Abstract
Translation initiation is a key step determining protein synthesis. Studies have uncovered a number of alternative translation initiation sites (TISs) in mammalian mRNAs and showed their roles in reshaping the proteome. However, the extent to which alternative TISs affect gene expression across plants remains largely unclear. Here, by profiling initiating ribosome positions, we globally identified in vivo TISs in tomato and Arabidopsis and found thousands of genes with more than one TIS. Of the identified TISs, >19% and >20% were located at unannotated AUG and non-AUG sites, respectively. CUG and ACG were the most frequently observed codons at non-AUG TISs, a phenomenon also found in mammals. In addition, although alternative TISs were usually found in both orthologous genes, the TIS sequences were not conserved, suggesting the conservation of alternative initiation mechanisms but flexibility in using TISs. Unlike upstream AUG TISs, the presence of upstream non-AUG TISs was not correlated with the translational repression of main open reading frames, a pattern observed across plants. Also, the generation of proteins with diverse N-terminal regions through the use of alternative TISs contributes to differential subcellular localization, as mutating alternative TISs resulted in the loss of organelle localization. Our findings uncovered the hidden coding potential of plant genomes and, importantly, the constraint and flexibility of translational initiation mechanisms in the regulation of gene expression across plant species.
Translation of mRNAs is a critical checkpoint in the control of gene expression, and translation initiation is the rate-limiting step determining when and where translation events start (Jackson et al. 2010). In eukaryotes the translation initiation process starts with 40S ribosomal subunits and eukaryotic initiation factors (eIFs), which form preinitiation complexes that bind to the 5′ cap and scan the 5′ untranslated regions (UTRs) of an mRNA for a proper translation initiation site (TIS) (Hinnebusch and Lorsch 2012; Hinnebusch 2014). It is commonly assumed that translation of a protein-coding gene starts with a universal AUG start codon and ends with one of three stop codons (TAA, TAG, and TGA). Yet, preinitiation complexes do not initiate at every AUG start site that they encounter. There are multiple cis- and trans-acting factors influencing the recognition of AUG start sites and consequently translation events (Jackson et al. 2010; Hinnebusch and Lorsch 2012). For example, TISs with high translation efficiency tend to have Kozak sequences (i.e., a purine [A or G] and a guanine at the −3 and +4 positions, respectively; +1 denotes the first base of the AUG start site) (Kozak 1984), and mammalian transcripts also have M (i.e., A or C), M and C at the −2, −4, and +5 positions, respectively (Noderer et al. 2014). Trans-acting factors such as eIF1 and eIF1A play critical roles in distinguishing AUG from non-AUG start codons (Takacs et al. 2011; Lind and Åqvist 2016).
Since the 1980s, studies on individual genes have reported that translation initiation can start with non-AUG codons, although the efficiency of translation initiated at these codons is lower than that initiated at AUG (Zitomer et al. 1984; Peabody 1987). Recent advances in combining ribosome profiling (i.e., the deep sequencing of ribosome-protected fragments [RPFs] to identify the position of ribosomes) and treatment with translation inhibitors that cause ribosomes to accumulate at initiation sites have enabled researchers to profile the positions of initiating ribosomes on transcripts and thus globally identify the in vivo TISs (Ingolia et al. 2011; Lee et al. 2012; Gao et al. 2015). Studies in mammalian cells have revealed the widespread presence of alternative TISs (i.e., a TIS different from the annotated AUG site) on transcripts, which tend to be located at AUG or near-cognate codons (i.e., codons one base different from AUG) (Ingolia et al. 2011; Fritsch et al. 2012; Lee et al. 2012; Gao et al. 2015). For example, nearly 50% of mammalian transcripts were found to have multiple TISs (Lee et al. 2012). After AUG codons, near-cognate codons, especially CUG, are the second major TIS codon group (Ingolia et al. 2009; Fritsch et al. 2012; Lee et al. 2012; Gao et al. 2015). These alternative AUG and non-AUG TISs were found to be evolutionarily conserved between humans and mice (Lee et al. 2012). Some alternative TISs modulate the translation of downstream main open reading frames (mORFs) (Ingolia et al. 2011; Lee et al. 2012; Gao et al. 2015), and others generate novel uncharacterized proteins or protein isoforms with diverse N-terminal ends (Ingolia et al. 2011; Lee et al. 2012). These findings in mammals reveal the biological impacts and the generality of alternative translation initiation mechanisms. They also show that the proteome diversity is much higher than previously expected and that it is not possible to unravel the entire proteome using current in silico prediction methods.
Based on the results of individual studies in the plants Arabidopsis thaliana and Nicotiana benthamiana, translation can also initiate at alternative AUGs or near-cognate codons in response to certain physiological conditions and stresses (Gordon et al. 1992; Riechmann et al. 1999; Kobayashi et al. 2002; Simpson et al. 2010; Willems et al. 2017). For example, upstream ORFs (uORFs) of the bZIP transcription factors, GDP-L-galactose phosphorylase and adenosylmethionine decarboxylase, which are involved in the sugar/polyamine/ascorbate-mediated translational repression of the mORFs, were reported to be translated with alternative AUG and ACG initiators (Wiese et al. 2004; Uchiyama-Kadokura et al. 2014; Laing et al. 2015). Translation of the RNA-binding protein FCA, which regulates flowering time, was found to initiate at a CUG codon in Arabidopsis (Simpson et al. 2010). In addition, an Arabidopsis N-terminal proteomics study revealed approximately 120 unique N-terminal peptides likely corresponding to translation events initiating at unannotated AUG and non-AUG TISs (Willems et al. 2017). These findings suggest that alternative translation initiation at AUG and non-AUG codons exists in plants. By globally profiling the positions of translating ribosomes, previous Arabidopsis and tomato studies have identified hundreds of upstream AUGs and some non-AUG start sites (Hsu et al. 2016; Willems et al. 2017; Wu et al. 2019) and their association with mORF translation (Liu et al. 2013; Wu et al. 2019). Nevertheless, the distribution of translating ribosomes across coding sequences (CDSs) and the high frequencies of non-AUG triplets on transcript sequences decreased the sensitivity and precision of pinpointing the in vivo AUG and non-AUG TISs. Thus, the central question of how general is the use of alternative AUG and non-AUG TISs for gene expression across plant species has yet to be addressed. Specifically, studies are still needed to determine to what extent alternative AUG initiation sites occur in genes and which non-AUG codons are favored as TISs. In addition, although sequence context is important for the recognition of AUG start sites, it remains unclear what sequence features facilitate the recognition of non-AUG start sites. The extent to which these alternative TISs affect protein expression and diversity remains to be determined. Lastly, whether the alternative TIS mechanism is a general feature across plant species remains to be explored.
To address the aforementioned questions, in this study we globally profiled the in vivo TISs in tomato by generating/analyzing data sets of initiating ribosome positions, and we also profiled the TISs in Arabidopsis by analyzing public data sets of initiating ribosome positions (Willems et al. 2017). We systematically identified the alternative TISs in genes and examined the preference for AUG and non-AUG TIS codons in both species and identified the sequence signatures for recognizing in vivo TISs in tomato. In addition, to evaluate the regulatory role of alternative translation initiation in gene expression, we investigated the translational efficiency/transcript abundance of genes with alternative TISs and also the organelle-specific targeting signals of genes with in-frame alternative TISs, which contribute to the generation of alternative N-terminal protein sequences. Finally, to further assess the biological significance of alternative TISs, we investigated the conservation of alternative TISs between Arabidopsis and tomato. This study uncovered the hidden AUG and non-AUG TISs in both plant genomes, and our findings highlight the evolutionary relationship between alternative TISs and gene expression in plants.
Results
Identification of in vivo translation initiation sites
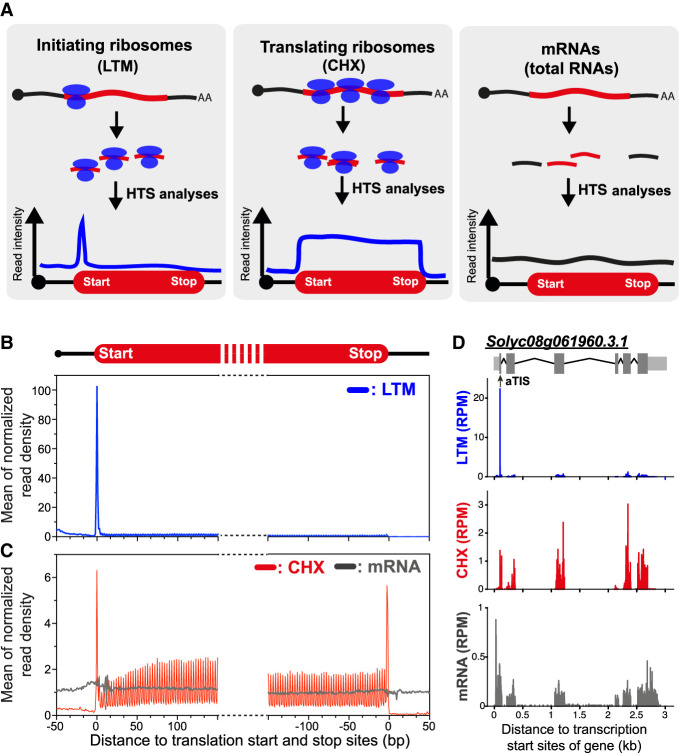
To globally characterize in vivo TISs, tomato leaves were treated with lactimidomycin (LTM; a translation inhibitor that blocks the very first round of elongation) to enrich for ribosomes positioned at start sites on mRNAs (Schneider-Poetsch et al. 2010; Lee et al. 2012). Polysome profiling analyses showed that, compared with the DMSO (mock) treatment, the LTM treatment led to stronger 80S (monosome) signals and decreased polysome signals (Supplemental Fig. S1A, black vs. blue). This result is in line with the effect of LTM in mammalian cells (Lee et al. 2012) and suggests that in tomato, LTM prevents the ribosomes from leaving the start sites on mRNAs and also allows the remaining elongating ribosomes to run off. Nevertheless, we consistently observed some residual signals in the polysome fractions of LTM-treated samples (Supplemental Fig. S1A, blue), implying the presence of residual elongating ribosomes on mRNAs. We therefore further performed an in vitro puromycin (PUR) treatment as described previously in mammalian studies to deplete the elongating ribosomes on mRNAs (Gao et al. 2015). Polysome profiling analysis revealed that, compared with the LTM-treated samples, the LTM plus PUR-treated samples had decreased polysome signals (Supplemental Fig. S1A, blue vs. orange), indicating that the PUR treatment could deplete elongating ribosomes on mRNAs and further enrich the initiating ribosomes. Thus, with this optimized protocol, we purified and analyzed the ribosome-protected fragments from LTM plus PUR-treated tomato samples using ribosome profiling (Ribo-seq); this data set is hereafter referred as the LTM data set (Methods) (Fig. 1A, left). In parallel, mRNAs isolated from tomato leaves treated with cycloheximide (CHX), which stabilizes the translating ribosomes and is used to infer the translated coding regions (Fig. 1A, middle), and total RNA isolated from untreated tomato leaves (used to evaluate the mRNA abundance) (Fig. 1A, right) were also sequenced using Ribo- and RNA-seq (Methods).
Figure 1.
Comparison of the densities of LTM- and CHX-associated ribosome-protected fragments (RPFs) in genes. (A) An illustration of high-throughput sequencing (HTS) read distribution on mRNAs from CHX- and LTM-treated samples, which induced ribosome stalling, and untreated (total mRNA) samples. (B,C) Metagene plots of mean normalized read densities in regions around translation start and stop sites of genes calculated for LTM-treated (B) and CHX-treated and untreated (mRNA) samples (C). Normalized read density of a gene was calculated by normalizing the read count per base to the average read density for the entire CDS. Only genes with 5′ UTRs and 3′ UTRs ≥ 20 nt were included. (D) As described in B and C, but for a single gene, Solyc08g061960, with a TIS peak identified at an annotated TIS (aTIS). (RPM) reads per million mapped reads. Within the gene model (top), light gray boxes indicate UTRs, dark gray boxes indicate annotated CDSs, thin lines indicate introns, and black arrow indicates aTIS.
For LTM treatment, the excised plant leaves were soaked in a solution for a period of time before sample collection (Methods), which might induce hypoxia and thus affect mRNA translation (Branco-Price et al. 2008; Juntawong et al. 2014). However, in contrast to previous observations of reduced polysome signals and a corresponding increase in 80S signals in response to hypoxia (Branco-Price et al. 2008; Juntawong et al. 2014), polysome profiling analyses revealed minor differences in 80S and polysome signals between freshly collected and solution-soaked leaf samples (Supplemental Fig. S1B). This result suggests that the LTM treatment performed in this study did not trigger a significant hypoxic response and likely affected the translation initiation of few, if any, transcripts.
With the obtained sequencing data sets, we mapped the reads and determined the LTM, CHX, and mRNA read densities per base along genes (Methods). We found that CHX read signals were located around the translation start and stop sites and were also distributed in coding regions (Fig. 1C, red), whereas mRNA signals were more evenly distributed across the 5′ UTRs, CDSs, and 3′ UTRs (Fig. 1C, gray), consistent with previous studies in yeast, mammals and Arabidopsis (Ingolia et al. 2009; Lee et al. 2012; Liu et al. 2013; Juntawong et al. 2014; Hsu et al. 2016). The LTM signals were primarily located at the annotated TIS (aTIS) and not in the rest of the CDS (Fig. 1B, blue). As an example, at the single-gene level, we observed an LTM peak at the annotated TIS of a ribosomal protein gene (Solyc08g061960) (Fig. 1D). These results reflect the stalling of initiating ribosomes caused by LTM (Fig. 1A, left) and also indicate that our data sets could capture the initiating ribosomes and be used to profile their positions in genes. In addition, side-by-side comparison of LTM and CHX signals showed the predominantly higher magnitude of LTM signals in annotated TISs compared with those of CHX (Fig. 1B,C). This feature allows us to minimize background and technical noise when identifying the in vivo TISs (Lee et al. 2012). Taken together, these results suggest that analyzing LTM data sets in parallel with CHX data sets could enable the identification of the in vivo TISs in tomato.
Prevalence of alternative AUG and non-AUG initiation sites in plant genes
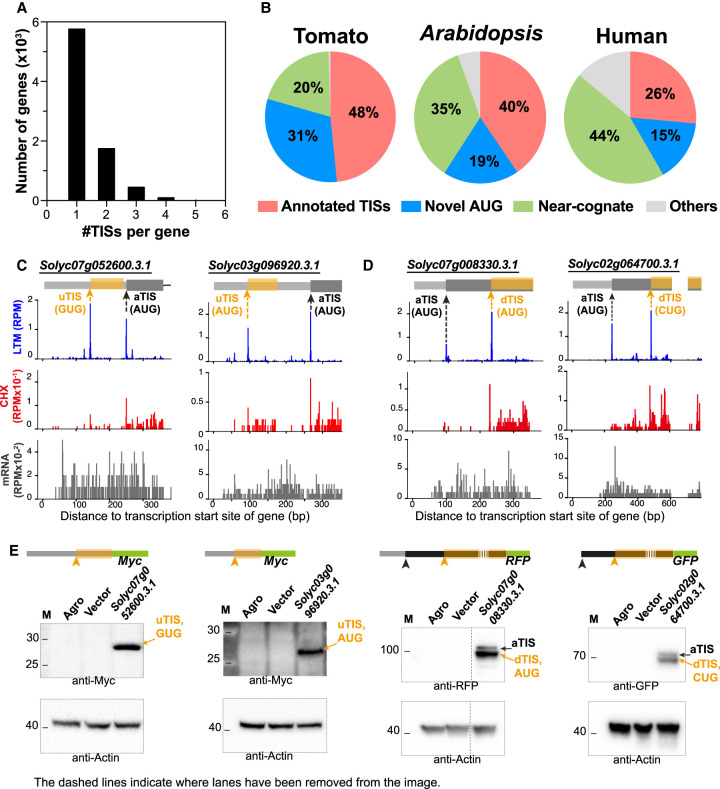
To globally characterize the in vivo TISs for tomato genes, we computed the differences in signal between the LTM and CHX samples per base in a given gene and identified the peaks with higher signals in the LTM sample (Methods). These peaks, called LTM peaks, represent the initiating ribosome positions and are inferred as in vivo TISs. There was a high correlation of LTM read densities between biological replicates (Spearman's rank correlation coefficient, rho = 0.89, P < 2.2 × 10−16) (Supplemental Fig. S2A; Supplemental Table S1; Methods), and we characterized the 11,488 TIS peaks that were found in both replicates. Of the 8212 genes with identified TISs, 71% had a single TIS peak, revealing that around one-third of genes had multiple TISs (Fig. 2A). Codon composition analyses further revealed that half of the experimentally derived TISs contained AUG codons and overlapped with annotated TISs (Fig. 2B, pink, left pie chart), thus validating a subset of in silico predicted TISs. This result also means that about half the in vivo TISs were not annotated; 31% of these unannotated TISs were AUG codons and 20% were near-cognate codons, which are one base different from AUG (Fig. 2B, blue and green, left pie chart). For example, in addition to LTM peaks at annotated AUGs, we found peaks at alternative start sites (i.e., TISs different from aTISs) in the 5′ UTR (referred to as upstream TIS [uTIS]) and CDS (referred to as downstream TIS [dTIS]) of genes; these alternative TISs included both AUG and non-AUG codons (Fig. 2C,D; Supplemental Fig. S3). These alternative uTISs and dTISs could be in frame or out of frame with annotated TISs and initiate the translation of separate, overlapping, or N-terminally extended/truncated ORFs (Supplemental Fig. S4A,B). For example, 83% of uTISs and 58% of dTISs initiate the translation of separate and N-terminally truncated ORFs, respectively (Supplemental Fig. S4C,D). Immunoblotting analyses further revealed the presence of protein bands corresponding to the expected sizes of alternative AUG- and non-AUG-initiated protein products (Fig. 2E), indicating that at least some of these alternative TISs can initiate mRNA translation. Taken together, our findings suggest the prevalence of alternative AUG and non-AUG TISs in tomato genes and indicate that, after AUG codons, near-cognate codons are the second most used for translation initiation. These findings are consistent with previous observations in mammals of strong TIS enrichment in both AUGs and near-cognate codons (Ingolia et al. 2011; Lee et al. 2012; Gao et al. 2015).
Figure 2.
Alternative initiation of translation at AUG and near-cognate codons in vivo. (A) Distribution of the number of identified in vivo TISs per gene in tomato. (B) Pie charts showing the proportion (%) of in vivo TISs in tomato (n = 11,488), Arabidopsis (n = 11,909), and humans (n = 7974) that are annotated TISs (pink) or novel TISs containing an AUG (blue), near-cognate (green), or other codons (gray). (C,D) Plots of read densities across genes as described in Figure 1D, but for genes with alternative in-frame (left) and out-of-frame (right) TISs in the 5′ UTR that initiate separate ORFs (C) and with alternative in-frame TISs in the CDS that lead to N-terminally truncated ORFs (D). Within the gene models (top), the orange arrows indicate upstream and downstream TISs (uTIS/dTIS) located in the 5′ UTR (light gray boxes) and within annotated CDSs (dark gray boxes), respectively, and the orange boxes are the alternative TIS-initiated ORFs. Examples of translation initiating at AUG, GUG, or CUG are shown. See Supplemental Fig. S3 for full-length gene models. (E) Immunoblotting analysis of proteins encoded by transcripts with annotated and/or alternative TISs indicated in C and D and transiently expressed in tobacco leaves. (Agro) tobacco leaves infiltrated with agrobacteria without expression plasmid; (Vector) tobacco leaves infiltrated with agrobacteria containing the expression vector (i.e., the plasmid without target gene sequences).
To further assess whether alternative translation initiation is a general feature in plants, we examined in vivo TISs using a public data set of Arabidopsis initiating ribosome positions (Willems et al. 2017). Similar to tomato, more than half of the Arabidopsis TISs were unannotated AUG and near-cognate start sites (Fig. 2B; Supplemental Table S1). For comparison, this pattern was not observed when analyzing the codon composition of coding regions of all protein-coding genes (Supplemental Fig. S5). These results showed that the failure to predict a significant portion of in vivo TISs through in silico sequence analyses is a general phenomenon and that translation initiation could occur at both AUG and near-cognate codons across plants.
Taken together, our findings showed that at least half of the in vivo TISs in genes have not been predicted via in silico analyses and are different from annotated TISs, suggesting the prevalence of alternative TISs. This prevalent alternative translation initiation occurs at AUG and non-AUG codons and is a phenomenon conserved in tomato, Arabidopsis, and humans.
Preference for near-cognate codons in translation initiation
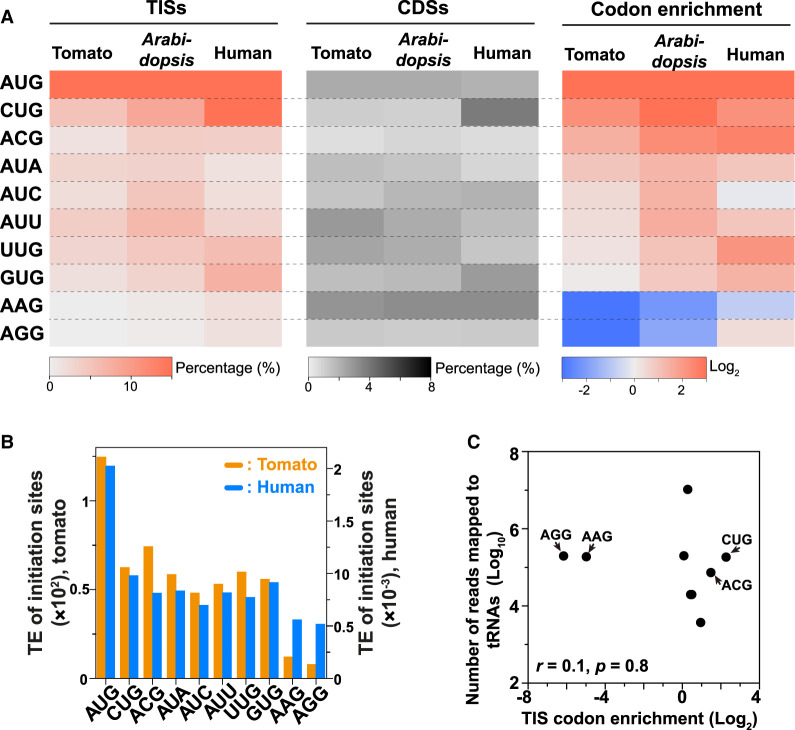
We showed that although AUG is the canonical and dominant TIS codon (Fig. 2B), near-cognate codons could serve as secondary translation initiation codons in plants (Fig. 2B). Because there are nine codons in the near-cognate group, this raises the question of which near-cognate codons serve more frequently as start codons across species. To address this, we examined the frequencies of each near-cognate codon among the identified TISs. In Arabidopsis and tomato, the percentage frequencies ranged from 0% to 8% (left, Fig. 3A). This result was in contrast to the previous observation of a specifically higher enrichment of CUG (∼18%) among human TISs (left Fig. 3A; Ingolia et al. 2011; Lee et al. 2012; Gao et al. 2015). These observations suggest that plants and humans may have different mechanisms for selecting near-cognate codons as start sites. Alternatively, the codon biases/abundances in the genome may influence the frequency with which preinitiating ribosomes encounter the codon in question and thus affect TIS codon preference. To assess these possibilities, we examined the codon abundances (%) in the CDSs of protein-coding genes in each species. More CUG codons (∼4%) are present in the CDSs of genes in humans than in tomato (∼1%) and Arabidopsis (∼1%) (Fig. 3A, middle), showing that there is a difference in overall codon usage between humans and plants, which likely affects the frequencies of TIS codons (Fig. 3A, left). Thus, to normalize the differences in codon usage across species, we calculated the TIS codon enrichment by taking the log2 ratio between the codon abundance among TISs (Fig. 3A, left) and that among CDSs (Fig. 3A, middle) for each codon. Among the nine near-cognate codons, CUG and ACG showed the highest enrichment, whereas AAG and AGG were the most depleted among TISs in tomato (Fig. 3A, right). The pattern of non-AUG codon usage was significantly correlated between tomato and Arabidopsis (Pearson correlation coefficient, r = 0.97, P = 1.7 × 10−5). Comparable results were also observed for tomato and humans (r = 0.67, P = 4.9 × 10−2) (Fig. 3A, right), showing that TIS codon preference among near-cognate codons is conserved across plants and mammals.
Figure 3.
Preference for near-cognate codons in translation initiation. (A) Heatmaps showing the percent frequencies of each type of codon among the identified TISs (left) and among annotated CDSs of all genes (middle), and the TIS codon enrichment (log2 ratio between the codon abundance among identified TISs and that among annotated CDSs; right). Enrichment values are shown for AUG and individual near-cognate codons in tomato, Arabidopsis, and humans. (B) The median translation initiation efficiency (TE) of TISs at AUG and individual near-cognate codons identified in tomato (orange) and humans (blue) (Methods). (C) The correlation between TIS codon enrichment and tRNA abundance in tomato. (r) Pearson correlation coefficient.
We also found that the degree of TIS codon enrichment was correlated with the magnitude of TIS translation initiation efficiency (i.e., the LTM read density of a given TIS vs. the mRNA read density of the gene with the TIS in question) (Methods) among near-cognate codons (r = 0.97 and 0.71; P = 2.5 × 10−5 and 3.2 × 10−2 in tomato and humans, respectively) (Fig. 3A,B), supporting our observation that specific near-cognate codons were favored as TISs. For example, CUG was enriched among near-cognate TISs and was also associated with higher TIS translation initiation efficiencies, whereas AAG was depleted among near-cognate TISs and also associated with lower TIS translation initiation efficiencies in tomato and humans (Fig. 3A,B). Our observations based on global analyses of in vivo near-cognate start sites were in agreement with the findings of previous studies using in vitro/in vivo reporter assay systems that AAG and AGG are generally poor start codons and that CUG is the best, followed by ACG and GUG, for initiating a noncanonical translation event (Peabody 1989; Gordon et al. 1992; Ivanov et al. 2010; Wei et al. 2013). The different magnitudes of translation initiation efficiencies at near-cognate codons between this study (Fig. 3B) and previous ones (de Arce et al. 2018; Kearse et al. 2019) may be attributable to flanking sequences around codon sites that affect their initiation efficiencies differentially (de Arce et al. 2018). In addition, only the identified in vivo TISs (i.e., TISs with significantly higher LTM signals) were included in Figure 3B, which may inflate the efficiency values of codons compared to those when considering all codon sites with/without translation initiation activities (de Arce et al. 2018).
During the process of translation, tRNAs with specific anticodon sequences form base pairs with the codon on the mRNA (Jackson et al. 2010). Thus, tRNA abundance has been considered to be one of the factors regulating TIS translation efficiency. To address the role of tRNA abundance in preference for near-cognate codons as TIS codons, we investigated the relationship between near-cognate TIS codon enrichment and tRNA abundance. No significant correlation was observed (r = 0.1, P = 0.8) (Fig. 3C), indicating that tRNA abundance is unlikely to play a major role in the selection of TIS codons. Other factors including trans-factors such as eukaryotic initiation factors (eIFs) and cis-factors such as RNA sequences, secondary structures, and codon-anticodon recognition may be involved in TIS codon usage (Kearse and Wilusz 2017). For example, eIF2A has been found to deliver tRNA elongators (e.g., Leu-tRNAs) to CUG and UUG (Starck et al. 2012; Kearse and Wilusz 2017). A purine in the central base pair of a codon greatly destabilizes complex formation between eIFs, tRNA initiators, 40S ribosomal subunits, and mRNAs (Kolitz et al. 2009), explaining the low usage of AAG and AGG as TIS codons.
Taken together, our findings show that, among the nine near-cognate codons, specific codons are favored as translation start sites in plants and mammals, suggesting biased utilization of specific near-cognate codons as TIS codons.
Characteristics of near-cognate start sites
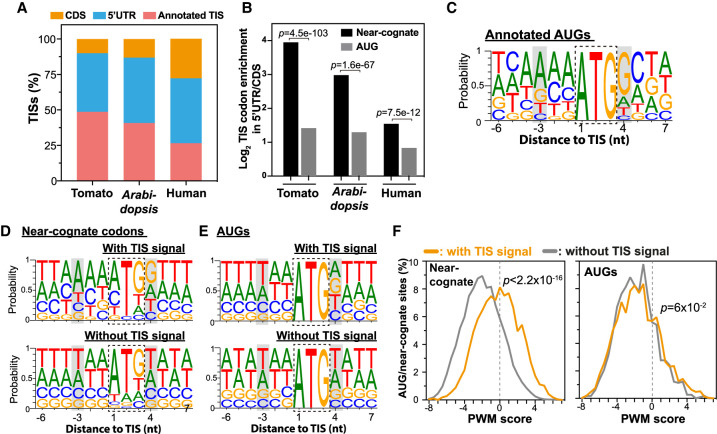
We showed that alternative TISs occurred in both 5′ UTR and CDS regions for several individual genes (Fig. 2C). In addition, global analyses of mammalian TISs revealed more in vivo TISs in 5′ UTRs than in CDSs (Lee et al. 2012; Gao et al. 2015). To determine whether this position bias is generally true for plant TISs, we calculated the relative abundance of TISs located in different regions of mRNAs. We found that although most TISs corresponded to annotated initiation sites, 5′ UTR regions had more alternative TISs than CDSs in both tomato and Arabidopsis (Fig. 4A, blue vs. orange), a pattern similar to that in humans (Fig. 4A; Lee et al. 2012; Gao et al. 2015). To further assess differences in the distribution bias between alternative AUG and near-cognate TISs, we calculated their enrichment in 5′ UTRs and CDSs. Although both AUG and near-cognate start sites were more likely to be located in 5′ UTR than in CDS regions in tomato (log2 TIS codon enrichment between the 5′ UTR and CDS = 1.4 and 3.9 for alternative AUG and near-cognate sites, respectively) (Fig. 4B), near-cognate start sites were significantly and highly enriched in 5′ UTRs (2.8-fold enrichment, P = 4.5 × 10−103, Fisher's exact test). The same result was also observed in Arabidopsis and humans (Fig. 4B), showing that the tendency for near-cognate start sites to be located in 5′ UTRs is evolutionarily conserved. This result possibly reflects the regulatory role of non-AUG codons in modulating the initiation of translation at annotated TISs, because near-cognate codons have lower initiation efficiency than annotated AUGs (Fig. 3B; Kearse and Wilusz 2017) and may be able to compete with annotated AUGs when located in 5′ UTRs but not in CDSs. The LTM read density at upstream non-AUG TISs was positively correlated with the CHX read density, but not the mRNA read density, in the corresponding TIS-initiated CDSs (Supplemental Fig. S6), indicating at least some of these alternative non-AUG TISs were used under the physiological conditions used in this study.
Figure 4.
Characteristics of alternative AUG and near-cognate start codons. (A) The relative distribution (%) of the identified TISs located in annotated TISs (pink), 5′ UTRs (blue), and CDSs (orange) in tomato, Arabidopsis, and humans. (B) The differential distribution of AUG (gray) and near-cognate (black) TISs between 5′ UTRs and CDSs in tomato, Arabidopsis, and humans. P-values are the statistical significance test of whether the enrichment of near-cognate codons between 5′ UTRs and CDSs differs from that of AUGs (Fisher's exact test). (C) The probability of occurrence of ATCG nucleotides in sequence regions around the annotated TISs in tomato. Gray boxes highlight the −3 and +4 positions of the Kozak sequence. (D,E) As in C, but for the upstream near-cognate (D) and AUG (E) codons with TIS signals (i.e., located at TISs, top) and without TIS signals (bottom). (F) Position-weight matrix (PWM) scores of codon sites with TIS signals (orange) and without TIS signals (gray) for near-cognate codons (left) and AUG codons (right). PWM score was used to represent the sequence similarity between the regions surrounding a given codon site and those surrounding annotated TISs (Methods). P-values are the test of whether the PWM scores generated based on the codon sites with TIS signals (orange) differ from those of the codon sites without TIS signals (gray) (Mann–Whitney U test).
Previous studies have shown that the presence of a “Kozak sequence” (i.e. [A/G]NNAUGG) is critical for AUG recognition (Kozak 1984); these sequences were also found in annotated TISs of tomato genes (Fig. 4C, gray). In addition, compared with annotated AUGs, the AUG start sites identified in 5′ UTRs tend to have weaker Kozak sequence contexts in Arabidopsis and tomato (Liu et al. 2013; Wu et al. 2019). Thus, to explore the mechanism for recognition of non-AUG start sites in plants, we analyzed the flanking sequence contexts of the identified TISs and their relationship with Kozak sequences in tomato. We found that, compared with the near-cognate sites without TIS signals (i.e., no LTM read signals), the near-cognate codons with TIS signals (i.e., detected TIS peaks) showed significantly stronger Kozak sequence contexts (P = 1.1 × 10−86 and 6.1 × 10−137 at the −3 and +4 positions, respectively, Fisher's exact test) (Fig. 4D, top vs. bottom). In contrast, the alternative AUG TISs showed weak Kozak sequence contexts (P = 1 and 0.32 at the −3 and +4 positions, respectively, Fisher's exact test) (Fig. 4E, top vs. bottom), in line with findings of previous studies in tomato and Arabidopsis (Liu et al. 2013; Wu et al. 2019). In addition, we noticed that, similar to annotated AUGs (Fig. 4C), near-cognate start sites with a TIS signal also had an A or C at the −4 and −2 positions (Fig. 4D, top). By summarizing the degree of flanking sequence similarity between the near-cognate start sites and the annotated AUG sites (shown as position-weight matrix [PWM] scores determined based on annotated TIS flanking sequences) (Methods), we found that the near-cognate sites with TIS signals (median PWM score = −0.14) had significantly higher PWM scores than the near-cognate sites without TIS signals (median PWM score = −2, Mann–Whitney U test, P < 2.2 × 10−16) (Fig. 4F, left). This pattern was not observed when alternative AUG sites were compared with annotated AUGs (median PWM = −1.7 and −1.9 for AUG sites with and without TIS signals, respectively, Mann–Whitney U test, P = 0.06) (Fig. 4F, right). Our findings support the importance of flanking sequence context for non-AUG recognition and are consistent with the previous observation that translation initiation in mammals is more dependent on sequence context for near-cognate start codons than for AUG codons (de Arce et al. 2018). Nevertheless, we should emphasize that the translation initiation efficiencies of alternative AUG and near-cognate start sites were lower than those of annotated AUG sites (Fig. 3B), reflecting their suboptimal sequence contexts compared with the annotated AUGs (Fig. 4C–E) and also indicating the regulatory role of alternative TISs in fine-tuning gene expression.
Taken together, these results suggest that there is a strong positional bias of near-cognate start sites toward the 5′ UTR region and that the Kozak and flanking sequence contexts play a more critical role in recognizing near-cognate TISs than alternative AUG TISs.
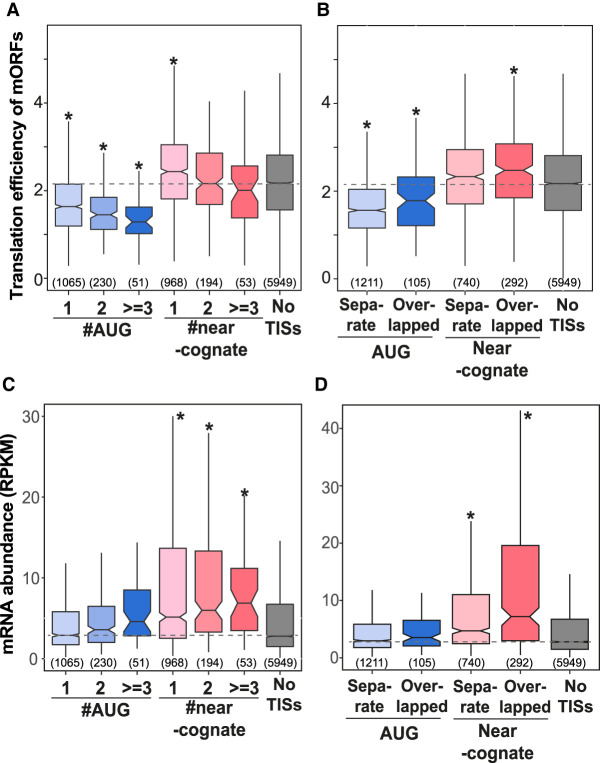
Upstream AUG and near-cognate TISs and translation efficiency of main ORFs
Our findings so far indicated that the majority of AUG and near-cognate TISs are located in 5′ UTRs, a phenomenon likely common among plants and mammals (Fig. 4). Upstream AUG initiators play a repressive role in the translation of mORFs in yeast and vertebrates including zebra fish, mouse, and human (Brar et al. 2012; Chew et al. 2016; Johnstone et al. 2016), whereas near-cognate TISs are positively associated with mORF translation in yeast (Brar et al. 2012; Spealman et al. 2018). In Arabidopsis, the mORFs of genes with uORFs initiated at AUG, but not CUG, tend to have lower translational efficiencies (Liu et al. 2013). To reveal to what extent these observations can be generalized to another plant species, we assessed the relationship between upstream AUG and near-cognate TISs and mORF translation efficiency (the CHX read density in the CDS vs. the mRNA read density in the CDS) (Methods) in tomato. Focusing on upstream AUG TISs first, we found that genes with increasing numbers of upstream AUG TISs had lower mORF translation efficiencies (median values ranging from 1.3 to 1.6) than genes with no AUG or near-cognate TISs in the 5′ UTR (median value = 2.2, Mann–Whitney U test, P < 7.6 × 10−14) (Fig. 5A), consistent with their general role in translational repression in Arabidopsis and vertebrates (Liu et al. 2013; Chew et al. 2016; Johnstone et al. 2016). In contrast, genes with increasing numbers of near-cognate TISs in the 5′ UTR (median values ranging from 2 to 2.4) showed minor differences from the genes with no TISs in the 5′ UTR (P < 0.5) (Fig. 5A). This observation was in line with the findings of a previous study in Arabidopsis (Liu et al. 2013) but different from the observation of a positive correlation between near-cognate TISs and mORF translation efficiencies in yeasts (Brar et al. 2012; Spealman et al. 2018), indicating that near-cognate codons may have a weaker influence on plant mORF translation.
Figure 5.
The correlation between alternative AUG/near-cognate TISs and translation efficiency/transcript abundance. (A) The translation efficiencies of main ORFs (mORFs) for genes with the indicated numbers of exclusively AUG-initiated TISs, exclusively near-cognate-initiated TISs, and without AUG/near-cognate-initiated TISs (“No TIS”) in the 5′ UTR. (B) The translation efficiencies of mORFs for genes with upstream AUG-initiated or near-cognate-initiated ORFs that are fully separate from or overlap and are out of frame with mORFs. (C,D) As described in A and B, but for steady-state mRNA abundances of genes. (*) P < 1 × 10−4. P-values are the test of whether the translation efficiency of mORFs and mRNA abundance determined for genes with AUG or near-cognate TISs differ from those of “No TIS” genes (Mann–Whitney U test). Dashed line shows the median value from the data for “No TIS” genes. Only genes with AUG or near-cognate sites in the 5′ UTR and with RPKM values ≥1 in the CDS region in both the CHX and mRNA RNA-seq data sets were included. The number of genes within a given group is shown in parentheses.
Depending on the position of the upstream TIS and the associated stop codon, a uORF could be fully separate from or overlap with the mORF (Supplemental Fig. S4A) and have different effects on mORF translation (Lee et al. 2012; Somers et al. 2013; Johnstone et al. 2016; Young and Wek 2016). For example, when an overlapping uORF is translated, the translation of the downstream mORF would be repressed and mORF initiation would solely depend on leaky scanning mechanisms. On the other hand, mORF translation would be favored in situations in which the uTIS of a separate ORF is bypassed via leaky scanning, or when the remaining 40S ribosomes, which complete the translation of separate uORFs, reinitiate at downstream start sites. Thus, we evaluated the respective contributions of these two uORF types to mORF translation repression. We found that gene groups with separate or overlapping AUG uORFs had significantly lower translational efficiencies than genes with no TISs in the 5′ UTR (P < 5 × 10−6) (Fig. 5B), and although these uORF gene groups had significantly different translational efficiencies, these values were not very different (median values = 1.6 and 1.8 for separate and overlapping uORFs, respectively, P = 0.02) (Fig. 5B, light blue vs. dark blue), suggesting that separate and overlapping AUG uORFs have similar repressive effects on mORF translation.
Our finding suggests that upstream AUG initiators play a general and conserved role in repressing downstream translation in plants and vertebrates. In addition, plant near-cognate initiators, distinct from those in vertebrates, likely play a minor role in modulating mORF translation.
Upstream AUG and near-cognate ORFs and steady-state mRNA levels
Our results thus far showed that plant upstream AUGs, but not near-cognate codons, are negatively associated with downstream mORF translation (Fig. 5A). In addition to functioning as translation repressors, the upstream AUG ORFs are globally associated with lower mRNA levels in vertebrates (Johnstone et al. 2016) and likely trigger decay of uORF-containing mRNA via the nonsense-mediated decay (NMD) pathway (Barbosa et al. 2013). In Arabidopsis, individual studies showed that ORFs with upstream AUGs could induce mRNA destabilization via NMD-dependent and NMD-independent pathways (Rayson et al. 2012; Uchiyama-Kadokura et al. 2014; Tanaka et al. 2016; Kurihara et al. 2018). Nevertheless, the global effects of uORFs on steady-state mRNA levels have not been evaluated systematically in any plant species. Employing our tomato total RNA data sets, we found that genes with increasing numbers of uAUG TISs did not show lower transcript abundances (median mRNA values ranging from 2.9 to 4.6) compared with genes with no upstream AUG/near-cognate TISs (median mRNA value = 2.8, Mann–Whitney U test, P < 0.1) (Fig. 5C), which is different from the observation of negative effects of uAUGs on mRNA abundance in vertebrates (Johnstone et al. 2016). To address whether this finding is true across plants, we retrieved the total RNA data sets of genes with AUG-initiated ORFs from Arabidopsis (Liu et al. 2013). Similar to tomato, no negative correlation was observed between uAUG TISs and transcript abundance in Arabidopsis (Supplemental Fig. S7A, blue vs. gray). Note that tomato and Arabidopsis total RNA samples were prepared using different mRNA enrichment methods (rRNA depletion and poly(A)+ RNA purification, respectively), so this observation is unlikely to be caused by technical bias. Our findings showed that unlike vertebrates, uAUG ORF translation may not globally trigger RNA degradation in plants and may possibly induce mRNA degradation of only a subset of transcripts or in specific processes (Uchiyama-Kadokura et al. 2014; Hou et al. 2016; Tanaka et al. 2016).
In parallel, we also assessed the effects of these upstream non-AUG-initiated ORFs on plant transcript abundance. Compared with genes with no upstream AUG/near-cognate TISs (median mRNA value = 2.8), genes with increasing numbers of near-cognate TISs had significantly higher transcript abundances in tomato (median mRNA values ranging from 5.1 to 6.9, P < 3.8 × 10−7) (Fig. 5C) and in Arabidopsis (Supplemental Fig. S7A, pink vs. gray), showing a positive correlation between initiation at upstream near-cognate codons and transcript abundance in plants. This pattern was also observed in both separate and overlapping non-AUG uORFs (Fig. 5D). However, the observed positive correlation might be a consequence of the higher probability of detecting near-cognate initiation sites, which generally have lower translation efficiencies (Fig. 3B), in high-abundance mRNAs.
Taken together, our findings show the conserved role of upstream AUGs in repressing mORF translation across species (Fig. 5A), but also imply that their function in down-regulating mRNA abundance has diverged between plants and mammals (Fig. 5C).
Relationship between alternative in-frame TISs and protein localization diversity
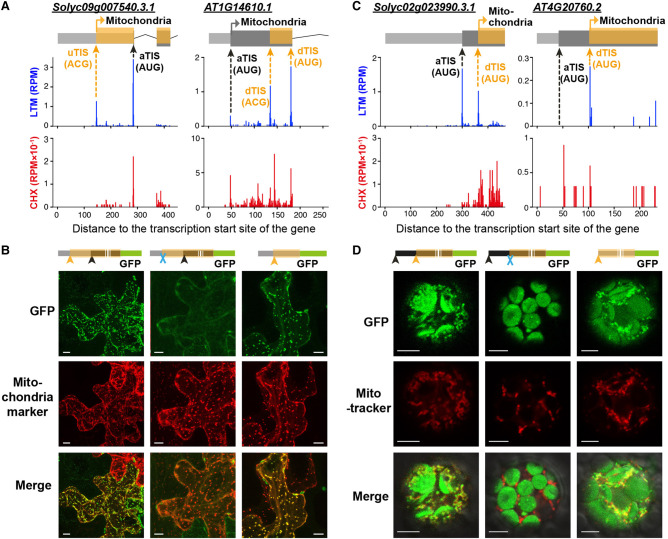
Alternative translation initiation serves as a mechanism to regulate the localization of proteins to different cellular compartments because the N terminus of the protein can function as a targeting signal (Mackenzie 2005; Yogev and Pines 2011; Carrie and Whelan 2013). In plants, analyses of individual genes have found that dual localized proteins can be encoded by a single gene with translation initiating from alternative in-frame AUG and non-AUG sites. For example, the Arabidopsis DNA polymerase γ2 uses a canonical AUG codon and an upstream CUG codon to produce proteins localized to the chloroplast and mitochondria, respectively (Christensen et al. 2005). However, the extent to which alternative translation initiation is involved in generating dual-targeted proteins and affects their localization remains to be systemically investigated. To address this, we first used the in vivo tomato TIS data sets and identified 179 and 668 alternative TISs that were in frame with annotated AUGs and led to N-terminally extended or truncated protein isoforms (Supplemental Table S2). Prediction of chloroplast and mitochondria localization further revealed that 27.4% and 16.8% of the proteins encoded by ORFs translated from alternative TISs gained and lost, respectively, targeting signals compared with the protein forms translated from annotated TISs (Supplemental Table S2). For example, Solyc09g007540, a valyl-tRNA synthetase gene whose ortholog in yeast is known to produce mitochondrial and cytosolic forms via alternative translation initiation (Chatton et al. 1988), was found to have an upstream and in-frame ACG TIS that could initiate translation of a mitochondria-localized protein (Fig. 6A, left; Supplemental Fig. S3). By introducing the 5′ UTR and CDS regions fused with GFP into tobacco leaves to reveal protein localization, we found that the region containing the 5′ UTR and CDS and the region containing only the 5′ UTR both produced GFP signals colocalizing with a mitochondria marker (Fig. 6B, left and right). In contrast, mutation of the ACG TIS resulted in cytosolic, but not mitochondria-localized, GFP signals (Fig. 6B, middle), supporting our hypothesis that the tomato valyl-tRNA synthetase transcript could produce both mitochondrial and cytosolic protein isoforms via alternative TIS mechanisms. The Arabidopsis orthologous gene also has multiple in vivo TISs, in which the annotated and alternative TISs generate mitochondrial and cytosolic protein isoforms, respectively (Fig. 6A, right; Supplemental Fig. S3). Another example is Solyc03g044470, which encodes a red chlorophyll catabolite reductase predicted to localize in chloroplasts (Supplemental Fig. S8A). This transcript has a downstream in-frame AUG TIS, which could lead to translation of a shorter protein isoform without a chloroplast localization signal (Supplemental Fig. S8A, left). We further found that when both the full-length CDS and the partial CDS region between the aTIS and dTIS were fused with GFP, GFP signals colocalized with chlorophyll (Supplemental Fig. S8C,D, left and right); however, mutation of the aTIS resulted in cytosolic GFP signals (Supplemental Fig. S8C,D, middle). Similarly, a downstream in-frame AUG TIS and a corresponding cytosolic protein isoform were also observed for the Arabidopsis orthologous gene (Supplemental Fig. S8B). Together, these results suggest that alternative translation initiation affects protein subcellular localization across plants.
Figure 6.
The differential localization of proteins encoded by genes with alternative in-frame TISs. (A,C) As indicated in Figure 1D, but for the orthologous gene pairs Solyc09g007540 and AT1G14610 (A) and Solyc02g023990 and AT4G20760 (C), which have identified alternative TISs (orange arrows) in frame with the annotated TIS (aTIS) that are either located upstream (uTIS) or downstream (dTIS) and encode N-terminally extended or truncated protein isoforms. The TIS of the protein isoform with predicted mitochondria targeting signals is indicated. See Supplemental Fig. S3 for full-length gene models. (B) Confocal images showing the localization of Solyc09g007540-GFP proteins with translation driven by the wild-type 5′ UTR and CDS (left), uTIS-mutated 5′ UTR and CDS (middle), and 5′ UTR region (right) in transiently transformed tobacco leaves. (Scale bar) 10 µm. The aTIS, uTISs/dTISs, and mutated TISs are indicated by a black arrow, orange arrow, and blue cross, respectively. (Mitochondria marker) CD3-992 (Nelson et al. 2007). (D) As described in B, but with the localization of Solyc02g023990-GFP proteins driven by the wild-type full-length CDS (left), dTIS-mutated full-length CDS (middle), and CDS region downstream of the dTIS (right) in transiently transformed Arabidopsis protoplasts. (MitoTracker) a mitochondria dye. (Scale bar) 5 µm.
In addition to organelle targeting signals present in the N terminus of the longest protein form (Fig. 6A; Supplemental Fig. S8), targeting signals could be embedded within CDS regions and become exposed when the shorter protein isoform is produced (Supplemental Table S2). For example, Solyc02g023990 has an in-frame dTIS, which generates a shorter protein isoform with predicted mitochondria localization (Fig. 6C, orange arrow in the left panel; Supplemental Fig. S3). A dTIS generating a mitochondrial protein isoform was also found in the Arabidopsis orthologous gene (Fig. 6C, right; Supplemental Fig. S3). Protein localization analyses further revealed that both the full-length CDS and dTIS-initiated protein forms of Solyc02g023990 colocalized with MitoTracker (a dye that stains mitochondria) signals (Fig. 6D, left and right), whereas mutation of dTIS yielded a protein that did not (Fig. 6D, middle). These results indicate that downstream alternative initiation is a strategy used in plants for achieving organ-specific targeting.
Together our findings suggest that alternative translation initiation is an evolutionarily conserved strategy that generates different protein isoforms with diverse N termini to achieve the removal or exposure of organelle targeting signals and consequently diversify the organelle proteome.
Conservation of alternative AUG and non-AUG TISs across species
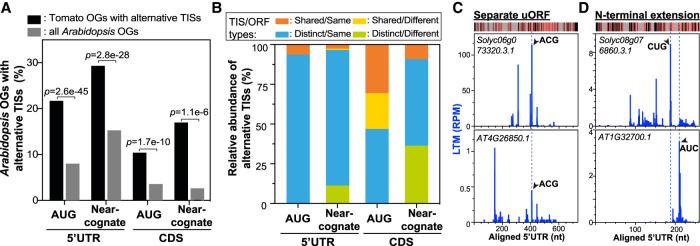
The prevalent alternative TISs could reshape proteome expression profiles by modulating translational efficiency or increasing protein diversity (Figs. 5, 6). To further assess the biological significance of alternative TIS mechanisms in plants, we next investigated the conservation of these mechanisms by examining whether the alternative TISs are present in both genes of an orthologous gene pair between Arabidopsis and tomato. Among the four alternative TIS groups (defined by whether translation is initiated from an AUG or near-cognate codon located in the 5′ UTR or CDS), 10%–29% of the tomato orthologs with an alternative TIS had an Arabidopsis ortholog with an alternative TIS; this is significantly higher than the percentage for “all Arabidopsis orthologs,” which served as a background data set (Fig. 7A). Among these alternative TISs, 3%–31% (depending on the alternative TIS group) were located at the same position and encoded the same type of ORF in tomato and Arabidopsis (Fig. 7B, orange; Supplemental Fig. S9A). For example, GDP-L-galactose phosphorylase, a key enzyme for the control of ascorbate biosynthesis, was reported to have an upstream near-cognate (ACG) initiator associated with a 65-aa uORF in Arabidopsis (Fig. 7C, bottom; Laing et al. 2015); this uORF was also found at the same position upstream of the tomato orthologous gene (Solyc06g073320) and encoded a peptide of 62 amino acids (Fig. 7C, top). The Solyc03g044470 and AT4G37000 orthologous genes, which both encode an N-terminally truncated protein (Supplemental Fig. S8A,B), use a downstream AUG TIS conserved between species (Supplemental Fig. S9B). These results show that some alternative TISs are shared across species and also support the findings of previous studies that uORFs are conserved across plants species and are predicted to be initiated from both AUG and non-AUG codons (Hayden and Jorgensen 2007; Jorgensen and Dorantes-Acosta 2012; Takahashi et al. 2012; Vaughn et al. 2012; van der Horst et al. 2019).
Figure 7.
The conservation of alternative TISs between tomato and Arabidopsis. (A) The proportion (%) of Arabidopsis orthologous genes (OGs) with alternative TISs when the tomato OG has an alternative TIS (black; n = 1320, 1089, 473, and 65, from left to right) and the proportion (%) of all Arabidopsis OGs with alternative TISs (gray; n = 9349), which were considered as a background data set. The alternative TISs were categorized into four groups depending on their location (5′ UTR or CDS) and the type of initiation codon (AUG or near-cognate codon). P-values are the test of whether the proportion of Arabidopsis OGs with alternative TISs differs between the black and gray data sets (Fisher's exact test). (B) The relative proportion (%) of alternative TISs detected in both genes of a given tomato-Arabidopsis OG pair. Pairs were categorized into four groups depending on (1) whether the TIS position and codon are identical between orthologs (i.e., “Shared”) or different (i.e., “Distinct”) and (2) whether the corresponding ORF type between orthologs is the same (i.e., “Same”) or different (i.e., “Different”) (Supplemental Fig. S9; Methods). (C) Plots of LTM read density along the aligned 5′ UTRs of an OG pair with alternative TISs (arrow) at the same position and corresponding to the same ORF type. The TIS codon and corresponding ORF type for each OG pair are shown beside the TIS peak and on the top left, respectively. The heatmap (top) shows the conserved (pink) and nonconserved (black) sequences and gapped regions (gray). (D) As defined in C, but for the OG pairs with alternative TISs located at different positions but producing the same ORF type.
In the majority (47%–94%) of orthologous genes pairs sharing alternative TISs, the TISs are located at different positions but encode the same type of ORF across species (Fig. 7B, blue; Supplemental Fig. S9A). For example, the two differentially located TISs in the 5′ UTRs of the Solyc08g076860 and AT1G32700 orthologous pair initiate the translation of the same type of ORF (i.e., the N-terminally extended form) (Fig. 7D). Similarly, the Solyc02g023990 and AT4G20760 orthologous genes, which both generate an N-terminally truncated protein (Fig. 6C), have downstream TISs located at different positions in the aligned CDS (Supplemental Fig. S9C). The most intriguing case is valyl-tRNA synthetase, in which both orthologous genes can generate N-terminal protein isoforms with different protein subcellular localizations, but the tomato gene uses an in-frame uTIS and the Arabidopsis gene uses an in-frame dTISs (Fig. 6A). These results suggest that compensatory TISs (i.e., alternative TISs located elsewhere in the mRNA) play a more dominant role in translational regulatory mechanisms across species than conserved TISs. A dominant role of compensatory TISs in generating the same ORF type was also observed between human and mouse cells (Lee et al. 2012). The use of multiple alternative TIS codons possibly reflects diversification in TISs used but maintenance of the translational regulation between plant species.
Taken together, our findings show the evolutionary flexibility of alternative TISs and the constraint on their associated ORFs; importantly, they suggest the functional significance of alternative initiation mechanisms in regulating gene expression across plants.
Discussion
Through genome-wide analysis of the initiating ribosome positions in tomato and Arabidopsis, we confirmed translation initiation from annotated TISs and further characterized novel and alternative TISs and showed their relevance to gene expression in plants. We found thousands of alternative AUG and near-cognate TISs located in 5′ UTRs and CDSs, which were more prevalent than previously appreciated (Figs. 2, 3). We further revealed that these alternative AUG TISs, but not near-cognate TISs, were tightly associated with mORF translation repression (Fig. 5); these alternative TISs also contribute to the expression and differential organelle localization of protein isoforms with different N-terminal ends (Fig. 6). These findings were observed both in tomato and Arabidopsis. Last, although the alternative TIS sequence diverged between Arabidopsis and tomato orthologous genes, they tended to be present in both orthologs and initiate the translation of same type of ORF (Figs. 6, 7). Together these findings suggest the biological significance and the conservation of the alternative translational initiation mechanism across plants. Several studies in plants have extensively characterized novel AUG and/or non-AUG codons initiating translational events in 5′ UTRs and noncoding RNAs by investigating the positions of translating ribosomes or conserved amino acid sequences (Hayden and Jorgensen 2007; Takahashi et al. 2012; Hsu et al. 2016; Bazin et al. 2017; van der Horst et al. 2019). Our approaches and findings complement these existing studies by providing an in-depth profile of in vivo TIS positions and further facilitate the investigation of novel translational events in the genome.
Nevertheless, although the characterized TISs may initiate a translation event, these TISs may differ in their biological functions, which include increasing proteome diversity or modulating gene expression. Some TIS-associated ORFs may contribute to the production of different protein isoforms and novel proteins/peptides that function in various processes (Simpson et al. 2010; Uchiyama-Kadokura et al. 2014; Yamashita et al. 2017). Others may represent cis-regulatory factors, in which the translation of a TIS-associated ORF, but not the associated protein product, affects the translation of other ORFs in the transcript (Tanaka et al. 2016; Ribone et al. 2017). Thus, the final protein products derived from ORFs in the latter group may not be generated or stably accumulate if translated. A global proteomic survey of small peptides and the genome-wide identification of the N-terminal proteome may facilitate the evaluation of these possibilities.
We found that alternative translation initiation at AUG and non-AUG codons is prevalent and is conserved in tomato, Arabidopsis, and humans (Fig. 2B). Nevertheless, there were significant differences in near-cognate TIS abundances between tomato and Arabidopsis/humans (20% vs. 35% and 44%, P < 2.8 × 10−151, Fisher's exact test) (Fig. 2B). This is possibly caused by the differential preference for TIS codons among species. An alternative explanation is the lower annotation quality in tomato compared with Arabidopsis and humans—50%, 15.5%, and 10% of genes in tomato, Arabidopsis, and humans, respectively, were without annotated 5′ UTRs, and the poorer 5′ UTR annotations in tomato may affect the identification of near-cognate start sites because the majority of these sites were located in 5′ UTRs (Fig. 4B). In addition, the higher abundance of novel and uncharacterized alternative AUG TISs in tomato than in Arabidopsis/humans (Fig. 2B, blue) may be, at least in part, because of misannotated start sites as shown for Solyc07g047850 (Supplemental Fig. S1D). Technical differences between studies may also have contributed to some of the differences across species.
In this study, we analyzed the initiating ribosome positions on mRNAs isolated from tomato leaf tissues and Arabidopsis suspension cells, using the annotated gene models to reveal translation initiation events from annotated TISs and identify novel and alternative TISs. Nevertheless, there are a few limitations to our approach. First, because only one tissue at a specific time point was used, the tissue- and condition-specific TISs were overlooked (Mustroph et al. 2009; Merchante et al. 2017). Second, studies on alternative transcription start sites and alternative mRNA splicing events have reported that a single gene may produce various mRNA isoforms with different 5′ UTR or CDS regions and thus lead to different TISs and/or coding regions being used for different protein isoforms (von Arnim et al. 2014; Brown et al. 2015; Mejía-Guerra et al. 2015; Kurihara et al. 2018). The genome-wide identification of transcription start sites of genes and the full-length transcript sequences will provide a better understanding of how alternative TISs are determined by alternative transcript start sites and alternative transcript processing. Last, the LTM treatment, in which the excised plant leaves were soaked in LTM solution before sample collection, may have triggered a minor hypoxic response (Supplemental Fig. S1B), induced excess accumulation of 40S ribosomal subunits (Supplemental Fig. S1A), or led to ribosome queuing upstream of stalled ribosomes (Juntawong et al. 2014; Ivanov et al. 2018; Kearse et al. 2019). These side effects may have led to the identification of TISs that are rarely used under normal physiological conditions. Despite these shortcomings, our approach uncovered alternative AUG and non-AUG initiators, expanding the coding potential of the tomato and Arabidopsis genomes and revealing the impact of alternative TISs on gene expression regulation. Our findings also shed light on the divergence of alternative start sites across plants. A comprehensive catalog of in vivo initiation sites and the corresponding ORFs in plant genomes is just the first step. Considering the biological significance of translational control in gene expression, which allows plants to quickly adapt to changing environmental conditions (Roy and von Arnim 2013; Merchante et al. 2017), further exploration will be needed to unveil the novel ORFs translationally expressed under a specific condition and reveal their functional or regulatory roles (Hellens et al. 2016).
Methods
Plant materials, chemical treatments, and RNA isolation for ribosome and total RNA profiling
Solanum lycopersicum cv CL5915 seeds were obtained from the World Vegetable Center and grown on soil in a growth chamber under a 12-h light (8:00–20:00, 150 μmol m−2 s−1)/12-h dark cycle at 25°C for 14 d before harvesting tomato leaves. All leaf samples, except for those treated with lactimidomycin (LTM; Merck), were excised and immediately frozen in liquid nitrogen. For the LTM treatment, the excised leaves were soaked in 30 µM LTM (dissolved in DMSO) solution at 25°C with gentle shaking for 30 min before freezing in liquid nitrogen. Two biological replicates (i.e., two separate sets of plants sampled on different days) were harvested.
To purify the ribosome-protected fragments (RPFs) for cycloheximide (CHX) sample sequencing, the polysome complexes were isolated by resuspending ground plant powder with polysome extraction buffer (20 mM HEPES, 100 mM KCl, 5 mM MgCl2) (Gao et al. 2015) containing 100 µg/mL CHX (Sigma-Aldrich), centrifuging at 13,000g for 5 min at 4°C and digesting with 1500 units of RNase I (Ambion, AM2295) per 40 µg of RNA at room temperature with gentle shaking for 40 min. The purified RPFs were further resolved in a 15% TBE-UREA polyacrylamide gel (Thermo Fisher Scientific), and the region of the gel corresponding to 26–32 nt was excised for construction of CHX-treated sample libraries as described previously (Hsu et al. 2016). To purify RNA samples for the total RNA data sets, total RNA was extracted from an aliquot of the aforementioned polysome extract using the RNA Clean Kit (Zymo), and then the Ribo-Zero rRNA depletion kit (Illumina) was used for rRNA removal. To purify the RPFs for LTM sample sequencing, the polysome complexes were isolated from the ground powder of the LTM-treated plants by extracting with polysome extraction buffer and centrifuging at 13,000g for 5 min at 4°C. The supernatant was then subjected to puromycin (PUR; Sigma-Aldrich) treatment based on a reported protocol (Gao et al. 2015) and digested with 3000 units of RNase I (Ambion, AM2295) per 54 µg of RNA at room temperature with gentle shaking for 40 min; the RPF purification was then performed as described above.
Polysome profiling analyses
For the DMSO-treated and the freshly collected leaf samples, the polysome extracts were purified from ground plant powder with polysome extraction buffer containing 100 μg/mL CHX. For the LTM-treated samples, the polysome complexes from LTM-treated samples were extracted with polysome extraction buffer, and for LTM plus PUR-treated samples, the extracts were further subjected to PUR treatment based on a reported protocol (Gao et al. 2015). The polysome extracts from fresh leaves and DMSO-, LTM-, and LTM plus PUR-treated samples were loaded onto a continuous sucrose gradient (10%–50% sucrose in polysome extraction buffer containing 50 μg/mL CHX and 100 μg/mL heparin [Sigma-Aldrich]) and spun at 35,000 rpm with a Beckman SW40 Ti rotor for 3.5 h at 4°C as performed previously (Liu et al. 2012). The distribution of the nucleic acids was examined by determining the UV254 absorbance profile (model UV-6, ISCO).
Sequencing data processing
Sequencing was performed with the Illumina HiSeq 2500 platform with single 75-nt end reads. The default parameters were used for the bioinformatics packages mentioned below unless otherwise specified. The raw sequencing reads from LTM, CHX, and total RNA samples in tomato were processed by quality filtering (parameter: -q20 -p85) and removing the first nucleotide from the 5′ end and the adaptor sequences (parameter: -a CTGTAGGCACCATCAAT) using the FASTX-Toolkit, and mapped to genes encoding ribosomal, transfer, small nucleolar, and small nuclear RNAs using Bowtie 2 (Langmead and Salzberg 2012) as suggested in a previous study (Ingolia et al. 2012). The unmapped reads were further aligned to the tomato genome (SL3.0) using STAR (parameters: ‐‐alignIntronMax 6756 ‐‐outFilterMismatchNmax 3) (Dobin et al. 2013) and BEDTools (Quinlan and Hall 2010) to obtain the regions of the genome aligning with the reads. The number of reads mapping along transcripts of a given gene based on the ITAG3.2 gene models was then determined using customized Python scripts (Supplemental Code). The P-site assignment for the reads in the LTM and CHX data sets was performed as described previously (Hsu et al. 2016) to determine the ribosome positioning. Briefly, the mapped reads in a data set were categorized into groups based on read length. In each group, the 5′ ends of the reads were first assigned to represent the aligned regions of a given read and used to generate metagene plots to reveal the offset between the peak with highest read intensity and the aTIS in a gene. The offset was then used to assign the P-site of a read. In parallel, the P-sites for the reads from total RNA data sets were assigned to the 12 and 11 nt downstream from the 5′ ends of reads in the first and second biological repeats, respectively, because a predominant fraction of RPF reads in the CHX data sets were assigned to those positions in the biological repeat in question. The S. lycopersicum genome sequences and gene models were based on the genome versions SL3.0 and ITAG3.2 (https://solgenomics.net). Because the rRNA, tRNA, snoRNA, and snRNA genes were not annotated in ITAG3.2, the gene model information for these noncoding genes was retrieved from the SL2.5 assembly in Ensembl Plants (https://plants.ensembl.org). Mapping statistics for the reads in each biological replicate are provided in Supplemental Table S3. Because the LTM read densities of the identified TISs, the CHX read densities of the CDSs, and the mRNA read densities of transcripts were highly correlated between replicates (Spearman's rank correlation coefficient > 0.9, P < 2.2 × 10−16) (Supplemental Fig. S2), the reads from replicates were combined together for the downstream analyses.
The Arabidopsis LTM and CHX sequencing data sets were retrieved from a published study (Willems et al. 2017) and analyzed using the same methods used for tomato. The Arabidopsis representative gene models were based on Araport11 (https://www.araport.org).
Identification of in vivo translation initiation sites
To identify LTM peaks representing in vivo TISs for genes in tomato and Arabidopsis, a given peak was required to meet the following criteria (pipeline modified from Lee et al. [2012] and Gao et al. [2015]): (1) the transcript has both LTM and CHX reads; (2) the position in question has ≥10 LTM reads and shows a local maximum of LTM read counts in a 31-nt window (−15, +15) flanking the position in question; (3) the difference between the normalized read densities (R) of LTM and CHX data is ≥ 0.1 (R was calculated as follows: R = [X/N]×10, where X is the number of reads mapping to the position in question and N is the total number of reads mapping to that transcript); and (4) the peak is located in the 5′ UTR or the first one-third of the CDS.
When AUGs or near-cognate codons were within 1 nt preceding or succeeding the codon corresponding to the identified TIS peak, the position of AUG or near-cognate codons was designated as an identified TIS peak (Lee et al. 2012; Gao et al. 2015). Only the TIS peaks present in both biological repeats were included in downstream analyses. To assess the false-positive and false-negative rates of our TIS identification pipeline, we used genes for which the number of CHX reads mapping within five codons downstream from the annotated TISs was in the top 10th percentile of read counts and genes with ≤5 CHX reads mapping within the same window as described previously (Lee et al. 2012). Of the genes in the top 10th percentile (n = 3244), 82.3% had a TIS peak call at the annotated TIS; the remaining 18% of genes with high CHX signals but without TIS peak calls at annotated TISs were regarded as false negatives. Of the 24,592 genes with ≤5 CHX reads, 4.1% had a detected TIS at an annotated TIS and were regarded as false positives. The AUG and near-cognate codons without translation initiation signals (Fig. 4D–F) were required to meet the following criteria: (1) located upstream of the most downstream TIS identified in the annotated 5′ UTR of the same gene; and (2) have mapped reads = 0 in the LTM sample and ≥1 in the mRNA sample.
The data sets of in vivo human TISs and expression levels of TIS/CDS/mRNAs in LTM/CHX/RNA samples are from a published study (Gao et al. 2015). The sequences of CDSs and cDNAs used to determine the overall codon composition and the presence of 5′ UTRs in genes in humans were retrieved from the Consensus Coding Sequence and Ensembl websites.
Determination of translation efficiency and tRNA abundance
The translation efficiency of the main ORF of a given gene (Fig. 5A,B; Supplemental Fig. S7B) was determined by normalizing the reads per kilobase per million mapped reads (RPKM) values for the CDS in the CHX sample to the RPKM values for the CDS in the mRNA sample. The translation initiation efficiencies at TISs (Fig. 3B) were determined by normalizing the RPKM values in a given TIS (a 5-nt window flanking the TIS) in the LTM sample to those in the transcribed regions of a gene with the TIS in question in the mRNA sample.
The transcript abundance of tRNAs was determined by mapping the sequencing reads from the total RNA data sets to the annotated S. lycopersicum tRNA loci retrieved from the SL2.5 version of the reference genome in Ensembl Plants (https://plants.ensembl.org) using Bowtie 2 (Langmead and Salzberg 2012) and are shown as the number of mapped reads as calculated with BEDTools (Quinlan and Hall 2010). Because multiple tRNA loci correspond to the same anticodon, the number of reads mapped to the tRNAs with the same anticodon was summed to represent the tRNA abundance for each codon.
Calculation of PWM scores for the flanking regions of TISs
The degree of sequence similarity between the flanking regions of alternative TISs and annotated TISs was summarized as position-weight matrix (PWM) scores (Fig. 4F) as described previously (Reuter et al. 2016). Briefly, a PWM matrix for a 13-nt window flanking the annotated TISs was computed by calculating the log2 ratio between nucleotide frequency of all annotated TISs and the background, that is, the nucleotide frequency of the entire 5′ UTR regions of all annotated genes. Thus, a positive value for a certain nucleotide at a given position indicates that the nucleotide is present more often in annotated TISs than in the background. A PWM score for an alternative TIS in question was then calculated by inputting the 13-nt sequences flanking the alternative TIS to the PWM matrix to obtain a PWM score (Reuter et al. 2016). Thus, a higher PWM score indicates a higher degree of sequence similarity between the input sequence and the sequence surrounding annotated TISs.
Prediction of chloroplast and mitochondria localization
Mitochondria and chloroplast localization predictions were performed using TargetP with specificity >0.9 (Emanuelsson et al. 2000) and LOCALIZER with default parameters (Sperschneider et al. 2017). Both TargetP and LOCALIZER show >60% sensitivity, >89% specificity, and >87% accuracy in predicting the chloroplast/mitochondria localization of experimentally supported proteins (Sperschneider et al. 2017). The organelle localization was assigned to the protein in question when at least one of the prediction tools indicated organelle localization.
Conservation of alternative TISs in orthologous gene pairs
To identify the orthologous gene (OG) pairs between tomato and Arabidopsis, an all-versus-all comparison of protein sequences was run on a combined set of S. lycopersicum and Arabidopsis genes using BLAST, and the OG pairs with the best reciprocal match between species were extracted. For each pair, protein sequences were aligned using MAFFT (Katoh et al. 2002), and the alignments were used to determine the Ks values using PAML (Yang 2007). Only the OGs with a Ks value <5 were extracted to generate a final set of 9349 OGs between S. lycopersicum and Arabidopsis.
For OGs with annotated 5′ UTRs and CDSs in both species, the 5′ UTR/CDS sequences were aligned with MUSCLE (Edgar 2004). A TIS identified in tomato was considered to be conserved (i.e., shared) if an Arabidopsis TIS peak was located at the same position in the aligned sequences and corresponded to the same codon (Fig. 7B; Supplemental Fig. S9A). To reveal whether the types of ORF generated by the TIS were the same between species (Fig. 7B; Supplemental Fig. S9A), ORFs were divided into different subtypes as previously performed in mammalian cell studies (Lee et al. 2012): uTISs were divided into two subtypes, “N-terminally extended” versus “overlapping” and “separate” (Supplemental Fig. S4A); dTISs were divided into two subtypes, “N-terminally truncated” and “separate” (Supplemental Fig. S4B).
Transient expression assay for protein expression and localization
Total RNAs isolated from tomato leaves using TRIzol reagent (Thermo Fisher Scientific) were used for the synthesis of cDNA with SuperScript II Reverse Transcriptase (Thermo Fisher Scientific). The 5′ UTR and CDS fragments encompassing the annotated and alternative TISs of the genes of interest (Figs. 2, 6; Supplemental Fig. S8) were amplified by PCR with the primers listed in Supplemental Table S4 and cloned into the pCambia1390-RFP, pGWB520-Myc, and pk7FWG2-eGFP Gateway destination vectors (Thermo Fisher Scientific). The site-directed PCR-mutagenesis of TISs in genes was performed using the primers listed in Supplemental Table S4 according to the manufacturer's instructions (Q5 Site-Directed Mutagenesis Kit, NEB).
To detect protein expression using immunoblotting, leaves of 3- to 4-wk-old N. benthamiana plants grown at 25°C under a 12-h light/12-h dark period were infiltrated with Agrobacterium tumefaciens strain LBA4404 carrying protein expression constructs. Infiltrated leaves were collected after 2–3 d for protein extraction and detection as described previously (Liu et al. 2013). The Myc-specific (GenScript A00173-40), GFP-specific (Roche 11814460001), RFP-specific (Rockland 600-401-379), and actin-specific (Sigma-Aldrich A0480) primary antibodies were used at concentrations of 1:3000, 1:5000, 1:3000, and 1:10,000, respectively. The anti-mouse (Promega W4021) and anti-rabbit (Promega W4011) HRP-coupled secondary antibodies were used at concentrations of 1:100,000 and 1:20,000, respectively, for detecting Myc/GFP/RFP fusion proteins via chemiluminescent detection (Millipore).
To reveal protein localization, A. thaliana protoplasts isolated from 2- to 3-wk-old leaves were transformed with GFP-tagged protein expression constructs. Images were acquired from protoplasts 16–24 h after transformation with a Zeiss LSM 780+ELYRA confocal microscope. Protoplasts were treated with MitoTracker Red CMXRos (200 nM, Thermo Fisher Scientific) to label mitochondria. Three- to 4-wk-old tobacco leaves were cotransformed with GFP-tagged protein expression constructs and the CD3-992 mitochondria-mCherry marker (Nelson et al. 2007) via agro-infiltration. Images were acquired from leaves 65–68 h after transformation with a laser scanning confocal (LSM710) imaging system. At least two independent transient expression assays were performed with consistent results.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE143311. Scripts for preforming analyses in this study are available as Supplemental Code.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank Mr. Bang-Chi Lin and Mr. Bo-Han Hou for technical support with data analyses, and Dr. Ho-Ming Chen, Dr. Kuan-Ju Lu, and Dr. Shin-Han Shiu for critical reading of the manuscript. This research was financially supported by grants from the Ministry of Science and Technology, Taiwan (MOST 106-2311-B-001-025; MOST 108-2628-B-001-005) and Academia Sinica to M.-J.L. We also thank Dr. Tzyy-Jen Chiou for sharing the pGWB520-Myc and pk7FWG2-eGFP plasmids; Dr. Shu-Hsing Wu for sharing the pCambia1390 plasmid; Dr. Stanton B. Gelvin for sharing the virD2-NLS-mRFP plasmid; Ms. Shu-Chen Shen and the ABRC/AS-BCST Confocal Microscopy Core Facilities for confocal image analyses and core services; the ABRC Plant Tech Core Facility for Arabidopsis protoplast transformation; and Dr. Melissa Lehti-Shiu for English editing of this article.
Author contributions: Y.-R.L. and M.-J.L. designed the research. Y.-R.L. performed the experiments. M.-J.L. analyzed the data. M.-J.L. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.261834.120.
References
- Barbosa C, Peixeiro I, Romão L. 2013. Gene expression regulation by upstream open reading frames and human disease. PLoS Genet 9: e1003529 10.1371/journal.pgen.1003529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin J, Baerenfaller K, Gosai SJ, Gregory BD, Crespi M, Bailey-Serres J. 2017. Global analysis of ribosome-associated noncoding RNAs unveils new modes of translational regulation. Proc Natl Acad Sci 114: E10018–E10027. 10.1073/pnas.1708433114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco-Price C, Kaiser KA, Jang CJ, Larive CK, Bailey-Serres J. 2008. Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J 56: 743–755. 10.1111/j.1365-313X.2008.03642.x [DOI] [PubMed] [Google Scholar]
- Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS. 2012. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335: 552–557. 10.1126/science.1215110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JWS, Simpson CG, Marquez Y, Gadd GM, Barta A, Kalyna M. 2015. Lost in translation: pitfalls in deciphering plant alternative splicing transcripts. Plant Cell 27: 2083–2087. 10.1105/tpc.15.00572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrie C, Whelan J. 2013. Widespread dual targeting of proteins in land plants: when, where, how and why. Plant Signal Behav 8: e25034 10.4161/psb.25034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatton B, Walter P, Ebel JP, Lacroute F, Fasiolo F. 1988. The yeast VAS1 gene encodes both mitochondrial and cytoplasmic valyl-tRNA synthetases. J Biol Chem 263: 52–57. [PubMed] [Google Scholar]
- Chew GL, Pauli A, Schier AF. 2016. Conservation of uORF repressiveness and sequence features in mouse, human and zebrafish. Nat Commun 7: 11663 10.1038/ncomms11663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AC, Lyznik A, Mohammed S, Elowsky CG, Elo A, Yule R, Mackenzie SA. 2005. Dual-domain, dual-targeting organellar protein presequences in Arabidopsis can use non-AUG start codons. Plant Cell 17: 2805–2816. 10.1105/tpc.105.035287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Arce AJD, Noderer WL, Wang CL. 2018. Complete motif analysis of sequence requirements for translation initiation at non-AUG start codons. Nucleic Acids Res 46: 985–994. 10.1093/nar/gkx1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016. 10.1006/jmbi.2000.3903 [DOI] [PubMed] [Google Scholar]
- Fritsch C, Herrmann A, Nothnagel M, Szafranski K, Huse K, Schumann F, Schreiber S, Platzer M, Krawczak M, Hampe J, et al. 2012. Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res 22: 2208–2218. 10.1101/gr.139568.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Wan J, Liu B, Ma M, Shen B, Qian SB. 2015. Quantitative profiling of initiating ribosomes in vivo. Nat Methods 12: 147–153. 10.1038/nmeth.3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon K, Fütterer J, Hohn T. 1992. Efficient initiation of translation at non-AUG triplets in plant cells. Plant J 2: 809–813. [PubMed] [Google Scholar]
- Hayden CA, Jorgensen RA. 2007. Identification of novel conserved peptide uORF homology groups in Arabidopsis and rice reveals ancient eukaryotic origin of select groups and preferential association with transcription factor-encoding genes. BMC Biol 5: 32 10.1186/1741-7007-5-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Brown CM, Chisnall MAW, Waterhouse PM, Macknight RC. 2016. The emerging world of small ORFs. Trends Plant Sci 21: 317–328. 10.1016/j.tplants.2015.11.005 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. 2014. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem 83: 779–812. 10.1146/annurev-biochem-060713-035802 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Lorsch JR. 2012. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol 4: a011544 10.1101/cshperspect.a011544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou CY, Lee WC, Chou HC, Chen AP, Chou SJ, Chen HM. 2016. Global analysis of truncated RNA ends reveals new insights into ribosome stalling in plants. Plant Cell 28: 2398–2416. 10.1105/tpc.16.00295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Calviello L, Wu HL, Li FW, Rothfels CJ, Ohler U, Benfey PN. 2016. Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. Proc Natl Acad Sci 113: E7126 10.1073/pnas.1614788113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802. 10.1016/j.cell.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. 2012. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc 7: 1534–1550. 10.1038/nprot.2012.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Loughran G, Sachs MS, Atkins JF. 2010. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1). Proc Natl Acad Sci 107: 18056–18060. 10.1073/pnas.1009269107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Shin BS, Loughran G, Tzani I, Young-Baird SK, Cao C, Atkins JF, Dever TE. 2018. Polyamine control of translation elongation regulates start site selection on antizyme inhibitor mRNA via ribosome queuing. Mol Cell 70: 254–264.e6. 10.1016/j.molcel.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127. 10.1038/nrm2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone TG, Bazzini AA, Giraldez AJ. 2016. Upstream ORFs are prevalent translational repressors in vertebrates. Embo J 35: 706–723. 10.15252/embj.201592759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen RA, Dorantes-Acosta AE. 2012. Conserved peptide upstream open reading frames are associated with regulatory genes in angiosperms. Front Plant Sci 3: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P, Girke T, Bazin J, Bailey-Serres J. 2014. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc Natl Acad Sci 111: E203–E212. 10.1073/pnas.1317811111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K-i, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066. 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse MG, Wilusz JE. 2017. Non-AUG translation: a new start for protein synthesis in eukaryotes. Genes Dev 31: 1717–1731. 10.1101/gad.305250.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse MG, Goldman DH, Choi J, Nwaezeapu C, Liang DM, Green KM, Goldstrohm AC, Todd PK, Green R, Wilusz JE. 2019. Ribosome queuing enables non-AUG translation to be resistant to multiple protein synthesis inhibitors. Gene Dev 33: 871–885. 10.1101/gad.324715.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Dokiya Y, Kumazawa Y, Sugita M. 2002. Non-AUG translation initiation of mRNA encoding plastid-targeted phage-type RNA polymerase in Nicotiana sylvestris. Biochem Biophys Res Commun 299: 57–61. 10.1016/S0006-291X(02)02579-2 [DOI] [PubMed] [Google Scholar]
- Kolitz SE, Takacs JE, Lorsch JR. 2009. Kinetic and thermodynamic analysis of the role of start codon/anticodon base pairing during eukaryotic translation initiation. RNA 15: 138–152. 10.1261/rna.1318509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1984. Point mutations close to the AUG initiator codon affect the efficiency of translation of rat preproinsulin in vivo. Nature 308: 241–246. 10.1038/308241a0 [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Makita Y, Kawashima M, Fujita T, Iwasaki S, Matsui M. 2018. Transcripts from downstream alternative transcription start sites evade uORF-mediated inhibition of gene expression in Arabidopsis. Proc Natl Acad Sci 115: 7831–7836. 10.1073/pnas.1804971115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing WA, Martínez-Sánchez M, Wright MA, Bulley SM, Brewster D, Dare AP, Rassam M, Wang D, Storey R, Macknight RC, et al. 2015. An upstream open reading frame is essential for feedback regulation of ascorbate biosynthesis in Arabidopsis. Plant Cell 27: 772–786. 10.1105/tpc.114.133777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB. 2012. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci 109: E2424–E2432. 10.1073/pnas.1207846109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind C, Åqvist J. 2016. Principles of start codon recognition in eukaryotic translation initiation. Nucleic Acids Res 44: 8425–8432. 10.1093/nar/gkw534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MJ, Wu SH, Chen HM, Wu SH. 2012. Widespread translational control contributes to the regulation of Arabidopsis photomorphogenesis. Mol Syst Biol 8: 566 10.1038/msb.2011.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MJ, Wu SH, Wu JF, Lin WD, Wu YC, Tsai TY, Tsai HL, Wu SH. 2013. Translational landscape of photomorphogenic Arabidopsis. Plant Cell 25: 3699–3710. 10.1105/tpc.113.114769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie SA. 2005. Plant organellar protein targeting: a traffic plan still under construction. Trends Cell Biol 15: 548–554. 10.1016/j.tcb.2005.08.007 [DOI] [PubMed] [Google Scholar]
- Mejía-Guerra MK, Li W, Galeano NF, Vidal M, Gray J, Doseff AI, Grotewold E. 2015. Core promoter plasticity between maize tissues and genotypes contrasts with predominance of sharp transcription initiation sites. Plant Cell 27: 3309–3320. 10.1105/tpc.15.00630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchante C, Stepanova AN, Alonso JM. 2017. Translation regulation in plants: an interesting past, an exciting present and a promising future. Plant J 90: 628–653. 10.1111/tpj.13520 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJH, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J. 2009. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci 106: 18843–18848. 10.1073/pnas.0906131106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. 2007. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136. 10.1111/j.1365-313X.2007.03212.x [DOI] [PubMed] [Google Scholar]
- Noderer WL, Flockhart RJ, Bhaduri A, de Arce AJD, Zhang JJ, Khavari PA, Wang CL. 2014. Quantitative analysis of mammalian translation initiation sites by FACS-seq. Mol Syst Biol 10: 748 10.15252/msb.20145136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody DS. 1987. Translation initiation at an ACG triplet in mammalian cells. J Biol Chem 262: 11847–11851. [PubMed] [Google Scholar]
- Peabody DS. 1989. Translation initiation at non-AUG triplets in mammalian cells. J Biol Chem 264: 5031–5035. [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayson S, Arciga-Reyes L, Wootton L, Zabala MD, Truman W, Graham N, Grant M, Davies B. 2012. A role for nonsense-mediated mRNA decay in plants: pathogen responses are induced in Arabidopsis thaliana NMD mutants. PLoS One 7: e31917 10.1371/journal.pone.0031917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter K, Biehl A, Koch L, Helms V. 2016. PreTIS: a tool to predict non-canonical 5′ UTR translational initiation sites in human and mouse. PLoS Comput Biol 12: e1005170 10.1371/journal.pcbi.1005170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribone PA, Capella M, Arce AL, Chan RL. 2017. A uORF represses the transcription factor AtHB1 in aerial tissues to avoid a deleterious phenotype. Plant Physiol 175: 1238–1253. 10.1104/pp.17.01060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Ito T, Meyerowitz EM. 1999. Non-AUG initiation of AGAMOUS mRNA translation in Arabidopsis thaliana. Mol Cell Biol 19: 8505–8512. 10.1128/MCB.19.12.8505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, von Arnim AG. 2013. Translational regulation of cytoplasmic mRNAs. Arabidopsis Book 11: e0165 10.1199/tab.0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO. 2010. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol 6: 209–217. 10.1038/nchembio.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG, Laurie RE, Dijkwel PP, Quesada V, Stockwell PA, Dean C, Macknight RC. 2010. Noncanonical translation initiation of the Arabidopsis flowering time and alternative polyadenylation regulator FCA. Plant Cell 22: 3764–3777. 10.1105/tpc.110.077990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers J, Pöyry T, Willis AE. 2013. A perspective on mammalian upstream open reading frame function. Int J Biochem Cell B 45: 1690–1700. 10.1016/j.biocel.2013.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spealman P, Naik AW, May GE, Kuersten S, Freeberg L, Murphy RF, McManus J. 2018. Conserved non-AUG uORFs revealed by a novel regression analysis of ribosome profiling data. Genome Res 28: 214–222. 10.1101/gr.221507.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperschneider J, Catanzariti AM, DeBoer K, Petre B, Gardiner DM, Singh KB, Dodds PN, Taylor JM. 2017. LOCALIZER: subcellular localization prediction of both plant and effector proteins in the plant cell. Sci Rep 7: 44598 10.1038/srep44598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck SR, Jiang VV, Pavon-Eternod M, Prasad S, McCarthy B, Pan T, Shastri N. 2012. Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science 336: 1719–1723. 10.1126/science.1220270 [DOI] [PubMed] [Google Scholar]
- Takacs JE, Neary TB, Ingolia NT, Saini AK, Martin-Marcos P, Pelletier J, Hinnebusch AG, Lorsch JR. 2011. Identification of compounds that decrease the fidelity of start codon recognition by the eukaryotic translational machinery. RNA 17: 439–452. 10.1261/rna.2475211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Takahashi A, Naito S, Onouchi H. 2012. BAIUCAS: a novel BLAST-based algorithm for the identification of upstream open reading frames with conserved amino acid sequences and its application to the Arabidopsis thaliana genome. Bioinformatics 28: 2231–2241. 10.1093/bioinformatics/bts303 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Sotta N, Yamazumi Y, Yamashita Y, Miwa K, Murota K, Chiba Y, Hirai MY, Akiyama T, Onouchi H, et al. 2016. The minimum open reading frame, AUG-stop, induces boron-dependent ribosome stalling and mRNA degradation. Plant Cell 28: 2830–2849. 10.1105/tpc.16.00481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama-Kadokura N, Murakami K, Takemoto M, Koyanagi N, Murota K, Naito S, Onouchi H. 2014. Polyamine-responsive ribosomal arrest at the stop codon of an upstream open reading frame of the AdoMetDC1 gene triggers nonsense-mediated mRNA decay in Arabidopsis thaliana. Plant Cell Physiol 55: 1556–1567. 10.1093/pcp/pcu086 [DOI] [PubMed] [Google Scholar]
- van der Horst S, Snel B, Hanson J, Smeekens S. 2019. Novel pipeline identifies new upstream ORFs and non-AUG initiating main ORFs with conserved amino acid sequences in the 5′ leader of mRNAs in Arabidopsis thaliana. RNA 25: 292–304. 10.1261/rna.067983.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn JN, Ellingson SR, Mignone F, von Arnim A. 2012. Known and novel post-transcriptional regulatory sequences are conserved across plant families. RNA 18: 368–384. 10.1261/rna.031179.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Jia QD, Vaughn JN. 2014. Regulation of plant translation by upstream open reading frames. Plant Sci 214: 1–12. 10.1016/j.plantsci.2013.09.006 [DOI] [PubMed] [Google Scholar]
- Wei J, Zhang Y, Ivanov IP, Sachs MS. 2013. The stringency of start codon selection in the filamentous fungus Neurospora crassa. J Biol Chem 288: 9549–9562. 10.1074/jbc.M112.447177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese A, Elzinga N, Wobbes B, Smeekens S. 2004. A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell 16: 1717–1729. 10.1105/tpc.019349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems P, Ndah E, Jonckheere V, Stael S, Sticker A, Martens L, Van Breusegem F, Gevaert K, Van Damme P. 2017. N-terminal proteomics assisted profiling of the unexplored translation initiation landscape in Arabidopsis thaliana. Mol Cell Proteomics 16: 1064–1080. 10.1074/mcp.M116.066662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HL, Song G, Walley JW, Hsu PY. 2019. The tomato translational landscape revealed by transcriptome assembly and ribosome profiling. Plant Physiol 181: 367–380. 10.1104/pp.19.00541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y, Takamatsu S, Glasbrenner M, Becker T, Naito S, Beckmann R. 2017. Sucrose sensing through nascent peptide-meditated ribosome stalling at the stop codon of Arabidopsis bZIP11 uORF2. Febs Lett 591: 1266–1277. 10.1002/1873-3468.12634 [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Yogev O, Pines O. 2011. Dual targeting of mitochondrial proteins: mechanism, regulation and function. Biochim Biophys Acta 1808: 1012–1020. 10.1016/j.bbamem.2010.07.004 [DOI] [PubMed] [Google Scholar]
- Young SK, Wek RC. 2016. Upstream open reading frames differentially regulate gene-specific translation in the integrated stress response. J Biol Chem 291: 16927–16935. 10.1074/jbc.R116.733899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitomer RS, Walthall DA, Rymond BC, Hollenberg CP. 1984. Saccharomyces cerevisiae ribosomes recognize non-AUG initiation codons. Mol Cell Biol 4: 1191–1197. 10.1128/MCB.4.7.1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.