remacle AG, Shiryaev SA, Strongin AY, Tsien RY, Wang Y. 2010. Simultaneous visualization of protumorigenic Src and MT1-MMP activities with fluorescence resonance energy transfer. Cancer Res 70: 2204–2212. 10.1158/0008-5472.CAN-09-3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang M, Wan R, Qin Q, Peng Q, Wang P, Wu J, Allen M, Shi Y, Laub S, Deng L, et al. 2019. Sensitive FRET biosensor reveals Fyn kinase regulation by submembrane localization. ACS Sens 4: 76–86. 10.1021/acssensors.8b00896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Lu S, Lei L, Lamberto I, Wang Y, Pasquale EB, Wang Y. 2019. Genetically encoded FRET biosensor for visualizing EphA4 activity in different compartments of the plasma membrane. ACS Sens 4: 294–300. 10.1021/acssensors.8b00465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SL, Buzzai A, Rautela J, Hor JL, Hochheiser K, Effern M, McBain N, Wagner T, Edwards J, McConville R, et al. 2019. Tissue-resident memory CD8+ T cells promote melanoma–immune equilibrium in skin. Nature 565: 366–371. 10.1038/s41586-018-0812-9 [DOI] [PubMed] [Google Scholar]
- Passaro D, Di Tullio A, Abarrategi A, Rouault-Pierre K, Foster K, Ariza-McNaughton L, Montaner B, Chakravarty P, Bhaw L, Diana G, et al. 2017. Increased vascular permeability in the bone marrow microenvironment contributes to disease progression and drug response in acute myeloid leukemia. Cancer Cell 32: 324–341.e6. 10.1016/j.ccell.2017.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippar U, Roussos ET, Oser M, Yamaguchi H, Kim HD, Giampieri S, Wang Y, Goswami S, Wyckoff JB, Lauffenburger DA, et al. 2008. A Mena invasion isoform potentiates EGF-induced carcinoma cell invasion and metastasis. Dev Cell 15: 813–828. 10.1016/j.devcel.2008.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickup MW, Mouw JK, Weaver VM. 2014. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep 15: 1243–1253. 10.15252/embr.201439246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinner S, Jordan P, Sharrock K, Bazley L, Collinson L, Marais R, Bonvin E, Goding C, Sahai E. 2009. Intravital imaging reveals transient changes in pigment production and Brn2 expression during metastatic melanoma dissemination. Cancer Res 69: 7969–7977. 10.1158/0008-5472.CAN-09-0781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piston DW, Kremers GJ. 2007. Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem Sci 32: 407–414. 10.1016/j.tibs.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Pittet MJ, Weissleder R. 2011. Intravital imaging. Cell 147: 983–991. 10.1016/j.cell.2011.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Progatzky F, Dallman MJ, Lo Celso C. 2013. From seeing to believing: labelling strategies for in vivo cell-tracking experiments. Interface Focus 3: 20130001 10.1098/rsfs.2013.0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S, Li H, Lu L, Qi Z, Liu L, Chen L, Shen G, Fu L, Luo Q, Zhang Z. 2016. Long-term intravital imaging of the multicolor-coded tumor microenvironment during combination immunotherapy. Elife 5: e14756 10.7554/eLife.14756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Hannezo E, Mangeat T, Liu C, Majumder P, Liu J, Choesmel-Cadamuro V, McDonald JA, Liu Y, Yi B, et al. 2018. A biochemical network controlling basal myosin oscillation. Nat Commun 9: 1210 10.1038/s41467-018-03574-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajoria S, Zhao L, Intes X, Barroso M. 2014. FLIM-FRET for cancer applications. Curr Mol Imaging 3: 144–161. 10.2174/2211555203666141117221111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves MQ, Kandyba E, Harris S, Del Rosario R, Balmain A. 2018. Multicolour lineage tracing reveals clonal dynamics of squamous carcinoma evolution from initiation to metastasis. Nat Cell Biol 20: 699–709. 10.1038/s41556-018-0109-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridder K, Keller S, Dams M, Rupp AK, Schlaudraff J, Del Turco D, Starmann J, Macas J, Karpova D, Devraj K, et al. 2014. Extracellular vesicle-mediated transfer of genetic information between the hematopoietic system and the brain in response to inflammation. PLoS Biol 12: e1001874 10.1371/journal.pbio.1001874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridder K, Sevko A, Heide J, Dams M, Rupp AK, Macas J, Starmann J, Tjwa M, Plate KH, Sültmann H, et al. 2015. Extracellular vesicle-mediated transfer of functional RNA in the tumor microenvironment. Oncoimmunology 4: e1008371 10.1080/2162402X.2015.1008371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritsma L, Steller EJ, Beerling E, Loomans CJ, Zomer A, Gerlach C, Vrisekoop N, Seinstra D, van Gurp L, Schäfer R, et al. 2012. Intravital microscopy through an abdominal imaging window reveals a pre-micrometastasis stage during liver metastasis. Sci Transl Med 4: 158ra145 10.1126/scitranslmed.3004394 [DOI] [PubMed] [Google Scholar]
- Ritsma L, Vrisekoop N, van Rheenen J. 2013. In vivo imaging and histochemistry are combined in the cryosection labelling and intravital microscopy technique. Nat Commun 4: 2366 10.1038/ncomms3366 [DOI] [PubMed] [Google Scholar]
- Roussos ET, Balsamo M, Alford SK, Wyckoff JB, Gligorijevic B, Wang Y, Pozzuto M, Stobezki R, Goswami S, Segall JE, et al. 2011. Mena invasive (MenaINV) promotes multicellular streaming motility and transendothelial migration in a mouse model of breast cancer. J Cell Sci 124: 2120–2131. 10.1242/jcs.086231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheele CL, Hannezo E, Muraro MJ, Zomer A, Langedijk NS, van Oudenaarden A, Simons BD, van Rheenen J. 2017. Identity and dynamics of mammary stem cells during branching morphogenesis. Nature 542: 313–317. 10.1038/nature21046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, Clevers H. 2012. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 337: 730–735. 10.1126/science.1224676 [DOI] [PubMed] [Google Scholar]
- Schindler SE, McCall JG, Yan P, Hyrc KL, Li M, Tucker CL, Lee JM, Bruchas MR, Diamond MI. 2015. Photo-activatable Cre recombinase regulates gene expression in vivo. Sci Rep 5: 13627 10.1038/srep13627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong J, Ouyang M, Kim T, Sun J, Wen PC, Lu S, Zhuo Y, Llewellyn NM, Schlaepfer DD, Guan JL, et al. 2011. Detection of focal adhesion kinase activation at membrane microdomains by fluorescence resonance energy transfer. Nat Commun 2: 406 10.1038/ncomms1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong J, Tajik A, Sun J, Guan JL, Humphries MJ, Craig SE, Shekaran A, García AJ, Lu S, Lin MZ, et al. 2013. Distinct biophysical mechanisms of focal adhesion kinase mechanoactivation by different extracellular matrix proteins. Proc Natl Acad Sci 110: 19372–19377. 10.1073/pnas.1307405110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong J, Huang M, Sim KM, Kim H, Wang Y. 2017. FRET-based visualization of PDGF receptor activation at membrane microdomains. Sci Rep 7: 1593 10.1038/s41598-017-01789-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova D, Baloban M, Emelyanov AV, Brenowitz M, Guo P, Verkhusha VV. 2016. Bright monomeric near-infrared fluorescent proteins as tags and biosensors for multiscale imaging. Nat Commun 7: 12405 10.1038/ncomms12405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh OA, Tanese D, Zampini V, Linghu C, Piatkevich K, Ronzitti E, Papagiakoumou E, Boyden ES, Emiliani V. 2017. Temporally precise single-cell-resolution optogenetics. Nat Neurosci 20: 1796–1806. 10.1038/s41593-017-0018-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, et al. 2010. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143: 134–144. 10.1016/j.cell.2010.09.016 [DOI] [PubMed] [Google Scholar]
- Spicer JD, McDonald B, Cools-Lartigue JJ, Chow SC, Giannias B, Kubes P, Ferri LE. 2012. Neutrophils promote liver metastasis via Mac-1-mediated interactions with circulating tumor cells. Cancer Res 72: 3919–3927. 10.1158/0008-5472.CAN-11-2393 [DOI] [PubMed] [Google Scholar]
- Stawarski M, Rutkowska-Wlodarczyk I, Zeug A, Bijata M, Madej H, Kaczmarek L, Wlodarczyk J. 2014. Genetically encoded FRET-based biosensor for imaging MMP-9 activity. Biomaterials 35: 1402–1410. 10.1016/j.biomaterials.2013.11.033 [DOI] [PubMed] [Google Scholar]
- Steeg PS. 2016. Targeting metastasis. Nat Rev Cancer 16: 201–218. 10.1038/nrc.2016.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbeek SC, Pham TV, de Ligt J, Zomer A, Knol JC, Piersma SR, Schelfhorst T, Huisjes R, Schiffelers RM, Cuppen E, et al. 2018. Cancer cells copy migratory behavior and exchange signaling networks via extracellular vesicles. EMBO J 37: e98357 10.15252/embj.201798357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steurer B, Turkyilmaz Y, van Toorn M, van Leeuwen W, Escudero-Ferruz P, Marteijn JA. 2019. Fluorescently-labelled CPD and 6-4PP photolyases: new tools for live-cell DNA damage quantification and laser-assisted repair. Nucleic Acids Res 47: 3536–3549. 10.1093/nar/gkz035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiers PJ, van Gastel N, Moermans K, Stockmans I, Carmeliet G. 2018. An ectopic imaging window for intravital imaging of engineered bone tissue. JBMR Plus 2: 92–102. 10.1002/jbm4.10028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subach OM, Patterson GH, Ting LM, Wang Y, Condeelis JS, Verkhusha VV. 2011. A photoswitchable orange-to-far-red fluorescent protein, PSmOrange. Nat Methods 8: 771–777. 10.1038/nmeth.1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M, Sakaue-Sawano A, Iimura T, Fukami K, Kitaguchi T, Kawakami K, Okamoto H, Higashijima S, Miyawaki A. 2009. Illuminating cell-cycle progression in the developing zebrafish embryo. Proc Natl Acad Sci 106: 20812–20817. 10.1073/pnas.0906464106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suijkerbuijk SJE, van Rheenen J. 2017. From good to bad: intravital imaging of the hijack of physiological processes by cancer cells. Dev Biol 428: 328–337. 10.1016/j.ydbio.2017.04.015 [DOI] [PubMed] [Google Scholar]
- Tan P, He L, Han G, Zhou Y. 2017. Optogenetic immunomodulation: shedding light on antitumor immunity. Trends Biotechnol 35: 215–226. 10.1016/j.tibtech.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanabalasuriar A, Neupane AS, Wang J, Krummel MF, Kubes P. 2016. iNKT cell emigration out of the lung vasculature requires neutrophils and monocyte-derived dendritic cells in inflammation. Cell Rep 16: 3260–3272. 10.1016/j.celrep.2016.07.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torcellan T, Stolp J, Chtanova T. 2017. In vivo imaging sheds light on immune cell migration and function in cancer. Front Immunol 8: 309 10.3389/fimmu.2017.00309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valon L, Marín-Llauradó A, Wyatt T, Charras G, Trepat X. 2017. Optogenetic control of cellular forces and mechanotransduction. Nat Commun 8: 14396 10.1038/ncomms14396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wal J, Habets R, Várnai P, Balla T, Jalink K. 2001. Monitoring agonist-induced phospholipase C activation in live cells by fluorescence resonance energy transfer. J Biol Chem 276: 15337–15344. 10.1074/jbc.M007194200 [DOI] [PubMed] [Google Scholar]
- van Huizen LMG, Kuzmin NV, Barbe E, van der Velde S, Te Velde EA, Groot ML. 2019. Second and third harmonic generation microscopy visualizes key structural components in fresh unprocessed healthy human breast tissue. J Biophotonics 12: e201800297 10.1002/jbio.201800297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennin C, Hermann D, Lucas MC, Timpson P. 2016. Intravital imaging reveals new ancillary mechanisms co-opted by cancer cells to drive tumor progression. F1000Res 5 10.12688/f1000research.8090.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennin C, Chin VT, Warren SC, Lucas MC, Herrmann D, Magenau A, Melenec P, Walters SN, del Monte-Nieto G, Conway JR, et al. 2017. Transient tissue priming via ROCK inhibition uncouples pancreatic cancer progression, sensitivity to chemotherapy, and metastasis. Sci Transl Med 9: eaai8504 10.1126/scitranslmed.aai8504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennin C, Murphy KJ, Morton JP, Cox TR, Pajic M, Timpson P. 2018. Reshaping the tumor stroma for treatment of pancreatic cancer. Gastroenterology 154: 820–838. 10.1053/j.gastro.2017.11.280 [DOI] [PubMed] [Google Scholar]
- Vera-Ramirez L, Vodnala SK, Nini R, Hunter KW, Green JE. 2018. Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat Commun 9: 1944 10.1038/s41467-018-04070-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R. 1839. Erlauterungstaflen zur physiologie und entwicklungsgeschichte. Leopold Voss, Leipzig, Germany. [Google Scholar]
- Wang Y, Botvinick EL, Zhao Y, Berns MW, Usami S, Tsien RY, Chien S. 2005. Visualizing the mechanical activation of Src. Nature 434: 1040–1045. 10.1038/nature03469 [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Xu ZP, Liu X, Roberts MS, Liang X. 2018. Anionic long-circulating quantum dots for long-term intravital vascular imaging. Pharmaceutics 10: 244 10.3390/pharmaceutics10040244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren SC, Nobis M, Magenau A, Mohammed YH, Herrmann D, Moran I, Vennin C, Conway JR, Mélénec P, Cox TR, et al. 2018. Removing physiological motion from intravital and clinical functional imaging data. Elife 7: e35800 10.7554/eLife.35800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelin B, Bakker GJ, Friedl P. 2012. Intravital third harmonic generation microscopy of collective melanoma cell invasion: principles of interface guidance and microvesicle dynamics. Intravital 1: 32–43. 10.4161/intv.21223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil S, Osswald M, Solecki G, Grosch J, Jung E, Lemke D, Ratliff M, Hänggi D, Wick W, Winkler F. 2017. Tumor microtubes convey resistance to surgical lesions and chemotherapy in gliomas. Neuro Oncol 19: 1316–1326. 10.1093/neuonc/nox070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissleder R. 2001. A clearer vision for in vivo imaging. Nat Biotechnol 19: 316–317. 10.1038/86684 [DOI] [PubMed] [Google Scholar]
- Wellenstein MD, de Visser KE. 2018. Cancer-cell-intrinsic mechanisms shaping the tumor immune landscape. Immunity 48: 399–416. 10.1016/j.immuni.2018.03.004 [DOI] [PubMed] [Google Scholar]
- Wilson WR, Hay MP. 2011. Targeting hypoxia in cancer therapy. Nat Rev Cancer 11: 393–410. 10.1038/nrc3064 [DOI] [PubMed] [Google Scholar]
- Wood S Jr. 1958. Pathogenesis of metastasis formation observed in vivo in the rabbit ear chamber. AMA Arch Pathol 66: 550–568. [PubMed] [Google Scholar]
- Wyckoff JB, Wang Y, Lin EY, Li JF, Goswami S, Stanley ER, Segall JE, Pollard JW, Condeelis J. 2007. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res 67: 2649–2656. 10.1158/0008-5472.CAN-06-1823 [DOI] [PubMed] [Google Scholar]
- Yang X, Liang X, Zheng M, Tang Y. 2018. Cellular phenotype plasticity in cancer dormancy and metastasis. Front Oncol 8: 505 10.3389/fonc.2018.00505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano S, Zhang Y, Miwa S, Tome Y, Hiroshima Y, Uehara F, Yamamoto M, Suetsugu A, Kishimoto H, Tazawa H, et al. 2014. Spatial–temporal FUCCI imaging of each cell in a tumor demonstrates locational dependence of cell cycle dynamics and chemoresponsiveness. Cell Cycle 13: 2110–2119. 10.4161/cc.29156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim M, Durr N, Ben-Yakar A. 2015. Tripling the maximum imaging depth with third-harmonic generation microscopy. J Biomed Opt 20: 096013 10.1117/1.JBO.20.9.096013 [DOI] [PubMed] [Google Scholar]
- You S, Tu H, Chaney EJ, Sun Y, Zhao Y, Bower AJ, Liu YZ, Marjanovic M, Sinha S, Pu Y, et al. 2018. Intravital imaging by simultaneous label-free autofluorescence-multiharmonic microscopy. Nat Commun 9: 2125 10.1038/s41467-018-04470-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chen L, Liu J, Dai B, Xu G, Shen G, Luo Q, Zhang Z. 2019. Immune modulation of liver sinusoidal endothelial cells by melittin nanoparticles suppresses liver metastasis. Nat Commun 10: 574 10.1038/s41467-019-08538-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukinaga H, Shionyu C, Hirata E, Ui-Tei K, Nagashima T, Kondo S, Okada-Hatakeyama M, Naoki H, Matsuda M. 2014. Fluctuation of Rac1 activity is associated with the phenotypic and transcriptional heterogeneity of glioma cells. J Cell Sci 127: 1805–1815. 10.1242/jcs.139733 [DOI] [PubMed] [Google Scholar]
- Zago G, Veith I, Singh MK, Fuhrmann L, De Beco S, Remorino A, Takaoka S, Palmeri M, Berger F, Brandon N, et al. 2018. RalB directly triggers invasion downstream Ras by mobilizing the Wave complex. Elife 7: e40474 10.7554/eLife.40474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Zhu X, Cui K, Mancuso J, Federley R, Fischer K, Teng G, Mittal V, Gao D, Zhao H, et al. 2016. In vivo visualization and characterization of epithelial-mesenchymal transition in breast tumors. Cancer Res 76: 2094–2104. 10.1158/0008-5472.CAN-15-2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS, Kalluri R. 2015. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 527: 525–530. 10.1038/nature16064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Wang J, Chen J, Qi Y, Nan D, Jin L, Qian X, Wang X, Chen Q, Liu X, et al. 2018. Optogenetic control of epithelial-mesenchymal transition in cancer cells. Sci Rep 8: 14098 10.1038/s41598-018-32539-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomer A, Ellenbroek SI, Ritsma L, Beerling E, Vrisekoop N, van Rheenen J. 2013. Intravital imaging of cancer stem cell plasticity in mammary tumors. Stem Cells 31: 602–606. 10.1002/stem.1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomer A, Maynard C, Verweij FJ, Kamermans A, Schäfer R, Beerling E, Schiffelers RM, de Wit E, Berenguer J, Ellenbroek SIJ, et al. 2015. In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell 161: 1046–1057. 10.1016/j.cell.2015.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]