Abstract
Rapid plant genome evolution is crucial to adapt to environmental changes. Chromosomal rearrangements and gene copy number variation (CNV) are two important tools for genome evolution and sources for the creation of new genes. However, their emergence takes many generations. In this study, we show that in Arabidopsis thaliana, a significant loss of ribosomal RNA (rRNA) genes with a past history of a mutation for the chromatin assembly factor 1 (CAF1) complex causes rapid changes in the genome structure. Using long-read sequencing and microscopic approaches, we have identified up to 15 independent large tandem duplications in direct orientation (TDDOs) ranging from 60 kb to 1.44 Mb. Our data suggest that these TDDOs appeared within a few generations, leading to the duplication of hundreds of genes. By subsequently focusing on a line only containing 20% of rRNA gene copies (20rDNA line), we investigated the impact of TDDOs on 3D genome organization, gene expression, and cytosine methylation. We found that duplicated genes often accumulate more transcripts. Among them, several are involved in plant–pathogen response, which could explain why the 20rDNA line is hyper-resistant to both bacterial and nematode infections. Finally, we show that the TDDOs create gene fusions and/or truncations and discuss their potential implications for the evolution of plant genomes.
In most eukaryotes, hundreds of ribosomal RNA (rRNA) genes compose the nucleolus organizer region (NOR). In Arabidopsis thaliana Columbia ecotype (Col-0), 375 tandem 45S rRNA gene copies are located at the top of both Chromosomes 2 (NOR2) and 4 (NOR4) (Copenhaver and Pikaard 1996). Only a portion of these copies is actively transcribed in the nucleolus to produce ribosomes. Most rRNA genes indeed remain transcriptionally inactive and accumulate repressive chromatin modification marks (Pontvianne et al. 2010, 2012, 2013; Grummt and Längst 2013). As in many species, rRNA gene copy numbers are highly variable among A. thaliana populations (Dopman and Hartl 2007; Kobayashi 2011; Gibbons et al. 2015; Rabanal et al. 2017). In natural inbred lines found in Sweden, rRNA copy number heterogeneity can account for up to 10% of genome size variation (Long et al. 2013). Worldwide, A. thaliana ecotypes can be found with a rRNA gene copy number ranging from 500 to 2500 in haploid cells (Long et al. 2013). Therefore, 500 copies could be considered as the lowest rRNA gene copy number found in natura so far (Rabanal et al. 2017). In budding yeast and in Drosophila, previous studies suggest that a minimum amount of inactive rRNA genes is necessary for global genome stability (Ide et al. 2010). One to two hundred rRNA gene units are usually found in budding yeast, but genome engineering allowed the creation of viable yeast lines with only 40 rRNA gene units (Takeuchi et al. 2003). Similarly, shifts in rRNA gene copy number affect genome-wide chromatin marks and alter gene expression in flies (Paredes and Maggert 2009; Paredes et al. 2011). In plants, neither the impact of this variability nor the consequences of having few copies of rRNA genes are known.
FASCIATA (FAS) 1 and 2 are part of the chromatin assembly factor (CAF) complex required for proper deposition of histones H3 and H4 upon DNA replication (Ramirez-Parra and Gutierrez 2007). In A. thaliana, their knockouts accumulate several signs of genomic instability, including double-stranded breaks (DSBs), telomere shortening, and drastic changes in rRNA gene copy number (Mozgová et al. 2010; Varas et al. 2017). Consequently, the intranuclear positioning of the NORs as well as their epigenetic state are modified in these mutants (Mozgová et al. 2010; Pontvianne et al. 2013). Crossing fas1-4 and fas2-4 mutants and subsequent inbreeding by self-fertilization led to the creation of A. thaliana wild-type segregant FAS genes lines with only 20% of the amount of rRNA gene copies in comparison to wild-type Col-0 (Fig. 1A; Pavlištová et al. 2016). This 20rDNA line (hereafter named 20rDNA L6) has a wild-type phenotype and retained a low amount of rRNA genes for five generations (referred as F5) (Pavlištová et al. 2016). In this study, we took advantage of this plant material to test the impact of a low amount of rRNA genes on plant genome stability during several generations. We found unexpected consequences on the genome structure and stability, as well as on its 3D genome organization. We also show the short-term consequences of gene copy number variation (CNV) on their expression and potentially their role in plant phenotypic traits such as pathogen responses.
Figure 1.
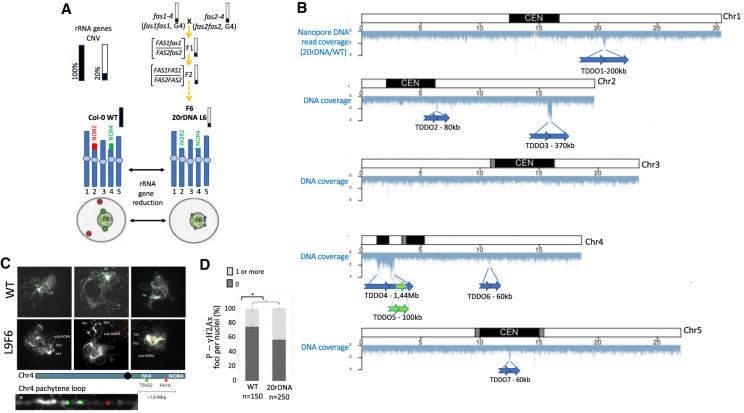
Genomic instability in the 20rDNA line L6F6. (A) Schematic representation of obtaining the 20rDNA L6F6 and its relative content in rRNA gene copies. NOR2 and NOR4 rRNA gene copies are both affected by the reduction, and according to the DNA-FISH experiment, it changes their nuclear distribution in the nucleus compared to wild-type Col-0 (WT), because all rRNA gene copies associate with the nucleolus (Pavlištová et al. 2016). (B) Distribution of the reads obtained by nanopore sequencing along all chromosomes in the 20rDNA L6F6 line compared to wild-type Col-0. The blue arrows represent the localization and the orientation of the TDDO identified in L6F6, except for TDDO5, which is represented by a green arrow. (C) DNA-FISH analyses of two loci present on the short arm of Chromosome 4 (kr4s), distant by 1.6 Mb: one present on the TDDO4 (BAC T5H22; green) and one located outside the TDDO4 (BAC F5I10; red). Three Pachytene chromosomes from WT Col-0 and in 20rDNA L6F6 are shown. A zoom of the unlooped kr4s from the middle panel of the 20rDNA line6 is presented at the bottom of the panel. (D) Histogram showing the percentage of nuclei displaying at least one P-γ-H2Ax foci in WT Col-0 versus L6F6.
Results
The 20rDNA L6 line accumulates features of genomic instability
The 20rDNA L6 contains only 20% of rRNA genes compared to the wild-type Col-0 and was obtained as a wild-type segregant from a cross between fas1-4 and fas2-4 mutant lines as described (Fig. 1A; Pavlištová et al. 2016). To confirm and precisely map the potential consequence of genomic instability in 20rDNA L6F6 (F6 for the sixth generation after F1), we performed long-read resequencing using nanopore technology. We obtained 6.4 Gb of total sequences with a midsize of 6 kb. We then analyzed the sequencing coverage against the TAIR10 A. thaliana Col-0 reference genome to identify the highly covered regions (Fig. 1B). We have detected seven large duplications, corresponding to tandem duplications in direct orientation (TDDO), named TDDO1 to TDDO7. The largest region, TDDO4, represents 1.44 Mb, spanning the heterochromatic knob on the short arm of Chromosome 4 (hk4s), a large heterochromatic region outside the pericentromeres, and a euchromatic region distal to the knob. Other TDDOs range in size from 60 to 370 kb long and are present on Chromosomes 1, 2, 4, and 5 (Fig. 1B). The absence/presence of TDDO4, the largest duplication, was also confirmed by DNA-fluorescence in situ hybridization (FISH) (Fig. 1C). We used two probes generated from BAC clones: one recognizing a portion of TDDO4 (hk4s–T5H22) and one recognizing an unduplicated genomic region located between TDDO4 and the NOR4 (F5I10). Different cell types were analyzed from vegetative as well as reproductive tissues: in both, more signals corresponding to TDDO4 were detected in the 20rDNA L6F6 nuclei compared to wild-type Col-0 cells (Supplemental Fig. S1). Analyses of pachytene chromosomes clearly showed that the additional signal actually belonged to the same chromosome, which confirms the duplication hypothesis (Fig. 1C).
The occurrence of duplication events is a sign of genomic instability. Thus, the chromosomal rearrangements observed in 20rDNA L6F6 could be the consequence of double-stranded breaks (DSBs). To test this hypothesis, we compared the amount of spontaneous DSBs between 20rDNA L6F6 and wild-type Col-0 cells by performing immunostaining of serine 139-phosphorylated H2Ax histone variant (P-γ-H2Ax), which is a marker of DSB (Charbonnel et al. 2010). P-γ-H2Ax foci were detected at a higher rate in 20rDNA L6F6 nuclei compared to wild-type nuclei from leaf tissues (Fig. 1D; Supplemental Fig. S2A). Accumulation of DSB foci can potentially be associated with a DNA repair defect. This hypothesis is supported by an increased susceptibility of the 20rDNA L6F6 line to a treatment with the genotoxin bleomycin (Supplemental Fig. S2B).
Appearance of the duplication events in the 20rDNA line
To show a potential link between low rDNA copies and TDDO appearance, it is crucial to know when these duplication events occurred. Like 20rDNA L6F6 line, 20rDNA L9F6 is an independent inbred line deriving from the cross between fas1-4 and fas2-4 mutants that both also display low amounts of rDNA copies (Supplemental Fig. S3; Mozgová et al. 2010; Pavlištová et al. 2016). We then performed long-read resequencing using nanopore technology and identified TDDO in the 20rDNA L9F6 line, as well as in the offspring of the parental lines fas1-4 and fas2-4 used to generate the initial cross (Fig. 2A).
Figure 2.
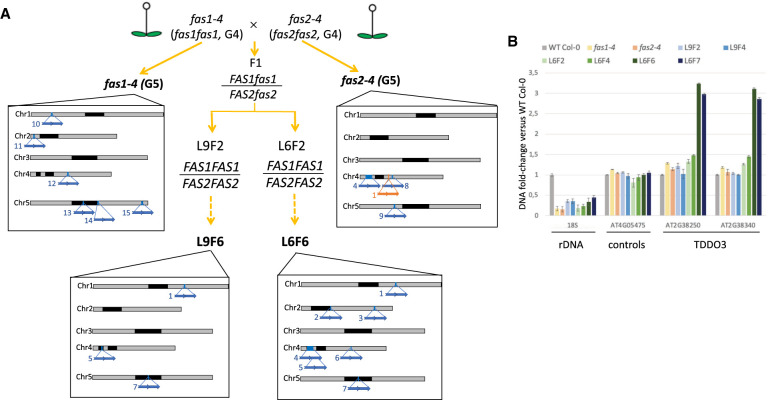
Identification of TDDO in the parental lines and in L9F6. (A) Identification of TDDO and deletion in the offspring of the parental lines fas1-4 and fas2-4 used to generate the L6 and L9 lines. Lines in bold were sequenced using nanopore technology. Relative distribution and names of each chromosome rearrangement identified are represented by blue (TDDO) and orange (deletion) arrows along chromosomes. Characteristics of each TDDO can be found in Supplemental Table S2. (B) CNV of genes present or not in TDDO3 and of rRNA genes were determined by quantitative PCR. Their relative enrichment was determined in WT Col-0, the parent lines fas1-4 and fas2-4, and in L6 and L9 at several generations. CNVs of rRNA genes were determined using probes amplifying the 18S, the loci AT4G05475 and AT4G16580 that are not duplicated are controls, and the loci AT4TE12140 and AT4G05030 allow the identification of the largest duplication of TDDO4.
In fas1-4, none of the seven TDDOs identified in 20rDNA L6F6 were detected, but we found six new TDDOs ranging from 57 to 175 kb long (Fig. 2A; Supplemental Fig. S4A). In fas2-4, only TDDO4 is present, as well as two additional duplications named TDDO8 (286 kb) and TDDO9 (106 kb) (Fig. 2A; Supplemental Fig. S4B). We also identified a deletion of 9.75 kb named DEL1 on Chromosome 4 (Fig. 2A; Supplemental Fig. S5). Analyses of 20rDNA L9F6 revealed that TDDOs 1, 5, and 7 are shared between L9F6 and L6F6, which suggests their presence in the F1 (Fig. 2A; Supplemental Fig. S4C). We could not find TDDO4, although this duplication is in one of the parents. Further analyses by quantitative PCR and DNA-FISH revealed that TDDO4 has been segregated out between the L9F2 and L9F4 (Supplemental Fig. S6). In parallel, the absence of TDDOs 2, 3, and 6 in the fas parents or in the L9 strongly suggests the appearance of these TDDOs between L6F2 and L6F6 (Supplemental Fig. S4D). This hypothesis is supported at least for TDDO3 by qPCR analyses (Fig. 2B). However, owing to a lack of long-read sequences obtained for L9F6, we are not able to determine the existence of novel TDDO.
In summary, our analyses identified 15 TDDOs that appeared independently, either in the parental line or in the two independent inbred lines resulting from the fas1-4 and fas2-4 cross (Fig. 2A; Supplemental Figs. S4A,D, S7). This hypothesis is supported by the absence of TDDO4 in all generation 1 (G1) mutants fas2-4 and fas2-5 analyzed by PCR (Supplemental Fig. S4E,F), suggesting the also very recent appearance of TDDO4 in the parental fas2-4 (G4) line.
Impact of low rDNA and TDDO on 3D genome organization
The nucleolus plays an important role in the spatial organization of the chromosomes (Bersaglieri and Santoro 2019; Pontvianne and Liu 2020; Santos et al. 2020). Nucleolus-associated chromatin domains (NADs), essentially composed of repressed chromatin domains, localize at the nucleolar periphery (Németh et al. 2010; van Koningsbruggen et al. 2010; Pontvianne et al. 2016b). Because rRNA gene nuclear distribution has a critical impact in NADs identity both in plant and animal cells (Quinodoz et al. 2018; Picart-Picolo et al. 2019, 2020), we analyzed NADs composition and 3D organization in 20rDNA L6F6. The fifth generation of the 20rDNA L6 (20rDNA L6F5) was transformed with a transgene ectopically expressing the FIBRILLARIN 2 nucleolar protein fused to the yellow fluorescent protein (FIB2:YFP). Using the FIB2:YFP nucleolar marker, we isolated nuclei and nucleoli from the transformants and identified NADs nuclear and nucleolar DNA sequences as previously described (Pontvianne et al. 2016a; Carpentier et al. 2018). Preceding studies have clearly shown that in wild-type Col-0 leaf cells, NOR4-derived rRNA genes are expressed and associate with the nucleolus. Conversely, NOR2 is excluded from the nucleolus, and NOR2-derived rRNA genes are silent (Pontvianne et al. 2013; Chandrasekhara et al. 2016). As a result, NADs are essentially distributed in the entire short arm of Chromosome 4 (kr4s), which juxtaposes the active NOR4 and associates with the nucleolus (Pontvianne et al. 2016b). Compared to the wild-type, NADs in 20rDNA L6F6 are enriched from genomic regions located on both Chromosomes 2 and 4 short arms (Fig. 3A; Supplemental Fig. S8). Among the 434 genes that gained nucleolar association, 144 belong to Chromosome 2 (33%) (Supplemental Table S1). In contrast, only 19 genes on Chromosome 4 gain nucleolar association. These results are consistent with the rDNA transcriptional state, as all leftover NOR2 and NOR4-derived rRNA genes are actively transcribed and associate with the nucleolus (Pavlištová et al. 2016). We also detected an enrichment of centromeric sequences associating with nucleoli in L6F6 compared to wild type. This type of reorganization was previously shown to associate with changes in NOR subnuclear organization (Pontvianne et al. 2016b; Pontvianne and Grob 2020). As in wild type, subtelomeric regions remain associated with the nucleolus in the 20rDNA L6F6 line (Fig. 3A; Supplemental Fig. S8). In summary, NAD identification in 20rDNA L6F6 revealed that 5.6 Mb of chromatin domains mainly enriched in silent epigenetic marks changed their subnuclear distribution, which suggests a substantial reorganization of the nuclear genome.
Figure 3.
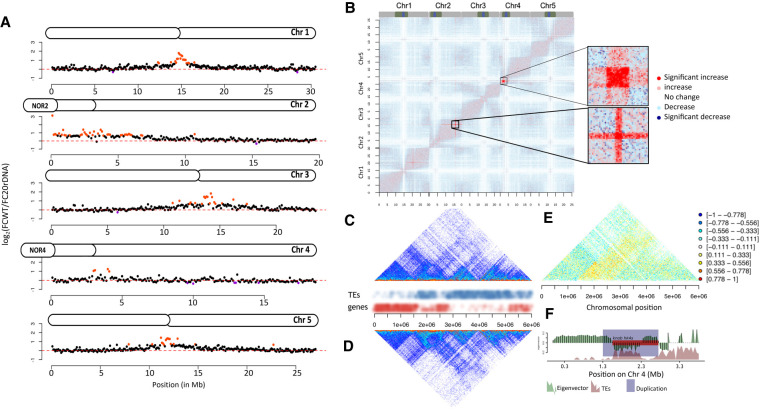
3D genome organization in L6F6. (A) Chromosome plots displaying the relative enrichment of a given genomic segment with the nucleolus. The y-axis displays the fold change nucleolus enrichment between wild-type Col-0 and the 20rDNA L6F6. Each dot represents a 100-kb window. Nucleolus-enriched genomic regions above the threshold are red, and depleted regions are violet. (B) Coverage-normalized t-test difference matrix (50-kb bins). The color of each pixel of the matrix is defined by the result of a t-test using the triplicate contact frequencies from wild-type and 20rDNA coverage-normalized Hi-C samples. The two magnified areas correspond to the two regions displaying the highest level of contact frequency changes. (C,D) Non-normalized Hi-C snapshot showing the contact frequencies on the short arm of Chromosome 4 in wild-type Col-0 (C) versus the 20rDNA L6F6 (D). TEs and genes are annotated to illustrate the occurrence of euchromatin and heterochromatin, respectively. (E) Ratio between Hi-C contact frequencies from wild-type and 20rDNA L6F6. Negative ratios correspond to more contacts in the wild type, whereas positive ratios correspond to more contacts in the 20rDNA L6F6. (F) Eigenvector of the wild-type Co-0 Hi-C data set and annotation of the TDDO4 affecting the knob hk4s. Note the central duplication breakpoint exactly coincides with a change between LSD and a CSD.
To get a global view of the chromatin 3D organization, we analyzed all chromatin–chromatin interactions using genome-wide chromosome-conformation capture (Hi-C). We generated triplicate Hi-C samples from both wild-type and 20rDNA L6F6 14-d-old seedlings (Fig. 3B; Supplemental Fig. S9). To assess differences between two given sets of Hi-C samples statistically, we took advantage of our triplicate Hi-C data sets and performed student t-tests on each contact frequency (pixel of the Hi-C matrix) and determined whether contact frequencies significantly changed between the wild-type and 20rDNA L6F6 (Fig. 3C,E). Contact frequencies assayed by Hi-C can be used to detect chromosomal rearrangements (Himmelbach et al. 2018). In our case, a duplication would lead to a twofold increase in coverage of the affected region, thus doubling of interaction frequencies at this region. We indeed found several regions displaying a significant (P < 0.01) increase of contact frequencies at several chromosomal locations, all corresponding to the previously described TDDO1 to TDDO7. Analyzing the genome-wide coverage using unpaired raw Hi-C sequencing reads confirmed the presence of significant increase in coverage of the affected regions (Supplemental Fig. S10). We subsequently normalized our Hi-C matrices for the assayed coverage. However, coverage-normalized Hi-C data showed that short-range contact frequencies within the duplicated regions are significantly depleted. Whether this depletion of contact frequencies is biologically significant or represents an artifact of the normalization procedure is extremely difficult to determine.
To further examine potential differences in 3D folding principles between wild-type Col-0 and 20rDNA L6F6, we performed a principal component analysis (PCA) to retrieve the eigenvector, which is characteristic of 3D folding patterns of a Hi-C data set (Grob et al. 2014; Lieberman-Aiden et al. 2009). Sign changes in the eigenvector delineate basic 3D folding domains, known as loose structural domains (LSDs) and closed structural domains (CSDs), which are analogous to animal A and B compartments (Lieberman-Aiden et al. 2009). We could not identify significant changes in the eigenvectors between wild-type Col-0 and the 20rDNA L6F6. Moreover, outside the duplicated regions, no changes in genomic bin contact frequencies could be observed. We therefore focused on the duplicated regions and analyzed duplication breakage points with the eigenvector obtained by the PCA analysis of the wild-type Col-0 Hi-C data (Supplemental Fig. S11). We observed that in a majority of the TDDOs in L6F6, at least one of the breakage points coincides with sign changes (CSDs to LSDs) or directional changes (valleys and peaks within a structural domain) in the eigenvector, with the exception of TDDO3. Hence, the changes in 3D conformation may have facilitated the occurrence of the TDDOs. This was most prominent for TDDO4, where the more central breakpoint exactly colocalizes with the change between the CSD and the LSD, which defines the ancient inversion breakpoint that gave rise to the knob (Fig. 3F; Zapata et al. 2016). This suggests the existence of continuously fragile chromosomal regions, the borders between structural domains being diagnostic for these regions.
Duplication events create chimeric genes
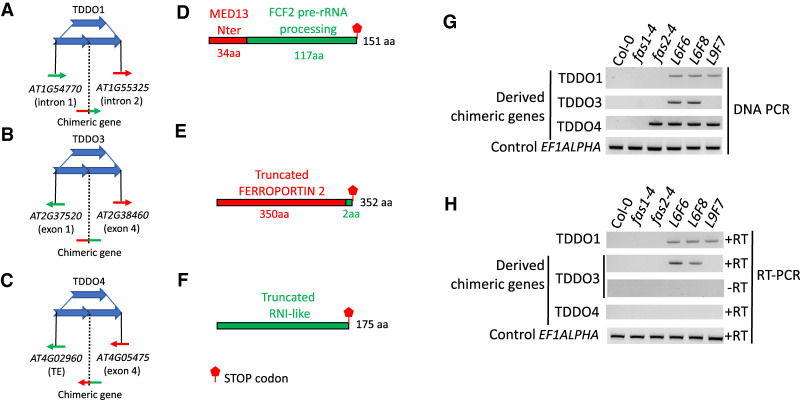
Most of the time, TDDOs keep genes intact and do not lead to gene loss. However, truncated genes can be generated at the breakpoint junction, while keeping intact genes on the edges of duplication (Newman et al. 2015). Besides, when breakpoints are located in two different genes in the same orientation, gene fusion can take place if the open reading frame (ORF) is preserved. In 20rDNA L6F6, we systematically analyzed the TDDO breakpoint junctions (Supplemental Figs. S7, S12). Of the seven cases of TDDO identified in this line, three potentially created fused or truncated proteins (Fig. 4A,F). On Chromosome 1, TDDO1 fused the first exon of gene AT1G55325 that encodes the N-terminal domain of the MEDIATOR 13-like with four of the five exons of AT1G54770 that encodes the FCF2 pre-rRNA processing factor. On Chromosome 2, although genes are in the opposite orientation, TDDO3 creates a shorter ORF of the AT2G38460 gene that potentially produces a truncated FERROPORTIN 1 protein. Finally, on Chromosome 4, TDDO4 fused the AT4G05475 gene to a transposable element (TE) (AT4G02960), leading to the potential expression of three new ORFs, including one that encodes a protein with two leucin rich repeats (LRR) (Fig. 4A,F). We then systematically analyzed the presence of these chimeric genes in the genome of the parental fas mutant lines and in 20rDNA L6 and L9, respectively (Fig. 4G). The TDDO1-derived chimeric gene can be detected in both L6 and L9, which confirm the appearance of TDDO1 after the cross between fas1-4 and fas2-4 (Figs. 2, 4G). The chimeric gene generated from TDDO3 was specifically detected in L6, whereas the chimeric gene generated by TDDO4 was detected in fas2-4, L6, and L9 plants, confirming the results obtained earlier (Fig. 2) but also suggesting that some generations of 20rDNA L9 inbreeds plants may still segregate TDDO4.
Figure 4.
TDDOs provoke chimeric genes formation. (A–C) Schematic representation of TDDO1, TDDO3, and TDDO4 that provoked TE and/or gene fusion in the 20rDNA L6F6. Genes or TEs present in the breaking junction and their orientation are shown. (D–F) Open reading frames (ORFs) potentially generated at the breaking points. TDDO1 provokes the fusion of the first exon of AT1G55325 that encodes for an ATPase motif of MEDIATOR 13 and the last four exons of AT1G54770 that contain an RNA processing domain (D). The chimeric gene created between AT2G37520 and AT2G38460 potentially encodes for a truncated FERROPORTIN protein (E). The breaking points at TDDO4 fuse the 5′ sequence of a TE (AT4G02960) with the second and last exon of the gene AT4G05475, which sequence encodes two Leucine Rich Repeats (LRR) (F). (G,H) PCR was performed with primers flanking the breaking junctions of TDDO1, TDDO3, and TDDO4 in the wild-type Col-0, the two mutants fas1-4 (G5) and fas2-4 (G5), and in 20rDNA lines L6 (generations F6 and F8) and L9 (generation F7). All PCR products were confirmed by Sanger sequencing. Genomic DNA (G) and cDNA (H) were used as templates. Amplicons from the locus encoding the elongation factor EF1ALPHA was used as a loading control.
We finally investigated whether these chimeric genes were transcribed. A first analysis of our RNA-seq data revealed that these genes were all able to accumulate transcripts. Using RT-qPCR, we confirmed the expression of the TDDO1- and TDDO3-derived chimeric genes, as well as the ability of the TDDO1-derived chimeric gene to be properly spliced (Supplemental Fig. S13). However, although reads could be detected in the RNA-seq data, we did not detect any signals for the TDDO4-derived chimeric gene by RT-PCR (Fig. 4H). In conclusion, our data show that TDDOs can promote the expression of chimeric genes.
Characterization and impact of duplication events on gene expression
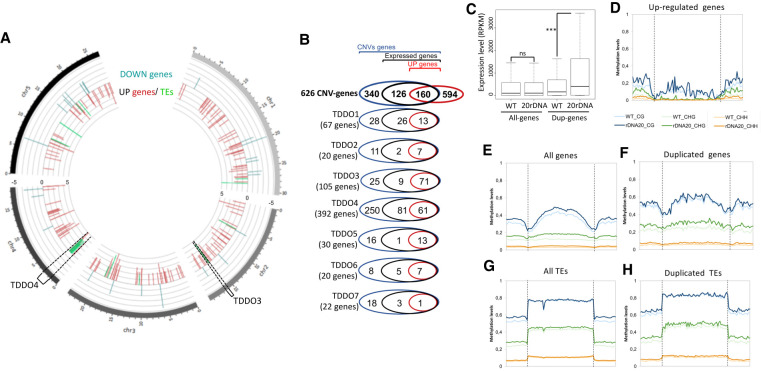
All TDDOs gained in 20rDNA L6F6 correspond to a gain of 2.31 Mb per haploid genome and induce CNVs of 626 genes and 851 transposable elements (TEs) (Supplemental Table S1). Changes in the 3D genome organization and CNVs can have an impact on chromatin marks and gene expression. We therefore analyzed the global gene expression pattern by poly(A)+ RNA-seq and the methylome by whole-genome bisulfite sequencing (WGBS) in wild-type Col-0 versus 20rDNA L6F6. We analyzed four replicates per samples by RNA-seq and identified differentially accumulating transcripts: 321 up-regulated genes and 14 up-regulated TEs, as well as 37 down-regulated genes but no down-regulated TEs in 20rDNA L6F6 compare to the wild-type Col-0 (with an adjusted P-value < 0.01 and log2[fold change] > 1.5 or <1.5) (Fig. 5A; Supplemental Table S1). We confirmed these results by quantitative RT-PCR (RT-qPCR) on nine randomly chosen genes and TEs (Supplemental Fig. S14). We did not find any correlation between differentially expressed genes and genes located in the newly arisen NADs of 20rDNA L6F6 (Supplemental Fig. S15A,C).
Figure 5.
Impact of TDDO on global gene expression and cytosine methylation in L6F6. (A) Representation of the chromosomal position of genes (red bars) and TEs (green bars) of differentially accumulated transcripts in the 20rDNA L6F6 versus WT Col-0 with an adjusted P-value < 0.01 and a log2(fold change) > 2. Data are displayed using Circos (Krzywinski et al. 2009). The brackets display the position of TDDO3 and TDDO4. (B) Venn diagrams representing the proportion of expressed genes (containing at least two reads/genes in wild-type) and up-regulated genes (P-value < 0.01, FC > 2) among all the duplicated genes or in each TDDO in L6F6. (C) Dot plot revealing the relative expression of all genes or duplicated (DUP) genes in leaves of 3-wk-old plants in WT Col-0 or in 20rDNA L6F6. (***) P-value = 0.0005 was calculated using a Wilcoxon test. (D–H) Global DNA methylation analyses from genome-wide bisulfite sequencing experiments in WT Col-0 versus the 20rDNA L6F6. Global CG, CHG, and CHH methylation are shown for up-regulated genes with an adjusted P-value < 0.01 and a log2(fold change) > 1.5 (D), all genes (E), duplicated genes (F), all TEs (G), and duplicated TEs (H).
However, we found that duplicated genes and TEs were significantly more expressed (Supplemental Fig. S15D,E). Of the up-regulated TEs, 57% (8) are also duplicated. If we consider the 321 up-regulated genes with a log2 fold change enrichment of 1.5, we found that 22% of these genes (71) belonged to duplicated genes, but the TDDOs only represent 2% of the genome. Conversely, no genes present in TDDO are down-regulated. Higher expression can only be observed from initially expressed genes in wild-type plants. Only 286 duplicated genes are actually expressed, and 160 of them are at least twice more expressed in 20rDNA L6F6 than in wild-type Col-0 (Fig. 5B). Depending on their genomic location, TDDOs perform differently. For instance, most of the TDDO3-derived genes produced at least twice as many transcripts in 20rDNA L6F6 (71 up-regulated genes of the 80 expressed genes), whereas genes present in TDDO4, enriched in genomic regions with heterochromatic features, were less up-regulated (61 fold change > 2 genes of the 142 expressed genes) (Fig. 5B). Finally, box-plot analyses of all genes versus the duplicated genes indeed revealed their overall ability to overaccumulate more transcripts in 20rDNA L6F6 (Fig. 5C). Thus, our data strongly suggest that gene duplication often leads to an increased expression, often higher than the twofold change expected in the hypothesis of additive expression.
To analyze the impact of CNVs at the DNA methylation level, we performed triplicate WGBS in wild-type Col-0 versus 20rDNA L6F6 lines. At the genome-scale, we observed a modest increase in CG, CHG, and CHH methylation in 20rDNA L6F6 at genes (Fig. 5E,F; Supplemental Fig. S16). However, methylation at TEs was affected in both CHG and CHH contexts, but not in the CG context (Fig. 5G,H; Supplemental Fig. S17). This observation is also true if we only analyze duplicated or up-regulated genes, with the exception of gene body methylation that is unaffected for up-regulated genes (Fig. 5D; Supplemental Fig. S16). Finally, differentially methylated regions (DMRs) identified in 20rDNA L6F6 compared to wild-type Col-0 did not show a potential overlap between up-regulated genes and hypomethylated regions.
Duplication events are linked to higher pathogen resistance
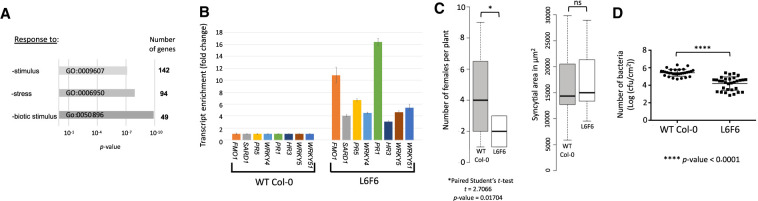
In the pool of up-regulated genes in 20rDNA L6F6, genes implicated in biotic and stress responses are particularly enriched (Fig. 6A). We performed RT-qPCR experiments and confirmed the overexpression of key genes involved in the plant–pathogen response (Fig. 6B). Among these genes are PATHOGENESIS-RELATED GENE 1 (PR1) and PATHOGENESIS-RELATED GENE 5 (PR5), whose higher expression levels are usually correlated with increased resistance against bacteria and nematodes (Wubben et al. 2008). Some of these genes were found in TDDO3 and TDDO4. Their higher expression rate could therefore be a consequence of the duplication events (Supplemental Fig. S17). Among them, ASYMETRIC LEAVES 1 (AS1), present in TDDO3, has an evolutionarily conserved role in plant–pathogen interactions (Yang et al. 2008). AS1 indeed acts as a positive regulator of extracellular defenses against bacterial pathogens in a salicylic acid-independent manner (Nurmberg et al. 2007). In addition, genes encoding four cysteine-rich receptor-like kinases (CRKs), located in TDDO4, also overaccumulate transcripts in the L6F6 (Supplemental Fig. S18). Among these genes is CRK36, whose overexpression is sufficient to enhance pattern-triggered immunity response and bacterial pathogen resistance (Yeh et al. 2015).
Figure 6.
Biotic stress genes are overexpressed in L6F6, which is more resistant to nematode and bacterial pathogens. (A) GO term enriched in the pool of up-regulated genes identified in 20rDNA L6F6. (B) Histogram displaying the relative transcript enrichment for eight genes implicated in the biotic stress response using quantitative RT-PCR in WT Col-0 versus 20rDNA L6F6. (C) Wild-type (WT) Col-0 and 20rDNA L6F7 were inoculated with the sugar beet cyst nematode (Heterodera schachtii). Four weeks after inoculation, the number of adult females per plant was determined. Data are the average number of adult females ± SE (n = 35 × 3). Data from the three independent experiments were pooled and are shown (left). The relative size of the syncytium cells was measured between both lines but no significant changes were noticed (right). (D) Wild-type (WT) Col-0 and 20rDNA L6F7 were inoculated with Pseudomonas strain DC3000 at 5 × 107 cfu/mL. Relative bacterial growth was determined 3 d after infection and is shown on the plot.
A. thaliana is susceptible to various pathogens, from prokaryotes to multicellular organisms. To test their resistance capabilities, we first infected both the wild-type Col-0 and 20rDNA L6F6 with the sugar beet cyst nematode Heterodera schachtii (Fig. 6C). We observed that only half the number of females was able to develop on 20rDNA L6F6 plants in comparison with wild-type Col-0 plants. However, we did not observe a change in the syncytium feeding site size, that is, the plant feeding structure induced by these nematodes (Fig. 6C). Secondly, we tested the ability of 20rDNA L6F6 to be infected by the virulent bacteria Pseudomonas syringae strain DC3000. Three days after inoculation, bacterial growth was significantly lower in 20rDNA L6F6 (Fig. 6D) than in wild-type Col-0. Single-nucleotide polymorphisms (SNPs) could also explain changes in plant–pathogen responses, but our analyses revealed that among the 196 SNPs found in genes in L6F6 compared to wild-type Col-0, none correspond to genes implicated in biotic stress response (Supplemental Fig. S19). Considering that fas2-4 mutant is hyper-resistant to P. syringae (Mozgová et al. 2015), and we identified TDDO4 in some lines of this mutant, one hypothesis is that the overexpression of pathogen response genes present in TDDO4 rather than FAS2 gene mutation is directly implicated in the resistance against P. syringae. However, we cannot exclude that fas2-4 and L6F6 pathogen resistance is mediated independently of TDDO4, which could also explain why very little overlap can be observed among the up-regulated genes in both lines (Supplemental Fig. S20).
In conclusion, we showed that higher accumulation of transcripts from genes implicated in the plant–pathogen response correlate with the plant's ability to resist against at least two types of distinct pathogens.
Discussion
Genomic structural variations shape animal and plant genomes (Krasileva 2019). Within a period of several millions of years, numerous rearrangements have occurred to shape the Arabidopsis thaliana genome, including duplications, translocations, inversions, and deletions (Blanc et al. 2000; Henry et al. 2006). Recently, genome analysis of seven accessions of A. thaliana revealed that they contain, on average, 15 Mb of rearranged sequences, generating CNVs for thousands of genes (Jiao and Schneeberger 2020). In this case, deletions, gain, or loss of copies are considered as important sources of CNVs and have potentially occurred in tens of thousands of years of evolution (Fulgione and Hancock 2018). CNVs occurring in the context of tandem duplication events represent between 3 and 4 Mb of genomic sequences in each of the seven accessions sequenced (Jiao and Schneeberger 2020). In our case, only a few generations were necessary to gain up to several megabases of genomic sequences by tandem duplications.
The rapid occurrence of these rearrangements is particularly intriguing. The relative sensitivity to genotoxic stress and the detection of a higher rate of spontaneous DSB in our 20rDNA lines is certainly one source of their appearance (Fig. 1D; Supplemental Fig. S2), but the precise mechanisms remain to be determined. One possibility is the implication of nonallelic homologous recombination (NAHR), usually responsible for TDDO (Zhang et al. 2013; Krasileva 2019). This mechanism can generate segmental duplications or deletions. In the 20rDNA L6F6, we detected duplications but no deletions, probably because of their deleterious effects.
Two other particular aspects of the detected TDDOs are their large sizes and locations, ranging from 57 kb to 1.44 Mb (Fig. 2; Supplemental Fig. S7). The TDDO borders do not share any genetic feature, and breakpoint junctions are not enriched in repetitive elements or particular genes. However, our Hi-C data revealed that sign changes in the eigenvector seem to be overrepresented at breaking junctions, suggesting a potential link between the 3D genome folding and the occurrence of TDDOs. The systematic identification and characterization of additional TDDOs would be necessary to strengthen this hypothesis.
It is also intriguing that more than half of the 15 TDDOs are located on NOR-bearing chromosomes (Fig. 2; Supplemental Fig. S7). Because of their tandemly repeated nature, NORs are indeed subjected to an inherent instability. Therefore, the existence of a sensing system monitoring their abundance has been proposed (Nelson et al. 2019), potentially via unequal sister chromatid exchange (Tartof 1974a,b). The 20rDNA lines derive from the cross between fas1 and fas2 mutants, whose mutations provoked a gradual loss of rRNA genes copies (Mozgová et al. 2010). Importantly, L6 and L9 are the only siblings in which the number of rRNA genes remained stable at a low level, whereas all other lineages quickly acquired rRNA genes (Pavlištová et al. 2016). However, our data actually show that rRNA gene copies are increasing progressively throughout the inbreeding of 20rDNA L6F6 (Supplemental Fig. S6), suggesting that the CNVs are found not only at the level of the TDDOs, but also at the level of the NORs. It remains to be elucidated whether a link between the rRNA gene gains and the appearance of TDDO exists and if the same mechanisms are involved. Nevertheless, a loss of rRNA gene copies also associates with genomic instability and hypersensitivity to DNA damage in cancer cells (Wang and Lemos 2017; Xu et al. 2017). Moreover, DNA damage sensitivity and rDNA replication defects also occur in budding yeast low rDNA copy strains (Ide et al. 2010).
Short-term consequences of gene duplications have been studied in animals, especially in cancer cells, where multiple de novo tandem duplication events induce gene CNVs (Quigley et al. 2018; Wee et al. 2018). The 20rDNA L6F6 line is an unprecedented opportunity to study the transcriptional behavior of newly duplicated genes. Globally, duplicated genes tend to be more expressed (Fig. 5C). Previous observations suggest that the expression of tandem genes recently duplicated is often greater than twofold (Loehlin and Carroll 2016). Although we cannot exclude that the detected transcripts come from only one of the duplicated genes, it is more likely that equivalent additive expression occurs for the duplicated genes. During evolution, duplicated gene expression can quickly lead to specialized expression patterns, often in a tissue-specific manner, although a significant number retain correlated transcriptional profiles (Blanc and Wolfe 2004; Guschanski et al. 2017). In our case, we were able to correlate this change in gene expression with the acquisition of increased resistance to different pathogens (Fig. 6). Analyzing gene expression in the future generation will allow us to evaluate if rapid transcriptional regulation occurs.
Plant genomes are rapidly evolving and their capacity to adapt to environmental changes is crucial. Like genome hybridization and TE mobilization, CNV is one important tool of genome evolution (Kondrashov 2012; Gabur et al. 2019; Quadrana et al. 2019). Together with previous observation, our data show the importance of systematically detecting CNVs. CNVs can indeed associate with adaptive traits (Kondrashov 2012; Gabur et al. 2019; Alonge et al. 2020). In our case, we found a potential link between CNV and pathogen resistance (Fig. 6). CNVs were already shown to be implicated in nematode resistance in soybean (Cook et al. 2012), but also in potato cultivar genome heterogeneity (Pham et al. 2017). We showed that the CNVs in the 20rDNA lines occurred only in a few generations in controlled growing conditions. This last point is particularly interesting in the context of plant breeding. In addition, TDDOs have the potential to create chimeric genes (Fig. 4). TDDO events can promote cancer cell formation, via the activation of oncogenes (Quigley et al. 2018). In that case, breaking junctions can affect the expression of an oncogene by modifying its regulation by enhancers, for example. In our study, the chimeric genes created are expressed and properly spliced. Although we do not have evidence concerning their potential ability to be translated or if the resultant protein would be functional, it is tempting to speculate that TDDO-mediated chimeric genes can lead to gene novelty as previously described (Chen et al. 2013). Studying the consequences of TDDOs in future generations will certainly shed light on their potential impact on genome evolution and plant adaptation.
Methods
Plant materials
Seeds corresponding to the fas1-4 (SAIL_662_D10) and fas2-4 (SALK_033228) were previously reported (Exner et al. 2006). All 20rDNA seeds that include fas1-4 and fas2-4 parental lines, as well as L6 and L9 lines used in this study correspond to stock previously reported (Pavlištová et al. 2016). For NADs identification, wild-type Col-0 expressing the FIB2:YFP fusion protein was described in Pontvianne et al. (2013). The 20rDNA L6F5 line was transformed by agroinfiltration to insert a transgene expressing FIB2:YFP fusion protein as described previously (Pontvianne et al. 2013).
Nanopore sequencing and data analyses
Genomic DNA preparation was performed as previously described (Debladis et al. 2017). After Qubit dosage (dsDNA High Sensitivity, Thermo Fisher Scientific), a second step of DNA purification was performed with the Genomic DNA Clean and Concentrator kit (Zymo Research) and precipitated. A last Qubit dosage was performed before library preparation using the 1D Genomic DNA by ligation kit SQK-LSK109 (Oxford Nanopore Technologies), following the manufacturer's instructions. The R9.5 ONT flow-cell FLO-MIN106D (Oxford Nanopore Technologies) was used. We obtained 6.4 Gb of sequences for L6F6, 0.7 Gb for L9F6, 5.9 Gb for fas1-4, and 11.4 Gb for fas2-4.
ONT reads were mapped on the TAIR10 reference genome using minimap2 with -a -Q -map-ont options (Li 2018). The alignment files were converted into BED files using BEDTools, and the coverage per 100-kb window was calculated using coverageBED (Quinlan and Hall 2010). For each 100-kb window, the ratio r = 20%rDNA coverage/wild-type Col-0 coverage was calculated. The mean (m) and standard error (SE) were calculated across the entire genome. Differentially covered regions in the 20%rDNA line were defined as regions for which r ≥ m+2SE or r ≤ m−2SE.
Additional methods can be found in the Supplemental Material.
Data access
All raw and processed sequencing data generated in this study have been submitted to the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena/browser/home) under accession number PRJEB35832. Code used to produce Hi-C figures is available as Supplemental Code.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank Rémy Merret and Michèle Laudié for Illumina sequencing and laboratory members for fruitful discussions. We also thank the flow cytometry facility, the microscopic facility, and the sequencing facility of Perpignan University Via Domitia Bioenvironnement (Perpignan, France). This work and A.P.-P.’s PhD fellowship are supported by the Agence Nationale de la Recherche (ANR), JCJC NucleoReg (ANR-15-CE12-0013-01) to F.P. F.P. was supported by the French Laboratory of Excellence project TULIP (ANR-10-LABX-41 and ANR-11-IDEX-0002-02). Work conducted at Iowa State University was supported by Hatch Act and State of Iowa funds. M.M. is a member of the European Training Network “EpiDiverse” that receives funding from the European Union Horizon 2020 program under Marie Skłodowska-Curie grant agreement No. 764965. M.D. and M.F. were supported by the Czech Science Foundation project 19-11880Y; by Ministry of Education, Youth and Sports of the Czech Republic INTER-COST (LTC18048); and by European Regional Development Fund, Project “SINGING PLANT” (CZ.02.1.01/0.0/0.0/16_026/0008446). A.P.-P., S.G., N.P., M.F., M.D., and F.P. are part of the European Cooperation in Science and Technology COST ACTION CA16212 INDEPTH.
Author contributions: T.J.B., V.P., and T.R.M. performed nematode infection assays. L.N. and T.H. performed Pseudomonas DC3000 infection assays. M.F. and M.D. performed DNA-FISH analyses. S.G. performed the Hi-C analyses. F.P. and A.P.-P. conceived and designed all other experiments. A.P.-P., S.G., C.L., E.J., J.D., and F.P. collected the data. P.Z. and M.M. contributed data or analysis tools. A.P.-P., N.P., M.M., and F.P. performed the analysis. F.P. wrote the paper. A.P.-P., S.G., and M.M. edited the paper. F.P. acquired main funding.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.261586.120.
Freely available online through the Genome Research Open Access option.
References
- Alonge M, Wang X, Benoit M, Soyk S, Pereira L, Zhang L, Suresh H, Ramakrishnan S, Maumus F, Ciren D, et al. 2020. Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell 182: 145–161.e23. 10.1016/j.cell.2020.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersaglieri C, Santoro R. 2019. Genome organization in and around the nucleolus. Cells 8: 579 10.3390/cells8060579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. 2004. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691. 10.1105/tpc.021410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Barakat A, Guyot R, Cooke R, Delseny M. 2000. Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12: 1093–1101. 10.1105/tpc.12.7.1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpentier MC, Picart-Picolo A, Pontvianne F. 2018. A method to identify nucleolus-associated chromatin domains (NADs). Methods Mol Biol 1675: 99–109. 10.1007/978-1-4939-7318-7_7 [DOI] [PubMed] [Google Scholar]
- Chandrasekhara C, Mohannath G, Blevins T, Pontvianne F, Pikaard CS. 2016. Chromosome-specific NOR inactivation explains selective rRNA gene silencing and dosage control in Arabidopsis. Genes Dev 30: 177–190. 10.1101/gad.273755.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonnel C, Gallego ME, White CI. 2010. Xrcc1-dependent and Ku-dependent DNA double-strand break repair kinetics in Arabidopsis plants. Plant J Cell Mol Biol 64: 280–290. 10.1111/j.1365-313X.2010.04331.x [DOI] [PubMed] [Google Scholar]
- Chen S, Krinsky BH, Long M. 2013. New genes as drivers of phenotypic evolution. Nat Rev Genet 14: 645–660. 10.1038/nrg3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DE, Lee TG, Guo X, Melito S, Wang K, Bayless AM, Wang J, Hughes TJ, Willis DK, Clemente TE, et al. 2012. Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338: 1206–1209. 10.1126/science.1228746 [DOI] [PubMed] [Google Scholar]
- Copenhaver GP, Pikaard CS. 1996. RFLP and physical mapping with an rDNA-specific endonuclease reveals that nucleolus organizer regions of Arabidopsis thaliana adjoin the telomeres on chromosomes 2 and 4. Plant J 9: 259–272. 10.1046/j.1365-313X.1996.09020259.x [DOI] [PubMed] [Google Scholar]
- Debladis E, Llauro C, Carpentier M-C, Mirouze M, Panaud O. 2017. Detection of active transposable elements in Arabidopsis thaliana using Oxford Nanopore Sequencing technology. BMC Genomics 18: 537 10.1186/s12864-017-3753-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopman EB, Hartl DL. 2007. A portrait of copy-number polymorphism in Drosophila melanogaster. Proc Natl Acad Sci 104: 19920–19925. 10.1073/pnas.0709888104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner V, Taranto P, Schonrock N, Gruissem W, Hennig L. 2006. Chromatin assembly factor CAF-1 is required for cellular differentiation during plant development. Dev Camb Engl 133: 4163–4172. 10.1242/dev.02599 [DOI] [PubMed] [Google Scholar]
- Fulgione A, Hancock AM. 2018. Archaic lineages broaden our view on the history of Arabidopsis thaliana. New Phytol 219: 1194–1198. 10.1111/nph.15244 [DOI] [PubMed] [Google Scholar]
- Gabur I, Chawla HS, Snowdon RJ, Parkin IAP. 2019. Connecting genome structural variation with complex traits in crop plants. Theor Appl Genet 132: 733–750. 10.1007/s00122-018-3233-0 [DOI] [PubMed] [Google Scholar]
- Gibbons JG, Branco AT, Godinho SA, Yu S, Lemos B. 2015. Concerted copy number variation balances ribosomal DNA dosage in human and mouse genomes. Proc Natl Acad Sci 112: 2485–2490. 10.1073/pnas.1416878112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob S, Schmid MW, Grossniklaus U. 2014. Hi-C analysis in Arabidopsis identifies the KNOT, a structure with similarities to the flamenco locus of Drosophila. Mol Cell 55: 678–693. 10.1016/j.molcel.2014.07.009 [DOI] [PubMed] [Google Scholar]
- Grummt I, Längst G. 2013. Epigenetic control of RNA polymerase I transcription in mammalian cells. Biochim Biophys Acta 1829: 393–404. 10.1016/j.bbagrm.2012.10.004 [DOI] [PubMed] [Google Scholar]
- Guschanski K, Warnefors M, Kaessmann H. 2017. The evolution of duplicate gene expression in mammalian organs. Genome Res 27: 1461–1474. 10.1101/gr.215566.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry Y, Bedhomme M, Blanc G. 2006. History, protohistory and prehistory of the Arabidopsis thaliana chromosome complement. Trends Plant Sci 11: 267–273. 10.1016/j.tplants.2006.04.002 [DOI] [PubMed] [Google Scholar]
- Himmelbach A, Ruban A, Walde I, Šimková H, Doležel J, Hastie A, Stein N, Mascher M. 2018. Discovery of multi-megabase polymorphic inversions by chromosome conformation capture sequencing in large-genome plant species. Plant J Cell Mol Biol 96: 1309–1316. 10.1111/tpj.14109 [DOI] [PubMed] [Google Scholar]
- Ide S, Miyazaki T, Maki H, Kobayashi T. 2010. Abundance of ribosomal RNA gene copies maintains genome integrity. Science 327: 693–696. 10.1126/science.1179044 [DOI] [PubMed] [Google Scholar]
- Jiao WB, Schneeberger K. 2020. Chromosome-level assemblies of multiple Arabidopsis genomes reveal hotspots of rearrangements with altered evolutionary dynamics. Nat Commun 11: 989 10.1038/s41467-020-14779-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T. 2011. Regulation of ribosomal RNA gene copy number and its role in modulating genome integrity and evolutionary adaptability in yeast. Cell Mol Life Sci 68: 1395–1403. 10.1007/s00018-010-0613-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA. 2012. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc Biol Sci 279: 5048–5057. 10.1098/rspb.2012.1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva KV. 2019. The role of transposable elements and DNA damage repair mechanisms in gene duplications and gene fusions in plant genomes. Curr Opin Plant Biol 48: 18–25. 10.1016/j.pbi.2019.01.004 [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: An information aesthetic for comparative genomics. Genome Res 19: 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34: 3094–3100. 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loehlin DW, Carroll SB. 2016. Expression of tandem gene duplicates is often greater than twofold. Proc Natl Acad Sci 113: 5988–5992. 10.1073/pnas.1605886113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q, Rabanal FA, Meng D, Huber CD, Farlow A, Platzer A, Zhang Q, Vilhjalmsson BJ, Korte A, Nizhynska V, et al. 2013. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet 45: 884–890. 10.1038/ng.2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozgová I, Mokroš P, Fajkus J. 2010. Dysfunction of Chromatin Assembly Factor 1 induces shortening of telomeres and loss of 45S rDNA in Arabidopsis thaliana. Plant Cell 22: 2768–2780. 10.1105/tpc.110.076182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozgová I, Wildhaber T, Liu Q, Abou-Mansour E, L'Haridon F, Métraux JP, Gruissem W, Hofius D, Hennig L. 2015. Chromatin assembly factor CAF-1 represses priming of plant defence response genes. Nat Plants 1: 15127 10.1038/nplants.2015.127 [DOI] [PubMed] [Google Scholar]
- Nelson JO, Watase GJ, Warsinger-Pepe N, Yamashita YM. 2019. Mechanisms of rDNA copy number maintenance. Trends Genet 35: 734–742. 10.1016/j.tig.2019.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Németh A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Peterfia B, Solovei I, Cremer T, Dopazo J, Langst G. 2010. Initial genomics of the human nucleolus. PLoS Genet 6: e1000889 10.1371/journal.pgen.1000889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman S, Hermetz KE, Weckselblatt B, Rudd MK. 2015. Next-generation sequencing of duplication CNVs reveals that most are tandem and some create fusion genes at breakpoints. Am J Hum Genet 96: 208–220. 10.1016/j.ajhg.2014.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurmberg PL, Knox KA, Yun BW, Morris PC, Shafiei R, Hudson A, Loake GJ. 2007. The developmental selector AS1 is an evolutionarily conserved regulator of the plant immune response. Proc Natl Acad Sci 104: 18795–18800. 10.1073/pnas.0705586104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes S, Maggert KA. 2009. Ribosomal DNA contributes to global chromatin regulation. Proc Natl Acad Sci 106: 17829–17834. 10.1073/pnas.0906811106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes S, Branco AT, Hartl DL, Maggert KA, Lemos B. 2011. Ribosomal DNA deletions modulate genome-wide gene expression: “rDNA-sensitive” genes and natural variation. PLoS Genet 7: e1001376 10.1371/journal.pgen.1001376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlištová V, Dvořáčková M, Jež M, Mozgová I, Mokroš P, Fajkus J. 2016. Phenotypic reversion in fas mutants of Arabidopsis thaliana by reintroduction of FAS genes: variable recovery of telomeres with major spatial rearrangements and transcriptional reprogramming of 45S rDNA genes. Plant J Cell Mol Biol 88: 411–424. 10.1111/tpj.13257 [DOI] [PubMed] [Google Scholar]
- Pham GM, Newton L, Wiegert-Rininger K, Vaillancourt B, Douches DS, Buell CR. 2017. Extensive genome heterogeneity leads to preferential allele expression and copy number-dependent expression in cultivated potato. Plant J Cell Mol Biol 92: 624–637. 10.1111/tpj.13706 [DOI] [PubMed] [Google Scholar]
- Picart-Picolo A, Picault N, Pontvianne F. 2019. Ribosomal RNA genes shape chromatin domains associating with the nucleolus. Nucleus 10: 67–72. 10.1080/19491034.2019.1591106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picart-Picolo A, Picart C, Picault N, Pontvianne F. 2020. Nucleolus-associated chromatin domains are maintained under heat stress, despite nucleolar reorganization in Arabidopsis thaliana. J Plant Res 133: 463–470. 10.1007/s10265-020-01201-3 [DOI] [PubMed] [Google Scholar]
- Pontvianne F, Grob S. 2020. Three-dimensional nuclear organization in Arabidopsis thaliana. J Plant Res 133: 479–488. 10.1007/s10265-020-01185-0 [DOI] [PubMed] [Google Scholar]
- Pontvianne F, Liu C. 2020. Chromatin domains in space and their functional implications. Curr Opin Plant Biol 54: 1–10. 10.1016/j.pbi.2019.11.005 [DOI] [PubMed] [Google Scholar]
- Pontvianne F, Abou-Ellail M, Douet J, Comella P, Matia I, Chandrasekhara C, Debures A, Blevins T, Cooke R, Medina FJ, et al. 2010. Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet 6: e1001225 10.1371/journal.pgen.1001225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Blevins T, Chandrasekhara C, Feng W, Stroud H, Jacobsen SE, Michaels SD, Pikaard CS. 2012. Histone methyltransferases regulating rRNA gene dose and dosage control in Arabidopsis. Genes Dev 26: 945–957. 10.1101/gad.182865.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Blevins T, Chandrasekhara C, Mozgova I, Hassel C, Pontes OMF, Tucker S, Mokros P, Muchova V, Fajkus J, et al. 2013. Subnuclear partitioning of rRNA genes between the nucleolus and nucleoplasm reflects alternative epiallelic states. Genes Dev 27: 1545–1550. 10.1101/gad.221648.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Boyer-Clavel M, Sáez-Vásquez J. 2016a. Fluorescence-activated nucleolus sorting in Arabidopsis. Methods Mol Biol 1455: 203–211. 10.1007/978-1-4939-3792-9_15 [DOI] [PubMed] [Google Scholar]
- Pontvianne F, Carpentier MC, Durut N, Pavlištová V, Jaške K, Schořová S, Parrinello H, Rohmer M, Pikaard CS, Fojtová M, et al. 2016b. Identification of nucleolus-associated chromatin domains reveals a role for the nucleolus in 3D organization of the A. thaliana genome. Cell Rep 16: 1574–1587. 10.1016/j.celrep.2016.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrana L, Etcheverry M, Gilly A, Caillieux E, Madoui M-A, Guy J, Bortolini Silveira A, Engelen S, Baillet V, Wincker P, et al. 2019. Transposition favors the generation of large effect mutations that may facilitate rapid adaption. Nat Commun 10: 3421 10.1038/s41467-019-11385-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley DA, Dang HX, Zhao SG, Lloyd P, Aggarwal R, Alumkal JJ, Foye A, Kothari V, Perry MD, Bailey AM, et al. 2018. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell 174: 758–769.e9. 10.1016/j.cell.2018.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinodoz SA, Ollikainen N, Tabak B, Palla A, Schmidt JM, Detmar E, Lai MM, Shishkin AA, Bhat P, Takei Y, et al. 2018. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 174: 744–757.e24. 10.1016/j.cell.2018.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabanal FA, Nizhynska V, Mandáková T, Novikova PY, Lysak MA, Mott R, Nordborg M. 2017. Unstable inheritance of 45S rRNA genes in Arabidopsis thaliana. G3 (Bethesda) 7: 1201–1209. 10.1534/g3.117.040204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Parra E, Gutierrez C. 2007. The many faces of chromatin assembly factor 1. Trends Plant Sci 12: 570–576. 10.1016/j.tplants.2007.10.002 [DOI] [PubMed] [Google Scholar]
- Santos AP, Gaudin V, Mozgová I, Pontvianne F, Schubert D, Tek AL, Dvořáčková M, Liu C, Fransz P, Rosa S, et al. 2020. Tidying-up the plant nuclear space: domains, function and dynamics. J Exp Bot 71: 5160−5179. 10.1093/jxb/eraa282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi Y, Horiuchi T, Kobayashi T. 2003. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev 17: 1497–1506. 10.1101/gad.1085403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartof KD. 1974a. Unequal mitotic sister chromatid exchange and disproportionate replication as mechanisms regulating ribosomal RNA gene redundancy. Cold Spring Harb Symp Quant Biol 38: 491–500. 10.1101/SQB.1974.038.01.053 [DOI] [PubMed] [Google Scholar]
- Tartof KD. 1974b. Unequal mitotic sister chromatin exchange as the mechanism of ribosomal RNA gene magnification. Proc Natl Acad Sci 71: 1272–1276. 10.1073/pnas.71.4.1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Koningsbruggen S, Gierliński M, Schofield P, Martin D, Barton GJ, Ariyurek Y, den Dunnen JT, Lamond AI. 2010. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell 21: 3735–3748. 10.1091/mbc.e10-06-0508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varas J, Santos JL, Pradillo M. 2017. The absence of the Arabidopsis chaperone complex CAF-1 produces mitotic chromosome abnormalities and changes in the expression profiles of genes involved in DNA repair. Front Plant Sci 8: 525 10.3389/fpls.2017.00525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Lemos B. 2017. Ribosomal DNA copy number amplification and loss in human cancers is linked to tumor genetic context, nucleolus activity, and proliferation. PLoS Genet 13: e1006994 10.1371/journal.pgen.1006994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee Y, Wang T, Liu Y, Li X, Zhao M. 2018. A pan-cancer study of copy number gain and up-regulation in human oncogenes. Life Sci 211: 206–214. 10.1016/j.lfs.2018.09.032 [DOI] [PubMed] [Google Scholar]
- Wubben MJE, Jin J, Baum TJ. 2008. Cyst nematode parasitism of Arabidopsis thaliana is inhibited by salicylic acid (SA) and elicits uncoupled SA-independent pathogenesis-related gene expression in roots. Mol Plant-Microbe Interact 21: 424–432. 10.1094/MPMI-21-4-0424 [DOI] [PubMed] [Google Scholar]
- Xu B, Li H, Perry JM, Singh VP, Unruh J, Yu Z, Zakari M, McDowell W, Li L, Gerton JL. 2017. Ribosomal DNA copy number loss and sequence variation in cancer. PLoS Genet 13: e1006771 10.1371/journal.pgen.1006771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Iwasaki M, Machida C, Machida Y, Zhou X, Chua NH. 2008. βC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev 22: 2564–2577. 10.1101/gad.1682208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh YH, Chang YH, Huang PY, Huang JB, Zimmerli L. 2015. Enhanced Arabidopsis pattern-triggered immunity by overexpression of cysteine-rich receptor-like kinases. Front Plant Sci 6: 322 10.3389/fpls.2015.00322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata L, Ding J, Willing EM, Hartwig B, Bezdan D, Jiao WB, Patel V, Velikkakam James G, Koornneef M, Ossowski S, et al. 2016. Chromosome-level assembly of Arabidopsis thaliana Ler reveals the extent of translocation and inversion polymorphisms. Proc Natl Acad Sci 113: E4052–E4060. 10.1073/pnas.1607532113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zuo T, Peterson T. 2013. Generation of tandem direct duplications by reversed-ends transposition of maize ac elements. PLoS Genet 9: e1003691 10.1371/journal.pgen.1003691 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.