Abstract
Mosquito control remains a central pillar of efforts to reduce malaria burden in sub-Saharan Africa. However, insecticide resistance is entrenched in malaria vector populations, and countries with a high malaria burden face a daunting challenge to sustain malaria control with a limited set of surveillance and intervention tools. Here we report on the second phase of a project to build an open resource of high-quality data on genome variation among natural populations of the major African malaria vector species Anopheles gambiae and Anopheles coluzzii. We analyzed whole genomes of 1142 individual mosquitoes sampled from the wild in 13 African countries, as well as a further 234 individuals comprising parents and progeny of 11 laboratory crosses. The data resource includes high-confidence single-nucleotide polymorphism (SNP) calls at 57 million variable sites, genome-wide copy number variation (CNV) calls, and haplotypes phased at biallelic SNPs. We use these data to analyze genetic population structure and characterize genetic diversity within and between populations. We illustrate the utility of these data by investigating species differences in isolation by distance, genetic variation within proposed gene drive target sequences, and patterns of resistance to pyrethroid insecticides. This data resource provides a foundation for developing new operational systems for molecular surveillance and for accelerating research and development of new vector control tools. It also provides a unique resource for the study of population genomics and evolutionary biology in eukaryotic species with high levels of genetic diversity under strong anthropogenic evolutionary pressures.
The 10 countries with the highest malaria burden in Africa account for 65% of all malaria cases globally, and attempts to reduce that burden further are stalling in the face of significant challenges (World Health Organization 2019). Not least among these, resistance to pyrethroid insecticides is widespread throughout African malaria mosquito populations, potentially compromising the efficacy of mosquito control interventions, which remain a core tenet of global malaria strategy (Hemingway et al. 2016; World Health Organization 2018). There is a broad consensus that further progress cannot be made if interventions are applied blindly, but must instead be guided by data from epidemiological and entomological surveillance (World Health Organization 2015). Genome sequencing technologies are considered to be a key component of future malaria surveillance systems, providing new insights into evolutionary and demographic events in mosquito and parasite populations (Ishengoma et al. 2019). Genomic surveillance systems cannot work in isolation, however, and depend on high-quality open genomic data resources, including baseline data on genome variation from multiple mosquito species and geographical locations, against which comparisons can be made and inferences regarding new events can be drawn.
Better surveillance can increase the impact and longevity of current mosquito control tools, but sustaining malaria control will also require the development of new tools (World Health Organization 2015). This includes repurposing existing insecticides from agriculture (Lees et al. 2019; Oxborough et al. 2019), developing entirely new insecticide classes, and developing tools that do not rely on insecticides, such as genetic modification of mosquito populations (Kyrou et al. 2018). The research and development of new mosquito control tools has been greatly facilitated by the availability of high-quality open genomic data resources, including genome assemblies (Holt et al. 2002; Sharakhova et al. 2007), annotations (Giraldo-Calderón et al. 2015), and, more recently, data on genetic variation in natural mosquito populations (The Anopheles gambiae 1000 Genomes Consortium 2017). Further expansion of these open data resources to incorporate unsampled mosquito populations and new types of genetic variation can provide new insights into a range of biological and ecological processes and help to further accelerate scientific discovery and applied research.
The Anopheles gambiae 1000 Genomes (Ag1000G) Project (https://www.malariagen.net/projects/ag1000g) was established in 2013 to build a large-scale open data resource on natural genetic variation in malaria mosquito populations. The Ag1000G Project forms part of the Malaria Genomic Epidemiology Network (MalariaGEN) (https://www.malariagen.net), a data-sharing community of researchers investigating how genetic variation in humans, mosquitoes, and malaria parasites can inform the biology, epidemiology, and control of malaria. The first phase of the Ag1000G Project released data from whole-genome Illumina deep sequencing of the major Afrotropical malaria vector species Anopheles gambiae and Anopheles coluzzii (The Anopheles gambiae 1000 Genomes Consortium 2017), two closely related siblings within the A. gambiae species complex (Coetzee et al. 2013). Mosquitoes were sampled in eight African countries from a broad geographical range, spanning Guinea-Bissau in the west to Kenya in the east. Genetic diversity was found to be high in most populations, and there were marked patterns of population structure, with some clear differences between populations in the magnitude and architecture of genetic diversity, indicating complex and varied demographic histories. However, many countries and ecological settings are not represented in the Ag1000G phase 1 resource. Also, only single-nucleotide polymorphisms (SNPs) were studied in phase 1, but other types of genetic variation are known to be important. In particular, copy number variation (CNV) has long been suspected to play a key role in insecticide resistance (Schimke et al. 1978; Devonshire and Field 1991; Weetman et al. 2015), but no previous attempts to call genome-wide CNVs have been made in these species. The Ag1000G Project also aims to provide a data resource of broad utility for the study of eukaryotic population genomics and evolutionary biology. In the first project phase, we found that nucleotide diversity among the mosquito populations we sampled was almost twice that reported for African populations of Drosophila melanogaster (Lack et al. 2016) and 10 times greater than modern human populations (Leffler et al. 2012). Among arthropods, the only data resource of comparable scale remains the Drosophila Genome Nexus, which has compiled and standardized genomic data from sampling of natural populations across the species ancestral range in sub-Saharan Africa, as well as naturalized populations in Europe and North America (Lack et al. 2016). Although Anopheles and Drosophila are both dipteran insects, there are fundamental differences in their biology and life histories, making a valuable comparison. Among other eukaryotes, comparable data resources exist only for humans (The 1000 Genomes Project Consortium 2015) and malaria parasites (MalariaGEN Plasmodium falciparum Community Project 2019), and thus, there remains an absence of open genomic data for studying demographic and evolutionary processes in natural populations. Furthermore, although many species have undoubtedly been exposed to new evolutionary pressures of anthropogenic origin, few species have been subject to such an intense and directed campaign of attack as malaria-transmitting mosquitoes. This year has seen the delivery of the 2 billionth insecticide-treated bed net in Africa, and programs of indoor residual spraying of insecticides protect more than 20 million people each year (Tangena et al. 2020). Thus whole-genome sequencing of Anopheles mosquitoes offers the opportunity to observe an evolutionary experiment on a continental scale.
This paper describes the data resource produced by the second phase of the Ag1000G Project. In this phase, sampling and sequencing were expanded to include wild-caught mosquitoes from five additional countries. This includes three new locations with A. coluzzii, providing greater power for genetic comparisons with A. gambiae, and two island populations, providing a useful reference point to compare against mainland populations. Seven new laboratory crosses are also included, providing a substantial resource for studying genome variation and recombination within known pedigrees. In this phase, we studied both SNPs and CNVs and rebuilt a haplotype reference panel using all wild-caught specimens. Here we describe the data resource and use it to re-evaluate major population divisions and characterize genetic diversity. We also illustrate the broad utility of the data by comparing geographical population structure between the two mosquito species to investigate evidence for differences in dispersal behavior, by analyzing genetic diversity within a gene in the sex-determination pathway currently targeted for gene drive development, and by providing some preliminary insights into the prevalence of different molecular mechanisms of pyrethroid resistance.
Results
Population sampling and sequencing
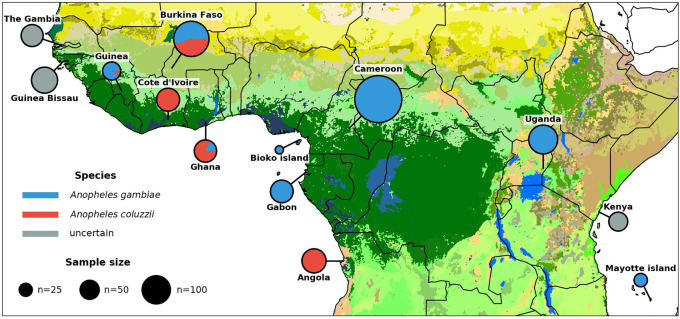
We performed whole-genome sequencing of 377 individual wild-caught mosquitoes, including three countries (The Gambia, Côte d'Ivoire, Ghana) and two oceanic islands (Bioko, Mayotte) not represented in the previous project phase. These individuals were collected using a variety of methods and at a different life stages (see Methods). We also sequenced 152 individuals comprising parents and progeny from seven laboratory crosses. We then combined these data with sequencing data from phase 1 of the project to create a total resource of data from 1142 wild-caught mosquitoes (1058 female, 84 male) from 13 countries (Fig. 1; Supplemental Table S1) and 234 mosquitoes from 11 laboratory crosses (Supplemental Table S2). All mosquitoes were sequenced individually on Illumina technology using 100-bp paired-end reads to a target depth of 30×. The median genome-wide coverage obtained across all 1142 wild-caught mosquitoes included in the final resource was 31×, with a minimum of 14×. Among these samples, on average 90% of the reference genome obtained at least 1× coverage, 82% at least 10×, and 68% at least 20×.
Figure 1.
Ag1000G phase 2 sampling locations. Color of circle denotes species, and area represents sample size. Species assignment is labeled as uncertain for samples from Guinea-Bissau, The Gambia, and Kenya because all individuals from those locations carry a mixture of A. gambiae and A. coluzzii ancestry informative markers (Supplemental Fig. S1). Map colors represent ecosystem classes; dark green designates forest ecosystems. For a compete color legend see Figure 9 in the work of Sayre (2013).
Genome variation
Sequence reads were aligned to the AgamP3 reference genome (Holt et al. 2002; Sharakhova et al. 2007), and SNPs were discovered using methods described previously (The Anopheles gambiae 1000 Genomes Consortium 2017). In total, we discovered 57,837,885 SNPs passing all variant quality filters, 11% of which were newly discovered in this project phase. Of these high-quality SNPs, 24% were multiallelic (three or more alleles). We also analyzed genome accessibility to identify all genomic positions where read alignments were of sufficient quality and consistency to support accurate SNP discovery and genotyping. Similar to the previous project phase, 61% (140 Mbp) of genome positions were accessible to SNP calling, including 91% (18 Mbp) of the exome and 58% (121 Mbp) of noncoding positions. Overall, we discovered an average of one variant allele every 1.9 bases of the accessible genome. We then used high-quality biallelic SNPs to construct a new haplotype reference panel including all 1142 wild-caught individuals, via a combination of read-backed phasing and statistical phasing as described previously (The Anopheles gambiae 1000 Genomes Consortium 2017).
In this project phase, we also performed a genome-wide CNV analysis, described in detail elsewhere (Lucas et al. 2019a). In brief, we called CNVs by analyzing data on depth of sequence read coverage in 300-bp genomic windows. We excluded windows with a high rate of ambiguous read mapping, leaving 77% (177 Mbp) of genomic windows accessible to CNV calling. For each individual mosquito, we fitted a hidden Markov model (HMM) to windowed depth of coverage values and then filtered the results to remove CNVs shorter than five contiguous windows (1.5 kbp) or with poor statistical support. We then compared calls between individuals to identify shared CNVs and compute population allele frequencies. The CNV call-set comprises 31,335 distinct CNVs, of which 7086 were found in more than one individual, and 1557 were above 5% frequency in one or more populations; 68 Mbp of the genome was overlapped by one or more CNVs, comprising 39% of genomic positions accessible to CNV calling, and 18 Mbp (10%) was affected by one or more nonsingleton CNVs. It is difficult to compare these results with other species owing to the few number of whole-genome studies and the differing methodologies and sample sizes. However, values in the range 1%–12% have been obtained in various studies of mammals (for review, see Locke et al. 2015), broadly comparable to the 10% we observed for nonsingleton CNVs. Although we applied a number of quality-filtering steps, it is likely that some false discoveries remain, particularly among CNVs only observed in a single individual. For our analysis of CNVs in insecticide-resistance genes, we therefore used only CNVs >5% frequency in at least one population. These high-frequency CNVs were significantly enriched in gene families associated with metabolic resistance to insecticides, with three loci in particular (two clusters of cytochrome P450 [CYP] genes Cyp6p/aa, Cyp9k1 and a cluster of glutathione S-transferase genes Gste) having a large number of distinct CNV alleles, multiple alleles at high population frequency, and evidence that CNVs are under positive selection (Lucas et al. 2019a). CNVs at these loci are thus likely to be playing an important role in adaptation to mosquito control interventions.
Species assignment
A. gambiae and A. coluzzii were originally defined as distinct species on the basis of fixed genetic differences (Coetzee et al. 2013). These sister species have overlapping ranges that span much of Sub-Saharan Africa, although A. coluzzii is absent from East Africa (Wiebe et al. 2017), and remain genetically distinct throughout much of this range, despite being often found in sympatry (Coetzee et al. 2013). Two molecular assays are widely used for differentiating these species, each of which reports the genotype at a single marker in the centromeric region of the X Chromosome (Fanello et al. 2002; Santolamazza et al. 2008). A single marker provides a restricted view, however, with a limited ability to detect some forms of hybridization or admixture or other complex patterns of population ancestry. In the previous project phase, we compared the results of these conventional assays with genotypes at 506 ancestry-informative SNPs distributed across all chromosome arms and found that, in some cases, the conventional assays were not concordant with species ancestry at other genome locations (The Anopheles gambiae 1000 Genomes Consortium 2017). In particular, all individuals from two sampling locations, Kenya and Guinea-Bissau, carried a mixture of A. gambiae and A. coluzzii alleles throughout their genomes, creating uncertainty regarding the appropriate species assignment. Among the new samples in phase 2, mosquitoes from The Gambia also carried a mixture of alleles from both species, in similar proportions to mosquitoes from Guinea-Bissau (Supplemental Fig. S1), confirming that populations with apparent mixed genomic ancestry are present in multiple countries in the far-western region. Previous studies using conventional assays have found that mosquitoes with heterozygous A. gambiae/A. coluzzii genotypes are common in several far-western countries, and have interpreted that as evidence for a recent breakdown of reproductive isolation between the species within that geographical area (Oliveira et al. 2008; Nwakanma et al. 2013). However, there are several possible explanations for those observations, including historical admixture and the presence of cryptic taxa that are ancestral to both A. gambiae and A. coluzzii that retain ancestral variation at species-diagnostic loci. Several studies have recently found evidence for cryptic taxa within the A. gambiae complex in other African regions (Riehle et al. 2011; Tennessen et al. 2020). Furthermore, our observations of apparent mixed ancestry in Kenyan mosquitoes, in East Africa where no mosquitoes identified as A. coluzzii have ever been observed (Wiebe et al. 2017), cannot be because of recent hybridization. A number of statistical methods are now available for use with genomic data that can test different hypotheses regarding the status and history of these populations, and work is ongoing within the Ag1000G Consortium to explore these fully. Until these questions are resolved, we regard the species assignment for individuals from Guinea-Bissau, The Gambia, and Kenya as uncertain.
In all other countries, genotypes at ancestry-informative SNPs were concordant with conventional assays, except on chromosome arm 2L where there has been a known introgression event carrying an insecticide-resistance allele from A. gambiae into A. coluzzii (Weill et al. 2000; Diabaté et al. 2004; Clarkson et al. 2014; Norris et al. 2015). We observed this introgression in A. coluzzii from both Burkina Faso and Angola in phase 1, and it was also present among A. coluzzii from Côte d'Ivoire, Ghana, and Guinea in the phase 2 cohort, confirming that A. coluzzii populations across a wide geographical range have been affected.
Population structure
We investigated genetic population structure within the wild-caught mosquitoes by performing dimensionality reduction analyses on the genome variation data, applying both UMAP (McInnes et al. 2018) and PCA (Patterson et al. 2006) to genotypes at biallelic SNPs from euchromatic regions of Chromosome 3 (Fig. 2; Supplemental Fig. S2) and applying PCA to CNVs from the whole genome (Supplemental Fig. S3). PCA has a direct genealogical interpretation (McVean 2009), but UMAP's nonlinear algorithm is able to represent a more complex structure in fewer dimensions, and hence, we compared both approaches. To complement these analyses, we fitted models of population structure and admixture to the SNP data (Supplemental Fig. S4; Frichot et al. 2014). We also used SNPs to compute two measures of genetic differentiation, average FST and rates of rare variant sharing, between pairs of populations defined by country of origin and species (Supplemental Fig. S5). From these analyses, three major groupings of individuals from multiple countries were evident: A. coluzzii from West Africa (Burkina Faso, Ghana, Côte d'Ivoire, Guinea), A. gambiae from west, central and near-east Africa (Burkina Faso, Ghana, Guinea, Cameroon, Uganda), and individuals with uncertain species status from far-west Africa (Guinea-Bissau, The Gambia). Within each of these groupings, samples clustered closely in all PCA and UMAP components and in admixture models for up to K = 5 ancestral populations, and differentiation between countries was weak, consistent with relatively unrestricted gene flow between countries. Each of the remaining clusters comprised samples from a single country and species (Angola A. coluzzii; Gabon A. gambiae, Mayotte A. gambiae; Bioko A. gambiae; individuals with uncertain species status from Kenya), and in general, each of these populations was more strongly differentiated from all other populations, consistent with a role for geographical factors limiting gene flow.
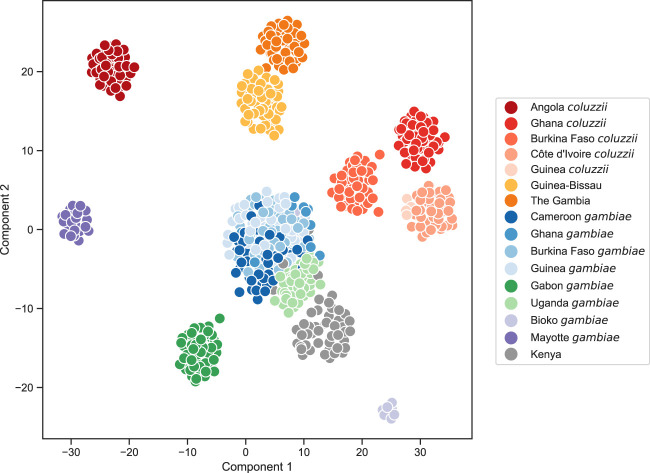
Figure 2.
Population structure analysis of the wild-caught mosquitoes using UMAP (McInnes et al. 2018). Genotype data at biallelic SNPs from euchromatic regions of Chromosome 3 were projected onto two components. Each marker represents an individual mosquito. Mosquitoes from each country and species were randomly down-sampled to at most 50 individuals.
All population subdivisions supported by the PCA analyses were also present in the UMAP analysis, although UMAP made all of these subdivisions apparent within only two components, providing a simpler visual summary. The SNP and CNV PCA results agreed in terms of identifying the same population subdivisions, but there were differences in the order in which divisions appeared. In the SNP PCA, PC1 largely divides populations by species, with the Guinea-Bissau and Gambia samples occurring with A. coluzzii populations and with the Kenya samples occurring with A. gambiae populations. In the CNV PCA, PC1 also appears to be largely driven by species, but the Guinea-Bissau, Gambia, and Kenya samples all group together. PC2 in the CNV PCA then splits out these three populations together, as distinct from populations with known A. gambiae and A. coluzzii status. Thus, the CNV PCA suggests the three populations with uncertain species status may share some common ancestry, although this would be surprising given the large geographical distance between them. These differences emphasize the need for further analyses to determine the ancestry and species status of these populations. The admixture analyses for Mayotte and Kenya modeled individuals from both populations as a mixture of multiple ancestral populations. This could represent some true admixture in these populations’ histories but could also be an artifact owing to strong genetic drift (Lawson et al. 2018). A comparison of the two A. gambiae island populations is interesting because Mayotte was highly differentiated from all other populations, but Bioko was more closely related to other West African A. gambiae, suggesting that Bioko may not be isolated from continental populations despite a physical separation of >30 km.
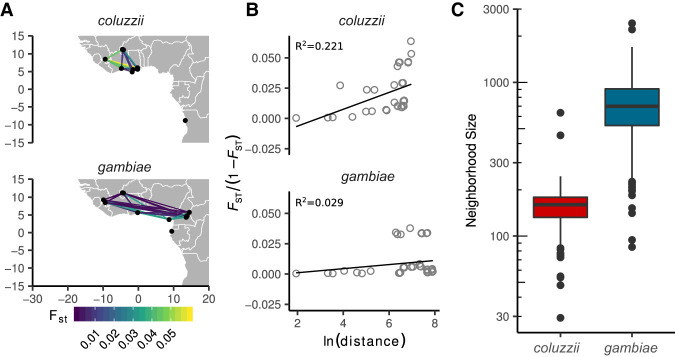
The new locations sampled in this project phase allow more comparisons to be made between A. gambiae and A. coluzzii, and there are many open questions regarding their behavior, ecology, and evolutionary history. For example, it would be valuable to know whether there are differences in long-range dispersal behavior (North et al. 2019) as have been suggested by recent studies in Sahelian regions (Dao et al. 2014; Huestis et al. 2019). Providing a comprehensive answer to this question is beyond the scope of this study, but we performed a preliminary analysis by estimating Wright's neighborhood size for each species (Wright 1946). This statistic is an approximation for the effective number of potential mates for an individual and can be viewed as a measurement of how genetic differentiation between populations correlates with the geographical distance between them (isolation by distance). We used Rousset's method for estimating neighborhood size based on a regression of normalized FST against the logarithm of geographical distance (Rousset 1997). To avoid any confounding effect of major ecological discontinuities, which may provide a natural barrier to gene flow (Lehmann et al. 2003; Pinto et al. 2013; The Anopheles gambiae 1000 Genomes Consortium 2017), we used only populations from West Africa and Central Africa north of the equatorial rainforest. We found that average neighborhood sizes are significantly lower in A. coluzzii than in A. gambiae (Wilcoxon, W = 1320, P < 2.2 × 10−16) (Fig. 3A–C), indicating stronger isolation by distance among A. coluzzii populations and suggesting a lower rate and/or range of dispersal. A recent kinship-analysis study of local dispersal in Malaysian Aedes aegypti mosquitoes estimated a neighborhood size of 268 (Jasper et al. 2019), between our estimates for A. gambiae and A. coluzzii, suggesting that our approach captures similar spatial dynamics as methods based on much denser spatial sampling. However, we do not have representation of both species at all sampling locations, and further sampling will be needed to confirm this result.
Figure 3.
Comparison of isolation by distance between A. coluzzii and A. gambiae populations from locations in West and Central Africa north of the equatorial rainforest. (A) Study region and pairwise FST. (B) Regressions of average genome-wide FST against geographic distance, following the method of Rousset (1997). Neighborhood size is estimated as the inverse slope of the regression line. Goodness-of-fit is reported as R2. (C) Difference in neighborhood size estimates by species. Box plots show medians and 95% confidence intervals of the distribution of estimates calculated in 200-kbp windows across the euchromatic regions of Chromosome 3.
Genetic diversity
The populations represented in Ag1000G phase 2 can serve as a reference point for comparisons with populations sampled by other studies at other times and locations. To facilitate population comparisons, we characterized genetic diversity within each of 16 populations in our cohort defined by country of origin and species by computing a variety of summary statistics using SNP data from the whole genome. These included nucleotide diversity (), the density of segregating sites (θW), Tajima's D, and site frequency spectra (SFS) (Fig. 4A,B; Supplemental Figs. S6, S7. We also estimated runs of homozygosity (ROH) within each individual and runs of identity by descent (IBD) between individuals, both of which provide information about haplotype sharing within populations (Fig. 4C,D).
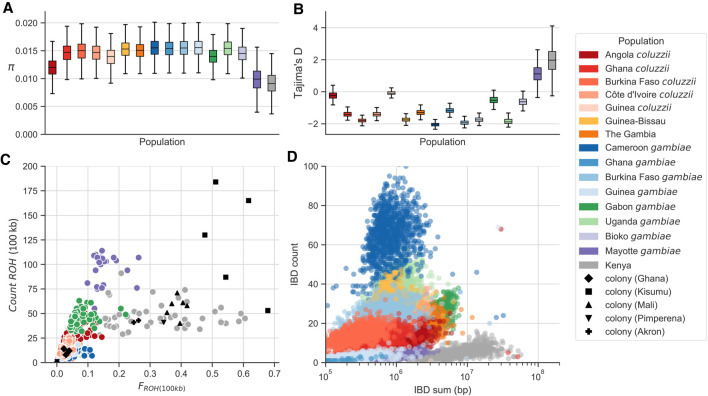
Figure 4.
Genetic diversity within populations. (A) Nucleotide diversity () calculated in nonoverlapping 20-kbp genomic windows using SNPs from euchromatic regions of Chromosome 3. (B) Tajima's D calculated in nonoverlapping 20-kbp genomic windows using SNPs from euchromatic regions of Chromosome 3. (C) Runs of homozygosity (ROH) in individual mosquitoes. Each marker represents an individual mosquito. (D) Runs of identity by descent (IBD) between individuals. Each marker represents a pair of individuals drawn from the same population.
The two easternmost populations (Kenya, Mayotte) were outliers in all statistics, with lower diversity, a deficit of rare variants relative to neutral expectation, and a higher degree of haplotype sharing within and between individuals. We previously described how the patterns of diversity in the Kenyan population were consistent with a severe and recent bottleneck (The Anopheles gambiae 1000 Genomes Consortium 2017). The similarities between Kenya and Mayotte suggest that Mayotte has also experienced a population bottleneck, which would be expected given that Mayotte is an oceanic island 310 km from Madagascar and 500 km from continental Africa, and may have been colonized by A. gambiae via small numbers of individuals. Although ROH and IBD were elevated in both populations, Mayotte individuals had a larger number of shorter tracts than Kenyan individuals, which may reflect differences in the timing and/or strength of a bottleneck.
Design of Cas9 gene drives
Nucleotide variation data from this resource are being used to inform the development of gene drives, a novel mosquito control technology using engineered selfish genetic elements to cause mosquito population suppression or modification (Burt 2003; Gantz et al. 2015; Hammond et al. 2016; Eckhoff et al. 2017; Kyrou et al. 2018). Gene drive target sequences need to be highly conserved, because any natural variation within the target sequence could inhibit association with the Cas9 guide RNA (Unckless et al. 2017). Data on natural genetic variation in multiple populations are therefore essential for identifying potential targets with low levels of nucleotide diversity and, hence, a low probability of encountering resistance. To facilitate the search for viable gene drive targets throughout the genome, we computed allele frequencies for all SNPs in all populations and included those data in the resource. We also compiled a table of all potential Cas9 target sites in the genome (23-bp regions with a protospacer-adjacent motif) that overlap a gene exon, including data for each target on the number of SNPs and nucleotide diversity in each of the populations we sampled.
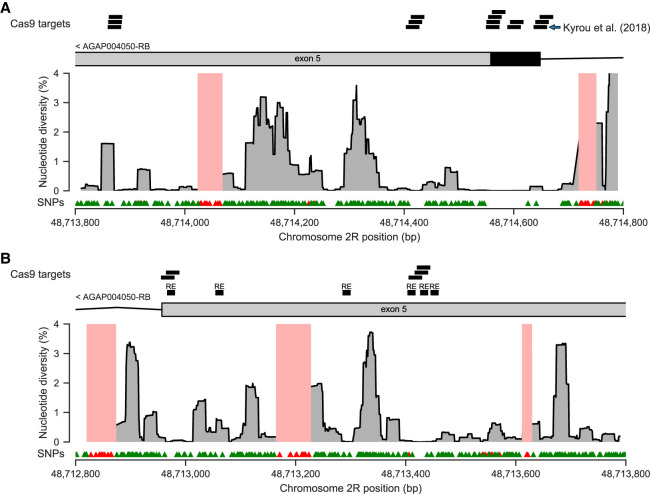
Promising results have been obtained in the laboratory with a Cas9 gene drive targeting the doublesex gene (dsx), a critical component of the sex determination pathway (Kyrou et al. 2018). This targets a sequence spanning the boundary of dsx exon 5, which is involved in sex-specific splicing, with exon 5 being included in the female transcript and excluded in the male transcript (Gempe and Beye 2011). Disruption of exon 5 causes sex determination to fail, producing sterile mosquitoes with an intersex phenotype (Kyrou et al. 2018). In addition to the Cas9 target studied by Kyrou et al. (2018), we found a further 19 Cas9 targets that overlap dsx exon 5, where the target sequence contains at most one SNP within the Ag1000G phase 2 cohort (Fig. 5A,B). Thus, there may be multiple viable targets for gene drives disrupting the sex determination pathway, providing opportunities to mitigate the impact of resistance owing to variation within any single target. The presence of multiple highly conserved regions within dsx also begs some interesting questions regarding the molecular biology of sex determination. The largest region of high conservation within dsx spanned the entire coding sequence of exon 5 and extended into 50 bp of noncoding sequence on either side (Fig. 5A). Such conservation of both coding and noncoding sites suggests that purifying selection is acting here on the nucleotide sequence. This in turn suggests that the nucleotide sequence may serve as an important target for factors that bind to DNA or pre-mRNA molecules and control differential splicing. However, this region is >1 kbp distant from the putative homologs of the regulatory factor binding sequences (dsxREs) that have been identified in D. melanogaster (Cline and and Meyer 1996; Scali et al. 2005; Gempe and Beye 2011). The upstream regulatory factors that control sex-specific splicing are not known in A. gambiae (Scali et al. 2005; Krzywinska et al. 2016), and our data add further evidence for fundamental differences in the sex determination pathway between Anopheles and Drosophila (Scali et al. 2005; Krzywinska et al. 2016).
Figure 5.
Nucleotide diversity within the female-specific exon 5 of the doublesex gene (dsx; AGAP004050), a key component of the sex determination pathway and a gene targeted for Cas9-based homing endonuclease gene drive (Kyrou et al. 2018). In both plots, the location of exon 5 within the female-specific isoform (AGAP004050-RB; AgamP4.12 gene set) is shown above (black indicates coding sequence; gray, untranslated region), with additional annotations above to show the location of Cas9 target sequences containing at most one SNP, and the putative exon splice enhancing sequences (“RE”) reported by Scali et al. (2005). The main region of the plot shows nucleotide diversity averaged across all Ag1000G phase 2 populations, computed in 23-bp moving windows. Regions shaded pale red indicate regions not accessible to SNP calling. Triangle markers below show the locations of SNPs discovered in Ag1000G phase 2 (green indicates passed variant filters; red, failed variant filters). (A) exon5/intron4 boundary. (B) exon5/intron6 boundary.
Resistance to pyrethroid insecticides
Malaria control in Africa depends heavily on mass distribution of long-lasting insecticidal nets (LLINs) impregnated with pyrethroid insecticides (Bhatt et al. 2015; Churcher et al. 2016; Ranson and Lissenden 2016). Entomological surveillance programs regularly test malaria vector populations for pyrethroid resistance using standardized bioassays, and these data have shown that pyrethroid resistance has become widespread in A. gambiae (Hemingway et al. 2016; World Health Organization 2018). However, pyrethroid resistance can be conferred by different molecular mechanisms, and it is not well understood which molecular mechanisms are responsible for resistance in which mosquito populations. The nucleotide variation data in this resource include 66 nonsynonymous SNPs within the Vgsc gene that encodes the binding target for pyrethroid insecticides, of which two SNPs (L995F, L995S) are known to confer a pyrethroid-resistance phenotype, and one SNP (N1570Y) has been shown to substantially increase pyrethroid resistance when present in combination with L995F (Jones et al. 2012). These SNPs can serve as markers of target-site resistance to pyrethroids, but knowledge of genetic markers of metabolic resistance in A. gambiae and A. coluzzii is currently limited (Mitchell et al. 2014; Weetman et al. 2018). Metabolic resistance to pyrethroids is mediated at least in part by increased expression of CYP enzymes (Kwiatkowska et al. 2013; Edi et al. 2014; Ngufor et al. 2015; Vontas et al. 2018), and we found CNV hot-spots at two loci containing Cyp genes (Lucas et al. 2019a). One of these loci occurs on chromosome arm 2R and overlaps a cluster of 10 Cyp genes, including Cyp6p3 previously shown to metabolize pyrethroids (Müller et al. 2008) and recently shown to confer pyrethroid resistance when expression is experimentally increased in A. gambiae (Adolfi et al. 2019). The second locus occurs on the X Chromosome and spans a single Cyp gene, Cyp9k1, which has also been shown to metabolize pyrethroids (Vontas et al. 2018). We also found CNVs at two other Cyp gene loci on chromosome arm 3R containing genes previously associated with pyrethroid resistance, Cyp6z1 (Nikou et al. 2003) and Cyp6m2 (Stevenson et al. 2011), although there was only a single CNV allele at each locus. Overexpression of Cyp6m2 has been shown to confer resistance to pyrethroids but increased susceptibility to the organophosphate malathion (Adolfi et al. 2019), and so, the selection pressures at this locus may be more complex. The precise phenotype of these CNVs remains to be characterized, but given the multiple lines of evidence showing that increased expression of genes at these loci confers pyrethroid resistance, it seems reasonable to assume that CNVs at these loci can serve as a molecular marker of CYP-mediated pyrethroid resistance.
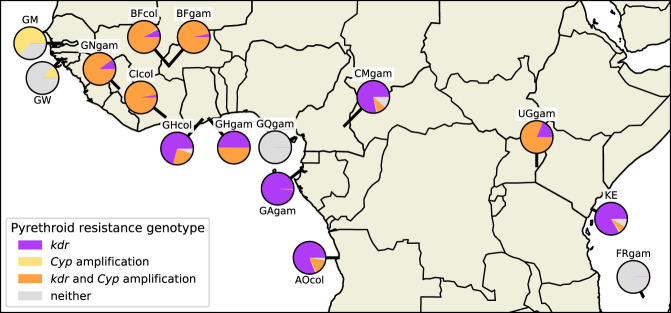
We constructed an overview of the prevalence of these two pyrethroid-resistance mechanisms—target-site resistance and CYP-mediated metabolic resistance—within the Ag1000G phase 2 cohort by combining the data on nucleotide and copy number variation (Fig. 6). The sampling of these populations was conducted at different times in different locations, and the geographical sampling is relatively sparse, so we cannot draw any general conclusions about the current distribution of resistance from our data. However, some patterns were evident. For example, West African populations of both species (Burkina Faso, Guinea, Côte d'Ivoire) all had >84% of individuals carrying both target-site and metabolic resistance markers. In Ghana, Cameroon, Gabon, and Angola, target-site resistance was nearly fixed, but metabolic resistance markers were at lower frequencies. Mosquitoes from Bioko carried no resistance markers at all but were collected in 2002, and so, the lack of resistance may be because sampling predated any major scale-up of vector control interventions (Vontas et al. 2018). However, the Gabon samples were collected in 2000 and show that high levels of target-site resistance were present in some populations at that time. In Guinea Bissau and The Gambia, target-site resistance was absent, but Cyp gene amplifications were present, and thus, surveillance using only molecular assays that detect target site resistance at those locations could be missing an important signal of metabolic resistance. In East Africa, both Kenya and Uganda had high frequencies of target-site resistance (88% and 100%, respectively). However, 81% of Uganda individuals also had Cyp gene amplifications, whereas only 4% of Kenyans carried these metabolic resistance markers. Denser spatiotemporal sampling will enable us to build a more complete picture of the prevalence and spread of these different resistance mechanisms and would be highly relevant to the design of insecticide-resistance management plans.
Figure 6.
Pyrethroid-resistance genotype frequencies. The geographical distribution of pyrethroid insecticide–resistance genotypes are shown by population. Pie chart colors represent resistance genotype frequencies: purple, these individuals were either homozygous or heterozygous for one of the two kdr pyrethroid target site resistance alleles Vgsc-L995F/S; yellow, these individuals carried a copy number amplification within any of the Cyp6p/aa, Cyp6m, Cyp6z, or Cyp9k gene clusters but no kdr alleles; orange, these individuals carried at least one kdr allele and one Cyp gene amplification; and gray, these individuals carried no known pyrethroid-resistance alleles (no kdr alleles or Cyp amplifications). The Guinea A. coluzzii population is omitted owing to small sample size.
Discussion
The Ag1000G phase 2 data resource provides a battery of new genetic markers that can be used to expand our capabilities for molecular surveillance of insecticide resistance. Insecticide-resistance management is a major challenge for malaria vector control, but the availability of new vector control products is opening up new possibilities. However, new products may be more expensive than products currently in use, so procurement decisions have to be justified and resources targeted to areas where they will have the greatest impact. For example, next-generation LLINs are now available that combine a pyrethroid insecticide with a synergist compound, piperonyl butoxide (PBO), which partially ameliorates metabolic resistance by inhibiting CYP enzyme activity in the mosquito. However, CYP-mediated metabolic resistance is only one of several possible mechanisms of pyrethroid resistance that may or may not be present in vector populations being targeted. It would therefore be valuable to survey mosquito populations and determine the prevalence of different pyrethroid-resistance mechanisms, both before and after any change in vector control strategy. Our data resource includes CNVs at four Cyp gene loci (Cyp6p/aa, Cyp6m, Cyp6z, and Cyp9k), which could serve as molecular markers of CYP-mediated metabolic resistance. Glutathione S-transferase enzymes are also associated with metabolic resistance to pyrethroids (Ochieng'Opondo et al. 2016; Adolfi et al. 2019) as well as other insecticide classes (Mitchell et al. 2014; Riveron et al. 2014; Pavlidi et al. 2018; Adolfi et al. 2019), and we found CNVs at the Gste locus that could serve as molecular markers of this alternative resistance mechanism, which is not inhibited by PBO. Gste CNVs were less prevalent in our data set than Cyp CNVs, and the geographical distribution also differed, suggesting they may be driven by different selection pressures (Supplemental Fig. S8).
To illustrate the potential for improved molecular surveillance of pyrethroid resistance, we combined the data on known SNP markers of target-site resistance and novel putative CNV markers of CYP-mediated metabolic resistance (Fig. 6). There are clear heterogeneities, with some populations at high frequency for both resistance mechanisms, particularly in West Africa. The presence of CYP-mediated pyrethroid resistance in a population suggests that PBO LLINs might provide some benefit over standard LLINs. However, if other resistance mechanisms are also at high frequency, the benefit of the PBO synergist might be diminished. Current WHO guidance states that PBO LLINs are recommended in regions with “intermediate levels” of pyrethroid resistance but not where resistance levels are high (World Health Organization 2017). This guidance is based on modeling of bioassay data and experimental hut trials, and it is not clear why PBO LLINs are predicted to provide diminishing returns at higher resistance levels, although high levels of resistance presumably correlate with the presence of multiple resistance mechanisms, including mechanisms not inhibited by PBO (Churcher et al. 2016). Without molecular data, however, this guidance is hard to interpret or improve upon.
Ideally, molecular data on insecticide-resistance mechanisms would be collected as part of routine entomological surveillance, as well as in field trials of new vector control products, alongside data from bioassays and other standard entomological variables. There are several options for scaling up molecular surveillance, including both whole-genome sequencing and targeted (amplicon) sequencing, with several choices of sequencing technology platform. Assays that target specific genetic loci are attractive because of lower cost and infrastructure requirements, and data from the Ag1000G Project have been used successfully to design multiplex assays for the Agena Biosciences iPLEX platform (Lucas et al. 2019b) and for Illumina amplicon sequencing (C Jacob, E Lucas, K Rockett, et al., in prep.). However, targeted assays need to be updated regularly to cover new forms of resistance as they emerge. To keep pace with evolving vector populations, regular whole-genome sequencing of contemporary populations from a well-chosen set of sentinel sites will be needed. None of the samples sequenced in this study were collected more recently than 2012, geographical sampling within each country was limited, and many countries are not yet represented in the resource; therefore, there remain important gaps to be filled. The next phase of the Ag1000G Project will expand the resource to cover 18 countries and will include another major malaria vector species, Anopheles arabiensis, and so will address some of these gaps. Looking beyond the Ag1000G Project, genomic surveillance of insecticide resistance will require new sampling frameworks that incorporate spatial and ecological modeling of vector distributions to guide sampling at appropriate spatial scales (Sedda et al. 2019). Fortunately, mosquitoes are easy to transport, and the costs of sequencing continue to decrease, so it is reasonable to consider a mixed strategy that includes both whole-genome sequencing and targeted assays.
These data also cast some new, and in some cases contrasting, light on the question of gene flow between malaria vector populations. The question is of practical interest, because gene flow is enabling the spread of insecticide resistance between species and across large geographical distances (The Anopheles gambiae 1000 Genomes Consortium 2017; Clarkson et al. 2018). Gene flow also needs to be quantified before new vector control interventions based on gene drive could be considered (North and Godfray 2018). We found evidence that isolation by distance is greater for A. coluzzii than for A. gambiae, at least within West Africa, suggesting that the effective rate of migration could be lower in A. coluzzii. A variety of anopheline species have recently been found to engage in long-distance wind-assisted migration, including A. coluzzii but not A. gambiae, which would appear to contradict our results, although the study was limited to a single location within the Sahelian region (Huestis et al. 2019). If A. coluzzii does have a lower rate and/or range of dispersal than A. gambiae, this is clearly not limiting the spread of insecticide-resistance adaptations between countries. For example, among the CNV alleles we discovered at the Cyp6p/aa, Cyp9k1, and Gste loci, 7/13 alleles found in A. coluzzii had spread to more than one country compared with 8/27 alleles in A. gambiae (Lucas et al. 2019a). There is also an interesting contrast between the spread of pyrethroid target-site and metabolic resistance alleles. We previously showed that target-site resistance has spread to countries spanning the equatorial rainforest and the Rift valley and has moved between A. gambiae and A. coluzzii (The Anopheles gambiae 1000 Genomes Consortium 2017; Clarkson et al. 2018). In the most extreme example, one haplotype (F1) has spread to countries as distant as Guinea and Angola. In contrast, although CNV alleles were commonly found in multiple countries, we did not observe any cases of CNV alleles crossing any of these ecological or biological boundaries, apart from a single allele found in both Gabon and Cameroon A. gambiae (Gste Dup5). This could be because of differences in the strength, timing, or spatial distribution of selective pressures or because of intrinsic factors such as differences in fitness costs in the absence of positive selection. Further work is required to investigate the selective forces affecting the spread of these different modes of adaptation to insecticide use.
The two island populations sampled in this project phase also provide an interesting contrast. Samples from Mayotte were highly differentiated from mainland A. gambiae, suggesting strong isolation, whereas Bioko samples were closely related to West African A. gambiae, suggesting ongoing gene flow. Bioko is part of Equatorial Guinea administratively, and there are frequent ferries to the mainland, which could provide opportunities for mosquito movement. However, there are no pyrethroid-resistance alleles in our Bioko samples, and these were collected in 2002, at a time when target-site resistance alleles were present in mainland populations, so the rate of contemporary migration between Bioko and mainland populations remains an open question. A recent study of A. gambiae populations on the Lake Victoria islands, separated from mainland Uganda by 4–50 km, found evidence for isolation between island and mainland populations, as well as between individual islands (Bergey et al. 2020). However, some selective sweeps at insecticide-resistance loci had spread through both mainland and island populations; thus, isolation is not complete, and some contemporary gene flow occurs. Some of these gene flow questions and apparent contradictions could, in principle, be resolved by inferring contemporary migration rates and population density from genomic data, but methodological improvements are needed in this area (Al-Asadi et al. 2019). The haplotype data we have generated provide a valuable resource to support the development of new statistical methods for demographic inference, and encourage application of these methods to nonmodel species.
Malaria has become a stubborn foe, frustrating global efforts toward elimination in both low and high burden settings. However, new vector control tools offer hope, as does the renewed focus on improving surveillance systems and using data to tailor interventions. The genomic data resource we have generated provides a platform from which to accelerate these efforts, showing the potential for data integration on a continental scale. It also provides a snapshot of populations in rapid evolutionary motion and, thus, an opportunity to study and understand the adaptive potential of genetically diverse eukaryotic species when subjected to strong selective pressures. Nevertheless, work remains to fill gaps in these data, by expanding geographical coverage, including other malaria vector species and integrating genomic data collection with routine surveillance of contemporary populations using quantitative sampling design. We hope that the MalariaGEN data-sharing community and framework for international collaboration can continue to serve as a model for coordinated action.
Methods
Population sampling
Ag1000G phase 2 mosquitoes were collected from natural populations at 33 sites in 13 sub-Saharan African countries (Fig. 1; Supplemental Table S1). Five of these countries and 18 of these collection sites were newly sampled in phase 2, and the remainder were previously sampled in phase 1. New samples in phase 2 comprised the following: A. coluzzii from Tiassalé, Côte d'Ivoire, collected as larvae from irrigated rice fields by dipping between May and September 2012; A. gambiae from Sacriba, Bioko Island, collected in September 2002 by overnight CDC light traps; A. gambiae from several sites on Mayotte Island, collected as larvae during March and April 2011 in temporary pools by dipping; mosquitoes from hamlets around Njabakunda, North Bank Region, The Gambia, collected between August and October 2011 by pyrethrum spray catch; A. gambiae and A. coluzzii from several sites in Ghana, collected as larvae from puddles near roads or farms between August and December 2012; and mosquitoes from Safim, Guinea-Bissau, collected in October 2010 using indoor CDC light traps. Further details of samples novel to phase 2 can be found in the Supplemental Material. Details of samples in phase 1 can be found in the supplementary information of The Anopheles gambiae 1000 Genomes Consortium (2017).
Laboratory crosses
Ag1000G phase 2 includes seven additional laboratory colony crosses: cross 18-5 (Ghana mother × Kisumu/G3 father, 20 offspring), 37-3 (Kisumu × Pimperena, 20 offspring), 45-1 (Mali × Kisumu, 20 offspring), 47-6 (Mali × Kisumu, 20 offspring), 73-2 (Akron × Ghana, 19 offspring), 78-2 (Mali × Kisumu/Ghana, 19 offspring), and 80-2 (Kisumu × Akron, 20 offspring). Colonies with two names, for example, “Kisumu/G3,” signify that the father is from one of these two colonies, but which one is unknown. Further details of crosses released in phase 1 can be found in the supplementary information of The Anopheles gambiae 1000 Genomes Consortium (2017), as well as methods for cross creation and processing that also apply to phase 2.
Whole-genome sequencing
Sequencing was performed on the Illumina HiSeq 2000 platform at the Wellcome Sanger Institute. Paired-end multiplex libraries were prepared using the manufacturer's protocol, with the exception that genomic DNA was fragmented using Covaris adaptive focused acoustics rather than nebulization. Multiplexes comprised 12 tagged individual mosquitoes, and three lanes of sequencing were generated for each multiplex to even out variations in yield between sequencing runs. Cluster generation and sequencing were undertaken per the manufacturer's protocol for paired-end 100-bp sequence reads with insert size in the range 100–200 bp. Target coverage was 30 × per individual. New sequencing data from this project phase were then analyzed in conjunction with sequencing data from phase 1 (The Anopheles gambiae 1000 Genomes Consortium 2017; PRJEB18691).
Genome accessibility
We constructed a map of the accessible genome, which identifies positions in the reference genome where we can confidently call nucleotide variation. For Ag1000G phase 2, we repeated the phase 1 genome accessibility analyses (The Anopheles gambiae 1000 Genomes Consortium 2017) but with 1142 samples and the additional Mendelian error information provided by the 11 crosses. We constructed annotations for each position in the reference genome based on data from sequence read alignments from all wild-caught samples, as well as additional data from repeat annotations. Annotations were then analyzed for association with rates of Mendelian errors in the crosses. Annotations and thresholds were chosen to remove classes of variants that were enriched for Mendelian errors. Following these analyses, it was apparent that the accessibility classifications used in phase 1 were also appropriate in application to phase 2. Reference genome positions were classified as accessible if the following were true: not repeat masked by DUST; no coverage ≤0.1% (at most one individual had zero coverage); ambiguous alignment ≤0.1% (at most one individual had ambiguous alignments); high coverage ≤2% (at most 20 individuals had more than twice their genome-wide average coverage); low coverage ≤10% (at most 114 individuals had less than half their genome-wide average coverage); and low mapping quality ≤10% (at most 114 individuals had average mapping quality below 30).
Sequence analysis and SNP calling
SNP calling methods were unchanged from phase 1 (The Anopheles gambiae 1000 Genomes Consortium 2017). Briefly, sequence reads were aligned to the AgamP3 reference genome (Holt et al. 2002; Sharakhova et al. 2007) using BWA version 0.6.2, duplicate reads marked (Li and Durbin 2009), reads realigned around putative indels, and SNPs discovered using GATK version 2.7.4 unified genotyper following best-practice recommendations (Van der Auwera et al. 2013).
Sample quality control
A total of 1285 individual mosquitoes were sequenced as part of Ag1000G phase 2 and included in the cohort for variant discovery. After variant discovery, quality-control (QC) steps using coverage and contamination filters alongside principal component analysis and metadata concordance were performed to exclude individuals with poor quality sequence and/or genotype data as detailed by The Anopheles gambiae 1000 Genomes Consortium (2017). A total of 143 individuals were excluded at this stage, retaining 1142 individuals for downstream analyses.
SNP filtering
Following Ag1000G phase 1 (The Anopheles gambiae 1000 Genomes Consortium 2017), we filtered any SNP that occurred at a genome position classified as inaccessible as described in the section on genome accessibility above, thus removing SNPs with evidence for excessively high or low coverage or ambiguous alignment. We then applied additional filters using variant annotations produced by GATK, filtering SNPs that failed any of the following criteria: QD < 5; FS > 100; ReadPosRankSum < −8; BaseQRankSum < −50.
Haplotype estimation
Haplotype estimation, also known as phasing, was performed on all phase 2 wild-caught individuals using methods from Ag1000G phase 1 (The Anopheles gambiae 1000 Genomes Consortium 2017). In short, SHAPEIT2 was used to perform statistical phasing with information from sequence reads (Delaneau et al. 2013).
CNV calling
Detailed methodology for detection and QC of CNVs was previously described (Lucas et al. 2019a). In brief, coverage was calculated for each individual in 300-bp windows and then normalized to account for bias owing to variation in (G + C) content. Windows were filtered to remove those with low mapping quality or extreme (G + C) content. To infer copy-number state in each window in each individual, we applied a Gaussian HMM to the individual's normalized filtered windowed coverage data. Putative CNVs were identified as sequences of five or more contiguous windows with a predicted copy number state greater than two (or greater than one for males on the X Chromosome). From this raw CNV call set, we created a quality-filtered call set by first removing samples with very high coverage variance and then removing CNV calls with poor statistical support (HMM likelihood ratio of predicted CNV state compared with the null hypothesis of no CNV < 1000). CNV calls were matched across samples, considering any two CNVs to be identical if the breakpoints predicted by their copy number state transitions were within one window of each-other. For the analysis of CNVs in metabolic insecticide-resistance genes, we characterized CNV alleles at five gene clusters (Cyp6aa1–Cyp6p2, Gstu4–Gste3, Cyp6m2–Cyp6m4, Cyp6z3–Cyp6z1, Cyp9k1) using unique patterns of discordant read pairs and split reads crossing the CNV breakpoint. Once the diagnostic reads were identified for a CNV allele, we recorded the presence of that allele in all samples with at least two supporting diagnostic reads.
Population structure
Ancestry informative marker (AIM), FST, doubleton sharing, and SNP PCA were conducted following methods previously defined (The Anopheles gambiae 1000 Genomes Consortium 2017). The PCA and UMAP analyses were performed on 131,679 SNPs from euchromatic regions of Chromosome 3 (3R: 1–37 Mbp; 3L: 15–41 Mbp) obtained from the full data set via random down-sampling to 100,000 nonsingleton SNPs from each chromosome arm and then performing LD-pruning. To generate the UMAP projection shown in Figure 2, each country and species was down-sampled to a maximum of 50 individuals to provide a projection that was less warped by differences in sample size. The UMAP analysis was also performed on the full set of individuals, which gave qualitatively identical results. UMAP was performed using the umap-learn Python package (McInnes et al. 2018) with the following parameter settings: ; ; spread = 5; metric = euclidean. Other parameter values for and were also performed, all producing qualitatively identical results. Guinea A. coluzzii (n = 4) was excluded from FST analysis, and Guinea A. coluzzii (n = 4), Bioko A. gambiae (n = 9), and Ghana A. gambiae (n = 12) were excluded from doubleton sharing analysis owing to small sample size. Unscaled CNV variation PCAs were built from the CNV presence/absence calls (Lucas et al. 2019a) using the prcomp function in R (R Core Team 2019).
Admixture models were fitted using LEA version 2.0 (Frichot and François 2015) in R version 3.6.1 (R Core Team 2019). Ten independent sets of SNPs were generated by selecting SNPs from euchromatic regions of Chromosome 3 with minor allele frequency >1%, randomly selecting 100,000 SNPs from each chromosome arm, and then applying the same LD pruning methodology as used for PCA. The resulting data were then analyzed using the snmf method (sparse nonnegative matrix factorization) (Frichot et al. 2014) to obtain ancestry estimates for each cluster (K) tested. We tested all K values from two to 15. Ten replicates of the analysis with snmf were run for each data set; thus, 100 runs were performed for each K. CLUMPAK (Kopelman et al. 2015) was used to summarize the results, identify the major and minor clustering solutions identified at each K (if they occurred), and estimate the average ancestry proportions for the major solution.
Genetic diversity
Analyses of genetic diversity were conducted following methods previously defined (The Anopheles gambiae 1000 Genomes Consortium 2017). In short, scikit-allel version 1.2.0 was used to calculate windowed averages of nucleotide diversity and Tajima's D (https://github.com/cggh/scikit-allel), IBDseq version r1206 (Browning and Browning 2015) was used to calculate IBD, and an HMM implemented in scikit-allel was used to calculate ROH.
Data access
The sequencing data and variation data generated in this study have been submitted to the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena/browser/home) under accession number PRJEB36277. Variation data from Ag1000G phase 2 can also be downloaded from the Ag1000G public FTP site via the MalariaGEN website (https://www.malariagen.net/resource/27).
The Anopheles gambiae 1000 Genomes Consortium
Please address correspondence to Alistair Miles (alistair.miles@bdi.ox.ac.uk) and Dominic Kwiatkowski (dominic@sanger.ac.uk).
Data analysis group
Chris S. Clarkson,2 Alistair Miles,2,3 Nicholas J. Harding,3 Eric R. Lucas,4 C.J. Battey,5 Jorge Edouardo Amaya-Romero,6,7 Andrew D. Kern,5 Michael C. Fontaine,6,7 Martin J. Donnelly,2,4 Mara K.N. Lawniczak,2 Dominic P. Kwiatkowski (chair),2,3
Partner working group
Martin J. Donnelly (chair), 2,3 Diego Ayala,6,8 Nora J. Besansky,9 Austin Burt,10 Beniamino Caputo,11 Alessandra della Torre,11 Michael C. Fontaine,6,7 H. Charles J. Godfray,12 Matthew W. Hahn,13 Andrew D. Kern,5 Dominic P. Kwiatkowski,2,3 Mara K.N. Lawniczak,2 Janet Midega,14 Samantha O'Loughlin,10 João Pinto,15 Michelle M. Riehle,16 Igor Sharakhov,17, 18 Daniel R. Schrider,19 Kenneth D. Vernick,20 David Weetman,4 Craig S. Wilding,21 Bradley J. White,22
Population sampling
Angola: Arlete D. Troco,23 João Pinto,15 Bioko: Jorge Cano,24 Burkina Faso: Abdoulaye Diabaté,25 Samantha O'Loughlin,10 Austin Burt,10 Cameroon: Carlo Costantini,6,26 Kyanne R. Rohatgi,9 Nora J. Besansky,9 Côte d'Ivoire: Edi Constant,27 David Weetman,4 Gabon: Nohal Elissa,28 João Pinto,15 Gambia: Davis C. Nwakanma,29 Musa Jawara,29 Ghana: John Essandoh,30 David Weetman,4 Guinea: Boubacar Coulibaly,31 Michelle M. Riehle,16 Kenneth D. Vernick,20 Guinea-Bissau: João Pinto,15 João Dinis,32 Kenya: Janet Midega,14 Charles Mbogo,14 Philip Bejon,14 Mayotte: Gilbert Le Goff,6 Vincent Robert,6 Uganda: Craig S. Wilding,21 David Weetman,4 Henry D. Mawejje,33 Martin J. Donnelly,4 Laboratory crosses: David Weetman,4 Craig S. Wilding,21 Martin J. Donnelly,4
Sequencing and data production
Jim Stalker,34 Kirk A. Rockett,3 Eleanor Drury,2 Daniel Mead,2 Anna E. Jeffreys,3 Christina Hubbart,3 Kate Rowlands,3 Alison T. Isaacs,4 Dushyanth Jyothi,35 Cinzia Malangone,35 Maryam Kamali,17,36
Project coordination
Victoria Simpson,3 Christa Henrichs,3 Dominic P. Kwiatkowski,2, 3
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank the staff of the Wellcome Sanger Institute Sample Logistics, Sequencing, and Informatics facilities for their contributions. The sequencing, analysis, informatics, and management of the Anopheles gambiae 1000 Genomes Project are supported by Wellcome through Sanger Institute core funding (098051), core funding to the Wellcome Centre for Human Genetics (203141/Z/16/Z), and a strategic award (090770/Z/09/Z) and by the MRC Centre for Genomics and Global Health, which is jointly funded by the Medical Research Council (MRC) and the Department for International Development (DFID; G0600718; M006212). M.K.N.L. was supported by MRC grant G1100339. S.O.’L. and A.B. were supported by a grant from the Foundation for the National Institutes of Health (NIH) through the Vector-Based Control of Transmission: Discovery Research (VCTR) program of the Grand Challenges in Global Health initiative of the Bill and Melinda Gates Foundation. D.W., C.S.W., H.D.M., and M.J.D. were supported by award numbers U19AI089674 and R01AI082734 from the National Institute of Allergy and Infectious Diseases (NIAID). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID or NIH.
Author contributions: C.S.C. and A.M. jointly led curation of the phase 2 data resource and wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.262790.120.
Freely available online through the Genome Research Open Access option.
Parasites and Microbes Programme, Wellcome Sanger Institute, Hinxton, Cambridge CB10 1SA, UK
MRC Centre for Genomics and Global Health, University of Oxford, Oxford OX3 7BN, UK
Department of Vector Biology, Liverpool School of Tropical Medicine, Liverpool L3 5QA, UK
Institute for Ecology and Evolution, University of Oregon, Eugene, OR 97403, USA
Laboratoire MIVEGEC (Université de Montpellier, CNRS 5290, IRD 229), Centre IRD de Montpellier, 34395 Montpellier Cedex 5, France
Groningen Institute for Evolutionary Life Sciences (GELIFES), University of Groningen, 9700 Groningen, The Netherlands
Unit d'Ecologie des Systèmes Vectoriels, Centre International de Recherches Médicales de Franceville, Franceville, Gabon
Eck Institute for Global Health, Department of Biological Sciences and University of Notre Dame, IN 46556, USA
Department of Life Sciences, Imperial College, Berkshire SL5 7PY, UK
Istituto Pasteur Italia–Fondazione Cenci Bolognetti, Dipartimento di Sanita Pubblica e Malattie Infettive, Università di Roma SAPIENZA, Rome, Italy
Department of Zoology, University of Oxford, Oxford OX1 3SZ, UK
Department of Biology and School of Informatics and Computing, Indiana University, Bloomington, IN 47405, USA
KEMRI-Wellcome Trust Research Programme, 80108 Kilifi, Kenya
Global Health and Tropical Medicine, GHTM, Instituto de Higiene e Medicina Tropical, IHMT, Universidade Nova de Lisboa, 1349-008 Lisbon, Portugal
Department of Microbiology and Immunology, Medical College of Wisconsin, Milwaukee, WI 53226, USA
Department of Entomology, Virginia Tech, Blacksburg, VA 24061, USA
Department of Cytology and Genetics, Tomsk State University, Tomsk 634050, Russia
Department of Genetics, University of North Carolina, Chapel Hill, NC 27599-7264, USA
Unit for Genetics and Genomics of Insect Vectors, Institut Pasteur, 75015 Paris, France
School of Biological and Environmental Sciences, Liverpool John Moores University, Liverpool L3 3AF, UK
Verily Life Sciences, South San Francisco, CA 94080, USA
Programa Nacional de Controle da Malária, Direcção Nacional de Saúde Pública, Ministério da Saúde, Luanda, Angola
London School of Hygiene and Tropical Medicine, London WC1E 7HT, UK
Institut de Recherche en Sciences de la Santé (IRSS), Bobo Dioulasso, B.P. 7192 Burkina Faso
Laboratoire de Recherche sur le Paludisme, Organisation de Coordination pour la lutte contre les Endémies en Afrique Centrale (OCEAC), B.P. 288 Yaoundé, Cameroon
Centre Suisse de Recherches Scientifiques. Yopougon, Abidjan - 01 BP 1303 Abidjan, Côte d'Ivoire
Institut Pasteur de Madagascar, Avaradoha, BP 1274, 101 Antananarivo, Madagascar
Medical Research Council Unit, The Gambia at the London School of Hygiene & Tropical Medicine (MRCG at LSHTM), Banjul, The Gambia
Department of Wildlife and Entomology, University of Cape Coast, Cape Coast, Ghana
Malaria Research and Training Centre, Faculty of Medicine and Dentistry, University of Mali, BP: E 423 Bamako-Mali
Instituto Nacional de Saaúde Paública, Ministaério da Saaúde Paública, Bissau, Guinaé-Bissau
Infectious Diseases Research Collaboration, Kampala, Uganda
Microbiotica Limited, Biodata, Innovation Centre, Wellcome Genome Campus, Cambridge CB10 1DR, UK
European Bioinformatics Institute, Hinxton, Cambridge CB10 1SA, UK
Department of Medical Entomology and Parasitology, Faculty of Medical Sciences, Tarbiat Modares University, Tehran, Iran
Contributor Information
Collaborators: Chris S. Clarkson, Alistair Miles, Nicholas J. Harding, Eric R. Lucas, C.J. Battey, Jorge Edouardo Amaya-Romero, Andrew D. Kern, Michael C. Fontaine, Martin J. Donnelly, Mara K.N. Lawniczak, Dominic P. Kwiatkowski, (chair), Martin J. Donnelly, (chair), Diego Ayala, Nora J. Besansky, Austin Burt, Beniamino Caputo, Alessandra della Torre, Michael C. Fontaine, H. Charles J. Godfray, Matthew W. Hahn, Andrew D. Kern, Dominic P. Kwiatkowski, Mara K.N. Lawniczak, Janet Midega, Samantha O'Loughlin, João Pinto, Michelle M. Riehle, Igor Sharakhov, Daniel R. Schrider, Kenneth D. Vernick, David Weetman, Craig S. Wilding, Bradley J. White, Angola: Arlete D. Troco, João Pinto, Bioko: Jorge Cano, Burkina Faso: Abdoulaye Diabaté, Samantha O'Loughlin, Austin Burt, Cameroon: Carlo Costantini, Kyanne R. Rohatgi, Nora J. Besansky, Côte d'Ivoire: Edi Constant, David Weetman, Gabon: Nohal Elissa, João Pinto, Gambia: Davis C. Nwakanma, Musa Jawara, Ghana: John Essandoh, David Weetman, Guinea: Boubacar Coulibaly, Michelle M. Riehle, Kenneth D. Vernick, Guinea-Bissau: João Pinto, João Dinis, Kenya: Janet Midega, Charles Mbogo, Philip Bejon, Mayotte: Gilbert Le Goff, Vincent Robert, Uganda: Craig S. Wilding, David Weetman, Henry D. Mawejje, Martin J. Donnelly, Laboratory crosses: David Weetman, Craig S. Wilding, Martin J. Donnelly, Jim Stalker, Kirk A. Rockett, Eleanor Drury, Daniel Mead, Anna E. Jeffreys, Christina Hubbart, Kate Rowlands, Alison T. Isaacs, Dushyanth Jyothi, Cinzia Malangone, Maryam Kamali, Victoria Simpson, Christa Henrichs, and Dominic P. Kwiatkowski
References
- The 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526: 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolfi A, Poulton B, Anthousi A, Macilwee S, Ranson H, Lycett GJ. 2019. Functional genetic validation of key genes conferring insecticide resistance in the major African malaria vector, Anopheles gambiae. Proc Natl Acad Sci. 116: 25764–25772. 10.1073/pnas.1914633116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Asadi H, Petkova D, Stephens M, Novembre J. 2019. Estimating recent migration and population-size surfaces. PLoS Genet 15: e1007908 10.1371/journal.pgen.1007908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Anopheles gambiae 1000 Genomes Consortium. 2017. Genetic diversity of the African malaria vector Anopheles gambiae. Nature 552: 96–100. 10.1038/nature24995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergey CM, Lukindu M, Wiltshire RM, Fontaine MC, Kayondo JK, Besansky NJ. 2020. Assessing connectivity despite high diversity in island populations of a malaria mosquito. Evol Appl 13: 417–431. 10.1111/eva.12878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S, Weiss DJ, Cameron E, Bisanzio D, Mappin B, Dalrymple U, Battle KE, Moyes CL, Henry A, Eckhoff PA, et al. 2015. The effect of malaria control on plasmodium falciparum in Africa between 2000 and 2015. Nature 526: 207–211. 10.1038/nature15535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. 2015. Accurate non-parametric estimation of recent effective population size from segments of identity by descent. Am J Hum Genet 97: 404–418. 10.1016/j.ajhg.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A. 2003. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. P Roy SocB-Biol Sci 270: 921–928. 10.1098/rspb.2002.2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churcher TS, Lissenden N, Griffin JT, Worrall E, Ranson H. 2016. The impact of pyrethroid resistance on the efficacy and effectiveness of bednets for malaria control in Africa. eLife 5: e16090 10.7554/eLife.16090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson CS, Weetman D, Essandoh J, Yawson AE, Maslen G, Manske M, Field SG, Webster M, Antão T, MacInnis B, et al. 2014. Adaptive introgression between Anopheles sibling species eliminates a major genomic island but not reproductive isolation. Nat Commun 5: 4248 10.1038/ncomms5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson CS, Miles A, Harding NJ, Weetman D, Kwiatkowski D, Donnelly M, The Anopheles gambiae 1000 Genomes Consortium. 2018. The genetic architecture of target-site resistance to pyrethroid insecticides in the African malaria vectors Anopheles gambiae and Anopheles coluzzii. bioRxiv 10.1101/323980 [DOI] [PMC free article] [PubMed]
- Cline TW, Meyer BJ. 1996. Vive la différence: males vs females in flies vs worms. Annu Rev Genet 30: 637–702. 10.1146/annurev.genet.30.1.637 [DOI] [PubMed] [Google Scholar]
- Coetzee M, Hunt RH, Wilkerson R, Della Torre A, Coulibaly MB, Besansky NJ. 2013. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa 3619: 246–274. 10.11646/zootaxa.3619.3.2 [DOI] [PubMed] [Google Scholar]
- Dao A, Yaro A, Diallo M, Timbiné S, Huestis D, Kassogué Y, Traoré A, Sanogo Z, Samaké D, Lehmann T. 2014. Signatures of aestivation and migration in Sahelian malaria mosquito populations. Nature 516: 387 10.1038/nature13987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Howie B, Cox AJ, Zagury JF, Marchini J. 2013. Haplotype estimation using sequencing reads. Am J Hum Genet 93: 687–696. 10.1016/j.ajhg.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devonshire AL, Field LM. 1991. Gene amplification and insecticide resistance. Annu Rev Entomol 36: 1–23. 10.1146/annurev.en.36.010191.000245 [DOI] [PubMed] [Google Scholar]
- Diabaté A, Brengues C, Baldet T, Dabire K, Hougard JM, Akogbeto M, Kengne P, Simard F, Guillet P, Hemingway J, et al. 2004. The spread of the Leu-Phe kdr mutation through Anopheles gambiae complex in Burkina Faso: genetic introgression and de novo phenomena. Trop Med Int Health 9: 1267–1273. 10.1111/j.1365-3156.2004.01336.x [DOI] [PubMed] [Google Scholar]
- Eckhoff PA, Wenger EA, Godfray HCJ, Burt A. 2017. Impact of mosquito gene drive on malaria elimination in a computational model with explicit spatial and temporal dynamics. Proc Natl Acad Sci 114: E255–E264. 10.1073/pnas.1611064114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edi CV, Djogbénou L, Jenkins AM, Regna K, Muskavitch MA, Poupardin R, Jones CM, Essandoh J, Kétoh GK, Paine MJ, et al. 2014. CYP6 p450 enzymes and ACE-1 duplication produce extreme and multiple insecticide resistance in the malaria mosquito Anopheles gambiae. PLoS Genet 10: e1004236 10.1371/journal.pgen.1004236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanello C, Santolamazza F, Della Torre A. 2002. Simultaneous identification of species and molecular forms of the Anopheles gambiae complex by PCR-RFLP. Med Vet Entomol 16: 461–464. 10.1046/j.1365-2915.2002.00393.x [DOI] [PubMed] [Google Scholar]
- Frichot E, François O. 2015. LEA: An R package for landscape and ecological association studies. Methods Ecol Evol 6: 925–929. 10.1111/2041-210X.12382 [DOI] [Google Scholar]
- Frichot E, Mathieu F, Trouillon T, Bouchard G, François O. 2014. Fast and efficient estimation of individual ancestry coefficients. Genetics 196: 973–983. 10.1534/genetics.113.160572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantz VM, Jasinskiene N, Tatarenkova O, Fazekas A, Macias VM, Bier E, James AA. 2015. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl Acad Sci 112: E6736–E6743. 10.1073/pnas.1521077112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gempe T, Beye M. 2011. Function and evolution of sex determination mechanisms, genes and pathways in insects. Bioessays 33: 52–60. 10.1002/bies.201000043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo-Calderón GI, Emrich SJ, MacCallum RM, Maslen G, Dialynas E, Topalis P, Ho N, Gesing S, the VectorBase Consortium, Madey G, et al. 2015. Vectorbase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res 43: D707–D713. 10.1093/nar/gku1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond A, Galizi R, Kyrou K, Simoni A, Siniscalchi C, Katsanos D, Gribble M, Baker D, Marois E, Russell S, et al. 2016. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat Biotechnol 34: 78–83. 10.1038/nbt.3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemingway J, Ranson H, Magill A, Kolaczinski J, Fornadel C, Gimnig J, Coetzee M, Simard F, Roch DK, Hinzoumbe CK, et al. 2016. Averting a malaria disaster: will insecticide resistance derail malaria control? Lancet 387: 1785–1788. 10.1016/S0140-6736(15)00417-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JMC, Wides R, et al. 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298: 129–149. 10.1126/science.1076181 [DOI] [PubMed] [Google Scholar]
- Huestis DL, Dao A, Diallo M, Sanogo ZL, Samake D, Yaro AS, Ousman Y, Linton YM, Krishna A, Veru L, et al. 2019. Windborne long-distance migration of malaria mosquitoes in the Sahel. Nature 574: 404–408. 10.1038/s41586-019-1622-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishengoma DS, Saidi Q, Roper C, Alifrangis M. 2019. Deployment and utilization of next-generation sequencing of Plasmodium falciparum to guide anti-malarial drug policy decisions in sub-Saharan Africa: opportunities and challenges. Malar J 18: 267 10.1186/s12936-019-2853-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasper M, Schmidt TL, Ahmad NW, Sinkins SP, Hoffmann AA. 2019. A genomic approach to inferring kinship reveals limited intergenerational dispersal in the yellow fever mosquito. Mol Ecol Resour 19: 1254–1264. 10.1111/1755-0998.13043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CM, Liyanapathirana M, Agossa FR, Weetman D, Ranson H, Donnelly MJ, Wilding CS. 2012. Footprints of positive selection associated with a mutation (N1575Y) in the voltage-gated sodium channel of Anopheles gambiae. Proc Natl Acad Sci 109: 6614–6619. 10.1073/pnas.1201475109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. 2015. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15: 1179–1191. 10.1111/1755-0998.12387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinska E, Dennison NJ, Lycett GJ, Krzywinski J. 2016. A maleness gene in the malaria mosquito Anopheles gambiae. Science 353: 67–69. 10.1126/science.aaf5605 [DOI] [PubMed] [Google Scholar]
- Kwiatkowska RM, Platt N, Poupardin R, Irving H, Dabire RK, Mitchell S, Jones CM, Diabaté A, Ranson H, Wondji CS. 2013. Dissecting the mechanisms responsible for the multiple insecticide resistance phenotype in Anopheles gambiae s.s., M form, from Vallée du Kou, Burkina Faso. Gene 519: 98–106. 10.1016/j.gene.2013.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrou K, Hammond AM, Galizi R, Kranjc N, Burt A, Beaghton AK, Nolan T, Crisanti A. 2018. A CRISPR–Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat Biotechnol 36: 1062–1066. 10.1038/nbt.4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lack JB, Lange JD, Tang AD, Corbett-Detig RB, Pool JE. 2016. A thousand fly genomes: an expanded Drosophila genome nexus. Mol Biol Evol 33: 3308–3313. 10.1093/molbev/msw195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson DJ, van Dorp L, Falush D. 2018. A tutorial on how not to over-interpret STRUCTURE and ADMIXTURE bar plots. Nat Commun 9: 3258 10.1038/s41467-018-05257-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees R, Praulins G, Davies R, Brown F, Parsons G, White A, Ranson H, Small G, Malone D. 2019. A testing cascade to identify repurposed insecticides for next-generation vector control tools: screening a panel of chemistries with novel modes of action against a malaria vector. Gates Open Research 3: 1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffler EM, Bullaughey K, Matute DR, Meyer WK, Ségurel L, Venkat A, Andolfatto P, Przeworski M. 2012. Revisiting an old riddle: what determines genetic diversity levels within species? PLoS Biol 10: e1001388 10.1371/journal.pbio.1001388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann T, Licht M, Elissa N, Maega B, Chimumbwa J, Watsenga F, Wondji C, Simard F, Hawley W. 2003. Population structure of Anopheles gambiae in Africa. J Hered 94: 133–147. 10.1093/jhered/esg024 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke MEO, Milojevic M, Eitutis ST, Patel N, Wishart AE, Daley M, Hill KA. 2015. Genomic copy number variation in Mus musculus. BMC Genomics 16: 497 10.1186/s12864-015-1713-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas ER, Miles A, Harding NJ, Clarkson CS, Lawniczak MK, Kwiatkowski DP, Weetman D, Donnelly MJ, and The Anopheles gambiae 1000 Genomes Consortium. 2019a. Whole-genome sequencing reveals high complexity of copy number variation at insecticide resistance loci in malaria mosquitoes. Genome Res 29: 1250–1261. 10.1101/gr.245795.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas ER, Rockett KA, Lynd A, Essandoh J, Grisales N, Kemei B, Njoroge H, Hubbart C, Rippon EJ, Morgan J, et al. 2019b. A high throughput multi-locus insecticide resistance marker panel for tracking resistance emergence and spread in Anopheles gambiae. Sci Rep 9: 13335 10.1038/s41598-018-37186-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MalariaGEN Plasmodium falciparum Community Project. 2019. An open dataset of Plasmodium falciparum genome variation in 7,000 worldwide samples. bioRxiv 10.1101/824730 [DOI] [PMC free article] [PubMed]
- McInnes L, Healy J, Melville J. 2018. UMAP: Uniform Manifold Approximation and Projection for dimension reduction. arXiv:1802.03426 [stat.ML].
- McVean G. 2009. A genealogical interpretation of principal components analysis. PLoS Genet 5: e1000686 10.1371/journal.pgen.1000686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SN, Rigden DJ, Dowd AJ, Lu F, Wilding CS, Weetman D, Dadzie S, Jenkins AM, Regna K, Boko P, et al. 2014. Metabolic and target-site mechanisms combine to confer strong DDT resistance in Anopheles gambiae. PLoS One 9: e92662 10.1371/journal.pone.0092662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P, Warr E, Stevenson BJ, Pignatelli PM, Morgan JC, Steven A, Yawson AE, Mitchell SN, Ranson H, Hemingway J, et al. 2008. Field-caught permethrin-resistant Anopheles gambiae overexpress CYP6P3, a P450 that metabolises pyrethroids. PLoS Genet 4: e1000286 10.1371/journal.pgen.1000286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngufor C, N'Guessan R, Fagbohoun J, Subramaniam K, Odjo A, Fongnikin A, Akogbeto M, Weetman D, Rowland M. 2015. Insecticide resistance profile of Anopheles gambiae from a phase II field station in Cové, southern Benin: implications for the evaluation of novel vector control products. Malar J 14: 464 10.1186/s12936-015-0981-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikou D, Ranson H, Hemingway J. 2003. An adult-specific CYP6 P450 gene is overexpressed in a pyrethroid-resistant strain of the malaria vector, Anopheles gambiae. Gene 318: 91–102. 10.1016/S0378-1119(03)00763-7 [DOI] [PubMed] [Google Scholar]
- Norris LC, Main BJ, Lee Y, Collier TC, Fofana A, Cornel AJ, Lanzaro GC. 2015. Adaptive introgression in an African malaria mosquito coincident with the increased usage of insecticide-treated bed nets. Proc Natl Acad Sci 112: 815–820. 10.1073/pnas.1418892112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North AR, Godfray HCJ. 2018. Modelling the persistence of mosquito vectors of malaria in Burkina Faso. Malar J 17: 140 10.1186/s12936-018-2288-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North AR, Burt A, Godfray HCJ. 2019. Modelling the potential of genetic control of malaria mosquitoes at national scale. BMC Biol 17: 26 10.1186/s12915-019-0645-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwakanma DC, Neafsey DE, Jawara M, Adiamoh M, Lund E, Rodrigues A, Loua KM, Konate L, Sy N, Dia I, et al. 2013. Breakdown in the process of incipient speciation in Anopheles gambiae. Genetics 193: 1221–1231. 10.1534/genetics.112.148718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochieng'Opondo K, Weetman D, Jawara M, Diatta M, Fofana A, Crombe F, Mwesigwa J, D'Alessandro U, Donnelly MJ. 2016. Does insecticide resistance contribute to heterogeneities in malaria transmission in The Gambia? Malar J 15: 166 10.1186/s12936-016-1203-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira E, Salgueiro P, Palsson K, Vicente J, Arez A, Jaenson T, Caccone A, Pinto J. 2008. High levels of hybridization between molecular forms of Anopheles gambiae from Guinea Bissau. J Med Entomol 45: 1057–1063. 10.1093/jmedent/45.6.1057 [DOI] [PubMed] [Google Scholar]
- Oxborough RM, Seyoum A, Yihdego Y, Dabire R, Gnanguenon V, Wat'senga F, Agossa FR, Yohannes G, Coleman S, Musa L, et al. 2019. Susceptibility testing of Anopheles malaria vectors with the neonicotinoid insecticide clothianidin; results from 16 African countries, in preparation for indoor residual spraying with new insecticide formulations. Malar J 18: 264 10.1186/s12936-019-2888-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. 2006. Population structure and eigenanalysis. PLoS Genet 2: e190 10.1371/journal.pgen.0020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidi N, Vontas J, Van Leeuwen T. 2018. The role of glutathione S-transferases (GSTs) in insecticide resistance in crop pests and disease vectors. Curr Opin Insect Sci 27: 97–102. 10.1016/j.cois.2018.04.007 [DOI] [PubMed] [Google Scholar]
- Pinto J, Egyir-Yawson A, Vicente J, Gomes B, Santolamazza F, Moreno M, Charlwood J, Simard F, Elissa N, Weetman D, et al. 2013. Geographic population structure of the African malaria vector Anopheles gambiae suggests a role for the forest-savannah biome transition as a barrier to gene flow. Evol Appl 6: 910–924. 10.1111/eva.12075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranson H, Lissenden N. 2016. Insecticide resistance in African Anopheles mosquitoes: a worsening situation that needs urgent action to maintain malaria control. Trends Parasitol 32: 187–196. 10.1016/j.pt.2015.11.010 [DOI] [PubMed] [Google Scholar]
- R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Riehle MM, Guelbeogo WM, Gneme A, Eiglmeier K, Holm I, Bischoff E, Garnier T, Snyder GM, Li X, Markianos K, et al. 2011. A cryptic subgroup of Anopheles gambiae is highly susceptible to human malaria parasites. Science 331: 596–598. 10.1126/science.1196759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveron JM, Yunta C, Ibrahim SS, Djouaka R, Irving H, Menze BD, Ismail HM, Hemingway J, Ranson H, Albert A, et al. 2014. A single mutation in the GSTe2 gene allows tracking of metabolically based insecticide resistance in a major malaria vector. Genome Biol 15: R27 10.1186/gb-2014-15-2-r27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F. 1997. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145: 1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santolamazza F, Mancini E, Simard F, Qi Y, Tu Z, della Torre A. 2008. Insertion polymorphisms of SINE200 retrotransposons within speciation islands of Anopheles gambiae molecular forms. Malar J 7: 163 10.1186/1475-2875-7-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayre RG. 2013. A new map of standardized terrestrial ecosystems of Africa. American Association of Geographers, Washington, DC. [Google Scholar]
- Scali C, Catteruccia F, Li Q, Crisanti A. 2005. Identification of sex-specific transcripts of the Anopheles gambiae doublesex gene. J Exp Biol 208: 3701–3709. 10.1242/jeb.01819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimke RT, Kaufman RJ, Alt FW, Kellems RF. 1978. Gene amplification and drug resistance in cultured murine cells. Science 202: 1051–1055. 10.1126/science.715457 [DOI] [PubMed] [Google Scholar]
- Sedda L, Lucas ER, Djogbénou LS, Edi AV, Egyir-Yawson A, Kabula BI, Midega J, Ochomo E, Weetman D, Donnelly MJ. 2019. Improved spatial ecological sampling using open data and standardization: an example from malaria mosquito surveillance. J R Soc Interface 16: 20180941 10.1098/rsif.2018.0941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharakhova MV, Hammond MP, Lobo NF, Krzywinski J, Unger MF, Hillenmeyer ME, Bruggner RV, Birney E, Collins FH. 2007. Update of the Anopheles gambiae PEST genome assembly. Genome Biol 8: R5 10.1186/gb-2007-8-1-r5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson BJ, Bibby J, Pignatelli P, Muangnoicharoen S, O'Neill PM, Lian LY, Müller P, Nikou D, Steven A, Hemingway J, et al. 2011. Cytochrome P450 6M2 from the malaria vector Anopheles gambiae metabolizes pyrethroids: sequential metabolism of deltamethrin revealed. Insect Biochem Mol Biol 41: 492–502. 10.1016/j.ibmb.2011.02.003 [DOI] [PubMed] [Google Scholar]
- Tangena JAA, Hendriks CMJ, Devine M, Tammaro M, Trett AE, Williams I, DePina AJ, Sisay A, Herizo R, Kafy HT, et al. 2020. Indoor residual spraying for malaria control in sub-saharan Africa 1997 to 2017: an adjusted retrospective analysis. Malar J 19 10.1186/s12936-020-03216-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Ingham VA, Toé KH, Guelbéogo WM, Sagnon N, Kuzma R, Ranson H, Neafsey DE. 2020. A population genomic unveiling of a new cryptic mosquito taxon within the malaria-transmitting Anopheles gambiae complex. bioRxiv 10.1101/116988 [DOI] [PMC free article] [PubMed]
- Unckless RL, Clark AG, Messer PW. 2017. Evolution of resistance against CRISPR/Cas9 gene drive. Genetics 205: 827–841. 10.1534/genetics.116.197285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Current Protoc Bioinformatics 43: 11.10.1–11.10.33. 10.1002/0471250953.bi1110s43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vontas J, Grigoraki L, Morgan J, Tsakireli D, Fuseini G, Segura L, de Carvalho J, Nguema R, Weetman D, Slotman MA, et al. 2018. Rapid selection of a pyrethroid metabolic enzyme CYP9K1 by operational malaria control activities. Proc Natl Acad Sci 115: 4619–4624. 10.1073/pnas.1719663115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weetman D, Mitchell SN, Wilding CS, Birks DP, Yawson AE, Essandoh J, Mawejje HD, Djogbenou LS, Steen K, Rippon EJ, et al. 2015. Contemporary evolution of resistance at the major insecticide target site gene Ace-1 by mutation and copy number variation in the malaria mosquito Anopheles gambiae. Mol Ecol 24: 2656–2672. 10.1111/mec.13197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weetman D, Wilding CS, Neafsey DE, Müller P, Ochomo E, Isaacs AT, Steen K, Rippon EJ, Morgan JC, Mawejje HD, et al. 2018. Candidate-gene based GWAS identifies reproducible DNA markers for metabolic pyrethroid resistance from standing genetic variation in East African Anopheles gambiae. Sci Rep 8: 2920 10.1038/s41598-018-21265-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill M, Chandre F, Brengues C, Manguin S, Akogbeto M, Pasteur N, Guillet P, Raymond M. 2000. The kdr mutation occurs in the Mopti form of Anopheles gambiae s.s. through introgression. Insect Mol Biol 9: 451–455. 10.1046/j.1365-2583.2000.00206.x [DOI] [PubMed] [Google Scholar]
- Wiebe A, Longbottom J, Gleave K, Shearer FM, Sinka ME, Massey NC, Cameron E, Bhatt S, Gething PW, Hemingway J, et al. 2017. Geographical distributions of African malaria vector sibling species and evidence for insecticide resistance. Malar J 16: 85 10.1186/s12936-017-1734-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. 2015. Global technical strategy for malaria 2016–2030. Technical report, World Health Organization, Geneva. [Google Scholar]
- World Health Organization. 2017. Conditions for deployment of mosquito nets treated with a pyrethroid and piperonyl butoxide. Technical report, World Health Organization, Geneva. [Google Scholar]
- World Health Organization. 2018. Global report on insecticide resistance in malaria vectors: 2010–2016. Technical report, World Health Organization, Geneva. [Google Scholar]
- World Health Organization. 2019. World malaria report 2019. Technical report, World Health Organization, Geneva. [Google Scholar]
- Wright S. 1946. Isolation by distance under diverse systems of mating. Genetics 31: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.