Abstract
To effectively analyze the increasing amounts of available genomic data, improved comparative analytical tools that are accessible to and applicable by a broad scientific community are essential. We built the “2-n-way” software suite to provide a fundamental and innovative processing framework for revealing and comparing inserted elements among various genomes. The suite comprises two user-friendly web-based modules. The 2-way module generates pairwise whole-genome alignments of target and query species. The resulting genome coordinates of blocks (matching sequences) and gaps (missing sequences) from multiple 2-ways are then transferred to the n-way module and sorted into projects, in which user-defined coordinates from reference species are projected to the block/gap coordinates of orthologous loci in query species to provide comparative information about presence (blocks) or absence (gaps) patterns of targeted elements over many entire genomes and phylogroups. Thus, the 2-n-way software suite is ideal for performing multidirectional, non-ascertainment-biased screenings to extract all possible presence/absence data of user-relevant elements in orthologous sequences. To highlight its applicability and versatility, we used 2-n-way to expose approximately 100 lost introns in vertebrates, analyzed thousands of potential phylogenetically informative bat and whale retrotransposons, and novel human exons as well as thousands of human polymorphic retrotransposons.
At any given time, evolutionary changes leave behind their traces in the genomes of all beings. To read out evolutionary signals from the past and compare them among a variety of living species enables us to understand processes of life and transitions in evolution. With the ever-accumulating amounts of available genomic data (Stephens et al. 2015), including the forthcoming Earth-Life Biome repository of more than a million new eukaryotic genome sequences (Lewin et al. 2018), improved tools and methods of analytical comparison are urgently required by a broad scientific community, including bioinformaticians as well as basic biologists and students.
With the first working draft of the human genome, the Santa Cruz Genomics Institute at the University of California Santa Cruz (UCSC) published their first graphical Genome Browser (Kent et al. 2002), which has become a comprehensive visual representation of the prevailing vertebrate genome assemblies (https://genome.ucsc.edu). Building on the Santa Cruz Genome Browser, we developed the graphical screening and extraction tool Genome Presence/Absence Compiler (GPAC). Based on open access UCSC multiway genome alignments and a table of user-defined reference genome coordinates, a simple graphical list of the presence or absence status of thousands of inserted elements (e.g., retrotransposons, numts, npcRNAs, and so forth) are depicted comparatively in a compilation of query species (Noll et al. 2015). A direct link from GPAC to the UCSC Genome Browser provides deposited annotation information of any specific locus. The tool was initially conceived and applied to find phylogenetically informative retrotransposon markers, activity patterns of retrotransposons, and other genomic insertions/deletions (Schmitz et al. 2016), and it was recently used to systematically screen for retrotransposon presence/absence homoplasy cases (Doronina et al. 2019). However, one significant limitation of GPAC is the restricted availability of suitable multiway genome alignments. Multiway genome alignments require powerful computational processing, currently restricted to a few specialists such as those in the UCSC Genome Bioinformatics Group. Applications are therefore critically constrained to the prefabricated collection of multiway alignments provided at the download area of the UCSC Genome Institute and are limited to only one-directional screenings from a fixed reference genome (first genome of a multiway alignment). To take advantage of multidirectional screenings that are indispensable for comparative evolutionary projects, the preparation of several multiway alignments with different reference genomes is mandatory.
We have now developed 2-n-way, a software suite that is applicable independently of expensive computational equipment or bioinformatics expertise, very flexible and fast, and based on freely selectable combinations of multiple 2-way genome alignments. We designed the tool for easy use as an extremely powerful, web-interactive platform. Herein we show the power of 2-n-way with some prime examples spanning the scope of genome architecture, functional genomics, population genomics, and phylogenomics.
Results
From a well-characterized target genome, we selected coordinates of interest, for example, from all UCSC deposited human introns, to obtain the corresponding information about the presence or absence of such introns in query genomes. 2-n-way is organized into two parts. First, in a web-based interface, the user uploads genome FASTA data of one or more target and associated query genomes to generate sets of 2-way alignments. After submitting the data to a server, the user obtains a unique ID, and a request to generate the alignments is placed in a queue. The status of processing is available via the ID number and displayed after processing (the last 10 requests are stored in the ID selection box). Second, by request, newly generated 2-way alignments can then be compiled into private or public projects and placed in the n-way interface. The web-based user interface of n-way accepts the coordinates of target (i.e., reference species) sequences of interest in direct screenings, or query coordinate data in reverse screenings, to search for and extract orthologous sequences for project-related query sequences. The interactive results table enables the user to sort and select interesting loci for subsequent MUSCLE (Edgar 2004) alignment optimization and to save all relevant information in an Excel/plain text format as well as FASTA sequence alignments for detailed examination. The 2-n-way processing is visualized in Figure 1, and the application plus examples are described in the Supplemental Materials. Details of n-way parameters are presented in Supplemental Note S1 and Supplemental Figure S1, and graphical tutorials about 2-way, n-way, and possible results are presented online together with the tool and as Supplemental Figures S2–S4. To show the usefulness of the 2-n-way tool, we conducted several example searches, which are described subsequently. Details of example materials and methods are provided in Supplemental Note S2. All of the examples are also deposited as individual projects in n-way.
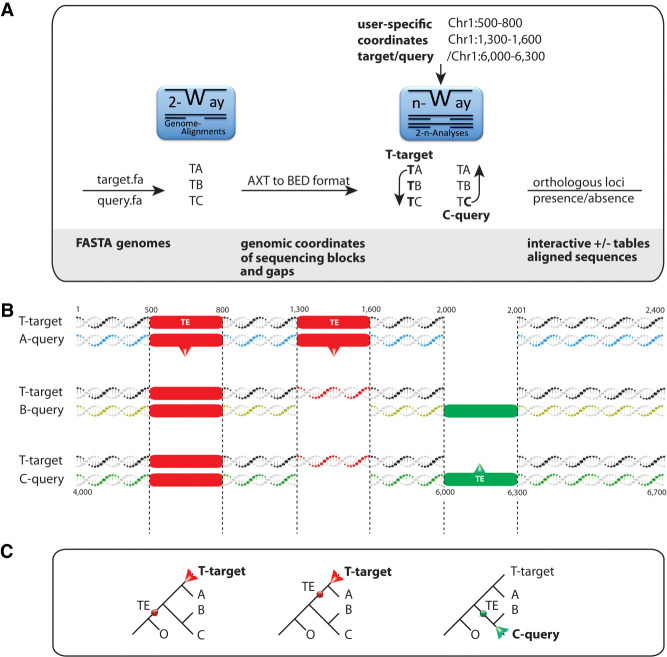
Figure 1.
The 2-n-way suite. (A) The workflow from genome data to 2-way genome alignments and extraction of presence/absence patterns. 2-way: Input for pairwise alignments consists of assembled genomes at the chromosome- or scaffold-level (FASTA genomes). We distinguish between target genome assemblies (T; target.fa) as leading sequences in the generation of 2-way alignments and the query genomes (A, B, C; query.fa). The 2-way module generates a list of files, including AXT-formatted (https://genome.ucsc.edu/FAQ/FAQformat.html) coordinates and sequences that will be transformed into BED-formatted coordinates that can then be transferred to the n-way module. N-way: In n-way, all combinations of 2-way alignments are screened for specific coordinates of sequences present in the target (direct search) or the query (reverse search). (B) Visualization of direct and reverse searches at the sequence level from pairwise aligned genomic regions with labeled coordinates of transposable elements. The green-labeled TE is not recognizable via the coordinates of the target T but is identified by the coordinates of query C, and its presence is subsequently screened for in query B (reverse search). (C) Phylogenetic representation of the two search strategies. The direct search for diagnostic presence/absence markers (red dots) via target T (red arrow) recognizes the first TE shared by T, A, B, and C and the second TE present only in the target T and query A from the RepeatMasker coordinate table derived from the target. The reverse screening starts from the presence of a TE from query C (green dot; green arrow) based on the RepeatMasker coordinate table of query C that would not be recognizable in direct search.
Carnivora: recovery of previously detected markers
To first verify its ability, thoroughness, and speed, we used 2-n-way to repeat a search for SINE and LINE phylogenetic markers to reconstruct the Arctoidea phylogeny (Doronina et al. 2015). Previously this search required months of semiautomatic screening and yielded 326 phylogenetically informative markers. 2-n-way enabled us to recover 323 of these in just 1 h. There were incomplete genome sequences at the loci of the three remaining cases.
Catarrhini: comparison of GPAC and 2-n-way
Currently, GPAC (Noll et al. 2015) is the only other tool available to systematically screen genome-wide for presence/absence loci. The crucial difference between GPAC and 2-n-way is that GPAC depends on complex multiway genome alignments, whereas 2-n-way generates multispecies comparisons via user-assigned combinations of 2-ways. In contrast to GPAC, 2-n-way allows flexible combinations of 2-way genome alignments to test all possible phylogenetic tree topologies. Furthermore, 2-n-way processes an unlimited number of input coordinates, whereas GPAC is restricted to about 160,000 coordinates. The processing time in 2-n-way is significantly shorter than in GPAC, for example, 2-n-way requires only 5 min (without MUSCLE-based optimization) for a data set of 100,000 coordinates compared to 4.5 h in GPAC. 2-n-way generates already aligned loci in FASTA format, whereas the output of GPAC is restricted to presence/absence tables. Only 2-n-way permits multiple, variable parameters to optimize screening procedures for phylogenetic and other markers such as exact intron losses. It also provides a MUSCLE realignment function to optimize presence/absence assignments and resultant sequences. In a direct comparison we used the genomes of human (hg38), chimpanzee (panTro5), and rhesus monkey (rheMac8) to detect diagnostic Alu insertions (39,583 loci in human, search for +−−/ ++−/+++). N-way re-found 94% of loci that were detected by GPAC (24,741 from 26,394) and found an additional 29% of the total loci that were not identified by GPAC (34,026 loci in n-way compared with 26,394 loci in GPAC). The 2-n-way process required 5 min (with MUSCLE-based optimization), while the GPAC search required 2 h.
Intron loss: abundance in mammals and birds
Eukaryotic intron loss (exact deletion of intronic sequences) is a well-known phenomenon at the genome level. As a consequence, the two neighboring exons merge, and intron splicing at this position is no longer possible. N-way is ideally suited to trace lost introns in query genomes compared to the presence of user-assigned introns in a target (reference) genome.
We performed n-way runs searching for introns that were lost in query species compared to 276,857 NCBI RefSeq introns located in human, 210,267 introns in mouse, 117,228 in cow, 13,326 in dog, 1545 in opossum, and 608,239 (Other RefSeq) introns in the zebra finch. This screening returned 315 potential intronless loci found from the human target (reference) genome, 130 from mouse, 133 from cow, 21 from dog, and eight from opossum. Manual checking of the orthology of the loci (Methods) revealed 87 clear cases of phylogenetically verified intron losses in mammals (Fig. 2, red balls; Supplemental Table S1; Supplemental Data S1).
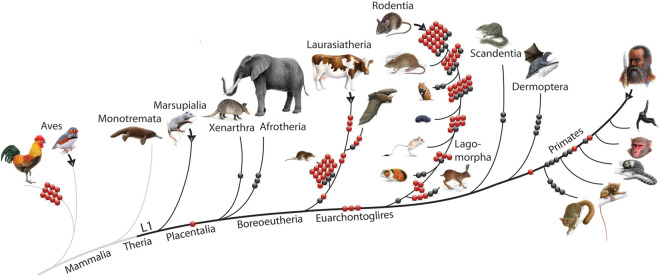
Figure 2.
2-n-way detected intron losses in representative mammals and birds. Red and black dots represent lost introns detected by 2-n-way. Red dots show introns whose loss was detected only once in evolution. Black dots represent the phylogenetic distribution of multiple intron losses in mammals. Black branches cohere with the activity of LINE-1 (L1) elements. Black arrows indicate the search directions starting in n-way from the coordinates for introns that are present and screening for intron loss in other lineages. The dog is not shown but was used to find intron losses in different taxa.
Of these loci, 35 were newly discovered cases, and 52 were previously detected (Coulombe-Huntington and Majewski 2007), but their exact phylogenetic positions were specified in the current research. We identified two prominent bursts of intron losses, in rodents and shrews, suggesting that a similar population dynamic/generation time exposed them both particularly frequently to losses of introns. Moreover, we detected 21 introns (three new and 18 reported previously) (Coulombe-Huntington and Majewski 2007), some from potential multifunctional moonlighting genes, that were lost by different animals more than once in mammalian evolution (Fig. 2, black balls; Supplemental Table S1), suggesting that some introns present a hotspot that can be especially easily lost, resulting today in a phylogenetic mosaic.
The n-way screening of zebra finch intron data sets revealed 533 potential intron losses. Of these, we verified 11 on the lineage leading to galliform birds (Supplemental Table S1; Supplemental Data S1) where intron loss has never been reported and known functional requirements for their loss are missing. The proposed classical mechanism of losing introns includes ectopic recombination between genes and intron-free retropseudogenes (Cohen et al. 2012). Retropseudogenes in therian mammals are processed by the LINE-1 (or L1) retrotransposition machinery that is largely absent in birds (Suh 2015). However, for several cases, we observed an ∼10-nt similarity between the 3′ ends of exons and introns that may have provided a hotspot for local recombination leading to intron loss. This finding may explain the unexpected number of intron losses in Galliformes in which just a few intronless retropseudogenes have been described (Hillier et al. 2004). A similar process referred to as microhomology-mediated intron loss has been illustrated for Drosophila and Caenorhabditis (van Schendel and Tijsterman 2013) but not yet described for amniotes.
2-n-way also enabled us to comparatively visualize intron loss between mouse and human. Intron losses were differently accumulated on chromosomes depending on their overall gene content; for example, many introns that are located on the gene-rich human Chromosome 19 were lost in the mouse lineage (Fig. 3).
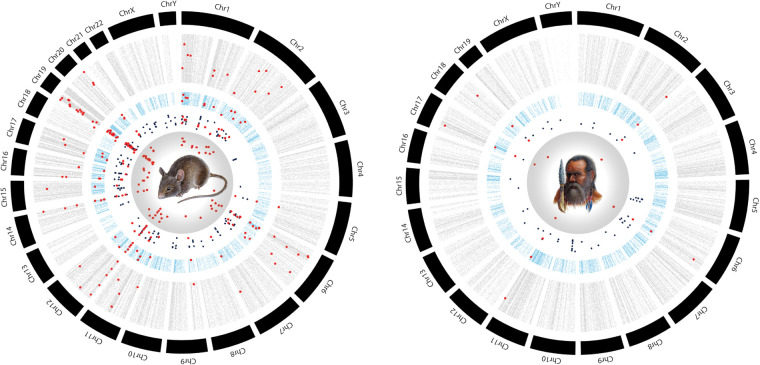
Figure 3.
Consecutive analysis steps for intron loss data sorted by human and mouse chromosomes (for methods, see Gu et al. 2014). The outer black shells show the chromosomal distributions of data points. The gray shells show all screened human introns for intron loss in mouse (left) or mouse introns for intron loss in humans (right). The blue shells represent potential intron losses extracted by 2-n-way under relaxed conditions (allowing perfect presence/absence, presence/presence, and semiperfect cases). The black dots in the white areas represent 2-n-way results from stringent selections of potential intron losses (perfect presence/absence cases). Red dots in the inner circles (containing the species illustrations) represent the manually verified candidate intron loss loci in mouse and human. The manually verified intron losses are highlighted in all the various processing areas.
Bats: ancestral lineage sorting and echolocation
Echolocating bats represent a paraphyletic group. The echolocating microchiropteran lineage Rhinolophoidea and the non-echolocating megachiropteran Pteropodidae merge in the monophyletic clade Yinpterochiroptera, whereas other echolocating microchiropteran bats form the monophyletic clade Yangochiroptera (Teeling et al. 2005). Analyses of 2083 individual gene trees (Hahn and Nakhleh 2016) revealed significant gene-tree/species-tree discordances (just ∼57% of analyzed gene trees supported the established species tree). The investigators suggested that the responsible noise in nucleotide sequence comparison stemmed from an ancestral period of intense incomplete lineage sorting (ILS).
To address this point, we used the 2-n-way tool to search for the presence/absence patterns of retrotransposed elements as phylogenetic markers. In a few hours, we analyzed 91,328 LINEs and 29,538 LTRs from the echolocating great roundleaf bat Hipposideros armiger (Rhinolophoidea, Yinpterochiroptera), 89,325 LINEs and 29,800 LTRs from the non-echolocating lesser dawn bat Eonycteris spelaea (Pteropodidae, Yinpterochiroptera), and 121,318 LINEs and 28,075 LTRs from the common vampire bat Desmodus rotundus (Phyllostomidae, Yangochiroptera). After manually rechecking the n-way results for orthology, we found 32 diagnostic markers supporting the Yinpterochiroptera monophyly (great roundleaf bat plus lesser dawn bat), one marker supporting a Pteropodidae-Yangochiroptera monophyly (lesser dawn bat plus common vampire bat), and no markers supporting the Rhinolophoidea-Yangochiroptera monophyly (great roundleaf bat plus common vampire bat) (Fig. 4A; Supplemental Table S2; Supplemental Data S2). The multidirectional KKSC test (Kuritzin et al. 2016) revealed significant support for the Yinpterochiroptera monophyly (32:1:0; P < 1.2 × 10−14). Thus, the 2-n-way algorithms revealed that 97% of the phylogenetic signals supported the current species tree, and only 3% of the markers supported a confounding tree topology.
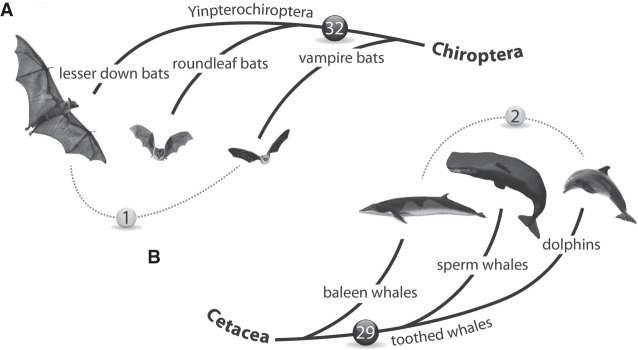
Figure 4.
Phylogenetic signals and ILS markers. (A) Thirty-two shared TE insertions in bat species support the monophyly of Yinpterochiroptera. A single conflicting insertion merging vampire bats with lesser down bats (dotted line) suggests the occurrence of ILS during the early evolution of bats. (B) Twenty-nine TE insertions in whale species support the monophyly of toothed whales. Nevertheless, two markers supporting the dolphin-baleen whale monophyly (dotted line) may indicate ancestral ILS or hybridization during the early evolution of whales.
Whales: incomplete lineage sorting at the root of the whale tree
Nikaido et al. (2007) investigated and divided ancestral diversifications of the whales into three lineages—baleen whales, sperm whales, and dolphins—using the SINE presence/absence marker system. Because they did not detect ILS in this relatively rapidly radiating group, they concluded that the fixation process of ancestral polymorphisms was prompted by the small size of ancestral whale populations. Using 2-n-way, we analyzed 1,396,816 CHR and CHRL SINEs (494,000 from bottlenose dolphin [Tursiops truncatus], 520,350 from sperm whale [Physeter macrocephalus], and 382,466 from baleen minke whale [Balaenoptera acutorostrata]). We extracted and manually checked loci with perfect presence/absence patterns from n-way and used a customized script to select loci with flanks nearly free of repetitive elements (<50 nt repeats). We found 29 diagnostic SINE markers supporting the monophyly of toothed whales (bottlenose dolphins plus sperm whale), two SINEs supporting the grouping of bottlenose dolphins plus minke whale, and zero supporting the sperm whale plus minke whale group (Fig. 4B; Supplemental Table S3; Supplemental Data S3). The multidirectional KKSC test (Kuritzin et al. 2016) significantly supported the monophyly of toothed whales (29:2:0; P < 3.1 × 10−12). Although the majority of the data from the lineages we investigated does support the monophyly of toothed whales (bottlenose dolphins plus sperm whales); nonetheless, in contrast to Nikaido et al. (2007), we did detect a small number of presence/absence markers supporting ancestral ILS that accompanied early whale speciation events following the relatively short diversification period of these whale groups.
Primates: novel exon gain
Exonization describes an evolutionary process of de novo exon formation from previously uninvolved genomic units such as TEs (Schrader and Schmitz 2019). Screening the human consensus CDS data (CCDS) (Pruitt et al. 2009) for the inclusion of Alu SINE TEs in introns of protein-coding genes, we detected 345 candidate exons to examine their TE origin and exonization history. In the coordinate-based n-way extraction, we analyzed these novel exons plus their TE-associated flanking regions from humans and compared them to other primate genomes to obtain a full recovery of the origin and evolutionary steps leading to novel exons and thereby significant novel variations in transcriptomes. Five as yet uncharacterized examples of primate loci (BIRC5, CCDC198, AZI2, BAHCC1, IFNAR2) with such possible exonizations are indicated in Supplemental Figure S5A (the alignments of the extracted loci are given in Supplemental Data S4). However, the phenotypic consequences of novel TE exon inclusions require further functional studies.
Human population: polymorphic TEs
Analyzing DNA polymorphism is the conventional gold standard in population genetic studies. The 2-n-way suite enables an additional track to visualize significant alternative variations in populations by screening for polymorphic patterns of unfixed TE insertions. For example, polymorphic TEs present in two human individuals at orthologous genomic positions, but absent in others, is a marker for their closer relationship. The increasing number of high-quality individual human genome assemblies derived from long-read sequencing technologies are the future of an additional mostly overlooked source of quick and reliable population genetic markers. The 2-n-way algorithm offers a way to analyze the frequency of polymorphic TE presence/absence patterns. Here, we uploaded two representative high-quality Pacific Biosciences (PacBio) and four Illumina HiSeq human genomes representing a worldwide distribution of polymorphic TE insertion patterns. From more than 100,000 Ape-specific AluY-TE insertion patterns, we detected 4093 human-specific insertions, 1665 of which were polymorphic. We also analyzed more than 5000 SINE-VNTR-Alu (SVA) retrotransposons, in which we identified 133 human-specific insertions and 33 polymorphic insertions. Furthermore, among approximately 175,000 primate-specific L1P element insertion patterns, 770 were human-specific and 161 were polymorphic. These examples strongly suggest that 2-n-way is a very powerful tool to extract genome-wide polymorphic TE markers for exhaustive population dynamic studies (Supplemental Fig. S5B; Supplemental Tables S4–S6).
Discussion
One obstacle in exploiting the unprecedented scale of information in depositories of genome data is that the amount and complexity of information is only fully accessible by highly experienced bioinformaticians and high-end computational equipment. With the 2-n-way suite, we provide a tool and the appropriate environment for any interested scientist to read, structure, extract, and visualize information of evolutionary changes encrypted in quasi-unlimited genomic space.
Initially, we designed the 2-n-way modules for phylogenomicists to access and visualize the diagnostic information of millions of genomic insertions/deletions. A preliminary offline version of 2-n-way had already proved exceptionally powerful at deciphering retrophylogenomic signals in the speciation network of the laurasiatherian orders Chiroptera, Perissodactyla, Cetartiodactyla, and Carnivora (Doronina et al. 2017).
Because parallel insertions of retrotransposons in two lineages at exactly the same genomic position are vanishingly rare, retroelement presence/absence markers are virtually homoplasy free (Doronina et al. 2019). Therefore, retroelement insertions as phylogenetic markers have the potential to build exceptionally reliable phylogenetic reconstructions. Nevertheless, discordant markers such as present in the laurasiatherian tree often lead to conflicting tree reconstructions afflicted by hemiplasy arising from ancestral ILS owing to ancestral polymorphic insertions. 2-n-way contains very versatile settings to recover the large numbers of presence/absence markers required, for example, for multidirectional coalescence-based analyses to reconstruct correct species trees (Springer et al. 2020). 2-n-way outshines all previous presence/absence screening methods. It showed nearly 100% success in a repeat search for TE markers previously described and required very little time. In an hour and with default options, we recovered, for example, 323 of 326 phylogenetically diagnostic markers from carnivore mammals that previously required months of semicomputational work (Doronina et al. 2015).
Compared with GPAC, 2-n-way represents an advanced and flexible tool independent of complex, predesigned, multiway alignments that enables the user to investigate individual and newly sequenced genomic data and requires only moderate computer power. Our comparative test of 2-n-way versus GPAC revealed that n-way was able to find 29% more Alu markers for human, chimpanzee, and rhesus monkey, and in a much shorter time than GPAC (5 min vs. 2 h).
We advocate that 2-n-way is ideally suited to straightforwardly test phylogenetic hypotheses using the virtually homoplasy-free insertion patterns of retrotransposed elements. As exemplified for bats, in a few hours we showed that homoplastic sequence data was the real reason for conflicting sequence-based phylogenetic reconstructions and not hemiplasy triggered by ancestral ILS as formerly predicted by Hahn and Nakhleh (2016). To confirm the expected absence of ILS in specific whale clades (Nikaido et al. 2007), we reconstructed, again in just hours, whale phylogeny based on TE presence/absence states and found two previously undiscovered ILS markers along with the 35 phylogenetically diagnostic loci; the latter significantly supporting the monophyly of toothed whales merging dolphins and sperm whales. The two conflicting markers clustering baleen whales and dolphins are probably an attribute of the expected short divergence time of the major lineages of extant whales and were only apparent from the high-throughput 2-n-way screening. On the other hand, 2-n-way easily detects polymorphic markers that should be excluded from phylogenetic investigations but do provide unique, valuable genome-wide data sets to investigate population structures. Preliminary screening of diagnostic polymorphic Alu, SVA, and L1P elements in human individuals revealed about 2000 of them distributed over the entire genomes. 2-n-way is not only excellently suited to detect the distribution of phylogenetic or population markers but also to extract unlimited numbers of orthologous genomic locations among organisms via a list of coordinates. The resulting FASTA sequences can be extracted as a reliable MUSCLE-based alignment.
To show the power of 2-n-way to analyze features of gene architecture, we screened the genomes of mammalian and bird species for lost introns. 2-n-way retrieved all previously detected and published cases of intron loss in selected mammalian species groups (Coulombe-Huntington and Majewski 2007) and discovered many more. Moreover, we were also able to detect the first reported incidences of intron loss in birds, an animal class that was thought to be free of retropseudogene–gene recombinations. Such recombinations are based on LINE-1 mobilized retropseudogenes (processed intron-less cDNA) and are considered to be responsible for intron loss in therian mammals. However, LINE-1s are absent or at most rare in birds. A possible explanation for this paradox might be microhomology-mediated intron losses (van Schendel and Tijsterman 2013).
We also used 2-n-way to produce the first comparative visualization of intron plasticity measured by intron loss for the mouse with its very short generation time and large effective population sizes versus human with long generations and small effective population size. Moreover, these intron losses were unequally distributed over chromosomes corresponding to their overall gene content (Fig. 3). Nevertheless, in addition to the effects of generation times and population sizes, the activity of L1 reverse transcriptase is an expected important mediator of intron losses (Coulombe-Huntington and Majewski 2007). However, the overall fast rate of genomic changes in mice might be decisive in the extreme difference seen in the data shown in Figure 3.
Novel exon gain via exonization of TEs (Schrader and Schmitz 2019) shows another application field that, with high-throughput n-way screenings, will shed new light on the emergence and evolution of novel protein-coding sequence regions (Supplemental Fig. S5A). The five selected cases represent two different processes of exonization: (1) that which took place immediately after insertion, in which all requirements for exon gain were already available (e.g., splice sites, polypyrimidine tract in a favorable position), and these are responsible for BIRC5, CCDC198, and AZL2; and (2) that which took place millions of years after insertion, in which not all required splice components were available at the time of insertion but did evolve subsequently. This is apparent in the case of BAHCC1 (insertion in Haplorrhini; exonization in Catarrhini) and FNAR2 (insertion in ancestral primates; exonization in Simians).
2-n-way can be used to compare any kind of defined genomic sequence type, including lncRNAs, tRNAs, snRNAs, rRNAs, snoRNAs, miRNA, viral components, and numts. In addition, 2-n-way provides a new analytical perspective for TE population-based studies. For example, initial analyses of 280,000 primate-specific TEs from six individual human genomes representing a worldwide distribution revealed more than 1850 polymorphic insertions ready for population dynamic studies. The nanopore sequencing technique with its ultralong but still error-prone sequence reads makes presence/absence analyses very efficient for phylogenetic population genomic studies, whereas the sequence-accuracy is less critical to determine diagnostic presence/absence loci.
The innovative concept of 2-n-way is that an enormous number of relatively simple to build individual components (2-ways) are combined to enable highly complex, multilocus, multispecies comparisons (n-way) based on moderate computer power. Users need no specialized knowledge of bioinformatics or data management and can select project-oriented, 2-way genome alignments to combine them with individual projects. A set of parameters enables one to fine-tune the detection process of presence/absence signals at orthologous loci. However, users should keep in mind that presence/absence tables and retrieved alignments may contain some noise produced by LASTZ or LAST (Harris 2007) as well as, albeit to a lesser extent, by MUSCLE (Edgar 2004). Therefore, it is essential that each determined locus be verified manually (Doronina et al. 2017).
2-n-way is also uniquely suited for phylogenomic projects involving DNA of extinct species. We have shown that ancient genomes are in principle well suited to analysis using the retrotransposon presence/absence strategy to derive highly reliable presence/absence markers for phylogeny (Feigin et al. 2018); 2-n-way provides the possibility to integrate medium- to high-quality ancient assemblies as targets as well as low-quality sequences as query genomes to derive the phylogenomic history of such species. Although most of the examples we present here are from highly complex genomes, it should be mentioned that 2-n-way is universally applicable for any grade of genome complexity.
Currently, the tool is running on several servers, and some of the operations/data are transferred to client computers. The 2-way whole-genome alignments are currently the most computationally intense and time-consuming steps handled in a queue system and, depending on the genome quality, can take some days. We will be continuously enhancing and updating the tool and developing novel strategies to reduce the running time on computer clusters.
Methods
A general obstacle to software design for molecular biological genome data is that it requires enormous resources for processing and storage on digital devices. Visualizing the results of analyzing such complex information is challenging and requires simplification. We optimized and fine-tuned the following processes to provide unique opportunities in searching for and sorting of evolutionarily informative units from an infinite multiple genome data space.
2-way whole-genome alignments
Comparative genome studies require reliable, fast-working tools to align large numbers of sequences. Interspersed repeats and low complexity regions of the target (alignment leading) reference genomes especially impede the alignment process and accidentally generate large numbers of nonhomologous alignments. Therefore, genomes must be initially masked (soft-masked repeats in lowercase letters), for example, via local repeat masking using an optimized species library of known repeats (http://www.repeatmasker.org/RMDownload.html). Basic tools of our 2-way aligner program are LASTZ version 1.04.00 (Harris 2007) and LAST (Kielbasa et al. 2011) that implement pairwise whole-genome alignments (https://lastz.github.io/lastz; March 2017; https://github.com/mcfrith/last-genome-alignments; July 2017). Typical INPUT is (1) chromosome or scaffold sequences of the soft-masked target genome, (2) chromosome or scaffold sequences of the optionally soft-masked query genome, and (3) an optional cutoff value of short sequences. Additional input parameters are (1) minimum block length (default 10 nt), (2) minimum gap size (default 30 nt), (3) maximum gap size (default 100,000 nt), and (4) maximum gap overlap (default 25 nt). The size distribution of genome data is visualized to customize cutoff values. Each request receives a personalized ID (in the left panel, Your Previous Runs/Views) (Supplemental Fig. S2), and data are retained for 2 mo. Public or non-public external soft-masked genomes of Targets and Queries can be accessed via pulldown menus or uploaded in FASTA format. A run can take from several hours up to a few days depending on the size, quality, and complexity of the investigated genomes. Highly fragmented target genome contigs containing more than 500,000 individual FASTA records (after, e.g., cutoff <100 nt) cannot currently be considered for generating 2-way alignments (2-ways) owing to their extremely long processing times. However, on request we can assist the user in processing such low-quality assemblies on our cluster of servers. The alignment progression is indicated in percentage, and a run can always be terminated by the user.
2-way output files
The following results generated by the 2-way application are downloadable: (1) target-query information file, (2) AXT-, (3) block-, (4) gap-BED files, (5) corresponding FASTA files for target and query, and (6) size files for target and query (for detailed definitions of AXT and BED, see https://genome.ucsc.edu/FAQ/FAQformat.html). The block-BED files contain all chromosomal coordinates of alignment blocks (sequence regions present in both genomes of a 2-way); the Gap-BED files contain all coordinates of gaps in alignments (sequence regions present only in one genome of a 2-way). An overview of the block and gap length distributions can be visualized. To use the newly generated 2-way alignments in n-way for 2-n-way combinatory analyses, a transfer of the block- and gap-BED files from 2-way to n-way is required and will be accomplished on user request.
N-way whole-genome alignment analysis
Because n-way alignments (n-ways) are conveyed in one step to the browser of the client computer, we recommend 8 GB RAM on client computers for very large data sets. Only the transfer of final FASTA sequences and RepeatMasker files requires additional transmissions from the server. Client computer analyses can undertake such tasks as sorting and selecting of investigated cases easily and independently from the repeated interchange with the remote server. Predesigned 2-ways can be selected from a public, target-directed, pulldown menu. Alternatively, the user can request to transfer their own 2-way alignments for public or personalized visualization in the n-way module.
In the next step, the coordinates of blocks and gaps (BED files) from the 2-ways generated above are transferred to the n-way part of the suite and sorted into projects. Each project contains 2-ways of at least one target and one or several target-associated queries. For example, the project “intron loss” (presented as an example) currently contains seven vertebrate genome-based targets and 47 target-associated queries. Target-associated queries (2-ways) become selectable for specific project groups (e.g., the target human in the “intron loss” project releases a list of associated primate and other mammalian queries/2-ways). The information about the genomes used (aliases of the species, Latin names, genome versions, and links) is provided in a “Species Overview” menu tab. Sequences for alignments are extracted via coordinate information from an established local genome database.
In n-way, two types of search strategies can be performed. In direct search, n-way searches for the presence or absence of given elements among the selected target genome coordinates. Consequently, the target species shows the presence (+) state, and queries can vary from presence (+), absence (−), or unidentified (N) (for further symbols of semiperfect results, see below). In reverse search, the screening is performed on selected coordinates of a chosen query, which in turn, represents the presence state, and the target is the absence state (target presence loci are excluded). The algorithm of reverse search implies that, based on the coordinates of a chosen query, the coordinates of targets are identified, and based on the target coordinates and the information of the length of the insert in the selected query, the presence or absence states in other queries are identified.
Any space- or tab-separated table of the structure—(1) name of species (alias; optional for direct search, obligatory for reverse search), (2) chromosome/scaffold number, (3) start coordinate, (4) end coordinate, and (5) optional information—can be uploaded for various genome coordinates in the field File (maximum file size 2 GB) or copied into the Reference genome coordinate field (maximum text size 2 MB). RepeatMasker report files can be uploaded to the File field without modifications. Pregenerated RepeatMasker reports stored on the n-way server are also available from the pulldown menu Server RM File. Uploaded RepeatMasker out-files will generate a selectable list of transposable element categories (e.g., SINE/Alu) and entered subfamilies (e.g., AluY, and so forth). During the process, n-way data are filtered for duplicated coordinates. Depending on the complexity of the search (roughly represented by the number of analyzed coordinates and species), the results are available in a couple of minutes or a few hours. For jobs in progress, a project can be restricted to specific IP addresses.
To improve critical misalignments that are occasionally caused in the LASTZ or LAST programs by short indels, we implemented a multiple sequence comparison by log-expectation (MUSCLE) (Edgar 2004) regional multiple sequences realigning option with subsequent reanalysis of the presence/absence state of sequences for highly accurate results of 1–1000 selected loci (the MUSCLE-based optimization option is applied after the initial n-way run). In addition, the user can select the MUSCLE-based optimization option for a specific size range of target/query sequences in the Muscle Based Parameter section for complete n-way runs. However, this will increase the n-way running time. In direct search, double symbols (+?, −?, +−, ++, −−) represent potentially informative semiperfect loci. The symbols (+?) or (−?) represent complicated cases at the boundary of the parameter range; (+−) represents incomplete presence/absence patterns; (++) indicates that the query sequence is significantly different in size than the target; and (−−) represents shorter query sequences aligned to BED-inserts that lack flanking sequences. To fine-tune n-way runs, a set of parameters is described in Supplemental Note S1 and Supplemental Figure S1. The tool can also be used to extract any other locus plus its flanking regions (e.g., genes present and conserved in all investigated species) from a suitable list of genome coordinates.
Gap search method
The preset-insert parameter denotes user-defined input coordinates of interest; for example, RepeatMasker coordinates from the Target (or Query) genome. BED-insert denotes the LASTZ- or LAST-generated insert in the derived 2-way alignment (one species sequence insert compared with a gap in the second species). Depending on the quality of alignments and the distribution of evolutionary changes, in ideal cases the preset-insert is identical to the BED-insert. We distinguish two search strategies. The Distance method uses stringent searches for minimal differences in boundary coordinates between target-insert and BED-insert regions, for example, to select the best candidates from large numbers of retrotransposon inserts, or exact intron/exon gain/loss detection. The Overlap method uses a relaxed search that allows some drift in boundary coordinates between target- and BED-inserts toward the center of the BED-insert region (by default maximal 30% from the BED-insert length). All parameters are described and presented graphically in the Parameter Tutorial (Supplemental Note S1; Supplemental Fig. S1).
N-way output files
Inspired by the user-friendly GPAC graphical interface (Noll et al. 2015), the n-way output is compiled in an interactive table with up to several thousand extractable orthologous loci with (1) the reference (target) name and coordinates, (2) the element screened for its presence/absence status, and (3) the list of species with + or − symbols for presence/absence status, respectively, of the focused elements. N represents unaligned regions in the corresponding query. With the option Display Perfect, only clear presence/absence patterns (+ or −) are visualized and can be sorted, for example, for (−), which rearranges n-way tables so that the lines with (−) are at the top. All patterns of interest can be labeled (Selected Rows) and extracted for further investigations. Up to 1000 selected rows can be automatically realigned by MUSCLE, and a corrected table will be displayed with bold red symbols for the changed presence/absence stages. For adjusted results, interpretations are based on the relative nucleotide density in gap regions of queries and targets. After correction, the symbols (+) and (−) indicate clear presence and absence patterns, respectively; double symbols (+? or −?) indicate possible presence or absence of a given element but with a different number of nucleotides in the BED-insert region of the corresponding queries. The table can be exported to Excel or displayed in a raw tabular format (Export to Excel/Download Table Data), and FASTA sequences/alignments of target, and all selected query species can be downloaded for subsequent processing (Download Fasta/MUSCLE). All sequences represent the same orientation.
Manual checking of presence/absence cases
After extracting hundreds of selected potentially diagnostic loci (duplicated regions of the target are automatically removed by n-way), it is essential and mandatory to make a final locus-by-locus check of the alignments and to inspect the loci to ascertain that true orthology, based on identical insertion sites, exists (e.g., Doronina et al. 2017). In particular in the intron loss searches described here, cases were only accepted as informative when the intron was completely absent in at least two closely related species (to avoid individual assembly errors) and the flanking exons were intact. For shared TE insertions, we checked for the same orientation and identical element types in orthologous positions and allowed no traces of the element in the absence state.
Software availability
The new web tool that includes the presented examples as individual projects for practice and reproduction may be found at http://retrogenomics.uni-muenster.de/tools/twoway (2-way) and http://retrogenomics.uni-muenster.de/tools/nway (n-way). All source codes are uploaded to the Supplemental Material as Supplemental Codes (twoway, nway).
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank Marsha Bundman for editorial assistance and Jon Baldur Hlidberg for the animal paintings; we also thank David Ray and his team for testing preliminary versions of 2-n-way. This work was supported by the Deutsche Forschungsgemeinschaft (DFG) grant number SCHM 1469/10-1 (to J.S.) and DFG grant number 281125614/GRK2220 to the Research Training Group Evolutionary Processes in Adaptation and Disease (EvoPAD).
Author contributions: J.S. and G.C. conceived the 2-n-way project. G.C. developed and optimized the n-way strategy. F.Z. processed and integrated the 2-way alignment procedure, and A.N. was responsible for the initial computerization of LASTZ-based alignments. N.G. assembled 2-n-way into the current web-interface and built the server-based interactive framework. L.D. and G.C. selected and performed the sample applications and intensively tested 2-n-way. W.M. provided the initial computer net for test-runs. J.S. and L.D. wrote the paper with input from all authors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.262261.120.
References
- Cohen NE, Shen R, Carmel L. 2012. The role of reverse transcriptase in intron gain and loss mechanisms. Mol Biol Evol 29: 179–186. 10.1093/molbev/msr192 [DOI] [PubMed] [Google Scholar]
- Coulombe-Huntington J, Majewski J. 2007. Characterization of intron loss events in mammals. Genome Res 17: 23–32. 10.1101/gr.5703406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doronina L, Churakov G, Shi J, Brosius J, Baertsch R, Clawson H, Schmitz J. 2015. Exploring massive incomplete lineage sorting in arctoids (Laurasiatheria, Carnivora). Mol Biol Evol 32: 3194–3204. 10.1093/molbev/msv188 [DOI] [PubMed] [Google Scholar]
- Doronina L, Churakov G, Kuritzin A, Shi J, Baertsch R, Clawson H, Schmitz J. 2017. Speciation network in Laurasiatheria: retrophylogenomic signals. Genome Res 27: 997–1003. 10.1101/gr.210948.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doronina L, Reising O, Clawson H, Ray DA, Schmitz J. 2019. True homoplasy of retrotransposon insertions in primates. Sys Biol 68: 482–493. 10.1093/sysbio/syy076 [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinf 5: 113 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigin CY, Newton AH, Doronina L, Schmitz J, Hipsley CA, Mitchell KJ, Gower G, Llamas B, Soubrier J, Heider TN, et al. 2018. Genome of the Tasmanian tiger provides insights into the evolution and demography of an extinct marsupial carnivore. Nat Ecol Evol 2: 182–192. 10.1038/s41559-017-0417-y [DOI] [PubMed] [Google Scholar]
- Gu Z, Gu L, Eils R, Schlesner M, Brors B. 2014. circlize implements and enhances circular visualization in R. Bioinformatics 30: 2811–2812. 10.1093/bioinformatics/btu393 [DOI] [PubMed] [Google Scholar]
- Hahn MW, Nakhleh L. 2016. Irrational exuberance for resolved species trees. Evolution; Int J Org Evolution 70: 7–17. 10.1111/evo.12832 [DOI] [PubMed] [Google Scholar]
- Harris RS. 2007. “Improved pairwise alignment of genomic DNA.” PhD thesis, Pennsylvania State University, University Park, PA. [Google Scholar]
- Hillier LW, Miller W, Birney E, Warren W, Hardison RC, Ponting CP, Bork P, Burt DW, Groenen MA, Delany ME, et al. 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432: 695–716. 10.1038/nature03154 [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The Human Genome Browser at UCSC. Genome Res 12: 996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielbasa S, Wan R, Sato K, Horton P, Frith MC. 2011. Adaptive seeds tame genomic sequence comparison. Genome Res 21: 487–493. 10.1101/gr.113985.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuritzin A, Kischka T, Schmitz J, Churakov G. 2016. Incomplete lineage sorting and hybridization statistics for large-scale retroposon insertion data. PLoS Comp Biol 12: e1004812 10.1371/journal.pcbi.1004812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin HA, Robinson GE, Kress WJ, Baker WJ, Coddington J, Crandall KA, Durbin R, Edwards SV, Forest F, Gilbert MTP, et al. 2018. Earth BioGenome project: sequencing life for the future of life. Proc Natl Acad Sci 115: 4325–4333. 10.1073/pnas.1720115115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido M, Piskurek O, Okada N. 2007. Toothed whale monophyly reassessed by SINE insertion analysis: the absence of lineage sorting effects suggests a small population of a common ancestral species. Mol Phylogenet Evol 43: 216–224. 10.1016/j.ympev.2006.08.005 [DOI] [PubMed] [Google Scholar]
- Noll A, Grundmann N, Churakov G, Brosius J, Makałowski W, Schmitz J. 2015. GPAC-genome presence/absence compiler: a web application to comparatively visualize multiple genome-level changes. Mol Biol Evol 32: 275–286. 10.1093/molbev/msu276 [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, et al. 2009. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res 19: 1316–1323. 10.1101/gr.080531.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J, Noll A, Raabe CA, Churakov G, Voss R, Kiefmann M, Rozhdestvensky T, Brosius J, Baertsch R, Clawson H, et al. 2016. Genome sequence of the basal haplorrhine primate Tarsius syrichta reveals unusual insertions. Nat Commun 7: 12997 10.1038/ncomms12997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader L, Schmitz J. 2019. The impact of transposable elements in adaptive evolution. Mol Ecol 28: 1537–1549. 10.1111/mec.14794 [DOI] [PubMed] [Google Scholar]
- Springer MS, Molloy EK, Sloan DB, Simmons MP, Gatesy J. 2020. ILS-aware analysis of low-homoplasy retroelement insertions: inference of species trees and introgression using quartets. J. Hered 111: 147–168. 10.1093/jhered/esz076 [DOI] [PubMed] [Google Scholar]
- Stephens ZD, Lee SY, Faghri F, Campbell RH, Zhai C, Efron MJ, Iyer R, Schatz MC, Sinha S, Robinson GE. 2015. Big data: astronomical or genomical? PLoS Biol 13: e1002195 10.1371/journal.pbio.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh A. 2015. The specific requirements for CR1 retrotransposition explain the scarcity of retrogenes in birds. J Mol Evol 81: 18–20. 10.1007/s00239-015-9692-x [DOI] [PubMed] [Google Scholar]
- Teeling EC, Springer MS, Madsen O, Bates P, O'Brien SJ, Murphy WJ. 2005. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 307: 580–584. 10.1126/science.1105113 [DOI] [PubMed] [Google Scholar]
- van Schendel R, Tijsterman M. 2013. Microhomology-mediated intron loss during metazoan evolution. Genome Biol Evol 5: 1212–1219. 10.1093/gbe/evt088 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.