Abstract
It is widely recognized that noncoding genetic variants play important roles in many human diseases, but there are multiple challenges that hinder the identification of functional disease-associated noncoding variants. The number of noncoding variants can be many times that of coding variants; many of them are not functional but in linkage disequilibrium with the functional ones; different variants can have epistatic effects; different variants can affect the same genes or pathways in different individuals; and some variants are related to each other not by affecting the same gene but by affecting the binding of the same upstream regulator. To overcome these difficulties, we propose a novel analysis framework that considers convergent impacts of different genetic variants on protein binding, which provides multiscale information about disease-associated perturbations of regulatory elements, genes, and pathways. Applying it to our whole-genome sequencing data of 918 short-segment Hirschsprung disease patients and matched controls, we identify various novel genes not detected by standard single-variant and region-based tests, functionally centering on neural crest migration and development. Our framework also identifies upstream regulators whose binding is influenced by the noncoding variants. Using human neural crest cells, we confirm cell stage–specific regulatory roles of three top novel regulatory elements on our list, respectively in the RET, RASGEF1A, and PIK3C2B loci. In the PIK3C2B regulatory element, we further show that a noncoding variant found only in the patients affects the binding of the gliogenesis regulator NFIA, with a corresponding up-regulation of multiple genes in the same topologically associating domain.
Hirschsprung (HSCR) disease is a rare, complex genetic disease characterized by missing enteric ganglia in various portions of the hindgut (Amiel et al. 2008). It is caused by failed migration, proliferation, differentiation, or colonization of enteric neural crest (NC) cells, which disrupts enteric nervous system (ENS) development (Badner et al. 1990; Chakravarti et al. 2006). Phenotypic severity of the disease is determined by the length of colonic aganglionosis and can be classified into short-segment HSCR (S-HSCR; 80% of cases), long-segment HSCR (L-HSCR; 15%–20% of cases), and total colonic aganglionosis (TCA; up to 5% of cases) (Udassin et al. 1981; Chakravarti et al. 2006).
HSCR is long recognized to be highly heritable (80%–97% for S-HSCR and ∼100% for L-HSCR), while around 80% of HSCR cases are sporadic (>95% for S-HSCR) (Badner et al. 1990; Tam et al. 2019). The incidence of HSCR varies across ethnic groups, from 1.4 to 2.8 in every 10,000 new births, with the highest incidence rate in Asia (Chakravarti et al. 2006; Amiel et al. 2008; Tam and Garcia-Barceló 2009). The genetic etiology of HSCR is multifactorial, involving rare and common, coding and noncoding variants in genes playing different roles in ENS development. Among the different subtypes of HSCR, S-HSCR is genetically most complex. Whereas L-HSCR and TCA cases are mostly autosomal dominant, the less severe S-HSCR subtype follows a complex, non-Mendelian inheritance pattern and has a male-to-female ratio of 4:1 (Badner et al. 1990; Emison et al. 2005), with the reason behind it not fully known.
Damaging rare variants at protein-coding sequences associated with HSCR have been found in many genes involved in ENS development. Among them, RET, encoding a receptor tyrosine kinase, is responsible for ∼50% of familial cases and 10%–20% of sporadic cases (Chakravarti et al. 2006). Other genes previously reported include BACE2, DNMT3B, ECE1, EDN3, EDNRB, FAT3, GDNF, GFRA1, KIFBP, L1CAM, NRG1, NRG3, NRTN, NTF3, NTRK3, PHOX2B, PROK1, PROKR1, PROKR2, PSPN, SEMA3A, SEMA3C, SEMA3D, SOX10, TCF4, and ZEB2 (Amiel et al. 1996; Garcia-Barcelo et al. 2009; Alves et al. 2013; Luzón-Toro et al. 2015; Tang et al. 2018; Tilghman et al. 2019), which together account for around 5% of all cases. These rare damaging variants often have incomplete penetrance and explain only a small portion of the phenotypic variance.
In contrast, the most well-known common variant, rs2435357, located in intron 1 of RET and affecting SOX10 binding to a RET enhancer, explains 10–20 times of the phenotypic variance as compared to the rare damaging RET variants (Emison et al. 2005, 2010; Chatterjee et al. 2016). Recently, some additional noncoding variants have also been found associated with HSCR. For example, two other variants in RET enhancers act synergistically with rs2435357 on HSCR risk (Chatterjee et al. 2016), while variants in four noncoding elements around the RET and SEMA3 loci were found in 48.4% of cases and 17.1% of controls, with the presence of five or more variants in these regions corresponding to a high disease risk (Tilghman et al. 2019). However, despite the ever-growing knowledge about HSCR, there is still a considerable portion of cases that cannot be explained by the cataloged coding or noncoding variants.
The emerging studies of HSCR-associated common variants in RET enhancers and other functional noncoding regions (Emison et al. 2010; Chatterjee et al. 2016; Tilghman et al. 2019) and the abundance of noncoding signals from genome-wide association studies of many human traits in general (Zhang and Lupski 2015) both suggest that noncoding regions would be a valuable territory to explore in the quest to explain the missing heritability in HSCR.
Existing methods for studying noncoding genetic variants can be broadly classified into site-based and region-based methods (Supplemental Materials). In general, both types of methods face several major challenges, namely (1) there is a large number of sites/regions to consider genome-wide, leading to low statistical power due to multiple hypothesis testing correction, (2) many genetic variants at noncoding regions do not have direct functional effects, such as those in linkage disequilibrium with the actual functional variants, (3) epistatic interactions can exist among genetic variants, making it unsuitable to consider each variant separately (Cheng et al. 2017), (4) genetic variants at different regulatory elements of the same gene/pathway may result in the same convergent effect, and therefore each of them may not have high recurrence in the study subjects, and (5) the loci of different genetic variants could be related to each other not by their genomic locations but by the upstream regulators that commonly bind them, which are more nontrivial to determine.
In this study, we propose a new noncoding variant prioritization framework, Multiscale Analysis of Regulatory Variants on the Epigenomic Landscape (MARVEL), that addresses these five challenges. Applying our framework to the whole-genome sequencing (WGS) data generated from an S-HSCR cohort (431 cases and 487 ethnically matched controls), we identify disease-associated enhancers, promoters, and genes based on epigenomic data generated from the human pluripotent stem cell (hPSC)-derived enteric NC-like cells (hNC). This is by far the largest WGS data set of S-HSCR with both cases and controls coming from a single population. By aggregating lower-level (genomic variants and TF binding motifs) information, marginally significant signals emerge as more significant associations at a higher level (enhancers, promoters, and genes) that converge to specific functional pathways and upstream regulators.
Results
A novel association framework for the noncoding genome
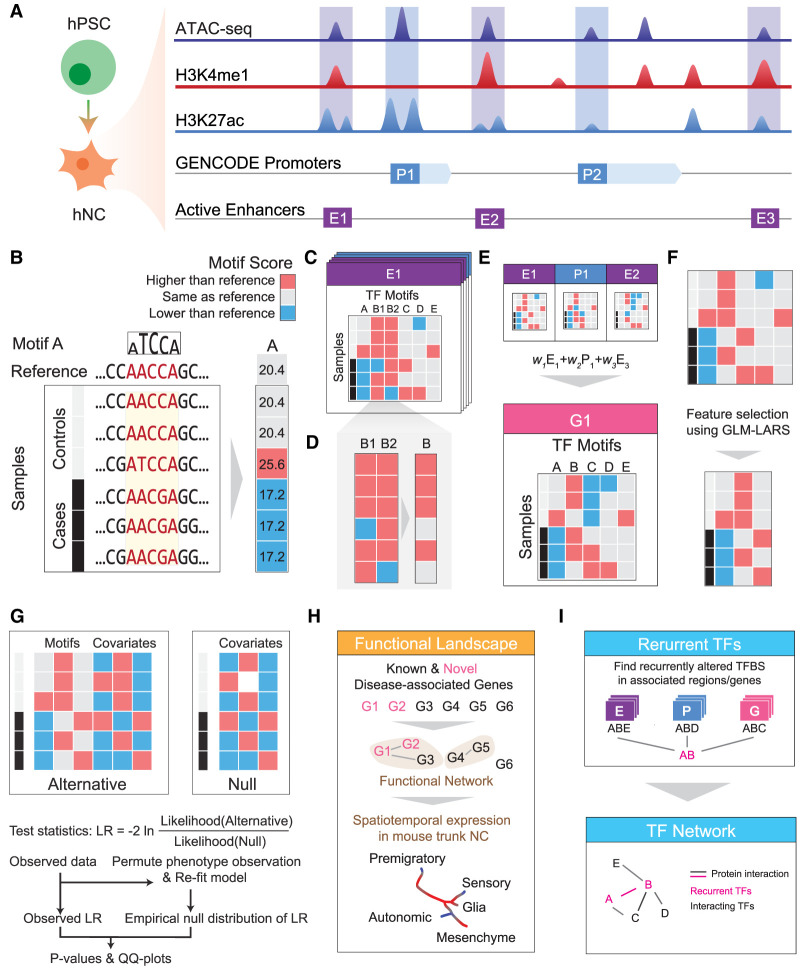
MARVEL is designed to address the five challenges listed above (Fig. 1; Methods; Supplemental Materials). First, we use cell type–specific epigenomic data to define a target set of active regulatory elements in the relevant cell types, in order to limit the number of statistical tests to be performed (Fig. 1A). Second, in these target elements, we use sequence motifs as a proxy to evaluate the functional potential of the genetic variants (Korhonen et al. 2017), which helps prioritize the genetic variants and their corresponding regulatory elements among those in linkage disequilibrium (Fig. 1B). Third, for each target element, we consider all genetic variants simultaneously to reconstruct the sample-specific sequence for evaluating their joint functional effects (Fig. 1C). For instance, if two genetic variants have opposite effects on the binding strength of a TF according to its sequence motif, the net expected effect would be smaller than the sum of their individual absolute effects. Fourth, our framework considers multiple occurrences of a motif in the same regulatory element (Fig. 1D) or different regulatory elements of the same gene (Fig. 1E) in a joint manner. As a result, a gene would be considered significantly perturbed when its regulatory elements have frequent motif-disrupting genetic variants even if in different subjects these regulatory elements contain different sets of genetic variants (Corradin et al. 2014; Chatterjee et al. 2016). The joint effect of multiple motifs is further captured by first selecting an important combination of the motifs using a feature selection procedure (Fig. 1F) followed by an association test (Fig. 1G), which together identify motifs whose match scores in these regulatory elements are significantly associated with the target phenotype, adjusted for covariates such as age and sex of the subjects. The identified phenotype-associated regulatory elements and genes are further investigated by additional analyses such as gene set enrichment and single-cell expression analyses (Fig. 1H). Fifth, by using sequence motifs as functional proxy, we also identify the potential upstream regulators whose binding to different target elements is frequently perturbed, which can be either loss or gain of binding. By identifying those whose binding motifs are commonly perturbed in the regulatory elements of different genes (Fig. 1I), we identify both the core regulators involved in the phenotype (Boyle et al. 2017) and genes potentially involved in the same pathways.
Figure 1.
Schematic overview of MARVEL. (A) Epigenomic data of relevant cell types (hNC in the case of HSCR) are integrated with a gene annotation set to identify the active regulatory elements relevant to the phenotype of interest. (B) In each regulatory element, the functional significance of genetic variants is evaluated by their perturbation to TF sequence motifs. (C) Since the perturbation effects of multiple genetic variants may not add up linearly, they are considered together to reconstruct the sample-specific sequences, based on which the overall change of TF motif match scores is determined. (D) For motifs with multiple appearances within the same regulatory element, their match scores are aggregated to give a single score. (E) At a higher level, if a gene involves multiple regulatory elements, the aggregated match scores of a motif in the different elements can be further aggregated into a single score. This is done in the gene-based analysis. (F,G) The aggregated match score matrix of all the motifs for a regulatory element/gene is used as the input of an association test, which selects a subset of the most informative motif features (F) and compares a model involving both these selected features and the covariates with a null model that involves only the covariates using likelihood ratio (LR) test (G). (H) The regulatory elements and genes identified to be significantly associated with the phenotype can be further studied by other downstream analyses, such as gene set enrichment and single-cell expression analyses. (I) TFs with recurrently perturbed match scores in different regulatory elements are collected to infer a network that highlights the phenotype-associated perturbations.
Using simulated data (Supplemental Materials), we verified that MARVEL is able to select the simulated informative motifs associated with the phenotype, including motifs whose match scores are affected by genetic variants at different allele frequencies, effect sizes, and correlations among each other (Supplemental Fig. S1). We also verified that, in the P-value and effect size distributions produced by MARVEL, the simulated disease-associated regions were clearly separated from the background (Supplemental Fig. S2).
Novel noncoding regulatory elements and gene loci associated with S-HSCR
We applied MARVEL to analyze our S-HSCR WGS data (Supplemental Materials). Briefly, target enhancers and promoters were defined using chromatin accessibility and histone modification data from hNC, which were, respectively, used in an enhancer-based and a promoter-based analysis. A gene-based analysis was also performed by considering at the same time all the promoters and enhancers that potentially regulate each gene, with the motif scores of the different regulatory elements weighted according to their expected chromatin contact frequencies with the transcription start site (TSS) in hPSCs (Supplemental Materials; Supplemental Table S4). This gene-based analysis allowed the detection of genes with frequent perturbations by multiple low-recurrence genetic variants at different regulatory elements. Association tests were then performed based on the motif scores in each of these three sets of target regions in turn, which contained 150,828 enhancers, 87,461 promoters, and 24,557 genes, respectively.
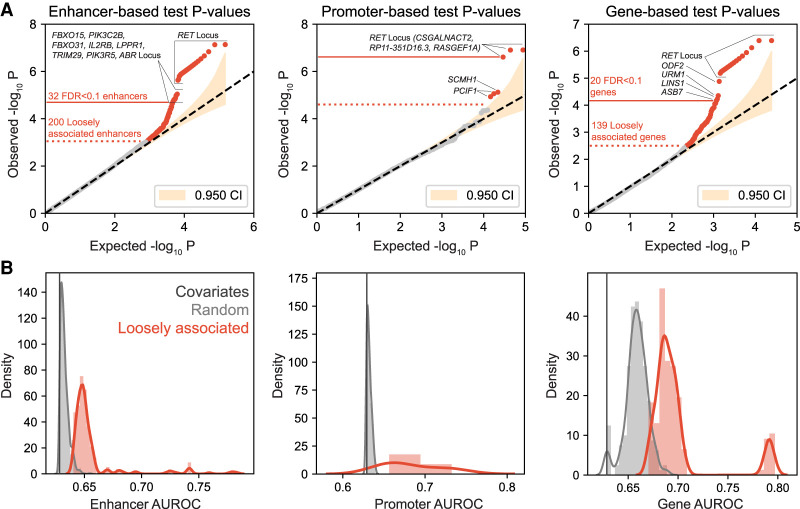
We defined all regions above the 95% confidence interval of the null in the quantile-quantile plot as loosely associated with S-HSCR, and all regions passing the FDR threshold of 0.1 (including a subset of the loosely associated regions) as significantly associated (Fig. 2A; Supplemental Table S1). Quantified by the area under the receiver operating characteristic (AUROC) (Methods), the loosely associated regions had generally larger effect sizes than background hNC enhancers, hNC promoters, and genes (Fig. 2B). Comparing the three sets of target regions, there were more enhancers and genes significantly associated with S-HSCR as compared to promoters. Noncoding variant association methods that focus on the promoter regions would have missed the many associated enhancers and aggregated regulatory regions of the genes. As a negative control, we repeated the procedures on a background set of enhancers that do not overlap the hNC enhancers (Methods) and found none of them either significantly or loosely associated with S-HSCR (Supplemental Fig. S3).
Figure 2.
Association test results. (A) Q-Q plots of association P-values in the enhancer-based, promoter-based, and gene-based tests. In each plot, the yellow shaded area shows the 95% confidence interval. The dotted red line marks the demarcation point of the loosely associated regions, above which all the regions are outside the 95% confidence interval. The solid red line marks the threshold for significantly associated regions, above which all regions have an FDR Q-value < 0.1. The significantly associated regions are a subset of the loosely associated regions. (B) Comparison of the AUROC value distributions of the models of the loosely associated regions with those of the models of the background regions and the covariate-only model.
As expected, all three analyses identified various regions near the RET locus as significantly associated with S-HSCR (Table 1; Supplemental Materials). Moreover, we also identified nine enhancers and four genes significantly associated with S-HSCR that are not close to RET (Table 1).
Table 1.
Significant S-HSCR-associated enhancers, promoters, and genes
Among the nine enhancers, the one on Chromosome 1 (Chr 1: 204,456,576–204,457,577) overlaps intron 10 of PIK3C2B. PIK3C2B is a phosphoinositide 3-kinase (PI3K) family protein, which is believed to have important roles in the signal integration and transduction in hNC (Frank and Tsai 2009). Binding sites of TWIST1 and NFIA are altered in this enhancer (Table 1), both with altered match scores in six cases but not in any controls. TWIST is an hNC specifier (Simoes-Costa and Bronner 2013) while NFIA has been shown to regulate gliogenesis, which is also crucial to a functional ENS (Kang et al. 2012; Glasgow et al. 2014; Tchieu et al. 2019). Another significantly associated enhancer, on Chromosome 17 (Chr 17: 8,915,658–8,916,659), is also close to two other PI3K family genes, PIK3R5 and PIK3R6. The two enhancers on Chromosomes 18 and 16 (Chr 18: 73,918,014–73,919,015 and Chr 16: 86,954,235–86,955,236) are within/close to F-box genes (FBXO15 and FBXO31, respectively). F-box proteins bind to CUL1 to form SCF (SKP-CUL1-F-box protein) E3 Ubiquitin Ligase complexes, which mediate ubiquitination of proteins that regulate the cell cycle and diverse neuronal activities (Liao et al. 2004; Frank and Tsai 2009). In particular, FBXO31 has been shown to regulate neural morphogenesis and migration (Vadhvani et al. 2013). Another enhancer on Chromosome 17 (Chr 17: 958,411–959,412) is close to ABR, which encodes a protein that is able to promote RHO activity and reduce CDC42 activity (Vaughan et al. 2011), both of which are important for actin regulation and cell polarization during migration (Vaughan et al. 2011; Cohen et al. 2018). The enhancer on Chromosome 9 (Chr 9: 100,937,755–100,938,756) is close to PLPPR1, which encodes a member of the plasticity-related gene (PRG) family. Members of the PRG family mediate lipid phosphate phosphatase activity in neurons and are known to be involved in neuronal plasticity (Savaskan et al. 2004; Trimbuch et al. 2009), potentially also involved in ENS development (Schäfer et al. 2009). The enhancer on Chromosome 11 (Chr 11: 120,020,704–120,021,705) is close to TRIM29, a gene that regulates epithelial-to-mesenchymal transition (EMT), which is an important process shared by both NC migration and cancer metastasis (Sun et al. 2019). Finally, the genes closest to the enhancer on Chromosome 22 (Chr 22: 37,128,477–37,129,478) are not clearly related to HSCR, which could represent new findings. In general, many of the top S-HSCR-associated enhancers identified by MARVEL cannot be identified by commonly used single-variant and region-based association (Supplemental Materials; Supplemental Fig. S4).
The four significantly associated genes not close to the RET locus come from two loci, respectively, on Chromosomes 9 and 15, with no loosely associated enhancers or promoters near them. Each of these genes was thus found to be significantly associated with S-HSCR due to weak association signals that distribute across multiple regulatory elements of it. Indeed, a significantly larger fraction of enhancers within 1 Mbp from the TSSs of these genes have P-values smaller than 0.1 as compared to the set of all enhancers (P = 0.025, Fisher's exact test). One of the genes in these loci is ODF2, which encodes the cenexin protein. Previous studies have demonstrated that, in various mammalian cell lines, cenexin controls centrosome positioning during directional cell migration (Hung et al. 2016). Since the centrosome acts as a steering machine for cell movement and stabilizes the migration direction (Ueda et al. 1997; Hung et al. 2016), ODF2 might help stabilize the direction of NC migration.
Loosely associated signals aggregate into functional pathways
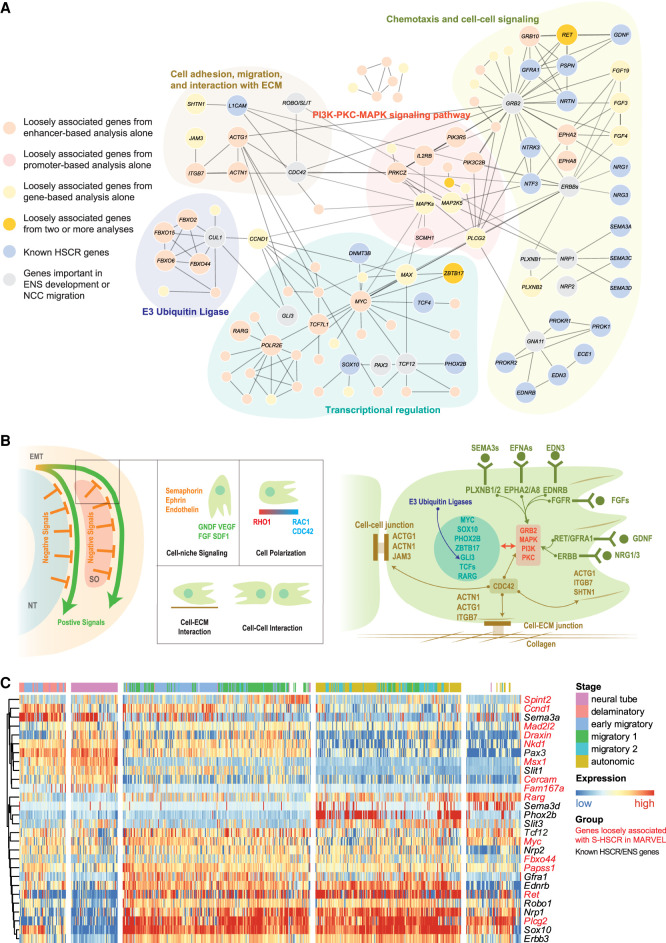
Having found that genetic variants weakly associated with S-HSCR at different regulatory elements aggregated to give stronger signals at the gene level, we next investigated whether association signals could be further aggregated into functional pathways. We performed this investigation by taking the 417 genes loosely associated with S-HSCR from our enhancer-based, promoter-based, and gene-based analyses and connected those with functional interactions in Reactome (Supplemental Materials; Wu and Haw 2017). A total of 69 genes were connected, by 94 functional interactions. This number of functional interactions was significantly larger than those of random gene sets of the same size (P = 0.032), suggesting that these S-HSCR genes were indeed functionally related.
To annotate the functions of these S-HSCR genes, we expanded this gene set by adding known HSCR genes and genes important in ENS development or NC migration (Methods) and found that the resulting genes formed five main clusters according to their functional interactions (Fig. 3A). These five clusters are, respectively, related to (1) chemotaxis and cell-cell signaling, (2) cell adhesion, migration, and interaction with extracellular matrix, (3) PI3K-PKC-MAPK signaling, (4) E3 Ubiquitin Ligase complexes, and (5) transcriptional regulatory factors. They are highly related to NC migration and its development (Fig. 3B), and many genes loosely associated with S-HSCR were involved in these functional clusters (Supplemental Table S2).
Figure 3.
Functional landscape of S-HSCR-associated genes. (A) Functional interactions among genes loosely associated with S-HSCR, known HSCR genes, and genes important in ENS functions or NC migration. Each node corresponds to a gene and each edge corresponds to a functional interaction cataloged in Reactome. Genes of particular interest are shown in bigger nodes, labeled with their names. (B) Schematic illustration of some biological processes and genes involved in NC migration (Szabó and Mayor 2018). Colors of gene names follow their functional categories in panel A. (NT) Neural tube, (SO) somites. (C) Spatiotemporal expression profiles of mouse trunk NCs. Each row corresponds to the stage-specific expression pattern of a mouse homolog of a human gene shown in panel A. Genes identified by MARVEL as loosely associated with S-HSCR are shown in red. Each column corresponds to a single cell with the predicted stage label taken from the original publication (Soldatov et al. 2019). Both the rows and the columns were clustered using hierarchical clustering with Pearson's correlation as the similarity measure, and the columns are divided into five partitions according to the clustering results.
Chemotaxis signaling is used to direct NC to target tissue (positive signaling) and works against the negative signals from the neural tube or somite to promote NC delamination and migration (Szabó and Mayor 2018). Additionally, cell polarization is critical during the directional migration of NC. Under the regulation of the RAC1-RHO1 gradient, actin is selectively polymerized on one side of the cell, extending the cell along the migration direction (Cohen et al. 2018; Szabó and Mayor 2018). Similarly, in rat axonal growth cones, under the regulation of CDC42/RAC1, SHTN1 is involved in the connection between F-actin and the cell adhesion molecule L1CAM (Kubo et al. 2015). The extracellular matrix (ECM) serves as the road of NC migration (Szabó and Mayor 2018), and collagens are basic structures in the ECM required for successful NC migration (Nagy et al. 2018). Cell-ECM interactions such as focal adhesion through integrins and vinculins provide both structural support and signaling sensor for migrating NC. In a previous study, mutations of vinculin in HSCR patients were shown to affect its function in focal adhesion (Lai et al. 2017).
Signals from cell-ECM interactions, cell-cell interactions, and chemotaxis are integrated inside NC through several signaling pathways including the PI3K, MAPK/ERK, and PKC pathways (Dinsmore and Soriano 2018). Through these pathways, signals from the environment are transformed into regulatory signals, controlling the activation and deactivation of genes.
An additional layer of regulation of these cellular signals was found at the protein level. For instance, E3 ubiquitin ligase is required by GLI3, a known HSCR-associated TF, to switch between its activator and repressor forms (Hsia et al. 2015), by which GLI regulates the differentiation and patterning of enteric NC (Liu et al. 2015).
Besides known HSCR TFs, we also found several novel TFs loosely associated with S-HSCR, such as MYC and ZBTB17 (Supplemental Table S2), which are known to regulate the size of the pool of premigratory NC (Kerosuo and Bronner 2016), and RARG, which is important in the retinoic acid signaling pathway, a pathway crucial for enteric NC migration (Uribe et al. 2018).
Overall, our results reveal an unprecedentedly broad linkage between noncoding genetic variants associated with S-HSCR and NC genes and clearly demonstrate that these genetic variants distribute across many different regulatory elements and genes that converge to key functional pathways.
As an additional way to explore the functions of the genes loosely associated with S-HSCR, we examined the spatiotemporal expression of their mouse homologs using a trunk NC single-cell RNA-seq (scRNA-seq) data set (Supplemental Materials; Soldatov et al. 2019). Some of the known HSCR genes and loosely associated genes display stage-specific expression (Fig. 3C; Supplemental Materials).
Upstream regulators of S-HSCR-associated genes
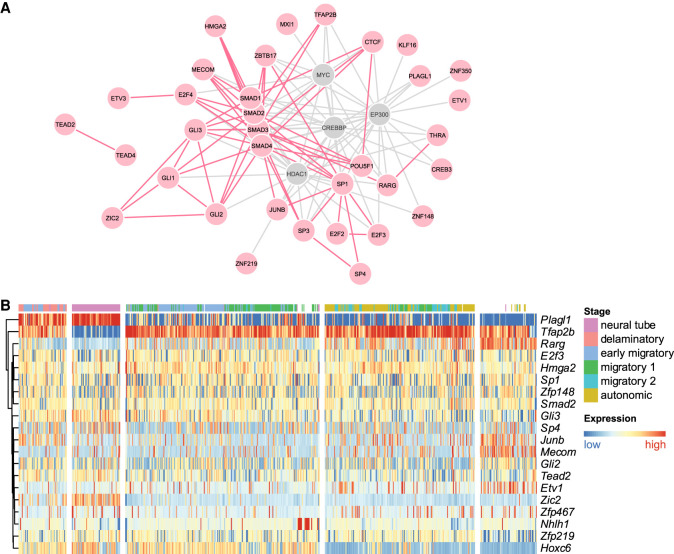
One major advantage of our framework is its ability to identify not only the disease-associated regulatory elements or genes but also their upstream transcriptional regulators whose binding sites are altered by the genetic variants. We developed a permutation test to identify TF binding motifs whose match scores are significantly more altered in the enhancers loosely associated with S-HSCR found in our enhancer-based analysis than the background set of all enhancers (Methods). This analysis identified 48 TFs that we will refer to as “recurrent TFs” (Supplemental Table S3). Among the top 100 promoters with the strongest association P-values with S-HSCR, 73 of them also had at least one of these recurrent TFs’ motifs selected by our procedure as a key motif, suggesting that, in NCs, these recurrent TFs regulate gene expression through both promoters and enhancers.
The recurrent TFs form an extensive network by way of physical protein-protein interactions (PPIs), with MYC and SMAD proteins acting as interaction hubs (Fig. 4A). The premigratory NC pool size regulators ZBTB17 and MYC and the retinoic acid receptor RARG discussed above in the functional pathway analysis are also contained in this network, showing that both the expression of these genes themselves and their downstream targets could be perturbed in S-HSCR. Some other interactions in this network are also known to have important functional roles. For example, MYC interacts with SMAD to activate expression of Snail, which are essential TFs that regulate EMT of NC (Smith et al. 2009; Kerosuo and Bronner 2016). The E2F proteins have been shown to regulate neuronal migration (McClellan et al. 2007). There are also reported HSCR genes in the network, such as GLI3 (Liu et al. 2015).
Figure 4.
Analysis of the recurrent TFs. (A) PPIs among the recurrent TFs (red) and several other proteins frequently interacting with them (gray). Direct interactions among the recurrent TFs are shown in red, while direct interactions involving the other proteins are shown in gray. Recurrent TFs that have no interactions with other proteins in this figure are excluded. (B) Spatiotemporal expression of recurrent TFs with stage-specific expression profiles in mouse trunk NCs. The heat map was produced in the same way as in Figure 3C, with the same order of columns.
Using the same mouse trunk NC scRNA-seq data described above, we found that the recurrent TFs with the strongest across-cell expression variance are Tfap2b and Plagl1 (Fig. 4B). TFAP2B (human homolog of Tfap2b) is a known migratory NC marker (Rabadán et al. 2013). Plagl1 is up-regulated in neural tube and in the delaminatory stage, which potentially regulates the composition of the ECM (Varrault et al. 2017) and thus potentially affects the migration environment of NC. One gene that has a similar expression pattern as Plagl1 is Zic2, which regulates the production of NC (Elms et al. 2003).
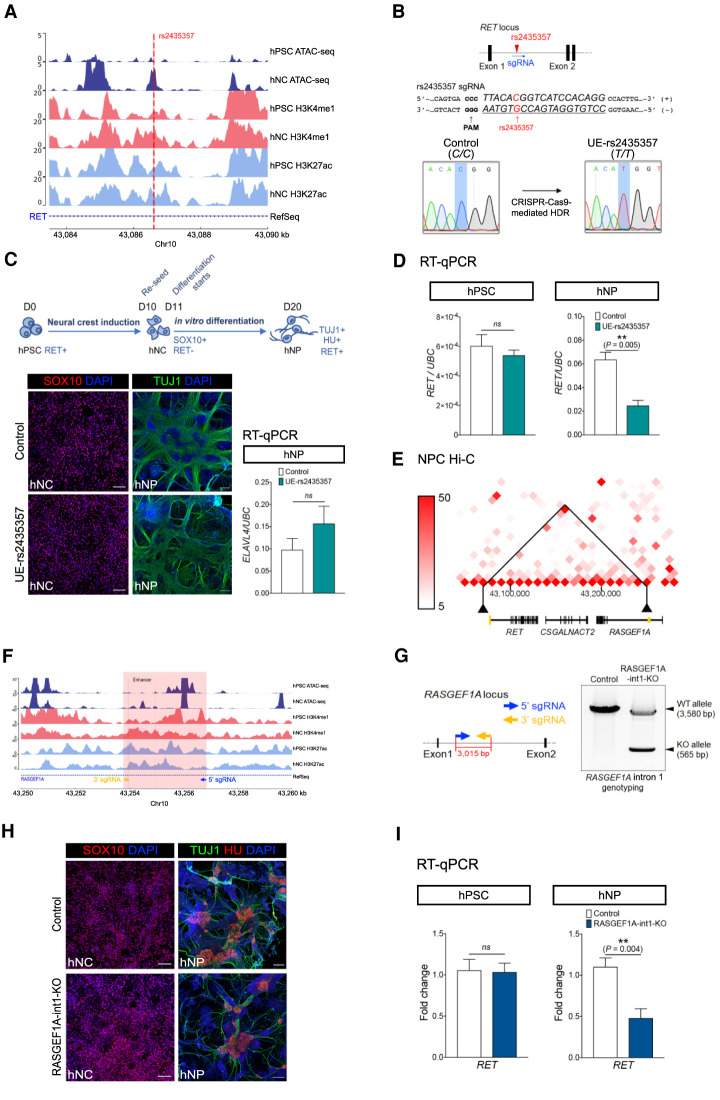
Enhancers significantly associated with S-HSCR confer cell stage–specific expression of RET
To delineate the potential biological roles of our newly identified S-HSCR-associated enhancers in disease susceptibility and pathogenesis, we employed hPSCs to model the development of the human ENS. We started with the most well-characterized HSCR-associated common SNP, rs2435357, where the risk allele T decreases the expression of RET and contributes to disease prevalence (Tang et al. 2016). As mentioned above, this SNP resides in an enhancer within intron 1 of RET that was found to be significantly associated with S-HSCR in our enhancer-based analysis (Table 1). It overlaps an hNC-specific ATAC-seq peak (Fig. 5A), suggesting that it may be implicated in the cell stage–specific expression of RET. To test this hypothesis, we introduced the risk allele T into a control hPSC line using a CRISPR-Cas9-mediated homology-directed repair (HDR) system with a specific single guide RNA (sgRNA) targeting the rs2435357(C/C) locus and single-stranded oligonucleotides (ssODNs) (Ran et al. 2013; Paquet et al. 2016; Kwart et al. 2017) containing the T allele (Fig. 5B). The control and mutant (UE-rs2435357) hPSC lines were then used to generate neural crest (hNC: SOX10+) and were subsequently directed to the neuronal lineage to make the early enteric neuronal progenitors (hNP: TUJ1+, ELAVL4+), recapitulating the two key developmental stages during the formation of the ENS as previously described (Fig. 5C; Fattahi et al. 2016; Lai et al. 2017). The single base (C > T) conversion alone did not affect hNC induction and the derivation of hNPs from hPSCs. We found that RET is highly expressed in the hNPs, while its expression remains at a low level in hNCs before committing to neuronal lineage, suggesting that the cell stage–specific enhancers are likely involved to mediate the dynamic expression of RET during the development of the ENS. In concordance with this observation, the C > T conversion in rs2435357 significantly reduced RET expression in hNPs derived from the UE-rs2435357 hPSC line, but it did not affect RET expression in hPSCs when compared to the isogenic control line (Fig. 5D). Our data suggest that the risk allele T in rs2435357 reduces RET expression in the early enteric neural progenitors (hNPs), contributing to HSCR pathogenesis.
Figure 5.
Functional impacts of a HSCR-associated SNP (rs2435357) and the deletion of a novel S-HSCR enhancer on RET expression. (A) ATAC-seq and ChIP-seq profiles of hPSC and hNC in intron 1 of RET show that rs2435357 is residing in a hNC-specific ATAC-seq peak. (B) Location of rs2435357 in the RET gene locus and in the sgRNA used for CRISPR-Cas9-mediated HDR for editing the C allele to the HSCR-associated risk allele T. The electrographs of Sanger sequencing show the successful introduction of the risk allele at rs2435357 in the UE-rs2435357 hPSC line. (C) Differentiation strategy to generate human neural crest (hNC) and neuronal progenitor (hNP). HU is encoded by the ELAVL4 gene. Immunostaining of SOX10 and TUJ1 in hNC and hNP of the control and the mutant (UE-rs2435357) lines. Scale bars: (hNC) 100 μm; (hNP) 200 μm. RT-qPCR analysis showing the comparable ELAVL4 expression level in hNP in the control (n = 5) and the mutant (UE-rs2435357) (n = 3) lines. t-test, (ns) not significant. (D) RT-qPCR analysis showing RET expression in the hPSC and hNP stages of the control (n = 5) and the mutant (UE-rs2435357) (n = 3). t-test, (ns) not significant. (E) Hi-C data from neural progenitor cells show that the enhancer in intron 1 of RASGEF1A (marked in yellow on the right) has physical interaction with the promoter of RET (marked in yellow on the left) at 10-kbp bin size. (F) ATAC-seq and ChIP-seq data from hPSC and hNC at the RASGEF1A intron 1 locus. (G) The design of sgRNAs used for the CRISPR-Cas9 system for deleting the DNA fragment in RASGEF1A intron 1. Genotyping reveals the specific deletion of RASGEF1A intron 1 in the UE-RASGEF1A-int1-KO hPSC line. (WT) Wild type, (KO) knockout. (H) Immunostaining of SOX10, TUJ1, and HU in hNC and hNP of the control and the mutant (RASGEF1A-int1-KO) lines, respectively. Scale bars: (hNC) 100 μm; (hNP) 200 μm. (I) RT-qPCR reveals the expression level of RET in the hPSC and hNP stages of the control (n = 4–5) and the mutant (RASGEF1A-int1-KO) (n = 6–7). t-test, (ns) not significant.
Next, we studied a novel enhancer (Chr 10: 43,253,960–43,254,961) significantly associated with S-HSCR within intron 1 of RASGEF1A that is located around 180 kbp downstream from the TSS of RET (Table 1). This putative enhancer is close to the TSS of RET in the three-dimensional (3D) genome architecture in neural progenitor cells based on previously published Hi-C data (Fig. 5E; Dixon et al. 2015). ATAC-seq data also revealed that there was an hNC-specific peak in this region, implying that this enhancer may also be implicated in ENS development (Fig. 5F). To investigate the potential effect of this novel enhancer in controlling RET expression, we generated a knockout (KO) mutant hPSC line (RASGEF1A-int1-KO) using a CRISPR-Cas9 system (Ran et al. 2013) with a pair of specific sgRNAs flanking the target region of intron 1 of RASGEF1A. Single hPSC colonies were isolated and genotyped to confirm the deletion of the target region (Fig. 5G). After neural induction and neuronal differentiation, we found that the differentiation potential of RASGEF1A-int1-KO hPSCs to make hNCs and hNPs was highly comparable to that of the control. Comparable numbers of SOX10-, TUJ1-, and HU-expressing cells were found in the control and the RASGEF1A-int1-KO groups (Fig. 5H). However, the deletion significantly reduced RET expression in hNPs, but not in hPSCs (Fig. 5I), implying that intron 1 of RASGEF1A contains a long-range regulator of RET in the early enteric progenitors.
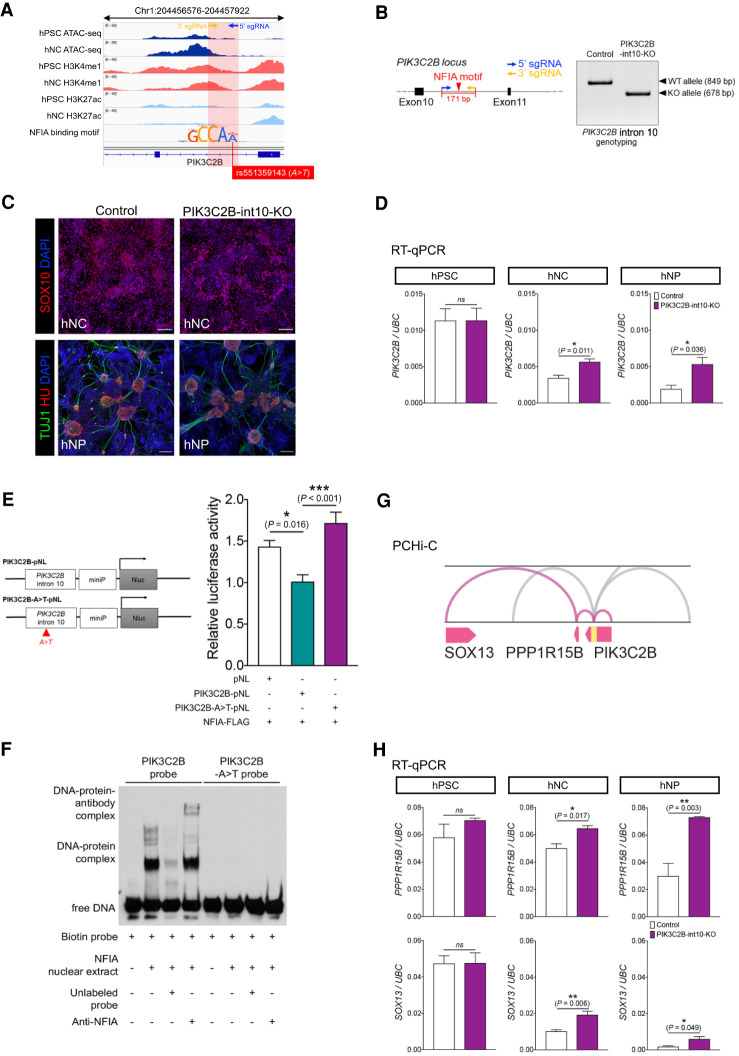
Disruption of NFIA binding in a novel S-HSCR-associated regulatory element in intron 10 of PIK3C2B interferes with the expression of PIK3C2B, PPP1R15B, and SOX13
We further experimentally studied the functional potential of another S-HSCR-associated regulatory element located in intron 10 of PIK3C2B (Chr 1: 204,456,576–204,457,577). This regulatory element was of particular interest because (1) it contained a strong hNC-associated ATAC-seq peak (Fig. 6A), (2) an A > T variant in this locus (rs551359143) was identified in six of our S-HSCR patients but not in any control subjects, and (3) the A > T conversion was predicted to disrupt the binding motif of NFIA (Table 1), which is a TF with a key role in regulating the differentiation of neural stem cells (Fig. 6A; Sanosaka et al. 2009; Piper et al. 2010; Kang et al. 2012; Tchieu et al. 2019). We generated another mutant hPSC line in which 171 bp of the intron 10 of PIK3C2B flanking the putative NFIA binding motif were deleted using a CRISPR-Cas9 system. The deletion of the target region was confirmed by genotyping in the PIK3C2B-int10-KO hPSC clone (Fig. 6B). The mutant hPSC clone was used to make hNCs and hNPs as described above. Similar to the other enhancers discussed above, the deletion of the fragment did not affect the differentiation potential of the hPSCs to make hNCs and hNPs (Fig. 6C). In terms of gene expression, our quantitative RT-PCR (RT-qPCR) revealed a sequential down-regulation of PIK3C2B along the enteric neuronal progenitor differentiation (hPSC-to-hNC-hNP). When the 171-bp fragment was deleted, the expression of PIK3C2B in hNCs and hNPs was significantly increased as compared to the isogenic control, implying that this 171-bp fragment probably serves as a negative regulatory element to mediate the sequential down-regulation of PIK3C2B along the differentiation path and the loss of it led to aberrant expression of PIK3C2B gene in hNCs and hNPs (Fig. 6D). We then further demonstrated the involvement of NFIA in mediating PIK3C2B expression using a luciferase assay. Our data showed that NFIA mainly acts as a repressor and the luciferase activity was significantly up-regulated when the NFIA binding motif was disrupted by introducing the A > T substitution (Fig. 6E). A direct binding of NFIA onto the intron 10 of PIK3C2B was demonstrated using a gel shift and supershift assay, in which NFIA binding was consistently abolished by the A > T conversion (Fig. 6F). All these results indicate that PIK3C2B was negatively regulated by NFIA and the HSCR-associated A > T variation would disrupt NFIA binding, leading to the increased expression of PIK3C2B.
Figure 6.
Characterization of a novel S-HSCR-associated regulatory element in intron 10 of PIK3C2B. (A) Overview of ATAC-seq and ChIP-seq profiles showing the putative hNC-specific regulatory element in PIK3C2B intron 10. The red shaded region indicates the location of the regulatory element and the line shows the A > T variant (rs551359143) found exclusively in the S-HSCR cases that disrupts the NFIA binding motif. The motif is not drawn to the same scale as the genomic signal tracks, with magnified characters. (B) Design of sgRNAs used for the CRISPR-Cas9 system for deleting the regulatory element. Genotyping reveals the specific deletion of the 171-bp fragment in intron 10 of PIK3C2B in the PIK3C2B-int10-KO hPSC line. (WT) Wild type, (KO) knockout. (C) Immunostaining shows that both the control and mutant (PIK3C2B-int1-KO) lines have comparable capability to make hNCs and hNPs. Scale bars: (hNC) 100 μm; (hNP) 200 μm. (D) RT-qPCR shows the changes in the expression of PIK3CB in different cell stages in the control and mutant lines. t-test, (ns) not significant. n = 3–4 per group. (E) Design of the constructs used for the luciferase assay. The bar chart shows the relative luciferase activities when the cells were transfected with different sets of constructs as indicated. Three independent assays were performed, each in triplicate. One-way ANOVA. (F) Gel mobility shift assays were performed with biotin-labeled probes containing the PIK3C2B intron 10 regulatory element with or without the A > T conversion and the nuclear extract from NFIA-overexpressing cells, in the presence of unlabeled probes or anti-NFIA antibody (0.1 µg). (G) Significant contacts (FDR < 0.05) in the promoter capture Hi-C data from GM12878 cells at the PIK3C2B locus. The putative regulatory element in intron 10 of PIK3C2B is marked in yellow. Contacts between the regulatory element and the TSSs of SOX13, PPP1R15B, and PIK3C2B are shown in purple curves, while contacts between the regulatory element and other promoters are shown in gray curves. Contacts that extend too far are trimmed. (H) RT-qPCR analysis shows the changes in the expression of PPP1R15B and SOX13 in the control and the mutant lines at different cell stages. t-test, (ns) not significant. n = 3–4 per group.
Long-range chromatin interactions form topologically associating domains (TADs) and the genes within a TAD are likely to be regulated by common enhancers (Chatterjee et al. 2016; Laugsch et al. 2019). A previous promoter capture Hi-C experiment suggests that chromatin loops can be formed between PIK3C2B intron 10 and the TSSs of PPP1R15B and SOX13 in GM12878 cells (Fig. 6G; Mifsud et al. 2015). This suggests that the gene expression of PPP1R15B and SOX13 may also be influenced by the S-HSCR-associated A > T variant in intron 10 of PIK3C2B. RT-qPCR revealed that the expression levels of PPP1R15B and SOX13, similar to that of PIK3C2B, were not affected in the hPSC stage but up-regulated in the hNC and hNP stages after the deletion of the 171-bp fragment (Fig. 6H). This result suggests that PIK3C2B intron 10 contains an element that can regulate the expression of genes within the same TAD and the usage of this regulatory element may be dynamic during the development of ENS. During neuronal differentiation, this element likely exerts negative control over gene expression, as the deletion of it consistently increased gene expression. In comparison, in the control line, all the genes regulated by this element were down-regulated when the cells differentiated into neuronal lineages, suggesting that up-regulation of these genes associated with the A > T variant may impose disease-causing risks during the later stage of enteric NC cell development.
Discussion
In this work, we have proposed a novel framework, MARVEL, for identifying noncoding genetic variants of potential functional significance. It was designed to overcome various issues in the study of noncoding variants. Our application of MARVEL to the analysis of WGS data of S-HSCR has clearly demonstrated the advantages of these designs. Starting from 37 million noncoding variants observed in the cases and controls, the epigenomic data reduced the number of genomic regions to study to the scale of tens of thousands (of genes and promoters) to hundreds of thousands (of enhancers), and the statistical procedures further selected no more than 10 key motifs from each of these regions among 771 of them. As a result, we were able to obtain some results other than those in the dominating RET locus at fairly strong significance levels, which would be impossible in the traditional approach of testing the disease association of each genetic variant separately since most noncoding variants are rare or have small effect sizes.
Comparing the enhancer-based and promoter-based analyses, we obtained more significant results from the former, showing that it is important to move beyond the immediate TSS-proximal regions when analyzing the noncoding variants. The designs of MARVEL made it possible to study genetic variants in distal enhancers in a computationally and statistically feasible manner.
The gene-based analysis led to findings in Chromosomes 9 and 15 not obtained from the enhancer-based or promoter-based analysis. We have found that around these gene loci there were more enhancers with marginally significant associations than other gene loci, suggesting that it was the aggregation of weak signals from multiple regulatory elements that helped identify these gene loci as significantly associated with S-HSCR. The importance of this aggregation idea was further demonstrated in the functional pathway analysis, in which some of the 417 genes loosely associated with S-HSCR were grouped into functional clusters that are highly related to NC migration.
Our analysis of upstream regulators has identified 48 recurrent TFs, and they form an extensive network through PPIs. Notably, some of these TFs also have their corresponding genes identified in the association tests, showing that the disruptions of both the expression of these TFs themselves and their binding at other genes’ regulatory regions are associated with S-HSCR, illustrating two main mechanisms by which genetic variants can exert their functional effects.
Making use of the CRISPR-Cas9 genome editing platform, we were able to generate various hPSC lines with isogenic background, such that any phenotypical differences observed should be solely due to the effects of the risk variants. This platform allowed us to study the functional impacts for the risk variants in the human genome, in contrast to previous studies that made use of other animal systems for enhancer studies, which usually focused on the highly conserved regions of the genome instead of the disease-associated variants.
The motif analysis in our new model is also capable of identifying the upstream transcription factors that regulate the HSCR-associated regulatory elements. For instance, we have found that NFIA may act as a negative regulator when binding to the regulatory element of intron 10 of PIK3C2B during neural differentiation. This finding is consistent with a previous report that suggests NFIA represses Hes1 during neural progenitor differentiation in mouse forebrain (Piper et al. 2010). A recent study from Qi et al. (2020) has found that the conditional knockout of Nfia in mouse neural crest cells (i.e., Wnt1-Cre; Nfiaf/f) would only disrupt the development of NPY2R-expressing neurons but not other neurons such as TH-expressing neurons in vivo (Qi et al. 2020). This suggests that NFIA is responsible for controlling the development of a specific neuronal subtype and may be involved in the later stage of neuronal development. Inhibition of PIK3C2B has been shown to improve function and prolong survival in myotubular myopathy through regulating the phosphoinositide (PIP) metabolism (Sabha et al. 2016). As the dysregulation of PIP metabolism may lead to neurodegenerative disease (Burke 2018), the up-regulation of PIK3C2B caused by the A > T SNP in intron 10 of PIK3C2B may affect the survival of enteric neurons.
Similar to PIK3C2B, up-regulation of PPP1R15B may induce cellular stress and affect cell survival through inducing protein misfolding conditions as seen in Huntington's disease (Krzyzosiak et al. 2018). On the other hand, it has been demonstrated that SOX13 can functionally replace SOX5 and SOX6 in governing oligodendrocyte development in mouse spinal cord (Baroti et al. 2016), suggesting that it modulates cell differentiation into glial lineages instead of neuronal lineages. Therefore, ectopic SOX13 expression may affect the cell fate determination of the enteric NC cells. Given that many of these genes may affect various stages of the ENS development, further studies are therefore required to unveil the functions of these differentially regulated genes in the development of ENS.
Overall, our work has produced a large number of novel regulatory elements, genes, and upstream regulators associated with S-HSCR for follow-up investigations. Due to the buffering of different regulatory elements and different genes, single perturbations may have little observable effects on cell differentiation, and as a result their roles in disease susceptibility and pathogenesis are subtle. In order to have a more complete understanding of the functional importance of the candidate regulatory elements, genes, and TFs identified in this study, it would be useful to perform large-scale genome editing assays to systematically perturb them individually and in combination and determine how they may interfere in various steps of ENS development.
Methods
Statistical procedures of the MARVEL framework
Here, we describe the statistical procedures used by the MARVEL framework. Additional details of the framework can be found in Supplemental Materials.
Identification of important motifs and phenotype-associated target regions
Suppose there are n subjects with their phenotype information recorded in the binary vector y, where yi = 1 if subject i is a case and yi = 0 if subject i is a control. For each target region, the standardized score profiles are represented by an n × m matrix X, where m is the number of motifs. Covariates of the n subjects are given in an n × v matrix C, where v is the number of covariates.
Based on X and y, generalized linear model-based least angle regression (GLM-LARS) (Tibshirani et al. 2004) is performed to select a subset of motifs whose match scores in this target region can best explain the phenotypes. We use glmpath (Park and Hastie 2007) (https://cran.r-project.org/web/packages/glmpath/index.html) to perform GLM-LARS with parameter settings min. lambda = 1e-2, max.steps = 10, and max.vars = 10 such that up to 10 motifs can be selected and the computations are efficient.
To quantify how well the target region's genetic variations are associated with the phenotype based on the scores of the selected motifs, we construct two logistic regression models
where is the logit function, X′ is a reduced version of X retaining only the columns corresponding to the selected motifs, (X′|C) is a matrix that concatenates the columns of X′ and C, and β0 and β1 are coefficients. The likelihood ratio between these two models is then used as a test statistic
The statistical significance of a target region is evaluated by comparing its LR with those from a permutation set, which involves all the target regions with 100 random permutations of y. For example, when we applied this procedure to perform association tests for the enhancers in the S-HSCR study, since there were 150,828 enhancers in total, each of these LR values were compared against the 15,082,800 values from the permutations to compute a P-value. The P-values of all the target regions are then corrected for multiple hypothesis testing using the Benjamini–Hochberg method (Benjamini and Hochberg 1995).
Estimation of effect size
We use AUROC to estimate the effect size of each target region. Specifically, after constructing the model g(y) = (X′|C)β1 for a target region, for each subject, this model is applied to compute a score that indicates how likely this subject has the phenotype. These phenotype scores together with the actual phenotype labels of the subjects are then used to produce the receiver operating characteristic, and the area under the curve is used as an estimate of the effect size.
Analysis of recurrent TFs
To identify the recurrent TFs, all motifs selected by GLM-LARS from the full set of 200 enhancers loosely associated with S-HSCR were collected. Ten thousand random sets of 200 enhancers were then formed by sampling from all enhancers, and GLM-LARS was applied to each of these sets. For each motif, its total model coefficients across all enhancers in a set were used as a test statistic, and a P-value was computed by comparing the test statistic obtained from the loosely associated enhancers with those from the random enhancer sets, to see if the former is significantly larger than the latter. These raw P-values were then corrected by the Benjamini–Hochberg method to control the FDR at 0.05.
The PPIs among the recurrent TFs were obtained from STRING (v11) (Szklarczyk et al. 2017). We included only the interactions based on manual curation or experimental evidence with a combined score >0.4.
Functional studies
Cell culture
A control hPSC line (UE02302) was established and maintained as previously described (Lau et al. 2019). Neural crest induction was performed according to a previously described protocol (Lai et al. 2017). In brief, hPSCs were plated on Matrigel-coated plates in a density of 5 × 104 cells cm−2 in ES cell medium containing 10 ng/mL fibroblast growth factor 2 (FGF2, Peprotech). The differentiation was started by replacing ES cell medium with KSR medium and gradually switched to N2 medium from day 4 to day 10. For hNC differentiation, the cells were treated with LDN193189, SB431542, CHIR99021, and retinoic acid.
For neuronal differentiation of hNCs to hNPs, hNC cells (2.5 × 104) were seeded as droplets on polyornithine/laminin/fibronectin-coated surface in N2 medium containing 10 ng/mL FGF2 and 3 μM CHIR99021. For the RET-associated study, p75NTR and HNK-1 double-positive hNC cells were sorted by a BD FACSAria III Cell Sorter; 5 × 104 sorted cells were seeded as droplets on a polyornithine/laminin/fibronectin-coated surface. Neuronal differentiation was initiated by replacing the medium with N2 medium containing BDNF, GDNF, NT-3, NGF, dibutyryl cAMP, and ascorbic acid for 9 d. Detailed procedures are described in Supplemental Materials.
Plasmid constructions
Human codon-optimized high fidelity Cas9 nuclease construct (Ran et al. 2013) was obtained from Addgene (48138). Oligos for sgRNA cloning are listed in Supplemental Table S5. For the luciferase assay, PIK3C2B intron 10 fragment was cloned into a NanoLuc luciferase reporter construct (pNL3.2[NlucP/minP]) (Promega N1041) to generate the PIK3C2B-pNL construct, while NFIA ORF was cloned into the pFLAG-CMV plasmid to generate an NFIA-FLAG expression construct. The A > T variant was introduced to the PIK3C2B-pNL construct by site-directed mutagenesis using a QuikChange Lightning Site-Directed Mutagenesis kit (Agilent). The cloning primers and mutagenesis primers are listed in Supplemental Table S5, and detailed procedures are described in Supplemental Materials.
Generation of new hPSC lines using the CRISPR-Cas9 system
For the generation of the mutant (UE-rs2435357) hPSC line, UE control hPSCs were transfected with sgRNA construct, ssODNs and pSpCas9(BB)-2A-GFP construct by the Human Stem Cell Nucleofector kit 2 (Lonza). For the generation of UE-RASGEF1A-int1-KO and PIK3C2B-int10-KO hPSC lines, UE control hPSCs were transfected with a pair of pSpCas9(BB)-2A-GFP constructs containing the specific sgRNAs using the P3 Primary Cell 4D-Nuclecfector X kit (Lonza). hPSCs expressing GFP were sorted as single cells and expanded for 2 wk. The single colonies were genotyped to confirm the site-specific conversion or the deletion of the target regions. Detailed methods are described in Supplemental Materials.
RT-qPCR
Total RNA from hPSCs, hNCs, and hNPs was extracted by an RNeasy Mini kit (Qiagen) and reverse-transcribed to cDNA using HiScript II Q RT SuperMix (Vazyme). The expression levels of the target genes were quantitated using real-time quantitative RT-PCR or droplet digital PCR (ddPCR). For real-time quantitative RT-PCR, target genes were amplified with Luna Universal Probe qPCR Master Mix (New England Biolabs) using a specific TaqMan Gene Expression Assay by ViiA 7 Real-Time PCR System (Thermo Fisher Scientific). ddPCR was used to measure RET expression in hNPs. In brief, cDNA samples were mixed with ddPCR Supermix for Probes (Bio-Rad 186-3010) and specific TaqMan probes and then subjected to droplet generation following the manufacturer's protocol. The reaction droplets were then subjected to thermal cycling, and the end-point fluorescence signals were measured by a QX200 Droplet Reader (Bio-Rad). Detailed procedures are described in Supplemental Materials.
Gel shift assay
The NFIA-FLAG expression construct was transfected to HeLa cells by FuGENE HD Transfection Reagent (Promega), and the nuclear extracts containing NFIA protein were then extracted using a nuclear and cytoplasmic extraction kit (Thermo Fisher Scientific). ssODNs derived from the intron 10 of PIK3C2B were biotin-labeled using a Biotin 3′-End DNA Labeling kit (Thermo Fisher Scientific) and then annealed with reverse complementary ssODNs to generate biotin-labeled probes. A gel shift assay was performed by mixing the nuclear extracts with biotin-labeled probes according to the manufacturer's protocol (LightShift Chemiluminescent EMSA kit; Thermo Fisher Scientific). For supershift assays, 0.1 µg anti-NFIA (Sigma-Aldrich HPA006111) was added to the mixture in the final step before incubation. Detailed methods are described in Supplemental Materials.
Luciferase assay
Control firefly luciferase construct (pGL3-control), NanoLuc luciferase constructs (pNL3.2[NlucP/minP] or PIK3C2B-pNL or PIK3C2B-A > T-pNL), and NFIA expression construct (NFIA-FLAG) were cotransfected into SH-SY5Y cells using jetPRIME transfection reagent (Polyplus Transfection) according to the manufacturer's protocol. Luciferase activities were detected with a Nano-Glo Dual-Luciferase Reporter Assay System (Promega) by the VICTOR Nivo Microplate Reader (PerkinElmer). Detailed procedures are described in Supplemental Materials.
Immunostaining
hNC cells and hNP-D9 cells were fixed in 4% PFA for 20 min at room temperature. Fixed cells were blocked with blocking solution (1% BSA, 0.1% Triton X-100 in PBS) at room temperature for 1 h. The blocked cells were then incubated with primary antibodies (mouse anti-SOX10 [1:500, R&D Systems MAB2864], rabbit anti-TUJ1 [1:1000, Abcam ab18207] and mouse anti-HU [1:1000, Thermo Fisher Scientific A-21271]) at 4°C overnight. After washing, the sections were incubated with respective secondary antibodies conjugated with Alexa Fluor 488/594 (Thermo Fisher Scientific 1:500) at room temperature for 2 h. The stained cells were mounted with ProLong Diamond Antifade Mountant with DAPI (Thermo Fisher Scientific). Fluorescence images were acquired by a Carl Zeiss LSM780 or LSM800 confocal microscope.
Chromatin interaction data
Hi-C data of NPC were obtained from the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) with accession number GSE52457.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE154972. The source code of MARVEL is provided as Supplemental Code S1 and on GitHub (https://github.com/fuxialexander/marvel).
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
ATAC-seq, ChIP-seq, and RNA-seq were performed in the Center for PanorOmic Sciences, The University of Hong Kong. Confocal imaging was performed with equipment maintained by the Li Ka Shing Faculty of Medicine Faculty Core Facility. This project is supported by Hong Kong Research Grants Council Theme-based Research Scheme T12C-714/14-R. K.Y.Y. is additionally supported by the Hong Kong Research Grants Council Collaborative Research Funds C4045-18WF, C4054-16G, C4057-18EF, and C7044-19G and General Research Funds 14170217 and 14203119, the Hong Kong Epigenomics Project (EpiHK), and the Chinese University of Hong Kong Young Researcher Award and Outstanding Fellowship. The work described in this paper is substantially supported by an HKSAR Department of Health, Health and Medical Research Fund (HMRF) grant (Project no.: 06173306) and a General Research Fund (HKU, 17108019) from the Department of Health and the Research Grants Council of Hong Kong Special Administrative Region, China Hong Kong, respectively, to E.S.W.N. K.N.C.L. is supported by the Hong Kong PhD Fellowship from the Research Grants Council of Hong Kong.
Author contributions: P.K.H.T., E.S.W.N., and K.Y.Y. conceived this study. A.X.F. and K.Y.Y. developed the MARVEL framework. A.X.F., C.S.M.T., M.M.G.B., P.C.S., and K.Y.Y. analyzed the sequencing data. R.K.N., F.P.L.L., S.T.L., and E.S.W.N. performed the epigenomics experiments. S.T.L. and Z.L. performed the RNA-seq analysis of hNC. A.X.F. and K.Y.Y. applied the MARVEL framework to the S-HSCR data. K.N.C.L. and E.S.W.N. performed the functional experiments and analyzed the data. A.X.F., K.N.C.L., E.S.W.N., and K.Y.Y. prepared the manuscript. P.C.S., P.K.H.T., E.S.W.N., and K.Y.Y. supervised the project.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.264473.120.
References
- Alves MM, Sribudiani Y, Brouwer RWW, Amiel J, Antiñolo G, Borrego S, Ceccherini I, Chakravarti A, Fernández RM, Garcia-Barcelo MM, et al. 2013. Contribution of rare and common variants determine complex diseases—Hirschsprung disease as a model. Dev Biol 382: 320–329. 10.1016/j.ydbio.2013.05.019 [DOI] [PubMed] [Google Scholar]
- Amiel J, Attié T, Jan D, Pelet A, Edery P, Bidaud C, Lacombe D, Tam P, Simeoni J, Flori E, et al. 1996. Heterozygous endothelin receptor B (EDNRB) mutations in isolated Hirschsprung disease. Hum Mol Genet 5: 355–357. 10.1093/hmg/5.3.355 [DOI] [PubMed] [Google Scholar]
- Amiel J, Sproat-Emison E, Garcia-Barcelo M, Lantieri F, Burzynski G, Borrego S, Pelet A, Arnold S, Miao X, Griseri P, et al. 2008. Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet 45: 1–14. 10.1136/jmg.2007.053959 [DOI] [PubMed] [Google Scholar]
- Badner JA, Sieber WK, Garver KL, Chakravarti A. 1990. A genetic study of Hirschsprung disease. Am J Hum Genet 46: 568–580. [PMC free article] [PubMed] [Google Scholar]
- Baroti T, Schillinger A, Wegner M, Stolt CC. 2016. Sox13 functionally complements the related Sox5 and Sox6 as important developmental modulators in mouse spinal cord oligodendrocytes. J Neurochem 136: 316–328. 10.1111/jnc.13414 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57: 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Boyle EA, Li YI, Pritchard JK. 2017. An expanded view of complex traits: from polygenic to omnigenic. Cell 169: 1177–1186. 10.1016/j.cell.2017.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JE. 2018. Structural basis for regulation of phosphoinositide kinases and their involvement in human disease. Mol Cell 71: 653–673. 10.1016/j.molcel.2018.08.005 [DOI] [PubMed] [Google Scholar]
- Chakravarti A, McCallion A, Lyonnet S. 2006. Multisystem inborn errors of development: Hirschsprung disease. In Scrivers’ online metabolic and molecular bases of inherited disease (ed. Valle DL, et al. ). McGraw-Hill Educ; https://ommbid.mhmedical.com/ 10.1036/ommbid.291 [DOI] [Google Scholar]
- Chatterjee S, Kapoor A, Akiyama JA, Auer DR, Lee D, Gabriel S, Berrios C, Pennacchio LA, Chakravarti A. 2016. Enhancer variants synergistically drive dysfunction of a gene regulatory network in Hirschsprung disease. Cell 167: 355–368.e10. 10.1016/j.cell.2016.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SJ, Shi FY, Liu H, Ding Y, Jiang S, Liang N, Gao G. 2017. Accurately annotate compound effects of genetic variants using a context-sensitive framework. Nucleic Acids Res 45: e82–e82. 10.1093/nar/gkx041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Kovari DT, Wei W, Keate R, Curtis JE, Nie S. 2018. Cdc42 regulates the cellular localization of Cdc42ep1 in controlling neural crest cell migration. J Mol Cell Biol 10: 376–387. 10.1093/jmcb/mjx044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradin O, Saiakhova A, Akhtar-Zaidi B, Myeroff L, Willis J, Cowper-Sallari R, Lupien M, Markowitz S, Scacheri PC. 2014. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res 24: 1–13. 10.1101/gr.164079.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinsmore CJ, Soriano P. 2018. MAPK and PI3K signaling: at the crossroads of neural crest development. Dev Biol 444: S79–S97. 10.1016/j.ydbio.2018.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. 2015. Chromatin architecture reorganization during stem cell differentiation. Nature 518: 331–336. 10.1038/nature14222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elms P, Siggers P, Napper D, Greenfield A, Arkell R. 2003. Zic2 is required for neural crest formation and hindbrain patterning during mouse development. Dev Biol 264: 391–406. 10.1016/j.ydbio.2003.09.005 [DOI] [PubMed] [Google Scholar]
- Emison ES, McCallion AS, Kashuk CS, Bush RT, Grice E, Lin S, Portnoy ME, Culler DJ, Green ED, Chakravarti A. 2005. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature 434: 857–863. 10.1038/nature03467 [DOI] [PubMed] [Google Scholar]
- Emison ES, Garcia-Barcelo M, Grice EA, Lantieri F, Amiel J, Burzynski G, Fernandez RM, Hao L, Kashuk C, West K, et al. 2010. Differential contributions of rare and common, coding and noncoding Ret mutations to multifactorial Hirschsprung disease liability. Am J Hum Genet 87: 60–74. 10.1016/j.ajhg.2010.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattahi F, Steinbeck JA, Kriks S, Tchieu J, Zimmer B, Kishinevsky S, Zeltner N, Mica Y, El-Nachef W, Zhao H, et al. 2016. Deriving human ENS lineages for cell therapy and drug discovery in Hirschsprung disease. Nature 531: 105–109. 10.1038/nature16951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank CL, Tsai LH. 2009. Alternative functions of core cell cycle regulators in neuronal migration, neuronal maturation, and synaptic plasticity. Neuron 62: 312–326. 10.1016/j.neuron.2009.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Barcelo M-M, Tang CS, Ngan ES, Lui VC, Chen Y, So M, Leon TY, Miao X, Shum CK, Liu F, et al. 2009. Genome-wide association study identifies NRG1 as a susceptibility locus for Hirschsprung's disease. Proc Natl Acad Sci 106: 2694–2699. 10.1073/pnas.0809630105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow SM, Zhu W, Stolt CC, Huang TW, Chen F, LoTurco JJ, Neul JL, Wegner M, Mohila C, Deneen B. 2014. Mutual antagonism between Sox10 and NFIA regulates diversification of glial lineages and glioma subtypes. Nat Neurosci 17: 1322–1329. 10.1038/nn.3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia EYC, Gui Y, Zheng X. 2015. Regulation of Hedgehog signaling by ubiquitination. Front Biol (Beijing) 10: 203–220. 10.1007/s11515-015-1343-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung HF, Hehnly H, Doxsey S. 2016. The mother centriole appendage protein cenexin modulates lumen formation through spindle orientation. Curr Biol 26: 793–801. 10.1016/j.cub.2016.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P, Lee HK, Glasgow SM, Finley M, Donti T, Gaber ZB, Graham BH, Foster AE, Novitch BG, Gronostajski RM, et al. 2012. Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron 74: 79–94. 10.1016/j.neuron.2012.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerosuo L, Bronner ME. 2016. Cmyc regulates the size of the premigratory neural crest stem cell pool. Cell Rep 17: 2648–2659. 10.1016/j.celrep.2016.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korhonen JH, Palin K, Taipale J, Ukkonen E. 2017. Fast motif matching revisited: high-order PWMs, SNPs and indels. Bioinformatics 33: 514–521. 10.1093/bioinformatics/btw683 [DOI] [PubMed] [Google Scholar]
- Krzyzosiak A, Sigurdardottir A, Luh L, Carrara M, Das I, Schneider K, Bertolotti A. 2018. Target-based discovery of an inhibitor of the regulatory phosphatase PPP1R15B. Cell 174: 1216–1228.e19. 10.1016/j.cell.2018.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo Y, Baba K, Toriyama M, Minegishi T, Sugiura T, Kozawa S, Ikeda K, Inagaki N. 2015. Shootin1–cortactin interaction mediates signal–force transduction for axon outgrowth. J Cell Biol 210: 663–676. 10.1083/jcb.201505011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwart D, Paquet D, Teo S, Tessier-Lavigne M. 2017. Precise and efficient scarless genome editing in stem cells using CORRECT. Nat Protoc 12: 329–354. 10.1038/nprot.2016.171 [DOI] [PubMed] [Google Scholar]
- Lai FP-L, Lau S-T, Wong JK-L, Gui H, Wang RX, Zhou T, Lai WH, Tse H-F, Tam PK-H, Garcia-Barcelo M-M, et al. 2017. Correction of Hirschsprung-associated mutations in human induced pluripotent stem cells via clustered regularly interspaced short palindromic repeats/Cas9, restores neural crest cell function. Gastroenterology 153: 139–153.e8. 10.1053/j.gastro.2017.03.014 [DOI] [PubMed] [Google Scholar]
- Lau S-T, Li Z, Pui-Ling Lai F, Nga-Chu Lui K, Li P, Munera JO, Pan G, Mahe MM, Hui C-C, Wells JM, et al. 2019. Activation of hedgehog signaling promotes development of mouse and human enteric neural crest cells, based on single-cell transcriptome analyses. Gastroenterology 157: 1556–1571.e5. 10.1053/j.gastro.2019.08.019 [DOI] [PubMed] [Google Scholar]
- Laugsch M, Bartusel M, Rehimi R, Alirzayeva H, Karaolidou A, Crispatzu G, Zentis P, Nikolic M, Bleckwehl T, Kolovos P, et al. 2019. Modeling the pathological long-range regulatory effects of human structural variation with patient-specific hiPSCs. Cell Stem Cell 24: 736–752.e12. 10.1016/j.stem.2019.03.004 [DOI] [PubMed] [Google Scholar]
- Liao EH, Hung W, Abrams B, Zhen M. 2004. An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature 430: 345–350. 10.1038/nature02647 [DOI] [PubMed] [Google Scholar]
- Liu JAJ, Lai FPL, Gui HS, Sham MH, Tam PKH, Garcia-Barcelo MM, Hui CC, Ngan ESW. 2015. Identification of GLI mutations in patients with Hirschsprung disease that disrupt enteric nervous system development in mice. Gastroenterology 149: 1837–1848.e5. 10.1053/j.gastro.2015.07.060 [DOI] [PubMed] [Google Scholar]
- Luzón-Toro B, Gui H, Ruiz-Ferrer M, Sze-Man Tang C, Fernández RM, Sham P-C, Torroglosa A, Kwong-Hang Tam P, Espino-Paisán L, Cherny SS, et al. 2015. Exome sequencing reveals a high genetic heterogeneity on familial Hirschsprung disease. Sci Rep 5: 16473 10.1038/srep16473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan KA, Ruzhynsky VA, Douda DN, Vanderluit JL, Ferguson KL, Chen D, Bremner R, Park DS, Leone G, Slack RS. 2007. Unique requirement for Rb/E2F3 in neuronal migration: Evidence for cell cycle-independent functions. Mol Cell Biol 27: 4825–4843. 10.1128/MCB.02100-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, Wingett SW, Andrews S, Grey W, Ewels PA, et al. 2015. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet 47: 598–606. 10.1038/ng.3286 [DOI] [PubMed] [Google Scholar]
- Nagy N, Barad C, Hotta R, Bhave S, Arciero E, Dora D, Goldstein AM. 2018. Collagen 18 and agrin are secreted by neural crest cells to remodel their microenvironment and regulate their migration during enteric nervous system development. Development 145: dev160317 10.1242/dev.160317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, Olsen KM, Gregg A, Noggle S, Tessier-Lavigne M. 2016. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533: 125–129. 10.1038/nature17664 [DOI] [PubMed] [Google Scholar]
- Park MY, Hastie T. 2007. L1-regularization path algorithm for generalized linear models. J R Stat Soc Ser B 69: 659–677. 10.1111/j.1467-9868.2007.00607.x [DOI] [Google Scholar]
- Piper M, Barry G, Hawkins J, Mason S, Lindwall C, Little E, Sarkar A, Smith AG, Moldrich RX, Boyle GM, et al. 2010. NFIA controls telencephalic progenitor cell differentiation through repression of the notch effector Hes1. J Neurosci 30: 9127–9139. 10.1523/JNEUROSCI.6167-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Yin G, Zhang Y, Tao Y, Wu X, Gronostajski RM, Qiu M, Liu Y. 2020. Nuclear factor I/A controls A-fiber nociceptor development. Neurosci Bull 36: 685–695. 10.1007/s12264-020-00486-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabadán MA, Usieto S, Lavarino C, Martí E. 2013. Identification of a putative transcriptome signature common to neuroblastoma and neural crest cells. Dev Neurobiol 73: 815–827. 10.1002/dneu.22099 [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8: 2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabha N, Volpatti JR, Gonorazky H, Reifler A, Davidson AE, Li X, Eltayeb NM, Dall'Armi C, Di Paolo G, Brooks SV, et al. 2016. PIK3C2B inhibition improves function and prolongs survival in myotubular myopathy animal models. J Clin Invest 126: 3613–3625. 10.1172/JCI86841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanosaka T, Namihira M, Nakashima K. 2009. Epigenetic mechanisms in sequential differentiation of neural stem cells. Epigenetics 4: 89–92. 10.4161/epi.4.2.8233 [DOI] [PubMed] [Google Scholar]
- Savaskan NE, Brauer AU, Nitsch R. 2004. Molecular cloning and expression regulation of PRG-3, a new member of the plasticity-related gene family. Eur J Neurosci 19: 212–220. 10.1046/j.1460-9568.2003.03078.x [DOI] [PubMed] [Google Scholar]
- Schäfer K-H, Van Ginneken C, Copray S. 2009. Plasticity and neural stem cells in the enteric nervous system. Anat Rec Adv Integr Anat Evol Biol 292: 1940–1952. 10.1002/ar.21033 [DOI] [PubMed] [Google Scholar]
- Simoes-Costa M, Bronner ME. 2013. Insights into neural crest development and evolution from genomic analysis. Genome Res 23: 1069–1080. 10.1101/gr.157586.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AP, Verrecchia A, Fagà G, Doni M, Perna D, Martinato F, Guccione E, Amati B. 2009. A positive role for Myc in TGFβ-induced snail transcription and epithelial-to-mesenchymal transition. Oncogene 28: 422–430. 10.1038/onc.2008.395 [DOI] [PubMed] [Google Scholar]
- Soldatov R, Kaucka M, Kastriti ME, Petersen J, Chontorotzea T, Englmaier L, Akkuratova N, Yang Y, Häring M, Dyachuk V, et al. 2019. Spatiotemporal structure of cell fate decisions in murine neural crest. Science 364: eaas9536 10.1126/science.aas9536 [DOI] [PubMed] [Google Scholar]
- Sun J, Zhang T, Cheng M, Hong L, Zhang C, Xie M, Sun P, Fan R, Wang Z, Wang L, et al. 2019. TRIM29 facilitates the epithelial-to-mesenchymal transition and the progression of colorectal cancer via the activation of the Wnt/β-catenin signaling pathway. J Exp Clin Cancer Res 38: 104 10.1186/s13046-019-1098-y [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Szabó A, Mayor R. 2018. Mechanisms of neural crest migration. Annu Rev Genet 52: 43–63. 10.1146/annurev-genet-120417-031559 [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. 2017. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 45: D362–D368. 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam PKH, Garcia-Barceló M. 2009. Genetic basis of Hirschsprung's disease. Pediatr Surg Int 25: 543–558. 10.1007/s00383-009-2402-2 [DOI] [PubMed] [Google Scholar]
- Tam PKH, Tang CSM, Garcia-Barceló M-M. 2019. Genetics of Hirschsprung's disease. In Hirschsprung's disease and allied disorders (ed. Holschneider AM, Puri P), pp. 121–131. Springer-Verlag, Berlin, Heidelberg. [Google Scholar]
- Tang CSM, Gui H, Kapoor A, Kim JH, Luzón-Toro B, Pelet A, Burzynski G, Lantieri F, So MT, Berrios C, et al. 2016. Trans-ethnic meta-analysis of genome-wide association studies for Hirschsprung disease. Hum Mol Genet 25: 5265–5275. 10.1093/hmg/ddw333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CSM, Li P, Lai FPL, Fu AX, Lau ST, So MT, Lui KNC, Li Z, Zhuang X, Yu M, et al. 2018. Identification of genes associated with Hirschsprung disease, based on whole-genome sequence analysis, and potential effects on enteric nervous system development. Gastroenterology 155: 1908–1922.e5. 10.1053/j.gastro.2018.09.012 [DOI] [PubMed] [Google Scholar]
- Tchieu J, Calder EL, Guttikonda SR, Gutzwiller EM, Aromolaran KA, Steinbeck JA, Goldstein PA, Studer L. 2019. NFIA is a gliogenic switch enabling rapid derivation of functional human astrocytes from pluripotent stem cells. Nat Biotechnol 37: 267–275. 10.1038/s41587-019-0035-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R, Johnstone I, Hastie T, Efron B. 2004. Least angle regression. Ann Stat 32: 407–499. 10.1214/009053604000000067 [DOI] [Google Scholar]
- Tilghman JM, Ling AY, Turner TN, Sosa MX, Krumm N, Chatterjee S, Kapoor A, Coe BP, Nguyen K-DH, Gupta N, et al. 2019. Molecular genetic anatomy and risk profile of Hirschsprung's disease. N Engl J Med 380: 1421–1432. 10.1056/NEJMoa1706594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimbuch T, Beed P, Vogt J, Schuchmann S, Maier N, Kintscher M, Breustedt J, Schuelke M, Streu N, Kieselmann O, et al. 2009. Synaptic PRG-1 modulates excitatory transmission via lipid phosphate-mediated signaling. Cell 138: 1222–1235. 10.1016/j.cell.2009.06.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udassin R, Nissan S, Lernau O, Hod G. 1981. The mild form of Hirschsprung's disease (short segment): fourteen years experience in diagnosis and treatment. Ann Surg 194: 767–770. 10.1097/00000658-198112000-00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M, Graf R, MacWilliams HK, Schliwa M, Euteneuer U. 1997. Centrosome positioning and directionality of cell movements. Proc Natl Acad Sci 94: 9674–9678. 10.1073/pnas.94.18.9674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe RA, Hong SS, Bronner ME. 2018. Retinoic acid temporally orchestrates colonization of the gut by vagal neural crest cells. Dev Biol 433: 17–32. 10.1016/j.ydbio.2017.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadhvani M, Schwedhelm-Domeyer N, Mukherjee C, Stegmüller J. 2013. The centrosomal E3 ubiquitin ligase FBXO31-SCF regulates neuronal morphogenesis and migration. PLoS One 8: e57530 10.1371/journal.pone.0057530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varrault A, Dantec C, Le Digarcher A, Chotard L, Bilanges B, Parrinello H, Dubois E, Rialle S, Severac D, Bouschet T, et al. 2017. Identification of Plagl1/Zac1 binding sites and target genes establishes its role in the regulation of extracellular matrix genes and the imprinted gene network. Nucleic Acids Res 45: 10466–10480. 10.1093/nar/gkx672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan EM, Miller AL, Yu H-YE, Bement WM. 2011. Control of local Rho GTPase crosstalk by Abr. Curr Biol 21: 270–277. 10.1016/j.cub.2011.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Haw R. 2017. Functional interaction network construction and analysis for disease discovery. Methods Mol Biol 1558: 235–253. 10.1007/978-1-4939-6783-4_11 [DOI] [PubMed] [Google Scholar]
- Zhang F, Lupski JR. 2015. Non-coding genetic variants in human disease. Hum Mol Genet 24: R102–R110. 10.1093/hmg/ddv259 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.